Abstract
Epigenetic remodeling is a molecular hallmark of gliomas, and it has been identified as a key mediator of glioma progression. Epigenetic dysregulation contributes to gliomagenesis, tumor progression, and responses to immunotherapies, as well as determining clinical features. This epigenetic remodeling includes changes in histone modifications, chromatin structure, and DNA methylation, all of which are driven by mutations in genes such as histone 3 genes (H3C1 and H3F3A), isocitrate dehydrogenase 1/2 (IDH1/2), α-thalassemia/mental retardation, X-linked (ATRX), and additional chromatin remodelers. Although much of the initial research primarily identified how the epigenetic aberrations impacted glioma progression by solely examining the glioma cells, recent studies have aimed at establishing the role of epigenetic alterations in shaping the tumor microenvironment (TME). In this review, we discuss the mechanisms by which these epigenetic phenomena in glioma remodel the TME and how current therapies targeting epigenetic dysregulation affect the glioma immune response and therapeutic outcomes. Understanding the link between epigenetic remodeling and the glioma TME provides insights into the implementation of epigenetic-targeting therapies to improve the antitumor immune response.
Introduction
Gliomas are the most common type of primary tumors originating in the central nervous system (CNS) (1). They are characterized as highly heterogeneous tumors, both biologically and morphologically. Gliomas are classified according to the WHO classification system in grades 1 to 4, indicating their growth rate and aggressiveness (2). While non-diffuse gliomas can be curable when resected entirely, diffuse gliomas, characterized by their infiltrative nature, are difficult to treat owing to the critical neural structures involved and the limited access of drugs to the brain. Thus complete surgical resection of diffuse gliomas is almost always impossible. For these reasons, recurrence of surgically removed diffuse gliomas is almost unavoidable, and patient outcomes have not improved significantly in recent decades, unlike with other cancers (3). Glioma progression and patient survival depend on the glioma location, the patient’s age, glioma cells’ genetic mutations, and epigenetic dysregulation.
We first describe the distinct adult and pediatric glioma microenvironments. Then we examine how epigenetic mechanisms modulate the glioma tumor microenvironment (TME) (Figure 1). We also cover the impact of epigenetic-targeting therapies on the immune response. This Review serves to discuss the literature on the impact of epigenetic dysregulation in glioma on several aspects of the glioma microenvironment. The mechanistic aspects of the epigenetic reprogramming have been reviewed in detail in refs. 4–6.
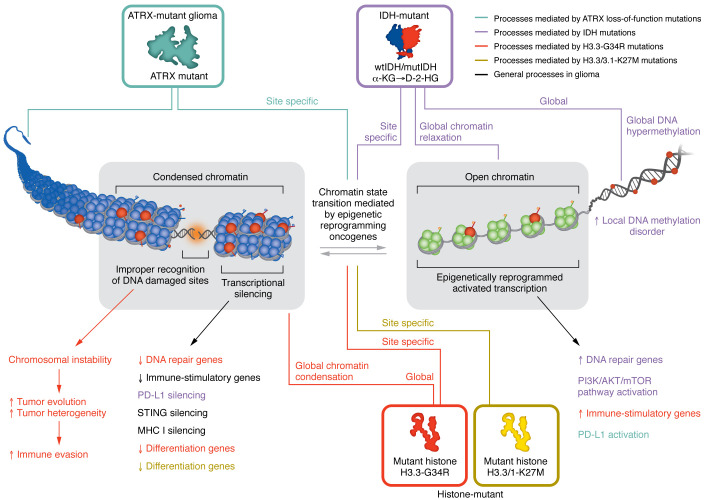
Figure 1. Effects of epigenetic mutations in molecular mechanisms that modulate immune cell activity within the TME.
The effects of epigenetic mutations on chromatin are indicated by color-coded arrows. The molecular effects of epigenetic changes that affect the immune TME are indicated below the chromatin.
Adult gliomas
It is well established that gliomas in adults generally differ both clinically and molecularly from pediatric gliomas. Adult gliomas have an incidence rate of about 5.0 per 100,000 people (7). The most prevalent subtype, glioblastoma (GBM), accounts for 57.7% of gliomas and has the most dismal median survival of approximately 15 months, even with standard-of-care treatments (8).
Adult diffuse gliomas comprise 3 types: astrocytoma, IDH-mutant; oligodendroglioma, IDH-mutant and 1p/19q-codeleted; and glioblastoma, IDH-wild-type — which includes the genetic modifiers, the mutation status of isocitrate dehydrogenase (IDH) genes, and codeletion of chromosome arms 1p and 19q (1p/19q codeletion) (2, 9, 10). Other molecular profiles that are characteristically altered in adult gliomas include ATRX, TP53, TERT promoter, CDKN2A/B, EGFR, and chromosomes 7 and 10. The most common mutation of IDH is in IDH gene 1, typically at arginine 132 to histidine (IDH1-R132H; mIDH1) (11). mIDH1 astrocytoma patients survive longer than patients with wild-type IDH (wtIDH) (12). In astrocytomas, mIDH occurs early in gliomagenesis and is accompanied by several other mutations, including ATRX, TP53, and CDKN2A/B (13, 14). In oligodendroglioma, the mIDH and 1p/19q codeletion are often accompanied by mutations in TERT promoter, FUBP1, and NOTCH1 (9, 11). Mutations in TERT promoter, EGFR, and chromosomes 7 and 10 are associated with GBM (2, 9). Molecular alterations are used in tandem with histopathological characteristics to determine the glioma grade.
Pediatric high-grade gliomas
Pediatric high-grade gliomas (pHGGs) constitute 8%–12% of all CNS pediatric tumors and have an incidence of approximately 0.85 per 100,000 children (15, 16). pHGG prognosis is dismal, with an overall median survival of approximately 10 to 18 months, and the 2-year survival rate is approximately 32% for hemispheric tumors versus approximately 10% to 22% for midline tumors (17). Despite intense research efforts, currently there are no effective treatment options. pHGG remains the leading cause of cancer-related death in children and adolescents under 19 (15, 18).
Many epigenetic-related mutations are seen more commonly (in some cases almost exclusively) in pediatric gliomas, particularly mutations in histone 3 (H3) genes. These mutations also include chromatin remodelers such as SETD2; α-thalassemia/mental retardation, X-linked (ATRX); and DAXX (5, 17, 19, 20). Midline diffuse gliomas typically harbor the H3.1-K27M or the H3.3-K27M histone mutation, while diffuse hemispheric pHGGs contain the H3.3-G34R/V mutation. ATRX and DAXX are often seen in pHGG and are associated with telomere lengthening (5, 20).
The unique microenvironment of glioma
Distinct immune-modulating features of the brain.
Compared with other tumors, gliomas have unique immune characteristics. The anatomical location within the brain as compared with anywhere else in the body mediates a host of distinct features that contribute to alterations in the immune response to the cancer cells. Distinct immune-modulating features of glioma include the blood-brain barrier (BBB) and the presence of neuronal and glial cells.
The BBB contributes to the homeostasis of the CNS via regulation of the influx and efflux of biological substances (21). Although the BBB generally serves to protect the brain from toxic substances in the blood while allowing for the passing of necessary nutrients, this semipermeable layer can also restrict the entry of peripheral leukocytes and therapeutic moieties (22). Inflammation within the brain has been shown to compromise BBB integrity (22, 23), allowing for increased immune cell infiltration, but by the time this occurs, gliomagenesis will likely have already escaped immune surveillance.
Neuronal activity has been shown to promote glioma growth and invasion (24, 25). Signaling molecules released by neurons, such as the synaptic protein neuroligin-3 (NLGN3), can influence glioma progression by activation of the PI3K/mTOR pathway (24, 25). Additionally, a positive-feedback mechanism promotes the expression of neurotransmitters in tumor cells and affects immune cell functions (25–29). The role of epigenetic dysregulation in glioma in altering neuronal behavior remains understudied. Recent independent studies by Venkatesh et al. (30) and Venkataramani et al. (31) demonstrated that glioma cells can establish NLGN3-dependent synaptic connections with neuronal cells in vivo.
Venkatesh et al. showed that oligodendroglial precursor cell–like (OPC-like) glioma cells express synaptic genes (30). Glioma cells can establish two different types of synapses with neurons, one mediated by AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid) receptors (a type of ionotropic glutamate receptor), reminiscent of a neuron-OPC synapse, and the other mediated by potassium currents, reminiscent of a neuron-astrocyte synapse. The authors showed that the expression of GluA2, an AMPA receptor, confers growth advantage to glioma, promoting a decreased survival in vivo, and that coculturing glioma cells with neurons in vitro promotes tumor cell growth, demonstrating the importance of neuron-to-glioma synaptic connections for glioma progression. Venkataramani et al. characterized AMPA receptor–mediated neuron-glioma synapses and demonstrated that these connections promote glioma invasion and proliferation (31). These studies highlight the importance of neurons and synaptic connections in promoting tumor growth and modulating glioma behavior.
