Abstract
Cell misuse and cross-contamination can affect the accuracy of cell research results and result in wasted time, manpower and material resources. Thus, cell line identification is important and necessary. At present, the commonly used cell line identification methods need cell staining and culturing. There is therefore a need to develop a new method for the rapid and automated identification of cell lines. Raman spectroscopy has become one of the emerging techniques in the field of microbial identification, with the advantages of being rapid and noninvasive and providing molecular information for biological samples, which is beneficial in the identification of cell lines. In this study, we built a library of Raman spectra for gastric mucosal epithelial cell lines GES-1 and gastric cancer cell lines, such as AGS, BGC-823, HGC-27, MKN-45, MKN-74 and SNU-16. Five spectral datasets were constructed using spectral data and included the full spectrum, fingerprint region, high-wavelength number region and Raman background of Raman spectra. A stacking ensemble learning model, SL-Raman, was built for different datasets, and gastric cancer cell identification was achieved. For the gastric cancer cells we studied, the differentiation accuracy of SL-Raman was 100% for one of the gastric cancer cells and 100% for six of the gastric cancer cells. Additionally, the separation accuracy for two gastric cancer cells with different degrees of differentiation was 100%. These results demonstrate that Raman spectroscopy combined with SL-Raman may be a new method for the rapid and accurate identification of gastric cancer. In addition, the accuracy of 94.38% for classifying Raman spectral background data using machine learning demonstrates that the Raman spectral background contains some useful spectral features. These data have been overlooked in previous studies.
Keywords: Raman spectroscopy, Cell line identification, Machine learning, Ensemble learning, Gastric cancer cell lines
Graphical Abstract
1. Introduction
As common materials in life sciences and clinical medicine research, cell lines are widely used in antitumour drug screening [1], proto-oncogenes and oncogenes analysis [2], monoclonal antibody preparation [3] and cytokine activity detection [4]. However, in experimental research, cell misuse or cross-contamination has always been a common but easily ignored problem for researchers. Using 'wrong' cells will affect the accuracy and reliability of the results and waste time, manpower and resources. In recent years, the National Institutes of Health (NIH) and other agencies have called on researchers to identify cells before experiments are performed [5]. There are various methods for cell identification, such as isozyme assays [6], cell-specific antibody staining [7], HLA typing [8], and short tandem repeat (STR) analysis [9]. These methods require complex operations such as the staining or culturing of cells. Therefore, we need a new method for cell line identification that is a convenient, label-free and noncontact approach.
The discovery of Raman spectroscopy can be traced to 1925, when an Indian scientist C. V. Raman developed Raman testing. Today, Raman spectroscopy is applied in various fields [10]. The Raman scattering that occurs between matter and photons can be reflected in the received Raman spectrum, and thus, the molecular movement of matter can be visualized. In recent years, Raman spectroscopy has been widely used in the fields of microbial identification, species identification, food authentication, microplastic identification, pharmaceutical analysis and tumour diagnosis [11], [12], [13], [14], [15], [16].
The Raman features associated with DNA, RNA, proteins, lipids, collagen and other biomolecules are mainly found in the range of 800–1800 cm−1 of the Raman spectrum, namely, the fingerprint region. Most studies have analysed the spectral data from the fingerprint region, and a few studies have investigated the effect of high wavenumber (HW) regions (2800–3800 cm−1) on spectral recognition. Relevant studies have shown that analysing the fingerprint region alone is superior to analysing the HW region alone, but analysing the fingerprint and HW regions together is optimal [17], [18]. In addition, in previous Raman spectroscopy studies, spectral baselines were removed in the spectral preprocessing stage using methods such as adaptive iteratively reweighted penalized least squares (airPLS) algorithm [19], [20], [21]. However, Tomasz et al. speculated in the study that there is a correlation between the Raman baseline and the fluorescence background of the sample [22]. At present, no one has explored whether the baseline of Raman spectroscopy can be used for spectral analysis. Some studies have also used Raman spectroscopy to study and differentiate the spectral compositions of tumour cells [23], [24], [25], but they use fewer cell lines and traditional spectral data analysis methods.
Due to the small characteristic differences among Raman spectra, computer methods such as machine learning and deep learning are often used to differentiate the spectra when analysing them. Algorithms such as the principal component analysis-linear discriminant analysis (PCA-LDA), K-nearest neighbour (KNN), partial least squares-discriminant analysis (PLS-DA), support vector machine (SVM), artificial neural network (ANN), and convolutional neural network (CNN) algorithms are widely used [20], [26], [27]. PCA can perform linear dimensionality reduction by orthogonal transformation [28] and reduce the difficulty of data analysis. However, reduction in variables might lose useful information and change the original patterns of the spectral data [21]. LDA and SVM are two popular methods for solving classification problems, but they encounter difficulties in setting boundaries for high-dimensional spectral data due to the curse of dimensionality [29]. ANN can be used in nonlinear calibration but has a tendency toward overfitting [21]. Model training for deep learning methods such as CNN often requires larger datasets [30].
Most of the existing Raman spectroscopy studies use classification models to identify and analyse individual spectral datasets. This data processing method cannot fully utilize the characteristics of different models and datasets. In data mining, to identify and analyse similarities among datasets, one of the commonly used combination-based methods is stacking ensemble learning [31], [32]. Ensemble learning combines multiple models in some way, so it is better than any individual model [33]. Marquis de Condorcet proved that if the probability of each voter being correct is greater than 0.5 and the voters are independent, then adding more voters increases the probability of the majority vote being correct [34]. Here, we use model fusion based on different datasets to enhance the traditional stacking ensemble learning algorithm.
In order to better evaluate the ability of Raman spectroscopy for cell line identification and overcome the shortcomings of existing methods for Raman data analysis, a self-built SL-Raman model was used to classify the Raman spectra of normal and gastric cancer cells. With fewer data samples, SL-Raman can execute several machine learning models for various dimensions spectrum datasets and provide quick recognition results through ensemble election. This not only allows for the full utilization of the spectra's information, but it also produces findings that are more accurate and representative of the data. We established a Raman spectral database of gastric cancer cell lines, constructed five different datasets through interception and recombination, and fully analysed the classification results for the different datasets. We distinguished the differences in biochemical composition between normal and gastric cancer cells, correctly identified gastric cancer cells, and correctly differentiated gastric cancer cells with different degrees of differentiation. This is the first time that a stacking ensemble learning model for different datasets has been proposed and applied for data processing. SL-Raman integrates the predictions from different underlying models, so it is more representative than the predictions from a single model. In addition, SL-Raman innovatively integrates 5 datasets, which enables the base model to obtain more comprehensive prediction results from different feature levels.
