Abstract
Background
To identify autophagy- and immune-related hub genes affecting the diagnosis and treatment of endometriosis.
Methods
Gene expression data were downloaded from the Gene Expression Omnibus (GEO) (GSE11691 and GSE120103 for training, and GSE7305 for validation). By overlapping the differentially expressed genes (DEGs), Weighted gene co-expression network analysis (WGCNA) module genes, and autophagy-related genes (ARGs), and immune-related genes (IRGs) separately, hub genes were identified using the least absolute shrinkage and selection operator (LASSO)and support vector machine recursive feature elimination (SVM-RFE). The hub genes were analyzed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. A hub gene-prediction model was constructed and assessed using five-fold cross-validation via five supervised machine-learning algorithms: random forest, the sequential minimal optimization (SMO), K-nearest neighbours (IBK), C4.5 decision tree (J48), and logistics regression. The area under the receiver operating characteristic curve (AUC) was adopted to assess the identification ability of characteristic genes.
Results
1,116 DEGs were obtained from the training cohort, and 22 endometriosis-related IRGs were identified by overlapping the 1,116 DEGs, 3,222 module genes, and 1,793 IRGs. Meanwhile, 45 endometriosis-related ARGs were obtained (1,928 ARGs). Subsequently, nine IRG hub genes (BST2, CCL13, CD86, CSF1, FAM3C, GREM1, ISG20, PSMB8, and S100A11) and nine ARG hub genes (GSK3A, HTR2B, RAB3GAP1, ARFIP2, BNIP3, CSF1, MAOA, PPP1R13L, and SH3GLB2) were obtained by LASSO and SVM-RFE. GO analysis indicated that the ARG hub genes responded to the regulation of autophagy and mitochondrial outer membrane permeabilization, and KEGG enrichment analysis involved serotonergic and dopaminergic synapses. GO analysis also indicated that the IRG hub genes responded to the regulation of leukocyte proliferation and mononuclear cell migration, and KEGG analysis showed enrichment involved in viral protein interaction with cytokines and cytokine receptors. The AUC of the random-forest algorithm of ARGs was 0.975 in the training cohort and 0.940 in the validation cohort, and the AUC of the SMO algorithm of IRGs was 0.907 in the training cohort and 0.8 in the validation cohort.
Conclusions
Seventeen hub genes are closely associated with endometriosis. These genes are potential autophagy- and immune-related biomarkers for diagnosis and treatment of endometriosis.
Keywords: Endometriosis, autophagy-related genes (ARGs), immune-related genes (IRGs), biomarker
Highlight box.
Key findings
• Seventeen hub genes were found to be closely associated with autophagy and immunity in endometriosis. These genes are potential autophagy-and immune-related biomarkers for diagnosis and treatment monitoring in endometriosis.
What is known and what is new?
• Autophagic abnormality and abnormal immune responses are involved in the pathogenesis of endometriosis. Autophagy inhibition can promote the expression of inflammatory cytokines, which underpins the pathogenesis of various autoimmune diseases.
• It preliminarily verified that autophagy and immune imbalance are involved in the pathogenesis of endometriosis. Seventeen hub genes were found to be closely associated with autophagy and immunity in endometriosis.
What is the implication, and what should change now?
• The present study aimed to evaluate the key points of ARGs and IRGs related to endometriosis, and provide reference for the development of therapeutic drugs and clinical treatment.
Introduction
Endometriosis refers to the abnormal relocation of endometrial cells outside the uterus, which is a chronic and debilitating disease that often causes pelvic pain and infertility (1,2). Superficial peritoneal lesions, deep-infiltrating endometriosis, and ovarian endometriotic cysts (endometriomas) are the most common pathological types of endometriosis (3). While, its pathogenic mechanisms remain to be elucidated genetics and the environment are key drivers, and the immune system is believed to play an important role in its pathophysiology and symptomatology (4). The theory of Samson (“retrograde menstruation”) is the highest acceptance. It says that the focus of endometriosis is the result of blood transfer to the peritoneal cavity through the fallopian tube (5). However, not all women with menstruation reflux will have endometriosis. Other factors may determine the survival and implantation of endometrial cells in peritoneal cavity (6). Conventional treatments for endometriosis include surgical removal of lesions and hormonotherapy. However, these therapies are often accompanied by multiple side effects, and the patients also have a high incidence of recurrence.
Globally, approximately 5–10% of child-bearing-age women are reported to have endometriosis, resulting in a huge economic burden (> US$22 billion in the United States alone) (7). Endometriosis is observed in 50–80% of women with pelvic pain and over 50% of those with infertility, and more than 176 million women worldwide have been diagnosed with endometriosis (1,4). Although definitive diagnosis is based on histology, endometriosis can be identified through visual diagnosis. However, visualization is challenging because the symptoms, signs, and lesion sites vary between patients (8-10). Given these conditions, diagnosis is often delayed (even for years), and about 65% of the patients have been misdiagnosed (11). Therefore, there is an urgent need to explore more accurate diagnostic methods.
Autophagy refers to physical programmed cell death in eukaryotic cells and plays a vital role in the metabolic process. It degrades cytoplasmic proteins and organelles into amino acids and fatty acids for energy generation and the recovery of metabolic wastes (12). Studies have shown that the conditional knockdown of autophagy-related genes (ARGs) in mice down-regulates the expression of inflammatory cytokines and suppresses autoreactive immune cells, thereby attenuating symptoms in the autoimmune disease model. Autophagy inhibition can promote the expression of inflammatory cytokines, which underpins the pathogenesis of various autoimmune diseases (13). Autophagy has been shown to play a role in antigen presentation, and the inhibition of autophagy may allow endometrial cells to escape from immune surveillance and facilitate intramyometrial implantation (14) and EMS. Therefore, the level of autophagy is most likely associated with the pathogenesis of EMS. More than 30 ARGs have been identified (15,16); yet, research into their association with endometriosis is lacking.
A study has demonstrated that chronic inflammation and abnormal immune responses are involved in the pathogenesis of endometriosis (17). Immune cells and proinflammatory cytokines can induce the proliferation and angiogenesis of ectopic endometrium by coordinating the immune system and endocrine system to form a specific local immune microenvironment of peritoneal fluid, so that it can escape the recognition and monitoring of immune cells and promote its implantation and growth, thus participating in the pathophysiological process of endometriosis. A study has confirmed that the probability of endometriosis patients with autoimmune diseases is significantly higher than that of normal people, and it is closely related to the pathogenesis, clinical symptoms, disease progress and outcome (18). Immunity-related genes (IRGs) play a crucial role in immune cell infiltration; however, there remains a lack of clear elucidation regarding the characteristics of IRG expression and their potential regulatory mechanisms in endometriosis.
