Abstract
MUC13, a transmembrane mucin glycoprotein, is overexpressed in colorectal cancer (CRC), however, its regulation and functions are not fully understood. It has been shown that MUC13 protects colonic epithelial cells from apoptosis. Therefore, studying MUC13 and MUC13-regulated pathways may reveal promising therapeutic approaches for CRC treatment. Growing evidence suggests that microRNAs (miRs) are involved in the development and progression of CRC. In the present study, the MUC13-miR-4647 axis was addressed in association with survival of patients. miR-4647 is predicted in silico to bind to the MUC13 gene and was analyzed by RT-qPCR in 187 tumors and their adjacent non-malignant mucosa of patients with CRC. The impact of previously mentioned genes on survival and migration abilities of cancer cells was validated in vitro. Significantly upregulated MUC13 (P=0.02) in was observed tumor tissues compared with non-malignant adjacent mucosa, while miR-4647 (P=0.05) showed an opposite trend. Higher expression levels of MUC13 (log-rank P=0.05) were associated with worse patient's survival. The ectopic overexpression of studied miR resulted in decreased migratory abilities and worse survival of cells. Attenuated MUC13 expression levels confirmed the suppression of colony forming of CRC cells. In summary, the present data suggested the essential role of MUC13-miR-4647 in patients' survival, and this axis may serve as a novel therapeutic target. It is anticipated MUC13 may hold significant potential in the screening, diagnosis and treatment of CRC.
Keywords: colorectal cancer risk and clinical outcomes, MUC13, microRNA, translation research
Introduction
Mucins are complex cell surface and secreted glycoproteins that provide protection and lubrication to the epithelial surface of mucosal tissues (1). The advantage of their expression in cancer cells is likely linked to their functions in healthy tissue, promoting epithelial resistance and resilience to toxic challenges at mucosal surfaces. Carcinoma cells derived from epithelia overexpress mucins to exploit their role in promoting proliferation, survival, migration and invasion of cancer cells. Mucins have thus been identified as markers of poor prognosis (2) and attractive therapeutic targets in numerous cancers (3).
The MUC13, cell surface mucin, is overexpressed in gastric (4), colorectal (CRC) (5–7), pancreatic (8,9), renal (10) and ovarian (11) cancers. Usually this protein is expressed on apical borders of epithelial cells of the intestine, with increased cytoplasmic expression observed in response to infection (12) and inflammation (13). MUC13 has a relatively short 151-amino acid extracellular domain compared with other cell surface mucins with three epidermal growth factor (EGF)-like domains, one sea urchin sperm protein enterokinase arginine domain within an extracellular component, followed by a short 23-amino acid transmembrane domain and a 69-amino acid cytoplasmic domain that includes eight serine and two tyrosine residues for potential phosphorylation. In addition, a protein kinase C consensus phosphorylation motif could play a role in cell signaling pathways and regulate proliferation and cell survival (7,13,14).
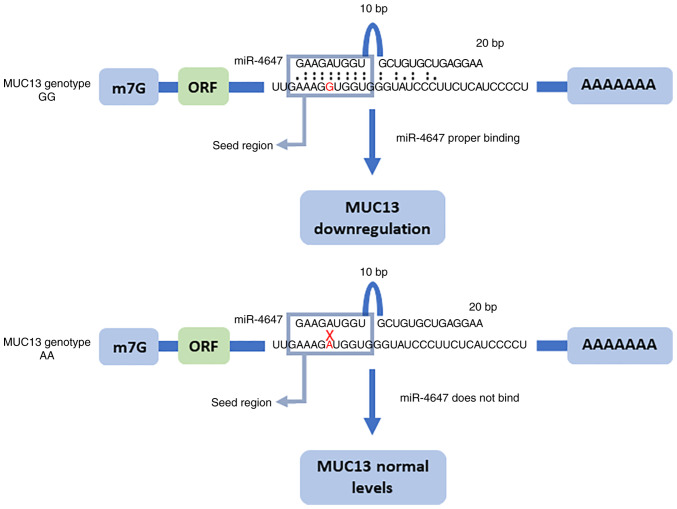
The elevated cytoplasmic MUC13 expression was reported in high-grade and metastatic CRC (6). In addition, MUC13 promotes activation of nuclear factor κB (NFκB), thus is anti-apoptotic (15), and pro-inflammatory (16). Gupta et al (6,7) suggested that MUC13 overexpression may influence colon cancer tumorigenesis and metastasis via multiple oncogenic proteins. It is considered that MUC13 may promote the survival of CRC cells under DNA damage via numerous mechanisms. Thus, it represents a potentially important new target for individualized therapy in advanced patients with CRC. CRC is a multifactorial disease resulting from individual genetic susceptibility (17,18), environmental factors (19), lifestyle, and inflammation (20). However, the molecular mechanisms responsible for the overexpression of MUC13 in CRC are poorly understood. It was hypothesized that one possibility is dysregulation by microRNAs (miRs) that regulate MUC13 mRNA. In our previous study on polymorphism in miR binding sites within mucin genes, a significant association was observed between homozygous variant genotype and decreased CRC risk for rs1532602 in MUC13 (21). The genetic variations in the 3′ untranslated regions (UTRs) of target genes may affect miRNA binding, ultimately imposing additional variability into the differential mRNA and protein expression levels. Aberrant miRNA expression and/or function are frequently observed in CRC. The present study thus provided important insights into the role of miR-4647 which is predicted to bind to polymorphic seed sequence site in MUC13, SNP spot rs1532602 (Fig. 1). The MUC13-miR-4647 axis was analyzed and pointed to their differential expression implication in survival of patients with CRC. CRC is a worldwide health burden, with nearly 1.2 million new cases expected each year globally (22). The five-year survival rate of patients with CRC in stages CRC I and II reaches 90%, while in advanced stages decreases to 14% (23). For newly diagnosed CRC patients, clarifying potential risk factors for prognosis and prediction of therapy response is paramount as it could have important clinical implications (24).
Figure 1.
Graphical representation of in silico predicted binding microRNA-4647 in the polymorphic seed sequence of MUC13 rs1532602.
The present results rendered MUC13-miR-4647 axis as a potential therapeutic target. At present, there is a great need to identify new diagnostic and prognostic markers, as well as to develop novel therapeutic strategies for the treatment of advanced CRC.
Materials and methods
Clinical samples
In total, 187 patients with sporadic CRC were recruited and underwent surgical resection between 2011 and 2015 at the General University Hospital (Prague, Czech Republic) and the Teaching Hospital and Medical School of Charles University (Pilsen, Czech Republic). All participants signed a written consent to participate in the study and approved using their biological samples for genetic analyses according to the Helsinki declaration. Ethics approval (approval nos. G 09-04-09 and G 14-08-67) was granted by the committees of the aforementioned hospitals. Written informed consent was provided by all patients. Study subjects provided information on their lifestyle habits, body mass index, diabetes, and family/personal history of cancer using a structured questionnaire to determine demographic characteristics and potential risk factors for CRC. The clinical characteristics of patients are presented in Table I.
Table I.
Clinical characteristics of the patients.
| Clinical characteristics | Total number (n=113) |
|---|---|
| Age, years (range) | 65±10 (37–86) |
| Sex | |
| Male | 69 |
| Female | 44 |
| SNP stratificationa | |
| G:G | 30 |
| A:G | 54 |
| A:A | 17 |
| CRC stratification | |
| Colon | 44 |
| Rectum | 69 |
| Ta | |
| 1 | 6 |
| 2 | 19 |
| 3 | 76 |
| 4 | 10 |
| Na | |
| 0 | 51 |
| >1 | 50 |
| Ma | |
| 0 | 87 |
| >1 | 21 |
| Gradea | |
| 1 | 19 |
| 2 | 77 |
| 3 | 8 |
| Local recidivea | |
| No | 79 |
| Yes | 33 |
| Neoadjuvant therapya | |
| No | 94 |
| Yes | 16 |
| Adjuvant therapya | |
| No | 65 |
| Yes | 42 |
| Living statusa | |
| Dead | 30 |
| Alive | 82 |
Numbers may not add up to 100% of available subjects because of missing data.
Tumor tissue and adjacent non-malignant mucosa tissue (5–10 cm distant from the tumor) were resected from each patient and deep frozen immediately after removal. The clinical stage at diagnosis was classified by the tumor-node-metastasis (TNM) system according to UICC (Union for International Cancer Control) (detail characteristic in Table I).