2. Materials and methods
2.1. Sample preparation
We used one normal cell line GES-1 and six gastric cancer cell lines in this study: AGS, BGC-823, HGC-27, MKN-45, MKN-74 and SNU-16. Cells were frozen and preserved in a − 80 °C freezer and were defrosted in a 37 °C water bath to promote melting before sample preparation. AGS cells were maintained in DMEM/F12 medium supplemented with 10% foetal bovine serum and 1% penicillin—streptomycin, while the other cell lines were cultured in RPMI-1640 medium with 10% foetal bovine serum and 1% penicillin—streptomycin. These cell lines were routinely cultured in a 5% CO2 humidified incubator at 37 °C. After culturing in a cell culture flask for 48 h, the cells were removed from the flask surface with 0.25% trypsin-EDTA and washed 3 times with phosphate buffered saline. The suspended cells were then collected via centrifugation and fixed in sterilized deionized water with 4% paraformaldehyde. Cells were then diluted to 10,000 per millilitre using deionized water, immobilized on a slide and air dried for Raman spectra measurements. The Raman detection sample of each cell line is about 7.5 μl (2.5 μl/drop * 3). The slide used in the experiment is a glass slide coated with 25 nm thick aluminum film. All reagents used in this process were purchased from Gibco Company, USA.
2.2. Raman measurements
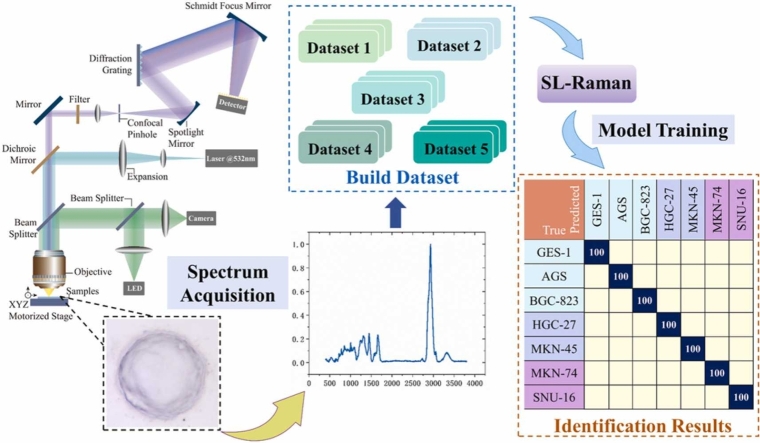
We collected the Raman spectra of gastric cancer cell lines using a Raman spectroscopy system (R300 (Objective lens: Olympus, 100 ×, NA=0.8), Hooke Instrument, Changchun, China) with a laser wavelength of 532 nm. To collect as much Raman data for gastric cancer cells as possible, we collected Raman spectra from five locations for each cell to create an average spectrum. In the subsequent spectral analyses, the average spectrum represents the overall spectral data of the unit. The conditions for collecting Raman spectra were a grating of 600, a 9 mW laser power and an 8 s integral time. Since the diameter of gastric cancer cells is about 20 µm, and the spot size of the laser is< 1 µm during the actual measurement, it is not possible to determine whether the specific location for detection is the nucleus, cytoplasm, or cell membrane when collecting spectra of gastric cancer cells. Therefore, we collected spectra from approximately 5 locations in each cell and used the average spectrum for spectroscopy in our analysis. We collected spectra from about 100 cells of each cell line for a total of about 3500 spectra and about 700 average spectra. A total of 3300 spectra and 660 average spectra were used for model training after signal-to-noise filtering.
2.3. Data preprocessing
Data preprocessing can effectively attenuate unnecessary spectral signal changes and interference caused by instruments and the environment [35]. The preprocessing process for Datasets 1–3 is: cosmic ray removal, filtering, background removal, and normalization. The preprocessing process for Dataset 4 is: cosmic ray removal, filtering, background removal, data before background removal minus data after background removal to obtain spectral background, normalization. Dataset 5 is composed of data sets 1–4 without additional data processing. By linear fitting the points around the singular values of the spectral data, we removed the cosmic rays. We filtered the data with a Savitzky—Golay filter, used the airPLS algorithm to gradually approximate the Raman spectral baseline and normalized the spectral data with min-max normalization [36], [37]. It is worth mentioning that to handle for minor differences in the x-axis among the different spectra, we processed the data using cubic spline interpolation. All processing was conducted in Python.
2.4. Composition of the dataset
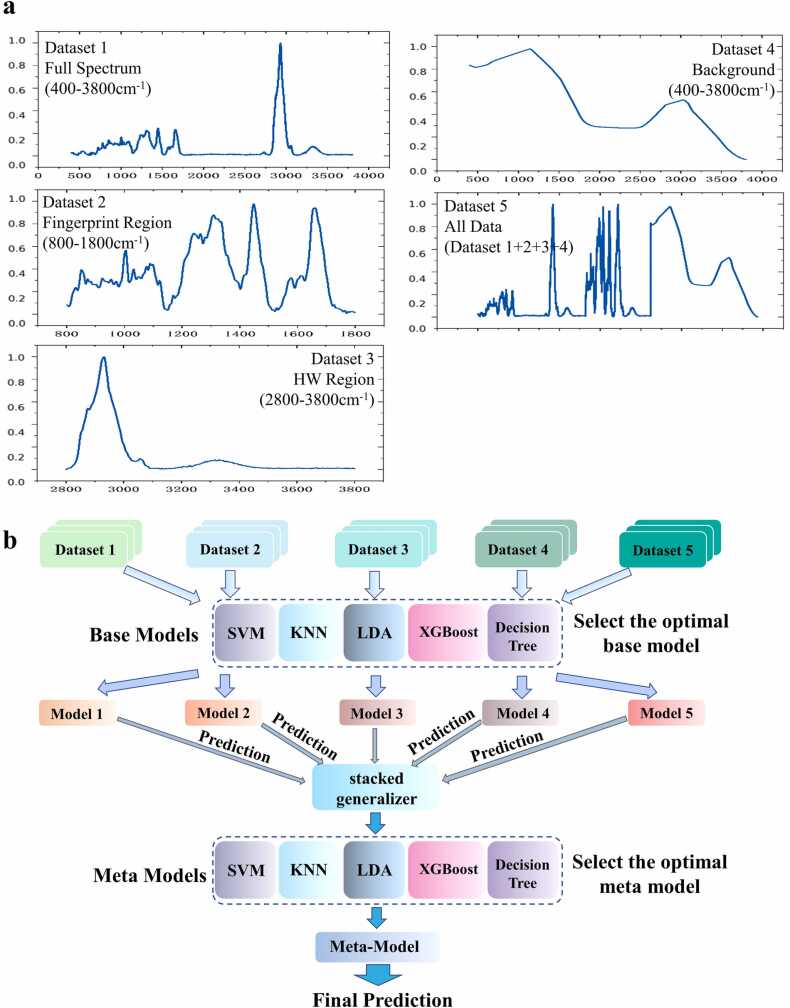
To fully utilize all the spectral information in Raman spectra, we divided the Raman data for gastric cancer cell lines into five datasets through data segmentation and merging. We used the spectral data from the 400–3800 cm−1 spectral range to form a dataset (Dataset 1, full spectrum dataset) and then selected the spectral data from the fingerprint region (800–1800 cm−1) and the HW number region (2800–3800 cm−1) as two separate datasets, namely, Dataset 2 (fingerprint region dataset) and Dataset 3 (HW region dataset). In addition, we merged the spectral backgrounds obtained with airPLS into a new dataset (Dataset 4, background dataset) and then merged Datasets 1–4 into a single dataset (Dataset 5, all data dataset). Dataset 5 exists to ensure that we have used all spectral information again.
