Abstract
Background
Immunotherapy has become the first-line treatment for advanced non-small-cell lung cancer (NSCLC), but most patients still fail to benefit or have disease progression following treatment. M2 phenotype tumor-associated macrophages (M2-TAMs) are important cellular components in the immunosuppressive microenvironment of NSCLC, but how they contribute to immunoresistance remains unclear. This study was conducted to investigate the role and mechanism of M2-TAMs in NSCLC immunoresistance.
Methods
We collected postoperative tumor samples for detection of M2-TAMs and other immune cells infiltration by immunofluorescence detection and flow cytometry. We then constructed a non-contact cell co-culture system using Transwell chambers. CCK-8, colony formation, wound healing and invasion assays were performed to evaluated the effect of M2-TAMs on the proliferation, migration and invasion abilities of lung adenocarcinoma (LUAD) cells in vitro. Xenograft model were performed to analysis the effect of M2-TAMs on the tumorigenesis and metastasis of LUAD cells in vivo.
Results
M2-TAMs were greatly increased in the tumor tissue of patients with immunoresistant LUAD. They could significantly promote the proliferation, invasion, and migration of LUAD cells, and improve their resistance to cytotoxic T lymphocytes (CTL) cytotoxicity. Further research showed M2-TAMs could considerably enhance the expression of METTL3 and total m6A RNA level in LUAD cells and interfering with METTL3 could significantly reverse the impairment of M2-TAMs on the efficacy of CTL in killing tumor cells.
Conclusions
In conclusion, M2-TAMs could promote LUAD immunoresistance by enhancing METTL3-mediated m6A methylation. Our results suggest METTL3 could be a potential therapeutic target for reversing immunoresistance and shed new light on the mechanism of M2-TAMs promoting LUAD immunoresistance.
Keywords: M2 phenotype tumor-associated macrophages (M2-TAMs), METTL3, non-small cell lung cancer (NSCLC), immunoresistance
Highlight box.
Key findings
• M2-TAMs could promote LUAD immunoresistance by enhancing METTL3-mediated m6A methylation.
What is known and what is new?
• As known, M2-TAMs are important cellular components in the immunosuppressive microenvironment of NSCLC.
• Our work first proposed the important role and therapeutic value of METTL3 in immunoresistance in NSCLC.
What is the implication, and what should change now?
• Our results shed new light on the mechanism of M2-TAMs in promoting immunoresistance. Further study is required to elucidate the mechanism of immunoresistance and find more potential intervention targets.
Introduction
Immune checkpoint inhibitors (ICIs), especially anti-PD-1/PD-L1 monoclonal antibodies (mAb), have become a standard of care treatment for lung cancer and many other tumors, but the general efficacy is limited. Due to primary, adaptive, and acquired resistance, most cancer patients still fail to benefit from immunotherapy (1), with a real-world study of advanced non-small cell lung cancer (NSCLC) showing the median overall survival with anti-PD-1 mAb treatment as approximately 9–11 months (2). Immune resistance has become the key limiting factor for overall survival of patients with advanced NSCLC. The immunosuppressive microenvironment is a critical facilitator for the formation of immune resistance, and consists of immunosuppressive cells, immunosuppressive cytokines, and tumor-promoting matrix proteins such as extracellular matrix secreted by carcinoma-associated fibroblasts CAFs (3). Therefore, it is of great value to study the operation mode of the immunosuppressive microenvironment for regulating the anti-tumor immune response and reversing immune tolerance.
Tumor-associated macrophages (TAMs) are the most abundant myeloid cell population in the tumor microenvironment and can be divided into an anti-tumorigenic M1 phenotype and pro-tumorigenic M2 phenotype according to their function and activity (4-6). M1-TAMs possess strong antigen-presenting ability and can produce proinflammatory cytokines, including IL-1β, IL-6, IL-12, IL-23, CXCL9, CXCL10, and TNF-α, which suppress tumor cells by activating type 1 immune responses (7). M2-TAMs mainly activate type 2 immunity and secrete anti-inflammatory cytokines including IL-10, TGF-β, CCL17, CCL18, CCL22, and CCL24, which promote tumorigenesis and development (7). At the time of tumor initiation, the recruited macrophages in lung adenocarcinoma (LUAD) switch to the M2 phenotype, which is the primary phenotype existing in LUAD (8), and a published study (9) showed M2-TAMs infiltration is increased in chemoresistant LUAD. However, their infiltration level and effect in immunoresistant LUAD require verification.
N6-methyladenosine (m6A) is the most abundant mRNA epigenetic modification in eukaryotes and plays an important role in the regulation of cell differentiation and cycles, biological metabolism, and responses to heat shock stress, cancers, and other diseases (10). A multicomponent m6A methyltransferase complex (MTC) composed of a METTL3-METTL14 heterodimeric core and other binding subunits is the major “writer” protein during m6A modification, and METTL3 is the only catalytic subunit using sadenosylmethionine (SAM) as a methyl donor (10,11). The m6A modification is dynamic and reversible, and can be removed by “eraser” proteins including FTO and ALKBH5 (12). “Reader” proteins such as YTHDC1/2 and YTHDF1/2/3 can specifically recognize and bind m6A-modified transcripts to regulate RNA splicing and nuclear export (12), and a recent study found various m6A-modified non-coding RNAs could play different roles in cancer stemness modulation (13).
Given these reported findings, we focused on the function and mechanism of M2-TAMs in the immunoresistance of LUAD, explored the relationship between them and m6A methylation modification. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-6104/rc).
