Summary
Novel therapeutics to manage bacterial infections are urgently needed as the impact and prevalence of antimicrobial resistance (AMR) grows. Antivirulence therapeutics are an alternative approach to antibiotics that aim to attenuate virulence rather than target bacterial essential functions, while minimizing microbiota perturbation and the risk of AMR development. Beyond known virulence factors, pathogen-associated genes (PAGs; genes found only in pathogens to date) may play an important role in virulence or host association. Many identified PAGs encode uncharacterized hypothetical proteins and represent an untapped wealth of novel drug targets. Here, we review current advances in antivirulence drug research and development, including PAG identification, and provide a comprehensive workflow from the discovery of antivirulence drug targets to drug discovery. We highlight the importance of integrating bioinformatic/genomic-based methods for novel virulence factor discovery, coupled with experimental characterization, into existing drug screening platforms to develop novel and effective antivirulence drugs.
Keywords: Antivirulence drug, Bacterial pathogens, Microbial genomics, Antimicrobial resistance, Infectious disease, Novel therapeutics
Introduction
Since Alexander Fleming's discovery of the first clinical antibiotic, penicillin, in 1928,1 antibiotics have drastically improved human health and expanded the average human lifespan.2 However, the frequent use of antibiotics accelerated the development of antimicrobial resistance (AMR) and contributed to a persistent and pressing global health crisis. According to the World Health Organization (WHO), AMR-related infections currently account for over 700,000 annual deaths and are projected to reach a catastrophic ten million annual deaths by 2050.3 The WHO declared AMR as one of the top ten global health threats in 2015 and published a list of priority AMR pathogens in urgent need of novel therapeutics in 2017.4, 5, 6 Antibiotic stewardship programs have been implemented to govern the clinical use of antibiotics but there is currently no conclusive evidence of these programs being effective in reducing AMR in hospital settings.7
The AMR crisis is further compounded by the limited innovative antibiotic classes that were successfully developed and approved for clinical use in the past few decades.8 The decline in development is attributed to the diminishing research funding devoted to antimicrobial drug discovery within the private sector. The inability to recoup the high cost of antibiotic development (estimated $1.5 billion USD) from sales revenue (estimated $46 million USD per year) has deterred many pharmaceutical and biotechnology companies from antimicrobial research.9 Additionally, limitations on sales of antibiotics due to careful stewardship from clinicians, the acute treatment duration, and the development of resistance by the pathogen further dissuades drug manufacturers from investing in developing new antibiotics.10 In the past few decades, most antibiotic development efforts have relied on the generation of derivatives of existing antibiotic classes to which bacteria already have or will likely develop broad resistance to. The WHO deems the current drug development pipeline incapable of tackling the global emergence and spread of AMR.11
Recent drug research and development efforts have increasingly turned to alternative therapeutic strategies which target bacterial pathogens through mechanisms distinct from existing antibiotics. There are three main premises for novel antimicrobial development strategies laid out by the WHO3: (1) reducing pathogen virulence or host damage, (2) minimizing resistance development in bacterial pathogens, and (3) addressing the lack of target specificity in broad-spectrum antibiotics. Precision antimicrobials are a recent and innovative therapeutic concept that aims to combat the rise of AMR pathogens by narrowing the activity to target pathogen-specific (virulence) components without disturbing the host microbiota.12, 13, 14 These antivirulence strategies aim to have specific activity against only the disease-causing bacterial pathogen and reduce selective pressures for resistance since they do not necessarily kill the pathogen and have minimal impact on the host's microbiota at large. Alternative antimicrobial approaches include the use of bacteriophages or phage-derived enzymes, immunomodulatory and antimicrobial peptides, quorum sensing inhibitors, biofilm disruptors, microbiota-modulating therapies, and antibody-drug conjugates.15 These are beyond the scope of this review and we recommend these other reviews by Ghosh et al.,15 Mookherjee et al.,16 and Nguyen et al.17 for more in-depth explorations of other alternative antimicrobial therapeutic approaches.
Here, we review current antivirulence drug research and development, with a focus on methodologies for genomics-based novel virulence factor identification and drug target prioritization. We present an integrated antivirulence drug discovery workflow that can be applicable towards any bacterial pathogen. Specifically, our proposed workflow aims to serve as a comprehensive guide, from the selection of pathogen-specific targets, functional characterization of pathogen-specific genes with unknown function, to the discovery of antivirulence compounds.
The antivirulence strategy against bacterial pathogens
The antivirulence strategy has been applied most notably in vaccine development. Many vaccines are in fact antivirulence agents since they target against key toxins and other virulence factor components by generating an immune response.18 However, for therapeutic agents (versus vaccines which are prophylactic agents) there has been more of a focus historically on antimicrobials. Only recently, as antimicrobial resistance becomes a significant threat, has interest shifted further to expand therapeutic approaches using an antivirulence approach.
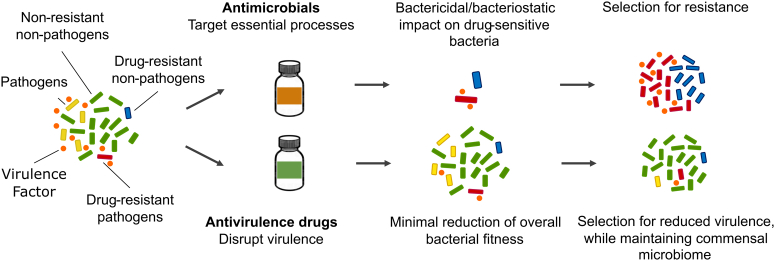
Unlike antimicrobials which focus on targeting essential bacterial processes, antivirulence therapeutic agents specifically target disease-causing components, or virulence factors, produced by bacterial pathogens—with minimal impact on their growth and survival (Fig. 1). While there are examples of antivirulence therapeutics that can impact growth,19 we propose that one of the major criterion in the development of novel antivirulence therapies is to ensure cell growth is not impacted. This reduces selective pressure for basic cellular survival and development of resistance to the therapeutic.20 As illustrated in part in Fig. 1, we postulate that antivirulence therapeutics can mitigate disease by diverse mechanisms (i.e., reducing host damage due to a virulence factor, while providing time for the host's immunity to clear the pathogen, and/or not causing damage to non-disease-causing microbiota, providing more competition for the potentially disadvantaged pathogenic bacteria).15, 16, 17, 18,20,21 Under the classic antibiotic regime in which most drug-sensitive bacteria are eliminated, some microbial species such as the opportunistic pathogen, Clostridioides difficile, can grow rapidly leading to microbial dysbiosis and secondary infections. In contrast, pathogen-specific monoclonal antibodies have been shown to preserve intestinal microbiome while targeting specific microorganisms in a mouse metagenomic study.21 Maintaining a normal microbiome, or supplementation with microbes that support the gut, has been shown to greatly limit the risk of some infections, such as by C. difficile, or help resolve such infections.22 Thus, precision antimicrobials and antivirulence therapeutics represent a promising approach in mitigating virulence of the disease-causing bacteria, preserving the host microbiota, and allowing subsequent pathogen clearance by the host immune system.