The glial cell population consists of three major subtypes: microglia, oligodendrocytes, and astrocytes. Microglia are brain-resident immune cells, and as such, they are discussed in detail in the myeloid cell section. Oligodendrocyte-lineage cells (OLCs), comprising oligodendrocytes and oligodendrocyte precursor cells, can increase the invasive activity, stemness, and chemoresistance of GBM cells by angiopoietin-2, FGF-1, and EGF secretion (32, 33). OLCs have been shown to modulate the immune response via the production of immune-mediating molecules, such as IL-18, IL-6, and CCL2 (34). Astrocytes are the most prevalent glial cells in the CNS (35), which activate and undergo transcriptomic reprogramming to become reactive astrocytes in response to CNS pathologies including glioma (36). In glioma, reactive astrocytes secrete protumor, antiinflammatory cytokines such as IL-6, TGF-β, and VEGF (36, 37).
Lymphoid cells.
Tumor-infiltrating lymphocytes (TILs) have strong antitumor immune functions and are key targets for immunotherapies. Although both B cells and T cells have been identified in gliomas (38), T cells are the predominant TILs and play a major role in glioma progression; thus the main body of glioma TIL research has been conducted on T cells. Regulatory T cells (Tregs), CD4+ T helper (Th) cells, and CD8+ T cells have all been shown to infiltrate the glioma TME (39). Tregs strongly suppress the functions of antitumor immune cells and support increased levels of other immunosuppressive cells (40, 41). Th cells, specifically Th1 cells, and CD8+ T cells contribute to the antitumor immune response by stimulating increased inflammation and tumor cell killing, respectively. Their antitumor effect is hindered by their low tumor infiltration and the immunosuppressive glioma TME (42, 43). Immune suppression is mediated primarily by glioma cells, myeloid-derived suppressor cells (MDSCs), Tregs, and tumor-associated macrophages and microglia (TAMMs) via the expression of immune checkpoint receptor ligands, such as PD-L1, and the secretion of immunosuppressive cytokines, such as TGF-β and IL-10 (44). These molecules, among others, mediate T cell dysfunction, i.e., anergy and exhaustion (45, 46). Exhausted Th cells and exhausted CD8+ T cells have reduced levels of proliferation and reduced effector cytokine production, i.e., IL-2, TNF-α, and IFN-γ (46, 47). T cell exhaustion is accompanied by increased chromatin accessibility and transcription in genes encoding immunosuppressive molecules (PDCD1, CTLA4, LAG3, ENTPD1) as well as decreased chromatin accessibility and transcription for genes important for cell differentiation (IL7R, TCF7, LEF1) (48, 49).
Myeloid cells.
Myeloid cells are the major immune population within the glioma TME, representing about 60% of all infiltrating immune cells (50–52). This population is composed primarily of brain-resident microglia, bone marrow–derived macrophages, and MDSCs (41, 53).
Despite the differences in their developmental origin, microglia and macrophages share several phenotypic properties. Microglia and macrophages can acquire antitumoral or protumoral phenotypes in the TME, but research suggests that the majority of TAMMs in the glioma TME have an immunosuppressive phenotype (54). In addition, TAMM characteristics have been shown to differ based on glioma cells’ mutations (55). For example, antitumoral macrophages are more abundant in wtIDH gliomas compared with mIDH1 gliomas (55).
MDSCs are a heterogeneous subgroup of immature myeloid cells with potent immunosuppressive properties. They promote proliferation and invasion of GBM cells (56). MDSCs are subdivided into polymorphonuclear MDSCs (PMN-MDSCs) and monocytic MDSCs (M-MDSCs), which exert different mechanisms to suppress immunity. PMN-MDSCs are suppressive via reactive oxygen species (ROS), peroxynitrite, arginase 1, and prostaglandin E2 production (57). M-MDSCs are suppressive via the expression of immunoregulatory molecules such as PD-L1 and by secreting nitric oxide, IL-10, and TGF-β (57).
Epigenetic mechanisms associated with TME modulation in glioma
Transcriptional reprogramming.
The most evident mechanism by which epigenetic alterations in glioma manipulate the tumor-infiltrating immune cells is via transcriptional modulation of genes associated with immune activation/suppression (Figure 2). For epigenetically mediated transcriptional modifications on the tumor cells to affect the immune cells, immune cells must sense a tumor-derived signal. The interaction between tumor cells’ surface ligands and immune cells’ receptors is an example of a direct mechanism. It was demonstrated that glioma TME-associated T cells are different from non-infiltrating T cells, and that glioma cells express ligands that can reprogram the T cell toward an exhausted phenotype (58). Our group and others demonstrated that mIDH1 expression mediates the transcriptional silencing of the PD-L1–encoding gene CD274 via DNA methylation (59, 60). The DNA methylation levels in the regulatory regions of CD274 decrease when mIDH1 glioma cells are treated with an mIDH1 inhibitor (59). PD-L1 expression by tumor cells supports T cell exhaustion, so mIDH1-harboring gliomas have less PD-L1 to suppress T cell activity. A recent study describes that loss of ATRX, another epigenetic modulator in both adult and pediatric glioma (20), epigenetically induces the expression of PD-L1 and several immunosuppressive cytokines, eliciting tolerogenic mechanisms in ATRX-mutant glioma (61).
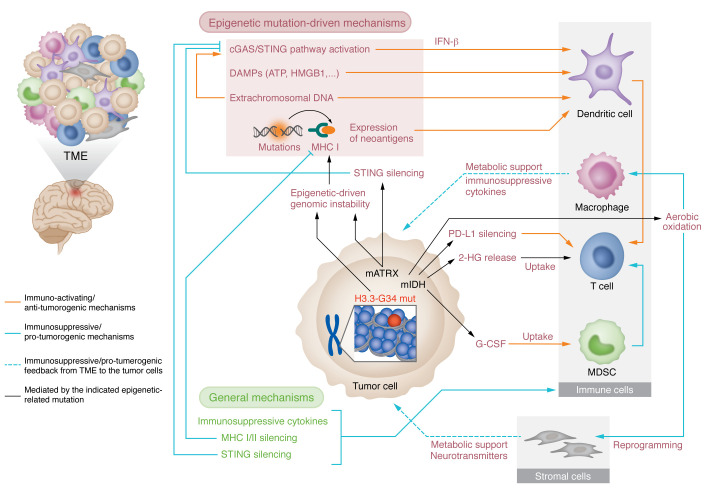
Figure 2. Epigenetic mechanism–mediated interactions between glioma cells and nontumoral cells that shape the TME.
Connections are indicated by arrows, and the color of the arrows indicates whether the interactions lead to immunosuppressive/protumoral or immune-activating/antitumoral mechanisms. The start of the black arrows indicates the mutations in the glioma cells that elicit the epigenetic mechanisms.
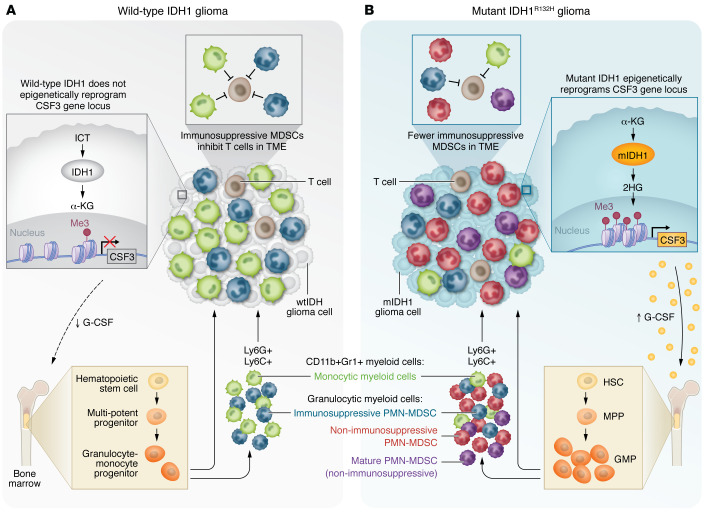
Tumor cells can also release cytokines that can be distally sensed by immune cells to activate or suppress the immune response. In particular, glioma cells secrete the immune-modulating cytokines IL-1β, IL-6, TGF-β, and IL-8 (62). Our group demonstrated that granulocyte colony-stimulating factor (G-CSF) expression is epigenetically activated by mIDH1 in glioma stem/progenitor-like cells, mediating reprogramming of myeloid cells within the mIDH1 glioma TME (63). This increased G-CSF prompts the expansion of pre-neutrophils and neutrophils, while reducing the immunosuppressive phenotype of PMN-MDSCs encountered in the mIDH1 TME (Figure 3) (63). Other potential mechanisms by which tumor cells can alter the immune cells distally involve the release of damage-associated molecular patterns (DAMPs). HMGB1 and extracellular ATP, for example, are released by glioma cells and promote inflammation in the TME (64, 65). Tumor cells also produce extracellular vesicles (EVs), which can mediate cell signaling. Glioma-derived exosomes can carry immunosuppressive molecules (66) and were shown to have functional suppressive activity on different immune cell types (66, 67). The role of epigenetic mutations in the regulation of the secretion of EVs and their cargo is an exciting, understudied field that could open opportunities for the development of tailored therapies.
