Abstract
Recent studies have reported that collagen type V alpha 2 (COL5A2) is a hub gene and associated with the prognosis of gastric cancer (GC) patients, playing an important role in GC. In this study, we aim to fathom out the biological roles of COL5A2 and its relevant mechanism in GC. Oncomine, gene expression profiling interactive analysis, and UALCAN were used to explore the effects of COL5A2 on GC. Cell counting kit-8 assay, colony formation assay, and transwell assay were conducted to investigate the biological behaviors of GC cell lines AGS and SGC-7901. Quantitative reverse transcription polymerase chain reaction and western blot were performed to determine gene and protein expressions. COL5A2 expression was up-regulated and negatively correlated with survival percentage of GC patients. COL5A2 expression was notably elevated in high stage and high grade of GC. Down-regulation of COL5A2 inhibited proliferation, migration, and invasion of AGS and SGC-7901 cells. COL5A2 induced epithelial–mesenchymal transition (EMT) by promoting the expressions of mesenchymal markers (SNAI1, SNAI2, TWIST, VIM, and MMP2), thereby facilitating the malignant phenotypes of GC. COL5A2 plays an oncogenic role in GC and has potential to predict the progression and prognosis of GC patients.
Keywords: collagen, gastric cancer, oncogenic, mesenchymal markers
1. Introduction
As a malignant tumor, gastric cancer (GC) is reported to be the second leading cause of cancer-related deaths globally, and there are 1,089,103 new cases (5.6% of total cases) and 768,793 deaths (7.7% of total cancer deaths) in 2020 globally [1]. China is the hardest-hit country, with 42.6% of the global incidence and 45% of all GC-related mortality [2]. Due to the lack of effective and safe screening methods and obvious signs at an early stage, patients with GC are already at an advanced stage when first diagnosed, and they face with high risks of metastasis and recurrence after treatment [2]. Although great efforts have been made in the treatment of GC, such as surgery and adjuvant chemotherapy, the prognosis of GC patients has been slightly improved [3,4]. The 5-year survival rate of patients at advanced stage is extremely low, only 20% with the median survival of less than half a year [4]. Therefore, much attention should be paid to the early diagnosis of GC. Recently, scientists have devoted to applying molecular markers in the improvement of cancer diagnosis and therapeutic strategies [5]. However, limited progress has been achieved in early diagnosis and effective therapy for GC. While many biomarkers for GC have been reported, such as carbohydrate antigen (CA) 72-4, alpha-fetoprotein, and CA12-5, carcinoembryonic antigen and CA19-9 are still the most frequently used biomarkers for GC in clinical practice [6,7]. Almost all of the patients with advanced GC cannot be treated with a targeted therapy. Currently, no diagnostic markers are available for secondary prevention. Thus, it is of great significance to dig out the GC-associated molecules and the underlying mechanisms for the management of this disease.
Collagen proteins are the major component of extracellular matrix (ECM) with the highest protein level in mammalian cells [8]. In vertebrates, at least 28 types of collagen are identified, which play various roles in scaffold, fibrosis, and adhesion of tissues [9]. Many studies have revealed that up-regulation and down-regulation of collagens are both involved in the progression of cancers by regulating the remodeling of ECM [10,11]. Collagen type V (COL5), a type of fibrillar collagens, is mainly expressed in bone, dermis, cornea, and placenta co-distributed with type I collagens [12]. COL5 includes three major subunits, including COL5A1, COL5A2, and COL5A3, which can bind to three different polypeptide α chains [12]. COL5 collagens play essential roles in tissue scaffold and cell adhesion via forming heterotrimer or homotrimer [13]. Recently, as one of the best-studied collagens, COL5A2 has been reported to be aberrantly expressed in various cancers, such as breast cancer [14], ovarian cancer [15], adenomas [16], and bladder cancer [17]. Of note, multiple bioinformatics analyses have revealed that COL5A2 is a hub gene involved in the prognosis of GC patients, with a vital role in GC [4,18–21]. In addition, COL5A2 level may be a risk factor for GC, and COL5A2 may act as a potential clinical biomarker for GC and renal metastasis [22]. However, there are no experimental data supporting this notion, and the relevant mechanism remains ambiguous.
Therefore, in this study, we aimed to explore biological roles of COL5A2 and the potential molecular mechanism in GC. We found that COL5A2 was highly expressed in GC tissues and cells. Additionally, the expression of COL5A2 was associated with grades and stages of GC as well as the survival percentage of patients. Loss-of-function assay revealed that COL5A2 played a promotive role in GC cell proliferation and mobility in vitro. Besides, we clarified the relevant mechanism via which COL5A2 could facilitate the protein expressions of pro-epithelial–mesenchymal transition (EMT)-related genes, including SNAI1, SNAI2, TWIST, VIM, and MMP2.
2. Materials and methods
2.1. Oncomine database analysis
The expression of COL5A2 mRNA was analyzed in Oncomine database (https://www.oncomine.org), a cancer microarray online database. Four reporters, including 2591643, 221730_at, ILMN_1729117, and IMAGE:429203, were used for the analysis of COL5A2 expression in GC.
2.2. Gene expression profiling interactive analysis (GEPIA) database
The mRNA expression of COL5A2 was also analyzed by a web-based tool GEPIA in 408 GC tissues and 211 normal tissues from the Cancer Genome Atlas (TCGA) database and in 408 GC tissues and 36 normal tissues from GTEx database. The correlation between COL5A2 expression and survival of GC patients was assessed by Kaplan–Meier (KM) curves plotted by GEPIA. Stage plot was generated to analyze COL5A2 expression in four stages of GC. The correlation between COL5A2 and metastasis-related genes (SNAI1, SNAI2, TWIST, VIM, and MMP2) was evaluated using GEPIA.