Fig. 1.
Comparison of traditional antimicrobials andantivirulence drugs. When exposed to an antimicrobial drug to treat an infection, the healthy microbiota and the pathogen are both impacted. Under such conditions, there is selection for antimicrobial resistance such that resistant microbes can potentially flourish, less impeded by the normal microbiota. When exposed to an antivirulence drug, the microbiota are relatively unaffected; in contrast, disease-causing pathogenic microbes compete with the healthy microbiota for resources while virulence is attenuated. Consequently, antivirulence approaches mitigate disease outcomes by neutralizing virulence factors, reducing the competitive advantage of the pathogen, aiding the immune system to effectively clear the pathogen, and/or selecting for commensalism—all with minimal disruption to the microbiota.
Antivirulence therapeutics developed to date
Antivirulence therapeutics to date largely focus on the inhibition of well-studied virulence factors associated with toxins, secretion systems and quorum sensing.23 For instance, several antivirulence drugs targeting bacterial toxins have been approved by the US Food and Health Administration (FDA) (Table 1). Raxibacumab, a monoclonal antibody against the Bacillus anthracis protective antigen, was one of the earliest antivirulence drugs approved in 2012.24 BabyBIG, approved in 2016, targets the botulinum toxin of Clostridium botulinum.25 Benzlotoxumab, FDA-approved in 2016, targets the C. difficile toxin B.26 Antivirulence drugs currently under clinical trials include: MEDI3902 which targets the Pseudomonas aeruginosa Type III secretion system toxin translocator PcrV and the biofilm exopolysaccharide Psl27; Shigamab which targets the Escherichia coli shiga toxin type 228; and MEDI4893 which targets the Staphylococcus aureus alpha toxin.29 Antivirulence compounds currently in use or under development target a variety of biological components from direct virulence factors (e.g., toxins, secretion system) to virulence-related proteins such as quorum-sensing molecules, regulators of virulence factor synthesis, and biofilm-associated proteins (Table 1).
Table 1.
Antivirulence drugs in development or approved, including natural products and repurposed drugs.
| Anti-virulence drug | Drug repur-posing | Target pathogen | Drug target | Effects | Drug discovery method | Target discovery method | Infection model | Status of the study | PMID | Publication year |
|---|---|---|---|---|---|---|---|---|---|---|
| Raxibacumab | No | Bacillus anthracis | protective antigen (PA) | Block receptor interaction of PA | Mab engineering | PA as chosen target | Rat, rabbit and monkey models | FDA-approved–2012 | 19587338 | 2009 |
| Obitoxaximab (ETI-204) | No | Bacillus anthracis | protective antigen (PA) | Block receptor interaction of PA | Mab engineering | PA as chosen target | Rabbit model | FDA-approved–2016 | 15664918 | 2006 |
| BabyBIG | No | Clostridium botulinum | Botulinum neurotoxins (BoNT) | Mitigates botulism intoxication | Human version of the equine botulism antitoxin | N/A | N/A | FDA-approved (2016) | 17484226 | 2007 |
| Botulism Antitoxin Heptavalent | No | Clostridium botulinum | BoNT serotype A-G | Mitigates botulism intoxication | N/A | BoTN as chosen targets | Guinea pig model | FDA-approved (2013) | 16452558 | 2006 |
| MEDI3902 | No | Pseudomonas aeruginosa | PsI & PcrV | Inhibits P. aeruginosa host cell attachment and promote opsonophagocytic killing | N/A | N/A | Murine model of pneumonia, bacteremia and thermal injury | Clinical–Phase II | 30107283 | 2018 |
| Shigamab | No | Escherichia coli | shiga toxin type 2 (Stx2) | Reduces renal damage in mice | Follow-up development after discovery of several Stx1 and Stx2-neutralizing antibodies | Stx2 as chosen target | Mouse model | Clinical–Phase I completed (NCT01252199) | 28337021 | 2013 |
| MEDI4893 | No | Staphylococcus aureus | alpha-toxin (AT) | Prevents AT-induced cell lysis | Development of an affinity-optimized, AT-neutralizing monoclonal antibody | AT as chosen target due to its suppression of adaptive and innate responses | Immunocompromised murine pneumonia model | Clinical––Phase II | 25987629 | 2015 |
| MAC-545496 (small molecule inhibitor) | No | MRSA | Glycopeptide-resistance-associated protein R (GraR) | Reverses β-lactam resistance in S. aureus | High-throughput synthetic small molecule screening for targocil antagonism | Transposon library screening | Wax moth larvae infection model | Preclinical | 31768032 | 2019 |
| Clotrimazole & miconazole (anti-fungal drugs) | Yes | Pseudomonas aeruginosa | PqsR | Reduces 2-alkyl-4-quinolone production | Screening of PHARMAKON library of 1600 FDA-approved drugs | docking assay | Wax moth larvae infection model | Preclinical | 30201815 | 2018 |
| Baicalin (Scutelleria) | Yes | Staphylococcus aureus | Sortase B | Reduces S. aureus adhesion and epithelial cell damage; attenuates inflammatory responses | N/A | SrtB as chosen target | Human alveolar epithelial cell model | Preclinical | 30555803 | 2018 |
| Azorella atacamensis (medicinal plant) | Yes | Pseudomonas aeruginosa | N-acyl homoserine lactones (AHLs) and/or hydroxy-alkyl quinolines (HAQs) | Decreases QS-molecule production–pyocyanin, elastase rhamnolipids production in P. aeruginosa | Screening o A. atacamensis phytochemicals for antivirulence properties | AHLs and HAQs as chosen target | C.elegans and human lung infection models | Preclinical | 33276611 | 2020 |
