Abstract
The act gene was identified and an act mutant as well as the pfl mutant was constructed in Streptococcus mutans. Pyruvate formate-lyase (PFL) activity was regenerated with the mixture of the respective cell extracts from these mutants by complementary reconstitution of the in vitro reactivating system. The S. mutans act gene encoded the sole enzyme able to activate the PFL protein in this organism.
Streptococcus mutans has been implicated as an important microbial agent in human dental caries (13). The regulation of acid production in this organism inhabiting dental plaque on tooth surfaces has been of particular interest because the resulting acid fermentation products from dietary sugars can demineralize the enamel tooth surfaces, resulting in dental caries.
In the natural environment of this organism, such as the interior of dental plaque, where anaerobic conditions exist, pyruvate formate-lyase (PFL) may play a more important role than lactate dehydrogenase (LDH) in pyruvate metabolism as a part of ATP synthesis and NAD+ and/or NADH recycling (2, 6, 27, 28). In addition, PFL of this organism is essential for anaerobic metabolism of sugar alcohols (e.g., sorbitol and mannitol) used as sweeteners for dental caries prevention (27). S. mutans produces various fermentation end products, including formate, acetate, and ethanol as well as lactic acid (27). Investigations of the enzymatically regulatory mechanisms controlling the production of fermentation end products have revealed that PFL plays a central role in fermentation in S. mutans (1, 2, 27). Therefore, this regulatory mechanism should be elucidated at the molecular level in this organism.
The Escherichia coli PFL has been identified as an enzyme containing a glycyl radical in its active structure (7, 8, 16, 22, 24) and can be converted from the reversibly inactive form to the active form by the PFL-activating enzyme, PFL activase (9, 10, 16, 23). The S. mutans PFL was also reported to be extremely dioxygen sensitive (2, 20, 29). We have cloned the S. mutans pfl gene encoding PFL (30), and the deduced amino acid sequence contains the putative glycyl radical at a similar position relative to the E. coli enzyme. Therefore, it is possible that S. mutans PFL could be activated by PFL activase or an equivalent enzyme.
In the present investigation, an act gene encoding PFL activase from S. mutans was cloned by PCR amplification. We describe the sequence analysis of the S. mutans act gene and present evidence that it is involved in the regulation of PFL.
Cloning and sequence analysis of the S. mutans act gene.
To isolate the act gene of S. mutans, we performed PCR amplification of an act internal region from S. mutans GS-5IS3 (30) chromosomal DNA. A 425-bp fragment was amplified by PCR using synthesized oligonucleotide primer sets consisting of act5′2 (5′ TATTGCCATAATCCGGAYACNTGG 3′ [forward]) and act3′3 (5′ GGAACCAAGACATRKCKDATCCA 3′ [reverse]) based upon the sequence of the act sequence of E. coli (14), Clostridium pasteurianum (25), and a putative act sequence of Streptococcus pyogenes (The University of Oklahoma's Advanced Center for Genome Technology [http:// www.genome.ou.edu/strep.html]). The three nucleotides at the 3′ ends of act5′2 and act3′3 were designed according to the unique tryptophan codon UGG. The nucleotide sequence of the 425-bp amplicon was determined and indicated that this amplicon was very likely a part of the act gene of S. mutans because of its similarity to the act genes from other organisms. To obtain the fragments upstream and downstream from the amplified act internal region, inverse PCR was carried out by using genome walking libraries of this organism. The nucleotide sequence of the 1.5-kb ScaI-HindIII region encompassing the entire S. mutans act gene region was then determined (accession number AB018417 [http://www.genome.ou.edu/strep.html]) by an automated DNA sequencer (model 373S; PE Biosystems, Foster City, Calif.). Nucleotide sequence analysis was carried out with the DNASIS-Mac program (Hitachi Software Engineering, Yokohama, Japan). The 789-bp open reading frame (ORF) encodes a 263-amino-acid protein with a calculated molecular weight of 30,148. A potential ribosome-binding sequence (AGGA) and a promoter-like sequence (TAGTCT-N18-TATAAT) could be identified immediately upstream from the putative initiation codon of the act gene. An inverted repeat sequence characteristic of transcription terminator could also be detected 11 bp downstream from the termination codon of the act gene. The formation of a putative stem-loop structure in this region of the mRNA corresponds to a free-energy change of 8.4 kJ per mol. Although random mutagenesis to isolate the act gene was initially attempted as previously reported for isolation of the pfl gene (30), act mutants were not isolated by this procedure. In fact, only one Sau3AI fragment capable of gene inactivation by this mutagenesis was detected, corresponding to a size of 156 bp based upon subsequent sequencing of the S. mutans act region. This size may have been too small for multiple integrational inactivations.
