Abstract
Introduction
Copy-number variations (CNVs) impacting on small DNA stretches and associated with language deficits provide a unique window to the role played by specific genes in language function.
Methods
We report in detail on the cognitive, language, and genetic features of a girl bearing a small deletion (0.186 Mb) in the 2p16.3 region, arr[hg19] 2p16.3(50761778_50947729)×1, affecting exons 3–7 of NRXN1, a neurexin-coding gene previously related to schizophrenia, autism (ASD), attention deficit hyperactivity disorder (ADHD), mood disorder, and intellectual disability (ID).
Results
The proband exhibits many of the features commonly found in subjects with deletions of NRXN1, like ASD-like traits (including ritualized behaviors, disordered sensory aspects, social disturbances, and impaired theory of mind), ADHD symptoms, moderate ID, and impaired speech and language. Regarding this latter aspect, we observed altered speech production, underdeveloped phonological awareness, minimal syntax, serious shortage of active vocabulary, impaired receptive language, and inappropriate pragmatic behavior (including lack of metapragmatic awareness and communicative use of gaze). Microarray analyses point to the dysregulation of several genes important for language function in the girl compared to her healthy parents.
Discussion
Although some basic cognitive deficit − such as the impairment of executive function − might contribute to the language problems exhibited by the proband, molecular evidence suggests that they might result, to a great extent, from the abnormal expression of genes directly related to language.
Keywords: 2p16.3 deletion, NRXN1, Cognitive impairment, Language deficits, Pragmatic impairment, Language-related gene networks
Introduction
Copy number variants (CNVs) found in individuals with language and pragmatic deficits provide a unique window to the genetic foundations of the human ability to learn and use languages. This is particularly true for CNVs affecting a reduced number of genes and resulting in a common phenotypic profile. In this paper, we report on a girl with a microdeletion in the 2p16.3 region affecting the NRXN1 gene. This gene encodes a neurexin [Rowen et al., 2002]. Neurexins are polymorphic cell surface proteins implicated in synapse development and maintenance [Missler and Südhof, 1998]. The NRXN1 protein has been shown to regulate synaptic activity, neuritogenesis, and neuronal network assembly during neocortical development [Südhof, 2008; Gjørlund et al., 2012; Jenkins et al., 2016]. Genomic losses involving NRXN1 have been associated with different neurocognitive conditions, including schizophrenia (SZ), autism spectrum disorders (ASD), attention deficit hyperactivity disorder (ADHD), seizures, mood disorder, or intellectual disability (ID) [for recent reviews, see Lowther et al., 2017; Hu et al., 2019; Castronovo et al., 2020; Tromp et al., 2021]. Whereas, as noted, the clinical profile of patients with deletions in this gene is pretty variable, language and communication deficits are commonly reported in most of them [Béna et al., 2013; Curran et al., 2013; Al Shehhi et al., 2019]. To date, the most detailed report on language and speech problems in subjects with deletions in NRXN1 is that of Brignell et al. [2018], who observed that children (aged 1.8–17 years) with NRXN1 deletions affecting exons 1–3 or 1–5 of the gene exhibited a pretty variable phenotype with regards to language deficits. Speech sound problems were frequent, but not severe, whereas many children also had difficulties with receptive language. Language use in social settings was impaired too.
The reported heterogeneity in the severity and the type of speech and language problems associated with deletions in this gene urges to find and characterize in detail the clinical presentation of new CNVs affecting the gene. Moreover, as genes work in a coordinated way, it is well possible that changes in the dosage of NRXN1 resulting from CNVs affect downstream genes with a more direct/profound impact on speech and language. Having all this in mind, in this paper we provide a detailed characterization of the speech, language, and communication deficits observed in our patient. Additionally, we build on our current knowledge of the genetics of language development and language evolution, as well as on the observed changes in the expression pattern of selected genes in the blood of our proband, for hypothesizing about the molecular causes of language dysfunction associated with NRXN1 deletions.