Figure 3. Model of aberrant granulocyte differentiation in mIDH1 tumors.
(A) In WT IDH1 tumors, tumor cells express low levels of G-CSF and the TME contains a high number of immunosuppressive MDSCs. (B) Through epigenetic reprogramming mediated by mIDH1-induced 2-HG accumulation, mIDH1 glioma cells express and secrete G-CSF. Circulating G-CSF has a direct effect on hematopoiesis in the bone marrow and spleen, promoting the expansion, differentiation, and mobilization of granulocytic myeloid cells. As a result, the granulocytes recruited to the TME are mainly neutrophils and preneutrophils, with inhibitory PMN-MDSCs, constituting a smaller fraction of the total granulocytes in the mIDH1 tumor. Figure adapted from Alghamri et al. (63).
The epigenetic modulation of tumor cells’ differentiation also has consequences for the TME. Glioma epigenetic mutations (in particular, H3 mutations in pediatric glioma and mIDH1 mutations in adults) have been shown to result in stalled differentiation, committing the cells to an undifferentiated, stem-like state. Cancer stem-like cells are less immunogenic, and can evade the immune system through various mechanisms, including major histocompatibility complex (MHC) transcriptional downregulation, induction of quiescence, and other mechanisms that promote immune tolerance. Thus, epigenetically mediated interruption of cell development in early neoplastic stages likely makes the initial tumor development possible by hampering the antitumoral immune response. A recent study shows that glioma stem cells (GSCs) selected to grow in immunocompetent hosts go through a process of epigenetic adaptation, which leads to secretion of immunosuppressive cytokines (68). Moreover, the adapted GSCs show upregulation of IRF8, a cytokine normally restricted to myeloid cells as it controls myeloid lineages and macrophage differentiation (68). This demonstrates that epigenetic mechanisms mediate reprogramming of glioma cells resulting in the modification of immune cells and induction of a pro-tumorigenic TME.
The role of histone mutations as modulators of the immune TME remains understudied. A recent study describes that the transcription factor RACK7 (ZMYND8) has increased affinity for the mutant histone H3.3-G34R (a driver mutation of pHGG). Increased binding of RACK7 to the mutant histone leads to transcriptional repression of selected genes, including CIITA, a master regulator of MHC class II expression (69).
Polycomb repressive complex 2 (PCR2) is a family of histone methyltransferases that controls epigenetic silencing, and whose function can be altered in glioma (70). PCR2 proteins’ expression levels were correlated with poor prognosis, and it was suggested that PCR2 chromatin silencing mediates immunosuppression by blocking the expression of immune-stimulatory cytokines in tumor cells (70). A better response to immune checkpoint inhibitors (ICIs) was recently reported for tumors with mutations in PCR2 (70). This opens possibilities to improve the response to immunotherapies by reverting the methylase activity of PCR2 via epigenetic pharmacological modulation (71).
The Notch pathway was shown to be epigenetically modulated in mIDH1 gliomas, through DNA methylation of CpG sites within the delta-like ligand 3 (DLL3) gene (72). DLL3 is an inhibitory Notch ligand, and its expression positively correlated with survival in mIDH1 gliomas. mIDH1 gliomas with high expression of DLL3 showed increased immune infiltration, suggesting an association between Notch signaling and immune activity in these tumors (72).
The role of noncoding RNAs in TME modulation.
In recent decades, noncoding RNAs (ncRNAs) like microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) were shown to play critical roles in glioma, including acting as epigenetic modulators. miRNAs are natural interference RNAs that act via inactivation of mRNA (73). Loss of miRNAs that naturally regulate critical mRNAs can be oncogenic, such as miR-31, which silences CDKN2A/B (74), and miR-34a, which controls EGFR levels (75). The expression of different miRNAs was observed in certain molecular subtypes, indicating that miRNAs can regulate the heterogeneity of glioma by mediating transcriptional subtype transitions (76).
LncRNAs are also involved in glioma biology, progression, and response to therapies (77). The lncRNA HOTAIRM1 acts as an epigenetic regulator by binding to transcription start sites and blocking the access of epigenetic modifiers to regulatory gene regions (78). Similarly, lncSNHG6 and lncRNA ZFAT-AS1 can promote epigenetic silencing by inducing H3K27me3 gene-specific deposition (79, 80).
One characteristic of ncRNAs is that they can function as intercellular signals (81). miRNA and lncRNA secretion by glioma cells can impact TME behavior. For example, lncRNA-ATB secreted by glioma cells can suppress miR-204-3p in astrocytes, which could promote migration of glioma cells (82). Other studies indicate that lncRNAs secreted via exosomes can have a paracrine effect, promoting adaptation to stress/hypoxia conditions and resistance (83, 84). The communication between glioma cells and the TME via ncRNA emerges as an area with great therapeutic potential, although the role of ncRNA in altering the epigenetic landscape of the TME, particularly the immune cells, remains understudied.
The epigenetic manipulation of metabolism within the TME.
Epigenetic activation of the PI3K/AKT/mTOR pathway is commonly observed in glioma, resulting in selection advantages such as increased metabolism, proliferation, stemness, and invasiveness. This excessive metabolism in glioma cells leads to the switch from oxidative phosphorylation (respiration) to the oxidation of pyruvate to lactate (aerobic glycolysis) (85). This phenomenon, typical of cancer cells, is called the Warburg effect. It was demonstrated that aerobic glycolysis can modulate immune cells’ functions. The release of lactic acid and the resulting hypoxia lead to the induction of an immunosuppressive TME by mechanisms that include increased secretion of TGF-β, inhibition of the monocyte differentiation to dendritic cells by lactic acid, and secretion of pro-tumorigenic cytokines (i.e., IL-23) (86). Additionally, the metabolic alteration of the TME can affect the function of astrocytes to promote the growth of glioma cells (via release of cholesterol) and the recruitment of immunosuppressive macrophages (87).
d-2-Hydroxyglutarate uptake by immune cells within the TME.
Glioma cells expressing mIDH1 produce increased levels of d-2-hydroxyglutarate (2-HG), resulting in epigenetic reprogramming of the tumor cells (88). The ability of 2-HG to affect the immune TME remains unclear. It has been reported that 2-HG can be internalized by T cells in vitro, and that T cells isolated from mIDH1 acute myeloid leukemia patients have high levels of 2-HG (89). One study found that 2-HG triggers HIF-1α protein destabilization, leading to metabolic skewing, oxidative phosphorylation, increased Treg frequency, and reduced Th17 polarization (89). A recent study found that exposure to 2-HG reduced proliferation of activated T cells, although a study from our group found no effects of 2-HG on T cell proliferation (59). Sodium-dependent dicarboxylate transporter 3 (SLC13A3) and organic anion transporter SLC22A6 were hypothesized to mediate 2-HG internalization by T cells (89, 90). The mechanisms mediating the internalization of 2-HG have yet to be elucidated for the glioma immune TME.
Genetic instability and immunity.
Besides the transcriptional alterations, epigenetic dysregulation can cause direct effects on the structure of the chromatin. In glioma, histone and ATRX mutations have been associated with genetic instability, which results from abnormal histone mark deposition in these cells (20).