For an in-depth and more exhaustive discussion of current therapeutics available and the development of novel antivirulence therapies, we recommend recent reviews from Allen et al.,20 Dickey et al.,23 and Mattar et al.30 Additionally, the “Antimicrobial Resistance Collaborators” have provided a recent, systematic review on global incidence of antimicrobial resistance and guidelines to reduce the associated public health and socioeconomic burden of AMR.31
Towards new therapeutic targets: expanding and integrating workflows
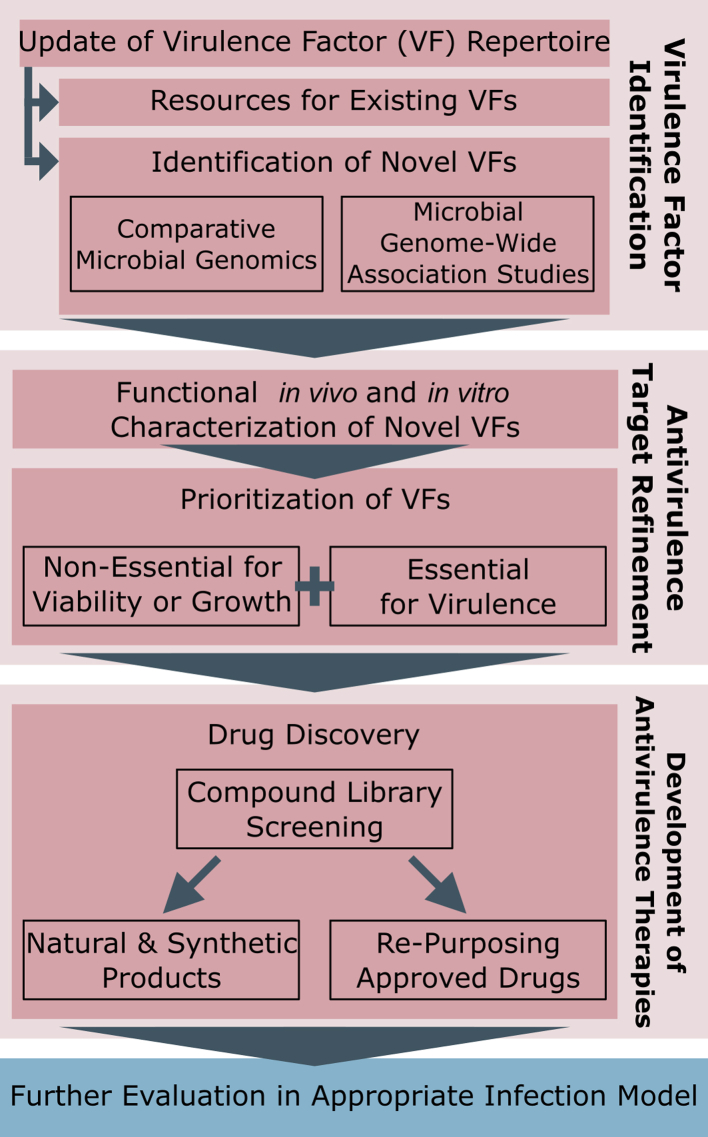
While there are a handful of new antivirulence therapies being developed, there is a need to increase the repertoire of possible drug targets and drug candidates. We can expand this repertoire by integrating workflows for the discovery of novel virulence factors together with the development of antivirulence therapeutics as follows (Fig. 2): (1) incorporating more genomics/bioinformatics approaches, (2) coupling this with more research on molecular mechanisms of virulence factors, and (3) incorporating into existing drug discovery pipelines with a focus on targeting microbial virulence rather than viability. This workflow here is intended to introduce omics approaches for discovery of novel antivirulence therapeutic targets, which can subsequently use existing drug development pipelines to identify antivirulence therapeutics. Though presented as a linear path in Fig. 2, we anticipate that Virulence Factor Identification and Antivirulence Target Refinement are likely iterative processes.
Fig. 2.
An integrated antivirulence drug development workflow from drug target to drug discovery. The flowchart provides a general guideline for how pathogen-associated genes (PAGs) analyses can be used to aid identification of new drug targets (red boxes), complementing/replacing existing antimicrobial target discovery approaches (Development of Antivirulence Therapies box). Candidate antivirulence compounds can then be incorporated into established drug development platforms (blue box).
Towards new antivirulence targets: databases of curated virulence factors and in silico virulence factor predictions
Initial development of antivirulence drugs focused mainly on targeting well-studied virulence factors with direct involvement in host interaction.23 These virulence factors, including microbial attachment and invasion factors (adhesins, pili, fimbriae), toxins (exotoxins and endotoxins), toxin trafficking systems (secretion system, transporters), and iron-binding factors (siderophores), are well-curated in knowledge databases such as the Virulence Factor Database (VFDB),32 the PAThosystems Resource Integration Center (PATRIC),33 and Victors.34 The expansion of genomics data has also enabled the development of genomics-based and metagenomics-based virulence factor predictors such as VFDB's VFanalyzer32 and PathoFact.35 These publicly available resources coupled with the wealth of microbial genomics data expedited the design and development of pathogen-specific antivirulence drugs.
Pathogen-specific genes as a novel avenue for antivirulence drug target discovery
To expand the druggable repertoire of bacterial virulence factors, more attention has been turned to virulence factors that do not directly interact with hosts but are involved in virulence-related pathways such as metabolic proteins for secondary metabolites important for virulence but not essential cell activities, regulators and bacterial signaling molecules. Better understanding of existing and novel virulence mechanisms deployed by pathogens under different environmental or clinical conditions may help to identify novel virulence factors that can be used as precision antivirulence drug targets. One promising source for pathogen-specific drug targets is by identifying pathogen-associated genes (PAGs). PAGs are defined as genes with homologs exclusively found in pathogens but not in non-pathogens. PAGs have been identified through the detection of genes unique to pathogens from large-scale comparative genome analyses of both pathogens and non-pathogens, as described by Sui et al.36 These pathogen-associated genes were found to be disproportionately enriched in more “offensive” functions such as host invasion, type III and type IV secretion systems, and toxins (versus more “defensive” functions that were found to be common to pathogens and non-pathogens).36,37 Virulence factors, including PAGs, have been found to also be over-represented in genomic islands (clusters of genes with probable horizontal origins) compared to other parts of the genome. Further assessing the phylogenetic distribution of pathogen-specific genes is of significant interest, as this may help prioritize those genes that truly may be more associated with the pathogen phenotype, versus simply be genes specific to a particular lineage. Such analyses may also help to fine tune the activity spectrum of antivirulence drugs based on how conserved the bacterial genes are among a species, genus or higher taxonomic rank. Within the current antivirulence development pipeline, most of antivirulence drug candidates are narrow-spectrum and are designed to target species-specific virulence mechanisms, such as biofilm formation and quorum-sensing in P. aeruginosa or botulinum toxin activity in C. botulinum (Table 1).