Materials and Methods
Linguistic, Cognitive, and Behavioral Assessment
The global developmental profile of the proband was assessed with the Spanish version of the Battelle Developmental Inventory [de la Cruz and González, 2011], which consists of 341 items aimed to evaluate the subject's personal/social development, adaptive capabilities, gross and fine motor abilities, receptive and expressive communication skills, and cognitive development. In addition, the proband's language features and communication skills were examined in detail through the analysis of a 10-min sample of an interaction between the child and her sister while playing in a room at her speech language therapist's place under her mother's surveillance. The conversation was video-recorded and then transcribed and coded using the CHAT (Codes for the Human Analysis of Transcripts) software, a tool of the CHILDES Project [MacWhinney, 2000]. CHAT allows to code for speech production phenomena on the main lines of the transcript, and for phonological, morphological, syntactic, and pragmatic phenomena on dependent lines. Information about prosody, dialectal articulation, communicative gestures, and communicative use of gaze can be included within square brackets in the main lines. Pauses are also coded in the main lines following CHAT conventions for estimated duration. We tagged every language error or anomaly with an indication of the structural components of language affected. We then evaluated their impact on communication and labelled them according to PREP-CORP (PRagmatic Evaluation Protocol for the analysis of oral CORPpora) [Fernández-Urquiza et al., 2017a]. This protocol has been previously used for outlining the pragmatic profiles of several neurodevelopmental disorders [e.g., Fernández-Urquiza et al., 2015, 2016, 2017b; Shiro et al., 2016, 2019; Diez-Itza et al., 2018, 2022].
Molecular Analyses
Microarrays for Whole-Genome CNV Search and Chromosome Aberration Analysis
Peripheral venous blood lymphocytes were grown following standard protocols and collected after 72 h. DNA from the patient and her parents was extracted from 100 μL of EDTA-anticoagulated whole blood using MagNA Pure (Roche Diagnostics, West Sussex, UK). The DNA was then hybridized on a CGH platform (Agilent Technologies). The derivative log ratio spread value was 0.13. The platform included 60.000 probes. Data were analyzed with the Agilent Genomic Workbench 7.0, and the ADM-2 algorithm (threshold = 6.0; aberrant regions had more than 5 consecutive probes).
Microarrays for Whole-Genome Expression Analysis
In order to determine the genes that could be differentially expressed in the proband compared to her healthy parents, microarray analyses of blood samples from the 3 of them were performed. Total RNA was extracted with the PAXgene Blood RNA Kit IVD (Cat. No./ID: 762164). RNA quality and integrity were confirmed with a Bioanalyzer RNA 6000 Nano. All samples had RNA integrity number values >9. An Affymetrix® Scanner 3000 7G was then used for analyzing transcriptome changes. The resulting raw data were processed with the Affymetrix® GeneChip® Command Console® 2.0 program. Next, *.CEL files were checked to certify the RNA integrity and the suitability of the labeling and the hybridization processes. Finally, the raw data from the different arrays were normalized with the SST (Signal Space Transformation)-RMA (Robust Microarray Analysis) tool [Irizarry et al., 2003]. Normalized data (*.CHP files) were subsequently used to search for differentially expressed genes in the proband compared to her parents. Statistical analyses were conducted with the LIMMA (Linear Models for Microarray Analysis) package of BioConductor, using the TAC 4.0 software.
In silico Analyses
In order to delve into the molecular causes of the speech and language problems exhibited by the girl, we used String 11.0 (www.string-db.org) for finding putative functional links between NRXN1 and the products of genes important for language. We focused on 2 types of genes. A first group of genes encompasses candidates for highly prevalent (specific) language disorders, in particular, developmental dyslexia (DD) and specific language impairment (SLI). For DD, we relied on the list provided by Paracchini et al. [2016], which includes genes resulting from candidate association studies, genome-wide association analyses (GWAs), quantitative GWAs, CNV studies, and next-generation sequencing (NGS) analyses; but we also surveyed the literature via PubMed (https://pubmed.ncbi.nlm.nih.gov/), looking for additional candidates. For SLI, we made use of the lists published by Pettigrew et al. [2016] and Chen et al. [2017], which include candidates resulting from linkage analyses, GWA studies, and NGS analyses; but we also surveyed the literature via PubMed for finding other strong candidates for SLI. A second group of genes encompasses candidates for language evolution, as discussed by Boeckx and Benítez-Burraco [2014a, b] and Benítez-Burraco and Boeckx [2015]. These genes are primarily expected to account for the recent globularization of the human skull/brain and ultimately, for the cognitive changes resulting in our species-specific ability for acquiring and using languages. These genes fulfil several of the following criteria: (1) they have changed (and/or interact with genes that have changed) after our separation from extinct hominins, particularly from Neanderthals, these changes including either changes in their coding regions, in their regulation, or in both; (2) they play a role in brain development, wiring, and/or function; and/or (3) they are candidates for language deficits in broader, highly prevalent cognitive disorders, particularly ASD and SZ [for further details, see Benítez-Burraco and Murphy, 2016; Murphy and Benítez-Burraco, 2016, 2017]. A potential limitation of this approach is that not all the genes within these 2 sets result from GWAs based on large samples. Both sets of genes are listed in online supplementary File 2 (for all online suppl. material, see www.karger.com/doi/10.1159/000524710). String 11.0 predicts physical and functional associations between proteins relying on different sources (genomic context, high-throughput experiments, conserved coexpression, and text mining) [for details, see Szklarczyk et al., 2015]. Because we were interested in finding robust functional links, we limited our search to protein-protein interaction databases and curated databases of protein interactions (Biocarta, BioCyc, GO, KEGG, and Reactome).