Current studies on the immune response, cellular autophagy, and endometriosis occurrence mainly focus on the following two aspects: (I) the effects of the immune response and autophagy on the pathological process of endometriosis (19,20); and (II) the efficacy of immune and autophagy inhibitors for the treatment of endometriosis (21). Although there have been reports of immune related gene in endometriosis, the relationship between immune related genes and endometriosis has not been comprehensively analyzed. The present study aimed to evaluate the key points of ARGs and IRGs related to endometriosis, and provide reference for the development of therapeutic drugs and clinical treatment. The hub genes identified in this study could also be autophagy-related and immune-related biomarkers for the diagnosis and treatment monitoring of endometriosis. We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5979/rc).
Methods
Data source
The gene expression data of GSE11691, GSE120103, and GSE7305 patients were collected from Gene Expression Omnibus (GEO) database (available from: https://www.ncbi.nlm.nih.gov/geo/). GSE11691 was in the GPL96 platform, [HG-U133A] Affymetrix Human Genome U133A Array, which included nine endometriosis samples and nine control samples. GSE120103 contained the Ribonucleic Acid (RNA) expression data annotated by GPL6480, which included 18 endometriosis samples and 18 normal tissue samples. GSE7305 was in the GPL570 platform, [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array, comprising 10 endometriosis and 10 control samples.
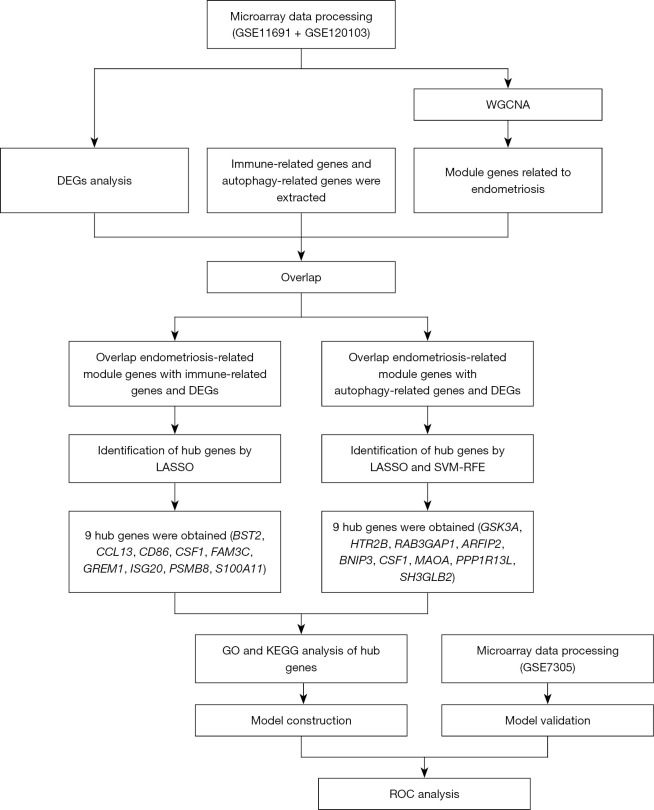
We downloaded the platform and series matrix files, as well as a list of IRGs, from the Immunology Database and Analysis Portal (ImmPort) (https://www.immport.org/shared/home), and assessed ARGs derived from Human Autophagy Database (HADb, http://www.autophagy.lu/index.html), the AUTOPHAGY DATABASE (http://www.tanpaku.org/autophagy/index.html), Index-Home-Human Autophagy Moderator Database (HAMdb, http://hamdb.scbdd.com/), Gene Set Enrichment Analysis Database (GSEA, http://www.gsea-msigdb.org/gsea/index.jsp) , and National Library of Medicine(NCBI, https://www.ncbi.nlm.nih.gov/). A flow diagram of the study is shown in Figure 1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Figure 1.
Flow diagram of the study. DEGs, differentially expressed genes; WGCNA, weighted gene co-expression network analysis; LASSO, least absolute shrinkage and selection operator; SVM-RFE, support vector machine recursive feature elimination; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Study design and data pre-processing
Original series matrix files of the GSE120103 and GSE11691 datasets were retrieved. A probe expression matrix of the raw data was extracted and normalized and subsequently converted into a gene expression matrix using the platform annotation file. The two gene matrices were merged using Perl script, and heterogeneity induced by various experimental batches and platforms was processed using the “sva” package of R software. A merging normalized gene expression matrix was obtained using the “limma” package of R for further analysis. The probe expression matrices were extracted sequentially using Perl script and converted into gene expression matrices. Analysis of the gene expression matrix was performed via the “limma” package of R. GSE7305 was downloaded in the form of a gene expression matrix.
Differential expression analysis
Differential expression analysis was conducted for endometriosis and normal samples using the “limma” package (22). Genes with P value <0.5 and |log2Fold Change (FC)| >1 were considered Differentially expressed genes (DEGs). Heat maps of the DEGs and volcano plots of all identified genes were plotted using the “pheatmap”, “ggplot2”, “dplyr”, and “ggrepel” packages.
Immune infiltration
CIBERSORT was used to evaluate the immune cell infiltration in the microenvironment, involving 547 biomarkers and 22 immune cells such as plasma, B cell, T cell, and myeloid cell subpopulations. Under the linear support vector regression principle, this tool was employed to conduct deconvolution analysis of the expression matrix of immune cells. The present study collected expression data from GSE120103 and GSE11691 and calculated the ratios of the 22 immune cells in each sample.
Weighted gene co-expression network analysis (WGCNA)
The “WGCNA” R package was applied to produce the co-expression networks (23) for genes in the GSE11691 and GSE120103 datasets, and abnormal samples were removed based on the cluster trees. A similarity matrix was formed by calculating the correlation coefficients of different gene pairs. A proper soft threshold was set for the transformation of the similarity matrix into an adjacency matrix to ensure scale-free network construction, and a topological overlap matrix (TOM) was subsequently provided to evaluate the mean network connectivity of each gene. Genes of similar expression profiles were grouped into different modules via the dynamic tree cutting method, and each module was depicted using a different color. Genes in gray modules were considered inapplicable to be assigned to any module.
The module eigengene (ME) was applied to represent the gene expression profile of each module, which was also used to assess the association between modules and phenotypes. The module with the highest absolute correlation coefficient value was selected as the key module and included in further analysis. Module membership (MM) reflected the correlation coefficient between the expression value of a gene and the ME of a corresponding module, while gene significance (GS) was used to present the correlation coefficient of a gene expression value with the phenotype.
Overlapping endometriosis-related module genes and DEGs with IRGs and ARGs
We overlapped the IRGs and ARGs with endometriosis-related module genes derived from WGCNA and DEGs, respectively. The detailed gene overlapping was presented using a Venn diagram.
