Abstract
Purpose:
Targeted cancer therapeutics have not significantly benefited patients with Ewing sarcoma with metastatic or relapsed disease. Understanding the molecular underpinnings of drug resistance can lead to biomarker-driven treatment selection.
Experimental Design:
Receptor tyrosine kinase (RTK) pathway activation was analyzed in tumor cells derived from a panel of Ewing sarcoma tumors, including primary and metastatic tumors from the same patient. Phospho-RTK arrays, Western blots, and IHC were used. Protein localization and the levels of key markers were determined using immunofluorescence. DNA damage tolerance was measured through PCNA ubiquitination levels and the DNA fiber assay. Effects of pharmacologic inhibition were assessed in vitro and key results validated in vivo using patient-derived xenografts.
Results:
Ewing sarcoma tumors fell into two groups. In one, IGF1R was predominantly nuclear (nIGF1R), DNA damage tolerance pathway was upregulated, and cells had low replication stress and RRM2B levels and high levels of WEE1 and RAD21. These tumors were relatively insensitive to IGF1R inhibition. The second group had high replication stress and RRM2B, low levels of WEE1 and RAD21, membrane-associated IGF1R (mIGF1R) signaling, and sensitivity to IGF1R or WEE1-targeted inhibitors. Moreover, the matched primary and metastatic tumors differed in IGF1R localization, levels of replication stress, and inhibitor sensitivity. In all instances, combined IGF1R and WEE1 inhibition led to tumor regression.
Conclusions:
IGF1R signaling mechanisms and replication stress levels can vary among Ewing sarcoma tumors (including in the same patient), influencing the effects of IGF1R and WEE1 treatment. These findings make the case for using biopsy-derived predictive biomarkers at multiple stages of Ewing sarcoma disease management.
Translational Relevance.
Patients with recurrent or metastatic Ewing sarcoma have few treatment options. Further, heterogeneity among tumors contributes to variations in response to treatment. Molecular predictors of response to targeted therapies will allow better patient–treatment matching. Here we show that Ewing sarcomas can be classified into two groups based on the subcellular localization of IGF1R and basal levels of replication stress. Nuclear-IGF1R promotes DNA-damage tolerance, thus permitting tumor cell survival and ameliorating replication stress. IGF1R inhibition retards growth of tumors with nuclear-IGF1R while arresting growth of tumors with membrane-associated IGF1R. WEE1 inhibition arrests tumor growth in both instances. A combination of IGF1R and WEE1 inhibition leads to tumor regression. This IGF1R/replication stress level–based classification does not correlate with stages of disease and could differ between primary and recurrent tumors in the same patient, making the case for biopsy-based evaluation at multiple stages of disease management.
Introduction
Ewing sarcoma (EwS), the second most prevalent sarcoma among adolescents and young adults, is an aggressive bone and soft-tissue tumor. Apart from the pathognomonic translocation that fuses the transactivation domain of EWSR1 with the DNA binding domain of FLI1 and occasional STAG2, p53, and CDKN2A mutations, the EwS genome is relatively silent. Despite this genetic homogeneity, the clinical presentation and the course of disease varies widely. Treatment consists of conventional chemotherapy, surgery, and radiation. With localized disease, the survival rate is around 50% to 70% (1–3), although long-term effects of treatment can be debilitating. Over 25% of patients present with metastatic disease, and the survival rate for patients with metastatic/relapsed disease can be as low as 10% to 15% (4–7). Salvage therapy regimens used to treat recurrent EwS are generally not curative and are limited by cumulative toxicity. Initial excitement over IGF1R inhibitors was followed by several clinical trials showing modest benefit (8), and numerous other well-tolerated tyrosine kinase inhibitors have not yet shown significant clinical benefit (9, 10).
EwS cells have high levels of replication stress (11, 12), as a result of hyper-proliferation and global upregulation of transcription by the EWS-FLI1 oncogene (12). EwS cell lines are remarkably sensitive to drugs targeting the replication stress response (RSR), and numerous clinical trials are exploring the utility of RSR-targeted therapeutics (12–15). However, a limitation of preclinical studies conducted using established EwS cell lines is that they provide little insight into the heterogeneity of replication stress responses among patient tumors, and in primary/metastatic/relapsed tumors from the same patient. Knowledge of intertumor variability in the RSR is relevant for patient-stratification when exploiting replication stress to therapeutic advantage.
In this study we profile 6 patient-derived tumors and cells, including a matched pair of tumors (primary and metastatic tumors from the same patient). Our data show that IGF1R signaling, and RSR pathways differ among patients and in different tumors from the same patient and are broadly classifiable as either tumors with high nuclear IGF1R (nIGF1R)-associated DNA damage-tolerance (DDT), or tumors with membrane-associated IGF1R (mIGF1R) and elevated replication stress levels. Sensitivity to IGF1R inhibitors differed among the samples analyzed and correlated with IGF1R localization, whereas a combination of IGF1R and WEE1 inhibition led to tumor regression. These observations raise the possibility that biopsy-derived biomarkers could aid selection of patient-specific therapeutic strategies and be informative at multiple stages of disease management.
Materials and Methods
EwS patient-derived xenografts (PDX), evaluation of pharmacologic agents
Biopsy samples were obtained with written informed patient consent at Cincinnati Children's Hospital Medical Center (CCHMC; samples are CCH1, CCH2, CCH4, CCH5) and Nationwide Children's Hospital [NCH; samples are NCH-EWS-2 (NCH2), and NCH-EWS-6 (NCH6)], and studies were conducted in accordance with institutional guidelines and the Declaration of Helsinki. Xenografts were established in NSG mice (6-week-old, female) by intramuscular injection of minced tumor fragments in Matrigel. Tumors were STR-profiled and used for experiments after the third implantation.
To evaluate the effect of pharmacologic agents, subcutaneous xenografts were used (4–5 mice per group, randomly assigned). When tumors reached ∼200 mm3 treatments (blinded treatment: oral, 5 days/week) were initiated with either vehicle (PBS, 10% DMSO), linsitinib (25 or 15 mg/kg), AZD1775 (40 or 80 mg/kg), or linsitinib + AZD1775. Experiments were terminated when control CCH1 tumors in the first set of experiments reached an average volume exceeding 2,000 mm3 (12 doses). For consistency, all other experiments report the same number of doses.
Animal experiments were performed in accordance with the recommendations of the Institutional Animal Care and Use Committee at CCHMC (IACUC2016–0019). Use of de-identified patient tumor tissue was approved by the Institutional Review Boards at CCHMC (IRB2008–0021) and NCH (IRB1100478). Details of all reagents, software, and instruments are provided in Supplementary Table S2. Isolation and in vitro culture of tumor cells in Supplementary Table S2. Experiments conducted between passages 3 to 7.
IHC DAB and immunofluorescence
After antigen-retrieval (citrate) on deparaffinized/rehydrated sections, peroxidase inactivation (for IHC-DAB, 3% H2O2) fixed samples were permeabilized and blocked, incubated in antibodies (Supplementary Table S2), washed with PBST (PBS, 0.1% Tween-20). For immunofluorescence-based detection, samples were incubated with Alexa Fluor-conjugated secondary antibody and counterstained with DAPI. For IHC-DAB, the ABC Kit was followed by DAB, counter-stained with fast-red nuclear stain. Samples were imaged and processed by ImageJ (NIH) and Adobe Photoshop.
Proliferation assays
WST-8 assay was used as described previously (16, 17). Cells were treated with indicated agents (48 hours), incubated with WST-8 reagent (2 hours), absorbance recorded at 450 nm.
DNA fiber assay
Cells were cultured in 12-well plates followed by 25 μmol/L IdU (30 minutes), and 250 μmol/L CldU (30 minutes), then trypsinized. Two microliters of 40,000 cells/mL was spotted on glass slides, partially dried, and treated with Tris 200 mmol/L, EDTA 50 mmol/L, and SDS 0.5%. Slides were placed at a 30° angle (2 hours), fixed with methanol/acetic acid, stored at −20°C, treated with 2.5M HCL, washed with PBS, blocked, incubated with anti-IdU and anti-BrdU antibodies, followed by anti-mouse-AF488 and anti-rat-AF594 secondary antibodies, and imaged. DNA fiber lengths in untangled areas of slides were measured by ImageJ and converted into total extension (2.59 kb/μm; refs. 18, 19). Speed of extension calculated as described in Henry-Mowatt and colleagues (20).
Phospho-RTK arrays
Cells were lysed, protein extract quantified by Bradford, and ∼200 to 300 μg protein lysate used in the human phospho-RTK array (R&D System) assay performed according to the manufacturer's protocol and visualized with ECL reagent using ChemiDoc.
SDS-PAGE and Western blotting
Lysates were run on SDS-PAGE, transferred onto PVDF membranes, blocked with 5% nonfat milk/TBS-T, incubated with primary antibody followed by HRP-conjugated secondary antibody, visualized with ECL, imaged on a BioRad imager. Bands quantified by Imagelab (v6.1.0). Quantifications and uncropped gels in Supplementary Data File S1 with membranes labeled to clearly indicate when the same load control panel (tubulin) applies to multiple antibodies in Western blot figures.
PI-based cell cycle and cell death determination
Cells were fixed in 70% ethanol and labeled with PI for 10 minutes and the percentage of cells in G1, S, and G2–M phases determined by flow cytometry. Cells treated in chambered slides were stained with PI and imaged on an inverted fluorescence microscope.
S-phase duration using dual EdU-BrdU pulse-chase labeling
S-phase duration was determined from asynchronous cells as described (previously 21). Cells were first labeled with EdU (10 μmol/L, 30 minutes), washed, labeled with BrdU (100 μmol/L) at 0, 2, 5, 8, and 14 hours. Except at the 0-hour time-point 20 mmol/L thymidine with 100 ng/mL Nocodazole was added in the medium and removed before BrdU labeling. Cells were fixed with 94% ethanol, permeabilized by HCL with Triton-X-100, EdU labeling detected by Click-iT reaction and BrdU by anti-BrdU-FITC. Data were acquired by flow cytometry and analyzed by FlowJo software. S-phase duration of each cell type was calculated using linear regression analysis trend-line equation.
Immunoprecipitation
Dynabeads magnetic separation was used. Nearly 250 μg total protein lysate was incubated with anti-IGF1R or anti-PCNA antibody-conjugated dynabeads overnight (4°C), pulled down using a magnetic separator, and immuno-blotted with the indicated antibodies.
