Abstract
Cytotoxic necrotizing factor type 1 (CNF1) is a 115-kDa toxin that activates Rho GTPases and is produced by uropathogenic Escherichia coli (UPEC). While both epidemiological studies that link CNF1 production by E. coli with urinary tract disease and the cytopathic effects of CNF1 on cultured urinary tract cells are suggestive of a role for the toxin as a UPEC virulence factor, few in vivo studies to test this possibility have been reported. Therefore, in this investigation, we evaluated the importance of CNF1 in a murine model of urinary tract infection (UTI) by comparing the degree of colonization and damage induced by three different CNF1-producing E. coli strains with isogenic CNF1-deficient derivatives. The data from single-strain challenge experiments with C3H/HeOuJ mice indicated a trend toward higher counts of the wild-type strains in the urine and bladders of these animals up to 3 days after challenge in two of three strain pairs. Furthermore, this difference was statistically significant at day 2 of infection with one strain pair, C189 and C189cnf1. To control for the animal-to-animal variability inherent in this model, we infected C3H/HeOuJ mice with a mixture of CNF1-positive and -negative isogenic derivatives of CP9. The CNF1-positive strain was recovered in higher numbers than the CNF1-negative strain in the urine, bladders, and kidneys of the mice up to 9 days postinfection. These striking coinfection findings, taken with the trends observed in single-strain infections, led us to conclude that CNF1-negative strains were generally attenuated compared to the wild type in the C3H/HeOuJ mouse model of UTI. Furthermore, histopathological examination of bladder specimens from mice infected with CNF1-positive strains consistently showed deeper, more extensive inflammation than in those infected with the isogenic mutants. Lastly, we found that CNF1-positive strain CP9 was better able to resist killing by fresh human neutrophils than were CP9cnf1 bacteria. From these data in aggregate, we propose that CNF1 production increases the capacity of UPEC strains to resist killing by neutrophils, which in turn permits these bacteria to gain access to deeper tissue and persist better in the lower urinary tract.
Acute infections of the bladder and kidneys in otherwise healthy individuals are among the most common types of bacterial infections in humans (20, 44). Indeed, there are an estimated 7 million episodes of acute cystitis and 250,000 cases of pyelonephritis in the United States every year (21, 39). This high incidence of disease disproportionately affects women, about 40% of whom experience cystitis in their lifetimes. Treatment of cystitis in women has been estimated to cost one billion dollars annually (44).
Escherichia coli is the most frequently isolated bacterial cause of uncomplicated urinary tract infection (UTI). Uropathogenic E. coli (UPEC) produces a number of virulence-associated factors that include P fimbriae, hemolysin, aerobactin, and cytotoxic necrotizing factor type 1 (CNF1) (29). CNF1 is a chromosomally encoded UPEC toxin that catalyzes the deamidation of the small GTPases RhoA, Rac, and Cdc42 (12, 14, 30, 42). Deamidation by CNF1 renders these GTPases constitutively active, which in most cells leads to the formation of actin stress fibers, lamellipodia, and filopodia. HEp-2 cells, which have been used as the prototypic cells for evaluation of CNF1 toxicity, not only display actin stress fibers but also become multinucleated (6, 10, 40). Additionally, CNF1 has been reported to (i) induce phagocytosis in epithelial cells (9, 11), (ii) reduce CR3-dependent phagocytosis in monocytes (5), (iii) increase the permeability of Caco-2 intestinal cell monolayers (14), (iv) efface the brush border of and decrease the transmigration of polymorphonuclear leukocytes (PMN) across a T84 monolayer (18), (v) inhibit wound repair in T24 bladder cells and Hs 738 fibroblast cells (24), and (vi) kill 5637 bladder cells through an apoptotic mechanism (34). Thus, CNF1 affects a variety of cellular functions in vitro, presumably through activation of the Rho GTPases. However, the role of this toxin in the pathogenesis of UTI remains to be delineated.
Several groups have reported an epidemiological link between the presence of the cnf1+ genotype or the production of CNF1 and E. coli strains that cause extraintestinal disease. Indeed, Yamamoto et al. showed that 61% of UTI isolates and 38% of bacteremia isolates harbored the cnf1+ gene, as opposed to only 10% of commensal fecal isolates (45). Furthermore, Mitsumori et al. reported that 64% of prostatitis isolates and 36% of pyelonephritis isolates were cnf1+ (35) and Andreu and colleagues reported that the percentages of cnf1+ prostatitis, pyelonephritis, and cystitis isolates were 63, 48, and 44%, respectively (3). Caprioli et al. found that 40% of UTI isolates tested positive for CNF1 expression, while only 0.9% of fecal isolates produced CNF1 (6). The common finding of cnf1+ in E. coli strains that cause extraintestinal infections is also consistent with the strong correlation between the presence of cnf1+ and the gene for hemolysin, hly, in these isolates. In fact, Yamamoto et al. reported that approximately 76% of isolates of E. coli from extraintestinal sites that were hly+ were also cnf1+, while 98% of cnf1+ strains were also hly+ (45).
In this study, we used a mouse model of ascending UTI to evaluate the role that CNF1 may play in the pathogenesis of UTI. We found that in two of three CNF1-positive UPEC strains tested, the bacterial counts in the bladders and urine of mice early in infection were higher than in those of animals inoculated with the respective isogenic cnf1 deletion mutant. In a few single-strain studies and in all mixed-inoculum studies, this difference was statistically significant. We also showed that CNF1 may alter the host response to infection, therefore allowing the bacteria to penetrate deeper into the muscular wall of the bladder and cause more severe and prolonged infection.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The media used were Luria Bertani (LB) broth, LB agar, and MacConkey agar (Difco, Becton-Dickinson, Sparks, Md.). When appropriate, ampicillin was used at 100 μg/ml of medium. Chloramphenicol was used at 30 μg/ml where necessary. Bacteria were routinely grown at 37°C.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain (serotype if relevant) or plasmid | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80 dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK−mK+) deoR thi-1 phoA supE44 λ−gyrA96 relA1 | Life Technologies, Inc., Gaithersburg, Md. |
| J96 (O4:H5:K6) | hly+cnf1+ | Pyelonephritis; 23 |
| CP9 (O4:H5:K54) | hly+cnf1+ | Blood; 41 |
| C85 (O2:H−) | hly+cnf1+ | Cystitis; 45 |
| C189 (O2:H−) | cnf1+ | Cystitis; 45 |
| J96cnf1 | hly+cnf1 | This study |
| CP9cnf1 | hly+cnf1 | This study |
| C85cnf1 | hly+cnf1 | This study |
| C189cnf1 | cnf1 | This study |
| CP9lacZ | hly+cnf1+lacZ | This study |
| Plasmids | ||
| pBluescript II SK(−) | Ampr | Stratagene, La Jolla, Calif. |
| pHLK102 | cnf1+ from J96 cloned in pBluescript II SK(−), Ampr | This study |
| pHLK116 | 4.4-kb cnf1+EcoRI-StuI fragment from 2CO2 cloned in pBC KS(−) | |
| pHLK120 | pHLK102 with internal BclI fragment of cnf1+ deleted; cnf1 Ampr | This study |
| pHLK125 | 2.0-kb BssHII fragment from pHLK120 cloned in pAM450; cnf1 | |
| 2CO2 | cnf1+ cosmid clone derived from J96 | 13, 43 |
| pSX34lacZα | Chlr; derived from pSC101 | S.-Y. Xu, New England Biolabs, Inc. |
| pHLK140 | cnf1+ from 2CO2 cloned in pSX34lacZα; Chlr | This study |
| pAM450 | Derived from pMAK705; sacB/R Ampr | 31 |
Cloning of cnf1+.