2.3. UALCAN database
The mRNA expression of COL5A2 based on sample types, individual cancer stages, and tumor grades were analyzed by UALCAN (http://ualcan.path.uab.edu), an interactive web resource.
2.4. Cell culture
GC cell lines including AGS and SGC-7901 and normal cell line GSE-1 were purchased from Chinese Academy of Medical Sciences cell bank (Shanghai, China). All the cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM), which was supplemented with 10% fetal bovine serum (FBS) and penicillin–streptomycin solution. Cells in logarithmic phase were inoculated into a six-well plate for further studies.
2.5. Transfection assay
COL5A2 expression was down-regulated by transfecting small interfering RNA (siRNA) targeting COL5A2 (si-COL5A2: si-COL5A2#1, 5′-CCATCCAGTGTACCACGTAAA-3′; si-COL5A2#2, 5′-CCAGGCTCCATAGGAATCAAA-3′) into cells with the help of Lipofectamine2000 (Thermo Scientific, USA) according to the manufacture’s guidance. The si-con (5′-ACGAGACACGAACGGAGAATT-3′) was used as the control. The two COL5A2 siRNAs and si-con were obtained from GenePharma (Shanghai, China). The overexpression plasmid of COL5A2 and empty vector purchased from GenePharma (Shanghai, China) were used to transfect AGS and SGC-7901 cells using Lipofectamine2000 (Thermo Scientific, USA) in strict accordance with the manufacturer’s guidance.
2.6. Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
The total RNA was isolated using TRIzol reagent (Thermo Scientific, USA) under the manufacturer’s instructions. Complementary DNAs (cDNAs) were reversely transcribed from extracted RNAs with RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA), followed by the qRT-PCR assay with TB Green Premix Ex Taqt II (TaKaRa, Japan). The 2–ΔΔCt method was applied to determine the level of mRNA, with normalization to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA expression. The primers used were listed as follows: COL5A2 (Forward: 5′-CAGGCTCCATAGGAATCAGAGG-3′, Reverse: 5′-CCAGCATTTCCTGCTTCTCCAG-3′) and GAPDH (Forward: 5′-GTCTCCTCTGACTTCAACAGCG-3′, Reverse: 5′-ACCACCCTGTTGCTGTAGCCAA-3′).
2.7. Western blot
Total proteins isolated from the cells were quantified by bicinchoninic acid protein assay kit (Beyotime, China), followed by the separation with sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Then the proteins were electrically transferred onto a polyvinylidene fluoride membrane, blocked with 5% skim milk, incubated with primary antibodies, washed with tris-buffered saline with tween-20 and maintained with appropriate secondary antibodies. Then BeyoECL Plus substrate (Beyotime, China) exploited to visualize the target protein band. The levels of proteins were analyzed on an ImageQuant LAS 4000 system (GE Healthcare) and normalized to GAPDH expression. The primary antibodies included those against COL5A2 (dilution ratio: 1:1,000, #PA5-14245; Invitrogen, Thermo Scientific, USA), SNAI1 (dilution ratio: 1:1,000, ab216347; Abcam, UK), SNAI2 (dilution ratio: 1:1,000, ab51772; Abcam, UK), TWIST (dilution ratio: 1:1,000, ab50887; Abcam, UK), VIM (dilution ratio: 1:1,000, ab92547, Abcam, UK), MMP2 (dilution ratio: 1:1,000, ab92536; Abcam, UK), and GAPDH (dilution ratio: 1:1,000, ab8245; Abcam, UK).
2.8. Cell counting kit-8 (CCK-8) assay and colony formation assay
Cell proliferation was detected by CCK-8 kit (Beyotime, China) and colony formation assay. For CCK-8 assay, we seeded the transfected cells in a six-well plate at a density of 1,000 cells/well. CCK-8 reagent was added for surveillance of cell viability at 0, 24, 48, and 72 h. After culture for another 1.5 h, the optical density (OD) of cells was determined at a wavelength of 450 nm with a microplate reader.
For colony formation assay, the transfected cells were first seeded in a 60 mm dish (400 cells/dish) and cultured at 37℃ for 1–2 weeks. When macroscopic colonies appeared, cell culture was terminated. After the cells were fixed and stained, the number of the colonies was counted under an optical microscope.
2.9. Transwell assay
After 48 h transfection, the cells were suspended in serum-free DMEM (1 × 104 cells/100 μL) and inoculated into the upper layer of transwell chamber. The lower layer was filled with medium containing 10% FBS. After maintenance at 37℃ overnight, the migrated cells were fixed and stained. The number of cells in five randomly selected microscopic vision fields was then counted and the cells were imaged under an Olympus BX41 light microscope (Olympus Corporation) at a magnification of ×200. For invasion assay, the upper layer of transwell chamber was coated with Matrigel.
2.10. Statistical analysis
SPSS 22.0 (IBM, USA) and GraphPad Prism7 (GraphPad, USA) were used to perform statistical analysis. Student’s t-test and one-way analysis of variance were utilized to compare the difference between the two groups and among the three or more groups, respectively. When P-value was less than 0.05, the difference was statistically significant.