RNA isolation, cDNA synthesis, and qPCR
Total RNA from cells was extracted under RNAase-free conditions using Pure-Link RNA Kit. cDNAs synthesized from using PrimeScript RT Reagent Kit. Real-time PCR was performed using primers:
WEE1 forward 5′-TGAAGAGGCTGGATGGATGC -3′, reverse 5′-TTCTGCCCACGCAGAGAAAT-3′ GAPDH forward 5′-TGCACCACCAACTGCTTAGC-3′, reverse 5′-GGCATGGACTGTGGTCATGAG-3′ in a QuantiStudio-3, Applied biosystem real-time-PCR machine; (95°C 5 minutes; 35 cycles of 95°C 30 seconds, 61°C 15 seconds, 72°C 30 seconds, extension 72°C, 5 minutes). CT (threshold cycle) of target gene was normalized to GAPDH (ΔCT). −ΔΔCT and 2−ΔΔCT values determined in two biological replicates.
Statistical analysis
Results presented as the mean ± SD. Statistical analyses were performed using Graphpad PRISM v9.3.1, www.graphpad.com (GraphPad Prism, RRID:SCR_002798). t test was used when two samples/conditions were compared and ANOVA for more than two groups. For tumor growth curves, we used ordinary one-way ANOVA with Tukey multiple comparison tests.
Data availability
All data generated or analyzed during this study are included in this manuscript and the associated supplementary files or will be made available on request from the corresponding author.
Results
Primary and metastatic tumor cells from the same patient have distinct RTK-signaling patterns
To evaluate changes in tumor cells through the course of disease, we used PDX from a matched pair of biopsies (Supplementary Table S1): CCH1 was acquired at original diagnosis from a primary tumor with no prior therapy (22) and CCH2 is a metastatic lesion in the femur biopsied at first relapse from the same patient. The patient never received RTK-targeted therapeutics. CCH1 cells were more elongated whereas CCH2 cells tended to aggregate. Both cells stained positive for the EwS cell-surface marker CD99 (Supplementary Figs. S1A and S1B).
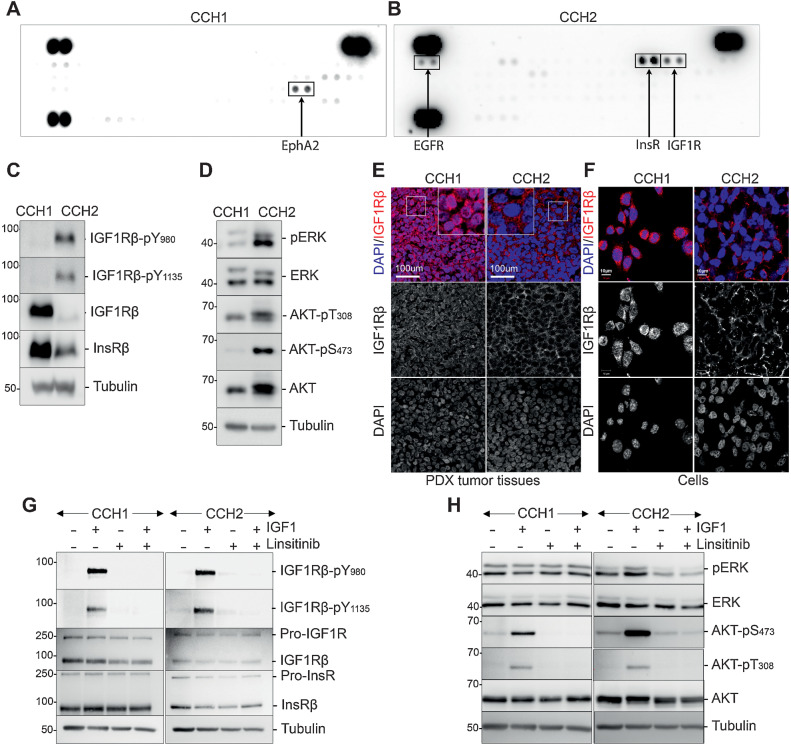
Phospho-RTK analysis of CCH1 and CCH2 cells revealed distinct RTK-activation patterns (Fig. 1A and B). The most prominently activated RTK in CCH1 cells was EPHA2, whereas the IGF receptor (IGF1R)/insulin receptor (InsR) family was the most activated in CCH2.
Figure 1.
RTK activation patterns differ between primary (CCH1) and metastatic (CCH2) tumor cells from the same patient. A, Phospho-RTK antibody arrays developed using lysates from CCH1 cells. The most prominently activated RTK EphA2 is boxed. B, Phospho-RTK antibody arrays developed using lysates from CCH2 cells. The most prominently activated RTKs (InsR, IGF1R, and EGFR) are boxed. C, Western blots on CCH1 and CCH2 cell lysates probed with the indicated antibodies, confirming differences in IGF1R activation between CCH1 and CCH2. D, Western blots on CCH1 and CCH2 cell lysates showing activation of AKT and ERK downstream of activated IGF1R only in CCH2 cells (right). E, Representative images of CCH1 and CCH2 patient-derived tumor xenograft tissues stained with antibody toward IGF1R (red) and DAPI. Individual channels are shown below the merged images. IGF1R is nuclear in CCH1 and membrane-associated in CCH2; 6×-magnified images are inset. F, Representative images of CCH1 and CCH2 cells cultured in vitro stained with antibody toward IGF1R (red) and DAPI. Individual channels are shown below the merged images. IGF1R is nuclear in CCH1 and membrane-associated in CCH2. G, Western blots showing the effect of either IGF1 stimulation (50 ng/mL, 10 minutes) or linsitinib treatment (1 μmol/L, 2 hours) on starved CCH1 and CCH2 cells. The blots were probed with the indicated antibodies and show that both CCH1 and CCH2 can be stimulated by high levels of exogenous IGF1. H, Western blots showing the effect of either IGF1 stimulation (50 ng/mL, 10 minutes) or linsitinib treatment (1 μmol/L, 2 hours) on starved CCH1 and CCH2 cells. The blots were probed with the indicated antibodies and show that exogenous IGF1 stimulates and linsitinib inhibits AKT phosphorylation in both CCH1 and CCH2, but ERK phosphorylation is only modulated in CCH2 cells.
Tumor cells were analyzed on Western blots using two phospho-IGF1Rß antibodies (pY1135/1136 and pY980). CCH2 cells showed strong IGF1Rß(Y980) phosphorylation and higher Y1135/1136 phosphorylation than CCH1 cells. Total IGF1Rß was significantly higher in CCH1 cells than in CCH2 cells (Fig. 1C).
In the canonical IGF1R pathway IGFs bind to the membrane-bound receptor leading to trans-autophosphorylation, followed by activation of the PI3K-AKT and the RAS–MAPK–ERK pathways. In Western blots, CCH2 cells exhibited higher phospho-ERK and phospho-Akt levels than CCH1 and had more total AKT than CCH1 cells, whereas the levels of total ERK were comparable in both samples (Fig. 1D).
EphA2 phosphorylation at Y772 (activation loop), S897 (ligand-independent mode of signaling), and total EphA2 levels were higher in CCH1 than in CCH2 cells (Supplementary Fig. S1C).
Distinct IGF1R signaling pathways in matched primary and metastatic tumor samples
The IGF1R pathway has been implicated in EwS tumor cell growth, hence we focused on IGF1R signaling. Immunostaining in tumor tissue revealed that IGF1R was localized to the membrane in CCH2 but was predominantly nuclear in CCH1 (Fig. 1E). Similar subcellular localization of IGF1R was seen in tumor cells cultured in vitro (Fig. 1F).
To evaluate the functioning of the IGF1R pathway, cells were serum-starved then stimulated with IGF1. IGF1 stimulated, and the IGF1R kinase inhibitor linsitinib (OSI-906) inhibited, IGF1R phosphorylation in both CCH1 and CCH2 (Fig. 1G). IGF1R remained predominantly nuclear upon either acute IGF1 or linsitinib treatment of CCH1 cells (Supplementary Fig. S1D). In CCH2 cells, acute IGF1 stimulation led to some internalization of IGF1R including to the nucleus, whereas IGF1R remained extra-nuclear in linsitinib treated CCH2 cells (Supplementary Fig. S1D).
AKT phosphorylation was stimulated by IGF1 and inhibited by linsitinib in both CCH1 and CCH2 (Fig. 1H) indicative of a functional IGF1 response. Basal levels of phospho-ERK were high in both CCH1 and CCH2 cells upon starvation, but they differed in their responses to IGF1 and linsitinib: IGF1 modestly upregulated pERK in CCH2 but not in CCH1 (Fig. 1H). Linsitinib lowered levels of pERK in CCH2, but there was no significant change in CCH1 cells.
CCH2 cells with membrane-associated IGF1R (mIGF1R) display higher levels of replication stress
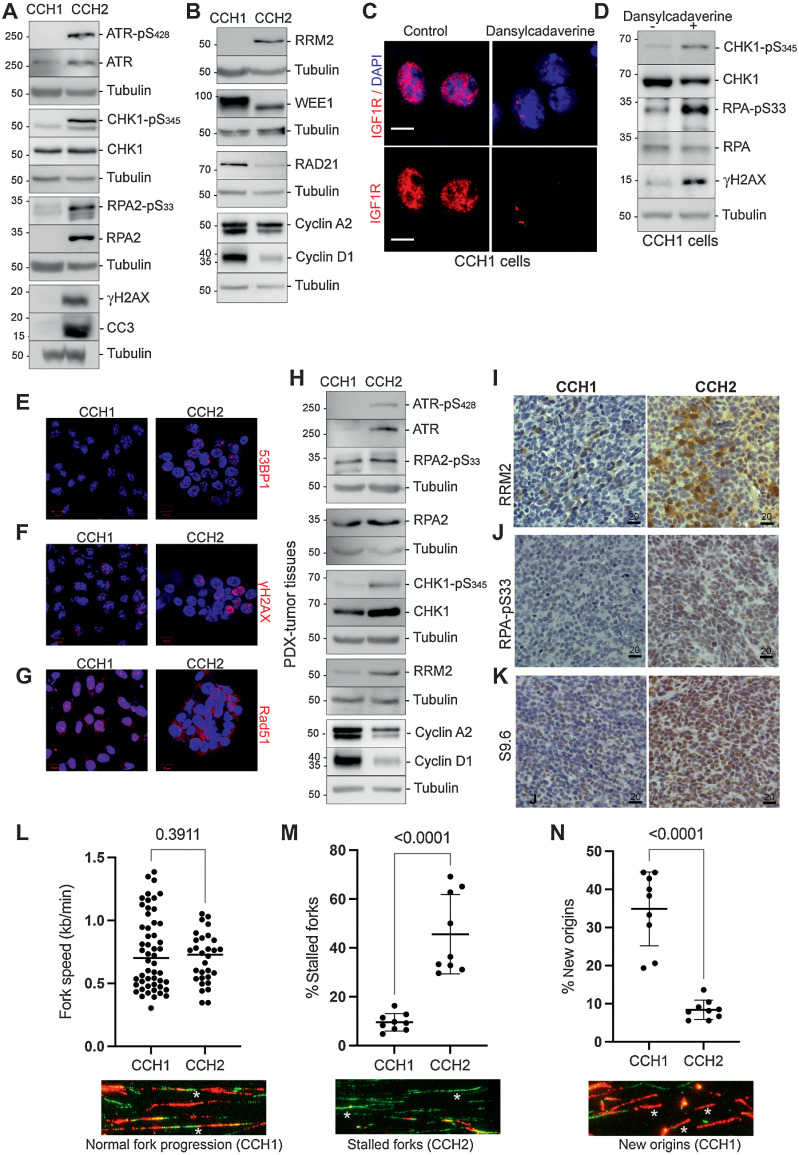
Because nIGF1R is known to protect cells from replication-fork stalling (23), we probed for markers of replication stress in CCH1 and CCH2 Western blots (Fig. 2A). Phospho-ATR and phospho-CHK1 levels were higher in CCH2 than in CCH1. Constitutive activation of the ATR–CHK1 pathway prevents the excessive accumulation of ssDNA and the consequent exhaustion of ssDNA binding protein replication protein A (RPA), which could lead to double-strand breaks and collapsed replication-forks (24). Ser33-phosphorylation of the RPA2 subunit (by ATR) minimizes ssDNA generation under conditions of replication stress by redirecting RPA activities from DNA-replication to DNA-repair (25, 26). CCH2 cells have higher levels of RPA2 (phospho-S33 and total) than CCH1. In addition, elevated levels of γH2AX and the apoptosis marker cleaved caspase-3 (CC3) were detected in CCH2 cells.