While we were conducting this study, the nucleotide sequences of 10 genes in the locus including the PFL activase gene (pflC) from S. mutans strain LT11 appeared in the GenBank nucleotide sequence database with the accession number AF051356, deposited by D. A. Boyd et al. (University of Manitoba, Winnipeg, Canada) (unpublished data). They reported that the pflC gene was located in the downstream region of the abcX gene, which encodes an ATP-binding cassette transporter protein. Our nucleotide sequence data of the act gene matched that of the pflC gene except for the 874th nucleotide, which was a T in our sequence. The amino acid sequence deduced from the act gene exhibited perfect consistency with that from the pflC gene.
All of the characterized bacterial act gene homologs are found downstream of the pfl gene homologs. For example, the E. coli and C. pasteurianum act genes are located downstream of their pfl genes (14, 16, 25). However, we did not detect any sequences homologous to the act gene in a similar region of the S. mutans chromosome (30). The act homologous sequence was not reported either upstream or downstream from the pfl gene in Lactococcus lactis or Streptococcus bovis (3, 4). The pfl gene was located on the NotI A fragment (433 kb) on the S. mutans chromosome (5). Meanwhile, by Southern hybridization analysis following pulsed-field electrophoresis we confirmed that the act probe hybridized with the NotI E fragment (223 kb) (data not shown). These results indicated that the act gene was not closely linked to the pfl gene on the S. mutans chromosome.
The amino acid sequences deduced from the act genes (S. mutans and E. coli) and a putative act gene (S. pyogenes) were aligned for comparison (Fig. 1) (14); The University of Oklahoma's Advanced Center for Genome Technology [http://www.genome.ou.edu/strep.html]). These three sequences have 42.1% identical and 79.3% conserved amino acid residues overall and have a conserved three-cysteine cluster, Cys-37, Cys-41, and Cys-44 (according to residue numbering in the S. mutans sequence), which corresponds to the catalytic site of the enzyme in E. coli (11, 26).
FIG. 1.
Multiple alignment of PFL activase sequences. The S. mutans PFL activase sequence was aligned with the S. pyogenes and E. coli sequences (allowing gaps [hyphens]). Identical and similar amino acid residues are indicated by asterisks and colons, respectively, in the consensus sequence. The region of the three-cysteine cluster, which is the catalytic site in the E. coli enzyme, is boxed.
Another ORF, designated act-5′orf, was detected on the same strand immediately upstream from the promoter-like sequence of the act gene. The putative transcription terminator sequence of the act-5′orf overlapped with the promoter-like sequence of the act gene. A part of this ORF exhibited similarity to a hemolysin gene (tlyC) from Haemophilis influenzae.
Characterization of the S. mutans act gene.