2.5. Machine learning classification methods
We used some popular machine learning classification models in this study: SVM, KNN, LDA, eXtreme gradient boosting (XGBoost), and decision tree (DT) [27] methods. We built a classification model using the sklearn machine learning library. Notably, we trained five classification models with five different datasets and compared the classification performance of different classifiers. 528 of the 660 spectra were used as the training set and 132 as the test set during model training. The 132 spectra used as the test set were not used in the training process. In addition to this, in the model selection phase of training 5 datasets using 5 base models, both 5-fold cross validation and grid search were used during training. At the end of training, the optimal model was used to predict 132 data from the test set.
2.6. Stacking ensemble learning
Stacking is a common ensemble learning framework in data mining. In general, this process involves training a learning structure with approximately two layers. In the first layer, N different base models are cross-validated to obtain the prediction results for each sample. These predictions are then combined into a new feature set and used as input to the next layer of the classifier. Each cross-validation consists of two processes: (i) training the model based on the training data and (ii) testing the model with test data. The predicted values for all samples are obtained after all the cross-validation steps are completed.
Unlike ordinary model stacking, we performed ensemble learning using different datasets from the same sample. In this way, a model with high classification accuracy based on all available spectral information was obtained. First, we used dataset 1 to train five base models and made subsequent predictions; then, we selected the most accurate model. By analogy, the optimal model for each dataset was finally obtained. The optimal model corresponding to each dataset is directly used to train and predict each dataset, and a 5-fold cross validation is used to predict each 132 data, and the final prediction results are obtained for all samples. We combined the predictions from the five best models for each of the five datasets and input them into the second layer of each model. Next, we compared the classification results. The optimal second layer was selected by comparing the prediction results, and the optimal two-layer ensemble learning model was identified for the selected sample. To ensure that the predictions based on different datasets were associated with the same sample, we decomposed the samples prior to training and ensured that the sample order remained the same in all subsequent training sessions. We call this ensemble learning model for Raman spectroscopy SL-Raman.
3. Results
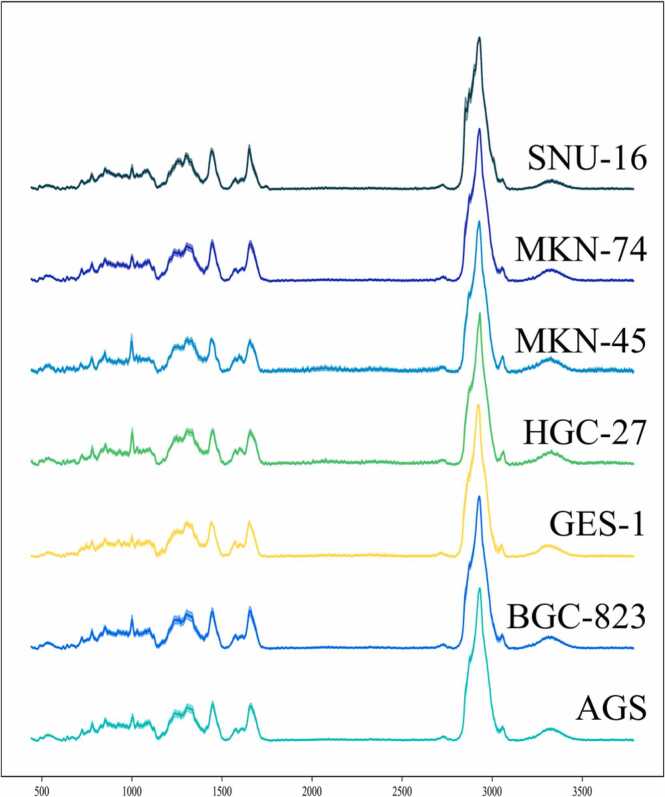
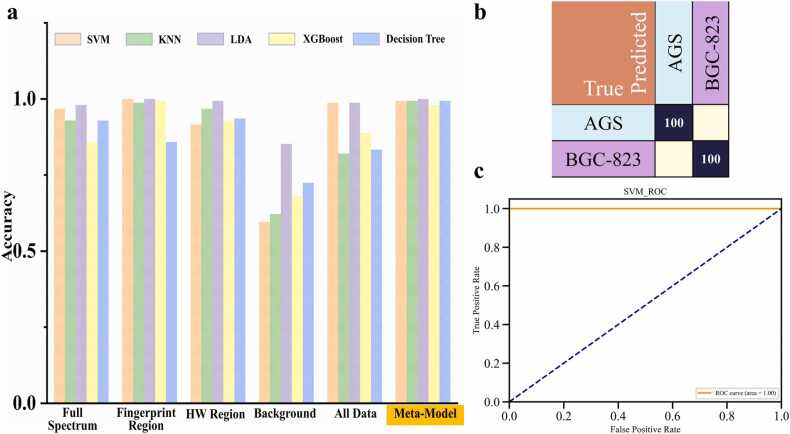
Previous Raman spectroscopy-based studies use machine learning or deep learning to classify and analyse the fingerprint region (800–1800 cm−1) of the Raman spectrum. First, the different datasets for the Raman spectra, spectral fingerprint regions, spectral HW regions and spectral backgrounds of normal and gastric cancer cells were obtained using five different machine learning methods. Second, the SL-Raman recognition model based on stacked ensemble learning for different datasets was successfully used to identify gastric cancer cells from different datasets. Finally, we analysed the differences between normal gastric epithelial cells and gastric cancer cells in the fingerprint area. Specifically, the composition of the five datasets is shown in Fig. 1.a, and the principle of the SL-Raman algorithm is shown in Fig. 1.b. Fig. 2 shows the average Raman spectra of normal gastric epithelial cells and all gastric cancer cells. The fingerprint region (800–1800 cm−1) involves most of the biomolecular vibrational modes within individual cells and exhibits unique observable features, the so-called Raman phenotype. The high wave number region (2800–3800 cm−1) is mainly associated with lipid and protein content.
Fig. 1.
The dataset composition and the algorithm used for the Raman spectrometer. a. Components of 5 datasets: Dataset 1, full spectrum dataset, 400–3800 cm−1; Dataset 2, fingerprint region dataset, 800–1800 cm−1; Dataset 3, HW region dataset, 2800–3800 cm−1; Dataset 4, background dataset; and Dataset 5, all data dataset, Datasets 1 + 2 + 3 + 4. b. Schematic diagram of the SL-Raman algorithm. The main process is as follows. (1) For each dataset, the model with the highest accuracy is selected as the base model. (2) The based model is trained using each dataset, and predictions are obtained; then, the predictions are combined into a new characteristic dataset through five-fold cross-validation. (3) The new feature dataset is input into each of the five meta-models, and the meta-model with the highest accuracy is selected for use with SL-Raman.
Fig. 2.