Commercially synthesized primers (Life Technologies, Inc., Gaithersburg, Md.) were designed to amplify cnf1+ from E. coli strain J96 (23) by PCR. The sequences of the primers were as follows: forward, 5′-TATTAATCTTCACAGAGGAG-3′; reverse, 5′-GGCCAATAAATAATTTCCCGAATC-3′ (8). Amplification reaction mixtures contained 1× PCR buffer (Perkin-Elmer, Foster City, Calif.), 0.2 mM each deoxynucleoside triphosphate (Perkin-Elmer), 1.0 μM each primer, 1.5 mM MgCl2, 10 to 20 ng of template DNA, and 2.5 U of Taq polymerase (Perkin-Elmer). Reaction mixtures were incubated in a thermocycler for 30 consecutive cycles of 1 min at 95°C, 1 min at 50°C, and 3 min at 72°C with a final extension at 72°C for 10 min. The PCR amplification product was purified (Wizard DNA Purification Kit; Promega Corp., Madison, Wis.) from low-melting-point agarose (SeaPlaque; BioWhittaker Molecular Applications, Rockland, Maine) and cloned in the SmaI site of pBluescript II SK(−) (Stratagene Cloning Systems, La Jolla, Calif.). The cnf1+ plasmid clone pHLK102 (Table 1) expressed CNF1 biological activity in the HEp-2 multinucleation assay and contained the restriction enzyme recognition sites predicted from the published cnf1+ DNA sequence (8).
A 4.4-kb cnf1+ EcoRI-StuI fragment from the cosmid 2CO2 (obtained from D. Berg [13, 43]) was cloned into pBC KS(−) (Stratagene Cloning Systems) to generate pHLK116. The cnf1+ fragment was reisolated following digestion of pHLK116 with SacI and SalI and subcloned into pSX34lacZα (obtained from S.-Y. Xu, New England Biolabs, Beverly, Mass.) to produce pHLK140 (Table 1).
Construction of cnf1 isogenic mutants.
An in-frame deletion mutation (pHLK120) was constructed by the removal of an internal 1.2-kb BclI cnf1+ fragment from pHLK102. The mutation (Δcnf-120) produced a truncated CNF1 protein that was detectable with CNF1-specific polyclonal antiserum (34), but strains carrying the Δcnf-120 mutation expressed no detectable CNF1 biological activity. A 2.0-kb BssHII fragment from pHLK120 that contained the Δcnf-120 mutation was ligated into the BssHII site in the suicide vector pAM450 (31) to yield pHLK125, which was subsequently used as previously described (16, 31) to introduce the Δcnf-120 mutation into different UPEC strains. For allelic exchange, bacteria were transformed with pHLK125 under selection for ampicillin resistance (Ampr) at 30°C. Transformants then underwent selection for Ampr at 42°C to isolate cointegrates that were subsequently plated at 30°C on LB agar without added NaCl and containing 5% sucrose to allow resolution of the plasmid. The resulting isogenic mutants were screened for loss of plasmid markers and loss of CNF1 activity. The replacement of the wild-type cnf1 gene with the Δcnf-120 allele was confirmed by PCR, Southern blot, and Western blot analyses. E. coli strains that carried the Δcnf-120 mutation were complemented in vitro with the wild-type cnf1 gene in pHLK102 or pHLK140 (Table 1). Growth curves of the wild-type and mutant strains were done in minimal-salts liquid containing glucose and in L broth over a 24-h period. The growth rates of the strain pairs appeared indistinguishable. Moreover, the plating efficiencies of the mutants on L agar and MacConkey agar were indistinguishable from those of the parental strains. All strains were tested for retention of the O antigen, capsular antigen (where present), type 1 pili, P fimbriae, hemolysin, and lipopolysaccharide by standard procedures.
Isolation of CP9lacZ.
Standard methods (32) were used to isolate and characterize a Lac− derivative of E. coli strain CP9, designated CP9lacZ (Table 1). Phenotypic and complementation analyses of CP9lacZ localized the mutation to lacZ, and the mutant was otherwise indistinguishable from strain CP9 in vitro or in vivo.
HEp-2 multinucleation assays.
Ninety-six-well tissue culture plates were seeded with 4 × 103 HEp-2 cells per well and incubated for 4 h at 37°C with 5% CO2. Overnight bacterial cultures were harvested by centrifugation, concentrated by resuspension in phosphate-buffered saline plus gentamicin (100 μg/ml), and disrupted by sonication. The resultant lysates were clarified by centrifugation at 4°C and sterilized by filtration. Serial dilutions of the lysates were applied to the HEp-2-seeded wells, and the microtiter plates were further incubated for 72 h before the wells were fixed and stained with Leukostat (Fisher Scientific, Pittsburgh, Pa.). Multinucleation of HEp-2 cells was assessed by microscopy.
Mouse UTI model.
Animal experiments were carried out in accordance with the principles outlined in the Guide for the Care and Use of Laboratory Animals (38). The mouse ascending UTI model was performed as previously described (15, 28, 36, 37). All of the E. coli strains evaluated in this model were first passed through mice as described below to ensure that the strains were not attenuated as a result of routine laboratory culture. The bacteria were isolated from infected kidneys at 2 to 5 days postinfection and stored at −70°C. The strains did not lose any virulence factors or CNF1 expression due to animal passage. To prepare inocula for mice, mouse-passaged bacteria were harvested from an LB agar plate after overnight incubation and resuspended in sterile, nonpyrogenic saline to an A600 of 1.12. Six milliliters of the bacterial suspension was pelleted and resuspended in 0.5 ml of saline to give a bacterial density of 109 CFU/ml. This suspension was adjusted by dilution so that 1 × 107 to 1 × 108 CFU were contained in 10 or 25 μl. The inoculum was pipetted into an adapter tube (6-in. Male/Female line; Surgimedics, The Woodlands, Tex.) connected to a 30-gauge, 0.5-in. sterile needle. The pipette tip was left in the tubing, and an electronic EDP pipettor (Rainin Instrument Co., Woburn, Mass.) was attached to the tip.
Four- to six-week-old C3H/HeOuJ and BALB/c female mice were obtained from Jackson Laboratory (Bar Harbor, Maine). C3H/HeOuJ mice were chosen for these experiments because they are easily infected by UPEC strains, and while they are congenic to C3H/HeJ mice, they do not express the lipopolysaccharide-hyporesponsive defect found in the latter strain. Mice were anesthetized with methoxyflurane (Metofane; Schering-Plough Animal Health Corp., Union, N.J.) or isoflurane (Forane; Anaquest, Madison, Wis.) and catheterized with a sterile 1-in. piece of tubing (inside diameter, 0.28 mm; outside diameter, 0.61 mm; Intramedic; Becton-Dickinson). Urine expressed from the bladder was collected on a sterile swab and streaked onto an LB plate to check for pre-existing bacteriuria. The needle connected to the inoculating apparatus was inserted into the catheter, and the Rainin EDP was used to deliver 10 or 25 μl over a 30-s period into the bladder of the anesthetized animal. After the inoculum was instilled, the catheter was removed from the mouse and the mouse was placed in a cage to recover from the anesthesia. Mice were given food and water ad libitum and were monitored over a period of 1 to 9 days for any signs of distress.
Mice were euthanatized with an overdose of methoxyflurane or isoflurane, and urine, bladders, and kidneys were collected aseptically. Urine was serially diluted in saline and plated on LB agar. Each organ was bisected; one half was homogenized for bacterial counts, and the other half was used to prepare histology sections. Tissue homogenates were diluted and plated on LB agar to quantitate bacterial CFU. In some cases, the remainder of the homogenate was inoculated into LB broth to recover bacteria present at numbers below the limit of detection by direct plating (approximately 10 CFU per specimen). Isolates from positive broth cultures were tested to confirm their identity. Specimens that were negative upon direct plating but which yielded positive broth cultures were assumed to contain 1 CFU. Isolates recovered from the mice were tested in the HEp-2 multinucleation assay for expression of functional CNF1 and were found in all cases to retain the phenotype of the strain that was inoculated.
Histopathology.
Organs were soaked in 10% phosphate-buffered formalin for a minimum of 4 h, embedded in paraffin, and cut into 5-μm sections. The slides were stained with hematoxylin and eosin and examined for depth of inflammation. Bladder sections were also stained with Brown-Brenn stain for bacteria. Sections were coded and viewed in a blinded fashion under ×600 magnification, and the area of greatest inflammation was located. Neutrophils and band forms were counted in that area. Severity of inflammation was scored according to the following scale: 0 to 25 neutrophils present, mild inflammation; 26 to 50 neutrophils present, moderate inflammation; greater than 50 neutrophils present, severe inflammation. No neutrophils were observed in saline-inoculated control animal sections.