Results
Clinical History
The proband was born by caesarean section after an uncomplicated pregnancy. She weighed 2.6 kg at birth and presented with congenital torticollis that required a year of rehabilitation. She was fed with artificial lactation, and the transition to solid feeding went smoothly. At 9 months of age, she was referred to the Paediatric Neurology Unit, because her parents reported that the girl's development was delayed compared to her older sister. She was diagnosed with global psychomotor developmental delay. At 13 months of age, a cranial MRI was performed. Results were suggestive of an increase in extra-axial spaces, supratentorial ventriculomegaly, and mega cisterna magna. At that age, the girl also began to attend day care. At 15 months of age, she seemed to evolve favourably despite her maturational delay. At 17 months of age, urine and metabolic studies were required, as the girl suffered from anemia. At 21 months of age, her verbal comprehension abilities seemed to have slightly improved, but her expressive impairment was still dramatic, as she was only able to produce a few single words. Regarding her family history, it is of interest that her paternal uncle suffered from SZ and ID, her father presented with motor tics, and her paternal great-grandmother suffered from cognitive and motor impairment.
Language and Cognitive Development
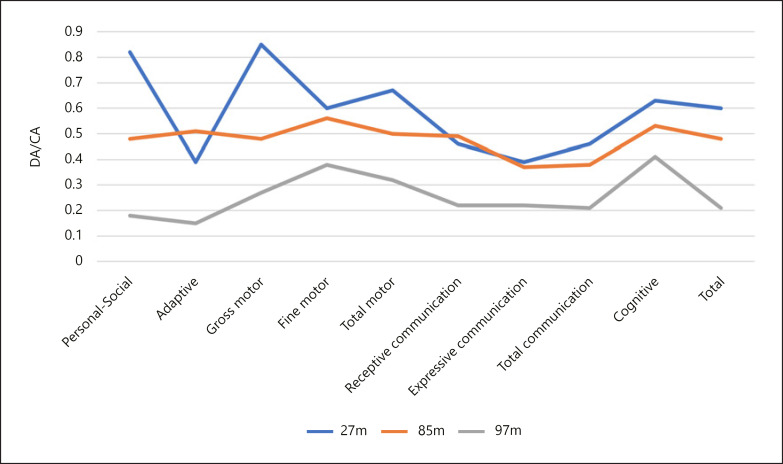
At 27 months of age, the proband was assessed with the Battelle Developmental Inventory (see Fig. 1). At this moment, the girl exhibited a 16-month delay in her adaptive abilities and her expressive communication skills, and a 14-month delay in her receptive communication abilities. She also presented with a 10-month cognitive delay. By contrast, her personal social skills and her gross motor abilities were less impaired.
Fig. 1.
Results of the assessment with the Battelle Developmental Inventory at 27, 85, and 97 months of age. DA/CA, developmental age/chronological age.
At 42 months of age, when the proband began to attend school, she was diagnosed with a severe developmental delay. Regarding her cognitive (dis)abilities, the girl showed scattered attention, problems with making associations, poor imitation capacity, absence of symbolic play, and difficulties with normal social interaction. She also experienced difficulties expressing and identifying emotions, although she did not present with behavioral problems. Concerning her language development, the girl exhibited a severe mixed language delay. She was unable to construct simple sentences and her speech was barely intelligible.
At 60 months of age, the proband's language abilities had improved only slightly. On the expressive domain, she occasionally uttered 2-word phrases, and her speech difficulties persisted. At that age, the girl started to receive speech therapy.