Figure 2.
CCH2 cells and tumor tissue have high levels of replication stress, whereas CCH1 cells have fewer stalled replication forks. A, Replication stress response pathway markers were evaluated using Western blots. Phosphorylation levels of ATR, CHK1, and RPA were negligible in CCH1 cells but high in CCH2 cells (20 μg protein lysates loaded). Further, γH2AX and cleaved caspase 3 (CC3) were also higher in CCH2 cells. B, Western blots show that RRM2 levels were high in CCH2 cells. In contrast, WEE1 and RAD21 were present at higher levels in CCH1 cells. CCH1 cells had higher levels of the proliferation markers cyclin A2 and cyclin D1. C, nIGF1R is depleted in CCH1 cells treated with 300 μmol/L dansylcadaverine for 4 hours. D, Levels of replication stress are elevated by nIGF1R depletion in CCH1 cells as measured by Western blots probed for phospho-CHK1, p-RPA, and γH2AX (60 μg protein lysates were loaded on the gel). E, Representative immunostaining for 53BP1 (red) in CCH1 and CCH2 cells shows more foci in CCH2. F, Representative immunostaining for γH2AX (red) in CCH1 and CCH2 cells. Twenty-eight percent (±7.3%) of CCH1 cells had more than 10 γH2AX foci per nucleus compared with 42.9% (±2.6%) for CCH2 cells. White asterisks indicate the pan-nuclear γH2AX seen in CCH2 cells. G, Immunostaining for RAD51 (red) in CCH1 and CCH2 cells shows nuclear RAD51 in CCH1 and cytoplasmic RAD51 in CCH2 cells. H, Western blot on CCH1 and CCH2 tumor tissue probed with antibodies toward the replication stress markers indicated show that CCH2 tumor tissue has elevated markers of replication stress and lower levels of proliferation markers (cyclins A2 and D1) than in CCH1. I, IHC on CCH1 and CCH2 tumor tissue stained with anti-RRM2 antibody shows elevated RRM2 in CCH2 present in discrete patches. J, IHC on CCH1 and CCH2 tumor tissue stained with anti-pRPA antibody shows elevated pRPA in CCH2. K, IHC on CCH1 and CCH2 tumor tissue stained with anti-S9.6 antibody shows more R-loops in CCH2. L, Fork speed measurement in the DNA fiber spreading assay indicate that CCH1 and CCH2 cells have comparable DNA polymerase progression. Speed of replication was estimated by measuring the lengths of fiber tracts (green, IdU; red, BrdU) and a reference of 2.59 kb per μmol/L. Data represent the mean ± SD, and significance was evaluated by an unpaired t test. A representative image of normal fork progression is shown below the graph and indicated by an asterisk. M, The relative frequency of stalled forks was determined as a percentage of all replication structures scored and was significantly higher in CCH2 cells. Data represent the mean ± SD, and significance was evaluated by an unpaired t test. A representative image of stalled forks (green tract only) is shown below the graph and indicated by asterisks; an additional region is shown in Supplementary Fig. S3A. N, The relative frequency of new origins was determined as a percentage of all replication structures scored and was significantly higher in CCH1 cells. Data represent the mean ± SD, and significance was evaluated by an unpaired t test. A representative image of new origins (red tract only) is shown below the graph and indicated by asterisks; an additional region is shown in Supplementary Fig. S3A.
We investigated the cell-cycle kinetics of CCH1 and CCH2 by probing for markers of proliferating cells and flow cytometry. CCH1 cells had a shorter doubling time than CCH2 cells (Supplementary Fig. S2). Levels of the proliferating cell markers cyclin D1 and cyclin A2 were higher in CCH1 than in CCH2 (Fig. 2B). The percentage of cells in S-phase measured using PI-staining was higher in CCH1 cells than in CCH2 (Supplementary Fig. S2). We also used double-staining with BrDU and EdU to establish the duration of the S-phase. CCH1 cells had shorter S-phase duration that CCH2 cells (Supplementary Fig. S2).
Recent studies implicated the IGF1–IGF1R pathway in transcriptional regulation of RRM2B [the ribonucleotide reductase (RNR) regulatory TP53 inducible subunit M2B, henceforth referred to as RRM2; ref. 27]. Accordingly, CCH2 cells with mIGF1R had higher levels of RRM2 than CCH1 (where RRM2 was barely detectable; Fig. 2B). Posttranscriptional regulation of RRM2 occurs through attenuation of RRM2 degradation by ATR–CHK1 activation and by the negative regulation of RRM2 accumulation by the WEE1 kinase (14). WEE1 is present at higher level in CCH1 cells than in CCH2 cells (Fig. 2B). Hence, multiple molecular mechanisms could contribute to the elevation of RRM2 in CCH2. CCH1 also has higher levels of RAD21 (Fig. 2B), consistent with a previous report showing that RAD21 dampens replication stress and promotes proliferation in EwS cells (28).
Supporting an association between nIGF1R and lower replication stress levels, treatment of CCH1 cells with the clathrin inhibitor dansylcadaverine (29) depletes nIGF1R (Fig. 2C) and increases levels of the replication stress markers pCHK1, pRPA, and γH2AX (Fig. 2D), while also reducing the percentage of cells in S-phase (Supplementary Fig. S2A).
CCH2 cells have discrete nuclear 53BP1 foci whereas CCH1 cells have rare 53BP1 foci (Fig. 2E). 53BP1 foci mark DNA-breaks, protecting DNA lesions generated by mitotic transmission of chromosomes under replication stress (30). The percentage of cells with nuclear γH2AX staining (either discrete foci or pan-nuclear) was higher in CCH2 than in CCH1 (Fig. 2F). CCH1 cells had prominent nuclear RAD51 foci, whereas RAD51 was cytoplasmic in CCH2 cells (Fig. 2G). RAD51 relocalizes into the nucleus upon DNA-damage (31) and contributes to the restart of stalled replication-forks (32).
pATR, pCHK1, pRPA, and RRM2 levels were also higher in CCH2 tumor tissue, while the proliferating cell markers CYCLIN A2 and D1 were higher in CCH1 tumor tissue (Fig. 2H). Further, immunostaining showed more RRM2 (Fig. 2I) and more pRPA (Fig. 2J) in CCH2 tumor tissue relative to CCH1. RRM2 staining in tissue samples was patchy suggestive of heterogeneity in RNR activity levels within the tumor.
High replication stress in EwS cells is accompanied by an accumulation of R-loops (DNA-RNA hybrid and displaced ssDNA that occur when replication-forks collide with the transcriptional machinery; ref. 12). CCH2 tissue was much more strongly positive for R-loops than CCH1 when probed with the RNA:DNA hybrid-specific antibody S9.6 (Fig. 2K).
To evaluate DNA replication dynamics in CCH1 and CCH2 cells, we used the DNA fiber assay, which measures nucleotide incorporation into nascent DNA during replication. CCH1 and CCH2 cells were sequentially incubated with IdU and CldU to allow incorporation at replication-forks. The speed of DNA-replication (ratio of lengths of nascent replication-tracts) was comparable between CCH1 and CCH2 cells (Fig. 2L). The percentage of stalled forks in CCH2 were greater than in CCH1 (Fig. 2M; Supplementary Fig. S3A) indicative of higher levels of replication stress, whereas CCH1 had a greater percentage of new origins firing (Fig. 2N; Supplementary Fig. S3A). Together this single-cell analysis reports higher levels of replication stress in CCH2 relative to CCH1.
Replication stress and IGF1R localization vary among EwS tumors
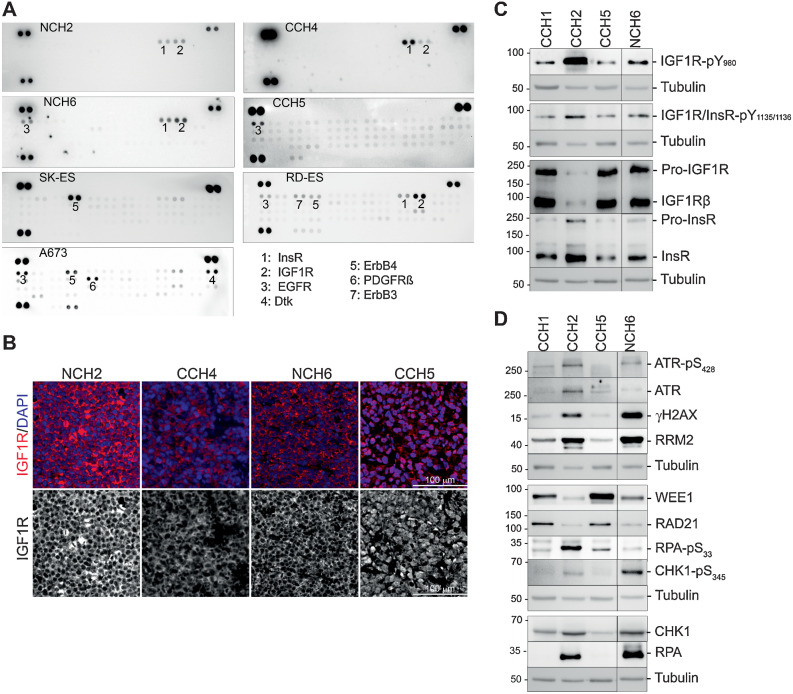
To more broadly sample IGF1R signaling and RSR pathways in EwS, we examined four additional PDX (Supplementary Table S1), and three well-studied EwS cell lines A673, RD-ES, and SK-ES. Cells cultured from the PDX stained positive for membranous CD99 (Supplementary Fig. S3). Phospho-RTK analysis (Fig. 3A) showed that the IGF1R/InsR family was the most frequently activated RTK (CCH4, NCH2, NCH6, RD-ES), followed by EGFR family RTKs (NCH6, CCH5, RD-ES, A673). In patient-derived samples, we only detected EGFR phosphorylation, whereas in the established cell lines phosphorylation of ErbB4 or ErbB3 was also detected. Interestingly, all three established cell lines had more phosphorylated RTKs than the patient-derived samples.