Average Raman spectra of normal and gastric cancer cells.
3.1. Distinguishing normal cells from single gastric cancer cells
A total of 3300 spectra and 660 average spectra were used for model training after signal-to-noise filtering. The specific number of each cell line is shown in Table 1. Raman spectral data for normal gastric mucosal epithelial cells and gastric cancer cells were distinguished using five machine learning algorithms: SVM, KNN, LDA, XGBoost and DT algorithms. Five different Raman spectral datasets were distinguished using these five classification algorithms, and the recognition accuracies are shown in Table 2. The accuracies in the table are based on the probabilities that the classification model correctly distinguished between normal GES-1 cells and each gastric cancer cell line. If we treat all the gastric cancer cell identification processes in Table 2 as 30 identification tasks and use the average accuracy as a measure of the recognition effectiveness of each machine learning model, we can conclude that the recognition accuracies of the SVM, KNN, LDA, XGBoost and DT algorithms are 98.66%, 97.73%, 99.20%, 93.61% and 95.51%, respectively. Thus, Raman spectroscopy-based and general machine learning models are effective for the identification of gastric cancer cells. Among them, LDA yielded the best identification effect.
Table 1.
The average number of spectra per cell line used for model training.
| Cell line name | Number of spectra |
|---|---|
| AGS | 98 |
| BGC-823 | 97 |
| GES-1 | 96 |
| HGC-27 | 84 |
| MKN-45 | 93 |
| MKN-74 | 89 |
| SNU-16 | 103 |
| Total | 660 |
Table 2.
Accuracy of machine learning models in identifying the Raman spectra of gastric cancer cells.
| Dataset | SVM | KNN | LDA | XGBoost | Decision Tree | |
|---|---|---|---|---|---|---|
| AGS | Full Spectrum | 100% | 99.36% | 100% | 92.95% | 100% |
| Fingerprint Region | 100% | 100% | 100% | 94.23% | 98.72% | |
| HW Region | 100% | 100% | 100% | 100% | 99.36% | |
| Background | 96.15% | 94.87% | 96.79% | 90.38% | 87.18% | |
| All Data | 97.44% | 96.79% | 99.36% | 92.95% | 88.46% | |
| BGC-823 | Full Spectrum | 100% | 100% | 100% | 94.84% | 100% |
| Fingerprint Region | 100% | 97.5% | 100% | 89.03% | 92.90% | |
| HW Region | 100% | 99.35% | 100% | 99.35% | 100% | |
| Background | 96.13% | 94.19% | 89.03% | 91.61% | 91.61% | |
| All Data | 96.77% | 97.5% | 100% | 94.84% | 100% | |
| HGC-27 | Full Spectrum | 100% | 100% | 100% | 95.14% | 94.44% |
| Fingerprint Region | 100% | 100% | 100% | 95.83% | 100% | |
| HW Region | 100% | 100% | 100% | 93.06% | 100% | |
| Background | 98.61% | 95.83% | 96.53% | 95.14% | 93.75% | |
| All Data | 100% | 98.61% | 100% | 95.14% | 97.22% | |
| MKN-45 | Full Spectrum | 97.14% | 95.39% | 99.34% | 87.5% | 94.08% |
| Fingerprint Region | 99.34% | 99.34% | 100% | 96.71% | 88.16% | |
| HW Region | 92.76% | 98.03% | 100% | 95.39% | 95.39% | |
| Background | 96.71% | 87.5% | 98.68% | 84.87% | 91.48% | |
| All Data | 98.03% | 90.13% | 99.34% | 87.5% | 91.48% | |
| MKN-74 | Full Spectrum | 100% | 100% | 100% | 93.24% | 100% |
| Fingerprint Region | 100% | 99.32% | 100% | 100% | 100% | |
| HW Region | 100% | 100% | 100% | 95.95% | 97.97% | |
| Background | 95.95% | 95.27% | 99.32% | 92.27% | 95.27% | |
| All Data | 96.62% | 95.95% | 100% | 93.34% | 94.59% | |
| SNU-16 | Full Spectrum | 100% | 100% | 100% | 91.88% | 96.25% |
| Fingerprint Region | 100% | 99.38% | 100% | 86.88% | 91.25% | |
| HW Region | 100% | 100% | 100% | 100% | 98.75% | |
| Background | 99.38% | 98.75% | 97.5% | 96.25% | 94.38% | |
| All Data | 98.75% | 98.75% | 100% | 91.88% | 92.5% |
We then compared the overall classification results of different models for the same dataset. The recognition accuracies of the different models for the full spectrum dataset, fingerprint region dataset, HW region dataset, background dataset and all data dataset were 97.72%, 97.62%, 98.85%, 94.38% and 96.13%, respectively. That is, in a binary classification task with high signal noise in Raman spectral data, the full spectrum, fingerprint region and HW region were all effectively used to identify the differences in spectral data. In this classification task, the highest classification accuracy was achieved with the HW region dataset. In addition, five classification models reached an accuracy of 94.38% in the task of identifying gastric cancer cell lines using the background dataset. This result suggests that the Raman spectral background is biologically informative [22].
When distinguishing AGS gastric cancer cells from GES-1 normal cells, we found that the LDA classification model yielded the highest recognition accuracy for different datasets. We then chose LDA as the base classification model for this round of SL-Raman modelling. Five predictions were made for each of the five datasets using five-fold cross-validation and LDA for each sample. The prediction results were combined into a new feature set and then input into the five classification models, namely, the SVM, KNN, LDA, XGBoost and DT models, and the classification accuracies were all 100%. Therefore, in this round of recognition tasks, we obtained particularly high accuracy values regardless of which meta-model we chose. The process for identifying other gastric cancer cells was similar. All identification results are shown in Table 3. After choosing the most effective machine learning model as the base model for SL-Raman, the fast and simple KNN meta-model was used to obtain good recognition results.
Table 3.
Accuracy of gastric cancer cell identification with SL-Raman and different meta-models.