All CRC cases were monitored until March 31st, 2021. Overall survival (OS) was defined as the time interval from diagnosis to death of any cause or the date of the last follow-up used for censoring.
Tumor and adjacent mucosal tissues were homogenized by MagNA Lyser (Roche Diagnostics). Genomic DNA, total RNA, and small RNA were isolated from tumor tissues and adjacent non-malignant tissue with miRVana isolation kit protocol according to the manufacturer's instructions (Thermo Fisher Scientific, Inc.) without small RNA enrichment. The concentration at 260 nm and purity of RNA using the 260/280 nm ratio were determined spectrophotometrically by measuring its optical density using Nanodrop (Thermo Fisher Scientific, Inc.). The integrity of total RNA was measured by Agilent 2100 Bioanalyzer with Agilent RNA 6000 Nano Kit (Agilent Technologies, Inc.).
Genotyping of human samples
Genetic polymorphism in the MUC13 gene, rs1532602, was analyzed with TaqMan allelic discrimination assay (Thermo Fisher Scientific, Inc.; Assay-on-demand, SNP genotyping products: C__11906718_1_). The TaqMan genotyping reaction was amplified on a 7500 Real-Time PCR system (Thermo Fisher Scientific, Inc.) as follows: 95°C for 10 min, 92°C for 15 sec, and 60°C for 1 min for 40 cycles.
The genotype screening was performed simultaneously for tumor and adjacent non-malignant tissues. The genotype correlation between the duplicate samples was >99%. The genotype call rate ranged between 97.0 and 99.5%.
Reverse transcription-quantitative (RT-q)PCR of the MUC13 gene
In the present study, the expression levels of MUC13 and miR-4647 were analyzed. For MUC13 gene detection (ID Hs00217230_m1, Table SI), cDNA was synthesized from 80 ng of total RNA in 10 µl RT reaction using TATAA GrandScript cDNA SuperMix kit (TATAA Biocenter AB), according to the manufacturer's protocol in C1000 PCR cycler (Bio-Rad Laboratories, Inc.). Before qPCR, cDNA was diluted 1:1 with nuclease-free water. A total of 10 µl of qPCR reaction contained 2 µl of diluted cDNA, 5 µl of Taqman universal mastermix with no UNG (Thermo Fisher Scientific, Inc.), 2.5 µl nuclease-free water and 0.5 µl of probe assay purchased from Thermo Fisher Scientific, Inc. The thermocycling conditions were as follows: 95°C 10 min, and 45 cycles of 15 sec at 95°C, 1 min at 60°C in CFX384 (Bio-Rad Laboratories, Inc.). The expression of MUC13 was normalized to the ACTB gene (Hs99999903_m1). ACTB was used as reference genes selected from TaqMan Endogenous Control assays (ACTB, GAPDH, and 18S, Thermo Fisher Scientific, Inc.) by Normfinder (GenEx Enterprise). All data were analyzed as a fold change of protein expression and by the 2−ΔΔCq method (25).
Immunohistochemistry (IHC) of MUC13 gene
Fixed specimens from the intestine wall (received from patients with CRC) were dehydrated and embedded in paraffin using routine procedures. Tissue blocks were cut into 5-µm sections, mounted on Super Frost slides coated with (3-aminopropyl)-triethoxysilane (Sigma Aldrich; Merck KGaA), deparaffinized using xylene, rehydrated using a descending ethanol series, and processed as follows: one section per block was stained with haematoxylin and eosin- and one section was stained with Verhoeff's haematoxylin and green trichrome to differentiate tissue components. Slides with a significant proportion of cancer tissue were selected for IHC staining. IHC was performed manually using the primary antibody MUCIN 13 (D-5) (1:100; cat. no. sc-373857; Santa Cruz Biotechnology, Inc.). Recommended negative and positive controls were employed.
Histological quantification was performed using stereological methods and Ellipse software (ViDiTo) as previously described (26,27). A sample of the human intestinal wall-ileum and colon-without pathological changes was used as a positive control (Figs. S1 and S2). The control colon sample was used from a patient enrolled in the present study. The result of IHC detection was in consistency with the manufacturer's datasheets. The negative control was a control sample of the human intestinal wall (colon; i.e. a sample containing the proven glycoprotein mucin 13) but the reaction with the primary antibody was left out during the IHC procedure. The preparation was negative (mucin 13 was inconclusive) and therefore a non-specific reaction of the detection system was ruled out.
Quantitative estimates were performed using stereological methods and the Ellipse software (ViDiTo) as established in the previous study on the abdominal aortic aneurysm (28). The method is based on counting intersections of detected structures with stereological grids randomly superposed on the micrographs and has been previously described (26). This point-counting method was used for estimating the area fraction of MUC13 within the intestine wall AA.
The microscope objective and magnification used for the quantitative assessment of MUC13 was at the lowest setting that permitted an exact and unambiguous identification of the counting events with respect to the IHC detection method. The number of counting events per sample is provided, and the resulting data are presented as arithmetic means calculated from stochastic methods. Quantification of MUC13 in the specimens was based on a total of 568 micrographs.
miRNA selection
miR-4647 is predicted to bind to MUC13 only in the presence of homozygous GG genotype of rs1532602 in MUC13 according to the freely available software: MicroSNiper http://epicenter.ie-freiburg.mpg.de/services/microsniper/). According to mirDIP (https://ophid.utoronto.ca/mirDIP/), Targetscan (https://www.targetscan.org/vert_80/), and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) miR-4647 binds to MUC13 with medium integrated score (mirDIP=0.18, Targetscan=−0.06, miRWalk=0.85).
RT of miR-4647
For miRNA detection (miR-4647 assay ID 461900_mat), total RNA was reversely transcribed into cDNA using a pool of gene-specific primers designed for miRNA analysis using TaqMan® MiR Reverse Transcription kit (Thermo Fisher Scientific, Inc.). A pool of 5X Taqman miR primers, each diluted at 1:100, contained all target primers plus spike control miR-39 and potential reference genes: 18S and small RNA assays: RNU6B, RNU44, and RNU48. RNU48 (ID 001006) was used as reference genes selected by Normfinder (GenEx Enterprise). A total of 10 µl RT reaction contained: 8 ng/µl of RNA, 4 µl of primer pool, 2 µl RTr enzyme, 0.2 µl of 100 mM dNTPs, 1 µl of 10X RT buffer, 0.13 µl of RNase inhibitor and 0.67 µl of RNAse-free water. The thermocycling conditions were as follows: 16°C for 30 min, 42°C for 30 min and 85°C for 5 min in C1000 (Bio-Rad Laboratories, Inc.).
cDNA preamplification of miR-4647
All samples were pre-amplified prior to use in the high-throughput quantitative real-time PCR instrument Biomark (Fluidigm Corporation). A preamplification pool was prepared from 20X TaqMan MiR Assays (Thermo Fisher Scientific, Inc.), each assay diluted at 1:100. A total of 10 µl of preamplification reaction contained: 2 µl of cDNA (not diluted), 1.5 µl of preamp pool, 5 µl of iQ Supermix (Bio-Rad Laboratories, Inc.) and 1.5 µl RNAse-free water. Preamplification was performed at C1000 (Bio-Rad Laboratories, Inc.) as follows: 95°C for 3 min, 18 cycles of 95°C for 15 sec and 59°C for 4 min. After preamplification, each reaction was diluted at 1:20.
High-throughput qPCR of miR-4647
MicR qPCR was performed using a high-throughput platform BioMark™ HD System (Fluidigm Corporation) and 48.48 GE Dynamic Arrays or 96.96 GE Dynamic Arrays. A total of 5 µl of sample premix contained: 1 µl of the sample (1:20 diluted preamp cDNA), 2.5 µl of Taqman Universal Mastermix without UNG (Thermo Fisher Scientific, Inc.), 0.25 µl of 20X GE sample loading reagent (Fluidigm Corporation) and 1.25 µl of water. A total of 5 µl of assay pre-mix contained 2.5 µl of 20X Taqman miR assays and 2.5 µl of 2X assay loading reagent (Fluidigm Corporation). The thermocycling conditions were as follows: 95°C for 10 min, 40 cycles of 95°C for 15 sec and 60°C for 1 min. All data were analyzed by the 2−ΔΔCq method (25).