Methods
Immunofluorescence staining and hematoxylin-eosin staining
LUAD tissues were acquired from the Third Affiliated Hospital of Soochow University from 2018 to 2019. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics committee of the Third Affiliated Hospital of Soochow University and informed consent was taken from all the patients. Collected clinical LUAD tissues sensitive or resistant to nivolumab were made into paraffin-embedded sections. The characteristics of LUAD patients were shown in Table 1. Sections were dewaxed in xylene and decreasing concentrations of alcohol, then repaired with a citric acid antigen repair solution. The cultured cells were directly made into cell slices and blocked with goat serum, and the primary antibody was diluted according to the manufacturer's instructions and incubated overnight. Slices were then washed in PBST and incubated with appropriate fluorescent secondary antibody for 1 hour. Images were acquired using an inverted fluorescence microscope (Olympus, CX53), and the mean fluorescence intensity was measured using ImageJ (NIH, Bethesda, MD, USA) software.
Table 1. Characteristics of LUAD patients.
| Characteristics | Data, n |
|---|---|
| Total patients | 16 |
| Sex | |
| Male | 13 |
| Female | 3 |
| Age at start of trial (years), mean [range] | 59.5 [25–71] |
| Lymph node biopsy | |
| Metastasis (+) | 8 |
| Metastasis (−) | 8 |
LUAD, lung adenocarcinoma.
Lung and mediastinal tissues of mice with lung metastases were completely removed and the number of lung metastases nodes was counted independently by two researchers. The tissues were then fixed, embedded, and made into paraffin sections, and the prepared slices were processed with a Hematoxylin and Eosin Staining Kit (Beyotime) according to the manufacturer’s instructions.
Flow cytometry
The collected fresh clinical tumor tissues were triturated, digested with 0.25% trypsin (Gibco) for 30 min at 37 ℃, and filtered through a 200-µm strainer to prepare single cell suspensions after the digestion was terminated. Anti-human antibodies used for incubation included antibodies against CD3 (Clone OKT3), CD8 (Clone SK1), CD161 (Clone HP-3G10), MHCII (Clone 6C6), CD11c (Clone Bu15), CD86 (Clone IT2.2), iNOS (Clone W16030C), CD206 (Clone 15-2), and CD163 (Clone S15049I). Data were acquired using a BD FACS Aria II flow cytometer and were analyzed using FlowJo software.
Cells, culture conditions, and cell coculture system
The human monocytic cell line Thp-1 was obtained from the American Type Culture Collection (ATCC, Manassas, VA), and human lung cancer A549 and H1975 cell lines were obtained from the Chinese Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were maintained in RPMI-1640 (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (P/S) solution at 37 ℃ with 5% CO2.
The upper and lower chambers of a Transwell chamber (4 µm pore inserts) were used to construct the co-culture system of M2-TAMs and lung cancer cells. LUAD cells (A549 and H1975) were cultured in the lower chambers and M2-TAMs were cultured in the upper chambers.
Polarization of M2-TAMs
Cultured human Thp-1 cells were resuspended and added to 100 ng/mL of PMA, and macrophages were obtained after 48 hours of induction. M2-TAMs were obtained after further induction with 10 ng/mL human IL-4 (ImmunoTools) and 10 ng/mL IL-6 (ImmunoTools) for 48 h.
Cell proliferation assay and colony formation
Cell proliferation ability was measured by CellTiter 96® Non-Radioactive Cell Proliferation Assay (CCK-8) kit (Dojindo) according to the manufacturer’s instructions. 1×103 cells/per-well were seeded into a 96-well plate and incubated for 2 h at 37 ℃ and 5% CO2. Absorbance was detected at 450 nm per 24 h for four times consecutively. All experiments were replicated in triplicate.
For colony formation assay, 1.5×103 suspended cells were seeded onto a coating of 0.8% low-melting agar in RPMI 1640 medium with 10% FBS, with each group set three repetitions. After a 21-day incubation at 37 ℃ and 5% CO2, the plates were stained with 0.1% crystal violet solution for 10 min and independently counted by three researchers.
Migration and invasion assays
Wound scrape assay was performed to examine the migration ability of M2-TAMs on LUAD cells as described previously (14). Image acquisition was performed using a phase contrast microscope (Leica, Leica Microsystems), and image analysis was performed using Image-Pro Plus software.
The evaluation of cell invasive ability was performed using a matrigel invasion assay to examine the role of M2-TAMs in the regulation of the LUAD cell invasion process as described previously (14). The average cell number was calculated from at least six fields of view (200×) independently by two researchers.
Mouse model
We purchased 5-week-old male BALB/c-nu nude mice from the SLAC Laboratory Animal Company (Shanghai, China) to use for xenograft models. The welfare and handling of laboratory animals followed the National Institutes of Health guide for the care and use of laboratory animals and the animal experiments were approved by the Institutional Animal Care and Use Committee of the Third Affiliated Hospital of Soochow University. A549 (3×106) cells in the co-culture or control group were subcutaneously inoculated into the murine left flank, respectively. Tumor volumes were monitored every 5 days, and the mice were sacrificed on the 30th day when tumors were excised, photographed, and weighed. The tumor volume was calculated as the following formula: V = Length × Width2/2.
For lung metastases formation assay, each mouse was injected with A549 (3×107) cells through the tail vein. The mice were sacrificed on the 28th day, and the lung and mediastinal tissues were completely removed. Lung metastasis nodes were counted independently by two researchers.
Colorimetric assay
Total intracellular RNA was extracted with TRIzol reagent (Thermo Fisher) according to the manufacturer’s instructions. An EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric, Epigentek) was then used to quantify the proportion of m6A in total RNA according to the manufacturer's instructions. Finally, the m6A content was quantified colorimetrically at 450 nm and calculated according to the standard curve.
Dot blot assay
Total intracellular RNA was extracted as described above. RNA was denatured at 65 ℃ for 5 minutes then immediately cooled on ice. Denatured RNA (2 µL) was dropped directly onto Immobilon-Ny+ Nylon membranes, which were then UV-crosslinked for 5 min and washed with PBST buffer. Membranes were incubated in 10 mL blocking buffer (5% defatted milk) for 1 hour at room temperature, then placed in 5 mL of primary antibody dilution buffer (1:250; 2 µg/mL) and incubated overnight at 4 ℃ with gentle shaking. Washed membranes were then placed in 5 mL of Goat anti rabbit IgG-HRP (1:10,000; 20 ng/mL) secondary antibody dilution buffer and incubated for 1 hour at room temperature with gentle shaking. The results were recorded with the Tanon fully automated chemiluminescence image analysis system.