3. Results
3.1. COL5A2 was highly expressed in GC patients
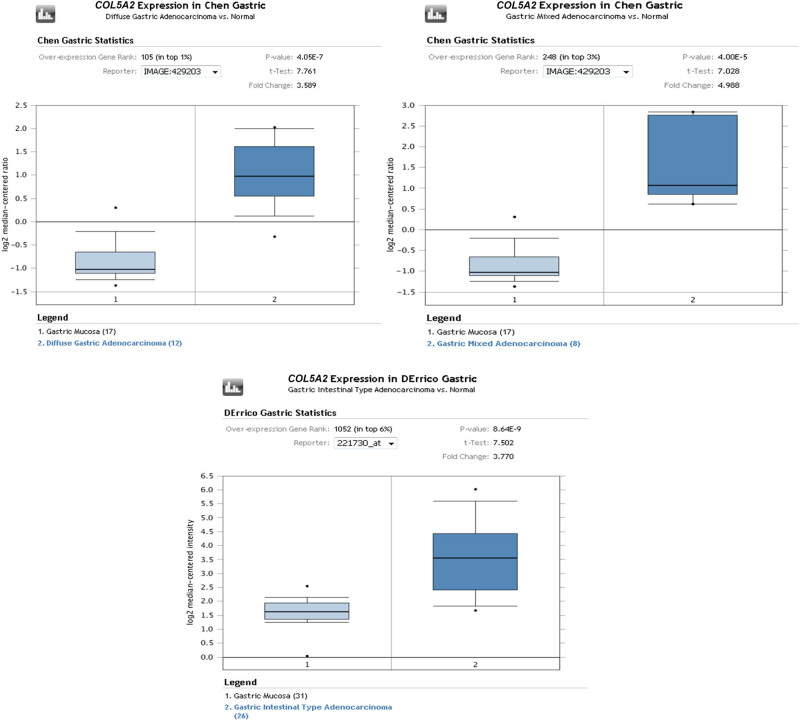
To figure out the role of COL5A2, we first analyzed the expression of COL5A2 using Oncomine database, GEPIA, and UALCAN. The results indicated that highly-expressed COL5A2 appeared in 18 of 20 tumor types, especially in breast, gastric, colorectal, and head and neck cancers (Figure 1a), which was consistent with the findings of several previous studies [18,19]. The mRNA expression of COL5A2 showed 2.294-fold (P = 2.52 × 10−15) increase in GC samples (n = 80) as compared with that in normal tissues (n = 80, Figure 1b). Besides, COL5A2 expression was elevated in three common histological subtypes of GC, including intestinal, diffused, and mixed gastric adenocarcinomas, as compared with that in gastric tissues or mucosa in Wang, Cho, Chen, and DErrico datasets (Figure 1b, Figures A1 and A2, P < 0.05).
Figure 1.
Oncomine analysis revealed that the mRNA expression of COL5A2 was up-regulated in GC. (a) mRNA expression of COL5A2 in a variety of cancers was analyzed, and COL5A2 expression was up-regulated in most tumor types. (b) mRNA expression of COL5A2 was evidently elevated in GC tissues.
Next, the mRNA level of COL5A2 was also analyzed by GEPIA with TCGA and GTEx databases. As depicted in Figure 2a, the expression of COL5A2 was determined to be higher in GC tissues than that in normal tissues (P < 0.05). Then, the survival information of 384 GC patients was classified into two groups according to the median expression level of COL5A2. The association between COL5A2 expression and overall survival of GC patients was evaluated by KM curves. Figure 2b revealed that the overall survival time was shorter in COL5A2 high expression group compared with that in COL5A2 low expression group (P < 0.01), and the cancer-specific survival time was shorter in COL5A2 high expression group as compared to that in COL5A2 low expression group (Figure 2c, P < 0.05). Besides, the mRNA expression of COL5A2 in four stages of GC was analyzed as well. The stage plot displayed that patients in stages II, III, and IV expressed higher level of COL5A2 than those in stage I (Figure 2d, P < 0.05).
Figure 2.
GEPIA analysis revealed that the mRNA expression of COL5A2 was up-regulated in GC. (a) mRNA expression of COL5A2 was analyzed in GEPIA combined with TCGA (left) or GTEx database (right); *P < 0.05. (b) KM survival curves of the overall survival for COL5A2 were plotted, and patients in COL5A2 high expression group had a shorter survival than those in COL5A2 low expression group. (c) KM survival curves of the GC-specific survival for COL5A2 was plotted, and patients in COL5A2 high expression group had a shorter survival than those in COL5A2 low expression group. (d) Association between COL5A2 expression and tumor stages of GC was analyzed by stage plot.
Further, we analyzed COL5A2 expression in GC using UALCAN based on sample types, cancer stages, and grades with TCGA database. The results further confirmed that COL5A2 expression was markedly up-regulated in 415 GC tissues relative to that in 34 normal tissues (Figure 3a, P < 0.05). When classified by cancer stages, COL5A2 expression level was notably higher in stages II, III, and IV subgroups than that in stage I and normal subgroups (Figure 3b, P < 0.05). Based on tumor grade, COL5A2 expression varied in grades I, II, and III subgroup patients (Figure 3c). Collectively, our results demonstrated that COL5A2 expression was up-regulated and associated with the prognosis and stages of GC patients, signifying the pivotal roles of COL5A2 in GC.
Figure 3.
UALCAN analysis revealed that the mRNA expression of COL5A2 was up-regulated in GC. (a–c) Expression of COL5A2 was analyzed based on sample types (a), cancer stages (b), and tumor grades (c); P < 0.05. Stomach adenocarcinoma (STAD). (d) mRNA expression of COL5A2 was detected in AGS, SGC-7901, and GES-1 cells. Data from three independent experiments were exhibited as mean ± standard deviation (SD). ∧∧∧ P < 0.001 vs GES-1.