Identification of hub genes
Hub gene identification was conducted using the “glmnet” package via the least absolute shrinkage and selection operator (LASSO) (24) and support vector machine recursive feature elimination (SVM-RFE). The latter is a vector-supporting machine learning algorithm capable of effectively deriving an informative gene subset to facilitate the identification of hub genes (25). Hub genes were screened using the “e1071” package with the 10-fold cross-validation method, and genes identified by LASSO and SVM-RFE were overlapped for determination.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of overlapped genes
GO and KEGG enrichment analysis was performed using the “clusterProfiler” package (26). P<0.05 indicated statistical significance. GO enrichment analysis aims to describe the gene-correlated biological processes (BP), cellular components (CC), and molecular functions (MF), while KEGG pathway analysis seeks to identify enriched biological pathways.
Module construction and validation
Weka 3.7.3 (www.cs.waikato.ac.nz/ml/weka) was applied for module construction, which takes into account multiple machine learning techniques such as decision trees (J48), the sequential minimal optimization (SMO), K-nearest neighbours (IBK), and logistic regression (LR). To optimize the performance of the selected technique, the algorithm performed an automatic search using five-fold cross-validation for the optimal parameter setting based on the best prediction results.
A receiver operating characteristic (ROC) curve was also plotted, and the area under the ROC curve (AUC) was calculated to assess the module accuracy. The area under the ROC curve (AUC) could be generated by plotting the sensitivity against (1-specificity) at different discrimination threshold settings. The AUC in the normalized units reveals the probability of a classifier ranking a randomly selected positive sample over a negative one.
Results
Identification of DEGs
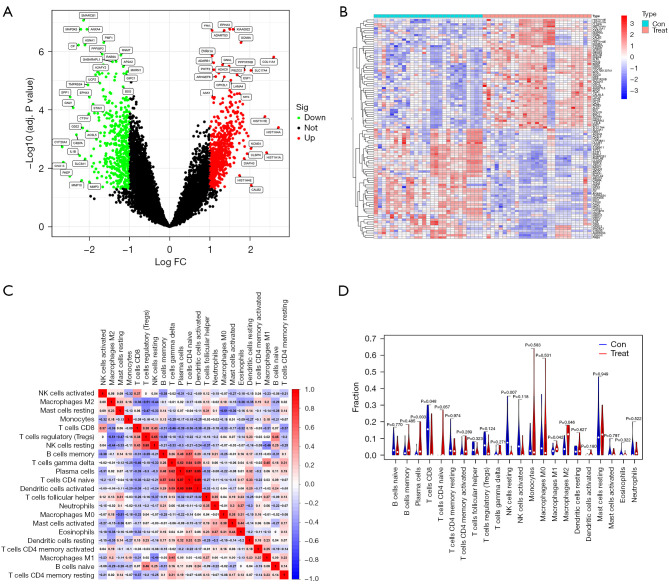
There were a total of 1,116 DEGs (660 up-regulated and 456 down-regulated) identified in the GSE11691 and GSE120103 datasets, and the top 50 DEGs were displayed in a heat map. Volcano plots showed the DEGs variation from the control in endometriosis (Figure 2A,2B).
Figure 2.
Differentially expressed genes in endometriosis. (A) Volcano map of differentially expressed genes in endometriosis compared with the controls from the GSE11691and GSE120013 datasets; (B) heat map of the top 50 differentially expressed genes. Immune infiltration between endometriosis samples and control samples. (C) The relative percentage of 22 immune cells in each sample. (D) Differences in immune infiltration between endometriosis samples and control samples. FC, fold change; Con, control.
Immune infiltration
In this part of the study, the proportion of 22 immune cells in 27 endometriosis samples and 27 control samples was estimated by the CIBERSORT algorithm, which can be seen in the corHeatmap (Figure 2C). The immune cell infiltration of endometriosis and endometriosis samples was compared in a vioplot (Figure 2D). The results showed that the endometriosis group had significantly higher proportions of Plasma cells (P=0.003), macrophages M1 (P=0.042), macrophages M2 (P=0.046) and lower proportions of NK cells resting (P=0.007), T cells CD8 (P=0.048) than the control group.
Weighted gene co-expression network construction and modules identification
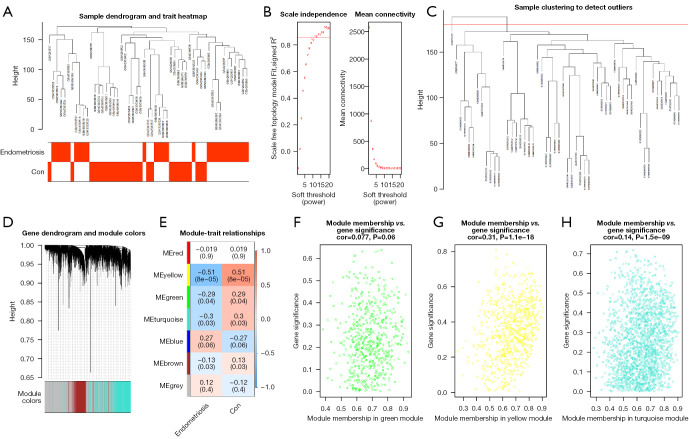
The soft threshold was set as a β-value of 10 (scale-free R2=0.85) to ensure scale-free networks (Figure 3A,3B). Genes with similar expression patterns were clustered into co-expression modules, which were differently colored. A total of six modules were identified (Figure 3C,3D).
Figure 3.
Construction and analysis of weighted gene co-expression network. (A) Clustering and phenotype in the combined data samples; the clustering is shown in the upper part of the figure, and the phenotype is shown in the lower part. The color stands for disease. Red square, the correlation between the disease or control group and the sample is 1; white square, the correlation between the disease or control group and the sample is 0. (B) Ascertainment of the soft threshold. (C) Merging of similar modules based on the dynamic cutting tree algorithm. (D) Clustering dendrogram of genes with various levels of similarity according to the topological overlap and the assigned module color. (E) Heat map for the association between various modules and clinical characteristics. The ordinate stands for various modules and the abscissa stands for different traits. Each block indicates a module’s and a trait’s correlation coefficient and P value. (F-H) Correlation between module membership and gene significance in the green module, yellow module and turquoise module. Con, control.
The association between each module and clinical information was displayed in the module-trait relationship (Figure 3E). There were 774 genes contained in the yellow-colored module, the majority of which were significantly associated with endometriosis (R=0.51, P=5×10-8), followed by 1,849 genes included in the turquoise module (R=0.3, P=0.03). The 599 genes in the green-colored module also exhibited an association with endometriosis (R=0.29, P=0.04). The genes in these three modules were included in further analysis.