| SVM | KNN | LDA | XGBoost | Decision Tree | |
|---|---|---|---|---|---|
| AGS | 100% | 100% | 100% | 100% | 100% |
| BGC-823 | 100% | 100% | 100% | 100% | 98.71% |
| HGC-27 | 100% | 100% | 100% | 100% | 100% |
| MKN-45 | 99.34% | 100% | 100% | 98.68% | 100% |
| MKN-74 | 100% | 100% | 100% | 100% | 100% |
| SNU-16 | 100% | 100% | 100% | 100% | 99.38% |
3.2. Distinguishing normal cells from multiple types of gastric cancer cells
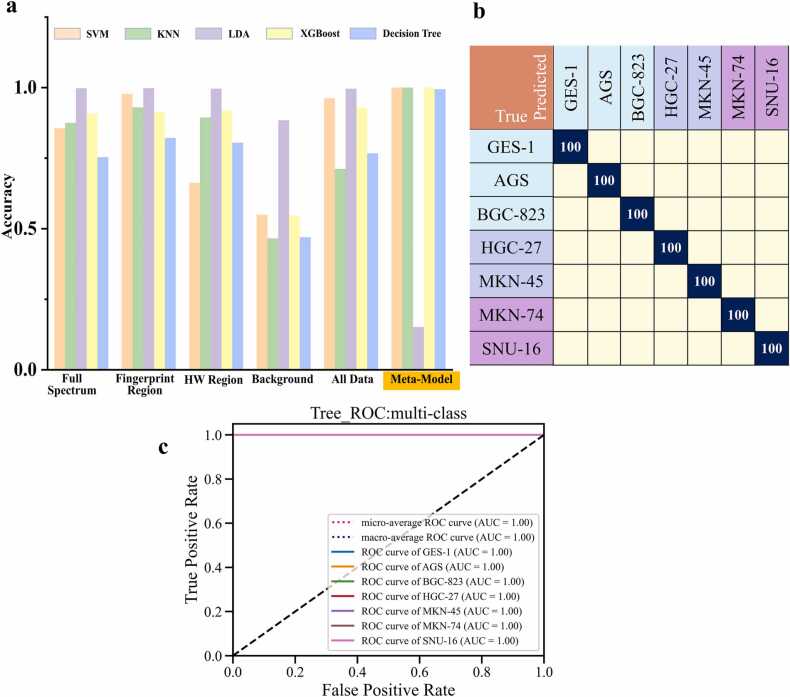
To validate the classification capability of SL-Raman in different cases, we trained the model on a total of seven cell line datasets for normal and gastric cancer. The resulting classification accuracy, confusion matrix and receiver operating characteristic (ROC) curve are shown in Fig. 3. The recognition accuracies of the SVM for the different datasets in Fig. 3a were 85.61%, 97.73%, 66.29%, 54.92% and 96.21%, respectively; those for the KNN approach were 87.5%, 92.99%, 89.39%, 46.59% and 71.21%, respectively; those for LDA were 99.81%, 99.81%, 99.62%, 88.45% and 99.62%, respectively; those for XGBoost were 90.72%, 91.29%, 91.86%, 54.73% and 92.80%, respectively; and those for the DT were75.38%, 82.20%, 80.49%, 46.97% and 76.70%, respectively. When LDA was used as the base model for SL-Raman, the SL-Raman accuracies for different meta-models were 100%, 100%, 15.15%, 100% and 99.43%, respectively. The final SL-Raman base model was LDA, and the meta-model was selected was the KNN, with a model accuracy of 100%.
Fig. 3.
Results of SL-Raman and different machine learning models in the identification of 7 cell lines. a. SL-Raman recognition accuracy for different models. b. Confusion matrix for the recognition of gastric cancer cells with SL-Raman. c. ROC curve for SL-Raman classification.
3.3. Distinguishing gastric cancer cells with different degrees of differentiation
To verify the ability of SL-Raman to identify gastric cancer cells with different degrees of differentiation, we constructed datasets for the gastric cancer cell lines AGS (highly differentiated) and BGC-823 (poorly differentiated). The classification accuracy, confusion matrix and ROC curve results are shown in Fig. 4. As shown in Fig. 4a, the recognition accuracies of the SVM for different datasets were 96.79%, 100%, 91.67%, 59.62% and 98.72%, respectively; those of the KNN were 92.95%, 98.72%, 96.79%, 62.18% and 82.05%, respectively; those of LDA were 98.08%, 100%, 99.36%, 85.26% and 98.72%, respectively; those of XGBoost were 85.90%, 99.36%, 92.95%, 67.95% and 88.90%, respectively; and those of the DT were 92.95%, 85.90%, 93.59%, 72.44% and 83.33%, respectively. When LDA was used as the base model for SL-Raman, the SL-Raman accuracies for different meta-models were 99.36%, 99.36%, 100%, 98.08% and 99.36%. The final SL-Raman base model and meta-model were both LDA, and the model accuracy was 100%.
Fig. 4.
Results of SL-Raman and different machine learning models in identifying gastric cancer cell lines with different degrees of differentiation. a. SL-Raman recognition accuracy for different models. b. Confusion matrix for the identification of gastric cancer cells with SL-Raman. c. ROC curve for SL-Raman classification.
3.4. Raman spectroscopy of gastric cancer cells
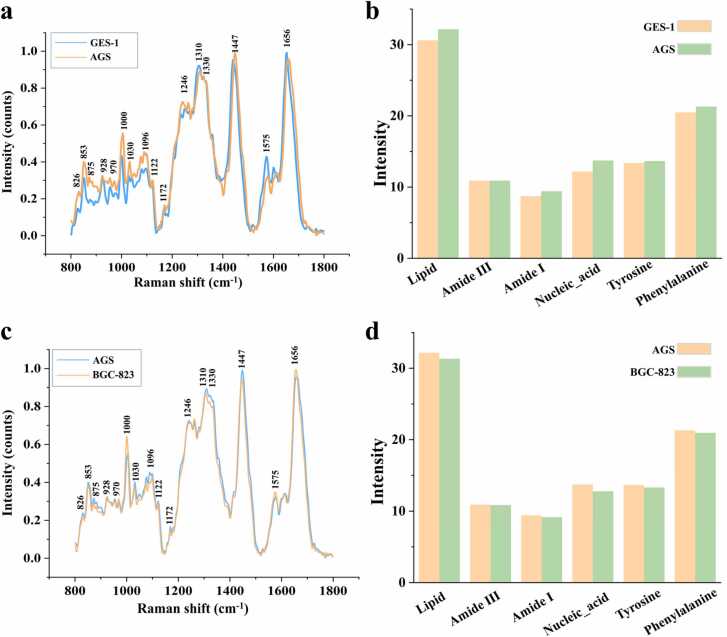
Raman spectroscopy can be used to identify cell lines and assess the biochemical composition of cells. As presented in Fig. 5a and Table 4, there was a significant difference in the spectra of normal and gastric cancer cells. The spectral intensity of gastric cancer cells was slightly higher than that of normal gastric epithelial cells in the range of 800–1300 cm−1. The Raman intensities at several distinct Raman peak positions, such as 853, 970, 1000, 1030, 1096, 1246, 1575 and 1656 cm−1, were also significantly different. We summed the intensity of the characteristic peak ranges to intuitively compare the overall variations in the different biochemical components. The spectral regions of the characteristic peaks were based on those suggested by Ye, J.[38]. The sums of the Raman characteristic peak intensities of six major biomolecular components, namely, lipids, amide III, amide I, nucleic acid, tyrosine and phenylalanine, for normal and gastric cancer cells are shown in Fig. 5.b. As the results show, the gastric cancer cells had more lipids, nucleic acids and phenylalanine. This finding also indicates that gastric cancer cells need more energy than non-cancer cells.
Fig. 5.
Raman spectroscopy analysis. a. Average Raman spectrum of the normal gastric mucosal epithelial cell line GES-1 compared to that for the gastric cancer cell line AGS. b. Sum of the Raman peak intensities of the major biomolecules of GES-1 and AGS. c. Comparison of the average Raman spectra for two gastric cancer cell lines with different degrees of differentiation: AGS and BGC-823. d. Sum of the Raman peak intensities of major biomolecules for AGS and BGC-823.