At the age of 72 months, when the girl entered the primary school, her cognitive and behavioral abilities were examined again. The girl was reported to exhibit moderate ID, emotional and behavioral deficits, as well as learning difficulties: for succesfully achieving and performing specific activities, she needed a very structured learning environment with constant monitoring and aid from adults. Overall, she showed a notable developmental delay compared to her peers.
At 85 months of age, the proband was assessed again with the Battelle Developmental Inventory. As shown in Figure 1, the gap between the girl's chronological age and her developmental age had increased in all the assessed areas, being expressive language the most impaired domain. At that moment, the proband presented with ID and significant problems in the areas of social interaction and verbal communication. She also exhibited ritualized and disordered behaviors, such as smelling objects, as well as high levels of impulsivity and attention deficit. Her broad independence and adaptive skills were also below expectations for her chronological age.
At 97 months of age, a third evaluation with the Battelle Developmental Inventory was carried out, which showed a significant setback with respect to the 2 previous assessments, the proband's personal/social and adaptive skills being the most affected domains, along with her expressive language (Fig. 1).
In addition, at that age, we also examined in detail the structural and functional features of the child's language, as well as her communication skills through the analysis of a naturally occurring interaction with her mother and sister. The full record can be found in online supplementary File 1. The proband's speech featured frequent omissions of initial consonants (e.g., siguiente > iguiente* “following”; quita > ita* “remove”; ventana > entana* “window”), as well as consonant clusters simplification (e.g., verde > vele* “green”; espejo > epejo* “mirror”; pastel > fafé* “cake”; armario > ababio* “closet”), and sound substitutions (e.g., ducha > lucha* “shower/fight”; roja > doja* “red”; armario > armadio* “closet”; aquí > adí* “here”; música > muzita* “music”; pastel > fafé* “cake”). Consonant frontalization was also frequently observed (e.g., aquí > adí* “here”; armario > ababio* “closet”; pastel > fafé* “cake”), particularly, of alveolar /s/ into interdental /θ/ (e.g., silla > zilla* “chair”; esto > ezto* “this”; eso > ezo* “that”; sí > zí* “yes”; música > muzita* “music”). Additionally, the mean length of utterance in words of the girl's discourse was very low, of 0.981. This roughly corresponds to a restricted holophrastic (i.e., one-word) language, which is typically found in children from 1 to 2 years of age. Although the proband did not produce truly syntactic structures, a hallmark of her discourse was the avoidance or the phonetic alteration of definite and indefinite articles (e.g., la ropa > e dopa* “the clothes”; las manos > a mano* “the hands”).
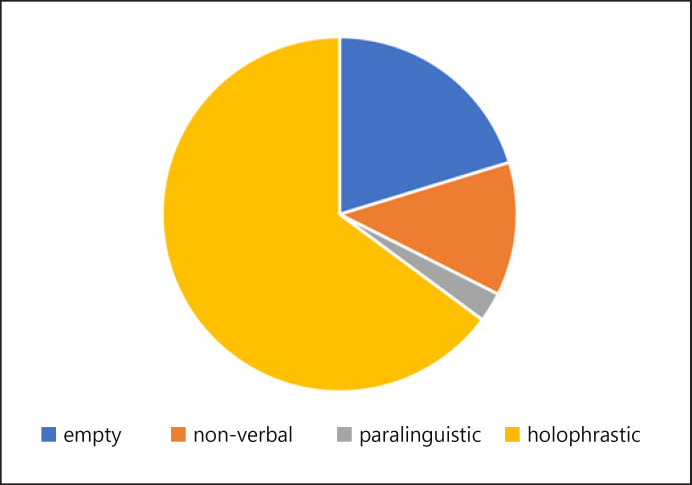
With regards to the pragmatic component, the child did not seem able to properly engage in conversation. Most of her turns at talk, even being verbal by nature (Fig. 2), were reactive, in response to her interlocutor's requests. Usually, the interlocutor needed to repeat the question twice or 3 times in order to obtain a minimal answer (verbal or nonverbal). In the infrequent cases when the child took the floor, she could generate echolalic utterances only.
Fig. 2.
Nature of the conversational turns provided by the child during a spontaneous conversation with a peer. Empty turns: the child provided no answer at all, ignoring her interlocutor and focusing on the objects she was playing with. Non-verbal turns: the child produced a gestural action in response to her interlocutor's request. Paralinguistic: the child tried to take the turn but was only able to produce fillers, such as non-verbal vocalizations. Holophrastic: the child generated single-word responses.