Figure 3.
A survey of RTK phosphorylation patterns and IGF1R localization in patient EwS tumor cells and commonly used EwS cell lines. A, Phospho-RTK antibody arrays developed using lysates from patient-derived NCH2, CCH4, NCH6, and CCH5 cells, and from the cell lines A673, RD-ES, and SK-ES. The most prominently activated RTKs are numbered. B, Immunofluorescence images of PDX tumor tissue stained with anti-IGF1R antibody (red) and DAPI. The IGF1R channel is shown below. Only CCH5 tissue has exclusively nuclear IGF1R. C, Two representatives each of EwS cells with nIGF1R (CCH1, CCH5) and mIGF1R (CCH2, NCH6) were selected for further analyses. Western blots show IGF1R phosphorylation patterns that match the phospho-RTK arrays. D, Evaluation of replication stress response markers in CCH1, CCH5, CCH2, and NCH6 cells confirms elevated replication stress in cells with mIGF1R.
We next used immunofluorescence to probe IGF1R subcellular localization in PDX tumor tissues: membranous IGF1Rß was present in NCH2, NCH6, and CCH4, whereas CCH5 had exclusively nuclear IGF1Rß (Fig. 3B) as previously seen in CCH1.
To further evaluate the correlation between IGF1R localization and replication stress, we selected CCH5 (with nIGF1R) and NCH6 (with mIGF1R) for comparison with CCH1 and CCH2. In Western blots pIGF1R (Fig. 3C) and key markers of replication stress (pATR, pCHK1, γH2AX, and RRM2) were elevated in cells with mIGF1R (Fig. 3D). However, although the level of RPA was higher in cells with mIGF1R, pRPA was not elevated in NCH6 cells. Cells with nIGF1R had higher levels of WEE1 and RAD21. CCH5 cells had a shorter doubling time, higher levels of the proliferation markers cyclin A2 and D1, more cells in S-phase, and shorter S-phase duration than NCH6 cells (Supplementary Fig. S2), paralleling previous observations made with CCH1 and CCH2.
None of the EwS cell lines (A673, RD-ES, SK-ES) had nIGF1R (Supplementary Fig. S4D). Total IGF1Rß was very low in A673, and only RD-ES showed some IGF1Rß phosphorylation (Supplementary Fig. S4A). The ATR–CHK1 pathway was activated in all cell-lines indicating some degree of replication stress, but pRPA was highest in RD-ES. γH2AX levels were higher in RD-ES and SK-ES. Comparable RRM2 was present in all cell-lines (Supplementary Figs. S4B and S4C). Overall, the cell-lines are most like CCH2 and NCH6 cells with classical IGF1R signaling and the presence of replication stress.
Nuclear IGF-1R (nIGF1R) signaling promotes ubiquitination of PCNA
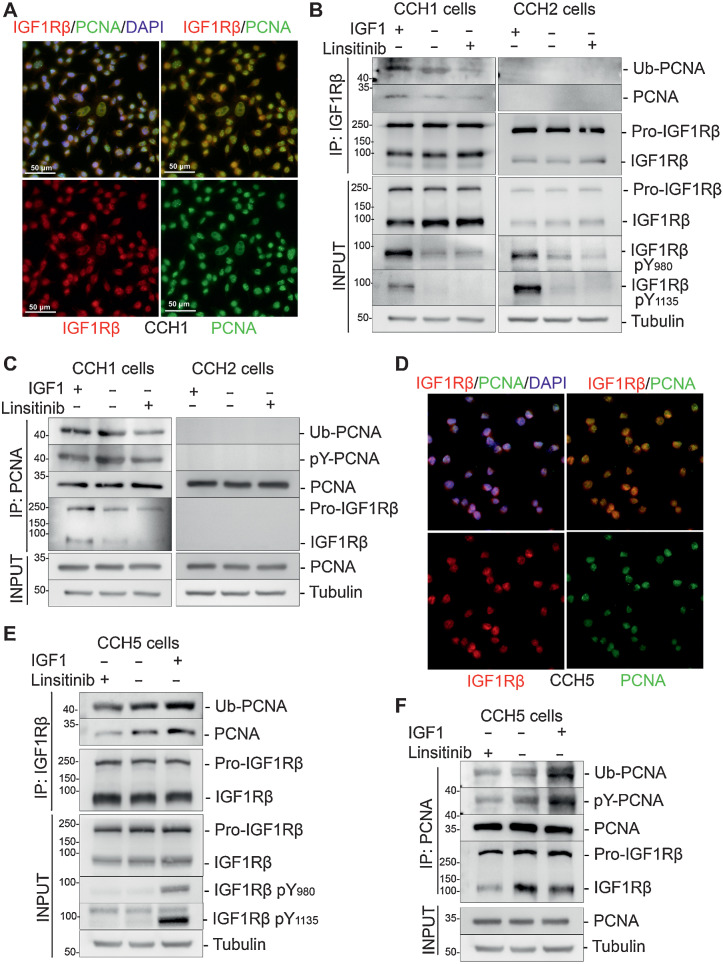
nIGF1R is known to promote DNA-Damage Tolerance (DDT), a process that facilitates genome duplication and protects from deleterious double-stranded DNA-breaks in the presence of replication perturbations (33). Mechanistically DDT involves ubiquitination of proliferating cell nuclear antigen (PCNA; ref. 23). IGF1R and PCNA co-localize in the nucleus in CCH1 cells (Fig. 4A). To determine whether nIGF1R in CCH1 cells correlates with PCNA mono-ubiquitination and phosphorylation, we treated cells with either IGF-1 or linsitinib, followed by immunoprecipitation of IGF1R (Fig. 4B). PCNA co-immunoprecipitated with IGF1R in CCH1 but not in CCH2 cells. Probing with anti-ubiquityl-PCNA and anti-phosphotyrosine showed that IGF-1 treatment increased the levels of PCNA ubiquitination and phosphorylation in CCH1 cells, whereasa there was no ubiquitinated-PCNA evident in CCH2 cells. Furthermore, linsitinib treatment inhibited PCNA–IGF1R interaction in CCH1 cells. Immunoprecipitation of PCNA followed by probing with anti-IGF1R and anti-ubiquitin antibodies similarly showed physical interaction between phospho-IGF1R and PCNA, as well as phosphorylation and ubiquitination of PCNA in CCH1, but not in CCH2, cells (Fig. 4C).
Figure 4.
Nuclear IGF1R in CCH1 and CCH5 cells binds to and promotes ubiquitination of PCNA. A, Nuclear colocalization of IGF1R (red) and PCNA (green) in CCH1 cells cultured in vitro. B, Interaction between IGF1R with PCNA was analyzed by co-immunoprecipitation of PCNA with immobilized IGF1R in control, 50 ng/mL IGF1-treated and 1 μmol/L linsitinib-treated (6 hours) CCH1 and CCH2 cells. Only CCH1 cells showed co-immunoprecipitation of IGF1R and PCNA. The blots were reprobed with anti-ubiquityl-PCNA(Lys164) antibody, which revealed a 43-kDa band corresponding to mono-ubiquitinated PCNA in the CCH1 blots. IGF1 stimulated, and linsitinib inhibited, PCNA:IGF1R interaction.C, Interaction between IGF1R with PCNA was analyzed by co-immunoprecipitation of IGF1R with immobilized PCNA in control, 50 ng/mL IGF1-treated, and 1 μmol/L linsitinib-treated (6 hours) CCH1 and CCH2 cells. Only CCH1 cells showed co-immunoprecipitation of IGF1R and PCNA. The blots were reprobed with anti-ubiquitin antibody, which revealed a 43-kDa band corresponding to mono-ubiquitinated PCNA in the CCH1 blots. IGF1 stimulated, and linsitinib inhibited, PCNA:IGF1R interaction. D, Nuclear colocalization of IGF1R (red) and PCNA (green) in CCH5 cells cultured in vitro. E, Co-immunoprecipitation of PCNA with immobilized IGF1R in CCH5 cells. Co-immunoprecipitated PCNA is mono-ubiquitinated and tyrosine phosphorylated as determined by probing with anti-ubiquityl-PCNA(Lys164) and anti-phospho-tyrosine antibodies, respectively. The input used for immunoprecipitations are shown below. F, Co-immunoprecipitation of IGF1R with immobilized PCNA in CCH5 cells. Immunoprecipitated PCNA is mono-ubiquitinated and tyrosine phosphorylated as determined by probing with anti-ubiquityl-PCNA(Lys164) and anti-phospho-tyrosine antibodies, respectively. The inputs used for immunoprecipitations are shown below.
IGF1R in CCH5 cells colocalized in the nucleus with PCNA (Fig. 4D), whereas PCNA and IGF1R do not colocalize in CCH2 and NCH6 cells (Supplementary Fig. S5). Pull-down experiments using either PCNA or IGF1Rß as bait confirmed a physical interaction between IGF1R and PCNA in CCH5 cells (Fig. 4E and F). PCNA was both phosphorylated and mono-ubiquitinated in the IGF1R–PCNA complex, and this was promoted by IGF1 treatment and inhibited by linsitinib treatment. Together these data support a model in which nIGF1R can potentiate DDT in CCH1 and CCH5 cells through interaction with ubiquitinated PCNA.