Mutant IDH was shown to epigenetically upregulate the DNA damage response (91). The genetic instability has many consequences, among them the emergence of extrachromosomal DNA in the cytoplasm. This phenomenon activates the cGAS/STING pathway in the tumor cells, leading to the activation of innate immune cells, such as dendritic cells (92). Additionally, epigenetically mediated genetic instability results in the accumulation of chromosomal alterations, promoting the expression of neoantigens arising from mutant proteins (93). These neoantigens can be recognized by adaptive immune cells, leading to immune activation or tolerance, depending on the tolerogenic properties of the TME. Notably, epigenetic regulation of STING (via STING promoter DNA methylation) has been proposed to modulate the immune response in glioma (94). Moreover, STING silencing in glioma can be reversed by DNA methyltransferase inhibition (95). A recent study from our group demonstrated that H3-G34 mutations, present in pHGG, confer genomic instability to these tumors (96). This results in activation of the cGAS/STING pathway, and promotes the activation of the immune system, improving the efficacy of DNA-damaging treatments (96).
Epigenetic reprogramming in tumor heterogeneity, evasion, and resistance.
Intratumoral heterogeneity in glioma was evidenced by the heterogeneous levels of expression of specific markers in biopsied tissue (97). Recent single-cell high-throughput analyses have helped to uncover the molecular basis of spatial and temporal heterogeneity (98, 99). The intratumoral heterogeneity can be based on genetic differences among the tumor cells, or due to epigenetic differences, which lead to different transcriptional profiles (100). Glioma cells were shown to transition between transcriptional states resembling mesenchymal (MES), astrocytic, neural precursor, and oligodendrocyte precursor lineages (98). A recent study aimed to characterize the interaction of these molecular programs in glioma cells and their interactions with the TME through the integration of spatial transcriptomics and scRNA-Seq from multiple glioma patients (101). The spatial transcriptomics uncovered that diverse molecular regions are recurrent in glioma. One of these niches encompasses tumor areas undergoing hypoxia and composed of MES-like cells. Glioma cells in these regions have increased genomic instability and are proposed as potential sources of adaptive evolution and development of resistance to therapies. Another niche is described as “reactive immune” and is characterized by increased immune infiltration, glioma cells with MES-like phenotype, and expression of immunosuppressive markers. The work also describes that the environment in which the tumor is growing (i.e., species and host age) can determine the molecular phenotypes adopted by glioma cells. This study demonstrated the impact of the intratumoral heterogeneity on the TME and reveals the potential of manipulating the TME to induce changes in the tumor cells.
Epigenetic heterogeneity can impose additional effects on the immune TME, as there can be differences among the immune-stimulating or immunosuppressive activities of different glioma cells according to the location of the cell and the stage of tumor development. As mentioned above, GSCs have immunosuppressive activities, and the epigenetic mutations commonly found in gliomas can affect the stemness of the tumor, as well as the identity of the cells (98). Heterogeneous expression of glioma markers mediated by epigenetic mechanisms has been mentioned as one factor limiting the success of chimeric antigen receptor (CAR) T cell therapies (102). Local DNA methylation disorder is another common dysregulation in glioma cells, particularly in those with mutations affecting epigenetic machinery (99, 103, 104). In response to stresses (i.e., hypoxia and irradiation), DNA methylation disorder increases, and it was speculated that this can provide a mechanism by which the plasticity of glioma cells increases to adapt to stressors.
Heterogeneity also plays a main role in the adaptation of gliomas to treatments, as it contributes to the generation of a larger population of phenotypes from which resistant cells will emerge. After the treatment, a fraction of the cells survive (i.e., resistant cells), leading to a transient reduction of tumor heterogeneity. Thus, a strategy recently proposed to treat glioma aims to target this window when heterogeneity is reduced, as the tumor cells have reduced plasticity to adapt to a second treatment.
Immune effects of therapies targeting epigenetic remodeling
The glioma epigenetic landscape revealed several epigenetic alterations that are mechanistically associated with tumor behavior (105, 106). Alterations in DNA methylation, histone methylation/acetylation, and IDH mutation status are frequent in gliomas and susceptible to targeting using epigenetic therapies. Strategies to target the glioma epigenome have been demonstrated to be effective in controlling tumor growth and represent a valuable alternative owing to the ability to reverse the epigenetic dysregulation that supports brain tumors (107). In fact, several DNA methyltransferase inhibitors, histone deacetylase inhibitors, PRC2-EZH2 methylase inhibitors, and mIDH1 inhibitors are currently being evaluated in clinical trials (108). Also, epigenetic processes in glioma cells can mediate the immune response. Thus, therapies targeting epigenetic mechanisms can be used to boost antitumor immunity (11).
DNA methyltransferase inhibition.
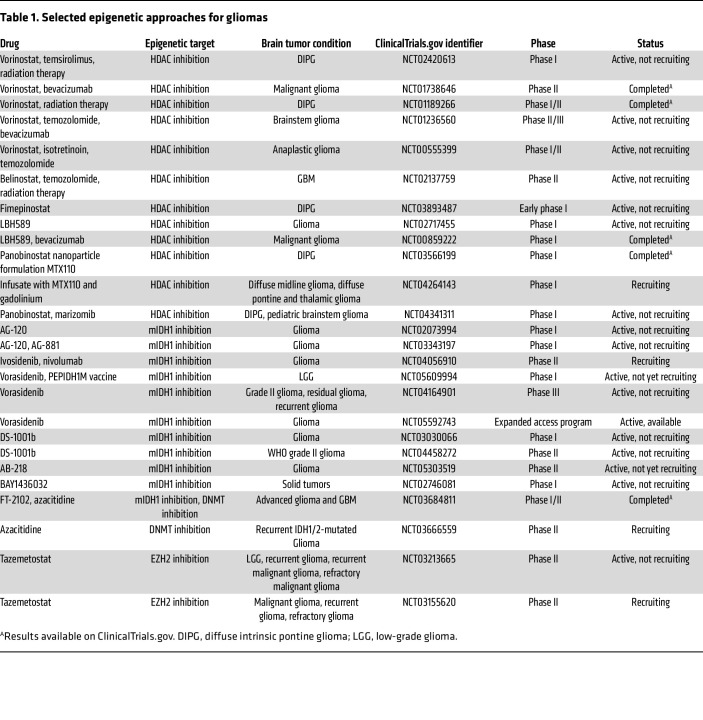
DNA methyltransferases (DNMTs) are enzymes that catalyze DNA methylation and regulate biological functions by modulating gene transcription (109). DNMTs catalyze the formation of 5-methylcytosine from cytosines in DNA CpG islands and ultimately suppress gene expression (109). Atypical DNMT functionality is often associated with tumor development via mechanisms leading to hypermethylation of tumor suppressor genes and increased genomic instability (109). DNMT inhibitors (DNMTis) can restore tumor suppressors’ activity by blocking DNA methylation, thereby reducing tumor cell proliferation and inducing apoptosis (110). Because DNA methylation processes are crucial for immune cell lineage progression and functionality (111), DNMTis can play a direct role in modulating antitumor immunity, yet the direct effects of DNMTis on immune cells remain unestablished. However, DNMTis can promote tumor-specific CD8+ T cell activation by upregulating MHC class I antigen presentation by glioma cells (112). Since T cell exhaustion is characterized by altered gene expression linked with alterations in DNA methylation by DNMTs (113), DNMTis may also promote antitumor immunity by preventing T cell exhaustion. Deleting a DNMT enzyme, DNMT3A, in CAR T cells was shown to prevent exhaustion and promote antitumor immunity (114). Therapies that include DNMTis, such as azacytidine and decitabine, are still in the early phases for safety and tolerability testing (Table 1). The ability of DNMTis to revert the chromatin structure, which is characteristic of T cell exhaustion, and enhance the efficacy of anti–PD-1 antibodies supports their use in combination with ICIs (115).
Table 1. Selected epigenetic approaches for gliomas.
Histone deacetylase inhibition.
Some common epigenetic changes in tumor cells are related to dysregulated histone mark deposition on regulatory regions in oncogenes and tumor suppressor genes (116). Aberrations in histone deacetylase (HDAC) expression in tumor cells cause altered cell cycle progression and can drive tumor development (116). Studies have linked HDAC expression with glioma grade (117, 118), i.e., lower expression of HDACs class II and IV was found in GBM compared with low-grade astrocytomas (117). Interestingly, HDAC1 was significantly overexpressed in several gliomas and is associated with dismal overall survival (118). While HDAC inhibitors (HDACis) have traditionally been investigated for their ability to target the aberrant epigenetic characteristics of tumor cells, they also induce changes in the antitumor immune response. HDACis enhance T cell chemokine expression, augment responses to PD-1–targeting immunotherapy, and upregulate PD-L1 and HLA-DR on tumor cells (119). This suggests that the combination of HDACis with ICIs could be a valuable therapeutic strategy for glioma patients. The HDACis vorinostat, belinostat, and fimepinostat are being evaluated in clinical trials for both adult and pediatric gliomas (Table 1). However, vorinostat exhibited toxicity and low effectiveness (120). This could be due to its poor BBB penetration (120). Combining HDACis with other therapies and improving HDACi BBB permeability may increase efficacy.