Modular genes in the co-expression network were characterized by high intramodular connectivity measured by GS and MM. Scatterplots of GS (y-axis) vs. MM (x-axis) are shown in Figure 3F-3H in different colors. WGCNA indicated that 3,222 genes were associated with endometriosis, and these genes were also included in the subsequent analyses.
Overlapping of endometriosis-related module genes and DEGs with IRGs and ARGs
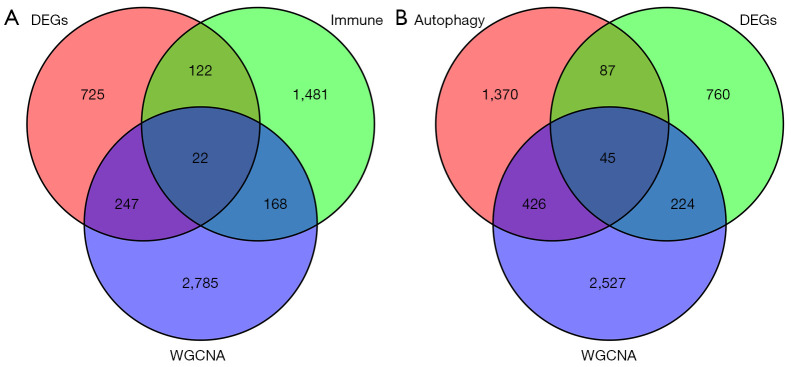
A total of 22 overlapping genes associated with immune-related module genes were retrieved by taking intersections of 1,116 DEGs, 3,222 module genes, and 1,793 IRGs, as shown in the Venn diagram (Figure 4A). At the same time, 45 overlapping genes associated with autophagy-related module genes were obtained by taking the intersections of the 1,116 DEGs, 3,222 module genes, and 1,928 ARGs, as shown in the Venn diagram (Figure 4B).
Figure 4.
Hub genes associated with endometriosis, including immune-related genes (A) and autophagy-related genes (B). WGCNA, weighted gene co-expression network analysis. DEGs, differentially expressed genes.
Identification of hub genes
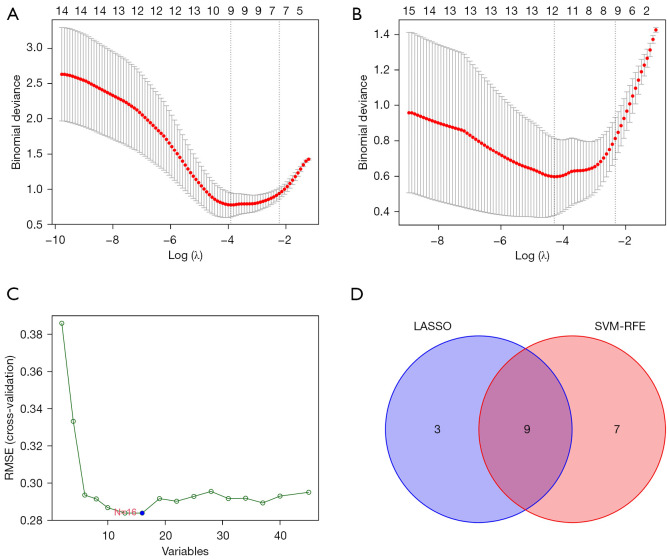
To construct the models, LASSO analysis was applied to obtain the hub genes in two groups of overlapped genes respectively. We obtained nine hub genes of IRGs (Figure 5A) and 12 hub genes of ARGs (Figure 5B). Next, 16 genes associated with endometriosis were obtained by SVM-RFE. Twelve hub genes of ARGs and 16 SVM-RFE genes were intersected again to obtain nine hub genes of ARGs (Figure 5C,5D). A total of 18 hub genes were finally included in the construction of the model.
Figure 5.
Identification of the hub genes of IRGs by LASSO (A); identification of the hub genes of ARGs by LASSO (B); SVM-RFE results of the differential genes in the GSE11691 and GSE120103 datasets (C); the intersection of the ARGs hub genes by LASSO and SVM-RFE shown in a Venn diagram (D). RMSE, root mean square error; LASSO, least absolute shrinkage and selection operator; SVM-RFE, support vector machine recursive feature elimination; IRGs, immune-related genes; ARGs, autophagy-related genes.
GO and KEGG analysis of hub genes
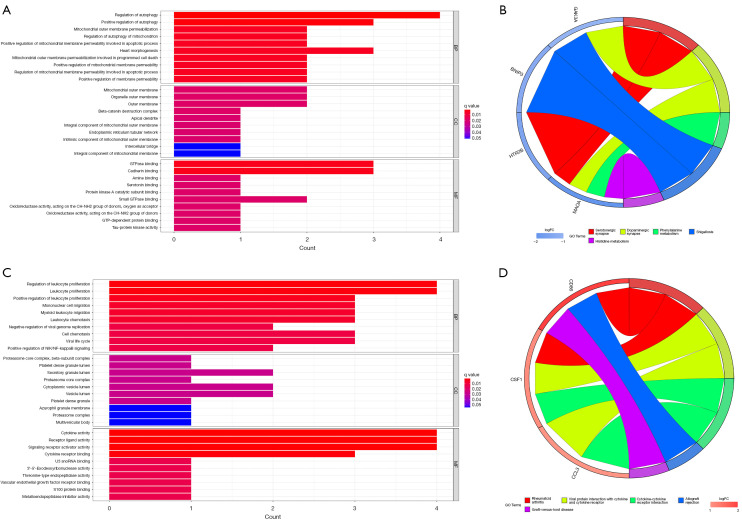
GO and KEGG functional enrichment analysis was performed on the ARGs-related hub genes to investigate the biological functions and pathways that these genes involved. A total of 452 GO entries (423 BP entries, eight CC entries, and 21 MF entries) and 12 KEGG pathways were enriched (Figure 6A,6B). On the other hand, the KEGG pathway analysis showed that these four genes were enriched in the serotonergic synapse, dopaminergic synapse, and phenylalanine metabolism.
Figure 6.
GO and KEGG enrichment of the hub genes. (A) GO results of the hub gene-related ARGs; (B) KEGG enrichment results of the hub gene-related ARGs; (C) GO results of the hub genes-related IRGs; (D) KEGG enrichment results of the hub gene-related IRGs. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; ARGs, autophagy-related genes; IRGs, immune-related genes; BP, biological process; CC, cellular component; MF, molecular function.
Enrichment analysis was also conducted for IRGs-related hub genes, and 358 GO entries (316 BP entries, seven CC entries, and 35 MF entries) and 11 KEGG pathways were enriched. These three genes were enriched in cytokine-cytokine receptor interaction and viral protein interaction with cytokines and cytokine receptors (Figure 6C,6D).