Neutrophil killing of bacteria in vitro
Overnight cultures of bacteria were washed and suspended in Hanks' balanced salt solution containing gelatin (1.0 mg/ml). Fresh human neutrophils were isolated from whole venous blood and suspended in Hanks' balanced salt solution–1.0 mg of gelatin per ml (4). Bacteria and neutrophils were mixed at ratios of two to five bacteria per neutrophil and incubated at 37°C with gentle tumbling. After 15 and 45 min of incubation, samples were plated on LB agar. The percentage of viable bacteria after incubation with neutrophils was calculated according to the following formula: % viable bacteria = (number of bacterial CFU in the mixtures at 15 or 45 min/number of bacterial CFU added to the mixtures at the start of the assay) × 100.
Statistical analysis.
The geometric mean number of CFU of bacteria from the urine, bladders, or kidneys of a group of mice infected with the wild-type strain was compared to the geometric mean number of CFU in mice infected with the mutant strain by using unpaired Student t tests. When the geometric mean numbers of CFU of a group of strains were compared, as in Fig. 1, an analysis of variance was used to evaluate the variance, followed by unpaired Student t tests to compare the means of two strains. The numbers of CFU from mice coinfected with two strains were compared by Student t tests of paired log10 numbers. A Student t test of paired samples was also used to examine the data obtained from the neutrophil-killing experiments.
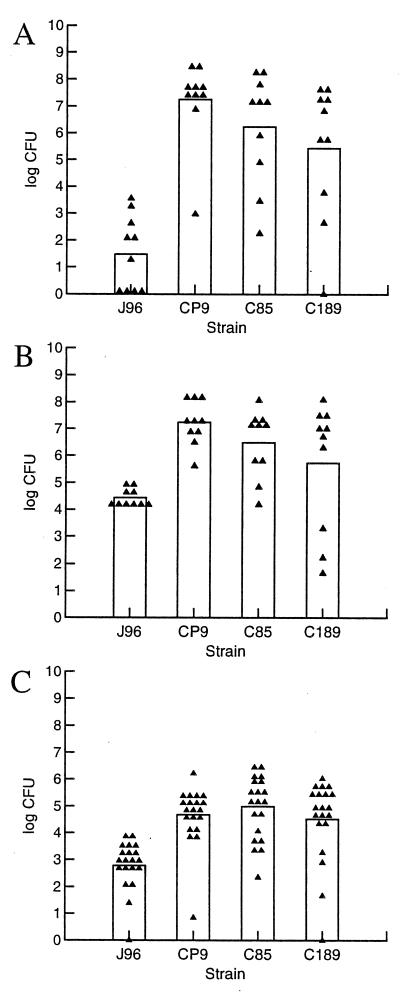
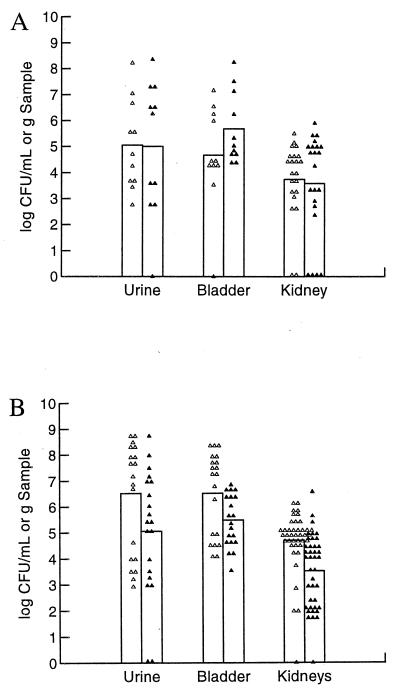
FIG. 1.
Comparison of different CNF1-positive UPEC strains in the C3H/HeOuJ mouse model of ascending UTI. Ten female C3H/HeOuJ mice per bacterial strain were inoculated with 1.0 × 108 CFU and sacrificed 3 days later. Urine, bladders, and kidneys were collected and homogenized, and serial dilutions were plated for colony counts. (A) Bacterial counts in urine. (B) Bacterial counts in bladders. (C) Bacterial counts in kidneys. Each point represents one sample. The height of the bar represents the mean for those samples. In all cases except C189 in the bladder, the counts for J96 were significantly lower than those of CP9, C85, and C189 (analysis of variance, P < 0.0001, followed by Student's t test, P < 0.05).
RESULTS
Rationale for selection of E. coli strains for mouse UTI studies.
Experiments were initially undertaken in the mouse UTI model with the well-characterized O4:H5:K6 UPEC strain J96. However, we concluded, on the basis of a set of experiments in which we varied a number of parameters to optimize the model, that the colonization levels of J96 were invariably too low to allow us to discern a difference between the wild-type and mutant J96cnf1 strains. Therefore, we sought to find CNF1-expressing UPEC strains that were more virulent (i.e., colonized the bladder and/or kidneys at higher levels) than J96 in the mouse model. For that purpose, we compared J96 with three independent CNF1-producing clinical isolates, CP9, C85, and C189 (Table 1) (41, 45). Colonization levels in the urine, bladder, and kidneys were significantly higher than those of J96 for all three strains (P < 0.05) except for C189 in the bladder (Fig. 1). Therefore, we proceeded to prepare isogenic cnf1 mutants of these strains (designated CP9cnf1, C85cnf1, and C189cnf1) by allelic exchange as we had done for J96. We then selected one strain pair, CP9 and CP9cnf1, to use in a series of studies in the mouse UTI model to assess the impact on relative bacterial counts of the time when samples were harvested after infection (kinetics of infection), the strain of mouse used as the host animal, and the challenge dose. When we evaluated the importance of these variables in the pairwise comparison, we also, on occasion, tested other CNF1-positive and CNF1-negative pairs.
Kinetics of infection of C3H/HeOuJ mice with CP9 and CP9cnf1 in the UTI model.
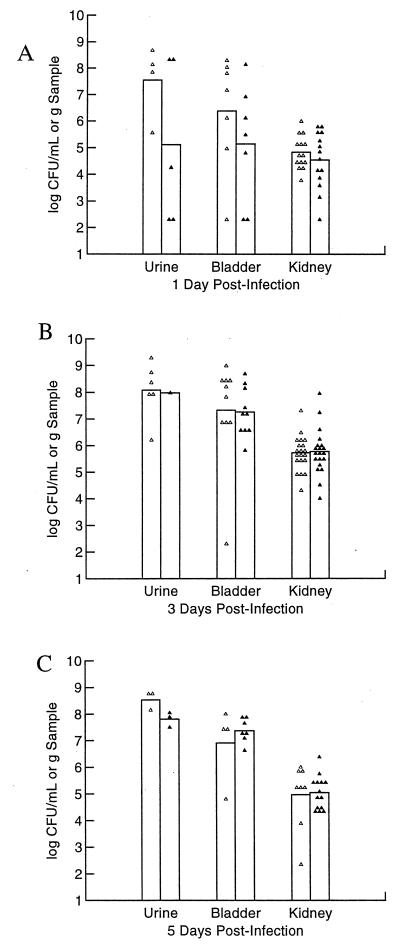
The goal of our first set of experiments with CP9 and CP9cnf1 was to define the optimal time for harvesting of infected urine, bladders, and kidneys for comparative enumeration. For this purpose, 30 mice per strain were inoculated with 1.9 × 107 CFU and 10 mice per strain were euthanatized at 1, 3, and 5 days postchallenge. At day 1 after inoculation, urine and bladder samples collected from mice infected with CP9 contained higher numbers of bacteria than did samples collected from CP9cnf1-infected mice (Fig. 2A). However, at day 3 postinfection, bacterial counts in the urine, bladders, and kidneys of mice infected with either strain were equivalent, although the absolute numbers for each type of sample were higher for both strains than on day 1 of infection (Fig. 2B). By 5 days after infection, mice challenged with CP9 again had somewhat higher bacterial numbers in their urine than did mice given CP9cnf1 but the bacterial numbers in the bladder slightly favored the mutant while equal numbers of bacteria of both strains were present in the kidney samples (Fig. 2C). Although none of the observed differences were statistically significant, the trend toward higher numbers of CNF1-positive bacteria in the urine and bladder on the first day following inoculation led us to focus on colonization in the first days of infection.
FIG. 2.
Kinetics of UTI of C3H/HeOuJ mice with CP9 and the cnf1 isogenic mutant CP9cnf1. Thirty female C3H/HeOuJ mice were inoculated transurethrally with 1.9 × 107 CFU of either CP9 or CP9cnf1 bacteria and sacrificed 1, 3, and 5 days later. Urine, bladder, and kidney samples were recovered, homogenized, and plated for colony counts. Urine samples were not obtained from every mouse, and mice that did not become infected were not included in the analysis. (A) Bacterial counts 1 day after infection. (B) Bacterial counts 3 days after infection. (C) Bacterial counts 5 days after infection. Each point represents one sample. The height of the bar represents the mean for those samples. Symbols: ▵, CP9; ▴, CP9cnf1.