Mutant IDH1 inhibition.
IDH1 mutation catalyzes the production of 2-HG, which is a competitive inhibitor of α-ketoglutarate–dependent dioxygenases including Jumonji-C domain–containing histone demethylases and the DNA demethylase TET2, generating a hypermethylated phenotype (121). Blocking 2-HG production can reverse DNA hypermethylation and promote differentiation in mIDH1 glioma cells (122). Several inhibitors of mIDH1 were shown to be effective in vitro and in vivo (59, 122, 123). The combination of current standard-of-care therapy (radiation and temozolomide) with mIDH1 inhibitor and PD-L1–blocking ICI increased tumor regression of mIDH1 glioma–bearing mice, decreased T cell exhaustion, and favored the generation of memory CD8+ T cells (59). Currently there are several clinical trials testing mIDH1 inhibitors (AG-120, AG-881, DS-1001b, BAY1436032) for glioma treatment, but these are still ongoing and in early phases of determining the safety profiles and ability to decrease 2-HG accumulation (Table 1) (124).
EZH2 inhibition.
Enhancer of zeste homolog 2 (EZH2) is a histone methyltransferase subunit of PCR2 that is altered in gliomas. This alteration leads to both gain- and loss-of-function activities (125). The aberrant expression of EZH2 impacts gene expression by binding to promoter regions and affecting methylation status, playing an oncogenic role in glioma (126). EZH2 controls the coordinated inactivation of several tumor suppressor genes, thereby promoting cancer growth, invasion, and drug resistance (126). EZH2 inhibitors (EZH2is) enhance p16 tumor suppressor gene expression, affecting glioma progression (125). Importantly, several studies suggest that EZH2 is a critical driver of immune response modulation by cancer cells, mediating immune evasion by downregulation of genes involved in immune activation, upregulation of immune checkpoints, and generation of an immunosuppressive TME (127). In addition, EZH2is increase T cell tumor infiltration, decrease tumor growth, and improve the therapeutic efficacy of ICIs in preclinical tumor models (128, 129). Several studies show that EZH2is decrease proliferation of glioma cells by halting cell cycle progression and altering the proinflammatory response (127, 130). In midline diffuse glioma, repression of EZH2 in microglia induces an antitumor phenotype resulting in decreased cancer cell invasion capability, increased phagocytosis by microglia, and tumor cell death (131). These studies suggest that EZH2is could improve glioma immunotherapy efficacy.
Challenges of epigenetic therapies.
As implied throughout this Review, epigenetic therapies have potential due to their antitumoral properties, but there are still challenges that must be overcome before epigenetic therapies become widely used. The main challenges are the induction of off-target effects, inability of drugs to cross the BBB, inability of drugs to penetrate into the tumor, and lack of efficacy given the heterogeneous nature of gliomas. Notably, the latter two problems are not specific to epigenetic therapies. Rather, they are common challenges among antiglioma therapies, including immunotherapies. Perhaps the largest hurdle more specific to epigenetic therapies is the chance for off-target effects. Epigenetic therapies target mechanisms either directly, e.g., mIDH1 inhibitors, or by broad epigenome reprogramming, e.g., HDACis and DNMTis. The direct mechanism is ideal for targeting specific mutations that contribute to alterations in epigenetic pathways. Contrarily, the broad epigenome-reprogramming therapies target general epigenetic mechanisms, which play a role in normal cellular processes in cancerous and non-cancerous cells alike. It is these broad epigenome-altering therapies that may result in more side effects. To limit the side effects of epigenetic therapies, it would be beneficial to reduce the volume of these drugs by optimizing the therapeutic window and combining them with other, more targeted treatments such as immunotherapies. Reduction of these off-target effects may also be accomplished through the use of targeted drug delivery systems.
Discussion
The ability of glioma cells to promote an immunosuppressive environment allows them to circumvent immune rejection. Glioma cells are naturally selected to exhibit molecular hallmarks that allow them to be ignored by the immune system. As gliomas are characterized by the occurrence of mutations that disrupt the epigenetic regulatory mechanisms, here we have discussed how epigenetic dysregulation provides an avenue for tumor evolution to occur. We have also discussed how the molecular intervention of these epigenetically driven mechanisms can be exploited to activate antitumoral immune activity.
An exciting recently uncovered field of study aims at exploiting signals emitted by DNA damage and genomic instability to promote an immune response (96). DNA-damaging therapies, such as radiation therapy and temozolomide, have historically been used for the treatment of gliomas, but these treatments are unable to overcome the immunosuppressive TME. This indicates that the mechanisms that connect DNA damage and innate immunity need to be further stimulated to elicit an effective immune response. In this sense, the direct stimulation of the cGAS/STING pathway via agonist small compounds emerges as an attractive therapeutic strategy (96, 132). STING epigenetic silencing can be an immune evasion mechanism in gliomas, so the epigenetic activation of STING expression is another possible therapeutic target. Likewise, inducing the expression of MHC class I and class II proteins, which are commonly silenced by epigenetic alterations in glioma, can promote antigen presentation and stimulate the immune system (133). In this sense, recent advances in the development of CRISPR/Cas9–based site-specific epigenetic editing systems allow gene-specific epigenetic therapies to be envisaged.
The epigenetically mediated chromosomal instability in some glioma subtypes is also a relevant mechanism that might allow for the selection of cells that have gained or lost genes that promote immune evasion or resistance. At the same time, the mutational burden imposed by evolutionary selection increases the amount of neoantigens expressed by tumor cells, providing opportunities to elicit adaptive antitumoral immune responses. The immunosuppressive TME does not normally allow for the neoantigens to prime an antitumor response, as the T cells become tolerized because of the lack of necessary costimulatory signals.
Intratumoral TME heterogeneity has been described in detail, in relation to tumor cells, stromal cells, and the immune compartment (98). A fraction of intratumoral heterogeneity cannot be explained by genetic differences among tumor cells, but rather has an epigenetic origin. Dysregulations observed in both adult and pediatric gliomas have been demonstrated to increase epigenetic plasticity and promote identity ambiguity in the tumor cells. Histone mutations lead to developmental stalling that promotes stem-like states that are more apt to induce a tolerogenic TME. Epigenetic intervention to promote differentiation of these stem cell populations within the tumor could reprogram the immune TME toward more antitumoral activity.
Understanding the mechanisms by which tumor cells induce an immunosuppressive TME should provide insights into how to manipulate the immune cells directly. We describe how aerobic oxidation (Warburg effect) in the tumor cells affects the immune cells, reprogramming the immune compartment to a tolerogenic environment. The blockade of pathways that sense metabolic stress in immune cells can be envisaged as a target to avoid this phenomenon. Likewise, the delivery of cytokines into the TME can reverse the immunosuppressive milieu mediated by tumor cells. Notably, diverse glioma mechanisms lead to the establishment of an immunosuppressive TME at the neoplastic stage of tumor development. At the time of treatment, the immunosuppressive TME is already established, and reverting it imposes a complex challenge. In summary, our improved understanding of the critical role that epigenetic processes play in shaping the TME and the immune response to glioma, together with the development of pharmacological agents to manipulate these processes, presents a new promising era of therapies that brings hope for the treatment of otherwise lethal tumors.
Acknowledgments
This work was supported by NIH/National Institute of Neurological Disorders and Stroke (NIH/NINDS) grants R37-NS094804, R01-NS105556, R01-NS122536, R01-NS124167, R01-NS122165, and R21-NS123879-01 as well as the Rogel Cancer Center Scholar Award to MGC; NIH/NINDS grants R01-NS076991, R01-NS082311, R01-NS096756, and R01NS122234 and NIH/National Cancer Institute grant R01-CA243916 to PRL; the Department of Neurosurgery, the Pediatric Brain Tumor Foundation, Leah’s Happy Hearts Foundation, Ian’s Friends Foundation, Chad Tough Foundation, and Smiles for Sophie Forever Foundation to MGC and PRL. BLM was supported by the University of Michigan Rackham Merit Fellowship and Research Training in Experimental Immunology training grant T32-AI007413.
Version 1. 01/17/2023
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2023, McClellan et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2023;133(2):e163450. https://doi.org/10.1172/JCI163450.