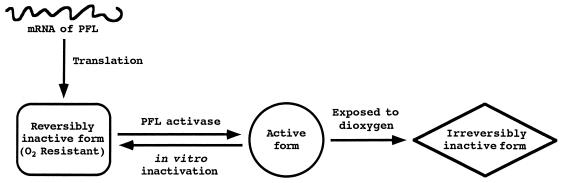
To confirm the function of the act gene, we constructed the S. mutans act mutant (YASC9YK2) by insertional inactivation with a suicide vector containing the tetracycline-resistant gene, following the transformation procedure routinely used in this laboratory (15, 30). The S. mutans act-5′orf mutant (YASCXYGEmr1) was also constructed by the same procedure with the erythromycin-resistant gene to determine whether or not the protein coded by act-5′orf gene is involved in pyruvate metabolism or its regulation. The predicted insertion of the vectors into the chromosome of parental strain (S. mutans GS-5IS3) was confirmed by Southern hybridization analysis (data not shown). The PFL and LDH activities with cell extracts from the parental strain and its mutants (act mutant:YASC9YK2; pfl mutant:SAKC5Y2C1 (30); act-5′orf mutant:YASCXYGEmr1) were spectrophotometrically determined by recording the change in absorbance at 340 nm. The parental strain was grown in medium D87M containing 1% glucose as carbon source (D87M-Glu). The act and pfl mutants and the act-5′orf mutant were grown in D87M-Glu containing 3 μg of tetracycline per ml and 10 μg of erythromycin per ml, respectively. Preparation of cell extracts from these cells at the earlier stages of the exponential growth phase and enzyme assays was performed throughout under strictly anaerobic conditions in a specially designed anaerobic chamber as described previously (2, 20, 21). This was necessary since the S. mutans PFL is very likely a glycyl radical enzyme that can be converted from the reversibly inactive form (R form) to the active form (A form) by PFL activase as indicated in Fig. 2 and also because the A form has been reported to be extremely dioxygen sensitive (2, 20, 27, 29). The protein concentrations in the cell extracts were determined by the biuret method (12). The PFL activities were not detected in the cell extracts from either act or pfl mutants, while the activities (2.11 ± 0.44 and 2.12 μmol/min/mg of protein) were almost identical in the extracts from the parental strain and the act-5′orf mutant (Table 1). Moreover, we did not detect formate as a fermentation end product in supernatants from the anaerobic cultures of both the act and pfl mutants by assaying as described previously (data not shown) (20). The LDH activities of the mutants were similar to that of the parental strain (Table 1). Inactivation of the act-5′orf gene did not induce any phenotypic changes concerning pyruvate metabolism, e.g., formate production, lactate production, and growth with various sugars. These results apparently indicate that the act mutant was not able to generate the A form of PFL and the act-5′orf is not involved in pyruvate metabolism.
FIG. 2.
Proposed model for interconversion of the reversibly inactive, active, and irreversibly inactive forms of the S. mutans PFL (21, 27).
TABLE 1.
PFL and LDH activities of S. mutans GS-5IS3 and its mutants
| S. mutans strain | Activitya (μmol/min/mg of protein)
|
|
|---|---|---|
| PFL | LDH | |
| GS-5IS3 | 2.11 ± 0.44 | 21.5 ± 2.0 |
| SAKC5Y2C1 | ND | 17.3 ± 0.45 |
| YASC9YK2 | ND | 16.4 ± 4.1 |
| YASCXYGEmr1 | 2.12 | 22.5 |
The PFL activity was measured at 30°C and the LDH activity was measured at 25°C. The enzyme activities are expressed as means ± standard deviations for triplicate cultures except for YASCXYGEmr1, and those of YASCXYGEmr1 are expressed as means for duplicate cultures. ND, not detected.
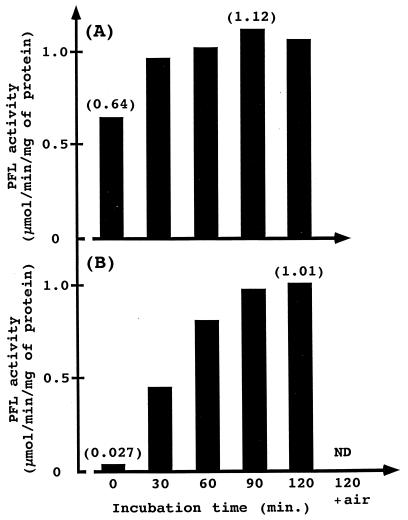
To detect PFL activating activity of the protein encoded by the act gene, we employed complementary reconstitution of the in vitro PFL reactivating system with mixtures of the respective cell extracts from the S. mutans act and pfl mutants. The act mutant would not be expected to express the A form of PFL but would retain the intact PFL protein as the R form. In addition, the pfl mutant should retain an enzyme functioning as the PFL activase. The R form of PFL was reactivated to the A form by the PFL activase in the cell extracts following incubation with 0.6 mM S-adenosyl-l-methionine, 10 mM sodium pyruvate, 2.85 mM dithiothreitol, 2 mM Fe(SO4)2(NH4)2, and 1 mM methylviologen in 100 mM potassium phosphate buffer (pH 6.5) for 90 min at 30°C under strictly anaerobic conditions (Fig. 2) (20, 21). To convert the A form of PFL in the cell extracts from parental strain GS-5IS3 to the R form, the extracts were incubated for 90 min at 35°C followed by storage at 4°C for several days under strictly anaerobic conditions (Fig. 2) (20, 21, 27). These extracts derived from parental strain containing the R form of PFL converted from the A form of PFL were used as a control experiment for complementary reconstitution of the in vitro PFL reactivating system with the pfl and act mutants, and reactivation of PFL by this reactivating system was determined with the parental strain (Fig. 3A). The PFL activity decreased to approximately 30% (0.64 μmol/min/mg of protein) of the value before inactivation (2.11 ± 0.44 μmol/min/mg of protein) and increased with the incubation time in reactivating mixtures containing the R form of PFL from these extracts (Table 1 and Fig. 3A). The maximum reactivation (1.12 μmol/min/mg of protein) was observed after 90 min of incubation, and the activity was approximately 1.8 times that before incubation. However, this activity value was not fully restored to that before inactivation by this in vitro reactivating system using methylviologen as a reductant.