3.2. COL5A2 expression was up-regulated in GC cells
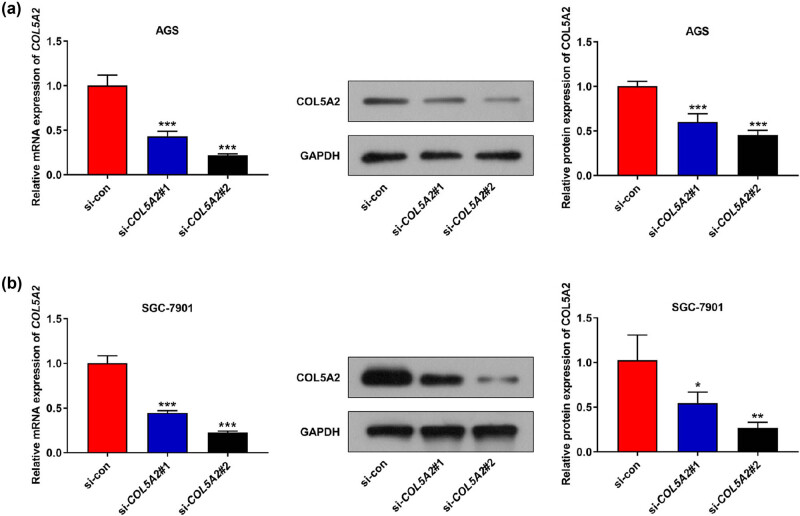
In order to investigate the roles of COL5A2 in GC in vitro, two GC cell lines, including AGS and SGC-7901, were singled out. qRT-PCR analyses revealed that the expression of COL5A2 in AGS and SGC-7901 cells was higher than that in the normal cell line GES-1 (Figure 3d, P < 0.01), which was consistent with the result of bioinformatics analysis. Next, we performed loss-of-function assay by knocking down COL5A2 level with si-COL5A2#1 and si-COL5A2#2 in GC cell lines. Figure 4a mirrored that the mRNA and protein expressions of COL5A2 were prominently decreased in AGS cells transfected with si-COL5A2#1 and si-COL5A2#2 as compared with those in cells transfected with si-con (P < 0.01). Moreover, the same results were obtained in SGC-7901 cells (Figure 4b, P < 0.01). Meanwhile, si-COL5A2#2 displayed better efficiency in AGS and SGC-7901 cells than in si-COL5A2#1 (Figure 4a and b, P < 0.01). Therefore, we selected si-COL5A2#2 to transfect GC cells for further analyses.
Figure 4.
COL5A2 expression was down-regulated in GC cells after transfected with si-COL5A2. (a) mRNA and protein expressions of COL5A2 were analyzed in si-COL5A2 (si-COL5A2#1 or si-COL5A2#2)- or si-con-transfected AGS cells. (b) mRNA and protein expressions of COL5A2 were observed in si-COL5A2 (si-COL5A2#1 or si-COL5A2#2) or si-con-transfected SGC-7901 cells. Data from three independent experiments were expressed as mean ± SD. *P < 0.05, **P < 0.01 and ***P <0.001 vs si-con group.
3.3. Knockdown of COL5A2 inhibited the proliferation, migration, and invasion of GC cells
Next, cell viability assay and transwell assay were conducted to explore the roles of COL5A2 in biological behavior of GC cells. As shown in Figure 5a–d, OD values and colony numbers of AGS and SGC-7901 cells were notably decreased in si-COL5A2 group, as compared with those in si-con group (P < 0.01). Besides, AGS and SGC-7901 cells transfected with si-COL5A2 appeared to decrease in migratory and invasive rates when compared with those transfected with si-con (Figure 6a and b, P < 0.01). These results suggested that COL5A2 positively regulated the proliferation, migration, and invasion of GC cells.
Figure 5.
Knockdown of COL5A2 inhibited proliferation of GC cells. (a and b) OD values in si-COL5A2- or si-con-transfected GC cell lines including AGS (a) and SGC-7901 (b) were determined by CCK-8 assay. (c and d) Colony numbers of si-COL5A2- or si-con-transfected GC cell lines including AGS (c) and SGC-7901 (d) were analyzed by colony formation assay. *P < 0.05, **P < 0.01 and ***P < 0.001 vs si-con group.
Figure 6.
Knockdown of COL5A2 inhibited the migratory and invasive rates of GC cells. (a and b) Numbers of migratory and invasive cells in si-COL5A2- or si-con-transfected AGS (a) and SGC-7901 (b) cells were analyzed by transwell assay. Data from three independent experiments were described as mean ± SD. *P < 0.05, **P < 0.01 and ***P < 0.001 vs si-con group.