Contributor Information
Brandon L. McClellan, Email: bmccle@umich.edu.
Santiago Haase, Email: sthaase@umich.edu.
Felipe J. Nunez, Email: felipej.nunez@gmail.com.
Ali A. Dabaja, Email: dabajaaa@umich.edu.
Maria G. Castro, Email: mariacas@med.umich.edu.
References
- 1.Ostrom QT, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2013-2017. Neuro Oncol. 2020;22(12 suppl 2):iv1–iv96. doi: 10.1093/neuonc/noaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weller M, et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18(6):e315–e329. doi: 10.1016/S1470-2045(17)30194-8. [DOI] [PubMed] [Google Scholar]
- 4.Uddin MS, et al. Epigenetics of glioblastoma multiforme: from molecular mechanisms to therapeutic approaches. Semin Cancer Biol. 2022;83:100–120. doi: 10.1016/j.semcancer.2020.12.015. [DOI] [PubMed] [Google Scholar]
- 5.Jones C, Baker SJ. Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer. 2014;14(10):10.1038/nrc3811. doi: 10.1038/nrc3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips RE, et al. Epigenomic reprogramming as a driver of malignant glioma. Cancer Cell. 2020;38(5):647–660. doi: 10.1016/j.ccell.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 8.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 9.Komori T. Grading of adult diffuse gliomas according to the 2021 WHO classification of tumors of the central nervous system. Lab Invest. 2022;102(2):126–133. doi: 10.1038/s41374-021-00667-6. [DOI] [PubMed] [Google Scholar]
- 10.Eckel-Passow JE, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Fabiani MB, et al. Genetic alterations in gliomas remodel the tumor immune microenvironment and impact immune-mediated therapies. Front Oncol. 2021;11:631037. doi: 10.3389/fonc.2021.631037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turkalp Z, et al. IDH mutation in glioma: new insights and promises for the future. JAMA Neurol. 2014;71(10):1319–1325. doi: 10.1001/jamaneurol.2014.1205. [DOI] [PubMed] [Google Scholar]
- 14.Mazor T, et al. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell. 2015;28(3):307–317. doi: 10.1016/j.ccell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostrom QT, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011-2015. Neuro Oncol. 2018;20(suppl 4):iv1–iv86. doi: 10.1093/neuonc/noy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones C, et al. Paediatric and adult malignant glioma: close relatives or distant cousins? Nat Rev Clin Oncol. 2012;9(7):400–413. doi: 10.1038/nrclinonc.2012.87. [DOI] [PubMed] [Google Scholar]
- 17.Mackay A, et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic pontine glioma. Cancer Cell. 2017;32(4):520–537. doi: 10.1016/j.ccell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sturm D, et al. Pediatric gliomas: current concepts on diagnosis, biology, and clinical management. J Clin Oncol. 2017;35(21):2370–2377. doi: 10.1200/JCO.2017.73.0242. [DOI] [PubMed] [Google Scholar]
- 19.Dobson THW, Gopalakrishnan V. Preclinical models of pediatric brain tumors—forging ahead. Bioengineering (Basel) 2018;5(4):81. doi: 10.3390/bioengineering5040081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haase S, et al. Mutant ATRX: uncovering a new therapeutic target for glioma. Expert Opin Ther Targets. 2018;22(7):599–613. doi: 10.1080/14728222.2018.1487953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arvanitis CD, et al. The blood-brain barrier and blood-tumour barrier in brain tumours and metastases. Nat Rev Cancer. 2020;20(1):26–41. doi: 10.1038/s41568-019-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SY, et al. Blood-brain barrier dysfunction-induced inflammatory signaling in brain pathology and epileptogenesis. Epilepsia. 2012;53(suppl 6):37–44. doi: 10.1111/j.1528-1167.2012.03701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchetti L, Engelhardt B. Immune cell trafficking across the blood-brain barrier in the absence and presence of neuroinflammation. Vasc Biol. 2020;2(1):H1–H18. doi: 10.1530/VB-19-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venkatesh HS, et al. Neuronal activity promotes glioma growth through neuroligin-3 secretion. Cell. 2015;161(4):803–816. doi: 10.1016/j.cell.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johung T, Monje M. Neuronal activity in the glioma microenvironment. Curr Opin Neurobiol. 2017;47:156–161. doi: 10.1016/j.conb.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, et al. Imipramine impedes glioma progression by inhibiting YAP as a Hippo pathway independent manner and synergizes with temozolomide. J Cell Mol Med. 2021;25(19):9350–9363. doi: 10.1111/jcmm.16874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belotti Y, et al. Prognostic neurotransmitter receptors genes are associated with immune response, inflammation and cancer hallmarks in brain tumors. Cancers (Basel) 2022;14(10):2544. doi: 10.3390/cancers14102544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodo TW, et al. Critical neurotransmitters in the neuroimmune network. Front Immunol. 2020;11:1869. doi: 10.3389/fimmu.2020.01869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yaman I, et al. Advances in understanding cancer-associated neurogenesis and its implications on the neuroimmune axis in cancer. Pharmacol Ther. 2022;239:108199. doi: 10.1016/j.pharmthera.2022.108199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venkatesh HS, et al. Electrical and synaptic integration of glioma into neural circuits. Nature. 2019;573(7775):539–545. doi: 10.1038/s41586-019-1563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venkataramani V, et al. Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature. 2019;573(7775):532–538. doi: 10.1038/s41586-019-1564-x. [DOI] [PubMed] [Google Scholar]
- 32.Kawashima T, et al. Oligodendrocytes up-regulate the invasive activity of glioblastoma cells via the angiopoietin-2 signaling pathway. Anticancer Res. 2019;39(2):577–584. doi: 10.21873/anticanres.13150. [DOI] [PubMed] [Google Scholar]
- 33.Hide T, et al. Oligodendrocyte progenitor cells and macrophages/microglia produce glioma stem cell niches at the tumor border. EBioMedicine. 2018;30:94–104. doi: 10.1016/j.ebiom.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peferoen L, et al. Oligodendrocyte-microglia cross-talk in the central nervous system. Immunology. 2014;141(3):302–313. doi: 10.1111/imm.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Placone AL, et al. The role of astrocytes in the progression of brain cancer: complicating the picture of the tumor microenvironment. Tumour Biol. 2016;37(1):61–69. doi: 10.1007/s13277-015-4242-0. [DOI] [PubMed] [Google Scholar]
- 36.Henrik Heiland D, et al. Tumor-associated reactive astrocytes aid the evolution of immunosuppressive environment in glioblastoma. Nat Commun. 2019;10(1):2541. doi: 10.1038/s41467-019-10493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan X, et al. Reactive astrocytes in glioblastoma multiforme. Mol Neurobiol. 2018;55(8):6927–6938. doi: 10.1007/s12035-018-0880-8. [DOI] [PubMed] [Google Scholar]
- 38.Lee-Chang C, et al. Myeloid-derived suppressive cells promote B cell-mediated immunosuppression via transfer of PD-L1 in glioblastoma. Cancer Immunol Res. 2019;7(12):1928–1943. doi: 10.1158/2326-6066.CIR-19-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Domingues P, et al. Tumor infiltrating immune cells in gliomas and meningiomas. Brain Behav Immun. 2016;53:1–15. doi: 10.1016/j.bbi.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 40.Humphries W, et al. The role of Tregs in glioma-mediated immunosuppression: potential target for intervention. Neurosurg Clin N Am. 2010;21(1):125–137. doi: 10.1016/j.nec.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z, Hambardzumyan D. Immune microenvironment in glioblastoma subtypes. Front Immunol. 2018;9:1004. doi: 10.3389/fimmu.2018.01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weenink B, et al. Low-grade glioma harbors few CD8 T cells, which is accompanied by decreased expression of chemo-attractants, not immunogenic antigens. Sci Rep. 2019;9(1):14643. doi: 10.1038/s41598-019-51063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohme M, Neidert MC. Tumor-specific T cell activation in malignant brain tumors. Front Immunol. 2020;11:205. doi: 10.3389/fimmu.2020.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grabowski MM, et al. Immune suppression in gliomas. J Neurooncol. 2021;151(1):3–12. doi: 10.1007/s11060-020-03483-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang H, et al. Different T-cell subsets in glioblastoma multiforme and targeted immunotherapy. Cancer Lett. 2021;496:134–143. doi: 10.1016/j.canlet.2020.09.028. [DOI] [PubMed] [Google Scholar]