Cell culture
Both CRC cell lines [HCT-116 (cat. no. CCL-247™) and DLD-1 (cat. no. CCL-221™; both from American Type Culture Collection) were initially obtained from ECACC (Sigma-Aldrich; Merck KGaA). Cell lines were cultured at 37°C in a humidified atmosphere of 5% CO2 in Dulbecco's Modified Eagle's Medium (DMEM; Sigma-Aldrich; Merck KGaA) and supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 1 mM L-Glutamine and 100 U/ml penicillin/streptomycin. Both cell lines were examined with MycoAlert (Lonza Group Ltd.) to exclude mycoplasma contamination.
Transient cell transfection
Cells were transfected 24 h after seeding with 10 nM hsa-miR-4647 (cat. no. 4464066; Sigma-Aldrich; Merck KGaA) mimics, and miRNA mimics negative control with no homology to the human genome (cat. no. HMC0003; Sigma-Aldrich; Merck KGaA) using Lipofectamine® RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. All the experiments were performed in three independent replicates.
The transfection efficiency, changes in miRs levels, and their impact on MUC13 mRNA levels were confirmed by real-time RT-qPCR as follows: Specific Silencer® Select small interfering (si)RNA for human MUC13 (am16708) and Silencer® Select negative control siRNA #1 (cat. no. 4464058) were purchased from Ambion; Thermo Fisher Scientific, Inc. and transfected with Lipofectamine® RNAiMAX Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Silencing efficiency was verified by RT-qPCR 48 h after transfection using the TaqMan® Gene Expression Assay (Thermo Fisher Scientific, Inc.) for MUC13 (ID Hs00217230_m1).
Isolation and RT-qPCR of RNA from cell culture samples
Total RNA, including miRNA, was extracted from cells using Qiagen miReasy Mini Kit (Qiagen GmbH) according to the manufacturer's protocol. The concentration of the total RNA was measured by Nanodrop™ 8000 Spectrophotometer (Thermo Fisher Scientific, Inc.), and the integrity of mRNA in each sample was measured by Agilent RNA 6000 Nano kit by Agilent Bioanalyzer 2100 (Agilent Technologies, Inc.). To measure levels of miR-4647 after transfection, RT of miRNAs was performed using TaqMan Small RNA Assay Protocols (Thermo Fisher Scientific, Inc.). cDNA for MUC13 gene detection after transfection was synthesized from 400 ng of total RNA in 20 µl RT reaction using High-Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol in MJ Research PTC-200 Thermal Cycler (Bio-Rad Laboratories, Inc.). cDNA was diluted 1:1 with nuclease-free water. The reaction contained 2 µl of the sample, 10 µl of Taqman universal mastermix with no UNG (Thermo Fisher Scientific, Inc.), 1 µl of assay and 7 µl of RNAse free water. The thermocycling conditions were as follows: 95°C 10 min, and 45 cycles of 15 sec at 95°C, 1 min at 60°C in 7500 Real Time PCR System (Thermo Fisher Scientific, Inc.). The expression of MUC13 was normalized to the ACTB gene. Expression of miR-4647 measured using TaqMan MiR Assays at 7500 Real Time PCR System (Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 sec and 60°C for 60 sec plus melting curve analysis. The expression of miRNAs was normalized to RNU48 as previously described (29). All data were analyzed by the 2−ΔΔCq method (25).
Genotyping and sequencing of cell lines
The assessment of the MUC13 genotype for rs1532602 in both cell lines was performed similarly as aforementioned for human samples. The genotype correlation between the duplicate samples was 100%. The genotype call rate ranged 100%. To determine which allele of SNP MUC13 rs1532602 is actively transcribed, total RNA was reversibly transcribed from both cell lines into cDNA by the aforementioned standard laboratory procedure. PCR with primers flanking the region of interest followed afterward. Primers 5′-CCATTGGAGGGATAGAAGCA3′ and 5′-CTTTTCCTGGTAGGGCAACA-3′ designed by online tool primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) produced 233-bp long amplicon. The final product was diluted at 1:100 in a subsequent, direct sequencing reaction that was performed using the BigDye Terminator v3.1 Cycle Sequencing kit (Thermo Fisher Scientific, Inc.) under standard conditions. DNA sequencing was performed on ABI PRISM 3130 Genetic Analyzer (Thermo Fisher Scientific, Inc.), and the results were evaluated by Mutation Surveyor software (SoftGenetics, LLC).
In vitro assays
For colony formation assay, transfected cells were placed on six-well plates (500 cells per well) 48 h after transfection. After 12 days, colonies were fixed at room temperature (RT) for 20 min with 3% formaldehyde and stained with 1% crystal violet at RT for 20 min. The number of colonies (formed from >50 cells) was counted manually. All measurements were repeated three times. Cell migration assays were performed 48 h after transfection using Transwell Permeable Supports 6.5-mm Insert (24 well plate; 8-µm pore size; Corning, Inc.). Transfected cells (1×104) were seeded in DMEM with 0.5% FBS on the top of the Transwell chamber. The lower chamber was filled with 20% FBS-DMEM. Cells were cultured for 24 h. After the time point, the migratory cells were fixed with cold 3% formaldehyde for 30 min, washed with PBS, stained at RT for 20 min with 1% crystal violet, and counted by light microscope in five random fields under ×200 magnification. All measurements were repeated three times.
The integrity of the plasma membrane was assessed using the ToxiLight assay (cat. LT17-217; Lonza Group Ltd.) according to the manufacturer's protocol. This assay measures the release of adenylate kinase in the extracellular space, which reflects the plasma membrane's integrity. Cancer cells were cultured in a 96-well plate (25,000 cells per well) and exposed to a hsa-miR-4647 (cat. no. 4464066; Sigma-Aldrich; Merck KGaA) miRNA mimic for 24 h. After incubation at 37°C, 20 µl supernatant of each well was transferred to a new 96-well plate. Then, 50 µl of assay buffer was added to each well. After incubation in the dark for 5 min, the luminescence was measured using a Spectramax iD3 (Molecular Devices, LLC).
Western blot analysis
Proteins (20 µg) were loaded and separated in 12% SDS-PAGE gels at 15 mA for 60 min, and the separated proteins were then transferred to 0.45-µm Amersham Protran Nitrocellulose Blotting Membrane (Cytiva) in methanol transfer buffer, using Mini Trans-Blot Cell (Bio-Rad Laboratories, Inc.). The membranes were blocked with 5% bovine serum albumin in Tris-buffered saline containing Tween 20 (TBST; 20 mM Tris-HCl at pH 7.4, 0.15 M NaCl and 0.1% Tween 20) for 1 h and incubated with anti-MUC13 (1:100; cat. no. ab235450; Abcam) and anti-GAPDH (1:500; cat. no. ab8245; Abcam) at 4°C overnight, followed by incubation for 1 h at RT with goat anti-rabbit secondary antibody (Abcam) conjugated with horseradish peroxidase. The membranes were developed with SupersignalWest Pico Chemiluminescent Substrate (Pierce; Thermo Fisher Scientific, Inc.) and visualized by Azure c600 (Azure Biosystems, Inc.).
Statistical analysis
Statistical analysis for expression levels was conducted by SPSS Statistics 20 (IBM Corp.). All qPCR reactions were run in triplicates. The expression level of each sample was normalized using pre-selected reference genes and then averaged. The expression of miR-4647 in patients with CRC was normalized with the expression of RNU48. The expression of MUC13 was normalized to the ACTB gene. Expression levels of all studied genes did not follow a normal distribution in the study population, as analyzed by the Kolmogorov-Smirnov test. Data were logarithmically transformed and the paired non-parametric Wilcoxon and non-parametric Man-Whitney tests were used for statistical analyses to compare medians. The relationships between the examined variables and survival were investigated using the Spearman's correlation, expressed by Spearman's rho (ρ). The survival analysis was performed using the log-rank test and Kaplan-Meier plots approach. For in vitro tests, the Man-Whitney test was used for comparing unpaired data, and the ANOVA test was used for comparing multiple groups with the Tukey's post hoc test. All statistical tests were conducted at a 95% confidence level and P<0.05 was considered to indicate a statistically significant difference.