Real-time PCR analysis
Total RNA was isolated using TRIzol reagent (Thermo Fisher), followed by cDNA synthesis with Superscript II (Invitrogen). Each cDNA synthesis reaction used 1µg of total RNA. RNA expression was measured by SYBR Green Universal Master Mix reagent (Vazyme Biotech, Nanjing, China) which operated on the Bio-Rad QX100 Droplet Digital PCR system (USA). The amount of relative mRNA was calculated by the 2ΔΔCt formula with the normalization to GAPDH. All primers were purchased from Tsingke Biological Technology (Beijng, China), and the primer sequences are shown in Table S1.
Western blot analysis
Total protein was extracted from tumor cells. An equal amount of protein sample was separated by 10% SDS-PAGE (GenScript, Nanjing, China) then transferred onto 0.45 µm PVDF membranes (Millipore, USA). Antibody incubation was conducted after blocking with Western blocking solution (P0023B), and anti-METTL3 antibody (Invitrogen) was used at a concentration of 1:2,000. GAPDH were selected as the loading control.
Luminex assay
After 5 days of cell culture, the cell supernatant was obtained by centrifugation, and cytokines detected using a Luminex assay (R&D System) kit according to the manufacturer’s instructions. Data were acquired with a Bio-Plex 200 reader (Bio-Rad) and analyzed using Bio-Plex Manager software (version 5.0). Results are represented as heatmaps.
siRNA knockdown
siRNA was obtained from GenePharma (Suzhou, Jiangsu, China), and its sequence was as follows: METTL3 siRNA: (5'-GAGATCCTAGAGCTATTAAA-3').
Statistical analysis
Statistical analyses were performed with GraphPad Prism 8.0 software. Two-tailed independent t-test or two-way ANOVA was performed to determine the statistical significance, with P<0.05 considered significant.
Results
Increased infiltration of M2-TAMs in nivolumab-resistant LUAD tumor tissue
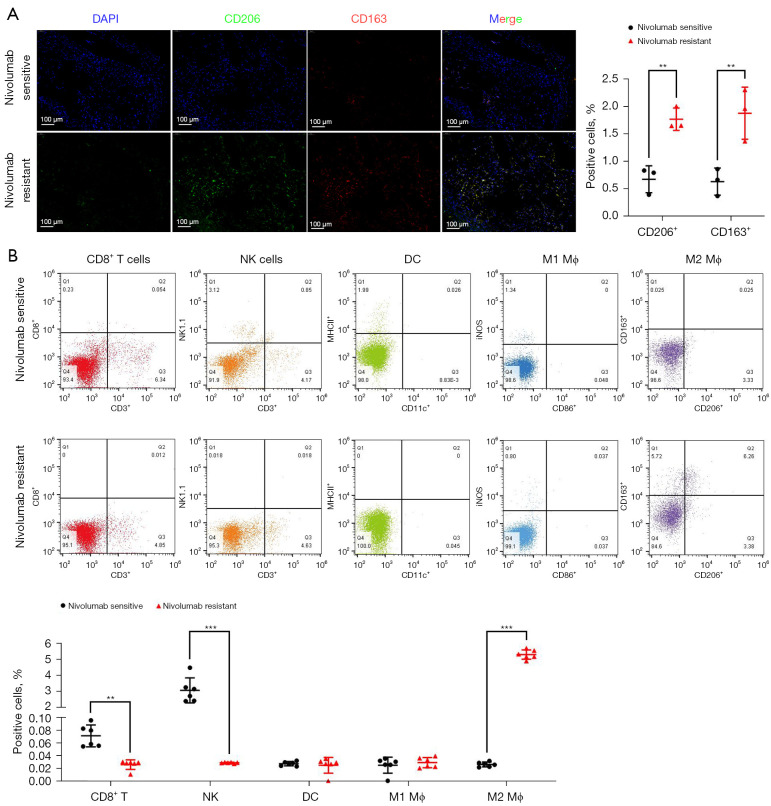
To determine whether the infiltration of M2-TAMs was increased in the tumor tissue of immunoresistant patients, we collected postoperative, fresh tumor tissue specimens from patients with stage I–III resectable LUAD who were diagnosed by pathology and imaging findings in the Third Affiliated Hospital of Soochow University from 01/01/2018 to 31/12/2019. A total of 16 patients were enrolled, including eight nivolumab-sensitive and eight nivolumab-resistant LUAD patients. All patients received nivolumab for the first time before surgery, and the course of treatment was more than four cycles, with no obvious immune-related adverse events of grade 3 or above observed. Using immunofluorescence analysis of the tumor tissue samples, we found the infiltration of M2-TAMs in tumor tissue of the nivolumab-resistant group was significantly increased (P<0.01) (Figure 1A). To further investigate whether this increase was accompanied by remodeling of the tumor microenvironment, the collected clinical samples were prepared into single cell suspension and analyzed by flow cytometry. The results showed the infiltration levels of CD8+ T cells (P=0.004) and NK cells (P<0.001) in the tumor tissues of the nivolumab-resistant group were significantly decreased, and the infiltration level of M2-TAMs (P<0.001) was significantly increased (Figure 1B). Collectively, these results suggested M2-TAMs might promote the immune resistance of LUAD by down-regulating the infiltration of CD8+ T cells and NK cells in the tumor microenvironment.
Figure 1.