FIG. 3.
Measurement of the reactivated PFL activities. Shown are results of reactivation of PFL with the cell extracts from the parental strain (S. mutans GS-5IS3) (A) and with the mixture of both cell extracts from the act and pfl mutants (B). The PFL activity was measured at 30°C, and bars represent means of enzyme activities resulting from two independent experiments. ND, not detected; air, reactivating mixture was exposed to air for 5 min after the 120-min incubation.
We expected that cell extracts prepared from the act and pfl mutants would complement each other for PFL activation, although both extracts did not independently exhibit PFL activities. The detected enzyme activities in the mixture of both extracts increased hyperbolically with incubation (Fig. 3B). To confirm that this activity was actually derived from the A form of PFL, the reactivating mixture was exposed to the air. The A form of PFL is irreversibly inactivated due to a cleavage of its peptide (I form) at the glycyl radical by dioxygen under aerobic conditions (24). In addition, other S. mutans enzymes converting pyruvate into acetyl coenzyme A such as pyruvate dehydrogenase complex are not inactivated by dioxygen. The restored enzyme activity completely disappeared when the reactivating mixture, after the 120-min incubation, was exposed to the air for 5 min (Fig. 3B). This complete abrogation of the activity apparently indicates that our cloned act gene encoded PFL activase. The highest reactivated PFL activity (1.01 μmol/min/mg of protein) observed after the 120-min incubation was approximately 90% of that detected in the control experiment with the extract from the parental strain (Fig. 3B).
Another dioxygen-sensitive anaerobic ribonucleotide reductase (ARNR) which catalyzes conversion of ribonucleotides to 2′ deoxy ribonucleotides was reported to be a glycyl radical enzyme in E. coli (18, 19). S. mutans ARNR utilizes ribonucleoside triphosphate as a substrate and mainly functions under strictly anaerobic conditions (N. Okada, personal communication). The active form of E. coli ARNR is similar to that of PFL and undergoes oxygenolytic cleavage of the polypeptide chain in vitro at the site of the glycyl radical (16, 18). The ARNR-activating enzyme exhibited significant similarity to PFL activase at the primary sequence level in E. coli (16, 17). Our finding that the act mutant did not exhibit PFL activity under anaerobic conditions excluded the possibility that the ARNR-activating enzyme of S. mutans might complement the function of the PFL activase. Together with the results indicated in Fig. 3, these results suggest that PFL activase encoded by the act gene was the sole enzyme able to activate PFL in S. mutans, although we cannot exclude the possibility that the PFL activase may activate ARNR.
Nucleotide sequence accession number. The act nucleotide sequence data appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases with the accession number AB018417.
Acknowledgments
We thank T. Yamada (School of Dentistry, Tohoku University) and H. K. Kuramitsu (State University of New York at Buffalo) for critical reading of the manuscript, Y. Yamashita (School of Dentistry, Kyushu University) and M. Kitamura (Himeji Institute of Technology) for their helpful comments, and Y. Mito (School of Dentistry, Tohoku University) and A. Gokan (Tokyo Dental College) for their kind assistance.
This investigation was supported in part by grants-in-aid for Encouragement of Young Scientists (no. 07771676, 08771623, and 09771547 to Y. Yamamoto) from the Japanese Ministry of Education, Science, Culture, and Sports and a grant-in-aid for Research supported by Tokyo Dental College.