3.4. Knockdown of COL5A2 inhibited biological behaviors of GC cells via regulating EMT-associated genes
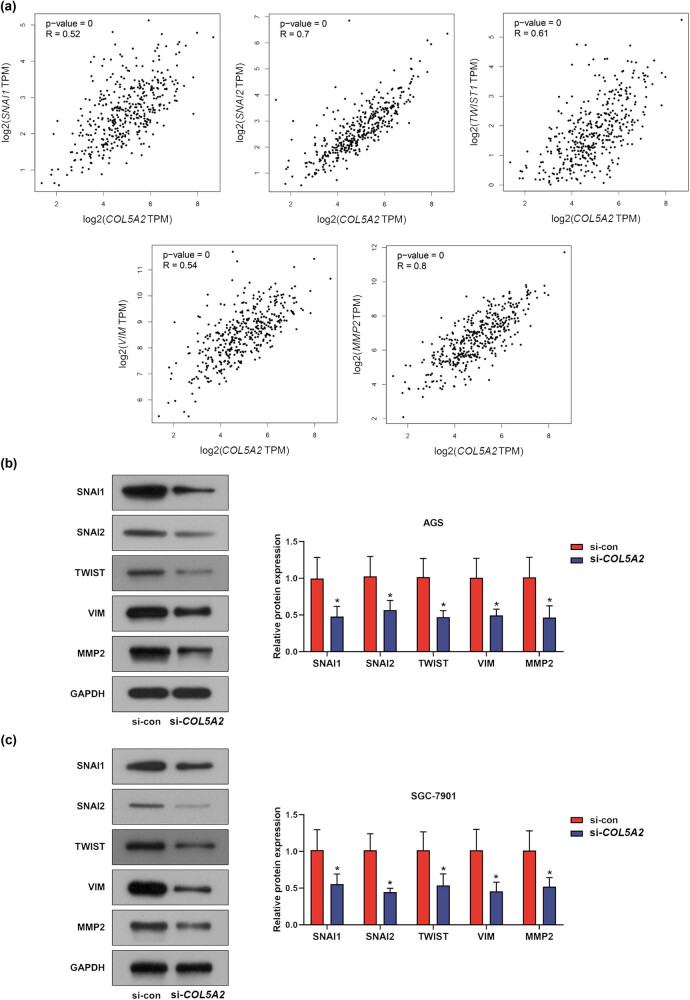
To find out the molecular mechanism on how COL5A2 regulates the viability, migration, and invasion of GC cells, we evaluated the correlation between COL5A2 expression and EMT-associated genes including SNAI1, SANI2, TWIST1, VIM, and MMP2 (Figure 7a). First, Spearman’s correlation test revealed that COL5A2 was positively correlated with the expressions of several mesenchymal markers including SNAI1 (P = 0, R = 0.52), SANI2 (P = 0, R = 0.7), TWIST1 (P = 0, R = 0.61), VIM (P = 0, R = 0.54), and MMP2 (P = 0, R = 0.8). Next, western blot was conducted to detect the protein expressions of the above-mentioned mesenchymal markers in COL5A2-silenced GC cells. As depicted in Figure 7b and c, the protein expressions of SNAI1, SANI2, TWIST1, VIM, and MMP2 were dramatically decreased in si-COL5A2 group when compared with those in si-con group (P < 0.01). These findings indicated that the knockdown of COL5A2 could inhibit the protein expressions of EMT-associated markers including SNAI1, SANI2, TWIST1, VIM, and MMP2 to suppress the EMT process of GC.
Figure 7.
Knockdown of COL5A2 inhibited biological behaviors of GC cells via regulating EMT-associated genes. (a) Correlation between COL5A2 expression and EMT-associated genes including VIM, SNAI1, SNAI2, VIM, and MMP2 was analyzed with GEPIA. (b and c) Protein expressions of VIM, SNAI1, SNAI2, VIM, and MMP2 in si-con- or si-COL5A2-transfected AGS (b) or SGC-7901 (c) cells were determined by western blot assay. Data from three independent experiments were presented as mean ± SD. *P < 0.05 vs si-con group.
3.5. Overexpression of COL5A2 promoted the proliferation, migration, invasion, and EMT-associated marker expressions in GC cells
In addition, overexpression of COL5A2 in the GC was also detected. As shown in Figure 8a–d, compared to those in the control group, the OD value and clone number were increased in the COL5A2 group (P < 0.01). Meanwhile, the migration and invasion were increased by overexpressed COL5A2 (Figure 9a and b, P < 0.05). The protein expressions of SNAI1, SANI2, TWIST1, VIM, and MMP2 were higher in COL5A2 group than those in control group (Figure 10a and b, P < 0.05). These findings indicated that overexpression of COL5A2 promoted the proliferation, migration, invasion, and EMT-associated marker expressions in GC.
Figure 8.
Overexpression of COL5A2 promoted the proliferation of GC cells. (a and b) After AGS and SGC-7901 cells were transfected with COL5A2 overexpression plasmid, the viability of AGS (a) and SGC-7901 cells (b) were analyzed by CCK-8 assay. (c and d) After AGS and SGC-7901 cells were transfected with COL5A2 overexpression plasmid, the colony number of AGS (c) and SGC-7901 cells (d) was analyzed by colony formation assay. **P < 0.01, ***P < 0.001 vs Con group.
Figure 9.
Overexpression of COL5A2 promoted the migration and invasion of GC cells. (a and b) After AGS and SGC-7901 cells were transfected with COL5A2 overexpression plasmid, the numbers of migratory and invasive cells in AGS (a) and SGC-7901 cells (b) were analyzed by transwell assay. Data from three independent experiments were exhibited by mean ± SD. *P < 0.05, **P < 0.01 vs Con group.
Figure 10.
Overexpression of COL5A2 promoted biological behaviors of GC cells via inducing EMT-associated genes. (a and b) After AGS and SGC-7901 cells were transfected with COL5A2 overexpression plasmid, the protein expressions of SNAI1, SNAI2, TWIST, VIM, and MMP2 in AGS (a) or SGC-7901 cells (b) were analyzed by western blot assay. Data from three independent experiments were exhibited by mean ± SD. *P < 0.05, **P < 0.01 vs Con group.