Differential infiltration of M2-TAMs and other immune cells in tumor tissues of nivolumab-sensitive and nivolumab-resistant LUAD patients. (A) Representative immunofluorescence images of M2-TAMs infiltration in LUAD tissues from nivolumab sensitive or resistant patients, n=3. (B) Representative flow cytometry plots of immune cell infiltration in LUAD tissues from nivolumab sensitive or resistant patients, including CD8+ T cells (CD3+/CD8+), NK cells (CD3-/NK1.1+), DC (CD11c+/MHCII+), M1-TAMs (CD86+/iNOS+), and M2-TAMs (CD206+/CD163+), n=6. Data are presented as mean ± SEM, **P<0.01, and ***P<0.001 according to two-tailed independent t-test. M2-TAMs, M2 phenotype tumor-associated macrophages; LUAD, lung adenocarcinoma.
M2-TAMs promote the proliferation, invasion, and migration of LUAD cells
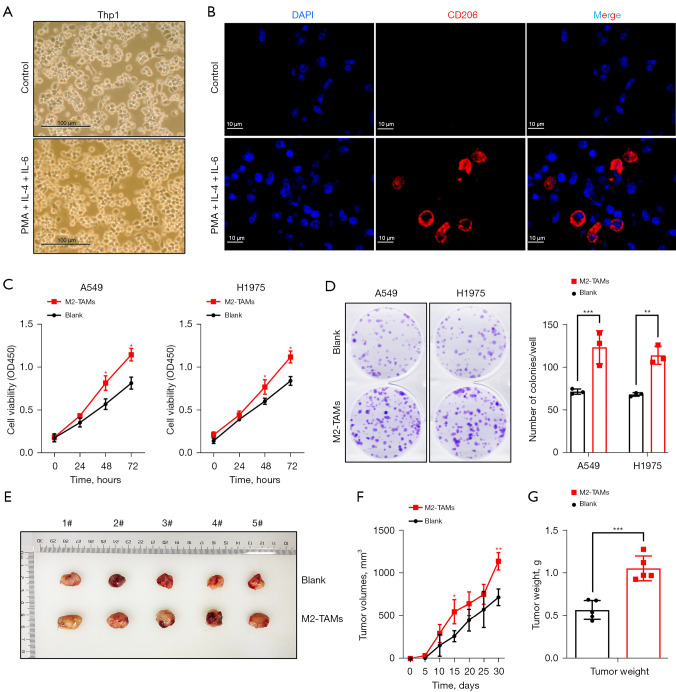
To examine the effect of elevated M2-TAMs in the tumor microenvironment of LUAD cells, we designed the following experiments. Firstly, the human monocytic cell Thp-1 was induced into M2-TAMs with PMA, IL-4, and IL-6 (Figure 2A,2B), and a co-culture system of M2-TAMs and LUAD cells (A549 and H1975) was constructed using Transwell chambers. LUAD cells cultured alone were used as a control group.
Figure 2.
M2-TAMs significantly promoted the proliferation of LUAD cells, both in vitro and in vivo. (A) Bright field images of Thp-1 cells by microscopy showed that Thp1 cells were induced to M2-TAMs by PMA, IL-4, and IL-6 after recovery. (B) Immunofluorescence was used to detect the induction of M2-TAMs (CD206+). (C) CCK8 assay detected the proliferation level of LUAD cells A549 and H1975 in M2-TAMs co-culture group or control group, n=3. (D) Colony formation assay and crystal violet staining of LUAD cells A549 and H1975 in M2-TAMs co-culture group or control group, n=3. (E) Assay of tumorigenicity in nude mice. A549 (3×106) cells in the co-culture or control group were subcutaneously inoculated into the murine left flank, n=5. (F) Volume of subcutaneous tumors in mice, n=5. (G) Weight of subcutaneous tumors in mice on day 30, n=5. Data are presented as mean ± SEM, *P<0.05, **P<0.01, and ***P<0.001 according to two-tailed independent t-test. M2-TAMs,M2 phenotype tumor-associated macrophages; LUAD, lung adenocarcinoma.
CCK-8 kits were used to investigate the effect of M2-TAMs on the proliferation ability of LUAD cells in the co-culture group or control group. This showed the proliferation ability of A549 and H1975 cells in the co-culture group was significantly higher than in the control group (P<0.05) (Figure 2C). A colony formation assay was then performed and showed the colony formation ability of A549 (P<0.001) and H1975 (P=0.002) cells was significantly improved after co-culture with M2-TAMs (Figure 2D). Finally, we performed nude mouse subcutaneous tumorigenic experiments, monitored the size of subcutaneous tumors every 5 days, and sacrificed the mice on day 30 and measured tumor weights (Figure 2E). We found the tumor volume of M2-TAMs in the co-culture group was observably larger than that of the control group on day 15 and day 30 (Figure 2F), with day 30 tumor weights in the former averaging 1.06 g compared with 0.57 g in the control group (P<0.001) (Figure 2G). Collectively, M2-TAMs markedly promoted the proliferation of LUAD cells in vitro and in vivo.