Module construction and validation
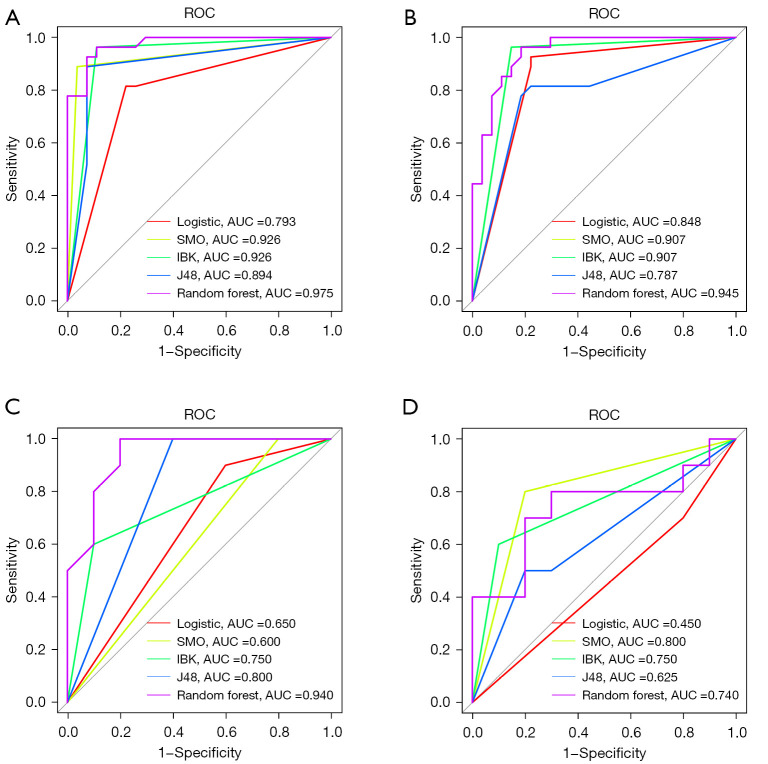
We utilized a variety of calculation methods to select the best result. In addition to the performance measures, a ROC curve was plotted and the AUC was calculated. All training cohort results were verified by the validation cohort (GSE7305). The performances of the classifiers are summarized in Tables 1-4. In terms of the ROC curve, SMO and IBK exhibited more or less similar results in the ARGs (0.926) and IRGs (0.907) training cohort. Random forest (0.975), J48 (0.894), and logistic regression (0.793) had a significantly lower discrimination ability in ARGS (Figure 7A). As for the IRGs results, random forest (0.945), logistic regression (0.848), and J48 (0.787) had a significantly lower identification ability in IRGs (Figure 7B).
Table 1. The performances of the classifiers (ARGs related training cohort).
| Algorithm | TP rate | FP rate | Precision | Recall | F-Measure | MCC | ROC area | PRC area |
|---|---|---|---|---|---|---|---|---|
| Random forest | 0.907 | 0.093 | 0.908 | 0.907 | 0.907 | 0.815 | 0.975 | 0.976 |
| J48 | 0.907 | 0.093 | 0.908 | 0.907 | 0.907 | 0.815 | 0.894 | 0.855 |
| IBK | 0.926 | 0.074 | 0.928 | 0.926 | 0.926 | 0.854 | 0.940 | 0.912 |
| SMO | 0.926 | 0.074 | 0.928 | 0.926 | 0.926 | 0.854 | 0.926 | 0.895 |
| Logistic regression | 0.778 | 0.222 | 0.779 | 0.778 | 0.777 | 0.557 | 0.849 | 0.815 |
ARGs, autophagy-related genes; TP, true positive; FP, false positive; MCC, Mathew correlation coefficient; ROC, receiver operating characteristic; PRC, precision recall curve; IBK, K-nearest neighbours; SMO, sequential minimal optimization.
Table 2. The performances of the classifiers (ARGs related validation cohort).
| Algorithm | TP rate | FP rate | Precision | Recall | F-Measure | MCC | ROC area | PRC area |
|---|---|---|---|---|---|---|---|---|
| Random forest | 0.650 | 0.350 | 0.794 | 0.650 | 0.601 | 0.420 | 0.940 | 0.939 |
| J48 | 0.550 | 0.450 | 0.763 | 0.550 | 0.436 | 0.229 | 0.810 | 0.753 |
| IBK | 0.800 | 0.200 | 0.857 | 0.800 | 0.792 | 0.655 | 0.800 | 0.757 |
| SMO | 0.600 | 0.400 | 0.778 | 0.600 | 0.524 | 0.333 | 0.600 | 0.578 |
| Logistic | 0.650 | 0.350 | 0.700 | 0.650 | 0.627 | 0.346 | 0.830 | 0.759 |
ARGs, autophagy-related genes; TP, true positive; FP, false positive; MCC, Mathew correlation coefficient; ROC, receiver operating characteristic; PRC, Precision Recall Curve; IBK, K-nearest neighbours; SMO, sequential minimal optimization.
Table 3. The performances of the classifiers (IRGs related training cohort).
| Algorithm | TP rate | FP rate | Precision | Recall | F-Measure | MCC | ROC area | PRC area |
|---|---|---|---|---|---|---|---|---|
| Random forest | 0.852 | 0.148 | 0.852 | 0.852 | 0.852 | 0.704 | 0.945 | 0.947 |
| J48 | 0.796 | 0.204 | 0.797 | 0.796 | 0.796 | 0.593 | 0.787 | 0.733 |
| IBK | 0.907 | 0.093 | 0.913 | 0.907 | 0.907 | 0.820 | 0.906 | 0.873 |
| SMO | 0.907 | 0.093 | 0.913 | 0.907 | 0.907 | 0.820 | 0.907 | 0.872 |
| Logistic | 0.852 | 0.148 | 0.860 | 0.852 | 0.851 | 0.712 | 0.853 | 0.810 |
IRGs, immune-related genes; TP, true positive; FP, false positive; MCC, Mathew correlation coefficient; ROC, receiver operating characteristic; PRC, precision recall curve; IBK, K-nearest neighbours; SMO, sequential minimal optimization.
Table 4. The performances of the classifiers (IRGs related validation cohort).
| Algorithm | TP rate | FP rate | Precision | Recall | F-Measure | MCC | ROC area | PRC area |
|---|---|---|---|---|---|---|---|---|
| Random forest | 0.750 | 0.250 | 0.753 | 0.750 | 0.749 | 0.503 | 0.740 | 0.759 |
| J48 | 0.650 | 0.350 | 0.665 | 0.650 | 0.642 | 0.314 | 0.625 | 0.589 |
| IBK | 0.750 | 0.250 | 0.775 | 0.750 | 0.744 | 0.524 | 0.750 | 0.694 |
| SMO | 0.800 | 0.200 | 0.800 | 0.800 | 0.800 | 0.600 | 0.800 | 0.740 |
| Logistic | 0.450 | 0.550 | 0.433 | 0.450 | 0.413 | −0.115 | 0.400 | 0.460 |
IRGs, immune-related genes; TP, true positive; FP, false positive; MCC, Mathew correlation coefficient; ROC, receiver operating characteristic; PRC, precision recall curve; IBK, K-nearest neighbours; SMO, sequential minimal optimization.