Comparison of CP9 and CP9cnf1 early in infection of C3H/HeOuJ or BALB/c mice in the UTI model and influence of challenge dose on infection of C3H/HeOuJ mice.
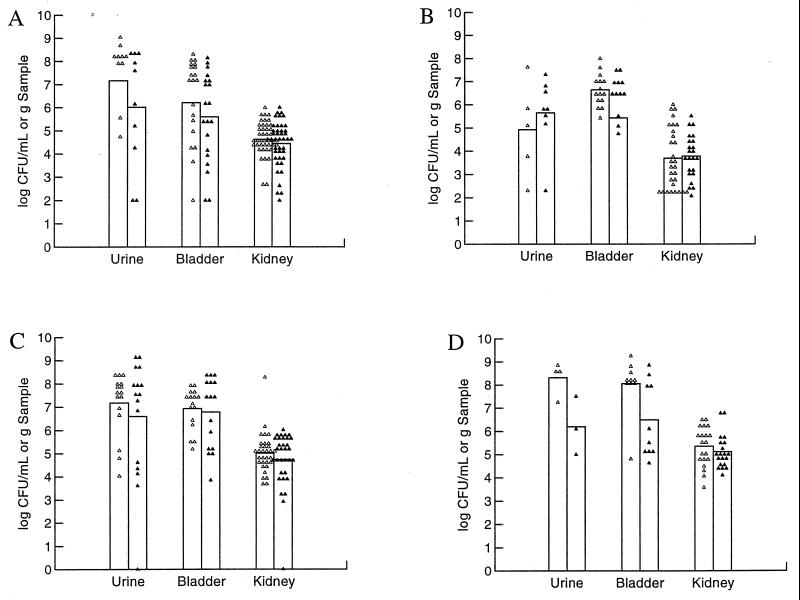
For the next series of kinetic studies, we again compared our prototypic strains, CP9 and CP9cnf1, but focused only on days 1 to 3 of infection. We also compared the influence of the mouse strain on the outcome at day 1 postchallenge. As with the previous kinetic experiments, 1 day after infection, C3H/HeOuJ mice challenged with strain CP9 had higher bacterial numbers in their urine and bladders than did mice challenged with the mutant (Fig. 3A). When identical experiments were done with BALB/c mice (Fig. 3B), the bladder samples had slightly higher numbers of wild-type CP9 bacteria while slightly higher numbers of CNF1-negative bacteria were present in the urine and kidney samples. Two days after infection, urine samples of C3H/HeOuJ mice infected with CP9 contained slightly more bacteria than did samples from CP9cnf1-infected mice (Fig. 3C). Bladders and kidneys were infected equally with the wild-type and mutant strains. BALB/c mice were not tested at this time point. From these data, we concluded that day 1 after challenge of C3H/HeOuJ permitted the best discrimination between the CNF1-postive CP9 strain and its isogenic mutant and that BALB/c mice did not display this difference across all sample types. Therefore, we elected to use C3H/HeOuJ mice in the remaining studies.
FIG. 3.
Comparison of CP9 and the cnf1 isogenic mutant CP9cnf1 at 1 to 3 days after inoculation of C3H/HeOuJ or BALB/c mice. (A) Twenty female C3H/HeOuJ mice were inoculated with 1.9 × 107 CFU and sacrificed 1 day later. Organ samples were collected, homogenized, serially diluted, and plated on LB agar. Symbols: ▵, CP9; ▴, CP9cnf1. (B) Fifteen female BALB/c mice were infected with 1.9 × 107 CFU and sacrificed for colony counts 1 day later. Symbols: ▵, CP9; ▴, CP9cnf1. (C) Fifteen female C3H/HeOuJ mice were infected with 1.9 × 107 CFU and sacrificed 2 days later. Symbols: ▵, CP9; ▴, CP9cnf1. (D) Ten female C3H/HeOuJ mice were infected with 1 × 108 CFU and euthanatized 3 days later. Counts of CP9 bacteria were significantly higher than those of CP9cnf1 in the urine and bladders (Student's t test, P < 0.05). Symbols: ▵, CP9; ▴, CP9cnf1. Each point represents one sample. The height of the bar represents the mean for those samples. Urine samples were not obtained from every mouse, and mice that did not become infected were not included in the analysis.
We also tested the effects of inoculum size in the early kinetic studies. In all previous studies, we used ≈2 × 107 bacteria as our challenge inoculum. In these experiments, we asked whether, if we raised the dose of the inoculum, we could discern a difference between the counts of CP9 and its isogenic mutant later in infection than day 1. We found that when we elevated the challenge inoculum to 108 bacteria, the answer was yes, at least for the urine and bladder samples (Fig. 3D). Thus, in this higher challenge dose experiment, significantly more CP9 than CP9cnf1 bacteria were present in the urine and bladder samples (P < 0.05) at day 3 of infection. The kidneys were colonized equally by both strains.
Evaluation of additional CNF1-postive and CNF1-negative isogenic pairs of UPEC strains early in the infection of C3H/HeOuJ mice in the UTI model.
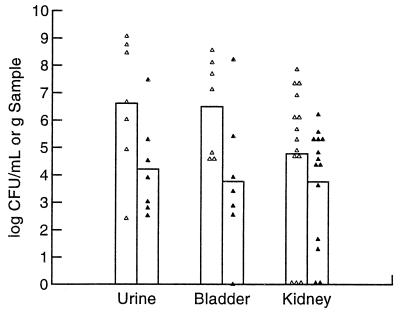
To determine whether our findings obtained with the CP9 and CP9cnf1 pair could be reproduced with other such pairs, we did comparative analyses of the numbers of C85 and C189 bacteria with those of their respective isogenic mutants at days 1 and 2 postinoculation. We found that the importance of CNF1 to the colonization levels of C85 and C189 differed. At 1 day postinoculation, C3H/HeOuJ mice infected with 1.9 × 107 C85 or C85cnf1 bacteria had equivalent numbers of CFU in the kidneys whereas the numbers of wild-type bacteria in the urine and bladders were slightly lower than those of mutant bacteria (data not shown). In contrast, when mice that were inoculated with 1.0 × 108 CFU of strains C189 and C189cnf1 were examined at 2 days postchallenge (day 1 was not tested), statistically significantly higher numbers of wild-type bacteria were found in the urine and bladders (Fig. 4). Colonization of the kidneys showed a trend toward higher numbers of C189 than C189cnf1 bacteria, but this trend was not statistically significant.
FIG. 4.
Single-strain infection comparing C189 with C189cnf1 2 days after inoculation. Seven female C3H/HeOuJ mice were inoculated with 1.0 × 108 CFU and sacrificed 2 days after challenge. Points represent individual samples, and the height of each bar represents the geometric mean of those samples. Counts of C189 were significantly higher than those of C189cnf1 in the urine and bladders (Student's t test, P < 0.05). Symbols: ▵, C189, ▴, C189 cnf1.
Although significantly higher levels of C189 than C189cnf1 bacteria were observed at 2 days postinoculation in the urine and bladders of infected mice and experiments done over a 3-day period with strain CP9 and its isogenic mutant showed a trend toward greater colonization of the urine and bladder by the wild-type strain, there was significant variability from experiment to experiment. We speculated that much of this variability was a consequence of the precision with which the inoculum was introduced intravesically into the animal and/or differences in the individual animal's response to infection. To control for experimental variation among animals, we next tested CP9 against CP9cnf1 in a mixed-infection model.
Mixed-strain infections.