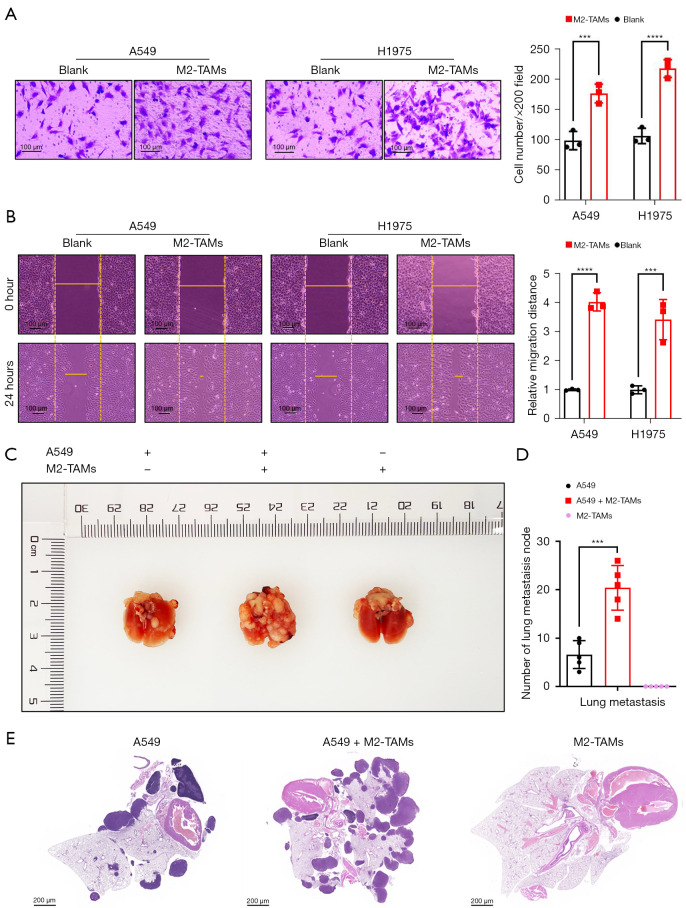
To explore the effect of M2-TAMs on the invasive ability of LUAD cells, we performed a matrigel invasion assay using a Transwell insert system, and after 24 hours of culture, the invasive ability of LUAD cells A549 (P<0.001) and H1975 (P<0.0001) was significantly enhanced (Figure 3A). Further, the results of wound scrape assay showed the migration ability of LUAD cells A549 (P<0.0001) and H1975 (P<0.001) in the M2-TAMs co-culture group was considerably higher than in the control group (Figure 3B). These results indicated M2-TAMs could significantly promote LUAD cell invasion and migration in vitro. We next established mouse lung metastasis models by injecting A549 cells from the co-culture group or the control group into nude mice via the tail vein (Figure 3C) and observed the A549 cells co-cultured with M2-TAMs formed more lung metastasis nodes than the control group (P<0.001) (Figure 3D). On day 28, the mice were sacrificed, and the lung and mediastinal tissue of mice with lung metastases was used to make representative HE staining slices (Figure 3E). Together, M2-TAMs significantly promoted LUAD cell proliferation, invasion, and migration in vivo and in vitro.
Figure 3.
M2-TAMs significantly promoted the invasion and migration of LUAD cells, both in vitro and in vivo. (A) Matrigel invasion assay and crystal violet staining of LUAD cells A549 and H1975 in M2-TAMs co-culture group or control group, n=3. (B) Wound scrape assay and bright field images of LUAD cells A549 and H1975 in M2-TAMs co-culture group or control group, n=3. (C) Mouse models of lung metastases formation via tail vein injection. (D)Number of lung metastasis node in mice, n=5. (E) Representative HE staining slices of lung and mediastinal tissues from mice with lung metastases. Data were presented as mean ± SEM, ***P<0.001, and ****P<0.0001 according to two-tailed independent t-test. M2-TAMs, M2 phenotype tumor-associated macrophages; LUAD, lung adenocarcinoma.
We have previously detected decreased infiltration levels of CD8+ T cells in LUAD specimens from nivolumab-resistant patients (Figure 1B). To determine whether the tumor-promoting effect of M2-TAMs on LUAD was related to CTLs, we performed T cell cytotoxicity assay in 96-well culture plates and found the cytotoxicity of CTLs to LUAD cells A549 and H1975 co-cultured with M2-TAMs was markedly weakened (Figure S1). This indicated that M2-TAMs enhanced the tolerance of LUAD cells to CTLs, which in turn facilitated the formation of immunoresistance.
M2-TAMs upregulated METTL3 and total m6A RNA levels in LUAD cells
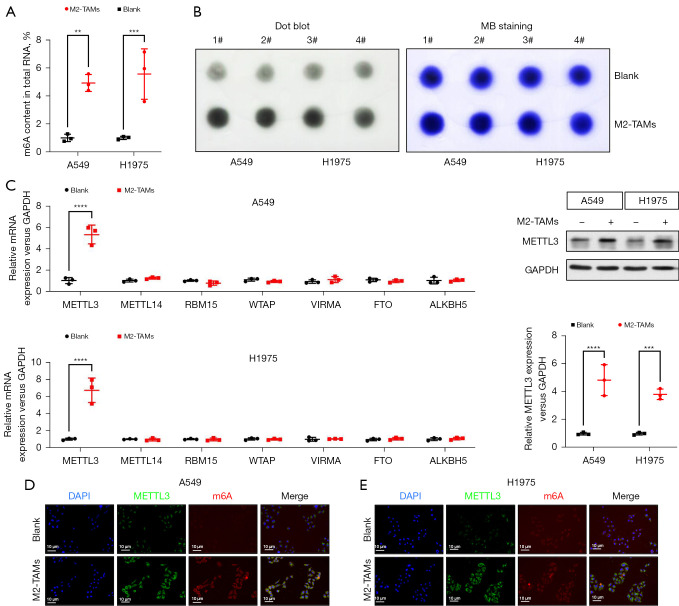
Mounting evidence suggests various m6A methylation-modified RNAs act as tumor-promoting factors to mediate drug resistance and metastasis in NSCLC (15-18). To examine whether the facilitating effect of M2-TAMs on LUAD was related to m6A methylation, we designed the following experiments. To begin with, we implemented a colorimetric assay to compare the m6A levels of total RNA in LUAD cells from the M2-TAMs co-culture group and the control group. The results showed the m6A levels of A549 (P<0.01) and H1975 (P<0.001) cells co-cultured with M2-TAMs were signally higher than those of the control group (Figure 4A). Dot Blot assay also proved M2-TAMs could observably increase m6A levels in LUAD cells (Figure 4B). To further determine which kind of m6A methyltransferase was involved in the M2-TAMs-mediated enhancement of m6A methylation in LUAD cells, we analyzed the expression levels of m6A methyltransferases in LUAD cells A549 and H1975 of the M2-TAMs co-culture group and control group by real-time PCR. The results indicated METTL3 was significantly upregulated in A549 (P<0.0001) and H1975 (P<0.0001) cells co-cultured with M2-TAMs (Figure 4C). Subsequent Western blot results also verified M2-TAMs could enhance the expression level of METTL3 in A549 (P<0.0001) and H1975 (P<0.001) cells (Figure 4D). Finally, we detected the expression of METTL3 and m6A in LUAD cells by immunofluorescence and found their levels in the M2-TAMs co-culture group were observably higher than in the control group (Figure 4E). Collectively, M2-TAMs significantly enhanced the expression of METTL3 in LUAD cells, increasing the level of total m6A modification.