Effects of IGF1R or WEE1 inhibition differ between EwS cells with nIGF1R and mIGF1R
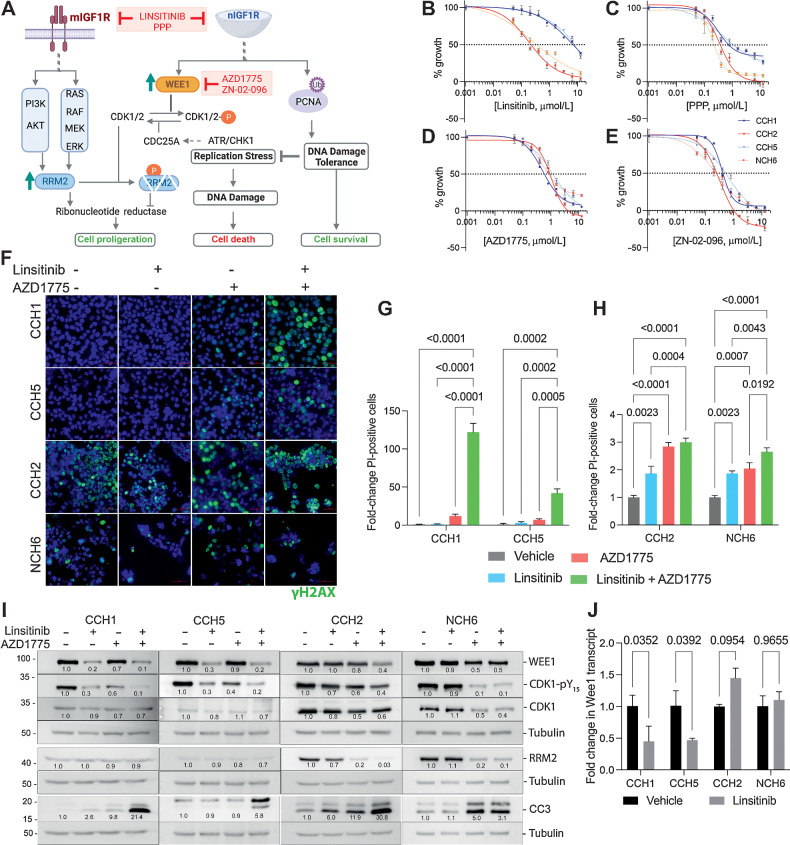
The data presented thus far are consistent with a model (schematized in Fig. 5A), in which cells with active DDT promoted by nIGF1R have low basal replication stress. On the other hand, cells with mIGF1R have high replication stress and high levels of RRM2, which could be a consequence of both classical IGF1R signaling (27) and activation of the ATR–CHK1 pathway.
Figure 5.
EwS cells can differ in their sensitivity to inhibitors of IGF1R signaling and WEE1. A, Schematic showing the proposed differences in the mechanisms through which EwS cells with either nuclear or membrane-associated IGF1R are regulated. Cells with mIGF1R activate the classical AKT and RAS pathways and upregulate RRM2 transcription. EwS cells with nIGF1R have higher levels of the WEE1 kinase, which in turn phosphorylates and inactivates CDK1, thus protecting RRM2 from degradation. High levels of RRM2 promote cell proliferation. nIGF1R also ubiquitinates and phosphorylates PCNA and promotes DNA damage tolerance (DDT), thus suppressing replication stress and the consequent DNA damage and cell death. Hence, nIGF1R can promote cell survival. Pharmacologic agents used in this study are indicated in red (schematic created with BioRender.com). B, Dose–response curves generated using CCH1 (blue), CCH5 (dotted blue), NCH6 (dotted red), and CCH2 (red) cells. The WST-8 assay was performed after 48 hours of linsitinib treatment. All experiments were conducted over three independent times with three technical replicates in each experiment. Representative data sets are shown here as the mean ± SD for each dose. Concentration of inhibitor that causes GI50 was calculated by plotting growth (%) versus drug concentration and using the formula 100 × (T − T0)/(C − T0), where optical density of the test well after 48 hours is T, optical density at time 0 is T0, and control (vehicle) optical density is C. GI50 values calculated using PRISM: CCH1 6.8 μmol/L, CCH2 0.2 μmol/L, CCH5 7.7 μmol/L, NCH6 0.3 μmol/L. C, Analyses conducted as in B for PPP. GI50 values calculated using PRISM: CCH1 0.8 μmol/L, CCH2 0.4 μmol/L, CCH5 1.1 μmol/L, NCH6 0.2 μmol/L. D, Analyses conducted as in B for AZD1775. GI50 values calculated using PRISM: CCH1 0.6 μmol/L, CCH2 0.9 μmol/L, CCH5 1.2 μmol/L, NCH6 0.8 μmol/L. E, Analyses conducted as in B for ZN-02–096. GI50 values calculated using PRISM: CCH1 0.4 μmol/L, CCH2 0.25 μmol/L, CCH5 0.5 μmol/L, NCH6 0.2 μmol/L. F, Immunofluorescence staining for γH2AX on CCH1, CCH5, CCH2, and NCH6 cells treated with linsitinib (5 μmol/L), AZD1775 (1 μmol/L), or a combination of linsitinib and AZD1775. CCH1 and CCH5 cells did not exhibit a large increase in DNA damage upon treatment with either agent alone. However, a combination of linsitinib and AZD1775 led to a dramatic induction of DNA damage. CCH2 and NCH6 cells had higher basal levels of γH2AX that remained at similar levels upon treatment with the indicated pharmacologic agents. G, Quantification of propidium-iodide positive cells with the linsitinib (5 μmol/L), AZD1775 (1 μmol/L), or linsitinib + AZD1775 treatments (24 hours) of CCH1 and CCH5 cells. Mean and SD are shown in each panel. H, Quantification of propidium iodide–positive cells with the linsitinib (5 μmol/L), AZD1775 (1 μmol/L), or linsitinib + AZD1775 treatments (48 hours) of CCH2 and NCH6 cells. Mean and SD are shown in each panel. I, Western blots showing the different effect of linsitinib (5 μmol/L), AZD1775 (5 μmol/L), or linsitinib + AZD1775 treatment for 24 hours on WEE1, RRM2, and cleaved caspase 3 (CC3) levels in EwS cells with nIGF1R (CCH1, CCH5) and mIGF1R (CCH2, NCH6) cells. J, qPCR analyses of Wee1 transcript levels upon linsitinib (5 μmol/L) treatment for 24 hours (two biological replicates) show a reduction in cells with nIGF1R.
We next evaluated the effect of IGF1R kinase inhibition on the proliferation and survival of representatives of both types of PDX cells: CCH5, CCH1 (nIGF1R), and CCH2, NCH6 (mIGF1R). Linsitinib was a more potent growth inhibitor of CCH2 and NCH6 cells than either CCH5 or CCH1 (Fig. 5B). Picropodophyllin (PPP) that selectively targets IGF1R over InsR (34) also failed to achieve total growth inhibition in EwS cells with nIGF1R (Fig. 5C).
Inhibition of IGF1R can induce replication stress by affecting RRM2 expression (via AKT, MEK/ERK and JUN; ref. 27). Further, the WEE1 kinase was identified as part of a “backup” pathway that protected cells from RRM2 depletion by preventing RRM2 degradation (27). Interestingly, the WEE1 kinase inhibitor AZD1775 (35) and the selective WEE1 degrader ZN-02–096 (36) exhibited similarly potent cell-growth inhibition in CCH1, CCH2, CCH5, and NCH6 (Fig. 5D and E).
Linsitinib caused negligible increase in γH2AX in CCH1 and CCH5, AZD1775 caused a small increase in γH2AX, and a combination of linsitinib and AZD1775 led to a significant increase in γH2AX-positive cells (Fig. 5F). In contrast, CCH2 and NCH6 cells had high basal levels of γH2AX, which did not increase substantially with any treatment (Fig. 5F). These results correlated with propidium-iodide (PI) and CC3 staining based evaluation of cell survival: linsitinib and AZD1775 were each independently able to induce significant cell-death in CCH2 and NCH6 cells, only slightly elevated by combination treatment (Fig. 5G–I). Linsitinib or AZD1775 had minimal effect on CCH1 or CCH5 cells, whereas the combination of linsitinib and AZD1775 led to a large increase in PI-positive cells. Flow cytometry analyses performed on CCH1 and CCH2 cells further confirmed these distinctions (Supplementary Fig. S6).
In Western blots, linsitinib treatment lowered the levels of WEE1 protein in CCH1 and CCH5 (but not in CCH2 or NCH6; Fig. 5I). WEE1 phosphorylates CDK1 at Y15 and the levels of CDK1–pY15 were similarly lowered by linsitinib treatment of CCH1 or CCH5, and by WEE1 inhibition with AZD1775 in all cells tested. Because this modulation of WEE1 levels by linsitinib appears to be restricted to cells with nIGF1R, and nIGF1R can regulate transcription (37, 38), we examined the effect of linsitinib treatment on the levels of WEE1 transcript. Lower levels of transcript were present in CCH1 and CCH5 cells treated with linsitinib, but no reduction in similarly treated CCH2 or NCH6 cells was observed (Fig. 5J).
Linsitinib treatment reduced RRM2 protein levels in CCH2 but had no effect on NCH6, CCH1, or CCH5 (Fig. 5I). The lack of effect in NCH6 was unanticipated given the known positive regulation of RRM2 by the classical IGF1R pathway (27). WEE1 kinase inhibition also lowered RRM2 levels in CCH2 and NCH6 cells, likely through increased degradation. Curiously, little effect was seen upon WEE1 inhibition on the very low basal levels of RRM2 in CCH1 or CCH5.