Results
Human-based research
Genes differentially expressed in CRC tissues
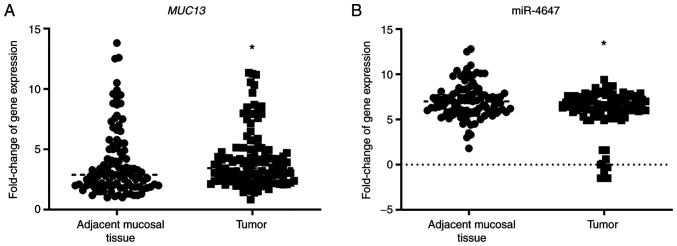
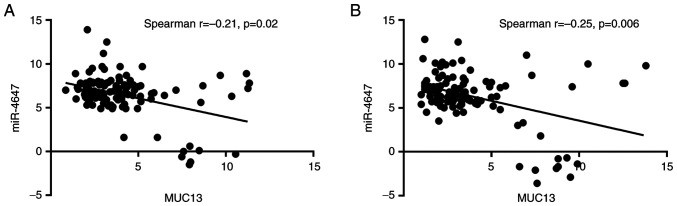
Expression levels of MUC13 and miR-4647 were successfully analyzed. Overall, MUC13 and miR-4647 were differentially expressed in the present study group: Significantly higher expression levels for MUC13 were observed in tumor tissues when compared with adjacent mucosal tissue while for miR-4647 an opposite trend was observed: Lower expression levels in tumor tissues (MUC13: 0.32-fold change-32% increase; P=0.02, Fig. 2A, miR-4647:0.17-fold change-17% decrease, P=0.05, respectively, Fig. 2B). A negative correlation between MUC13 and miR-4647 expression levels was observed in tumor tissue as well as in adjacent non-malignant mucosa (Spearman correlation r=−0.21, P=0.02 and r=−0.25, P=0.006, respectively; Fig. 3A and B). Significantly higher miR-4647 expression levels were observed in women compared with men (0.20-fold change-20% increase; P=0.009). Patients not receiving neoadjuvant therapy evinced significantly lower expression levels of miR-4647 when compared with those receiving neoadjuvant treatment (0.53-fold change-53% decrease; P=0.05, respectively). No other associations with clinicopathological data were observed. All data are summarized in Table II and Fig. S3A-F.
Figure 2.
Expression of MUC13 and related miRNA in patients with colorectal cancer. (A) Expression of MUC13 in tumor and non-malignant tissue. (B) Expression of miR-4647 in tumor and non-malignant tissue (Total n=187). *P<0.05. miR, microRNA.
Figure 3.
Expression correlation of analyzed genes. (A and B) Negative correlation of MUC13 and miR-4647 in (A) tumor tissue and (B) in non-malignant tissue (Total n=187). miR, microRNA.
Table II.
Fold change difference in expression of log2 scale of analyzed genes by non-parametric Wilcoxon test and Mann-Whitney test.
| MUC13 | microRNA-4647 | |||
|---|---|---|---|---|
|
|
|
|||
| Covariate | Fold change | P-value | Fold change | P-value |
| Relative expression (tumor vs. normal mucosa) | 0.32 | 0.02 | −0.17 | 0.05 |
| Sex (female vs. male) | 0.85 | 0.23 | 0.20 | 0.009 |
| Stratification (colon vs. rectum) | 0.58 | 0.13 | −0.29 | 0.81 |
| Neoadjuvant therapy (no vs. yes) | −0.04 | 0.64 | −0.53 | 0.05 |
miR-4647 is differentially expressed in CRC tissues
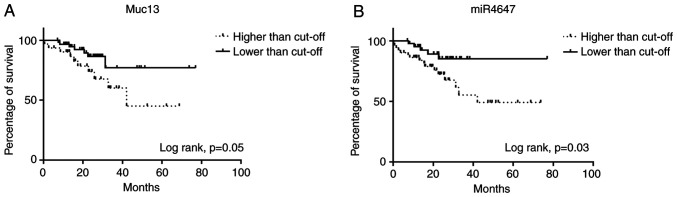
To further support the potential importance of analyzing miRNAs and MUC13 in CRC, their relation to OS was analyzed. It was observed that higher expression levels of MUC13 (Fig. 4A, log-rank P=0.05) and miR-4647 (Fig. 4B, log-rank P=0.03) were associated with worse survival of patients.
Figure 4.
Kaplan-Meier overall survival curves stratified for high and low expression for MUC13 and miR-4647. (A) MUC13. (B) miR-4647 (Total n=187). Survival analysis was performed using the log-rank test and Kaplan-Meier plot approach. miR, microRNA.
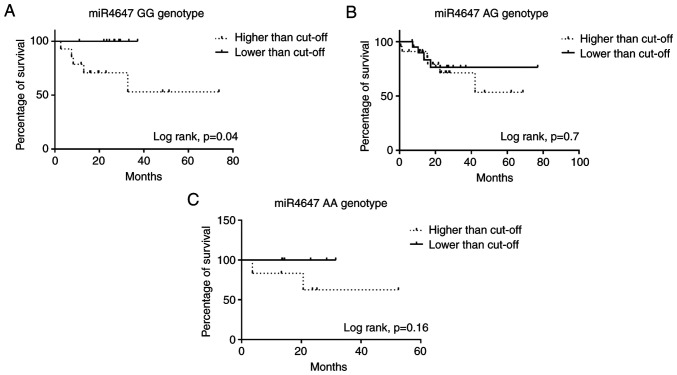
In silico, miR-4647 is predicted to bind to the homozygous GG genotype for rs1532602 in the MUC13 gene. However, in the presence of the variant homozygous AA genotype, miR-4647 does not bind to the MUC13 gene (Fig. 1). Previously, it has been observed that the homozygous variant AA genotype of rs1532602 in the miRNA binding site of MUC13 was associated with a decreased risk of CRC (21). Notably, patients carrying the homozygous GG genotype for rs1532602 in MUC13, together with higher levels of miR-4647, displayed worse OS (log-rank P=0.04, Fig. 5A). This could explain the controversial observation of higher miR-4647 expression levels and worse survival of patients. No other significant association with patient's survival was observed for heterozygous or variant genotypes (Fig. 5B and C). Additionally, survival analyses were also performed dividing patients according to the miRSNP rs1532602. No significant association with prognosis of patients was observed (data not shown).
Figure 5.
Kaplan-Meier overall survival curves stratified for rs1532602 in MUC13 in CRC patients with different genotypes. (A) Homozygous GG genotype (P=0.04), (B) heterozygous GA genotype, (C) homozygous variant AA genotype. Survival analysis was performed using the log-rank test and Kaplan-Meier plot approach.
IHC
In total, samples from 44 patients included in the study were analyzed for MUC13 positivity in tumor and adjacent non-malignant tissue. However, this procedure was not commercially available for miR-4647.
Control specimens
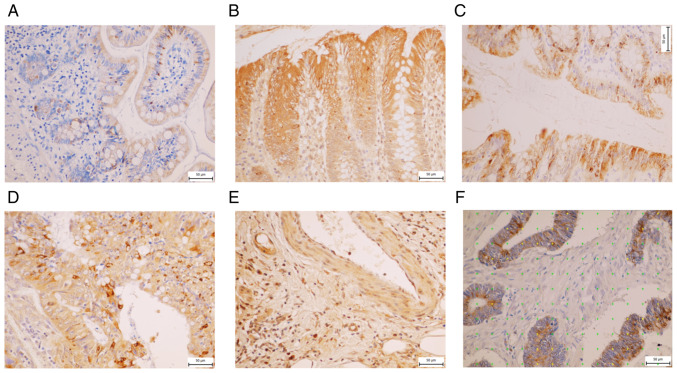
The control specimens of non-malignant human ileum, colon ascending and rectum were analyzed. In all specimens, IHC revealed the MUC13 positivity in columnar epithelial cells (enterocytes) and goblet cells (base of cells). MUC13 was localized in the apex of enterocytes and on their surface. In the non-malignant tunica mucosa of ileum, MUC13 was detected at Lieberküns' crypts base predominantly, on the top of villi of ileum in the cytoplasm of enterocytes and also in the base of goblet cells with intense MUC 13 positivity (Fig. 6A). In the non-malignant colon tissue, a change in positivity for MUC13 was observed. The expression of MUC13 was more evenly distributed in the crypts and, on the contrary, was more pronounced in enterocytes close to intestinal lumen (Fig. 6B). In the rectum, more pronounced positivity was observed in the crypts and the enterocytes (Fig. 6C).