Figure 4.
M2-TAMs significantly upregulated METTL3 and total m6A RNA in LUAD cells. (A) Colorimetric assay to detect total m6A RNA proportion of LUAD cells A549 and H1975 in M2-TAMs co-culture group or control group, n=3. (B) Dot blot assay verified M2-TAMs could significantly upregulate the level of total m6A RNA in LUAD cells A549 and H1975. Methylene blue staining was used as the internal reference, n=2. (C) Real-time PCR detected the expression levels of m6A methyltransferases in A549 and H1975 cells from M2-TAMs co-culture group or control group, n=3. (D) Western blot detection of METTL3 expression in A549 and H1975 cells from M2-TAMs co-culture group or control group, n=3. (E) Representative immunofluorescence images of METTL3 and m6A expression in A549 and H1975 cells from M2-TAMs co-culture group or control group, n=3. Data were presented as mean ± SEM. **P<0.01, ***P<0.001, and ****P<0.0001 according to two-tailed independent t-test. M2-TAMs, M2 phenotype tumor-associated macrophages; LUAD, lung adenocarcinoma.
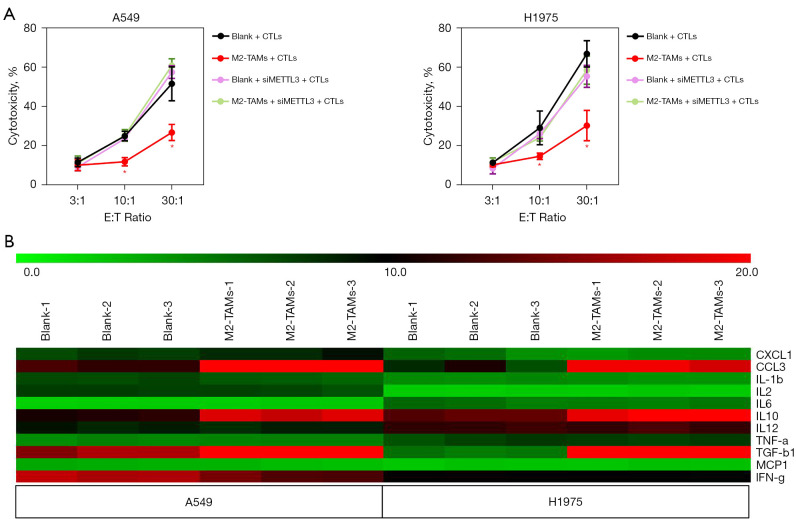
M2-TAMs may enhance the resistance of LUAD cells to CD8+ T cells by upregulating METTL3
We have verified that M2-TAMs can significantly impair the cytotoxicity of CTLs against LUAD cells (Figure S1) and that this effect was accompanied by the elevation of METTL3 in tumor cells (Figure 4C-4E). To further determine the effect of M2-TAMs-induced METTL3 elevation on antitumor immunity, we designed a T cell cytotoxicity assay that silenced METTL3 by siRNA transfection, and the results showed METTL3 interference significantly reversed the impairment of M2-TAMs on the efficacy of CTL in killing LUAD cells (Figure 5A). This suggests M2-TAMs might attenuate the antitumor immune response of CD8+ T cells by upregulating the levels of METTL3 and m6A RNA in LUAD cells, promoting immunoresistance and tumor progression. In the co-culture system of M2-TAMs and LUAD cells constructed by Transwell chamber, M2-TAMs did not directly contact tumor cells, but indirectly affected their function via effector molecules. To determine the cytokines through which M2-TAMs affect the function of LUAD cells, we used a Luminex liquid-chip to analyze the content of various cytokines in the supernatant of the M2-TAMs co-culture group and the control group, and the results showed the contents of M2 markers IL-10, TGF-β1, and CCL3 were significantly increased in the supernatant of co-culture group (Figure 5B). IL-10 and TGF-β1 are both well-known anti-inflammatory cytokines, and M2-TAMs-derived TGF-β1 has been shown to promote lung cancer angiogenesis (19), liver cancer, and colorectal cancer migration and invasion (20). CCL3 is closely related to immune surveillance and tolerance and has become a prognostic biomarker for a variety of malignant tumors (21).
Figure 5.
M2-TAMs-induced METTL3 may be involved in the regulation of LUAD cell sensitivity to CD8+ T cell cytotoxicity. (A) T cell cytotoxicity assay to detect the effects of M2-TAMs and METTL3 interference on LUAD cell survival at different CTL/tumor cell ratios, n=3. (B) Luminex liquid-chip to detect the cytokine content in supernatant of M2-TAMs co-culture group and control group, n=3. Data are presented as mean ± SEM. M2-TAMs, M2 phenotype tumor-associated macrophages; LUAD, lung adenocarcinoma.
Discussion
Lung cancer is the malignant tumor with the highest mortality and second highest morbidity, and its 5-year relative survival rate is only 22% (22). NSCLC is the most common pathological type of lung cancer, accounting for approximately 85% of total cases. For advanced NSCLC patients without sensitive gene mutations or who are resistant to targeted therapy, immunotherapy holds the most potential. Although several immune checkpoint inhibitors have been approved for the first-line treatment of advanced NSCLC, many offer no benefit to patients due to primary and acquired resistance. Immunoresistance presents an urgent problem that limits the overall survival of advanced NSCLC. Recent research suggests quiescent cancer cells resist T cell attack by forming an immunosuppressive niche (23), and M2-TAMs are important components of the immunosuppressive microenvironment.