REFERENCES
- 1.Abbe K, Carlsson J, Takahashi Abbe S, Yamada T. Oxygen and the sugar metabolism in oral streptococci. Proc Finn Dent Soc. 1991;87:477–487. [PubMed] [Google Scholar]
- 2.Abbe K, Takahashi S, Yamada T. Involvement of oxygen-sensitive pyruvate formate-lyase in mixed-acid fermentation by Streptococcus mutans under strictly anaerobic conditions. J Bacteriol. 1982;152:175–182. doi: 10.1128/jb.152.1.175-182.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnau J, Jorgensen F, Madsen S M, Vrang A, Israelsen H. Cloning, expression, and characterization of the Lactococcus lactis pfl gene, encoding pyruvate formate-lyase. J Bacteriol. 1997;179:5884–5891. doi: 10.1128/jb.179.18.5884-5891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asanuma N, Iwamoto M, Hino T. Structure and transcriptional regulation of the gene encoding pyruvate formate-lyase of a ruminal bacterium, Streptococcus bovis. Microbiology. 1999;145:151–157. doi: 10.1099/13500872-145-1-151. [DOI] [PubMed] [Google Scholar]
- 5.Cappiello M G, Hantman M J, Zuccon F M, Peruzzi F, Amjad M, Piggot P J, Daneo-Moore L. Physical and genetic map of Streptococcus mutans GS-5 and localization of five rRNA operons. Oral Microbiol Immunol. 1999;14:225–232. doi: 10.1034/j.1399-302x.1999.140405.x. [DOI] [PubMed] [Google Scholar]
- 6.Carlsson J, Griffith C J. Fermentation products and bacterial yields in glucose-limited and nitrogen-limited cultures of streptococci. Arch Oral Biol. 1974;19:1105–1109. doi: 10.1016/0003-9969(74)90238-6. [DOI] [PubMed] [Google Scholar]
- 7.Frey M, Rothe M, Wagner A F, Knappe J. Adenosylmethionine-dependent synthesis of the glycyl radical in pyruvate formate-lyase by abstraction of the glycine C-2 pro-S hydrogen atom. Studies of [2H]glycine-substituted enzyme and peptides homologous to the glycine 734 site. J Biol Chem. 1994;269:12432–12437. [PubMed] [Google Scholar]
- 8.Knappe J, Elbert S, Frey M, Wagner A F. Pyruvate formate-lyase mechanism involving the protein-based glycyl radical. Biochem Soc Trans. 1993;21:731–734. doi: 10.1042/bst0210731. [DOI] [PubMed] [Google Scholar]
- 9.Knappe J, Neugebauer F A, Blaschkowski H P, Ganzler M. Post-translational activation introduces a free radical into pyruvate formate-lyase. Proc Natl Acad Sci USA. 1984;81:1332–1335. doi: 10.1073/pnas.81.5.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knappe J, Wagner A F. Glycyl free radical in pyruvate formate-lyase: synthesis, structure characteristics, and involvement in catalysis. Methods Enzymol. 1995;258:343–362. doi: 10.1016/0076-6879(95)58055-7. [DOI] [PubMed] [Google Scholar]
- 11.Kulzer R, Pils T, Kappl R, Huttermann J, Knappe J. Reconstitution and characterization of the polynuclear iron-sulfur cluster in pyruvate formate-lyase-activating enzyme. Molecular properties of the holoenzyme form. J Biol Chem. 1998;273:4897–4903. doi: 10.1074/jbc.273.9.4897. [DOI] [PubMed] [Google Scholar]
- 12.Layne E. Spectrophotometric and turbidimetric methods for measuring proteins. Methods Enzymol. 1957;3:447–454. [Google Scholar]
- 13.Loesche W J. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodel W, Plaga W, Frank R, Knappe J. Primary structures of Escherichia coli pyruvate formate-lyase and pyruvate-formate-lyase-activating enzyme deduced from the DNA nucleotide sequences. Eur J Biochem. 1988;177:153–158. doi: 10.1111/j.1432-1033.1988.tb14356.x. [DOI] [PubMed] [Google Scholar]
- 15.Sato Y, Yamamoto Y, Kizaki H, Kuramitsu H K. Isolation, characterization and sequence analysis of the scrK gene encoding fructokinase of Streptococcus mutans. J Gen Microbiol. 1993;139:921–927. doi: 10.1099/00221287-139-5-921. [DOI] [PubMed] [Google Scholar]
- 16.Sawers G, Watson G. A glycyl radical solution: oxygen-dependent interconversion of pyruvate formate-lyase. Mol Microbiol. 1998;29:945–954. doi: 10.1046/j.1365-2958.1998.00941.x. [DOI] [PubMed] [Google Scholar]