Figure 6.
Immunohistochemical staining of MUC13 in different parts of colon. (A) Non-malignant ileum, (B) non-malignant colon, (C) non-malignant rectum, (D) cytoplasm of cancer cells and (E) endothelial cancer cells. (F) Figure from Ellipse software of cancer cells using a point-counting method where yellow points stand for positivity for mucin 13, green points for negative.
CRC tissue and its stroma
Higher MUC13 expression intensity was observed in cancer tissue compared with its adjacent non-malignant mucosa (32.57 vs. 5.04%). In the majority, MUC13 positivity was detected in the cytoplasm of the cancer cells and in some cells in the stroma as well (Fig. 6D). In certain cancer tissue samples, positive vessels for MUC13 were also detected. Endothelial cells, layer on top of the luminal part of endothelium and leiomyocytes in larger vessel wall were positive for MUC13, too (Fig. 6E). Mucinous tumors expressed MUC13, but at a lower level, based on the lower staining intensity than adenocarcinomas without mucinous components (25 vs. 34%).
To investigate whether there is a differential distribution in MUC13 expression in tumor tissue and non-malignant mucosa, the population was stratified for the rs1532602 genotype of the MUC13 gene. No significant distribution of MUC13 expression was observed. However, the patients with homozygous AA variant genotype evinced lower MUC13 expression levels both in tumor tissue and non-malignant mucosa when compared with homozygous GG genotype (Fig. S4A and B).
MUC13 relation to OS was analyzed to support further the potential importance of MUC13 expression levels in CRC. Although it was observed that higher expression levels of MUC13 (Fig. S4C, log-rank P=0.5) were associated with worse survival of patients, this association was not significant.
Characteristics of histological sections and microscopic image fields representing each colorectal tumor sample used for estimating the parameters are presented in Tables SII and SIII, and Fig. 6F.
Cell culture-based research
Determination of MUC13 genotype status
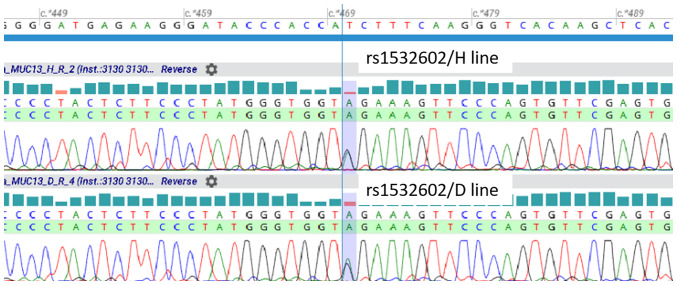
To assess the MUC13 genotype for rs1532602 in both cell lines, genotype screening by TaqMan allelic discrimination assay was performed. Both cell lines, HCT-116 and DLD-1, revealed heterozygous GA genotype for MUC13 rs1532602. To ascertain whether cell lines transcribe both alleles, the corresponding MUC13 cDNA was sequenced. It was confirmed that both cell lines expressed both alleles (G and A allele) of MUC13 rs1532602 in the same concentration (Fig. 7).
Figure 7.
Determination of MUC13 genotype status in colorectal cancer cell lines. HCT-116 (H line) and DLD-1 (D line) evinced heterozygous GA genotype for MUC13 rs1532602.
Determination of transfection efficiency
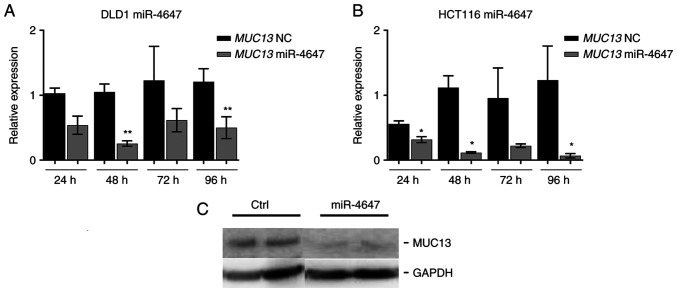
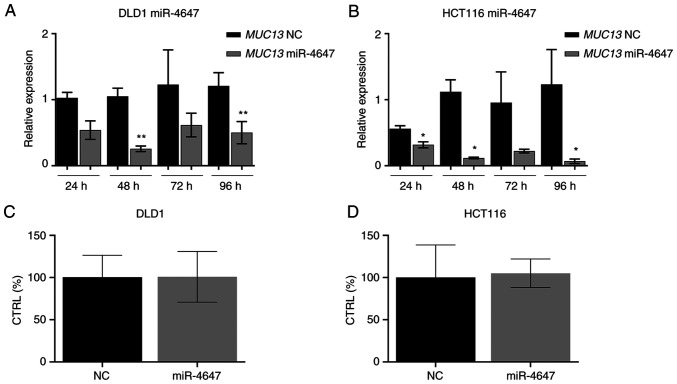
To analyze the effects of miR4647 on cell migration and colony forming, the transfection process of corresponding miRs precursors was first optimized. HCT-116 and DLD-1 cells were transfected using Lipofectamine RNAiMAX and efficiency was subsequently determined by RT-qPCR at 24, 48, 72 and 96 h after transfection (Fig. 8A and B). Significantly increased levels of tumor suppressive miR-4647 were recorded (more than 1,000-fold increase was achieved in both cell lines, n=3). In the majority, the most eminent effect was observed 24 h after transfection. The silenced MUC13 protein expression by miR-4647 was confirmed by western blot analysis (Fig. 8C).
Figure 8.
Transfection efficacy. (A and B) Transfection efficacy by miR-4647 mimics in (A) DLD1 and (B) HCT-116 cells. (C) Western blotting of silenced MUC13 by miR-4647. All presented results are average of 3 independent experiments (Man-Whitney test). *P<0.05 and **P<0.01. miR, microRNA; NC, negative control.
To further examine the MUC13-miR4647 axis, CRC cell lines were transfected with miR-4647 to detect the MUC13 mRNA and protein expression levels. The data revealed that overexpression of miR-4647 inhibited MUC13 mRNA expression compared with the negative control in the DLD-1 (Fig. 9A, P=0.05) and HCT-116 cells (Fig. 9B, P=0.003).
Figure 9.
Expression levels of MUC13 after overexpression of miR-4647. (A and B) Expression of MUC13 decreased after miR-4647 overexpression in (A) DLD1 and (B) HCT-116 cells. (C and D) The adenylate kinase levels after miR-4647 overexpression in (C) DLD1 and (D) HCT-116 cells. All presented results are average of 3 independent experiments (Man-Whitney test). *P<0.05 and **P<0.01. miR, microRNA; NC, negative control.
To determine whether analyzed miR-4647 affects cell homeostasis, the HCT-116 and DLD-1 cells were transfected with miR-4647 precursor and adenylate kinase enzyme in cell medium was measured 48 h after transfection. It was observed that the upregulated expression of miR-4647 did not affect the cellular homeostatic network (Fig. 9C and D).
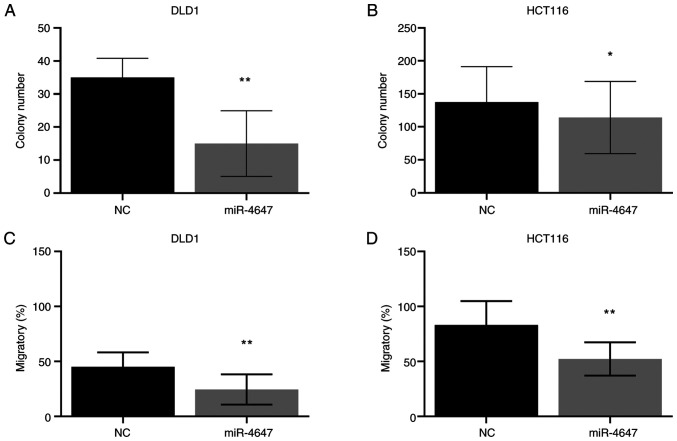
miR-4647 inhibits colony formation and migration of CRC cells
To determine the effect of miRs on cell survival, the anchorage dependence of proliferation using colony formation assay was assessed. After 12 days, HCT-116 and DLD-1 cells transfected with tested miR formed significantly fewer colonies than cells transfected with control oligonucleotide (miR-4647: HCT-116 with 20% decrease, P=0.03; DLD-1 with 57% decrease, P=0.05, respectively; Figs. 10A and B and S5A).