Table 4.
Peak positions and tentative assignments of the major Raman bands from biological samples.
| Peak position, cm−1 | Major Assignments |
|---|---|
| 826[39] | O-P-O stretch DNA |
| 853[40] | Ring breathing mode of tyrosine and C-C stretch of proline ring |
| 875[41] | Antisymmetric stretch vibration of choline group N + (CH3)3, characteristic for phospholipids Phosphatidylcholine, sphingomyelin |
| 928[42] | ν(C-C), stretching-probably in amino acids proline & valine (protein band) |
| 970[43] | Phosphate monoester groups of phosphorylated proteins & cellular nucleic acids |
| 1000[44] | Phenylalanine Bound & free NADH |
| 1030[45] | Phenylalanine of collagen |
| 1096[46] | O-P-O (stretching PO2) symmetric (Phosphate II) of phosphodiesters |
| 1122[47] | νsym(C-O-C) (polysaccharides, cellulose) |
| 1172[48] | δ(C-H), tyrosine (protein assignment) |
| 1246[45] | Amide III (of collagen) |
| 1310[49] | CH3CH2 twisting mode of collagen/lipids |
| 1330[44], [50], [51] | Typical phospholipids Region associated with DNA & phospholipids Collagen Nucleic acids and phosphates |
| 1447[52] | CH2 bending mode of proteins. CH2 (overlapping) asymmetric CH3 bending, and CH2 scissoring (associated with elastin, collagen, and phospholipids) |
| 1575[53] | Ring breathing modes in the DNA bases G, A (ring breathing modes of the DNA/RNA bases) |
| 1656[52] | Amide I (C = O stretching mode of proteins, a-helix conformation)/C = C lipid stretch |
Fig. 5c and d show the Raman spectra of two gastric cancer cells with different degrees of differentiation: AGS (highly differentiated) and BGC-823 (poorly differentiated). Highly differentiated AGS cells contained more nucleic acid and lipids.
4. Discussion
Raman spectroscopy can be used for the identification of pathogenic bacteria and malignant tumours. Lihao Zhang et al. used Raman spectroscopy to identify breast cancer cell lines with an accuracy of 97% [23]. However, they downscaled the Raman spectral data before analysis, which resulted in the loss of many spectral features. In this study, instead of downscaling the spectral data, we constructed multiple datasets and implemented operations to expand data features so that the recognition model could better identify spectral features. SL-Raman integrates features from datasets with different spectral ranges and machine learning classification models that yield good classification results to obtain precise, accurate and stable recognition results.
We not only compared the classification ability of different classification models for the fingerprint region of Raman spectra but also verified the ability of machine learning models to classify the HW region of these spectra, the full spectra, and the Raman background. The accuracies of the SVM, KNN, LDA, XGBoost and DT algorithms in distinguishing gastric cancer cells from normal cells were 98.66%, 97.73%, 99.20%, 93.61% and 95.51%, respectively. The classification accuracy of Raman background data also reached 94.38%. The accuracy of SL-Raman based on stacking-ensemble learning for normal cells and gastric cancer cells from different datasets was 100%. The accuracy of SL-Raman in differentiating seven cell types was 100%, and the accuracy of differentiating two different gastric cancer cell lines was 100%.
The high accuracy obtained by SL-Raman in identifying the Raman spectra of gastric cancer cells also reflects the differences in chemical composition among different cells. This differentiation was the main objective of this study, and a powerful cell line identification technique for classifying Raman spectroscopy data was established. Overall, SL-Raman is more accurate than most machine learning algorithms and is able to fully utilize all the characteristic information in Raman spectral data.
Our current work is still based on Raman spectroscopy detection in pure samples, but this is only at the method exploration stage. We are figuring out the ratio of mixed samples, sample handling process, and spectral acquisition conditions. The use of Raman spectroscopy for cell identification in mixed samples is the next research we are working on. In addition, one of the main advantages of Raman spectroscopy is the ability to directly detect samples in liquids, and using Raman spectroscopy in combination with optical tweezers and microfluidics for liquid detection of biological samples is one of our next research efforts.
5. Conclusions
Raman spectroscopy can be used to identify differences in the biotic component of biological samples. Raman spectroscopy combined with SL-Raman enables the identification of normal gastric epithelial cells and gastric cancer cells. SL-Raman achieves 100% accuracy in distinguishing one gastric cancer cell line from normal cells, 100% accuracy in distinguishing six gastric cancer cell lines from normal cells and 100% accuracy in distinguishing two gastric cancer cell lines with different degrees of differentiation. The findings show that SL-Raman successfully integrates the benefits of various machine learning techniques with the data from a multidimensional spectral dataset. The technique offers a novel way to analyze Raman spectrum data since it can achieve high recognition accuracy with a small amount of data. In addition, the accuracy of classifying the Raman spectral background using machine learning is 94.38%. These results suggest that the Raman spectral background also contains some useful features that may be related to the fluorescent background of the biological samples. In subsequent research, we will continue to build a Raman spectral database that includes other human cells so that Raman spectroscopy can be used in new ways for cell lineage identification.
Funding
The work was supported by the National key R&D Program of China (2021YFA1302100 to Q.Z); the National Natural Science Foundation of China (82172861 to Q.Z); Guangdong Basic and Applied Basic Research Foundation (2021A1515011743 to Q.Z).
CRediT authorship contribution statement
Kunxiang Liu: Formal analysis, Writing – original draft, Visualization, Implementation. Bo Liu: Methodology. Yuhong Zhang: Experiment. Qinian Wu: Experiment. Ming Zhong: Experiment. Lindong Shang: Writing – original draft. Yu Wang: Formal analysis. Peng Liang: Formal analysis. Weiguo Wang: Writing – review & editing. Qi Zhao: Writing – review & editing, Funding acquisition. Bei Li: Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Qi Zhao, Email: zhaoqi@sysucc.org.cn.
Bei Li, Email: beili@ciomp.ac.cn.