The mixed-infection model allows direct comparison of two strains in a single animal and eliminates the variation between animals that is inherent in the single-infection model system. Therefore, a Lac-negative derivative of CP9 was generated by UV irradiation for use in such an experiment. This CNF1-producing strain, designated CP9lacZ, could be differentiated from Lac-positive CP9cnf1 on MacConkey agar. To confirm that generation of the Lac− phenotype did not alter the virulence of CP9lacZ, that strain was tested against the parental strain in a mixed infection. The two strains were mixed in a 1:1 ratio and used to inoculate C3H/HeOuJ mice intravesically (Fig. 5A). Urine and kidney samples from mice euthanatized 2 days postinoculation contained equal numbers of each strain. Slightly higher numbers of wild-type CP9 bacteria were found in the bladders, but the difference in the numbers of each strain was not significant. Based on these data, it was determined that CP9lacZ could infect mice as well as CP9. CP9lacZ was then tested against CP9cnf1 in the mixed-infection model. The strains were mixed in equal numbers and used at an inoculum of 108 CFU per strain, a dose similar to that previously shown to discriminate well between CP9 and CP9cnf1 in single-dose experiments at 3 days postinfection (Fig. 3C). This mixture was used to infect C3H/HeOuJ mice (Fig. 5B). Two days postinfection, CNF1-positive CP9lacZ had grown to significantly higher numbers than CP9cnf1 in the urine, bladders, and kidneys of the mice tested (P < 0.05; Fig. 5B). Next, a time course study was done with the same mixture of CP9lacZ and CP9cnf1 as in the previous experiment. Mice were infected intravesically with the mixture and sacrificed 6 h, 2 days, 4 days, 7 days, and 9 days postinoculation (Fig. 6). In the urine, the numbers of CFU of both strains increased between 6 h and 2 days. Between 2 and 9 days, the number of CFU of each strain decreased but the decrease in the number of CP9cnf1 bacteria was more rapid than the decrease in the number of CP9lacZ bacteria (Fig. 6A). In the bladder, the number of CFU of CP9lacZ bacteria was stable throughout the experiment while the number of CP9cnf1 CFU decreased (Fig. 6B). CP9cnf1 and CP9lacZ numbers decreased in the kidneys during the 9-day period, with the average number of CP9lacZ CFU decreasing by 1.5 log units (Fig. 6C). In contrast, CP9cnf1 was nearly cleared from the kidneys after 9 days. The average numbers of CFU of the cnf1 isogenic mutant decreased by 5.8 log units (Fig. 6C), and in 100% (six of six) of the infected kidneys, bacteria were only detectable after enrichment culture. Results of the mixed-infection time course study indicate that the wild-type strain consistently colonized the mice at higher levels than did the cnf1 isogenic mutant strain. In addition, there was more rapid clearing of the mutant strain from the urinary tracts of the mice.
FIG. 5.
Mixed infections of C3H/HeOuJ mice. Two strains were mixed in equal numbers and inoculated into female mice. Mice received 2.0 × 108 CFU and were sacrificed 2 days after infection. (A) Mixed infections comparing CP9 (CNF1+ Lac+) with CP9lacZ (CNF1+ Lac−). Eleven female C3H/HeOuJ mice were inoculated. Symbols: ▵, CP9; ▴, CP9lacZ. (B) Mixed infections comparing CP9lacZ with CP9cnf1. Twenty female C3H/HeOuJ mice were inoculated. Counts of CP9lacZ were significantly higher than those of CP9cnf1 in the urine, bladders, and kidneys (Student's t test, P < 0.05). Symbols: ▵, CP9lacZ, ▴, CP9cnf1. Points represent bacterial numbers in individual samples, and bars represent the means of those samples.
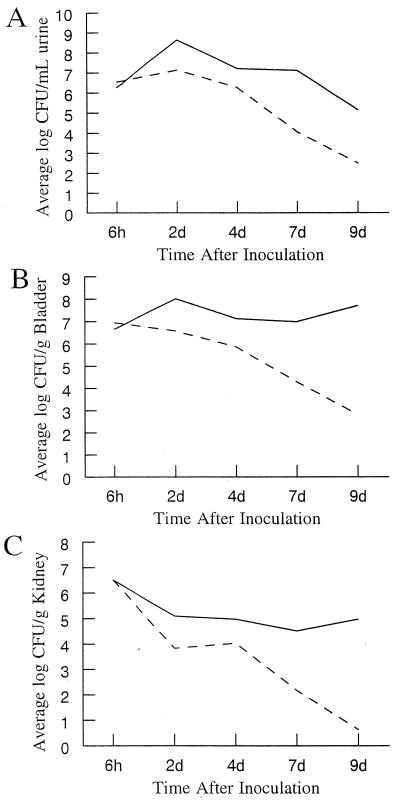
FIG. 6.
CNF1 contributes to bacterial survival in a mixed infection. Time courses of mixed infections with CP9lacZ and CP9cnf1 are shown. The bacterial strains were mixed in equal numbers, and C3H/HeOuJ mice received 2.0 × 108 CFU. Mice were sacrificed 6 h, 2 days, 4 days, 7 days, and 9 days after infection, and colony counts were performed on collected samples. (A) Mean bacterial numbers in the urine of infected mice. (B) Mean bacterial numbers in the bladders of infected mice. (C) Mean bacterial numbers in the kidneys of infected mice. ——, CP9lacZ; –––, CP9cnf1. Each point represents the mean number of CFU per sample for three mice.
Complementation of CP9cnf1 with a cloned cnf1+ gene.
The results obtained with the mixed-infection model indicated higher colonization of the urinary tract by the wild-type strain. To confirm our assumption that the defect of CP9cnf1 was, in fact, a consequence of the absence of CNF1 expression, we attempted in vivo complementation studies. The cnf1+ gene was cloned from cosmid 2CO2 into pSX34lacZα, a low-copy-number vector derived from pSC101. The resulting construct, designated pHLK140, was transformed into CP9cnf1 and expressed CNF1 at a level that was five times higher than wild-type expression in CP9. However, we found that pHLK140 was not stable in vitro, either in the presence or in the absence of antibiotic selection, perhaps due to the toxicity of overexpressed CNF1. When tested in the mouse model, the vector alone was stable in the absence of antibiotic selection. Similar to our findings with in vitro passage of transformants harboring pHLK140, this plasmid was not completely stable in vivo in the absence of selection. Loss of the plasmid was detected in the urine and kidney samples but not in the bladder samples. In mixed complementation infection experiments with CP9lacZ(pSX34lacZα) (Lac negative, CNF1 positive, vector only) and CP9cnf1(pHLK140) (Lac positive, CNF1 positive), there were no differences in the numbers of bacterial CFU recovered from the bladders. pHLK140 was retained by 83% of CP9cnf1 in the urine and 73% of CP9cnf1 in the kidneys, and half as many of these bacteria were recovered from either site as were CNF1-positive CP9lacZ(pSX34lacZα) bacteria. Thus, functional complementation appears to have occurred in the bladders, indicating some selective pressure for retention of the plasmid in the bladders, but not in the urine or kidneys, of the mice. We cannot preclude the possibility that overexpression of CNF1 in the kidneys impaired the survival of the mutant at that site. These results, taken together with the results of the single- and mixed-infection experiments, indicate that production of CNF1 allows better infection of the bladder by UPEC.
Histological analysis of bladder samples and light microscopic examination of urine from infected mice.
Single and mixed infections, as well as complementation experiments, showed that in the mouse model of UTI, CNF1 conferred an advantage on CP9 in the bladder. Therefore, we focused our histological evaluations on formalin-fixed bladder tissue from mice infected with that strain or CP9cnf1. Microscopic examination of infected bladder sections stained with hematoxylin and eosin revealed that both wild-type and mutant strains elicited an acute inflammatory response from the host that consisted of an influx of neutrophils and edema. The location of the influx of neutrophils ranged from the epithelium only in some bladders through the muscularis in others. The percentage of bladders infected with CP9 that exhibited moderate-to-severe inflammation (Table 2) was higher than in the CP9cnf1-infected bladders (100% versus 20%). These comparative data from isogenic strains that differ only in CNF1 production suggest that inflammation of the submucosa and muscularis may be more severe in the bladders of animals infected with CNF1-positive bacteria. In support of this premise, bacteria were visible (by Brown-Brenn staining) within the areas of inflammation (data not shown).
TABLE 2.
Severity of neutrophil influx in mouse bladders 1 day after infection with CNF1-positive CP9 or its cnf1 isogenic mutant
| Straina | No. of CFU/gb | Avg no. of neutrophils (range)c | Severity of inflammationd |
|---|---|---|---|
| CP9 (6) | 104–107 | 95 (39–181) | 100 |
| CP9 cnf1 (5) | 105–107 | 39 (7–129) | 20 |
Numbers in parentheses are the numbers of mice tested.
Range of bacterial counts present in the bladders examined.