Figure 10.
Effect of overexpression of miR-4647 on growth and invasion behaviour of CRC cells. (A and B) The effect of miR overexpression on the proliferation of (A) DLD1 and (B) HCT-116 cells. (C and D) Effect of miR overexpression on the migratory ability of (C) DLD1 and (D) HCT-116 cells. All presented results are average of 3 independent experiments (Man-Whitney test). *P<0.05 and **P<0.01. miR, microRNA; NC, negative control.
Migration assay was performed to investigate the effect of miR on the invasive behavior of cancer cells. The migratory abilities of CRC cells after transfection with miR-4647 were reduced in both cell lines (miR-4647: HCT-116 with 37% decrease, P=0.002; DLD-1 with 46% decrease, P=0.001, respectively; Figs. 10C and D, and S5B).
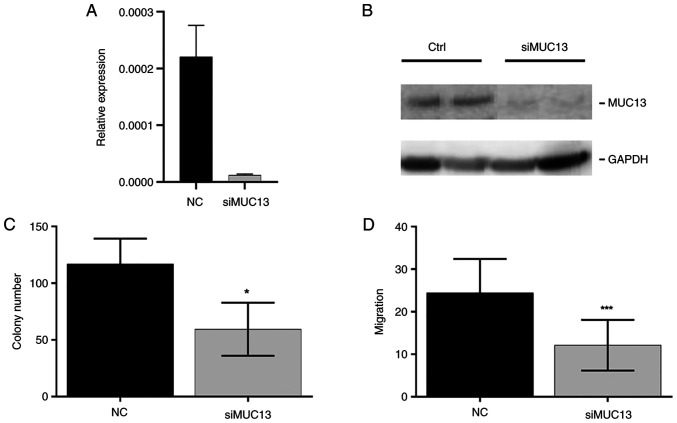
Silencing of MUC13 influences colony formation and migration of CRC cells
To evaluate whether the downregulation of MUC13 is responsible for the observed effects in CRC cells, MUC13 was silenced by siRNA in HCT-116 cells. The silenced MUC13 mRNA levels and protein expression in HTC-116 cells were confirmed by RT-qPCR and western blot analysis (Figs. 11A and B, and S5C). Lower expression levels of MUC13 confirmed the suppression of colony formation (33% decrease, P=0.02, Fig. 11C) and cell migration (64% decrease, P=0.0003, Fig. 11D). This effect was also observed after artificial overexpression of miR-4647, thus these data may indicate the role of MUC13 in CRC cells.
Figure 11.
Effect of silenced MUC13 on HCT-116 cell proliferation and migration. (A) The control expression of MUC13 after silencing. (B) The western blot analysis of silenced MUC13. (C) The effect of MUC13 silencing on the proliferation of CRC cells. (D) Migratory behaviour of CRC cells after MUC13 silencing. All presented results are average of 3 independent experiments (Man-Whitney test). *P<0.05 and ***P<0.001. NC, negative control; si-, small interfering.
Discussion
Recently, it has been observed that the homozygous variant AA genotype of rs1532602 in the miR binding site of MUC13 was associated with a decreased risk of CRC (21). Mucinous CRCs have been found to have worse survival and a higher TNM stage at diagnosis. These results inspired the authors to investigate the role of miRNAs regulation of MUC13. The in silico predicted MUC13-miR-4647 axis was analyzed and the association of their differential expression in survival of patients was pointed out.
Firstly, it was confirmed that MUC13 expression levels decreased after ectopic overexpression of miR-4647 by RT-qPCR. Further, the decreased miR-4647 expression was observed in human CRC tissues compared with matching adjacent non-malignant tissue, while MUC13 was found to be overexpressed in CRC tumors.
The main result of the present study is the observation of higher expression levels of MUC13 in association with poor survival of patients with CRC. In vitro functional tests confirmed this finding where CRC cells with attenuated MUC13 expression levels evinced decreased ability to form colonies. Besides, CRC cells with overexpressed miR-4647 formed significantly fewer colonies and had a reduced ability to migrate. MiR-4647 is predicted to bind to the G allele in polymorphic seed sequence site in MUC13, SNP spot rs1532602. This observation pointed out that the poorer survival of CRC cells may be linked to the MUC13 gene and not to miR-4647 overexpression. Higher expression levels of miR-4647 in CRC tumors also increased the risk of death in patients with the homozygous GG genotype of rs1532602 in MUC13. To the best of our knowledge, the effect of miR-4647 expression levels in patients with CRC as well as on patient's survival has not been analyzed in available literature yet. However, this miRNA was recently presented at ESMO Annals of Oncology and identified as a biomarker for the selection of patients with chronic myeloid leukemia that can qualify for achieving deep molecular response and can be considered for imatinib discontinuation trial (30). Similarly, the analysis of MUC13 in relation to association with relevant miRNA is, to the best of our knowledge, performed in the present study for the first time.
The presence of SNPs could also contribute to a different combination of miRNAs interacting in the region. This fact may additionally affect the modulation of post-transcriptional regulation mediated by a single SNP. Therefore, at present, it cannot be excluded that the observed clinical phenotypes may be the result of different combinations of miRNAs binding to one of such predicted SNP. Moreover, a recent study (31) suggested that variations in gene regions other than 3′UTRs may also affect the binding of miRNAs.
Notably, higher expression levels of miR-4647 were observed in association with poor survival of patients with CRC. This controversial association with the present data should be considered with caution. One of the hypotheses could be the fact that according to Targetscan, the other target for miR-4647 is the tumor necrosis factor receptor superfamily, member 13C (TNFRSF13C), which plays an important role in B cell homeostasis, immune system processes, adaptive immune response and the tumor necrosis factor-mediated signaling pathway. MiRNAs work to fine-tuning translation through specific mRNA binding. Negative regulation of TNFRSF13C gene expression by miR-4647 inhibits its translation and causes degradation of target mRNA. The association of poorer survival with higher levels of miR-4647 could be, thus, an explanation of the observed effect. Notably, Sheng et al observed that MUC13 promoted TNF-induced NF-κB activation by interacting with TNFR1 and the E3 ligase, cIAP1, which increased the ubiquitination of RIPK1 (15). However, patients carrying the homozygous GG genotype for rs1532602 in MUC13 together with higher levels of miR-4647, displayed worse OS. This could explain the observation of higher miR-4647 expression levels and worse patients' survival.
In a healthy colon, MUC13 was detected as a thin layer on the apical surface of glands. In cancer tissue, higher MUC13 expression intensity was observed compared with their adjacent non-malignant mucosa. MUC13 positivity was detected in the cytoplasm of the cancer cells. On the other hand, in our study, mucinous tumors expressed MUC13, but at a lower level, based on the lower staining intensity than adenocarcinomas (25 vs. 34%). In the present study, we have observed the overexpression of MUC13 in tumors compared with an adjacent colon. Further studies are required to clarify the relationships between MUC13 expression and colon cancer stages and prognosis.
Dysregulated expression of MUC13 has been shown in ovarian, pancreatic, gastric and colorectal cancers (4–6,9,11). We have confirmed that CRC patients with higher expression levels of MUC13 in tumor tissue displayed worse survival than those with lower expression levels. These observations agree with the studies of Gupta et al (6,7). On the contrary, Packer et al reported that the mRNA level of MUC13 was decreased in colon cancer; however, this was a small study with only 23 samples of colon cancer and 6 non-malignant colon tissue samples (32). MUC13 mRNA was also detected in the blood of CRC patients; however, MUC13 mRNA was also identified in the blood of healthy individuals (33). Sheng et al (13) observed that MUC13 may protect epithelial cells in the colon from apoptosis and through targeting MUC13 and MUC13-regulated pathways sensitize cancer cells to death and therefore may present an attractive target for cancer treatment (15).