Combined IGF1R and WEE1 inhibition leads to tumor regression
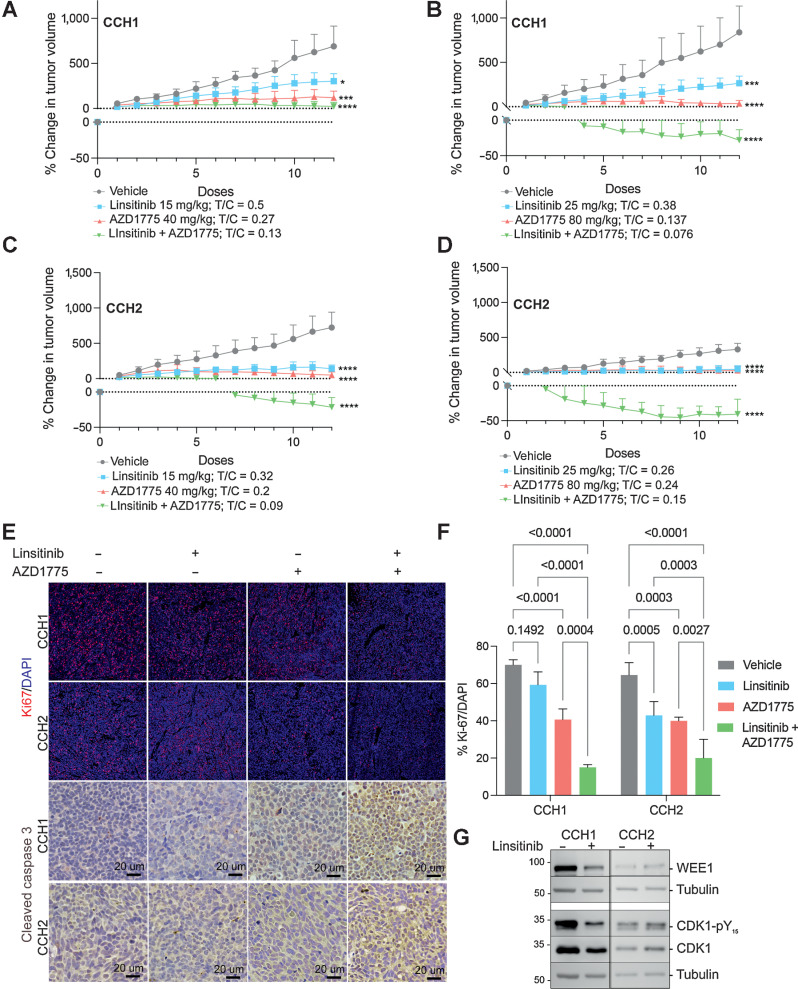
To further evaluate sensitivity to IGF1R and WEE1 inhibition, we used CCH1 (Fig. 6A and B) and CCH2 (Fig. 6C and D) subcutaneous xenografts (Fig. 6). Tumors were treated 5 days a week with linsitinib [either 15 mg/kg (Fig. 6A and C) or 25 mg/kg (Fig. 6B and D)], starting when tumor volume reached approximately 200 mm3; 25 mg/kg is known to inhibit tumor growth without elevating blood glucose levels in other xenograft models (39). At both doses, CCH2 tumors were more sensitive to treatment than CCH1 tumors. In parallel experiments, we assessed the effect of the WEE1 inhibitor AZD1775 at either 80 or 40 mg/kg 5 days a week. WEE1 inhibition largely arrested tumor growth of both CCH1 and CCH2 tumors. Treatment with a combination of linsitinib and AZD1775 led to significant tumor regression for both CCH1 and CCH2 xenografts.
Figure 6.
Effectiveness of linsitinib, AZD1775, and a combination of linsitinib and AZD1775 in nIGF1R and mIGF1R PDXs. A and B, Growth rates of CCH1 tumors treated with either vehicle or the indicated doses of linsitinib and AZD1775 are shown by plotting % change in tumor volume over time. Tumors were treated daily for 5 days/week. The T/C value (mean volume of treated tumor after 12 doses/mean volume of vehicle-treated tumor after 12 doses) is shown. Each data point is the mean and SD of 4 to 5 mice per group. C and D, Growth rates of CCH2 tumors treated with either vehicle or the indicated doses of linsitinib and AZD1775 are shown by plotting % change in tumor volume over time. Tumors were treated daily for 5 days/week. The T/C value (mean volume of treated tumor after 12 doses/mean volume of vehicle-treated tumor after 12 doses) is shown. Each data point is the mean and SD of 4 to 5 mice per group. E, Tumor sections stained for the proliferation marker Ki67 (red) and the apoptosis marker CC3 (DAB-brown). F, Quantification of Ki67-positive cells per tumor section. The data are shown as the mean and SD. Each data point represents the average of four sections per mouse and 3 to 4 mice per treatment group. G, Western blot analysis of tumor tissue confirms that linsitinib treatment lowers WEE1 and pCDK1 levels in CCH1 tumors but not in CCH2 tumors.
An insignificant decrease in cell proliferation upon linsitinib treatment of CCH1 tumors was observed (Ki-67 positive cells; Fig. 6E and F), whereas CCH2 tumors displayed an over 20% decrease in Ki67-positive cells. AZD1775 alone and in combination with linsitinib were similarly effective in reducing tumor cell proliferation in both tumors. Staining for the apoptotic marker cleaved caspase-3 showed elevated cell-death in CCH1 and CCH2 tumors with combination treatment (Fig. 6E).
In Western blot analysis of tumor tissues (Fig. 6G), linsitinib treatment reduced levels of WEE1 in CCH1 cells but not in CCH2 cells. The level of the downstream effector CDK1-pY was also lowered upon linsitinib-treatment of CCH1 but not CCH2, corroborating in vitro findings.
In summary, in vivo studies demonstrate that a tumor with nIGF1R is less sensitive to linsitinib-treatment compared with a mIGF1R-containing tumor, and that a combination of IGF1R and WEE1 inhibition can cause tumor regression in both classes of EwS tumors.
Discussion
Herein we demonstrate that IGF1R-signaling and the replication stress response pathway can be utilized to place EwS tumors into two groups. In one, IGF1R is largely nuclear, replication stress and RRM2 levels are low, whereas WEE1 and RAD21 levels are high. The other group has predominantly membrane-associated IGF1R signaling, high replication stress and RRM2, and low WEE1 and RAD21. These distinctions between EwS tumors could contribute to distinct therapeutic vulnerabilities and lay the groundwork for multitargeted-agent treatment strategies based upon histopathologic biomarkers.
In the primary-metastatic pair of tumors examined, the expression level of IGF1R went from being high and largely nuclear (primary tumor), to being low, phosphorylated, and primarily membrane-associated (CCH2 metastatic tumor). In the wider panel of PDX-derived samples, there was no apparent correlation between IGF1R localization and disease stage. Some studies suggest that high nIGF1R predicts good prognoses and treatment responsiveness (40, 41), whereas others predict a worse outcome (42, 43). nIGF1R is linked to increased IGF-induced proliferation, and enhanced tumorigenicity (44–46), can promote both transcription and DDT (23). DDT in turn permits the completion of replication without fork-stalling, thus supporting the survival of highly proliferative tumor cells (Fig. 5A). CCH1 and CCH5 cells have nIGF1R and higher basal levels of ubiquitinated-PCNA, consistent with the existence of a mechanism to bypass the development of replication stress. These cells with nIGF1R also have higher levels of RAD21 a suppressor of replication stress in EwS (28). Accordingly, these highly proliferative cells have low levels of basal replication stress. However, although inhibition of IGF1R-kinase activity with linsitinib inhibits DDT, it did not induce either DNA-damage or cell-death in cells with nIGF1R. In contrast, DNA-damage and cell-death result from linsitinib-treatment of cells with classical IGF1R signaling. Similarly, relative insensitivity to linsitinib is seen in vivo towards CCH1 (nIGF1R) xenografts versus CCH2.
Replication stress is a widely recognized hallmark of EwS (11, 12) and other cancer cells (47, 48). In EwS it is attributed to elevation of transcription and accelerated entry into S-phase driven by the EWS–FLI1 fusion protein, and the formation of R-loops (12). Indeed, the capability to resolve R-loops and reduce replication stress is a prerequisite for the stable expression of EWSR1–FLI1 in primary cells (49). Therapeutically targeting replication stress is an area of active investigation and can take two routes: depleting cellular resources essential for DNA replication and/or by over-riding checkpoints to push cells with unresolved replication stress into the cell-cycle, thus inducing catastrophic DNA-damage (50, 51). Most functional studies rely on established EwS cell-lines because of the rarity of the disease and the difficulty in establishing EwS PDX. Moreover, differences in tumor cells through the course of disease are rarely studied since repeat biopsies on patients with EwS are uncommon. Our results show that tumors from the same patient at different stages of disease can have distinct therapeutic vulnerabilities and highlight the inherent challenge in selecting targeted therapeutics based on biomarkers derived from diagnostic biopsies.
In the matched primary-metastatic pair of tumors analyzed here, the metastatic tumor (CCH2) had higher levels of replication stress and low DDT. Although there is some precedent for the observation that the DDT mechanism might be lost in metastatic lesions, and in tumors with poor response to neoadjuvant treatment (52), our data set does not clearly link primary tumors with DDT and low replication stress.
Cell survival under conditions of replication stress is supported by a large pool of nucleotides. The rate-limiting step in deoxyribonucleotide synthesis is catalyzed by RNR (composed of subunits RRM1 and RRM2). RRM2 expression is associated with metastatic disease at EwS diagnosis, occurrence of metastatic/local relapse, and correlates with poor patient outcome (53). RRM2 levels are higher in CCH2 and NCH6 than in CCH1 or CCH5. Two mechanisms, both active in CCH2 and NCH6, could contribute to this: IGF1R/AKT/ERK signaling upregulates RRM2 transcription (27), and the ATR/CHK1/WEE1 pathway prevents RRM2 degradation (Fig. 5A; ref. 14). Curiously levels of WEE1 are higher in cells with nIGF1R, whereas the ATR/CHK1 pathway is activated in CCH2 and NCH6. Further, IGF1R inhibition appears to lower the levels of Wee1 transcript in cells with nIGF1R suggesting that nIGF1R could exert transcriptional control. nIGF1R plays a role in transcription via DNA binding and RNAPol2 recruitment (37) and through association with the transcription factor LEF1 (38). However, these have not previously been linked with IGF1R kinase activity. Although the observation that linsitinib modulates Wee1 transcription is intriguing and warrants further investigation, the possibility that increased WEE1-degradation contributes to the reduction in protein levels upon linsitinib treatment cannot be ruled out.
Inhibition of WEE1 with AZD1775 reduced RRM2 levels in mIGF1R-containing tumor cells but not in those with nIGF1R (which have low levels of RRM2). However, WEE1 also triggers G2–M arrest through inhibitory phosphorylation of CDK1 giving cells the opportunity to repair damaged DNA. Hence regardless of the effect on RRM2 levels, AZD1775 can also lead to high CDK1 activity allowing cells to progress through the cell cycle with unrepaired DNA damage thus leading to mitotic catastrophe (54). This mechanism likely contributes to the ability of AZD1775 to induce some cell-death of nIGF1R-containing cells. Preclinical studies using EwS cell-lines corroborate the effectiveness of WEE1 inhibition (55).
Combined WEE1- and IGF1R-inhibition is effective in both nIGF1R- and mIGF1R-containing cells, but likely through distinct mechanisms. In cells with nIGF1R, linsitinib inhibits DDT and lowers WEE1 levels. Consequently, cells could become more dependent on residual WEE1 and unable to bypass DNA-damage caused by AZD1775. In mIGF1R-containing cells WEE1 and IGF1- targeted treatments appear to have an additive effect accompanied by RRM2 depletion, CDK1 activation, and induction of DNA-damage. These observations are in keeping with the recent report of replication stress and RRM2 downregulation upon co-inhibition of IGF1R and WEE1 in breast cancer cell-lines (56). Our observations demonstrate the effectiveness of this multiple-targeted agent strategy in EwS and suggest that the combination could provide greater benefit in tumors with nIGF1R.