4. Discussion
Multiple bioinformatics analyses have manifested an oncogenic role of COL5A2 in GC. It has been reported that COL5A2 was an overexpressed hub gene in GC and associated with the outcome of GC patients [23]. Consistently, our study found that COL5A2 expression was up-regulated and negatively correlated with the survival percentage of GC. Besides, there was a notable elevation of COL5A2 expression in stages II, III, and IV compared to that in stage I. Meanwhile, the expression of COL5A2 was higher in GC in grade III than that in grade I. These results implied that COL5A2 may play a critical role in the progression of GC.
With the in-depth study of GC, several oncogenes of GC have been found. For example, a high level of legumain was correlated with worse prognosis and peritoneal metastasis in GC patients [24]. C-Maf-inducing protein is an oncogene in human GC cells [25]. Besides, cadherin-6 was highly expressed in GC, and its high expression was correlated with tumor progression and poor prognosis of patients with GC [26]. Additionally, the mRNA expression of FBXO50 was increased in GC cell lines, and positively correlated with levels of ITGA5, ITGB1, MMP2, MSN, COL5A2, GNG11, and WNT5A [27]. In this study, we discovered that COL5A2 was an oncogenic factor in GC, and silencing of COL5A2 could repress the proliferation, migration, and invasion of GC cells.
ECM is a key mediator of extracellular microenvironment, which plays an essential role in cancer development by regulating cell transformation, mobility, tumor initiation, and metastasis [28]. Deposited and fibrotic collagens could provide a linearized “tunnel” for cancer cells to migrate and invade [29]. For example, Choi et al. have revealed that weak expression of lysyl oxidase supports the growth and metastasis of breast cancer via regulating the production and remodeling of collagens I and IV [30]. Importantly, COL5A3 has been reported to enhance proliferative potential in breast cancer cells via binding a membrane proteoglycan GPC-1 [31]. Similarly, in this present study, COL5A2 potentiated the proliferation, migration, and invasion of GC cells.
As the basic component of ECM, collagens are reported to be involved in the process of EMT [32]. Wei et al. have demonstrated that the accumulation of fibrotic collagens facilitates tumor metastasis by initiating EMT [33]. Besides, collagen type I could augment SNAI1- and LEF-1-mediated EMT in breast cancer [34]. EMT is a cellular process that allows epithelial cells to gain mesenchymal characteristics and entails their mobility via loss of cell adhesion [35]. In this way, EMT facilitates the tumorigenesis, metastasis, and growth of cancers [36]. Recently, accumulating evidence has expounded that there is an intimate correlation among aberrant EMT, cell invasion, and spread of GC [37,38]. EMT is a reversible process involved in two major hallmark proteins, namely adhesion molecules and mesenchymal markers, the former ones contribute to the assembly of epithelial cells and the latter ones are helpful to cancer cell invasion and metastasis [39]. In GC, EMT is induced along with alternation in expressions of various mesenchymal proteins [40]. VIM, a cytoskeletal protein, could promote cell mobility by remodeling cytoskeleton in the process of EMT [41]. SNAI1, SNAI2, and TWIST are three main transcriptional factors regulating EMT [41,42]. Miyoshi et al. have pointed out that SNAI1 could facilitate the invasion of liver cancer cells by elevating the expressions of MMPs [43]. These EMT-associated genes also have been investigated in GC. For example, Zhang et al. have demonstrated that NUB1 blocks EMT process in GC by suppressing the expressions of EMT-related proteins including N-cadherin, VIM, and MMP-2 [44]. AOC1 induces EMT by elevating the expressions of mesenchymal markers including N-cadherin, SNAI1, and SNAI2 and facilitates the progression of GC [45]. Interestingly, similar results were obtained in our study that the protein expressions of VIM, SNAI1, SNAI2, MMP2, and TWIST were decreased by the inhibition of COL5A2, suggesting that COL5A2 could trigger EMT process to regulate the malignant phenotypes of GC. Of course, the effect of COL5A2 on GC needs to be further elucidated in animal experiments in vivo.
In summary, our study reveals that COL5A2 expression is up-regulated and associated with the dismal outcome of GC patients. Besides, COL5A2 may be correlated with the higher stages and grades of this disease. COL5A2 promoted cell growth and mobility of GC. Collectively, COL5A2 facilitates malignant phenotypes of GC through inducing EMT via promoting the protein expressions of SNAI1, SNAI2, TWIST, VIM, and MMP2, demonstrating the oncogenic role of COL5A2 in GC and providing a novel clue for the therapeutic application of COL5A2 in GC.
Acknowledgement
Not applicable.
Appendix
Figure A1.
Oncomine analysis revealed that the mRNA expression of COL5A2 was up-regulated in GC.
Figure A2.
The mRNA expression of COL5A2 was evidently elevated in three common histological subtypes of gastric cancer including intestinal, diffused and mixed gastric adenocarcinomas in Chen and DErrico datasets.
Footnotes
Funding information: Not applicable.
Conflict of interest: The authors report no conflict of interest.
Data availability statement: The analytical datasets generated during the study can be made available from the corresponding author on reasonable request.