Figure 7.
ROC curve of the hub genes associated with endometriosis, including autophagy-related genes (A) and immune-related genes (B). (B) The yellow line show up because it’s a duplicate of the green and they both have the same result. ROC curve of testing set (GSE7305) genes, including autophagy-related genes (C) and immune-related genes (D). ROC, receiver operating characteristic; AUC, area under the ROC curve; SMO, sequential minimal optimization; lBK, K-nearest neighbours.
The validation cohort ROC curves of the five algorithms were as follows: random forest (0.940), J48 (0.800), IBK (0.750), logistic regression (0.650), and SMO (0.600) had the lowest verification result ability in the ARGs (Figure 7C). Similarly, SMO (0.800), IBK (0.750), random forest (0.740), J48 (0.625), and logistic regression (0.450) had the lowest verification result ability in the IRGs (Figure 7D). These results indicated overfitting in the application of the random-forest classifier.
Discussion
The occurrence of endometriosis is primarily attributed to the abnormal spread and growth of endometrioid tissue or the relocation and metaplasia of endometrial tissue outside the uterus, which is characterized by the functional response of uterine glands and stroma to stimulation by local, endogenous, and exogenous hormones. The clinical symptoms of endometriosis mainly include dysmenorrhea, pelvic pain, infertility, and sexual intercourse difficulty (27,28), and the lack of sensitive biomarkers causes delayed diagnosis. Therefore, there is a pressing need to identify accurate diagnostic molecular biomarkers. In this study, we investigated the diagnostic effects of IRGs and ARGs for endometriosis.
Endometriosis is a chronic inflammatory disease associated with local and systemic immune responses. In this study, immune cell infiltrations were further explored. The result showed significant differences in multiple immune cells between the endometriosis and the control groups. Plasma cells, macrophages M1 and macrophages M2 significantly increased in endometriosis. The change of peritoneal microenvironment in patients with endometriosis can promote the M2 polarization and M1 proinflammatory phenotype of macrophages, and the abnormal phenotype can lead to abnormal gene expression in ectopic endometrium, which is related to the pathophysiology of endometriosis and the reproductive outcome of patients (29). The activity of B cells is regulated by many factors. Macrophages can produce B lymphocyte stimulator and induce B cells to differentiate into plasma cells, which is necessary for the development of normal B cells. However, the level of B lymphocyte stimulator in ectopic lesions of endometriosis patients is significantly higher than that in normal endometriosis patients (30).
Endometriosis-associated model genes were identified through WGCNA, which could provide directions for exploring the pathogenesis of endometriosis to improve its clinical management. Through the endometriosis-associated key modules, we determined that genes in the yellow-, turquoise-, and green-colored modules exert different effects on endometriosis. We also obtained the DEGs in the training set, which further support our results.
Studies have indicated that immune-dysfunction and cellular autophagy are involved in the pathogenesis and progress of endometriosis (31,32), and these two processes are closely correlated (33). Components related to the innate immune response, such as conventional pattern recognition receptors (PRR) and inflammasomes, are integrated with autophagy. Abnormal cellular autophagy triggers inflammatory and immune responses, leading to autoimmune injury. However, the role of the immune response and autophagy in mediating endometriosis remains unclear. Thus, exploring the underlying mechanisms of the immune response and autophagy in endometriosis onset is a direction for the development of therapeutic agents for endometriosis as well as other similar diseases (19).
Nine hub genes were identified through our study, including BST2, CCL13, CD86, CSF1, FAM3C, GREM1, ISG20, PSMB8, and S100A11. These genes may be involved in the abnormal immune response of endometriosis. Moreover, nine hub genes were determined to be involved in autophagy in endometriosis, including GSK3A, HTR2B, RAB3GAP1, ARFIP2, BNIP3, CSF1, MAOA, PPP1R13L, and SH3GLB2. These findings could provide a reference for early identification and drug development in endometriosis.
The activated inflammatory cascade induces the proliferation, adhesion, and migration of endometrial cells, thereby forming the basis of ectopic implantation (34). Augmented macrophage function is also observed in endometriotic lesions (35). Bacci et al. found that macrophages are pro-inflammatory and facilitate endometriotic lesion growth and vascularization (36). The function of the IRGs identified in this study is mostly related to macrophages. Chemokines serve as vital molecules regulating the recruitment and migration of specific leukocytes, with which ISG20 expression is associated. CCL13 belongs to the CC chemokine family and is involved in CCR2 and CCR3, the migration of monocytes/macrophages, T-lymphocytes, and eosinophils via its functional ligands (37). CD86, a pro-inflammatory indicator for peritoneal macrophages, plays a crucial role in anti-endometriosis (38). CSF-1 is released by endometriotic macrophages and is involved in the formation of early endometriotic lesions through the enhancement of cell proliferation as well as via the attachment and invasion of endometrial cells (39). S100A11 is highly expressed and correlated to tumor promotion and progression in most cancers; however, its role in endometriosis is unclear. BST2 is essential in the antiviral immune response and is capable of activating the Nuclear Factor Kappa-Β (NF-kB) signaling pathway and subsequently the innate immune responses to infection (40). NF-kβ is important in the pathogenesis of endometriosis and is directly associated with endometrial cell invasion (41). PSMB8 refers to an immunoproteasome, and its inhibition induces cell apoptosis and migration/invasion blocking via the Phosphatidylinositol 3-kinase (PI3K)/Akt pathway in glioblastoma (42).
FAM3C is involved in Transforming growth factor β (TGF-β) -induced epithelial-mesenchymal transition (EMT); the knockdown of FAM3C inhibits TGF-β-induced EMT (43). GRHL2 belongs to the grainy head-like transcription family and plays an essential role in the inhibition of TGF-β mediated activation of Smad2/3 (44). TGF-β-induced EMT plays a key role in endometriosis. Autophagy is involved in numerous physiological processes, such as the elimination of misfolded or aggregated proteins, starvation, anti-aging, cell growth, and innate immunity (45). The ARGs identified in this study participate in the occurrence of diseases through multiple pathways. GSK3, as a member of the Wnt/β-catenin pathway, is involved in a wide range of cellular processes, such as apoptosis, mitosis, and proliferation. The Wnt/β-Catenin pathway is closely related to endometriosis (46). HTR2B is related to neovascularization and immunomodulation as well as neurotransmission (47), indicating that it could be involved in the pathogenesis of endometriosis. MAOA is a putative endometrial receptivity biomarker, which is also linked to endometriosis (48). PPP1R13L is a member of the apoptosis-stimulating proteins of the p53 (ASPP) family and was identified as a novel RelA-associated inhibitor (RAI) that binds to the NF-κB subunit p65/RelA and inhibits its transcriptional activity (49). SH3GLB2 is a TGF-β-induced anti-apoptotic factor (TIAF1), which is responsive to TGF-β1-induced polymerization (50). CSF1 is a co-gene of two pathways, which can be focused on in future endometriosis research.