References
- 1.Fera B.D., Liu H., Tian F. Abstract 1648: Checkpoint molecule profiling in tumor cell lines and immune cell lines for application in immuno-oncology drug screening. Cancer Res. 2021;81(13_Supplement) 1648-1648. [Google Scholar]
- 2.Linnenbach A.J., Huebner K., Reddy E.P., Herlyn M., Parmiter A.H., Nowell P.C., Koprowski H. Structural alteration in the MYB protooncogene and deletion within the gene encoding alpha-type protein kinase C in human melanoma cell lines. Proc Natl Acad Sci U S A. 1988;85(1):74–78. doi: 10.1073/pnas.85.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrara S.C., Ulitzka M., Grzeschik J., Kornmann H., Hock B., Kolmar H. From cell line development to the formulated drug product: The art of manufacturing therapeutic monoclonal antibodies. Int J Pharm. 2021;594 doi: 10.1016/j.ijpharm.2020.120164. [DOI] [PubMed] [Google Scholar]
- 4.Gurgul-Convey E., Mehmeti I., Plotz T., Jorns A., Lenzen S. Sensitivity profile of the human EndoC-betaH1 beta cell line to proinflammatory cytokines. Diabetologia. 2016;59(10):2125–2133. doi: 10.1007/s00125-016-4060-y. [DOI] [PubMed] [Google Scholar]
- 5.Barallon R., Bauer S.R., Butler J., Capes-Davis A., Dirks W.G., Elmore E., Furtado M., Kline M.C., Kohara A., Los G.V., MacLeod R.A., Masters J.R., Nardone M., Nardone R.M., Nims R.W., Price P.J., Reid Y.A., Shewale J., Sykes G., Steuer A.F., Storts D.R., Thomson J., Taraporewala Z., Alston-Roberts C., Kerrigan L. Recommendation of short tandem repeat profiling for authenticating human cell lines, stem cells, and tissues. In Vitro Cell Dev Biol Anim. 2010;46(9):727–732. doi: 10.1007/s11626-010-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nims R.W., Shoemaker A.P., Bauernschub M.A., Rec L.J., Harbell J.W. Sensitivity of isoenzyme analysis for the detection of interspecies cell line cross-contamination. In Vitro Cell Dev Biol Anim. 1998;34(1):35–39. doi: 10.1007/s11626-998-0050-9. [DOI] [PubMed] [Google Scholar]
- 7.Poppema S., Bhan A.K., Reinherz E.L., McCluskey R.T., Schlossman S.F. Distribution of T cell subsets in human lymph nodes. J Exp Med. 1981;153(1):30–41. doi: 10.1084/jem.153.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim T.O., Greenwood M., Eagar T., Despotovic J.M. Novel HLA Typing Method Identifies HLA Alleles Associated with Pediatric ITP. Blood. 2019;134(Supplement_1) 1067-1067. [Google Scholar]
- 9.Edwards A., Civitello A., Hammond H.A., Caskey C.T. DNA typing and genetic mapping with trimeric and tetrameric tandem repeats. Am J Hum Genet. 1991;49(4):746. [PMC free article] [PubMed] [Google Scholar]
- 10.Raman C.V., Krishnan K.S., New A. Type of secondary radiation. Nature. 1928;121(3048):501–502. [Google Scholar]
- 11.Araujo C.F., Nolasco M.M., Ribeiro A.M.P., Ribeiro-Claro P.J.A. Identification of microplastics using Raman spectroscopy: Latest developments and future prospects. Water Res. 2018;142:426–440. doi: 10.1016/j.watres.2018.05.060. [DOI] [PubMed] [Google Scholar]
- 12.Pahlow S., Meisel S., Cialla-May D., Weber K., Rosch P., Popp J. Isolation and identification of bacteria by means of Raman spectroscopy. Adv Drug Deliv Rev. 2015;89:105–120. doi: 10.1016/j.addr.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Saletnik A., Saletnik B., Puchalski C. Raman Method in Identification of Species and Varieties, Assessment of Plant Maturity and Crop Quality-A Review. Molecules. 2022;27(14):4454. doi: 10.3390/molecules27144454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W.T., Zhang H., Yuan Y., Guo Y., He S.X. Research progress of Raman spectroscopy in drug analysis. AAPS PharmSciTech. 2018;19(7):2921–2928. doi: 10.1208/s12249-018-1135-8. [DOI] [PubMed] [Google Scholar]
- 15.Xu Y., Zhong P., Jiang A., Shen X., Li X., Xu Z., Shen Y., Sun Y., Lei H. Raman spectroscopy coupled with chemometrics for food authentication: A review. TrAC Trends Analytical Chem. 2020;131 [Google Scholar]
- 16.Shang L., Xu L., Wang Y., Liu K., Liang P., Zhou S., Chen F., Peng H., Zhou C., Lu Z.-M., Li B. Rapid detection of beer spoilage bacteria based on label-free SERS technology. Anal Methods. 2022 doi: 10.1039/d2ay01221a. [DOI] [PubMed] [Google Scholar]
- 17.Lin K., Wang J., Zheng W., Ho K.Y., Teh M., Yeoh K.G., Huang Z. Rapid Fiber-optic Raman Spectroscopy for Real-Time In Vivo Detection of Gastric Intestinal Metaplasia during Clinical Gastroscopy. Cancer Prev Res ((Phila)) 2016;9(6):476–483. doi: 10.1158/1940-6207.CAPR-15-0213. [DOI] [PubMed] [Google Scholar]
- 18.Zhou X., Dai J., Chen Y., Duan G., Liu Y., Zhang H., Wu H., Peng G. Evaluation of the diagnostic potential of ex vivo Raman spectroscopy in gastric cancers: fingerprint versus high wavenumber. J Biomed Opt. 2016;21(10) doi: 10.1117/1.JBO.21.10.105002. [DOI] [PubMed] [Google Scholar]
- 19.Bocklitz T., Walter A., Hartmann K., Rosch P., Popp J. How to pre-process Raman spectra for reliable and stable models? Anal Chim Acta 704( 2011;(1–2):47–56. doi: 10.1016/j.aca.2011.06.043. [DOI] [PubMed] [Google Scholar]
- 20.Liu K., Zhao Q., Li B., Zhao X. Raman spectroscopy: a novel technology for gastric cancer diagnosis. Front Bioeng Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.856591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J., Xu J., Zhang X., Wu C., Lin T., Ying Y. Deep learning for vibrational spectral analysis: Recent progress and a practical guide. Anal Chim Acta. 2019;1081:6–17. doi: 10.1016/j.aca.2019.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Buchwald T., Buchwald Z., Daktera-Micker A. The fluorescence background in Raman spectra of sound enamel. Vibrational Spectrosc. 2021;115 [Google Scholar]
- 23.Zhang L., Li C., Peng D., Yi X., He S., Liu F., Zheng X., Huang W.E., Zhao L., Huang X. Raman spectroscopy and machine learning for the classification of breast cancers. Spectrochim Acta A Mol Biomol Spectrosc. 2022;264 doi: 10.1016/j.saa.2021.120300. [DOI] [PubMed] [Google Scholar]
- 24.Crow P., Barrass B., Kendall C., Hart-Prieto M., Wright M., Persad R., Stone N. The use of Raman spectroscopy to differentiate between different prostatic adenocarcinoma cell lines. Br J Cancer. 2005;92(12):2166–2170. doi: 10.1038/sj.bjc.6602638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talari A.C.S., Evans C.A., Holen I., Coleman R.E., Rehman I.U. Raman spectroscopic analysis differentiates between breast cancer cell lines. J Raman Spectrosc. 2015;46(5):421–427. [Google Scholar]
- 26.Liu B., Liu K., Wang N., Ta K., Liang P., Yin H., Li B. Laser tweezers Raman spectroscopy combined with deep learning to classify marine bacteria. Talanta. 2022;244 doi: 10.1016/j.talanta.2022.123383. [DOI] [PubMed] [Google Scholar]
- 27.Ralbovsky N.M., Lednev I.K. Towards development of a novel universal medical diagnostic method: Raman spectroscopy and machine learning. Chem Soc Rev. 2020;49(20):7428–7453. doi: 10.1039/d0cs01019g. [DOI] [PubMed] [Google Scholar]
- 28.Wold S., Esbensen K., Geladi P. Principal component analysis. Chemometr Intell Lab Syst. 1987;2(1–3):37–52. [Google Scholar]
- 29.Szymańska E., Gerretzen J., Engel J., Geurts B., Blanchet L., Buydens L.M. Chemometrics and qualitative analysis have a vibrant relationship. TrAC Trends Anal Chem. 2015;69:34–51. [Google Scholar]
- 30.Riaz S., Arshad A., Jiao L., Semi-Supervised A., With C.N.N. Fuzzy Rough C-Mean for Image Classification. IEEE Access. 2019;7:49641–49652. [Google Scholar]
- 31.Cao H., Gu Y., Fang J., Hu Y., Ding W., He H., Chen G. Application of stacking ensemble learning model in quantitative analysis of biomaterial activity. Microchem J. 2022;183 [Google Scholar]
- 32.Haghighi F., Omranpour H. Stacking ensemble model of deep learning and its application to Persian/Arabic handwritten digits recognition. Knowledge-Based Syst. 2021;220 [Google Scholar]
- 33.Ganaie M.A., Hu M., Malik A.K., Tanveer M., Suganthan P.N. Ensemble deep learning: A review. Eng Appl Artificial Intelligence. 2022;115 [Google Scholar]
- 34.L. Hansen, P. Salamon, Neural network ensembles IEEE Transactions on Pattern Analysis and M achine Intellidence, 1990.