Neutrophils were counted within the tissue in the area of greatest inflammation, and PMN in blood vessels were excluded (magnification, ×600). Slides were coded and examined in a blinded fashion by a pathologist. All slides from CP9-infected animals were reported to contain many more areas of inflammation than the slides from animals infected with the CNF1-negative mutant. Inflammation was scored as described in Materials and Methods.
Percentage of animals with moderate to severe inflammation.
Killing of bacteria by neutrophils.
The finding of increased inflammation within the bladder tissue of wild-type-infected mice compared to mutant-infected animals prompted us to examine the interaction of neutrophils with the bacteria. We examined the ability of freshly isolated human neutrophils to kill CP9 and CP9cnf1 after incubation of the bacteria and neutrophils together for 15 and 45 min. Despite day-to-day variation in neutrophil viability and bactericidal activity, isolated neutrophils were less effective in killing wild-type CP9 than in killing CP9cnf1 (data not shown). In seven of eight samples, the viability of CP9 after 15 or 45 min of exposure to neutrophils was significantly greater than that of CP9cnf1 (P < 0.05). Lysates prepared from the CP9-neutrophil mixture after incubation expressed CNF1, as demonstrated by the multinucleation of HEp-2 cells. This activity was not present in the CP9cnf1-neutrophil incubation mixture. These data indicated that human neutrophils kill CNF1-negative UPEC more effectively than they kill isogenic CNF1-positive UPEC.
DISCUSSION
In this investigation, we sought to test the hypothesis that CNF1 contributes to the virulence of UPEC through the use of a mouse model of ascending UTI. Prior to this report, the evidence in favor of such a proposal was primarily based on epidemiological findings (3, 6, 35, 45) and, more indirectly, the cytopathic effects of CNF1 on a human bladder epithelial cell line, 5637 (34). The evidence against such a theory was recently presented by Johnson and colleagues (26). Those investigators used the same general approach that we employed in our investigation: a comparison of the net growth and histological damage induced by a CNF1-positive UPEC strain to those of its isogenic cnf1 mutant in a mouse model of ascending UTI. Although we do not dispute the conclusions of those researchers that CNF1 did not contribute to the virulence of the UPEC strain that they tested, our results, for the reasons described below, led us to the opposite conclusion. Indeed, we generated four lines of evidence to support our original tenet. First, in single-challenge inoculation experiments, we showed that up to 3 days after single-strain intravesical challenge, two of the three CNF1-producing strains tested achieved greater numbers in the urine and bladders of mice than did their respective cnf1 isogenic mutants. On days 1, 2, and 3 after challenge, the average numbers of CFU of CP9 were higher than the average numbers of CFU of CP9cnf1 in the urine or bladders. Strain C85 was only tested at day 1, and it was present in slightly lower numbers in the urine and bladders than its cnf1 isogenic mutant at that point of infection. C189 and its isogenic mutant were tested only on day 2 of infection, and at that time, the wild-type strain was present in higher numbers in the urine and bladders than C189cnf1. Second, in mixed-inoculation studies, CP9 consistently outgrew CP9cnf1 in the urine and tissues of the urinary tract. Third, CNF1-positive strains CP9 and C85 induced a greater degree of inflammation and appeared to traverse deeper into the bladder tissue at day 1 of infection than did CP9cnf1 and C85cnf1, respectively, even though the latter strain was present at slightly higher numbers than its wild-type parent. Fourth, CP9 survived better in the presence of human neutrophils than did CP9cnf1.
That the conclusions of this study and the report of Johnson et al. (13, 26) are contradictory may be explained, in part, on the basis of differences in certain experimental variables used by the two groups. The parameters that are known to alter the severity and time course of single-strain challenge studies in the mouse UTI model include the following. First, the type of human isolate influences the infection, i.e., strains from a patient with cystitis or pyelonephritis (reference 27 and Fig. 1 of this study). Indeed, Johnson and colleagues previously reported that cystitis isolates colonize the mouse bladder and urine better than do pyelonephritis isolates whereas pyelonephritis isolates colonize the kidneys at higher levels. The strain used by Johnson et al. for their analysis of the contribution of CNF1 to the urovirulence of E. coli in the mouse model was F11, an isolate from the urine of a patient with symptoms of cystitis (26). In our experiments, we primarily examined CP9, a strain isolated from the blood of a patient (41). We also tested C85 and C189, both of which are cystitis isolates. Second, the strain of mouse used in the model may also affect the pattern of infection, as demonstrated by Hopkins et al. (22). Those investigators examined the time course and host response of various mouse strains to a single E. coli strain and concluded that mice of various strains differ in their initial susceptibility to infection and in the ability to resolve an infection. In our studies, infection of BALB/c mice with CP9 and CP9cnf1 resulted in higher numbers of the mutant strain in the urine and kidneys of the mice (but not the bladder) at 1 day of infection. Conversely, when C3H/HeOuJ mice were infected with the same dose and the same strains, there were higher numbers of the wild-type strain than the mutant in the urine and bladders of the mice on that first day of infection. In addition, the overall numbers of bacteria in the urine and tissues of C3H/HeOuJ mice were higher than the numbers in the comparable specimens from BALB/c mice. These differences could indicate that for the BALB/c infections, the time of sampling was not optimal for observation of an effect of CNF1 on the infection process. In contrast, the data could also indicate that CNF1 does not play a role in the infection of BALB/c mice. Since Johnson et al. used CBA/J/Hsd mice (26), perhaps the absence of a demonstrable effect of CNF1 on the urovirulence of F11 could reflect their choice of mouse strains for these experiments. Third, the time in infection at which the comparison between a CNF1-positive and a CNF1-negative strain was made had an impact in our studies of mice that were inoculated with single bacterial strains (earlier was generally better at a comparable dose). We focused our single-strain infection experiments on days 1 to 3; Johnson et al. focused most of their experiments on a 7-day infection period. Fourth, the challenge dose might have an impact on the outcome of a differential analysis between the wild type and an isogenic cnf1 mutant. We generally used an inoculum of 2 × 107 CFU per mouse in single-strain studies, while Johnson and colleagues used a dose of 2 × 109 CFU. The higher dose might obscure the type of trends that we observed between CNF1-postive and CNF1-negative isogenic mutants in single-strain inoculation experiments. Finally, the spectrum of additional virulence determinants expressed by a particular UPEC strain (e.g., hemolysin, cytolethal distending toxin) might alter or mask the influence that CNF1 has on the pathogenesis of infection. In fact, in our study, the only strain pair in which a statistically significant difference in colony counts (rather than a trend) was observed between the wild-type and mutant strains in single-strain infection studies was with the hemolysin-negative isolate C189.
Taken together, the general trends of our single-strain challenge studies with C3H/HeOuJ mice suggested that CNF1 contributed to UTI early in the infection process in the mouse model in two of the three strains tested. However, the variation in bacterial numbers in each animal test group made it difficult to draw definitive conclusions supported by statistically significant differences in the geometric mean numbers of CFU. This wide range of counts within experimental groups has been previously demonstrated (26, 37) and may reflect differences between individual animals' responses to infection. Some individual animals may clear the initial infection faster than others because of their immune system efficiency or the speed with which the animals urinate after infection. Regardless of the reason for the variability, when such variation among animals was minimal or experimentally controlled for by utilizing mixed infections (see below), there were statistically significant differences between the numbers of CFU of strains CP9 and CP9cnf1 recovered from the urine.
A mixed-infection model was utilized to control for the variation in severity of infection from animal to animal. To our knowledge, this is the first published study comparing a CNF1-positive strain to an isogenic CNF1-negative mutant in a mixed-infection model. We generated a Lac-negative mutant of strain CP9 (CP9lacZ) to differentiate the parent from the isogenic cnf1 mutant. When equal numbers of CP9lacZ and CP9cnf1 bacteria were inoculated into mice, CP9lacZ was recovered in significantly higher numbers from the urinary tract than CP9cnf1. The capacity of CP9lacZ to outgrow CP9cnf1 in the urinary tract was not due to a difference between the growth rates of the bacteria, as demonstrated in vitro. Rather, CNF1 appeared to confer an advantage on the bacterium in the urinary tract. In fact, time course studies of mixed infections showed that the CNF1-positive strain was able to persist in the bladder and kidneys whereas CP9cnf1 was cleared more rapidly from the urinary tract. In a previous report, Miller and Creaghe described mixed-infection studies with two unrelated E. coli strains isolated from a patient with pyelonephritis (33). When they inoculated a mixed culture directly into the kidney, they found stable cocolonization of the kidney, but when inoculation was by a retrograde infection, a serogroup O8 strain was found to infect the kidneys to the exclusion of a serogroup O75 strain. The investigators concluded that bacterial interference played some part in infection of the kidney by a single strain. The influence of virulence factors was not discussed in that report, but another explanation for these results may be that the O8 strain possessed virulence determinants that gave that strain an advantage over the other strain. Our experimental design differs from that of Miller and Creaghe in that we used isogenic strains isolated from blood that varied only in CNF1 production and lactose utilization. We concluded that the capacity of CP9lacZ to colonize the urinary tract in higher numbers than CP9cnf1 is due to the presence of CNF1.