In this study, we utilized Weka machine learning software to develop prediction models for the evaluation of hub genes and applied a variety of calculation methods to select the best result. These algorithms have an overall satisfactory accuracy and identification ability, outperforming SMO, IBK, J48, and logistic regression.
Our study identified several multi-molecule biomarkers for the diagnosis and treatment of endometriosis. However, some limitations should be noted. Firstly, there was a lack of relevant clinical information, such as disease course and medication history. Secondly, the potential immune- and autophagy-related biomarkers identified in our analysis require further validation by more studies. Lastly, the related genes were collected from an electronic database; despite its continuous updates, there are more genes to be discovered.
Conclusions
We identified nine hub genes that are closely associated with the immune response in endometriosis. At the same time, nine hub genes of autophagy-related genes in endometriosis were also identified. Our model validation results were good. Thus, these genes could be potential biomarkers for disease diagnosis and therapeutic monitoring.
Supplementary
The article’s supplementary files as
Acknowledgments
We would like to thank the researchers and study participants for their contributions.
Funding: This work was supported by the National Natural Science Foundation of China (No. 82260949), the Scientific Research and Innovation Fund Project of Gansu University of Traditional Chinese Medicine (No. 2022KCZD-5), and the Gansu Provincial University Innovation Fund Project (No. 2021A-088).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5979/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5979/coif). The authors have no conflicts of interest to declare.
(English Language Editor: A. Kassem)
References
- 1.Giudice LC. Clinical practice. Endometriosis. N Engl J Med 2010;362:2389-98. 10.1056/NEJMcp1000274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macer ML, Taylor HS. Endometriosis and infertility: a review of the pathogenesis and treatment of endometriosis-associated infertility. Obstet Gynecol Clin North Am 2012;39:535-49. 10.1016/j.ogc.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bulun SE. Endometriosis. N Engl J Med 2009;360:268-79. 10.1056/NEJMra0804690 [DOI] [PubMed] [Google Scholar]
- 4.Zondervan KT, Becker CM, Koga K, et al. Endometriosis. Nat Rev Dis Primers 2018;4:9 . 10.1038/s41572-018-0008-5 [DOI] [PubMed] [Google Scholar]
- 5.Lebovic DI, Mueller MD, Taylor RN. Immunobiology of endometriosis. Fertil Steril 2001;75:1-10. 10.1016/s0015-0282(00)01630-7 [DOI] [PubMed] [Google Scholar]
- 6.Witz CA, Schenken RS. Pathogenesis. Semin Reprod Endocrinol 1997;15:199-208. 10.1055/s-2008-1068749 [DOI] [PubMed] [Google Scholar]
- 7.Simoens S, Dunselman G, Dirksen C, et al. The burden of endometriosis: costs and quality of life of women with endometriosis and treated in referral centres. Hum Reprod 2012;27:1292-9. 10.1093/humrep/des073 [DOI] [PubMed] [Google Scholar]
- 8.Wykes CB, Clark TJ, Khan KS. Accuracy of laparoscopy in the diagnosis of endometriosis: a systematic quantitative review. BJOG 2004;111:1204-12. 10.1111/j.1471-0528.2004.00433.x [DOI] [PubMed] [Google Scholar]
- 9.Nisolle M, Paindaveine B, Bourdon A, et al. Histologic study of peritoneal endometriosis in infertile women. Fertil Steril 1990;53:984-8. 10.1016/s0015-0282(16)53571-7 [DOI] [PubMed] [Google Scholar]
- 10.Fauconnier A, Fritel X, Chapron C. Endometriosis and pelvic pain: epidemiological evidence of the relationship and implications. Gynecol Obstet Fertil 2009;37:57-69. 10.1016/j.gyobfe.2008.08.016 [DOI] [PubMed] [Google Scholar]
- 11.Greene R, Stratton P, Cleary SD, et al. Diagnostic experience among 4,334 women reporting surgically diagnosed endometriosis. Fertil Steril 2009;91:32-9. 10.1016/j.fertnstert.2007.11.020 [DOI] [PubMed] [Google Scholar]
- 12.Cătană CS, Atanasov AG, Berindan-Neagoe I. Natural products with anti-aging potential: Affected targets and molecular mechanisms. Biotechnol Adv 2018;36:1649-56. 10.1016/j.biotechadv.2018.03.012 [DOI] [PubMed] [Google Scholar]
- 13.Yin H, Wu H, Chen Y, et al. The Therapeutic and Pathogenic Role of Autophagy in Autoimmune Diseases. Front Immunol 2018;9:1512 . 10.3389/fimmu.2018.01512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren Y, Mu L, Ding X, et al. Decreased expression of Beclin 1 in eutopic endometrium of women with adenomyosis. Arch Gynecol Obstet 2010;282:401-6. 10.1007/s00404-009-1280-0 [DOI] [PubMed] [Google Scholar]
- 15.Díaz M, García C, Sebastiani G, et al. Placental and Cord Blood Methylation of Genes Involved in Energy Homeostasis: Association With Fetal Growth and Neonatal Body Composition. Diabetes 2017;66:779-84. 10.2337/db16-0776 [DOI] [PubMed] [Google Scholar]
- 16.Wildenberg ME, Koelink PJ, Diederen K, et al. The ATG16L1 risk allele associated with Crohn's disease results in a Rac1-dependent defect in dendritic cell migration that is corrected by thiopurines. Mucosal Immunol 2017;10:352-60. 10.1038/mi.2016.65 [DOI] [PubMed] [Google Scholar]
- 17.Ritchie ME, Silver J, Oshlack A, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics 2007;23:2700-7. 10.1093/bioinformatics/btm412 [DOI] [PubMed] [Google Scholar]
- 18.Suryawanshi S, Huang X, Elishaev E, et al. Complement pathway is frequently altered in endometriosis and endometriosis-associated ovarian cancer. Clin Cancer Res 2014;20:6163-74. 10.1158/1078-0432.CCR-14-1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang HL, Mei J, Chang KK, et al. Autophagy in endometriosis. Am J Transl Res 2017;9:4707-25. [PMC free article] [PubMed] [Google Scholar]
- 20.Yu JJ, Sun HT, Zhang ZF, et al. IL15 promotes growth and invasion of endometrial stromal cells and inhibits killing activity of NK cells in endometriosis. Reproduction 2016;152:151-60. 10.1530/REP-16-0089 [DOI] [PubMed] [Google Scholar]