Chauhan et al (9) showed that MUC13 expression increased the expression of HER2 in multiple cell types and MUC13 knockdown resulted in the downregulation of HER2 expression. HER2 belongs to the human EGF receptor (EGFR/ErbB) family of receptor tyrosine kinases that have been widely implicated in human cancers (34) and cancer pathogenesis (35). Although it is understood that receptor activation requires interactions between their specific ligands, so far, no soluble ligand has been identified for HER2. However, previous studies suggested that activation of ErbB receptors can be potentiated by proteins such as mucins, like MUC1 and MUC4 (36). Expression of MUC13 on the surface of CRC cells may also influence the growth characteristics via interactions with c-erbB growth factor receptors, modulate adhesion and interfere with immune recognition. Duan et al (37) suggested that MUC13 expressed on platelets in rats is involved in the interaction of platelets with endothelial cells, and, possibly by the same mechanism, MUC13 on cancer cells interacts with endothelial ligands during metastasis (14). MUC13 released from the surface of cancer cells may be a helpful serum diagnostic target in patients with gastrointestinal cancers.
There is a significant clinical need for biomarkers to provide an early indication of CRC. Such markers could act as an adjunct in CRC detection and allow a suitable use of chemotherapy and monitoring response to treatment (38). Investigations to identify biomarkers can also identify novel targets for therapy. MiRNAs not only contribute to diverse biological processes but are also implicated in the progression and metastasis of human cancers (39). Falzone et al (40) identified that miR-183-5p, miR-21-5p, miR-195-5p and miR-497-5p are directly related to CRC through the interaction with the mismatch repair pathway. In another recent study, clusters miR-17/92a-1, miR-106a/363, miR-106b/93/25 and miR-183/96/182 showed the strongest association with metastasis occurrence and poor patient survival (41). Similarly, Wang et al (42) observed that the miR-99a-HS3ST2-miR-100 axis, a novel miRNA-mRNA network, was associated with the presence of lymph node metastasis in CRC. Insights into the roles of miRNAs in cancer have established them as targets for novel therapeutic approaches. Previously, several miRNA-targeted therapeutics have reached clinical development (43). Consistent with the aforementioned findings, in the present study, miR-4647 suppressed proliferation in vitro.
Significantly higher miR-4647 expression levels were observed in women in comparison with men. Male and female patients have different endocrine backgrounds. It can be hypothesized that the changes observed in the expression levels of miR-4647 may be thus related to sex of patients instead of MUC13 or CRC.
There are certain limitations to the present study, such as the absence of non-cancerous cell lines as a control. However, as the aim of the study was to identify prognostic markers as well as to disclose novel therapeutic strategies for the treatment of advanced CRC, the exclusion of non-cancerous cell lines was not of utmost importance.
There is proof-of-principle evidence that miRNA SNPs can play a critical role in predicting cancer risk, treatment response and outcome. Understanding the factors contributing to cancer risk can represent a powerful future tool for clinicians and genetic counselors, as well as in advancing our understanding of cancer biology. If one risk allele or a signature of alleles were identified, clinicians could advise a specific group of patients to begin earlier, more frequent, and intensive screening, or even stronger preventative measures, in hopes of preventing disease or diagnosing it at an earlier and more treatable stage. More notably, as miRNAs are stimulated by external stimuli, it is also possible to manage patients with such SNPs by modifying lifestyle factors to maintain homeostasis of their inherited differences. This path of active research may prove the most promising. While assessing an individual's risk can be a valuable tool for diagnosing cancer at an earlier stage, the question of the best treatment for individual patients remains open.
In summary, the present study investigated a MUC13-miR-4647axis in CRC. The present data revealed the essential role of MUC13 in survival of patients and the MUC13-miR-4647axis pathway may provide novel therapeutic approaches. It is expected that MUC13 may hold significant potential in cancer screening, diagnosis and treatment.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Ondrej Daum (Department of Pathology, Faculty Hospital and Faculty of Medicine in Pilsen, Charles University in Prague, Czech Republic) for the consultation and help, and Dr Marketa Slajerova and Mrs Jaroslava Berankova (both from the Department of Histology and Embryology, Faculty of Medicine in Pilsen, Charles University in Prague, Czech Republic) for their technical help.
Funding Statement
The present study was supported by the National Science Foundation (grant no. 22-05942S), the Czech Health Research council of the Ministry of Health of the Czech Republic (grant no. NV19-09-00237), the Cooperation Program, research area ‘Oncology and Haematology’ (grant no. LX22NPO5102), the National institute for cancer research and the Cooperation Program, research area SURG and research area MED/DIAG.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
VV and PV conceptualized the present study. LS, AO, LB, JH, MU, VKr, JB and OK developed methodology. VKo, VN, JS, LV and VV conducted formal analysis. LS, JB, VL, MS and JS provided resources. LS and VV prepared the original draft. LS, AO, VKo, VN, VKr, JB, VL, MS, LV, MU, JS, PV and VV wrote, reviewed and edited the manuscript. PV, VL and JS supervised the study. VV, VL, LV, JS, MS and PV conceived the study. VV, JB, VL, LV, PV and VKr acquired funding. All authors read and approved the final version of the manuscript. AO and VKo confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Ethics approval (approval nos. G 09-04-09 and G 14-08-67) was granted by the committees of the General University Hospital (Prague; Czech Republic) and the Teaching Hospital and Medical School of Charles University (Pilsen; Czech Republic). Written informed consent was provided by all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 2.Jonckheere N, Skrypek N, Van Seuningen I. Mucins and tumor resistance to chemotherapeutic drugs. Biochim Biophys Acta. 2014;1846:142–151. doi: 10.1016/j.bbcan.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Kufe DW. Mucins in cancer: Function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimamura T, Ito H, Shibahara J, Watanabe A, Hippo Y, Taniguchi H, Chen Y, Kashima T, Ohtomo T, Tanioka F, et al. Overexpression of MUC13 is associated with intestinal-type gastric cancer. Cancer Sci. 2005;96:265–273. doi: 10.1111/j.1349-7006.2005.00043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh MD, Young JP, Leggett BA, Williams SH, Jass JR, McGuckin MA. The MUC13 cell surface mucin is highly expressed by human colorectal carcinomas. Hum Pathol. 2007;38:883–892. doi: 10.1016/j.humpath.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 6.Gupta BK, Maher DM, Ebeling MC, Sundram V, Koch MD, Lynch DW, Bohlmeyer T, Watanabe A, Aburatani H, Puumala SE, et al. Increased expression and aberrant localization of mucin 13 in metastatic colon cancer. J Histochem Cytochem. 2012;60:822–831. doi: 10.1369/0022155412460678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta BK, Maher DM, Ebeling MC, Stephenson PD, Puumala SE, Koch MR, Aburatani H, Jaggi M, Chauhan SC. Functions and regulation of MUC13 mucin in colon cancer cells. J Gastroenterol. 2014;49:1378–1391. doi: 10.1007/s00535-013-0885-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan S, Ebeling MC, Zaman MS, Sikander M, Yallapu MM, Chauhan N, Yacoubian AM, Behrman SW, Zafar N, Kumar D, et al. MicroRNA-145 targets MUC13 and suppresses growth and invasion of pancreatic cancer. Oncotarget. 2014;5:7599–7609. doi: 10.18632/oncotarget.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chauhan SC, Ebeling MC, Maher DM, Koch MD, Watanabe A, Aburatani H, Lio Y, Jaggi M. MUC13 mucin augments pancreatic tumorigenesis. Mol Cancer Ther. 2012;11:24–33. doi: 10.1158/1535-7163.MCT-11-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheng Y, Ng CP, Lourie R, Shah ET, He Y, Wong KY, Seim I, Oancea I, Morais C, Jeffery PL, et al. MUC13 overexpression in renal cell carcinoma plays a central role in tumor progression and drug resistance. Int J Cancer. 2017;140:2351–2363. doi: 10.1002/ijc.30651. [DOI] [PubMed] [Google Scholar]