Furthermore, the pragmatic skills needed for understanding the logic of conversation as described by Grice [1975] were seriously impaired. Specifically, transgressions of the maxim of relation (i.e., say things relevant to the topic under discussion) were pervasive (e.g., when asked to name a music player, she might say “dance”; when asked about a picture, she might reply “paper”; or when asked about some windows in the room, she might reply by counting them: “one, two, etc.”), as well as transgressions of the maxim of quantity (i.e., provide the right amount of information), so that the child might refer to the majority of the things around by means of demonstrative pronouns like “this” and “that”, which are not informative enough. Finally, her communicative use of gaze was also impaired. Usually, she did not establish visual contact with her conversational partner, and she experienced difficulties with joint attention when asked to name an object. No compensatory strategies aimed to improve her communication deficits were observed (e.g., asking for help when she was unable to find a word).
To sum up, our proband exhibits severe speech and language deficits, with her expressive skills being dramatically affected, to the point that she is unable to pronounce words correctly, to build up simple sentences, and to name familiar things around her. These deficits hinder the intelligibility of her discourse and preclude from successfully communicating. Language comprehension is also seriously impaired, as well as pragmatic abilities: she shows no metapragmatic awareness (i.e., she is unable to realize which of her utterances are problematic or failed, and she is unable to generate compensatory strategies aimed to improve communicative success); she frequently produces non-related, tangential and/or insufficiently informative answers; she is not able to properly engage in conversation according to the turn-taking rules [Sacks et al., 1974], and she makes no communicative use of gaze.
Cytogenetic and Molecular Analyses
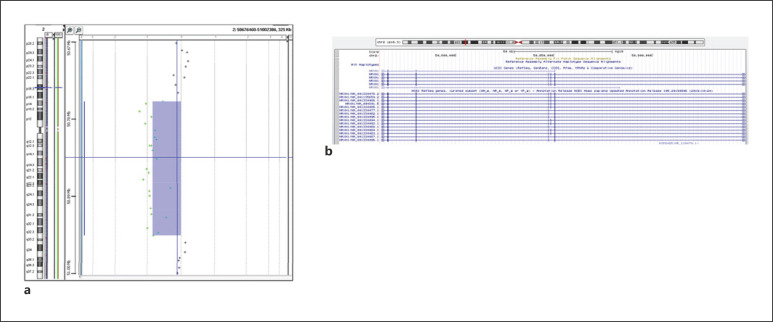
An array comparative genomic hybridization (array-CGH) was performed when the proband was 5 years and 1 month old. The array confirmed the presence of a deletion of 0.186 Mb in the 2p16.3 region (arr[hg19] 2p16.3(50761778_50947729)×1), predicted to be pathological, and affecting exons 3–7 of the NRXN1 gene (Fig. 3a, b). The deletion was inherited from the proband's healthy father.
Fig. 3.
Chromosomal alterations found in the proband. a Screen capture of the array-CGH of the proband's chromosome 2 showing the microdeletion at 2p16.3. b Screen capture of the UCSC Genome Browser (https://genome.ucsc.edu/) showing the extension of the deletion in NRXN1.