- 21.Hung SW, Zhang R, Tan Z, et al. Pharmaceuticals targeting signaling pathways of endometriosis as potential new medical treatment: A review. Med Res Rev 2021;41:2489-564. 10.1002/med.21802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47 . 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559 . 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen KA, Chalpe A, Eyster KM. Management of endometriosis-associated pain. Clin Obstet Gynecol 2010;53:439-48. 10.1097/GRF.0b013e3181dbda06 [DOI] [PubMed] [Google Scholar]
- 25.Huang ML, Hung YH, Lee WM, et al. SVM-RFE based feature selection and Taguchi parameters optimization for multiclass SVM classifier. ScientificWorldJournal 2014;2014:795624 . 10.1155/2014/795624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peiris AN, Chaljub E, Medlock D. Endometriosis. JAMA 2018;320:2608 . 10.1001/jama.2018.17953 [DOI] [PubMed] [Google Scholar]
- 28.Zondervan KT, Becker CM, Missmer SA. Endometriosis. N Engl J Med 2020;382:1244-56. 10.1056/NEJMra1810764 [DOI] [PubMed] [Google Scholar]
- 29.Vallvé-Juanico J, Santamaria X, Vo KC, et al. Macrophages display proinflammatory phenotypes in the eutopic endometrium of women with endometriosis with relevance to an infectious etiology of the disease. Fertil Steril 2019;112:1118-28. 10.1016/j.fertnstert.2019.08.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore PA, Belvedere O, Orr A, et al. BLyS: member of the tumor necrosis factor family and B lymphocyte stimulator. Science 1999;285:260-3. 10.1126/science.285.5425.260 [DOI] [PubMed] [Google Scholar]
- 31.Ren XU, Wang Y, Xu G, et al. Effect of rapamycin on endometriosis in mice. Exp Ther Med 2016;12:101-6. 10.3892/etm.2016.3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen HH, Zhang T, Yang HL, et al. Ovarian hormones-autophagy-immunity axis in menstruation and endometriosis. Theranostics 2021;11:3512-26. 10.7150/thno.55241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deretic V. Autophagy in inflammation, infection, and immunometabolism. Immunity 2021;54:437-53. 10.1016/j.immuni.2021.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Famularo G, Procopio A, Giacomelli R, et al. Soluble interleukin-2 receptor, interleukin-2 and interleukin-4 in sera and supernatants from patients with progressive systemic sclerosis. Clin Exp Immunol 1990;81:368-72. 10.1111/j.1365-2249.1990.tb05340.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capobianco A, Rovere-Querini P. Endometriosis, a disease of the macrophage. Front Immunol 2013;4:9 . 10.3389/fimmu.2013.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bacci M, Capobianco A, Monno A, et al. Macrophages are alternatively activated in patients with endometriosis and required for growth and vascularization of lesions in a mouse model of disease. Am J Pathol 2009;175:547-56. 10.2353/ajpath.2009.081011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Zepeda EA, Combadiere C, Rothenberg ME, et al. Human monocyte chemoattractant protein (MCP)-4 is a novel CC chemokine with activities on monocytes, eosinophils, and basophils induced in allergic and nonallergic inflammation that signals through the CC chemokine receptors (CCR)-2 and -3. J Immunol 1996;157:5613-26. [PubMed] [Google Scholar]
- 38.Krasnyi AM, Sadekova AA, Smolnova TY, et al. The Levels of Ghrelin, Glucagon, Visfatin and Glp-1 Are Decreased in the Peritoneal Fluid of Women with Endometriosis along with the Increased Expression of the CD10 Protease by the Macrophages. Int J Mol Sci 2022;23:10361 . 10.3390/ijms231810361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Budrys NM, Nair HB, Liu YG, et al. Increased expression of macrophage colony-stimulating factor and its receptor in patients with endometriosis. Fertil Steril 2012;97:1129-35.e1. 10.1016/j.fertnstert.2012.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tokarev A, Suarez M, Kwan W, et al. Stimulation of NF-κB activity by the HIV restriction factor BST2. J Virol 2013;87:2046-57. 10.1128/JVI.02272-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nasiri N, Babaei S, Moini A, et al. Controlling Semi-Invasive Activity of Human Endometrial Stromal Cells by Inhibiting NF-kB Signaling Pathway Using Aloe-emodin and Aspirin. J Reprod Infertil 2021;22:227-40. 10.18502/jri.v22i4.7648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang BY, Song JW, Sun HZ, et al. PSMB8 regulates glioma cell migration, proliferation, and apoptosis through modulating ERK1/2 and PI3K/AKT signaling pathways. Biomed Pharmacother 2018;100:205-12. 10.1016/j.biopha.2018.01.170 [DOI] [PubMed] [Google Scholar]
- 43.Waerner T, Alacakaptan M, Tamir I, et al. ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell 2006;10:227-39. 10.1016/j.ccr.2006.07.020 [DOI] [PubMed] [Google Scholar]
- 44.Cieply B, Riley P, 4th, Pifer PM, et al. Suppression of the epithelial-mesenchymal transition by Grainyhead-like-2. Cancer Res 2012;72:2440-53. 10.1158/0008-5472.CAN-11-4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siracusa R, D'Amico R, Impellizzeri D, et al. Autophagy and Mitophagy Promotion in a Rat Model of Endometriosis. Int J Mol Sci 2021;22:5074 . 10.3390/ijms22105074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat Rev 2018;62:50-60. 10.1016/j.ctrv.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi WG, Choi W, Oh TJ, et al. Inhibiting serotonin signaling through HTR2B in visceral adipose tissue improves obesity-related insulin resistance. J Clin Invest 2021;131:e145331 . 10.1172/JCI145331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vargas E, García-Moreno E, Aghajanova L, et al. The mid-secretory endometrial transcriptomic landscape in endometriosis: a meta-analysis. Hum Reprod Open 2022;2022:hoac016 . 10.1093/hropen/hoac016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang JP, Hori M, Sanda T, et al. Identification of a novel inhibitor of nuclear factor-kappaB, RelA-associated inhibitor. J Biol Chem 1999;274:15662-70. 10.1074/jbc.274.22.15662 [DOI] [PubMed] [Google Scholar]
- 50.Lee MH, Lin SR, Chang JY, et al. TGF-β induces TIAF1 self-aggregation via type II receptor-independent signaling that leads to generation of amyloid β plaques in Alzheimer's disease. Cell Death Dis 2010;1:e110 . 10.1038/cddis.2010.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as