We examined the basis for the apparent advantage that production of CNF1 conferred on UPEC strains in this model. Histological analysis of bladder samples taken from mice 1 day after infection indicated that CNF1-positive bacteria caused severe inflammation more often than the isogenic mutants (Table 2). Functional complementation of the cnf1 mutant also was evident in the bladder, a result consistent with a role for CNF1 in UPEC pathogenesis at that site. One possible benefit of CNF1 production by bacteria in bladder tissues is that the toxin induces epithelial cells to take up the bacteria and promote their transcytosis across the mucosal barrier. In fact, Falzano et al. reported that CNF1 induced phagocyte-like activity in human epithelial cells, allowing the uptake of noninvasive bacteria (9). Alternatively, cell death caused by CNF1 (34) or tissue damage due to a more intense inflammatory response to CNF1-positive strains may allow the infecting strain to gain access to deeper tissues. Infiltration of PMN was found more often in the mucosa and submucosa of bladders infected with CNF1-positive strains. Abnormal multinucleation was not evident in any of the bladder sections examined, a finding consistent with a recent report on the differences in the effects of CNF1 on human bladder 5637 cells (cytotoxicity) and HEp-2 cells (multinucleation) (34).
That CNF1 production may increase the inflammatory response of the host is suggested by our observations and the work of others. Specifically, Elliott et al. reported that CNF1 evokes edema and necrosis and is associated with inflammation in the intestines of rabbits in a diarrhea model of infection (7). Additionally, Fournout et al. found that germfree piglets infected orally with a CNF1-positive E. coli strain developed pulmonary inflammation more frequently than those infected with an isogenic CNF1-negative mutant (13). In that study, CNF1-positive and CNF1-negative bacteria were both disseminated in the lungs of the infected pigs. Hofman et al. recently reported that treatment of isolated PMN with purified CNF1 for 16 h, followed by stimulation with zymosan, resulted in an increase in superoxide generation and adherence of PMN to T84 cells; however, the phagocytic function of CNF1-treated PMN was decreased (19). Our observation that human neutrophils are less effective at killing CNF1-positive bacteria than CNF1-negative bacteria may result from decreased phagocytosis of the bacteria by the neutrophils.
If CNF1 gives bacteria an advantage in persistence in the lower urinary tract, how does that happen in the face of the host responses to bacterial entry into the bladder? The defensive responses of the host include the washout flow of urine, shedding of the uroepithelium, and production of an acute inflammatory response, with the resultant production of host cytokines, such as interleukins 6 and 8, that increase the influx of polymorphonuclear lymphocytes into the area (1, 2, 17). Our results show that CNF1 production protects UPEC from killing by neutrophils. That result should be viewed in the context of the fact that CNF1-producing bacteria have been reported to evoke greater granulocyte colony-stimulating factor levels in patients than E. coli strains that do not produce CNF1 (25). Granulocyte colony-stimulating factor plays a role in maintaining the normal blood neutrophil count and determining the neutrophilic response to infection in the human host. Thus, in spite of an apparent capacity of CNF1-positive bacteria to recruit neutrophils to the site of infection, the toxin acts to protect the bacteria from neutrophilic attack.
ACKNOWLEDGMENTS
This research was supported by National Institutes of Health grant AI38281-05.
We thank Raghav Wusirika for help with the neutrophil experiments and Rebecca Gillespie and Beth Baker for assistance with the animal experiments. We are also grateful to Karen Meysick for critical evaluation of the manuscript and many helpful suggestions.
REFERENCES
- 1.Agace W, Connell H, Svanborg C. Host resistance to urinary tract infections. In: Mobley H L T, Warren J W, editors. Urinary tract infections: molecular pathogenesis and clinical management. Washington, D.C.: American Society for Microbiology; 1996. pp. 221–243. [Google Scholar]
- 2.Agace W, Hedges S, Ceske M, Svanborg C. IL-8 and the neutrophil response to mucosal gram negative infection. J Clin Investig. 1993;92:780–785. doi: 10.1172/JCI116650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreu A, Stapleton A E, Fennell C, Lockman H A, Xercavins M, Fernandez F, Stamm W E. Urovirulence determinants in Escherichia coli strains causing prostatitis. J Infect Dis. 1997;176:464–469. doi: 10.1086/514065. [DOI] [PubMed] [Google Scholar]
- 4.Campbell D E, Douglas S D. Phagocytic cell functions. I. Oxidation and chemotaxis. In: Rose N R, de Macario E C, Folds J D, Lane H C, Nakamura R M, editors. Manual of clinical laboratory immunology. 5th ed. Washington, D.C.: ASM Press; 1997. pp. 320–328. [Google Scholar]
- 5.Capo C, Meconi S, Sanguedolce M-V, Bardin N, Flatau G, Boquet P, Mege J-L. Effect of cytotoxic necrotizing factor-1 on actin cytoskeleton in human monocytes: role in the regulation of integrin-dependent phagocytosis. J Immunol. 1998;161:4301–4308. [PubMed] [Google Scholar]
- 6.Caprioli A, Falbo V, Ruggeri F M, Baldassarri L, Bisicchia R, Ippolito G, Romoli E, Donelli G. Cytotoxic necrotizing factor production by hemolytic strains of Escherichia coli causing extraintestinal infections. J Clin Microbiol. 1987;25:146–149. doi: 10.1128/jcm.25.1.146-149.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott S J, Srinivas S, Albert M J, Alam K, Robins-Browne R M, Gunzburg S T, Mee B J, Chang B J. Characterization of the roles of hemolysin and other toxins in enteropathy caused by alpha-hemolytic Escherichia coli linked to human diarrhea. Infect Immun. 1998;66:2040–2051. doi: 10.1128/iai.66.5.2040-2051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falbo V, Pace T, Picci L, Pizzi E, Caprioli A. Isolation and nucleotide sequence of the gene encoding cytotoxic necrotizing factor 1 of Escherichia coli. Infect Immun. 1993;61:4909–4914. doi: 10.1128/iai.61.11.4909-4914.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falzano L, Fiorentini C, Donelli G, Michel E, Kocks C, Cossart P, Cabanié L, Oswald E, Boquet P. Induction of phagocytic behavior in human epithelial cells by Escherichia coli cytotoxic necrotizing factor type 1. Mol Microbiol. 1993;9:1247–1254. doi: 10.1111/j.1365-2958.1993.tb01254.x. [DOI] [PubMed] [Google Scholar]
- 10.Fiorentini C, Arancia G, Caprioli A, Falbo V, Ruggeri F M, Donelli G. Cytoskeletal changes induced in HEp-2 cells by the cytotoxic necrotizing factor of Escherichia coli. Toxicon. 1988;26:1047–1056. doi: 10.1016/0041-0101(88)90203-6. [DOI] [PubMed] [Google Scholar]
- 11.Fiorentini C, Fabbri A, Matarrese P, Falzano L, Boquet P, Malorni W. Hinderance of apoptosis and phagocytic behaviour induced by Escherichia coli cytotoxic necrotizing factor 1: two related activities in epithelial cells. Biochem Biophys Res Commun. 1997;241:341–346. doi: 10.1006/bbrc.1997.7723. [DOI] [PubMed] [Google Scholar]