- 17.Sun X, Eliasson R, Pontis E, Andersson J, Buist G, Sjoberg B M, Reichard P. Generation of the glycyl radical of the anaerobic Escherichia coli ribonucleotide reductase requires a specific activating enzyme. J Biol Chem. 1995;270:2443–2446. doi: 10.1074/jbc.270.6.2443. [DOI] [PubMed] [Google Scholar]
- 18.Sun X, Harder J, Krook M, Jornvall H, Sjoberg B M, Reichard P. A possible glycine radical in anaerobic ribonucleotide reductase from Escherichia coli: nucleotide sequence of the cloned nrdD gene. Proc Natl Acad Sci USA. 1993;90:577–581. doi: 10.1073/pnas.90.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun X, Ollagnier S, Schmidt P P, Atta M, Mulliez E, Lepape L, Eliasson R, Graslund A, Fontecave M, Reichard P, Sjoberg B M. The free radical of the anaerobic ribonucleotide reductase from Escherichia coli is at glycine 681. J Biol Chem. 1996;271:6827–6831. [PubMed] [Google Scholar]
- 20.Takahashi N, Abbe K, Takahashi Abbe S, Yamada T. Oxygen sensitivity of sugar metabolism and interconversion of pyruvate formate-lyase in intact cells of Streptococcus mutans and Streptococcus sanguis. Infect Immun. 1987;55:652–656. doi: 10.1128/iai.55.3.652-656.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi S, Abbe K, Yamada T. Purification of pyruvate formate-lyase from Streptococcus mutans and its regulatory properties. J Bacteriol. 1982;149:1034–1040. doi: 10.1128/jb.149.3.1034-1040.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unkrig V, Neugebauer F A, Knappe J. The free radical of pyruvate formate-lyase. Characterization by EPR spectroscopy and involvement in catalysis as studied with the substrate-analogue hypophosphite. Eur J Biochem. 1989;184:723–728. doi: 10.1111/j.1432-1033.1989.tb15072.x. [DOI] [PubMed] [Google Scholar]
- 23.Wagner A F, Demand J, Schilling G, Pils T, Knappe J. A dehydroalanyl residue can capture the 5′-deoxyadenosyl radical generated from S-adenosylmethionine by pyruvate formate-lyase-activating enzyme. Biochem Biophys Res Commun. 1999;254:306–310. doi: 10.1006/bbrc.1998.9931. [DOI] [PubMed] [Google Scholar]
- 24.Wagner A F, Frey M, Neugebauer F A, Schafer W, Knappe J. The free radical in pyruvate formate-lyase is located on glycine-734. Proc Natl Acad Sci USA. 1992;89:996–1000. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weidner G, Sawers G. Molecular characterization of the genes encoding pyruvate formate-lyase and its activating enzyme of Clostridium pasteurianum. J Bacteriol. 1996;178:2440–2444. doi: 10.1128/jb.178.8.2440-2444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong K K, Murray B W, Lewisch S A, Baxter M K, Ridky T W, Ulissi DeMario L, Kozarich J W. Molecular properties of pyruvate formate-lyase activating enzyme. Biochemistry. 1993;32:14102–14110. doi: 10.1021/bi00214a005. [DOI] [PubMed] [Google Scholar]
- 27.Yamada T. Regulation of glycolysis in streptococci. In: Reizer J, Peterkofsky A, editors. Sugar transport and metabolism in Gram-positive bacteria. Chichester, United Kingdom: Ellis Horwood Limited; 1987. pp. 69–93. [Google Scholar]
- 28.Yamada T, Carlsson J. Regulation of lactate dehydrogenase and change of fermentation products in streptococci. J Bacteriol. 1975;124:55–61. doi: 10.1128/jb.124.1.55-61.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamada T, Takahashi Abbe S, Abbe K. Effects of oxygen on pyruvate formate-lyase in situ and sugar metabolism of Streptococcus mutans and Streptococcus sanguis. Infect Immun. 1985;47:129–134. doi: 10.1128/iai.47.1.129-134.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto Y, Sato Y, Takahashi Abbe S, Abbe K, Yamada T, Kizaki H. Cloning and sequence analysis of the pfl gene encoding pyruvate formate-lyase from Streptococcus mutans. Infect Immun. 1996;64:385–391. doi: 10.1128/iai.64.2.385-391.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]