Here we had the unique opportunity to evaluate a matched pair of tumors from the same patient and identified differences with relevance to therapy choice. It is possible that the replication stress response and the DDT capacities of CCH1 and CCH2 evolved in a linear fashion influenced by either treatment or microenvironment. But another possibility is that they could have distinct clonal origins. Genomic analyses of matched primary-metastatic EwS tumors reported that the ancestral EwS cell could clonally diverge years before diagnosis via catastrophic genomic events (chromoplexy), and then undergo parallel evolution (57, 58). Our inability to correlate disease-stage with either IGF1R-localization or basal replication stress levels supports the possibility that the tumors evaluated here were a result of early clonal divergence.
Two conclusions with clinical relevance derive from this study. First, nuclear localization of IGF1R correlates with poor response to IGF1R-inhibition, but a combination of IGF1R- and WEE1-inhibition leads to tumor-regression regardless of IGF1R-localization. Second, given the variability among tumors and the likelihood of independent clonal evolution, biomarker analysis performed on each tumor incarnation (primary, metastatic, relapsed) will be valuable in predicting response to therapeutics. The results presented here could form the basis of a future clinical trial using the subcellular localization of IGF1R as a biomarker of response and are likely to be relevant in tumors other than EwS.
Supplementary Material
Patient tumor information
Reagents, instruments, tumor cell culture details
Western blots and quantifications
A. Bright field images of CCH1 and CCH2 cells B. CCH1 and CCH2 cells cells stained with antibody towards CD99 (green) and DAPI (blue) C. Western blots on CCH1 and CCH2 cell lysates probed with the indicated antibodies, confirming differences in EphA2 phosphorylation between CCH1 and CCH2. D. Immunofluorescence imaging of IGF1R-stained CCH1 and CCH2 cells starved, treated with IGF1 or treated with linsitinib for 2 hours. Individual channels are shown below the merged images. Nuclear IGF1R in CCH1 cells does not change localization with either treatment. IGF1 treatment induces slight internalization of IGF1R in CCH2 cells and this is inhibited by linsitinib.
Cell cycle analyses A. Dansylcadaverine-mediated nIGF1R depletion arrested CCH1 cells at the G1-phase and reduced the percentage of cells in the S-phase. B. Western blots show that Cyclin A2 and Cyclin D1 levels were high in CCH5 cells with nIGF1R relative to NCH6 with mIGF1R. C. Cell cycle analyses shows higher percentages of CCH1 and CCH5 cells in S-phase than CCH2 and NCH6 cells. D. CCH5 cells with nIGF1R divided faster, had higher percentages of S-phase cells with shorter S-phase duration than NCH6 with mIGF1R. E. EdU/BrdU double labelling-based S-phase duration analysis was carried out. Top panel shows that cells showed EdU-AF55+ and BrdU-FITC+ signals in comparison to unstained cells. F. Percentages of EdU+BrdU+ cells (CCH1, CCH5, CCH2 and NCH6 cells), at indicated time points, are shown. G.Timings of S-phase exit (red) for all cells were calculated using linear regression.
A. Representative images for DNA fiber experiments conducted on CCH1 and CCH2 cells. In each image asterisk (*) indicates a normal fork (red-green), “N” indicates a new fork (red only), and “S” indicates a stalled fork (green only).B. Brightfield and CD99-stained (green) images of patient-derived cells used in the studies reported here.
Characterization of Ewing sarcoma cell lines A673, RD-ES and SK-ESA.Western blot analysis of the IGF1R signaling pathway B. Western blot analysis of markers associated with the Replication Stress Response C. Western blot analysis of RPA phosphorylation and apoptosis measured by cleaved caspase 3 (CC3 levels).Cells stained with antibody towards IGF1R (red) and DAPI (blue) shows membranous staining in RD-ES and SK-ES cells, and very low levels in A673 cells.
Immunofluorescence images of CCH2 and NCH6 cells stained with antibodies towards IGF1R (red) and PCNA (green).
Flow cytometry analysis for propidium-iodide positive cells with the indicated treatments (48 hours). The percentage of PI-positive cells are shown in each panel.
Acknowledgments
This work was supported by NIH grant RO1 CA207068 (awarded to R.S. Hegde) and a grant from the Jeff Gordon Children's Foundation (awarded to R.S. Hegde). We thank Dr. David Milewski for providing a frozen PDX tissue sample for CCH2.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
J.G. Pressey reports other support from Zentalis, Cellectar, Gradalis, Epizyme, Eisai, Springworks, and Novartis and personal fees from General Dynamics outside the submitted work. R.S. Hegde reports grants from NIH and Jeff Gordon Foundation during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
U.K. Soni: Data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. Y. Wang: Data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. R.N. Pandey: Investigation, methodology, writing–review and editing. R. Roberts: Resources, writing–review and editing. J.G. Pressey: Resources, writing–review and editing. R.S. Hegde: Conceptualization, resources, formal analysis, supervision, funding acquisition, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Womer RB, West DC, Krailo MD, Dickman PS, Pawel BR, Grier HE, et al. Randomized controlled trial of interval-compressed chemotherapy for the treatment of localized Ewing sarcoma: a report from the Children's Oncology Group. J Clin Oncol 2012;30:4148–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Longhi A, Ferrari S, Tamburini A, Luksch R, Fagioli F, Bacci G, et al. Late effects of chemotherapy and radiotherapy in osteosarcoma and Ewing sarcoma patients: the Italian Sarcoma Group Experience (1983–2006). Cancer 2012;118:5050–9. [DOI] [PubMed] [Google Scholar]
- 3. Nagarajan R, Clohisy DR, Neglia JP, Yasui Y, Mitby PA, Sklar C, et al. Function and quality-of-life of survivors of pelvic and lower extremity osteosarcoma and Ewing's sarcoma: the Childhood Cancer Survivor Study. Br J Cancer 2004;91:1858–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leavey PJ, Mascarenhas L, Marina N, Chen Z, Krailo M, Miser J, et al. Prognostic factors for patients with Ewing sarcoma (EWS) at first recurrence following multi-modality therapy: a report from the Children's Oncology Group. Pediatr Blood Cancer 2008;51:334–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stahl M, Ranft A, Paulussen M, Bölling T, Vieth V, Bielack S, et al. Risk of recurrence and survival after relapse in patients with Ewing sarcoma. Pediatr Blood Cancer 2011;57:549–53. [DOI] [PubMed] [Google Scholar]
- 6. Shankar AG, Ashley S, Craft AW, Pinkerton CR. Outcome after relapse in an unselected cohort of children and adolescents with Ewing sarcoma. Med Pediatr Oncol 2003;40:141–7. [DOI] [PubMed] [Google Scholar]
- 7. Van Mater D, Wagner L. Management of recurrent Ewing sarcoma: challenges and approaches. Onco Targets Ther 2019;12:2279–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wagner LM, Fouladi M, Ahmed A, Krailo MD, Weigel B, DuBois SG, et al. Phase II study of cixutumumab in combination with temsirolimus in pediatric patients and young adults with recurrent or refractory sarcoma: a report from the Children's Oncology Group. Pediatr Blood Cancer 2015;62:440–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Just MA, Van Mater D, Wagner LM. Receptor tyrosine kinase inhibitors for the treatment of osteosarcoma and Ewing sarcoma. Pediatr Blood Cancer 2021;68:e29084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fleuren ED, Versleijen-Jonkers YM, Boerman OC, van der Graaf WT. Targeting receptor tyrosine kinases in osteosarcoma and Ewing sarcoma: current hurdles and future perspectives. Biochim Biophys Acta 2014;1845:266–76 [DOI] [PubMed] [Google Scholar]
- 11. Brenner JC, Feng FY, Han S, Patel S, Goyal SV, Bou-Maroun LM, et al. PARP-1 inhibition as a targeted strategy to treat Ewing's sarcoma. Cancer Res 2012;72:1608–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gorthi A, Romero JC, Loranc E, Cao L, Lawrence LA, Goodale E, et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature 2018;555:387–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nieto-Soler M, Morgado-Palacin I, Lafarga V, Lecona E, Murga M, Callen E, et al. Efficacy of ATR inhibitors as single agents in Ewing sarcoma. Oncotarget 2016;7:58759–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koppenhafer SL, Goss KL, Terry WW, Gordon DJ. Inhibition of the ATR-CHK1 pathway in Ewing sarcoma cells causes DNA damage and apoptosis via the CDK2-mediated degradation of RRM2. Mol Cancer Res 2020;18:91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goss KL, Koppenhafer SL, Harmoney KM, Terry WW, Gordon DJ. Inhibition of CHK1 sensitizes Ewing sarcoma cells to the ribonucleotide reductase inhibitor gemcitabine. Oncotarget 2017;8:87016–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pandey RN, Wang TS, Tadjuidje E, McDonald MG, Rettie AE, Hegde RS. Structure-activity relationships of benzbromarone metabolites and derivatives as EYA inhibitory Anti-angiogenic agents. PLoS One 2013;8:e84582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tadjuidje E, Wang TS, Pandey RN, Sumanas S, Lang RA, Hegde RS. The EYA tyrosine phosphatase activity is Pro-angiogenic and is inhibited by benzbromarone. PLoS One 2012;7:e34806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 1998;140:1285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Quinet A, Carvajal-Maldonado D, Lemacon D, Vindigni A. DNA fiber analysis: Mind the Gap! Methods Enzymol 2017;591:55–82. [DOI] [PubMed] [Google Scholar]
- 20. Henry-Mowatt J, Jackson D, Masson J-Y, Johnson PA, Clements PM, Benson FE, et al. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol Cell 2003;11:1109–17. [DOI] [PubMed] [Google Scholar]
- 21. Bialic M, Al Ahmad Nachar B, Kozlak M, Coulon V, Schwob E. Measuring S-phase duration from asynchronous cells using dual EdU-BrdU Pulse-chase labeling flow cytometry. Genes (Basel) 2022;13:408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Y, Pandey RN, Roychoudhury K, Milewski D, Kalin TV, Szabo S, et al. Targeting Eya3 in ewing sarcoma retards tumor growth and angiogenesis. Mol Cancer Ther 2021;20:803–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Waraky A, Lin Y, Warsito D, Haglund F, Aleem E, Larsson O. Nuclear insulin-like growth factor 1 receptor phosphorylates proliferating cell nuclear antigen and rescues stalled replication forks after DNA damage. J Biol Chem 2017;292:18227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Toledo LI, Altmeyer M, Rask M-B, Lukas C, Larsen DH, Povlsen LK, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 2013;155:1088–103. [DOI] [PubMed] [Google Scholar]