- 12.Flatau G, Lemichez E, Gauthier M, Chardin P, Paris S, Fiorentini C, Boquet P. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–733. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- 13.Fournout S, Dozois C M, Odin M, Desautels C, Pérès S, Hérault F, Daigle F, Segafredo S, Laffitte J, Oswald E, Fairbrother J M, Oswald I P. Lack of a role of cytotoxic necrotizing factor 1 toxin from Escherichia coli in bacterial pathogenicity and host cytokine response in infected germfree piglets. Infect Immun. 2000;68:839–847. doi: 10.1128/iai.68.2.839-847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerhard R, Schmidt G, Hofmann F, Aktories K. Activation of Rho GTPases by Escherichia coli cytotoxic necrotizing factor 1 increases intestinal permeability in Caco-2 cells. Infect Immun. 1998;66:5125–5131. doi: 10.1128/iai.66.11.5125-5131.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagberg L, Engberg I, Freter R, Lam J, Olling S, Svanborg Eden C. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect Immun. 1983;40:273–283. doi: 10.1128/iai.40.1.273-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedlund M, Svensson M, Nilsson A, Duan R-D, Svanborg C. Role of the ceramide-signaling pathway in cytokine responses to P-fimbriated Escherichia coli. J Exp Med. 1996;183:1037–1044. doi: 10.1084/jem.183.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofman P, Flatau G, Selva E, Gauthier M, Le Negrate G, Fiorentini C, Rossi B, Boquet P. Escherichia coli cytotoxic necrotizing factor 1 effaces microvilli and decreases transmigration of polymorphonuclear leukocytes in intestinal T84 epithelial cell monolayers. Infect Immun. 1998;66:2494–2500. doi: 10.1128/iai.66.6.2494-2500.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofman P, Le Negrate G, Mograbi B, Hofman V, Brest P, Alliana-Schmid A, Flatau G, Boquet P, Rossi B. Escherichia coli cytotoxic necrotizing factor-1 (CNF-1) increases the adherence to epithelia and the oxidative burst of human polymorphonuclear leukocytes but decreases bacterial phagocytosis. J Leukoc Biol. 2000;68:522–528. [PubMed] [Google Scholar]
- 20.Hooton T M, Scholes D, Hughes J P, Winter C, Roberts P L, Stapleton A E, Stergachis A, Stamm W E. A prospective study of risk factors for symptomatic urinary tract infection in young women. N Engl J Med. 1996;335:468–474. doi: 10.1056/NEJM199608153350703. [DOI] [PubMed] [Google Scholar]
- 21.Hooton T M, Stamm W E. Diagnosis and treatment of uncomplicated urinary tract infection. Infect Dis Clin N Am. 1997;11:551–582. doi: 10.1016/s0891-5520(05)70373-1. [DOI] [PubMed] [Google Scholar]
- 22.Hopkins W J, Gendron-Fitzpatrick A, Balish E, Uehling D T. Time course and host responses to Escherichia coli urinary tract infection in genetically distinct mouse strains. Infect Immun. 1998;66:2798–2802. doi: 10.1128/iai.66.6.2798-2802.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hull R A, Gill R E, Hsu P, Minshew B H, Falkow S. Construction and expression of recombinant plasmids encoding type 1 or d-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect Immun. 1981;33:933–938. doi: 10.1128/iai.33.3.933-938.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Island D M, Cui X, Warren J W. Effect of Escherichia coli cytotoxic necrotizing factor 1 on repair of human bladder cell monolayers in vitro. Infect Immun. 1999;67:3657–3661. doi: 10.1128/iai.67.7.3657-3661.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson S H, Lu Y, Brauner A. Soluble interleukin-6 receptor, interleukin-10 and granulocyte colony-stimulating factor in acute pyelonephritis: relationship to markers of bacterial virulence and renal function. Nephron. 1998;80:401–407. doi: 10.1159/000045211. [DOI] [PubMed] [Google Scholar]
- 26.Johnson D E, Drachenberg C, Lockatell C V, Island M D, Warren J W, Donnenberg M S. The role of cytotoxic necrotizing factor-1 in colonization and tissue injury in a murine model of urinary tract infection. FEMS Immunol Lett. 2000;28:37–41. doi: 10.1111/j.1574-695X.2000.tb01454.x. [DOI] [PubMed] [Google Scholar]
- 27.Johnson D E, Lockatell C V, Russell R G, Hebel J R, Island M D, Stapleton A, Stamm W E, Warren J W. Comparison of Escherichia coli strains recovered from human cystitis and pyelonephritis infections in transurethrally challenged mice. Infect Immun. 1998;66:3059–3065. doi: 10.1128/iai.66.7.3059-3065.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson D E, Russell R G, Lockatell C V, Zulty J C, Warren J W. Urethral obstruction of 6 hours or less causes bacteriuria, bacteremia, and pyelonephritis in mice challenged with “nonuropathogenic” Escherichia coli. Infect Immun. 1993;61:3422–3428. doi: 10.1128/iai.61.8.3422-3428.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson J R. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev. 1991;4:80–128. doi: 10.1128/cmr.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lerm M, Schmidt G, Goehring U-M, Schirmer J, Aktories K. Identification of the region of Rho involved in substrate recognition by Escherichia coli cytotoxic necrotizing factor 1 (CNF1) J Biol Chem. 1999;274:28999–29004. doi: 10.1074/jbc.274.41.28999. [DOI] [PubMed] [Google Scholar]
- 31.McKee M L, Melton-Celsa A R, Moxley R A, Francis D H, O'Brien A D. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect Immun. 1995;63:3739–3744. doi: 10.1128/iai.63.9.3739-3744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 33.Miller T E, Creaghe E. Bacterial interference as a factor in renal infection. J Lab Clin Med. 1976;87:792–803. [PubMed] [Google Scholar]
- 34.Mills M, Meysick K C, O'Brien A D. Cytotoxic necrotizing factor type 1 of uropathogenic Escherichia coli kills cultured human uroepithelial 5637 cells by an apoptotic mechanism. Infect Immun. 2000;68:5869–5880. doi: 10.1128/iai.68.10.5869-5880.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitsumori K, Terai A, Yamamoto S, Ishitoya S, Yoshida O. Virulence characteristics of Escherichia coli in acute bacterial prostatitis. J Infect Dis. 1999;180:1378–1381. doi: 10.1086/314976. [DOI] [PubMed] [Google Scholar]
- 36.Mobley H L, Green D M, Trifillis A L, Johnson D E, Chippendale G R, Lockatell C V, Jones B D, Warren J W. Pyelonephritic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun. 1990;58:1281–1289. doi: 10.1128/iai.58.5.1281-1289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mobley H L, Jarvis K G, Elwood J P, Whittle D I, Lockatell C V, Russell R G, Johnson D E, Donnenberg M S, Warren J W. Isogenic P-fimbrial deletion mutants of pyelonephritic Escherichia coli: the role of alpha Gal(1–4) beta Gal binding in virulence of a wild-type strain. Mol Microbiol. 1993;10:143–155. doi: 10.1111/j.1365-2958.1993.tb00911.x. [DOI] [PubMed] [Google Scholar]
- 38.National Institutes of Health. Guide for the care and use of laboratory animals. Publication no. 85-23. Bethesda, Md: National Institutes of Health; 1985. [Google Scholar]
- 39.Neu H C. Urinary tract infections. Am J Med. 1992;92:63S–70S. doi: 10.1016/0002-9343(92)90312-y. [DOI] [PubMed] [Google Scholar]
- 40.Ruggeri F M, Fiorentini C, Caprioli A, Arancia G, Falbo V, Donelli G. HEp-2 cell multinucleation induced by an E. coli cytotoxic factor. IRCS Med Sci. 1986;14:833–834. [Google Scholar]
- 41.Russo T, Guenther J E, Wenderoth S, Frank M M. Generation of isogenic K54 capsule-deficient Escherichia coli strains through TnphoA-mediated gene disruption. Mol Microbiol. 1993;9:357–364. doi: 10.1111/j.1365-2958.1993.tb01696.x. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- 43.Swenson D L, Bukanov N O, Berg D E, Welch R A. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect Immun. 1996;64:3736–3743. doi: 10.1128/iai.64.9.3736-3743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warren J W. Clinical presentations and epidemiology of urinary tract infections. In: Mobley H L T, Warren J W, editors. Urinary tract infections: molecular pathogenesis and clinical management. Washington, D.C.: ASM Press; 1996. pp. 3–27. [Google Scholar]
- 45.Yamamoto S, Tsukamoto T, Terai A, Kurazono H, Takeda Y, Yoshida O. Distribution of virulence factors in Escherichia coli isolated from urine of cystitis patients. Microbiol Immunol. 1995;39:401–404. doi: 10.1111/j.1348-0421.1995.tb02219.x. [DOI] [PubMed] [Google Scholar]