Bacterial genome-wide association studies (GWAS)
Bacterial genome-wide association studies (GWAS)38 and genotype-phenotype studies enable the discovery of novel virulence-related genes through the analysis of widely available, high-quality bacterial genomes isolated from clinical infections. A recent comparative genomics study of a collection of P. aeruginosa strains, examined further using a lab-based infection model, found genetic elements in the accessory genome that have novel associations with virulence.39 Another recent study applying GWAS to clinical data was able to identify multiple single nucleotide polymorphisms (SNPs) in S. aureus associated with worse outcome in bacteremia (predicting greater patient mortality).40 Genetic knockout studies in lab-based isolates plus large-scale genomic analyses of populations of bacterial pathogens from clinical and environmental sources with associated real-world data both represent powerful tools for the identification and characterization of novel virulence factors.
Characterizing virulence-associated genes with unknown function
Genes encoding hypothetical proteins can comprise a significant number of genes predicted in a bacterial genome.40,41 Interestingly, a large fraction of PAGs is annotated as hypothetical proteins or genes with unknown function. Currently, computational annotation of hypothetical proteins can enable high-throughput characterization of novel genes, and benefits from a diverse combination of various in silico methods supported by experimental data. These computational functional analyses include sequence and structural-based methods such as BLAST-based ortholog detection,42 protein domain analysis with Pfam,43 structural modeling with Phyre2,44 and AlphaFold,45 transmembrane domain prediction with TMHMM,46 and protein subcellular localization prediction using PSORTb.47 Additionally, BLAST-based genome annotation tools such as Prokka48 can further aid identification of potential virulence roles for hypothetical proteins identified in the PAG analysis, though caution must be exercised when direct annotation transfer approaches are used, due to the contextual nature of virulence (i.e., what may be a virulence factor in one pathogen may not be in another depending on its context).
Bioinformatic approaches described above are powerful tools for finding and distinguishing candidate virulence factors among hypothetical proteins but alone are not sufficient to characterize virulence factors. Experimental validation is required to determine whether candidate PAGs have a role in virulence. The two major criteria to distinguish PAGs from other hypothetical proteins are: (1) deletion analysis showing that the PAG has an impact on virulence and (2) with minimal impact on bacterial cell growth. Use of in vivo infection models and in vitro tissue culture will be necessary to characterize potential virulence functions of PAGs and whether these PAGs can serve as targets for subsequent antivirulence drug development.
The potential role a gene may have for virulence can be validated using methods of gene deletion mutagenesis. Loss of a gene abrogating virulence and improving survival of an infected model organism is a powerful experimental result for characterizing novel virulence factors.49, 50, 51 Specific phenotypic traits such as biofilm formation, motility, growth under varying nutrient conditions, and even antimicrobial resistance may also provide insight into the functional importance of a gene for virulence in a bacterial pathogen.52 Justifying this interest in genes encoding hypothetical proteins, it should be emphasized that many previously undefined bacterial genes have been found to be important for virulence, reflecting our still limited understanding of all the players in complex disease phenotypes. In a Streptococcus pneumoniae knockout study, 44 genes were identified to impact virulence and included conserved hypothetical proteins, essential genes, known virulence factors, and transcriptional regulators.45
Among the newly identified pneumococcal virulence factors that do not affect bacterial growth rate is LytB which was found to prevent phagocytosis in host cell.45 Knockouts of LytA, Spr0084, and Spr0930 attenuated virulence in a murine sepsis model but their exact functional roles are yet to be determined. Another study in Mycobacterium tuberculosis H37Rv found that 27% of encoded proteins are functionally unknown.53 However, through a series of in silico functional analyses including secondary structure-based alignment using SSEalign, functional enrichment by PANTHER, and virulence factor identification using VFDB and VICMpred, RpoS and PspA are identified from the pool of hypothetical proteins to be involved in the mycobacterial stress response pathway important for survival in the host.53 As the knowledge of virulence factors expands, there is also a need to expand existing virulence factor databases. Expanded publicly available and centralized information on virulence-related genes, including the context under which they play a role in virulence, would enable more efficient target selection for antivirulence drug discovery.
Prioritization of “accessible” targets for antivirulence drugs
Antimicrobial drug development usually needs to factor in drug delivery across the bacterial cell wall.54 Gram-negative bacterial pathogens are particularly noted for low membrane permeability and active drug efflux, which collectively hinders drug action on intracellular targets.55 As such, there has been an interest in prioritizing more accessible cell-surface or secreted virulence factors as antivirulence drug targets. Bioinformatic tools such as PSORTb,47,56 or BUSCA,57 (see PSORT.org for a list) can aid identification of such targets through prediction of bacterial protein subcellular localization—with PSORTb the first tool to exceed the accuracy of high-throughput proteomic laboratory methods for protein localization determination.47,56,58 Such tools have recently been reviewed well by Imai and Nakai59 but some tools have notable limitations such as BUSCA which does not predict periplasmic proteins (critical for differentiation from outer membrane proteins in gram-negative bacteria, to avoid over predicting cell surface proteins). All the widely used computational predictors have web-based interfaces and PSORTb can additionally be run locally or in a command–line interface as an open-source software. Pre-computed and regularly updated PSORTb predictions also available via PSORTdb.54 While such prioritization is not essential, it can simplify drug discovery efforts by not having to address the need to cross the cell envelope. By focusing more on drug action outside of the bacterial cell—targeting external virulence factors versus the cell itself—this may more likely lead to drug discovery which has only antivirulence activity and lessens impact on bacterial cell viability.
Application of the antibiotic drug discovery platform towards antivirulence drug development
During the “golden age of antibiotics” between 1940s and 1960s, many modern antibiotics were discovered from natural compounds produced by the soil-derived actinomycete.2 The natural product discovery pipeline eventually dried up when novel classes of antibiotics became increasingly scarce to find without the constant re-discovery of existing compounds. Since antivirulence drug targets differ from traditional antimicrobial drug targets that are often parts of essential bacterial processes, the development of antivirulence drugs can potentially revive and build upon existing antimicrobial drug discovery platforms while incorporating the concept of precision antimicrobial therapeutics to address the global health concern of AMR as well as the technical challenges of producing novel drugs against bacteria. The design of antivirulence drugs also satisfies the WHO's innovation criteria for a novel antimicrobial therapeutic as they: (1) target specific virulence factors or virulence-related proteins rather than essential genes (novel target and mode of action), (2) do not affect bacterial survival and thus reduce selective pressure for drug resistance (minimizes cross-resistance), and (3) are of distinct chemical classes from antibiotics and act on different cellular processes.60
Most of the approved antivirulence drugs and drug candidates under development were discovered through high-throughput screenings of existing compound libraries against specific bacterial targets including well characterized and known virulence factors such as quorum-sensing molecules and toxins (Table 1). The following subsections outline the success of antivirulence drug discovery using natural product libraries, small molecules libraries and approved drug libraries that were originally designed for classic antibiotic discovery. These studies illustrate that existing antibiotic drug development platforms including drug library screening methods, may be adapted for antivirulence drug discovery.