- 35.Liu Y.J., Kyne M., Wang C., Yu X.Y. Data mining in Raman imaging in a cellular biological system. Comput Struct Biotechnol J. 2020;18:2920–2930. doi: 10.1016/j.csbj.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorry P.A. General least-squares smoothing and differentiation by the convolution (Savitzky-Golay) method. Analytical Chem. 2002;62(6):570–573. [Google Scholar]
- 37.Zhang Z.M., Chen S., Liang Y.Z. Baseline correction using adaptive iteratively reweighted penalized least squares. Analyst. 2010;135(5):1138–1146. doi: 10.1039/b922045c. [DOI] [PubMed] [Google Scholar]
- 38.Ye J., Yeh Y.T., Xue Y., Wang Z., Zhang N., Liu H., Zhang K., Ricker R., Yu Z., Roder A., Perea Lopez N., Organtini L., Greene W., Hafenstein S., Lu H., Ghedin E., Terrones M., Huang S., Huang S.X. Accurate virus identification with interpretable Raman signatures by machine learning. Proc Natl Acad Sci U S A. 2022;119(23) doi: 10.1073/pnas.2118836119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stone N., Kendall C., Smith J., Crow P., Barr H. Raman spectroscopy for identification of epithelial cancers. Faraday Discuss. 2004;126:141–157. doi: 10.1039/b304992b. discussion 169-83. [DOI] [PubMed] [Google Scholar]
- 40.Stone N., Kendall C., Shepherd N., Crow P., Barr H. Near-infrared Raman spectroscopy for the classification of epithelial pre-cancers and cancers. J Raman Spectrosc. 2002;33(7):564–573. [Google Scholar]
- 41.Krafft C., Neudert L., Simat T., Salzer R. Near infrared Raman spectra of human brain lipids. Spectrochim Acta A Mol Biomol Spectrosc. 2005;61(7):1529–1535. doi: 10.1016/j.saa.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 42.Lau D.P., Huang Z., Lui H., Man C.S., Berean K., Morrison M.D., Zeng H. Raman spectroscopy for optical diagnosis in normal and cancerous tissue of the nasopharynx-preliminary findings. Lasers Surg Med. 2003;32(3):210–214. doi: 10.1002/lsm.10084. [DOI] [PubMed] [Google Scholar]
- 43.R.K. Dukor, Vibrational Spectroscopy in the Detection of Cancer, Handbook of Vibrational Spectroscopy, 2006.
- 44.Malini R., Venkatakrishna K., Kurien J., Pai K.M., Rao L., Kartha V.B., Krishna C.M. Discrimination of normal, inflammatory, premalignant, and malignant oral tissue: a Raman spectroscopy study. Biopolymers. 2006;81(3):179–193. doi: 10.1002/bip.20398. [DOI] [PubMed] [Google Scholar]
- 45.Cheng W.T., Liu M.T., Liu H.N., Lin S.Y. Micro-Raman spectroscopy used to identify and grade human skin pilomatrixoma. Microsc Res Tech. 2005;68(2):75–79. doi: 10.1002/jemt.20229. [DOI] [PubMed] [Google Scholar]
- 46.Liu C.H., Zhou Y., Sun Y., Li J.Y., Zhou L.X., Boydston-White S., Masilamani V., Zhu K., Pu Y., Alfano R.R. Resonance Raman and Raman spectroscopy for breast cancer detection. Technol Cancer Res Treat. 2013;12(4):371–382. doi: 10.7785/tcrt.2012.500325. [DOI] [PubMed] [Google Scholar]
- 47.Shetty G., Kendall C., Shepherd N., Stone N., Barr H. Raman spectroscopy: elucidation of biochemical changes in carcinogenesis of oesophagus. Br J Cancer. 2006;94(10):1460–1464. doi: 10.1038/sj.bjc.6603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang Z., McWilliams A., Lui H., McLean D.I., Lam S., Zeng H. Near-infrared Raman spectroscopy for optical diagnosis of lung cancer. Int J Cancer. 2003;107(6):1047–1052. doi: 10.1002/ijc.11500. [DOI] [PubMed] [Google Scholar]
- 49.Qiu S., Huang Q., Huang L., Lin J., Lu J., Lin D., Cao G., Chen C., Pan J., Chen R. Label-free discrimination of different stage nasopharyngeal carcinoma tissue based on Raman spectroscopy. Oncol Lett. 2016;11(4):2590–2594. doi: 10.3892/ol.2016.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrus P.G., Strickland R.D. Cancer grading by Fourier transform infrared spectroscopy. Biospectroscopy. 1998;4(1):37–46. doi: 10.1002/(sici)1520-6343(1998)4:1<37::aid-bspy4>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 51.Utzinger U., Heintzelman D.L., Mahadevan-Jansen A., Malpica A., Follen M., Richards-Kortum R. Near-Infrared Raman Spectroscopy for in vivo Detection of Cervical Precancers. Applied Spectroscopy. 2016;55(8):955–959. [Google Scholar]
- 52.Surmacki J., Musial J., Kordek R., Abramczyk H. Raman imaging at biological interfaces: applications in breast cancer diagnosis. Mol Cancer. 2013;12(1):48. doi: 10.1186/1476-4598-12-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan J.W., Taylor D.S., Zwerdling T., Lane S.M., Ihara K., Huser T. Micro-Raman spectroscopy detects individual neoplastic and normal hematopoietic cells. Biophys J. 2006;90(2):648–656. doi: 10.1529/biophysj.105.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]