- 11.Chauhan SC, Vannatta K, Ebeling MC, Vinayek N, Watanabe A, Pandey KK, Bell MC, Koch MD, Aburatani H, Lio Y, Jaggi M. Expression and functions of transmembrane mucin MUC13 in ovarian cancer. Cancer Res. 2009;69:765–774. doi: 10.1158/0008-5472.CAN-08-0587. [DOI] [PubMed] [Google Scholar]
- 12.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1:183–197. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheng YH, Lourie R, Linden SK, Jeffery PL, Roche D, Tran TV, Png CW, Waterhouse N, Sutton P, Florin TH, McGuckin MA. The MUC13 cell-surface mucin protects against intestinal inflammation by inhibiting epithelial cell apoptosis. Gut. 2011;60:1661–1670. doi: 10.1136/gut.2011.239194. [DOI] [PubMed] [Google Scholar]
- 14.Williams SJ, Wreschner DH, Tran M, Eyre HJ, Sutherland GR, McGuckin MA. Muc13, a novel human cell surface mucin expressed by epithelial and hemopoietic cells. J Biol Chem. 2001;276:18327–18336. doi: 10.1074/jbc.M008850200. [DOI] [PubMed] [Google Scholar]
- 15.Sheng YH, He Y, Hasnain SZ, Wang R, Tong H, Clarke DT, Lourie R, Oancea I, Wong KY, Lumley JW, et al. MUC13 protects colorectal cancer cells from death by activating the NF-κB pathway and is a potential therapeutic target. Oncogene. 2017;36:700–713. doi: 10.1038/onc.2016.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheng YH, Triyana S, Wang R, Das I, Gerloff K, Florin TH, Sutton P, McGuckin MA. MUC1 and MUC13 differentially regulate epithelial inflammation in response to inflammatory and infectious stimuli. Mucosal Immunol. 2013;6:557–568. doi: 10.1038/mi.2012.98. [DOI] [PubMed] [Google Scholar]
- 17.Naccarati A, Pardini B, Hemminki K, Vodicka P. Sporadic colorectal cancer and individual susceptibility: A review of the association studies investigating the role of DNA repair genetic polymorphisms. Mutat Res. 2007;635:118–145. doi: 10.1016/j.mrrev.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Tomlinson IP, Dunlop M, Campbell H, Zanke B, Gallinger S, Hudson T, Koessler T, Pharoah PD, Niittymäki I, Tuupanen S, et al. COGENT (COlorectal cancer GENeTics): An international consortium to study the role of polymorphic variation on the risk of colorectal cancer. Br J Cancer. 2010;102:447–454. doi: 10.1038/sj.bjc.6605518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rattray NJW, Charkoftaki G, Rattray Z, Hansen JE, Vasiliou V, Johnson CH. Environmental influences in the etiology of colorectal cancer: The premise of metabolomics. Curr Pharmacol Rep. 2017;3:114–125. doi: 10.1007/s40495-017-0088-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, Zhang W, Huo M, Wang P, Liu X, Wang Y, Li Y, Zhou Z, Xu N, Zhu H. XBP1 regulates the protumoral function of tumor-associated macrophages in human colorectal cancer. Signal Transduct Target Ther. 2021;6:357. doi: 10.1038/s41392-021-00761-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vymetalkova V, Pardini B, Rosa F, Jiraskova K, Di Gaetano C, Bendova P, Levy M, Veskrnova V, Buchler T, Vodickova L, et al. Polymorphisms in microRNA binding sites of mucin genes as predictors of clinical outcome in colorectal cancer patients. Carcinogenesis. 2017;38:28–39. doi: 10.1093/carcin/bgw114. [DOI] [PubMed] [Google Scholar]
- 22.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 23.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 24.Hao M, Wang K, Ding Y, Li H, Liu Y, Ding L. Which patients are prone to suffer liver metastasis? A review of risk factors of metachronous liver metastasis of colorectal cancer. Eur J Med Res. 2022;27:130. doi: 10.1186/s40001-022-00759-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Krizkova V, Dubova M, Susova S, Vycital O, Bruha J, Skala M, Liska V, Daum O, Soucek P. Protein expression of ATP-binding cassette transporters ABCC10 and ABCC11 associates with survival of colorectal cancer patients. Cancer Chemother Pharmacol. 2016;78:595–603. doi: 10.1007/s00280-016-3114-7. [DOI] [PubMed] [Google Scholar]
- 27.Witter K, Tonar Z, Matejka VM, Martinca T, Jonák M, Rokosný S, Pirk J. Tissue reaction to three different types of tissue glues in an experimental aorta dissection model: A quantitative approach. Histochem Cell Biol. 2010;133:241–259. doi: 10.1007/s00418-009-0656-3. [DOI] [PubMed] [Google Scholar]
- 28.Eberlová L, Tonar Z, Witter K, Křížková V, Nedorost L, Korabečná M, Tolinger P, Kočová J, Boudová L, Třeška V, et al. Asymptomatic abdominal aortic aneurysms show histological signs of progression: A quantitative histochemical analysis. Pathobiology. 2013;80:11–23. doi: 10.1159/000339304. [DOI] [PubMed] [Google Scholar]
- 29.Cervena K, Novosadova V, Pardini B, Naccarati A, Opattova A, Horak J, Vodenkova S, Buchler T, Skrobanek P, Levy M, et al. Analysis of MicroRNA expression changes during the course of therapy in rectal cancer patients. Front Oncol. 2021;11:702258. doi: 10.3389/fonc.2021.702258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guru SA, Sumi MP, Najar IA, Mir AR, Saxena A. MO10-6 miR-4647 an early biomarker of outcome in chronic myeloid leukaemia patients. Ann Oncol. 2022;33((Suppl 6)):S488. doi: 10.1016/j.annonc.2022.05.124. [DOI] [Google Scholar]
- 31.Liu C, Rennie WA, Carmack CS, Kanoria S, Cheng J, Lu J, Ding Y. Effects of genetic variations on microRNA: Target interactions. Nucleic Acids Res. 2014;42:9543–9552. doi: 10.1093/nar/gku675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Packer LM, Williams SJ, Callaghan S, Gotley DC, McGuckin MA. Expression of the cell surface mucin gene family in adenocarcinomas. Int J Oncol. 2004;25:1119–1126. [PubMed] [Google Scholar]
- 33.Lauriola M, Ugolini G, Rosati G, Zanotti S, Montroni I, Manaresi A, Zattoni D, Rivetti S, Mattei G, Coppola D, et al. Identification by a digital gene expression displayer (DGED) and test by RT-PCR analysis of new mRNA candidate markers for colorectal cancer in peripheral blood. Int J Oncol. 2010;37:519–525. doi: 10.3892/ijo_00000701. [DOI] [PubMed] [Google Scholar]
- 34.Settleman J. Predicting response to HER2 kinase inhibition. Oncotarget. 2015;6:588–589. doi: 10.18632/oncotarget.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaturvedi P, Singh AP, Chakraborty S, Chauhan SC, Bafna S, Meza JL, Singh PK, Hollingsworth MA, Mehta PP, Batra SK. MUC4 mucin interacts with and stabilizes the HER2 oncoprotein in human pancreatic cancer cells. Cancer Res. 2008;68:2065–2070. doi: 10.1158/0008-5472.CAN-07-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Senapati S, Das S, Batra SK. Mucin-interacting proteins: From function to therapeutics. Trends Biochem Sci. 2010;35:236–245. doi: 10.1016/j.tibs.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan Y, Naruse T, Nakamura M, Yamaguchi Y, Kawashima T, Morikawa Y, Kitamura T, Suda T. Expression and functional analysis of a hemopoietic progenitor antigen, NJ-1 (114/A10), in the megakaryocytic lineage. Biochem Biophys Res Commun. 1998;253:401–406. doi: 10.1006/bbrc.1998.9700. [DOI] [PubMed] [Google Scholar]
- 38.Zhu J, Xu Y, Liu S, Qiao L, Sun J, Zhao Q. MicroRNAs associated with colon cancer: New potential prognostic markers and targets for therapy. Front Bioeng Biotechnol. 2020;8:176. doi: 10.3389/fbioe.2020.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Detassis S, Grasso M, Del Vescovo V, Denti MA. microRNAs make the call in cancer personalized medicine. Front Cell Dev Biol. 2017;5:86. doi: 10.3389/fcell.2017.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falzone L, Scola L, Zanghi A, Biondi A, Di Cataldo A, Libra M, Candido S. Integrated analysis of colorectal cancer microRNA datasets: Identification of microRNAs associated with tumor development. Aging (Albany NY) 2018;10:1000–1014. doi: 10.18632/aging.101444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pidíková P, Herichová I. miRNA clusters with up-regulated expression in colorectal cancer. Cancers (Basel) 2021;13:2979. doi: 10.3390/cancers13122979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Gao G, Chen Z, Chen Z, Han M, Xie X, Jin Q, Du H, Cao Z, Zhang H. Identification of the miRNA signature and key genes in colorectal cancer lymph node metastasis. Cancer Cell Int. 2021;21:358. doi: 10.1186/s12935-021-02058-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.