References
- [1].Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed]
- [2].Wang FH, Shen L, Li J, Zhou ZW, Liang H, Zhang XT, et al. The Chinese Society of Clinical Oncology (CSCO): clinical guidelines for the diagnosis and treatment of gastric cancer. Cancer Commun (Lond). 2019;39(1):10. [DOI] [PMC free article] [PubMed]
- [3].Japanese gastric cancer treatment guidelines 2014 (ver. 4). Gastric Cancer. 2017;20(1):1–19. [DOI] [PMC free article] [PubMed]
- [4].Shen H, Wang L, Chen Q, Xu J, Zhang J, Fang L, et al. The prognostic value of COL3A1/FBN1/COL5A2/SPARC-mir-29a-3p-H19 associated ceRNA network in gastric cancer through bioinformatic exploration. J Cancer. 2020;11(17):4933–46. [DOI] [PMC free article] [PubMed]
- [5].Wu HH, Lin WC, Tsai KW. Advances in molecular biomarkers for gastric cancer: miRNAs as emerging novel cancer markers. Expert Rev Mol Med. 2014;16:e1. [DOI] [PMC free article] [PubMed]
- [6].Asao T, Fukuda T, Yazawa S, Nagamachi Y. Carcinoembryonic antigen levels in peritoneal washings can predict peritoneal recurrence after curative resection of gastric cancer. Cancer. 1991;68(1):44–7. [DOI] [PubMed]
- [7].Shimada H, Noie T, Ohashi M, Oba K, Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer. 2014;17(1):26–33. [DOI] [PubMed]
- [8].Wahyudi H, Reynolds AA, Li Y, Owen SC, Yu SM. Targeting collagen for diagnostic imaging and therapeutic delivery. J Control Rel. 2016;240:323–31. [DOI] [PMC free article] [PubMed]
- [9].Fang M, Yuan J, Peng C, Li Y. Collagen as a double-edged sword in tumor progression. Tumour Biol. 2014;35(4):2871–82. [DOI] [PMC free article] [PubMed]
- [10].Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139(5):891–906. [DOI] [PMC free article] [PubMed]
- [11].Arnold SA, Rivera LB, Miller AF, Carbon JG, Dineen SP, Xie Y, et al. Lack of host SPARC enhances vascular function and tumor spread in an orthotopic murine model of pancreatic carcinoma. Dis Model Mech. 2010;3(1–2):57–72. [DOI] [PMC free article] [PubMed]
- [12].Myllyharju J, Kivirikko KI. Collagens and collagen-related diseases. Ann Med. 2001;33(1):7–21. [DOI] [PubMed]
- [13].Shoulders MD, Raines RT. Collagen structure and stability. Annu Rev Biochem. 2009;78:929–58. [DOI] [PMC free article] [PubMed]
- [14].Vargas AC, McCart Reed AE, Waddell N, Lane A, Reid LE, Smart CE, et al. Gene expression profiling of tumour epithelial and stromal compartments during breast cancer progression. Breast Cancer Res Treat. 2012;135(1):153–65. [DOI] [PubMed]
- [15].Yue H, Wang J, Chen R, Hou X, Li J. Gene signature characteristic of elevated stromal infiltration and activation is associated with increased risk of hematogenous and lymphatic metastasis in serous ovarian cancer. BMC Cancer. 2019;19(1):1266. [DOI] [PMC free article] [PubMed]
- [16].Guillen-Ahlers H, Buechler SA, Suckow MA, Castellino FJ, Ploplis VA. Sulindac treatment alters collagen and matrilysin expression in adenomas of ApcMin/+ mice. Carcinogenesis. 2008;29(7):1421–7. [DOI] [PMC free article] [PubMed]
- [17].Zeng XT, Liu XP, Liu TZ, Wang XH. The clinical significance of COL5A2 in patients with bladder cancer: a retrospective analysis of bladder cancer gene expression data. Medicine (Baltimore). 2018;97(10):e0091. [DOI] [PMC free article] [PubMed]
- [18].Li Z, Liu Z, Shao Z, Li C, Li Y, Liu Q, et al. Identifying multiple collagen gene family members as potential gastric cancer biomarkers using integrated bioinformatics analysis. PeerJ. 2020;8:e9123. [DOI] [PMC free article] [PubMed]
- [19].Wang Y. Transcriptional regulatory network analysis for gastric cancer based on mRNA microarray. Pathol Oncol Res. 2017;23(4):785–91. [DOI] [PubMed]
- [20].Liu Q, Zhang W. Construction of a circular RNA-microRNA-messengerRNA regulatory network in stomach adenocarcinoma. J Cell Biochem. 2020;121(2):1317–31. [DOI] [PubMed]
- [21].Tan Y, Chen Q, Xing Y, Zhang C, Pan S, An W, et al. High expression of COL5A2, a member of COL5 family, indicates the poor survival and facilitates cell migration in gastric cancer. Biosci Rep. 2021;41(4):BSR20204293. [DOI] [PMC free article] [PubMed]
- [22].Ding YL, Sun SF, Zhao GL. COL5A2 as a potential clinical biomarker for gastric cancer and renal metastasis. Medicine (Baltimore). 2021;100(7):e24561. [DOI] [PMC free article] [PubMed]
- [23].Cao L, Chen Y, Zhang M, Xu DQ, Liu Y, Liu T, et al. Identification of hub genes and potential molecular mechanisms in gastric cancer by integrated bioinformatics analysis. PeerJ. 2018;6:e5180. [DOI] [PMC free article] [PubMed]