- 46.Woroniecka K, Fecci PE. T-cell exhaustion in glioblastoma. Oncotarget. 2018;9(82):35287–35288. doi: 10.18632/oncotarget.26228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bengsch B, et al. Epigenomic-guided mass cytometry profiling reveals disease-specific features of exhausted CD8 T cells. Immunity. 2018;48(5):1029–1045.e5. doi: 10.1016/j.immuni.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan O, et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature. 2019;571(7764):211–218. doi: 10.1038/s41586-019-1325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simonds EF, et al. Deep immune profiling reveals targetable mechanisms of immune evasion in immune checkpoint inhibitor-refractory glioblastoma. J Immunother Cancer. 2021;9(6):e002181. doi: 10.1136/jitc-2020-002181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang AL, et al. CCL2 produced by the glioma microenvironment is essential for the recruitment of regulatory T cells and myeloid-derived suppressor cells. Cancer Res. 2016;76(19):5671–5682. doi: 10.1158/0008-5472.CAN-16-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamran N, et al. Immunosuppressive myeloid cells’ blockade in the glioma microenvironment enhances the efficacy of immune-stimulatory gene therapy. Mol Ther. 2017;25(1):232–248. doi: 10.1016/j.ymthe.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown NF, et al. Harnessing the immune system in glioblastoma. Br J Cancer. 2018;119(10):1171–1181. doi: 10.1038/s41416-018-0258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kennedy BC, et al. Tumor-associated macrophages in glioma: friend or foe? J Oncol. 2013;2013:486912. doi: 10.1155/2013/486912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehani B, et al. Immune cell gene expression signatures in diffuse glioma are associated with IDH mutation status, patient outcome and malignant cell state, and highlight the importance of specific cell subsets in glioma biology. Acta Neuropathol Commun. 2022;10(1):19. doi: 10.1186/s40478-022-01323-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lakshmanachetty S, et al. New insights into the multifaceted role of myeloid-derived suppressor cells (MDSCs) in high-grade gliomas: from metabolic reprograming, immunosuppression, and therapeutic resistance to current strategies for targeting MDSCs. Cells. 2021;10(4):893. doi: 10.3390/cells10040893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Veglia F, et al. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol. 2021;21(8):485–498. doi: 10.1038/s41577-020-00490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woroniecka K, et al. T-cell exhaustion signatures vary with tumor type and are severe in glioblastoma. Clin Cancer Res. 2018;24(17):4175–4186. doi: 10.1158/1078-0432.CCR-17-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kadiyala P, et al. Inhibition of 2-hydroxyglutarate elicits metabolic reprogramming and mutant IDH1 glioma immunity in mice. J Clin Invest. 2021;131(4):e139542. doi: 10.1172/JCI139542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mu L, et al. The IDH1 mutation-induced oncometabolite, 2-hydroxyglutarate, may affect DNA methylation and expression of PD-L1 in gliomas. Front Mol Neurosci. 2018;11:82. doi: 10.3389/fnmol.2018.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu C, et al. ATRX loss promotes immunosuppressive mechanisms in IDH1 mutant glioma. Neuro Oncol. 2022;24(6):888–900. doi: 10.1093/neuonc/noab292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeung YT, et al. Interleukins in glioblastoma pathophysiology: implications for therapy. Br J Pharmacol. 2013;168(3):591–606. doi: 10.1111/bph.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alghamri MS, et al. G-CSF secreted by mutant IDH1 glioma stem cells abolishes myeloid cell immunosuppression and enhances the efficacy of immunotherapy. Sci Adv. 2021;7(40):eabh3243. doi: 10.1126/sciadv.abh3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Candolfi M, et al. Release of HMGB1 in response to proapoptotic glioma killing strategies: efficacy and neurotoxicity. Clin Cancer Res. 2009;15(13):4401–4414. doi: 10.1158/1078-0432.CCR-09-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen R, et al. The mechanism of HMGB1 secretion and release. Exp Mol Med. 2022;54(2):91–102. doi: 10.1038/s12276-022-00736-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Azambuja JH, et al. Molecular profiles and immunomodulatory activities of glioblastoma-derived exosomes. Neurooncol Adv. 2020;2(1):vdaa056. doi: 10.1093/noajnl/vdaa056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Vrij J, et al. Glioblastoma-derived extracellular vesicles modify the phenotype of monocytic cells. Int J Cancer. 2015;137(7):1630–1642. doi: 10.1002/ijc.29521. [DOI] [PubMed] [Google Scholar]
- 68.Gangoso E, et al. Glioblastomas acquire myeloid-affiliated transcriptional programs via epigenetic immunoediting to elicit immune evasion. Cell. 2021;184(9):2454–2470.e26. doi: 10.1016/j.cell.2021.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jiao F, et al. RACK7 recognizes H3.3G34R mutation to suppress expression of MHC class II complex components and their delivery pathway in pediatric glioblastoma. Sci Adv. 2020;6(29):eaba2113. doi: 10.1126/sciadv.aba2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vlachostergios PJ. Polycomb repressive complex 2 mutations predict survival benefit in advanced cancer patients treated with immune checkpoint inhibitors. Immunooncol Technol. 2021;10:100035. doi: 10.1016/j.iotech.2021.100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Covre A, et al. Antitumor activity of epigenetic immunomodulation combined with CTLA-4 blockade in syngeneic mouse models. Oncoimmunology. 2015;4(8):e1019978. doi: 10.1080/2162402X.2015.1019978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Noor H, et al. DLL3 expression and methylation are associated with lower-grade glioma immune microenvironment and prognosis. Genomics. 2022;114(2):110289. doi: 10.1016/j.ygeno.2022.110289. [DOI] [PubMed] [Google Scholar]
- 73.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 74.Rajbhandari R, et al. Loss of tumor suppressive microRNA-31 enhances TRADD/NF-κB signaling in glioblastoma. Oncotarget. 2015;6(19):17805–17816. doi: 10.18632/oncotarget.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69(19):7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang T, et al. A regulatory circuit of miR-125b/miR-20b and Wnt signalling controls glioblastoma phenotypes through FZD6-modulated pathways. Nat Commun. 2016;7:12885. doi: 10.1038/ncomms12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peng Z, et al. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer. 2018;17(1):61. doi: 10.1186/s12943-018-0812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Q, et al. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer Res. 2018;37(1):265. doi: 10.1186/s13046-018-0941-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li X, et al. NCBP3/SNHG6 inhibits GBX2 transcription in a histone modification manner to facilitate the malignant biological behaviour of glioma cells. RNA Biol. 2021;18(1):47–63. doi: 10.1080/15476286.2020.1790140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang F, et al. DGCR8/ZFAT-AS1 promotes CDX2 transcription in a PRC2 complex-dependent manner to facilitate the malignant biological behavior of glioma cells. Mol Ther. 2020;28(2):613–630. doi: 10.1016/j.ymthe.2019.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cheng J, et al. Exosomal noncoding RNAs in glioma: biological functions and potential clinical applications. Mol Cancer. 2020;19(1):66. doi: 10.1186/s12943-020-01189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bian EB, et al. Exosomal lncRNA‑ATB activates astrocytes that promote glioma cell invasion. Int J Oncol. 2019;54(2):713–721. doi: 10.3892/ijo.2018.4644. [DOI] [PubMed] [Google Scholar]
- 83.Dai X, et al. AHIF promotes glioblastoma progression and radioresistance via exosomes. Int J Oncol. 2019;54(1):261–270. doi: 10.3892/ijo.2018.4621. [DOI] [PubMed] [Google Scholar]
- 84.Li J, et al. Hypoxic glioma stem cell-derived exosomes containing Linc01060 promote progression of glioma by regulating the MZF1/c-Myc/HIF1α axis. Cancer Res. 2021;81(1):114–128. doi: 10.1158/0008-5472.CAN-20-2270. [DOI] [PubMed] [Google Scholar]
- 85.Kanwore K, et al. Cancer metabolism: the role of immune cells epigenetic alteration in tumorigenesis, progression, and metastasis of Glioma. Front Immunol. 2022;13:831636. doi: 10.3389/fimmu.2022.831636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kesarwani P, et al. The interplay between metabolic remodeling and immune regulation in glioblastoma. Neuro Oncol. 2017;19(10):1308–1315. doi: 10.1093/neuonc/nox079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perelroizen R, et al. Astrocyte immunometabolic regulation of the tumour microenvironment drives glioblastoma pathogenicity. Brain. 2022;145(9):3288–3307. doi: 10.1093/brain/awac222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465(7300):966. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Böttcher M, et al. D-2-hydroxyglutarate interferes with HIF-1α stability skewing T-cell metabolism towards oxidative phosphorylation and impairing Th17 polarization. Oncoimmunology. 2018;7(7):e1445454. doi: 10.1080/2162402X.2018.1445454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bunse L, et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat Med. 2018;24(8):1192–1203. doi: 10.1038/s41591-018-0095-6. [DOI] [PubMed] [Google Scholar]