In this study, we reported that compared with nivolumab-sensitive LUAD patients, nivolumab-resistant LUAD patients had higher level of M2-TAMs infiltration and lower levels of CD8+ T and NK cell infiltration in primary tumor tissues. One study showed M2-TAMs could produce functional cross-talk with a variety of NSCLC cells via CCR2 and CX3CR1 (24). Therefore, we constructed a non-contact cell co-culture system using Transwell chambers and demonstrated that M2-TAMs could promote the proliferation, invasion, and migration of LUAD cells by secreting a variety of cytokines, including IL-10, TGF-β1, and CCL3. We also found M2-TAMs could significantly enhance METTL3 expression in LUAD cells, which increased their total m6A RNA content, and interfering with METTL3 could significantly reverse the impairment of M2-TAMs on the efficacy of CTL in killing LUAD cells. These results indicated M2-TAMs might facilitate the immunoresistance of LUAD cells by enhancing METTL3-mediated m6A modification, and blockade of METTL3 could reverse the established immunosuppressive microenvironment and enhance immunotherapy efficacy.
Tumor cells and M2-TAMs have been proven to promote each other in several cancers. For instance, M2-TAMs-derived IL-6 regulated the EMT process and enhanced the migration and invasion of CRC cells in colorectal cancer, and could promote CCL2 secretion from CRC cells via the JAK2/STAT3 pathway, promoting macrophage recruitment (25). In breast cancer, M2-TAMs-derived lncRNA PCAT6 promoted tumorigenesis and angiogenesis in triple-negative breast cancer by regulating VEGFR2 (26), and inflammatory breast cancer was shown to promote the development of M2-TAMs through a complex chemokine network (27). In lung cancer, tumor cell-derived exosomes could be internalized by M0 macrophages and promoted the differentiation of M0 macrophages to the immunosuppressive M2 phenotype (28). Published studies have focused on the molecular mechanism of macrophage polarization toward M2-TAMs in lung cancer (29-32), and our work filled the gap on how they promote its metastasis, progression, and immunoresistance.
METTL3 is a key catalytic subunit of m6A methylation proven to play multiple roles in NSCLC. METTL3‑mediated m6A modification of Bcl‑2 mRNA promoted NSCLC progression (33), and METTL3-induced lncRNA ABHD11-AS1 facilitated the Warburg effect of NSCLC (34). METTL3 induced chemoresistance and metastasis in NSCLC by regulating the MALAT1-miR-1914-3p-YAP axis (15), and METTL3-mediated autophagy played an important role in reversing gefitinib resistance by β-elemene in NSCLC cells (35). Our work first proposed the important role and therapeutic value of METTL3 in immunoresistance in NSCLC.
Conclusions
Our results shed new light on the mechanism of M2-TAMs in promoting immunoresistance and identified METTL3 as a potential therapeutic target. However, the following limitations to this research exist: (I) our results did not reveal the specific molecular mechanism by which M2-TAMs upregulate METTL3 expression in LUAD cells; (II) this study did not further reveal which m6A RNA promoted LUAD immunoresistance and its specific molecular mechanism. Further study is required to elucidate the mechanism of immunoresistance and find more potential intervention targets.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank all patients involved in this study.
Funding: This work was supported by the National Natural Science Foundation of China (No. 82072561); Major Science and Technology Project of Changzhou Health Commission (Nos. ZD202004, ZD202007, and ZD202201); and Young Talent Development Plan of Changzhou Health Commission (No. CZQM2020024).
Ethical Statement: All authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics committee of the Third Affiliated Hospital of Soochow University and informed consent was taken from all the patients. Animal experiments were approved by the Institutional Animal Care and Use Committee of the Third Affiliated Hospital of Soochow University. The welfare and handling of laboratory animals followed the National Institutes of Health guide for the care and use of laboratory animals.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-6104/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-6104/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-6104/coif). The authors have no conflicts of interest to declare.
(English Language Editor: B. Draper)
References
- 1.Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017;168:707-23. 10.1016/j.cell.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.La J, Cheng D, Brophy MT, et al. Real-World Outcomes for Patients Treated With Immune Checkpoint Inhibitors in the Veterans Affairs System. JCO Clin Cancer Inform 2020;4:918-28. 10.1200/CCI.20.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biffi G, Tuveson DA. Diversity and Biology of Cancer-Associated Fibroblasts. Physiol Rev 2021;101:147-76. 10.1152/physrev.00048.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity 2014;41:49-61. 10.1016/j.immuni.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mills CD, Kincaid K, Alt JM, et al. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol 2000;164:6166-73. 10.4049/jimmunol.164.12.6166 [DOI] [PubMed] [Google Scholar]
- 6.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol 2010;11:889-96. 10.1038/ni.1937 [DOI] [PubMed] [Google Scholar]
- 7.Wu K, Lin K, Li X, et al. Redefining Tumor-Associated Macrophage Subpopulations and Functions in the Tumor Microenvironment. Front Immunol 2020;11:1731. 10.3389/fimmu.2020.01731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Redente EF, Dwyer-Nield LD, Merrick DT, et al. Tumor progression stage and anatomical site regulate tumor-associated macrophage and bone marrow-derived monocyte polarization. Am J Pathol 2010;176:2972-85. 10.2353/ajpath.2010.090879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baghdadi M, Wada H, Nakanishi S, et al. Chemotherapy-Induced IL34 Enhances Immunosuppression by Tumor-Associated Macrophages and Mediates Survival of Chemoresistant Lung Cancer Cells. Cancer Res 2016;76:6030-42. 10.1158/0008-5472.CAN-16-1170 [DOI] [PubMed] [Google Scholar]