Natural product screening
Plants represent a diverse reservoir of secondary metabolites, or bioactive phytochemicals, accumulated by co-evolution with other organisms in competitive or hostile environments. Medicinal plants are known to confer therapeutic effects against many ailments including bacterial infections.61 Between 2006 and 2020, 100 plants globally have been reported to show antimicrobial activities against the highly virulent and antimicrobial resistant ESKAPE (Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter spp) pathogens.62 Approximately 5,000 of the ∼45,000 African plant species and more than 2,000 Chinese herbal compounds have documented medicinal properties with many also conferring antivirulence properties.63 For instance, anti-quorum-sensing activity has already been reported in several Chinese medicinal plant extracts, Poria cum Radix pini, Angelica dahurica, Rhizoma cibotii, and Schizonepeta tenuifolia, as well as the South African plant Sclerocarya birrea.63,64 Likewise, diterpenoids, phytochemicals found in the Chilean medicinal plant Azorella atacamensis, can interfere with the production of quorum-sensing molecules in P. aeruginosa.65 These studies tap into two approaches to characterize bioactive compounds, one is to begin with whole plant extracts that have not been previously characterized and purify novel secondary metabolites. The other is to consult traditional knowledge for known naturally occurring curatives and isolate specific compounds. Both approaches then screen for activity against bacterial factors. The exact activity depending on the antivirulence activity sought. Significant success has been demonstrated in developing anti-quorum-sensing therapeutics as described in studies mentioned immediately above and in Table 1.
These recent investigations for antivirulence drug candidates have demonstrated the potential revival of the natural product-based drug discovery programs previously developed for antibiotic discovery. Further screening of medicinal plants for activity against existing and novel bacterial virulence factors can enable the discovery of phytochemicals as promising antivirulence compounds.
Small molecule screening
Besides natural product screening, the use of high-throughput screening (HTS) of synthetic, small molecules has accelerated the drug development process in different fields of medicines. Screening thousands of small molecules drastically accelerates the “hit to lead” generation, the process at which a compound with target binding affinity and potential therapeutic properties is identified and optimized.66 HTS assays are now being adopted in the antivirulence drug development process. For example, the small molecule MAC-545496 inhibits the cell-envelope stress regulator GraR resulting in reduced biofilm formation, abated intracellular survival in macrophages, and reversed β-lactam resistance in methicillin-resistant S. aureus.67 In addition to traditional HTS, the fragment-based lead discovery method is a more recent approach for large-scale small molecule screening that utilizes high-quality chemical fragment libraries with thousand-fold fewer molecules but covers a larger chemical space.68 This method identifies fragments with binding affinity to target and structurally expand the fragment into a drug lead with optimized binding and biological activity. Both methods have been widely adopted in many drug discovery platforms and can also be applied towards novel antivirulence drug development. Such screening approaches will benefit from the development of more robust protein structure prediction methods such as AlphaFold,45 though some of the very novel protein-based drug targets will require additional experimental structural data to more accurately identify their small molecular interactions.
Repurposing approved drugs
Traditional drug discovery process can take more than ten years to bring a novel drug to the market. This has inspired the more cost- and time-effective strategy of finding novel indications of already-available drugs, known as drug repositioning or repurposing. This approach has reportedly accounted for roughly 30% of newly approved drugs by the US Food and Drug Administration in recent years.64 Since safety and efficacy information of existing drugs are readily available, the time to develop an approved drug for an alternative indication may effectively be shortened. This is particularly crucial in the realm of infectious disease where novel therapeutics are urgently needed to combat the continual rise of AMR. Antimicrobial activities have been reported in a variety of drug classes including anthelmintics, anticancer drugs, anti-inflammatory drugs and antipsychotic drugs.69 Similar screening methods with the addition of a non-bactericidal and non-bacteriostatic criterion would help to identify existing drugs with potential antivirulence properties. For example, the FDA-approved antifungal drugs clotrimazole and miconazole as well as the antipsychotic drug pimozide targets the P. aeruginosa transcriptional regulator PqsR to inhibit the Pseudomonas Quinolone Signal (PQS) quorum-sensing system.70
In addition, the combination of pathogen-associated genes analysis and in silico docking assay identified a repurposing potential for the FDA-approved osteoporosis drug Raloxifene. Raloxifene was predicted to be an antivirulence agent against P. aeruginosa, specifically targeting the pathogen-associated PhzB2 protein involved in pyocyanin biosynthesis. Subsequent in vivo studies confirmed an antivirulence activity of the drug without impacting bacterial growth.71
Diagnostics as an important component of an antivirulence therapeutic strategy
The effective use of pathogen-specific drugs, including narrow-spectrum antibiotics and antivirulence drugs, relies on rapid and accurate diagnosis of the infection/infecting pathogen. This is increasingly becoming possible, as more rapid approaches are developed in addition to traditional laboratory culturing. Identification via protein markers through use of MALDI-TOF and targeted genomic sequencing such as multi-locus sequence typing are becoming routine clinical practice. Rapid, very specific, diagnostics are becoming more achievable with the lowering cost of DNA/genomic-based rapid-screening methods and the continual advancement of public health genomics platforms for pathogen identification.72,73
Discussion
Though antibiotics have been critical for bacterial infectious disease control, the overuse and over-reliance on antibiotics have also exacerbated the public health crisis of antimicrobial resistance. Novel antimicrobial drug discovery efforts need to focus on innovative therapeutics that are less prone to resistance. Antivirulence therapeutics are a promising approach to combat this global public health threat by aiming to attenuate bacterial virulence rather than to negatively impact the survival of specific bacterial pathogens—potentially selecting more for drug resistance. Revitalizing and applying the antibiotic drug discovery pipeline, including the use of existing natural product and small molecule libraries, for the discovery of novel antivirulence therapeutics may be a promising strategy in mitigating AMR-related morbidities and mortalities. Likewise, as more bacterial genomes are sequenced, novel PAGs with little to no sequence similarity to well characterized genes represent an unexplored source of potentially novel antivirulence drug targets. Additionally, identifying PAGs and their evolutionary conservation across different taxonomic levels can help fine-tune the antimicrobial spectrum of novel drugs and create precision medicine against specific pathogens without affecting the host's established, non-disease-causing microbiota. This approach, especially if directed at PAGs shared across diverse genera, may also identify drugs that could target more broadly conserved themes in pathogenicity.