- 91.Nunez FJ, et al. IDH1-R132H acts as a tumor suppressor in glioma via epigenetic up-regulation of the DNA damage response. Sci Transl Med. 2019;11(479):eaaq1427. doi: 10.1126/scitranslmed.aaq1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kwon J, Bakhoum SF. The cytosolic DNA-sensing cGAS-STING pathway in cancer. Cancer Discov. 2020;10(1):26–39. doi: 10.1158/2159-8290.CD-19-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mardis ER. Neoantigens and genome instability: impact on immunogenomic phenotypes and immunotherapy response. Genome Med. 2019;11(1):71. doi: 10.1186/s13073-019-0684-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Qiu L, et al. STING cg16983159 methylation: a key factor for glioblastoma immunosuppression. Signal Transduct Target Ther. 2022;7(1):228. doi: 10.1038/s41392-022-01093-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Low JT, et al. Epigenetic STING silencing is developmentally conserved in gliomas and can be rescued by methyltransferase inhibition. Cancer Cell. 2022;40(5):439–440. doi: 10.1016/j.ccell.2022.04.009. [DOI] [PubMed] [Google Scholar]
- 96.Haase S, et al. H3.3-G34 mutations impair DNA repair and promote cGAS/STING-mediated immune responses in pediatric high-grade glioma models. J Clin Invest. 2022;132(22):e154229. doi: 10.1172/JCI154229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Strommer K, et al. Cellular and tumoural heterogeneity of EGFR gene amplification in human malignant gliomas. Acta Neurochir (Wien) 1990;107(3–4):82–87. doi: 10.1007/BF01405784. [DOI] [PubMed] [Google Scholar]
- 98.Neftel C, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849.e21. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Johnson KC, et al. Single-cell multimodal glioma analyses identify epigenetic regulators of cellular plasticity and environmental stress response. Nat Genet. 2021;53(10):1456–1468. doi: 10.1038/s41588-021-00926-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hoffman M, et al. Intratumoral genetic and functional heterogeneity in pediatric glioblastoma. Cancer Res. 2019;79(9):2111–2123. doi: 10.1158/0008-5472.CAN-18-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ravi VM, et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell. 2022;40(6):639–655.e13. doi: 10.1016/j.ccell.2022.05.009. [DOI] [PubMed] [Google Scholar]
- 102.Antonucci L, et al. CAR-T therapy for pediatric high-grade gliomas: peculiarities, current investigations and future strategies. Front Immunol. 2022;13:867154. doi: 10.3389/fimmu.2022.867154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Flavahan WA, et al. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357(6348):eaal2380. doi: 10.1126/science.aal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Unruh D, et al. Methylation and transcription patterns are distinct in IDH mutant gliomas compared to other IDH mutant cancers. Sci Rep. 2019;9(1):8946. doi: 10.1038/s41598-019-45346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Masui K, et al. Metabolic reprogramming in the pathogenesis of glioma: update. Neuropathology. 2019;39(1):3–13. doi: 10.1111/neup.12535. [DOI] [PubMed] [Google Scholar]
- 106.Nicholson JG, Fine HA. Diffuse glioma heterogeneity and its therapeutic implications. Cancer Discov. 2021;11(3):575–590. doi: 10.1158/2159-8290.CD-20-1474. [DOI] [PubMed] [Google Scholar]
- 107.Romani M, et al. Epigenetic targeting of glioblastoma. Front Oncol. 2018;8:448. doi: 10.3389/fonc.2018.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kukreja L, et al. Emerging epigenetic therapies for brain tumors. Neuromolecular Med. 2022;24(1):41–49. doi: 10.1007/s12017-021-08691-x. [DOI] [PubMed] [Google Scholar]
- 109.Zhang J, et al. DNA methyltransferases in cancer: biology, paradox, aberrations, and targeted therapy. Cancers (Basel) 2020;12(8):2123. doi: 10.3390/cancers12082123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Park JW, Han J-W. Targeting epigenetics for cancer therapy. Arch Pharm Res. 2019;42(2):159–170. doi: 10.1007/s12272-019-01126-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15(5):763–774. doi: 10.1016/S1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 112.Natsume A, et al. The DNA demethylating agent 5-aza-2’-deoxycytidine activates NY-ESO-1 antigenicity in orthotopic human glioma. Int J Cancer. 2008;122(11):2542–2553. doi: 10.1002/ijc.23407. [DOI] [PubMed] [Google Scholar]
- 113.Scharer CD, et al. Cutting edge: chromatin accessibility programs CD8 T cell memory. J Immunol. 2017;198(6):2238–2243. doi: 10.4049/jimmunol.1602086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Prinzing B, et al. Deleting DNMT3A in CAR T cells prevents exhaustion and enhances antitumor activity. Sci Transl Med. 2021;13(620):eabh0272. doi: 10.1126/scitranslmed.abh0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ghoneim HE, et al. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell. 2017;170(1):142–157.e19. doi: 10.1016/j.cell.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bojang P, Ramos KS. The promise and failures of epigenetic therapies for cancer treatment. Cancer Treat Rev. 2014;40(1):153–169. doi: 10.1016/j.ctrv.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lucio-Eterovic AK, et al. Differential expression of 12 histone deacetylase (HDAC) genes in astrocytomas and normal brain tissue: class II and IV are hypoexpressed in glioblastomas. BMC Cancer. 2008;8:243. doi: 10.1186/1471-2407-8-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fan Y, et al. Comprehensive analysis of HDAC family identifies HDAC1 as a prognostic and immune infiltration indicator and HDAC1-related signature for prognosis in glioma. Front Mol Biosci. 2021;8:720020. doi: 10.3389/fmolb.2021.720020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zheng H, et al. HDAC inhibitors enhance T-cell chemokine expression and augment response to PD-1 immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2016;22(16):4119–4132. doi: 10.1158/1078-0432.CCR-15-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee P, et al. Mechanisms and clinical significance of histone deacetylase inhibitors: epigenetic glioblastoma therapy. Anticancer Res. 2015;35(2):615–625. [PMC free article] [PubMed] [Google Scholar]
- 121.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rohle D, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang F, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340(6132):622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 124.Karpel-Massler G, et al. Novel IDH1-targeted glioma therapies. CNS Drugs. 2019;33(12):1155–1166. doi: 10.1007/s40263-019-00684-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med. 2016;22(2):128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Paskeh MDA, et al. EZH2 as a new therapeutic target in brain tumors: molecular landscape, therapeutic targeting and future prospects. Biomed Pharmacother. 2022;146:112532. doi: 10.1016/j.biopha.2021.112532. [DOI] [PubMed] [Google Scholar]
- 127.Stazi G, et al. Dissecting the role of novel EZH2 inhibitors in primary glioblastoma cell cultures: effects on proliferation, epithelial-mesenchymal transition, migration, and on the pro-inflammatory phenotype. Clin Epigenetics. 2019;11(1):173. doi: 10.1186/s13148-019-0763-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 128.Peng D, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527(7577):249–253. doi: 10.1038/nature15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zingg D, et al. The histone methyltransferase Ezh2 controls mechanisms of adaptive resistance to tumor immunotherapy. Cell Rep. 2017;20(4):854–867. doi: 10.1016/j.celrep.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 130.Kang N, et al. EZH2 inhibition: a promising strategy to prevent cancer immune editing. Epigenomics. 2020;12(16):1457–1476. doi: 10.2217/epi-2020-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Keane L, et al. Inhibition of microglial EZH2 leads to anti-tumoral effects in pediatric diffuse midline gliomas. Neurooncol Adv. 2021;3(1):vdab096. doi: 10.1093/noajnl/vdab096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Boudreau CE, et al. Intratumoral delivery of STING agonist results in clinical responses in canine glioblastoma. Clin Cancer Res. 2021;27(20):5528–5535. doi: 10.1158/1078-0432.CCR-21-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sun T, et al. Histone deacetylase inhibition up-regulates MHC class I to facilitate cytotoxic T lymphocyte-mediated tumor cell killing in glioma cells. J Cancer. 2019;10(23):5638–5645. doi: 10.7150/jca.34471. [DOI] [PMC free article] [PubMed] [Google Scholar]