- 10.Zhou H, Yin K, Zhang Y, et al. The RNA m6A writer METTL14 in cancers: Roles, structures, and applications. Biochim Biophys Acta Rev Cancer 2021;1876:188609. 10.1016/j.bbcan.2021.188609 [DOI] [PubMed] [Google Scholar]
- 11.Zeng C, Huang W, Li Y, et al. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol 2020;13:117. 10.1186/s13045-020-00951-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 2019;20:608-24. 10.1038/s41580-019-0168-5 [DOI] [PubMed] [Google Scholar]
- 13.Qin S, Mao Y, Wang H, et al. The interplay between m6A modification and non-coding RNA in cancer stemness modulation: mechanisms, signaling pathways, and clinical implications. Int J Biol Sci 2021;17:2718-36. 10.7150/ijbs.60641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lan J, Sun L, Xu F, et al. M2 Macrophage-Derived Exosomes Promote Cell Migration and Invasion in Colon Cancer. Cancer Res 2019;79:146-58. 10.1158/0008-5472.CAN-18-0014 [DOI] [PubMed] [Google Scholar]
- 15.Jin D, Guo J, Wu Y, et al. m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol 2019;12:135. 10.1186/s13045-019-0830-6 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Li B, Zhu L, Lu C, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun 2021;12:295. 10.1038/s41467-020-20527-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin H, Chen L, Piao S, et al. M6A RNA methylation-mediated RMRP stability renders proliferation and progression of non-small cell lung cancer through regulating TGFBR1/SMAD2/SMAD3 pathway. Cell Death Differ. 2021; 10.1038/s41418-021-00888-8. Correction appears in Cell Death Differ 2022. [DOI]
- 18.Liu Z, Wang T, She Y, et al. N6-methyladenosine-modified circIGF2BP3 inhibits CD8+ T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol Cancer 2021;20:105. 10.1186/s12943-021-01398-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong X, Bu J, Chen J, et al. PIGF and Flt-1 on the surface of macrophages induces the production of TGF-β1 by polarized tumor-associated macrophages to promote lung cancer angiogenesis. Eur J Pharmacol 2021;912:174550. 10.1016/j.ejphar.2021.174550 [DOI] [PubMed] [Google Scholar]
- 20.Liu M, Zhong YB, Shao J, et al. Tumor-associated macrophages promote human hepatoma Huh-7 cell migration and invasion through the Gli2/IGF-II/ERK1/2 axis by secreting TGF-β1. Cancer Biol Ther 2020;21:1041-50. 10.1080/15384047.2020.1824478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ntanasis-Stathopoulos I, Fotiou D, Terpos E. CCL3 Signaling in the Tumor Microenvironment. Adv Exp Med Biol 2020;1231:13-21. 10.1007/978-3-030-36667-4_2 [DOI] [PubMed] [Google Scholar]
- 22.Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 23.Baldominos P, Barbera-Mourelle A, Barreiro O, et al. Quiescent cancer cells resist T cell attack by forming an immunosuppressive niche. Cell 2022;185:1694-1708.e19. 10.1016/j.cell.2022.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmall A, Al-Tamari HM, Herold S, et al. Macrophage and cancer cell cross-talk via CCR2 and CX3CR1 is a fundamental mechanism driving lung cancer. Am J Respir Crit Care Med 2015;191:437-47. 10.1164/rccm.201406-1137OC [DOI] [PubMed] [Google Scholar]
- 25.Wei C, Yang C, Wang S, et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer 2019;18:64. 10.1186/s12943-019-0976-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong F, Ruan S, Wang J, et al. M2 macrophage-induced lncRNA PCAT6 facilitates tumorigenesis and angiogenesis of triple-negative breast cancer through modulation of VEGFR2. Cell Death Dis 2020;11:728. 10.1038/s41419-020-02926-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valeta-Magara A, Gadi A, Volta V, et al. Inflammatory Breast Cancer Promotes Development of M2 Tumor-Associated Macrophages and Cancer Mesenchymal Cells through a Complex Chemokine Network. Cancer Res 2019;79:3360-71. 10.1158/0008-5472.CAN-17-2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard A, Tousif S, Wang Y, et al. Lung Tumor Cell-Derived Exosomes Promote M2 Macrophage Polarization. Cells 2020;9:1303. 10.3390/cells9051303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren W, Hou J, Yang C, et al. Extracellular vesicles secreted by hypoxia pre-challenged mesenchymal stem cells promote non-small cell lung cancer cell growth and mobility as well as macrophage M2 polarization via miR-21-5p delivery. J Exp Clin Cancer Res 2019;38:62. 10.1186/s13046-019-1027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu JY, Huang TW, Hsieh YT, et al. Cancer-Derived Succinate Promotes Macrophage Polarization and Cancer Metastasis via Succinate Receptor. Mol Cell 2020;77:213-227.e5. 10.1016/j.molcel.2019.10.023 [DOI] [PubMed] [Google Scholar]
- 31.Li Z, Feng C, Guo J, et al. GNAS-AS1/miR-4319/NECAB3 axis promotes migration and invasion of non-small cell lung cancer cells by altering macrophage polarization. Funct Integr Genomics 2020;20:17-28. 10.1007/s10142-019-00696-x [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, Wang SQ, Hang L, et al. GRP78 facilitates M2 macrophage polarization and tumour progression. Cell Mol Life Sci 2021;78:7709-32. 10.1007/s00018-021-03997-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Liu S, Zhao T, et al. METTL3-mediated m6A modification of Bcl-2 mRNA promotes non-small cell lung cancer progression. Oncol Rep 2021;46:163. 10.3892/or.2021.8114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue L, Li J, Lin Y, et al. m6 A transferase METTL3-induced lncRNA ABHD11-AS1 promotes the Warburg effect of non-small-cell lung cancer. J Cell Physiol 2021;236:2649-58. 10.1002/jcp.30023 [DOI] [PubMed] [Google Scholar]
- 35.Liu S, Li Q, Li G, et al. The mechanism of m6A methyltransferase METTL3-mediated autophagy in reversing gefitinib resistance in NSCLC cells by β-elemene. Cell Death Dis 2020;11:969. 10.1038/s41419-020-03148-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as