Although antivirulence drugs theoretically do not directly impact bacterial growth and survival, resistance may still emerge depending on the biological role and dependence on the chosen virulence factor target and the treatment environment.20 Allen et al. hypothesized that it is possible to design an evolutionary robust drug that selects against resistance if the antivirulence drug target (virulence factor) damages the host but confers no survival advantage to the pathogen at the site of infection.20 Expanding the knowledge base of bacterial virulence factors and specifically their functional roles would therefore increase the selection of appropriate antivirulence drug targets that are ideally both pathogen specific and confer negligible impact on pathogen fitness. This would also help to expand the antivirulence drug options and improve their clinical lifespan against existing AMR-related infections.
Adoption of antivirulence drugs as a part of precision antimicrobial therapeutics would benefit from more accurate clinical diagnoses of infections and the disease-causing agent, including sequence-based approaches for diagnostics that have been prioritized.74,75 It is important to note that while genomics and bioinformatics have advanced our knowledge of bacterial virulence as well as accelerated the drug prediction workflows, the integration of both computational and laboratory analyses into virulence drug target, and drug discovery pipelines is crucial for the successful discovery of novel bacterial targets and antivirulence therapeutics.
Outstanding questions
In recent years, high-throughput omics approaches have provided an improved understanding of bacterial virulence mechanisms that can help to advance antivirulence drug discoveries; however, there are still some significant questions that need to be addressed:
1) DNA sequencing technologies have generated massive amounts of high-quality genomic data but in silico gene functional annotation and laboratory validation remains a challenge in the characterization of novel, pathogen-specific genes with limited sequence similarity to known genes. GWAS and genotype-phenotype association studies improve our confidence in the discovery of novel virulence factors, but more specific laboratory methods that enable a deeper understanding of pathogen virulence and disease processes are needed. Such in-depth characterization of virulence factors of interest is a rate-limiting factor for anti-virulence drug discovery.
2) It is predicted that antivirulence therapies would less strongly select for resistance versus antimicrobial therapies that directly target pathogen survival; instead, antivirulence therapies may select for a non-pathogen phenotype. However, we need more investigation of how pathogens evolve under antivirulence selective pressure versus direct survival. More investigation of the potential for antivirulence drug resistance, using combined computational and laboratory experimentation is needed.
Search strategy and selection criteria
For this review we identified literature by searches through PubMed from August 2020 to August 2022 using the search terms “antivirulence compounds/drugs/therapies”, “antimicrobial resistance”, “quorum sensing inhibitors”, “phage therapy”, “pathogen-specific”, “novel antimicrobial therapeutics”, and “bacterial pathogens”. For sections on antivirulence drug discovery workflow and compound screening combinations of keywords “antivirulence” with “drug discovery”, “compound screens”, “novel therapeutics”, “natural sources therapeutics”, “bioinformatic/computational approaches”, “workflow”, and “pipelines” were utilized for literature searches. Only peer-reviewed articles in English, that were published up to August 2022, were included.
Contributors
Conceptualization: WYVL, PKT, FSLB, AHL; literature search: WYVL, PKT, FSLB, AHL; data analysis: WYVL, PKT; data interpretation: WYVL, PKT, original draft: WYVL, PKT; figures and visualization: WYVL, PKT, FSLB; review and editing: WYVL, PKT, FSLB, AHL. All authors read and approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Acknowledgements
AHL acknowledges the support of a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC), Project Grant from the Canadian Institutes of Health Research (CIHR) and a Banting Research Foundation Discovery Award. PKT acknowledges the support of an Omics Data Science Initiative (ODSI) Fellowship from Simon Fraser University. WYVL was the recipient of a CIHR Scholarship and FSLB received additional support from Genome Canada and CIHR. Funders had no role in the design or writing of this manuscript.
Contributor Information
Fiona S.L. Brinkman, Email: brinkman@sfu.ca.
Amy H.Y. Lee, Email: amy_lee_10@sfu.ca.
References
- 1.Fleming A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Bull World Health Organ. 2001;79:780–790. [PMC free article] [PubMed] [Google Scholar]
- 2.Hutchings M.I., Truman A.W., Wilkinson B. Antibiotics: past, present and future. Curr Opin Microbiol. 2019;51:72–80. doi: 10.1016/j.mib.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 3.World Hsealth Organization New report calls for urgent action to avert antimicrobial resistance crisis. 2019. https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis (WHO Joint News Release). Available from:
- 4.World Health Organization, Food and Agriculture Organization of the United Nations, World Organisation for Animal Health Monitoring and evaluation of the global action plan on antimicrobial resistance: framework and recommended indicators. 2019. https://apps.who.int/iris/handle/10665/325006 (WHO Document). Available from:
- 5.World Health Organization Global action plan on antimicrobial resistance. https://apps.who.int/iris/handle/10665/193736 (WHO Document). Available from: [DOI] [PubMed]
- 6.Tacconelli E., Carrara E., Savoldi A., et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 7.Bertollo L.G., Lutkemeyer D.S., Levin A.S. Are antimicrobial stewardship programs effective strategies for preventing antibiotic resistance? a systematic review. Am J Infect Control. 2018;46:824–836. doi: 10.1016/j.ajic.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Theuretzbacher U., Outterson K., Engel A., Karlen A. The global preclinical antibacterial pipeline. Nat Rev Microbiol. 2020;18:275–285. doi: 10.1038/s41579-019-0288-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plackett B. Why big pharma has abandoned antibiotics. Nature. 2020;586:S50–S52. [Google Scholar]