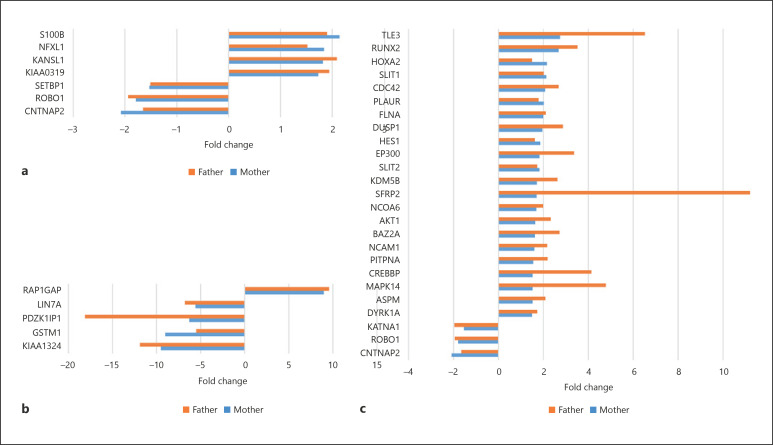
We then used String 11.0 (https://www.string-db.org) for finding functional links between NRXN1 and the products of genes important for language, specifically, candidates for DD and SLI, but also candidates for language evolution. Because String did not predict any functional link of interest between NRXN1 and the products of these genes (data not shown), we conducted additional in vitro analyses. Specifically, we performed microarray analyses of blood samples from the proband to determine whether she exhibited altered patterns of gene expression that may account for the observed deficits. We used her healthy carrier father and her healthy non-carrier mother as controls. The results of the microarrays are shown in online supplementary File 3. We found that NRXN1 was only slightly upregulated in the proband (1.27-fold change [FC] compared to her healthy non-carrier mother; 1.1 FC compared to her healthy carrier father). Additionally, we checked whether the candidate genes for language disorders and/or evolution used in our in silico analyses were differentially expressed in the girl compared to her parents. For most of the candidates for language disorders, we found no significant differences (i.e., FC > 1.5 compared to both healthy parents). However, some of them (CNTNAP2, ROBO1, and SETBP1) were significantly downregulated in the proband, whereas some others (KIAA0319, KANSL1, NFXL1, and S100B) were significantly upregulated (Fig. 4a). Regarding candidates for language evolution, we found that most genes that were differentially expressed genes (FC > 1.5 compared to both healthy parents) were upregulated in the proband (including DYRK1A, ASPM, NCAM1, BAZ2A, SLIT2, DUSP1, FLNA, PLAUR, CDC42, SLIT1, RUNX2, and TLE3) (Fig. 4c). Finally, we searched for additional candidates for the proband's language deficits, looking for genes exhibiting the strongest FC in our subject (i.e., FC > 5 compared to both unaffected parents). We found that several genes of interest were strongly downregulated (KIAA1324, GSTM1, PDZK1IP1, and LIN7A), whereas only 1 gene (RAP1GAP) was strongly upregulated (Fig. 4b).
Fig. 4.
Variation in the expression levels of genes of interest in the proband's blood compared to her healthy parents. a Fold changes in the expression levels of candidates for language disorders. b Genes exhibiting fold changes >5 in the proband compared to both healthy parents. c Fold changes in the expression levels of candidates for language evolution.
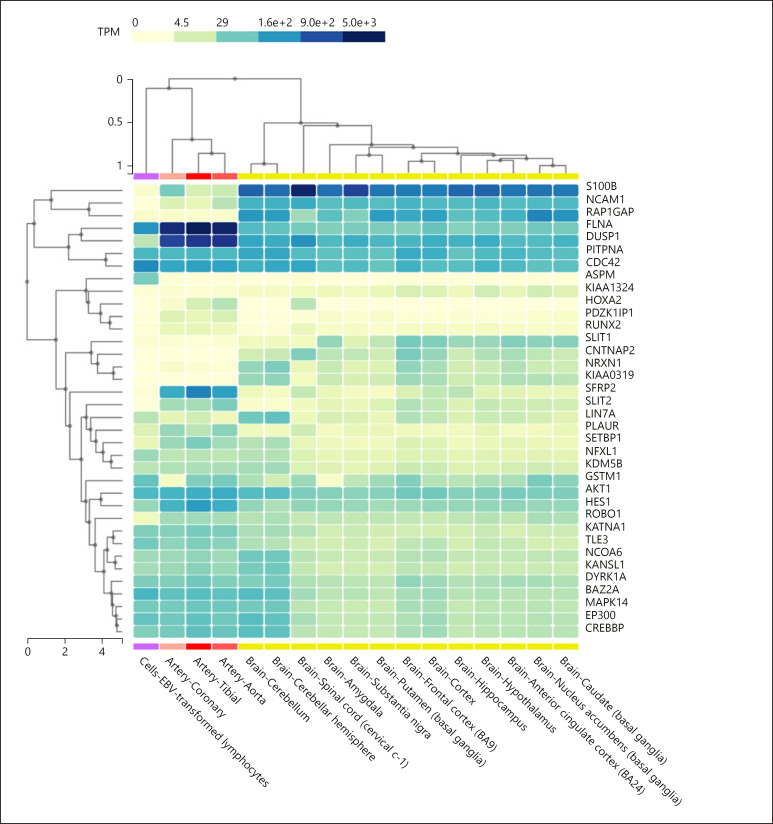
We then checked the expression patterns in the brain of genes of interest using the Genotype-Tissue Expression (GTEx) project interface (https://www.gtexportal.org/home/). Our results are summarized in Figure 5. With regards to the candidates for language disorders and/or evolution that are significantly down- or upregulated in the blood of our proband, we found that around 50% are more expressed in the brain than in the blood. These include S100B (strongly expressed in the spinal cord, the substantia nigra, the hypothalamus, and the hippocampus), NCAM1 (highly expressed across the whole brain), SLIT1 (expressed throughout the brain), CNTNAP2 (more expressed in the cortex), and KIAA0319 (highly expressed in the cortex and parts of the basal ganglia). Finally, with regards to the genes that are strongly downregulated in the proband, we found that LIN7A is expressed at similar levels in the nervous system and in the blood, although it is upregulated in the cerebellum. PDZK1IP1 is expressed at lower levels in the brain than in the blood. Transcripts of this gene are preferentially found in the spinal cord. GSTM1 is expressed at similar levels in the blood and in the brain, where it is mostly expressed in the frontal cortex and the basal ganglia. KIAA1324 is expressed at higher levels in the brain, particularly in the cortex, the hypothalamus and parts of the basal ganglia. Lastly, RAP1GAP is expressed at higher levels in the brain than in the blood, particularly in the cortex and the basal ganglia.