- 25. Vassin VM, Anantha RW, Sokolova E, Kanner S, Borowiec JA. Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J Cell Sci 2009;122:4070–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marechal A, Zou L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res 2015;25:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rieunier G, Wu X, Harris LE, Mills JV, Nandakumar A, Colling L, et al. Targeting IGF perturbs global replication through ribonucleotide reductase dysfunction. Cancer Res 2021;81:2128–41. [DOI] [PubMed] [Google Scholar]
- 28. Su XA, Ma D, Parsons JV, Replogle JM, Amatruda JF, Whittaker CA, et al. RAD21 is a driver of chromosome 8 gain in Ewing sarcoma to mitigate replication stress. Genes Dev 2021;35:556–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aleksic T, Chitnis MM, Perestenko OV, Gao S, Thomas PH, Turner GD, et al. Type 1 insulin-like growth factor receptor translocates to the nucleus of human tumor cells. Cancer Res 2010;70:6412–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Sølvhøj Pedersen R, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 2011;13:243–53. [DOI] [PubMed] [Google Scholar]
- 31. Gildemeister OS, Sage JM, Knight KL. Cellular redistribution of Rad51 in response to DNA damage: novel role for Rad51C. J Biol Chem 2009;284:31945–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene 2003;22:1115–23. [DOI] [PubMed] [Google Scholar]
- 33. Branzei D, Psakhye I. DNA damage tolerance. Curr Opin Cell Biol 2016;40:137–44. [DOI] [PubMed] [Google Scholar]
- 34. Girnita A, Girnita L, Prete FD, Bartolazzi A, Larsson O, Axelson M. Cyclolignans as inhibitors of the insulin-like growth factor-1 receptor and malignant cell growth. Cancer Res 2004;64:236–42. [DOI] [PubMed] [Google Scholar]
- 35. Fu S, Wang Y, Keyomarsi K, Meric-Bernstam F, Meric-Bernstein F. Strategic development of AZD1775, a wee1 kinase inhibitor, for cancer therapy. Expert Opin Investig Drugs 2018;27:741–51. [DOI] [PubMed] [Google Scholar]
- 36. Li Z, Pinch BJ, Olson CM, Donovan KA, Nowak RP, Mills CE, et al. Development and characterization of a wee1 kinase degrader. Cell Chem Biol 2020;27:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aleksic T, Gray N, Wu X, Rieunier G, Osher E, Mills J, et al. Nuclear IGF1R interacts with regulatory regions of chromatin to promote RNA polymerase II recruitment and gene expression associated with advanced tumor stage. Cancer Res 2018;78:3497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Warsito D, Sjostrom S, Andersson S, Larsson O, Sehat B. Nuclear IGF1R is a transcriptional co-activator of LEF1/TCF. EMBO Rep 2012;13:244–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mulvihill MJ, Cooke A, Rosenfeld-Franklin M, Buck E, Foreman K, Landfair D, et al. Discovery of OSI-906: a selective and orally efficacious dual inhibitor of the IGF-1 receptor and insulin receptor. Future Med Chem 2009;1:1153–71. [DOI] [PubMed] [Google Scholar]
- 40. Scotlandi K, Manara MC, Serra M, Marino MT, Ventura S, Garofalo C, et al. Expression of insulin-like growth factor system components in Ewing's sarcoma and their association with survival. Eur J Cancer 2011;47:1258–66. [DOI] [PubMed] [Google Scholar]
- 41. Asmane I¨N, Watkin E, Alberti L, Duc A, Marec-Berard P, Ray-Coquard I, et al. Insulin-like growth factor type 1 receptor (IGF-1R) exclusive nuclear staining: a predictive biomarker for IGF-1R monoclonal antibody (Ab) therapy in sarcomas. Eur J Cancer 2012;48:3027–35. [DOI] [PubMed] [Google Scholar]
- 42. van de Luijtgaarden ACM, Versleijen-Jonkers YMH, Roeffen MHS, Schreuder HWB, Flucke UE, van der Graaf WTA. Prognostic and therapeutic relevance of the IGF pathway in Ewing's sarcoma patients. Target Oncol 2013;8:253–60. [DOI] [PubMed] [Google Scholar]
- 43. Gonzalez E, Bui M, Ahmed AA. IGF1R immunohistochemistry in Ewing's sarcoma as predictor of response to targeted therapy. Int J Health Sci (Qassim) 2020;14:17–21 [PMC free article] [PubMed] [Google Scholar]
- 44. Bodzin AS, Wei Z, Hurtt R, Gu T, Doria C. Gefitinib resistance in HCC mahlavu cells: upregulation of CD133 expression, activation of IGF-1R signaling pathway, and enhancement of IGF-1R nuclear translocation. J Cell Physiol 2012;227:2947–52. [DOI] [PubMed] [Google Scholar]
- 45. Aslam MI, Hettmer S, Abraham J, LaTocha D, Soundararajan A, Huang ET, et al. Dynamic and nuclear expression of PDGFRalpha and IGF-1R in alveolar Rhabdomyosarcoma. Mol Cancer Res 2013;11:1303–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lin Y, Liu H, Waraky A, Haglund F, Agarwal P, Jernberg-Wiklund H, et al. SUMO-modified insulin-like growth factor 1 receptor (IGF-1R) increases cell cycle progression and cell proliferation. J Cell Physiol 2017;232:2722–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Toledo LI, Murga M, Zur R, Soria R, Rodriguez A, Martinez S, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol 2011;18:721–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montaña MF, et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol 2011;18:1331–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Miller HE, Gorthi A, Lawrence LA, Iskra BS, Bishop AJR. Reconstruction of Ewing sarcoma developmental context from mass-scale transcriptomics reveals characteristics of EWSR1-FLI1 permissibility. Cancers (Basel) 2020;12:948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ubhi T, Brown GW. Exploiting DNA Replication Stress for Cancer Treatment. Cancer Res 2019;79:1730–9. [DOI] [PubMed] [Google Scholar]
- 51. Zhu H, Swami U, Preet R, Zhang J. Harnessing DNA replication stress for novel cancer therapy. Genes (Basel) 2020;11:990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang C, Zhang Y, Chen Y, Ragaller F, Liu M, Corvigno S, et al. Nuclear IGF1R interact with PCNA to preserve DNA replication after DNA-damage in a variety of human cancers. PLoS One 2020;15:e0236291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ohmura S, Marchetto A, Orth MF, Li J, Jabar S, Ranft A, et al. Translational evidence for RRM2 as a prognostic biomarker and therapeutic target in Ewing sarcoma. Mol Cancer 2021;20:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moiseeva TN, Qian C, Sugitani N, Osmanbeyoglu HU, Bakkenist CJ. WEE1 kinase inhibitor AZD1775 induces CDK1 kinase-dependent origin firing in unperturbed G1- and S-phase cells. Proc Nat Acad Sci USA 2019;116:23891–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martin JC, Hoegel TJ, Lynch ML, Woloszynska A, Melendy T, Ohm JE. Exploiting replication stress as a novel therapeutic intervention. Mol Cancer Res 2021;19:192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu X, Seraia E, Hatch SB, Wan X, Ebner DV, Aroldi F, et al. CHK1 inhibition exacerbates replication stress induced by IGF blockade. Oncogene 2022;41:476–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Anderson ND, de Borja R, Young MD, Fuligni F, Rosic A, Roberts ND, et al. Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumors. Science 2018;361:eaam8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov 2014;4:1326–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Patient tumor information
Reagents, instruments, tumor cell culture details
Western blots and quantifications
A. Bright field images of CCH1 and CCH2 cells B. CCH1 and CCH2 cells cells stained with antibody towards CD99 (green) and DAPI (blue) C. Western blots on CCH1 and CCH2 cell lysates probed with the indicated antibodies, confirming differences in EphA2 phosphorylation between CCH1 and CCH2. D. Immunofluorescence imaging of IGF1R-stained CCH1 and CCH2 cells starved, treated with IGF1 or treated with linsitinib for 2 hours. Individual channels are shown below the merged images. Nuclear IGF1R in CCH1 cells does not change localization with either treatment. IGF1 treatment induces slight internalization of IGF1R in CCH2 cells and this is inhibited by linsitinib.
Cell cycle analyses A. Dansylcadaverine-mediated nIGF1R depletion arrested CCH1 cells at the G1-phase and reduced the percentage of cells in the S-phase. B. Western blots show that Cyclin A2 and Cyclin D1 levels were high in CCH5 cells with nIGF1R relative to NCH6 with mIGF1R. C. Cell cycle analyses shows higher percentages of CCH1 and CCH5 cells in S-phase than CCH2 and NCH6 cells. D. CCH5 cells with nIGF1R divided faster, had higher percentages of S-phase cells with shorter S-phase duration than NCH6 with mIGF1R. E. EdU/BrdU double labelling-based S-phase duration analysis was carried out. Top panel shows that cells showed EdU-AF55+ and BrdU-FITC+ signals in comparison to unstained cells. F. Percentages of EdU+BrdU+ cells (CCH1, CCH5, CCH2 and NCH6 cells), at indicated time points, are shown. G.Timings of S-phase exit (red) for all cells were calculated using linear regression.
A. Representative images for DNA fiber experiments conducted on CCH1 and CCH2 cells. In each image asterisk (*) indicates a normal fork (red-green), “N” indicates a new fork (red only), and “S” indicates a stalled fork (green only).B. Brightfield and CD99-stained (green) images of patient-derived cells used in the studies reported here.
Characterization of Ewing sarcoma cell lines A673, RD-ES and SK-ESA.Western blot analysis of the IGF1R signaling pathway B. Western blot analysis of markers associated with the Replication Stress Response C. Western blot analysis of RPA phosphorylation and apoptosis measured by cleaved caspase 3 (CC3 levels).Cells stained with antibody towards IGF1R (red) and DAPI (blue) shows membranous staining in RD-ES and SK-ES cells, and very low levels in A673 cells.
Immunofluorescence images of CCH2 and NCH6 cells stained with antibodies towards IGF1R (red) and PCNA (green).
Flow cytometry analysis for propidium-iodide positive cells with the indicated treatments (48 hours). The percentage of PI-positive cells are shown in each panel.
Data Availability Statement
All data generated or analyzed during this study are included in this manuscript and the associated supplementary files or will be made available on request from the corresponding author.