- [24].Wang Y, Zhang S, Wang H, Cui Y, Wang Z, Cheng X, et al. High level of legumain was correlated with worse prognosis and peritoneal metastasis in gastric cancer patients. Front Oncol. 2020;10:966. [DOI] [PMC free article] [PubMed]
- [25].Zhang J, Huang J, Wang X, Chen W, Tang Q, Fang M, et al. CMIP is oncogenic in human gastric cancer cells. Mol Med Rep. 2017;16(5):7277–86. [DOI] [PMC free article] [PubMed]
- [26].Zhao Z, Li S, Li S, Wang J, Lin H, Fu W. High expression of oncogene cadherin-6 correlates with tumor progression and a poor prognosis in gastric cancer. Cancer Cell Int. 2021;21(1):493. [DOI] [PMC free article] [PubMed]
- [27].Miwa T, Kanda M, Tanaka H, Tanaka C, Kobayashi D, Umeda S, et al. FBXO50 enhances the malignant behavior of gastric cancer cells. Ann Surg Oncol. 2017;24(12):3771–9. [DOI] [PubMed]
- [28].Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196(4):395–406. [DOI] [PMC free article] [PubMed]
- [29].Giese A, Kluwe L, Laube B, Meissner H, Berens ME, Westphal M. Migration of human glioma cells on myelin. Neurosurgery. 1996;38(4):755–64. [PubMed]
- [30].Choi SK, Kim HS, Jin T, Moon WK. LOXL4 knockdown enhances tumor growth and lung metastasis through collagen-dependent extracellular matrix changes in triple-negative breast cancer. Oncotarget. 2017;8(7):11977–89. [DOI] [PMC free article] [PubMed]
- [31].Huang G, Ge G, Izzi V, Greenspan DS. α3 Chains of type V collagen regulate breast tumour growth via glypican-1. Nat Commun. 2017;8:14351. [DOI] [PMC free article] [PubMed]
- [32].Tzanakakis G, Kavasi R-M, Voudouri K, Berdiaki A, Spyridaki I, Tsatsakis A, et al. Role of the extracellular matrix in cancer-associated epithelial to mesenchymal transition phenomenon. Dev Dyn. 2018;247(3):368–81. [DOI] [PubMed]
- [33].Wei SC, Fattet L, Tsai JH, Guo Y, Pai VH, Majeski HE, et al. Matrix stiffness drives epithelial–mesenchymal transition and tumour metastasis through a TWIST1–G3BP2 mechanotransduction pathway. Nat Cell Biol. 2015;17(5):678–88. [DOI] [PMC free article] [PubMed]
- [34].Medici D, Nawshad A. Type I collagen promotes epithelial–mesenchymal transition through ILK-dependent activation of NF-kappaB and LEF-1. Matrix Biol. 2010;29(3):161–5. [DOI] [PMC free article] [PubMed]
- [35].Lu H, Zhang Q, Sun Y, Wu D, Liu L. LINC00689 induces gastric cancer progression via modulating the miR-338-3p/HOXA3 axis. J Gene Med. 2020;22(12):e3275. [DOI] [PubMed]
- [36].Xu Q, Deng F, Qin Y, Zhao Z, Wu Z, Xing Z, et al. Long non-coding RNA regulation of epithelial–mesenchymal transition in cancer metastasis. Cell Death Dis. 2016;7(6):e2254. [DOI] [PMC free article] [PubMed]
- [37].Zhang H, Wu X, Xiao Y, Wu L, Peng Y, Tang W, et al. Coexpression of FOXK1 and vimentin promotes EMT, migration, and invasion in gastric cancer cells. J Mol Med (Berl). 2019;97(2):163–76. [DOI] [PubMed]
- [38].Sun S, Hang T, Zhang B, Zhu L, Wu Y, Lv X, et al. miRNA-708 functions as a tumor suppressor in colorectal cancer by targeting ZEB1 through Akt/mTOR signaling pathway. Am J Transl Res. 2019;11(9):5338–56. [PMC free article] [PubMed]
- [39].Santamaria PG, Moreno-Bueno G, Portillo F, Cano A. EMT: present and future in clinical oncology. Mol Oncol. 2017;11(7):718–38. [DOI] [PMC free article] [PubMed]
- [40].Peng Z, Wang CX, Fang EH, Wang GB, Tong Q. Role of epithelial–mesenchymal transition in gastric cancer initiation and progression. World J Gastroenterol. 2014;20(18):5403–10. [DOI] [PMC free article] [PubMed]
- [41].Kim B, Sohn EJ, Jung JH, Shin EA, You OH, Im J, et al. Inhibition of ZNF746 suppresses invasion and epithelial to mesenchymal transition in H460 non-small cell lung cancer cells. Oncol Rep. 2014;31(1):73–8. [DOI] [PubMed]
- [42].Jagadish N, Parashar D, Gupta N, Agarwal S, Purohit S, Kumar V, et al. A-kinase anchor protein 4 (AKAP4) a promising therapeutic target of colorectal cancer. J Exp Clin Cancer Res. 2015;34:142. [DOI] [PMC free article] [PubMed]
- [43].Miyoshi A, Kitajima Y, Sumi K, Sato K, Hagiwara A, Koga Y, et al. Snail and SIP1 increase cancer invasion by upregulating MMP family in hepatocellular carcinoma cells. Br J Cancer. 2004;90(6):1265–73. [DOI] [PMC free article] [PubMed]
- [44].Zhang D, Wu P, Zhang Z, An W, Zhang C, Pan S, et al. Overexpression of negative regulator of ubiquitin-like proteins 1 (NUB1) inhibits proliferation and invasion of gastric cancer cells through upregulation of p27Kip1 and inhibition of epithelial–mesenchymal transition. Pathol Res Pract. 2020;216(8):153002. [DOI] [PubMed]
- [45].Xu F, Xu Y, Xiong JH, Zhang JH, Wu J, Luo J, et al. AOC1 contributes to tumor progression by promoting the AKT and EMT pathways in gastric cancer. Cancer Manag Res. 2020;12:1789–98. [DOI] [PMC free article] [PubMed]