Fig. 5.
Expression pattern in the blood and the brain of genes of interest. Expression levels of the genes were retrieved from the Genotype-Tissue Expression (GTEx) project (https://www.gtexportal.org/home/). Statistical analysis and data interpretation were performed by the GTEx Consortium Analysis Working Group using the GTEx Multi Gene Query tool. TPM, transcripts per million.
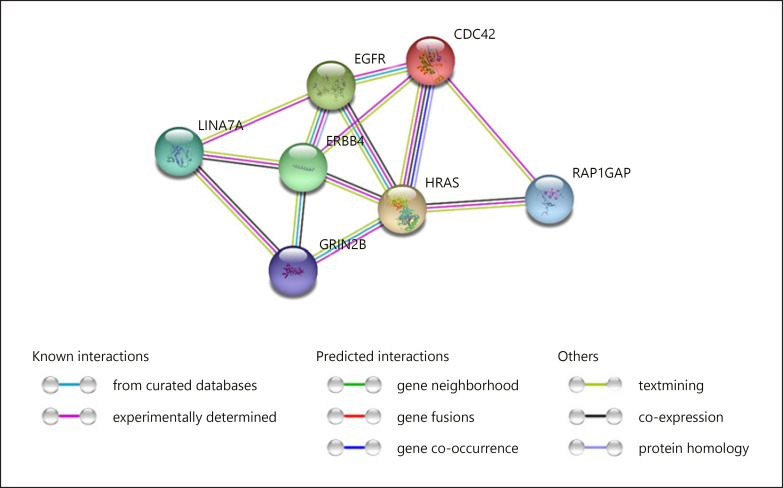
Finally, we used String 11.0 for knowing more about functional interactions of interest between the proteins encoded by the genes found strongly dysregulated in the proband and the proteins encoded by candidate genes for language disorders and/or evolution. We found predicted functional links between LIN7A and GRIN2B, ERBB4, and EGFR, as well as between RAP1GAP and CDC42 and HRAS (Fig. 6).
Fig. 6.
Interaction network of genes found strongly dysregulated in the proband and the proteins encoded by candidate genes for language disorders and/or evolution. The network was drawn with String, version 11.0 [Szklarczyk et al., 2015] license-free software (http://string-db.org/), using the molecular action visualization. Colored nodes symbolize genes/proteins included in the query. The color of the lines represents different kind of evidence, as shown in the legend. Stronger associations between proteins are represented by thicker lines. The medium confidence value was 0.0400 (a 40% probability that a predicted link exists in a particular database). The diagram only represents the potential connectivity between the involved proteins, which has to be mapped onto particular biochemical networks, signaling pathways, cellular properties, aspects of neuronal function, or cell types of interest.
Discussion
The widespread application of NGS facilities, particularly array-CGH, to people with conditions featuring cognitive and language deficits has resulted in an increasing list of candidate genes for these problems. Nonetheless, it is usually difficult to obtain robust genotype-to-phenotype links, particularly in the case of complex, multifactorial traits like language. In this paper, we provide a detailed characterization of the language problems exhibited by a girl with a microdeletion in 2p16.3 affecting part of the NRXN1 gene. Mutations or genomic losses in this gene have been associated with diverse cognitive and behavioral problems, including language and communication deficits [Béna et al., 2013; Curran et al., 2013; Brignell et al., 2018; Al Shehhi et al., 2019], although we still lack a mechanistic account of these problems.