- 10.Cama J., Leszczynski R., Tang P.K., et al. To Push or To Pull? In a post-COVID world, supporting and incentivizing antimicrobial drug development must become a governmental priority. ACS Infect Dis. 2021;7:2029–2042. doi: 10.1021/acsinfecdis.0c00681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler M.S., Gigante V., Sati H., et al. Analysis of the clinical pipeline of treatments for drug-resistant bacterial infections: despite progress, more action is needed. Antimicrob Agents Chemother. 2022;66 doi: 10.1128/aac.01991-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paharik A.E., Schreiber H.L.I.V., Spaulding C.N., Dodson K.W., Hultgren S.J. Narrowing the spectrum: the new frontier of precision antimicrobials. Genome Med. 2017;9:110. doi: 10.1186/s13073-017-0504-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rakesh K.P., Marichannegowda M.H., Srivastava S., et al. Combating a master manipulator: Staphylococcus aureus immunomodulatory molecules as targets for combinatorial drug discovery. ACS Comb Sci. 2018;20:681–693. doi: 10.1021/acscombsci.8b00088. [DOI] [PubMed] [Google Scholar]
- 14.Spaulding C.N., Klein R.D., Schreiber H.L.I.V., Janetka J.W., Hultgren S.J. Precision antimicrobial therapeutics: the path of least resistance? NPJ Biofilms Microbiomes. 2018;4:4. doi: 10.1038/s41522-018-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh C., Sarkar P., Issa R., Haldar J. Alternatives to conventional antibiotics in the era of antimicrobial resistance. Trends Microbiol. 2019;27:323–338. doi: 10.1016/j.tim.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 16.Mookherjee N., Anderson M.A., Haagsman H.P., Davidson D.J. Antimicrobial host defence peptides: functions and clinical potential. Nat Rev Drug Discov. 2020;19:311–332. doi: 10.1038/s41573-019-0058-8. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen J., Pepin D.M., Tropini C. Cause or effect? the spatial organization of pathogens and the gut microbiota in disease. Microbes Infect. 2021;23 doi: 10.1016/j.micinf.2021.104815. [DOI] [PubMed] [Google Scholar]
- 18.Micoli F., Bagnoli F., Rappuoli R., Serruto D. The role of vaccines in combatting antimicrobial resistance. Nat Rev Microbiol. 2021;19:287–302. doi: 10.1038/s41579-020-00506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.García-Contreras R., Peréz-Eretza B., Jasso-Chávez R., et al. High variability in quorum quenching and growth inhibition by furanone C-30 in Pseudomonas aeruginosa clinical isolates from cystic fibrosis patients. Pathog Dis. 2015;73 doi: 10.1093/femspd/ftv040. [DOI] [PubMed] [Google Scholar]
- 20.Allen R.C., Popat R., Diggle S.P., Brown S.P. Targeting virulence: can we make evolution-proof drugs? Nat Rev Microbiol. 2014;12:300–308. doi: 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- 21.Jones-Nelson O., Tovchigrechko A., Glover M.S., et al. Antibacterial monoclonal antibodies do not disrupt the intestinal microbiome or its function. Antimicrob Agents Chemother. 2020;64 doi: 10.1128/AAC.02347-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrof E.O., Gloor G.B., Vanner S.J., et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome. 2013;1:3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickey S.W., Cheung G.Y.C., Otto M. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat Rev Drug Discov. 2017;16:457–471. doi: 10.1038/nrd.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skoura N., Wang-Jairaj J., Della Pasqua O., et al. Effect of raxibacumab on immunogenicity of anthrax vaccine adsorbed: a phase 4, open-label, parallel-group, randomised non-inferiority study. Lancet Infect Dis. 2020;20:983–991. doi: 10.1016/S1473-3099(20)30069-4. [DOI] [PubMed] [Google Scholar]
- 25.Payne J.R., Khouri J.M., Jewell N.P., Arnon S.S. Efficacy of human botulism immune globulin for the treatment of infant botulism: the first 12 years post licensure. J Pediatr. 2018;193:172–177. doi: 10.1016/j.jpeds.2017.10.035. [DOI] [PubMed] [Google Scholar]
- 26.Orth P., Xiao L., Hernandez L.D., et al. Mechanism of action and epitopes of Clostridium difficile toxin B-neutralizing antibody bezlotoxumab revealed by X-ray crystallography. J Biol Chem. 2014;289:18008–18021. doi: 10.1074/jbc.M114.560748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ali S.O., Yu X.Q., Robbie G.J., et al. Phase 1 study of MEDI3902, an investigational anti-Pseudomonas aeruginosa PcrV and Psl bispecific human monoclonal antibody, in healthy adults. Clin Microbiol Infect. 2019;25:629.e1–629.e6. doi: 10.1016/j.cmi.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Krautz-Peterson G., Chapman-Bonofiglio S., Boisvert K., et al. Intracellular neutralization of shiga toxin 2 by an a subunit-specific human monoclonal antibody. Infect Immun. 2008;76:1931–1939. doi: 10.1128/IAI.01282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua L., Cohen T.S., Shi Y., et al. MEDI4893∗ promotes survival and extends the antibiotic treatment window in a Staphylococcus aureus immunocompromised pneumonia model. Antimicrob Agents Chemother. 2015;59:4526–4532. doi: 10.1128/AAC.00510-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattar C., Edwards S., Baraldi E., Hood J. An overview of the global antimicrobial resistance research and development hub and the current landscape. Curr Opin Microbiol. 2020;57:56–61. doi: 10.1016/j.mib.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 31.Murray C.J., Ikuta K.S., Sharara F., et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu B., Zheng D., Jin Q., Chen L., Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47:D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis J.J., Wattam A.R., Aziz R.K., et al. The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res. 2020;48:D606–D612. doi: 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sayers S., Li L., Ong E., et al. Victors: a web-based knowledge base of virulence factors in human and animal pathogens. Nucleic Acids Res. 2019;47:D693–D700. doi: 10.1093/nar/gky999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Nies L., Lopes S., Busi S.B., et al. PathoFact: a pipeline for the prediction of virulence factors and antimicrobial resistance genes in metagenomic data. Microbiome. 2021;9:49. doi: 10.1186/s40168-020-00993-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho Sui S.J., Fedynak A., Hsiao W.W., Langille M.G., Brinkman F.S. The association of virulence factors with genomic islands. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertelli C., Laird M.R., Williams K.P., et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saber M.M., Shapiro B.J. Benchmarking bacterial genome-wide association study methods using simulated genomes and phenotypes. Microb Genom. 2020;6 doi: 10.1099/mgen.0.000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vasquez-Rifo A., Veksler-Lublinsky I., Cheng Z., Ausubel F.M., Ambros V. The Pseudomonas aeruginosa accessory genome elements influence virulence towards Caenorhabditis elegans. Genome Biol. 2019;20:270. doi: 10.1186/s13059-019-1890-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lobb B., Tremblay B.J., Moreno-Hagelsieb G., Doxey A.C. An assessment of genome annotation coverage across the bacterial tree of life. Microb Genom. 2020;6 doi: 10.1099/mgen.0.000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsiao W.W., Ung K., Aeschliman D., Bryan J., Finlay B.B., Brinkman F.S. Evidence of a large novel gene pool associated with prokaryotic genomic islands. PLoS Genet. 2005;1:e62. doi: 10.1371/journal.pgen.0010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 43.Mistry J., Chuguransky S., Williams L., et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jumper J., Evans R., Pritzel A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 47.Yu N.Y., Wagner J.R., Laird M.R., et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 49.Seol-Hee K., Shin-Young P., Yun-Jeong H., You-Hee C. Drosophila melanogaster-based screening for multihost virulence factors of Pseudomonas aeruginosa PA14 and identification of a virulence-attenuating factor, HudA. Infect Immun. 2008;76:4152–4162. doi: 10.1128/IAI.01637-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paczosa M.K., Silver R.J., McCabe A.L., et al. Transposon mutagenesis screen of Klebsiella pneumoniae identifies multiple genes important for resisting antimicrobial activities of neutrophils in mice. Infect Immun. 2020;88 doi: 10.1128/IAI.00034-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falkow S. Molecular Koch's postulates applied to microbial pathogenicity. Rev Infect Dis. 1988;10:S274–S276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- 52.Harrison F. Bacterial cooperation in the wild and in the clinic: are pathogen social behaviours relevant outside the laboratory? Bioessays. 2013;35:108–112. doi: 10.1002/bies.201200154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Z., Zeng X., Tsui S.K.W. Investigating function roles of hypothetical proteins encoded by the Mycobacterium tuberculosis H37Rv genome. BMC Genomics. 2019;20:394. doi: 10.1186/s12864-019-5746-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pontefract B.A., Ho H.T., Crain A., Kharel M.K., Nybo S.E. Drugs for Gram-negative bugs from 2010-2019: a decade in review. Open Forum Infect Dis. 2020;7 doi: 10.1093/ofid/ofaa276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharma D., Misba L., Khan A.U. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Control. 2019;8:76. doi: 10.1186/s13756-019-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lau W.Y.V., Hoad G.R., Jin V., et al. PSORTdb 4.0: expanded and redesigned bacterial and archaeal protein subcellular localization database incorporating new secondary localizations. Nucleic Acids Res. 2021;49:D803–D808. doi: 10.1093/nar/gkaa1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Savojardo C., Martelli P.L., Fariselli P., Profiti G., Casadio R. BUSCA: an integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018;46:W459–W466. doi: 10.1093/nar/gky320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rey S., Gardy J.L., Brinkman F.S.L. Assessing the precision of high-throughput computational and laboratory approaches for the genome-wide identification of protein subcellular localization in bacteria. BMC Genomics. 2005;6:162. doi: 10.1186/1471-2164-6-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Imai K., Nakai K. Tools for the recognition of sorting signals and the prediction of subcellular localization of proteins from their amino acid sequences. Front Genet. 2020;11 doi: 10.3389/fgene.2020.607812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.World Health Organization Antibacterial agents in clinical development: an analysis of the antibacterial clinical development pipeline. 2019. https://www.who.int/publications/i/item/9789240000193 (WHO Technical Document). Available from:
- 61.Chassagne F., Samarakoon T., Porras G., et al. A systematic review of plants with antibacterial activities: a taxonomic and phylogenetic perspective. Front Pharmacol. 2020;11 doi: 10.3389/fphar.2020.586548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shahbaaz M., Bisetty K., Ahmad F., Hassan M.I. Current advances in the identification and characterization of putative drug and vaccine targets in the bacterial genomes. Curr Top Med Chem. 2016;16:1040–1069. doi: 10.2174/1568026615666150825143307. [DOI] [PubMed] [Google Scholar]
- 63.Mahavy C.E., Duez P., El Jaziri M., Rasamiravaka T. African plant-based natural products with antivirulence activities to the rescue of antibiotics. Antibiotics (Basel) 2020;9:830. doi: 10.3390/antibiotics9110830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chong Y.M., How K.Y., Yin W.F., Chan K.G. The effects of Chinese herbal medicines on the quorum sensing-regulated virulence in Pseudomonas aeruginosa PAO1. Molecules. 2018;23:972. doi: 10.3390/molecules23040972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Azuama O.C., Ortiz S., Quiros-Guerrero L., et al. Tackling Pseudomonas aeruginosa virulence by mulinane-like diterpenoids from Azorella atacamensis. Biomolecules. 2020;10:1626. doi: 10.3390/biom10121626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hevener K.E., Pesavento R., Ren J., Lee H., Ratia K., Johnson M.E. Hit-to-Lead: hit validation and assessment. Methods Enzymol. 2018;610:265–309. doi: 10.1016/bs.mie.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 67.El-Halfawy O.M., Czarny T.L., Flannagan R.S., et al. Discovery of an antivirulence compound that reverses β-lactam resistance in MRSA. Nat Chem Biol. 2020;16:143–149. doi: 10.1038/s41589-019-0401-8. [DOI] [PubMed] [Google Scholar]
- 68.Lamoree B., Hubbard R.E. Using fragment-based approaches to discover new antibiotics. SLAS Discov. 2018;23:495–510. doi: 10.1177/2472555218773034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miro-Canturri A., Ayerbe-Algaba R., Smani Y. Drug repurposing for the treatment of bacterial and fungal infections. Front Microbiol. 2019;10:41. doi: 10.3389/fmicb.2019.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.D’Angelo F., Baldelli V., Halliday N., et al. Identification of FDA-approved drugs as antivirulence agents targeting the pqs quorum-sensing system of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2018;62 doi: 10.1128/AAC.01296-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ho Sui S.J., Lo R., Fernandes A.R., et al. Raloxifene attenuates Pseudomonas aeruginosa pyocyanin production and virulence. Int J Antimicrob Agents. 2012;40:246–251. doi: 10.1016/j.ijantimicag.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maxson T., Mitchell D.A. Targeted treatment for bacterial infections: prospects for pathogen-specific antibiotics coupled with rapid diagnostics. Tetrahedron. 2016;72:3609–3624. doi: 10.1016/j.tet.2015.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dozois C.M., Daigle F., Curtiss R. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc Natl Acad Sci U S A. 2003;100:247–252. doi: 10.1073/pnas.232686799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miao Q., Ma Y., Wang Q., et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018;67:S231–S240. doi: 10.1093/cid/ciy693. [DOI] [PubMed] [Google Scholar]
- 75.Simner P.J., Miller S., Carroll K.C. Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin Infect Dis. 2018;66:778–788. doi: 10.1093/cid/cix881. [DOI] [PMC free article] [PubMed] [Google Scholar]