Our proband exhibits most of the cognitive, behavioral, and neurological anomalies observed in patients with losses of NRXN1. This is specifically true for her speech, language, and communication (dis)abilities (Table 1).
Table 1.
Summary table with the most relevant clinical features of our proband compared to patients with deletions of NRXN1 according to Brignell et al., 2018 and DECIPHER
| Cognitive/behavioural features | |
|---|---|
| Intellectual disability | + |
| Autism spectrum disorder | + |
| Attention deficit hyperactivity disorder | + |
| Schizophrenia | |
| Anxiety | |
| Speech impairment | + |
| Expressive language impairment | + |
| Receptive language impairment | + |
| Impaired pragmatics (social communication difficulties) | + |
| Brain/neurological findings | |
| Childhood apraxia of speech | + |
| Dysarthria | |
| Hypotonia | + |
| Epilepsy | |
| Complex neurodevelopmental disorder | + |
+, features present in our proband.
From our detailed analysis of the proband's speech problems, we conclude that they are indicative of childhood apraxia of speech (CAS), a condition characterized by inconsistent errors on consonants and vowels in repeated productions of words and inappropriate prosody. Nonetheless, we have also found evidence of impaired phonological awareness. Our results are thus in line with findings by Brignell et al. [2018], who suggest that articulation and phonological problems co-occur in children with NRXN1 deletions.
We also found that besides her severe expressive language delay, the receptive skills of our proband are also notably affected. This finding is also in line with previous research [e.g., Ching et al., 2010; Gregor et al., 2011; Béna et al., 2013; Curran et al., 2013; Dabell et al., 2013].
Finally, we report for the first time on the pragmatic impairment associated with deletions of NRXN1. This pragmatic deficit consists of an inability to properly engage in turn-taking during conversation together with a reduced metapragmatic awareness, which impedes perceiving the violation of the logic and the rules of conversation. All this results in different abnormal behaviors during conversation, like producing tangential or perseverating answers to new questions, or interpreting utterances in a literal way. These pragmatic problems seemingly become exacerbated by other co-occurring problems, like her reduced vocabulary, her altered expressive language/grammatical skills, her attention deficit, and her ASD-like features.
Overall, this language and communication profile partially overlaps with the deficits found in other neurodevelopmental disorders, such as ASD, ADHD, and moderate to severe ID, which are commonly comorbid conditions in children with NRXN1 deletions (ASD: 43–65% [Schaaf et al., 2012; Béna et al., 2013; Dabell et al., 2013; González-Mantilla et al., 2016]; ADHD: 9–41% [Schaaf et al., 2012; Lowther et al., 2017]; ID: 77–92% [Schaaf et al., 2012; Béna et al., 2013; Dabell et al., 2013]). In fact, as pointed out by Lowther et al. [2017], around 46% of individuals with NRXN1 deletions present with dual neurodevelopmental disorder diagnoses, as it is the case of our proband, who was diagnosed with a moderate ID, but who also exhibits ADHD features, as well as ASD-like traits, including problems with social interactions, some ritualized behaviors, and altered sensory experience. Brignell et al. [2018] have suggested that the speech and language problems of individuals with NRXN1 deletions could result from their ID and ASD features, a possibility that is supported by several studies pointing to a correlation between ID, ASD, and language difficulties [Levy et al., 2010; Liao et al., 2015].
For this reason, besides providing a more detailed profile of the problems with language exhibited by our proband, as well as some hints about their cognitive causes, we also aimed to advance an alternative hypothesis about the causes of the language problems resulting from losses of the NRXN1 gene. The known roles of NRXN1 in synapse development and maintenance, and neuron network assembly during cortical development [Missler and Südhof, 1998; Südhof, 2008; Gjørlund et al., 2012; Jenkins et al., 2016], are compatible with a putative role in the development and maintenance, specifically, of brain circuits underlying language function. This possibility is reinforced by the fact that the gene is expressed in areas involved in language processing, particularly, the cerebellum, which contributes significantly to motor planning, but also to many aspects of language processing [Vias and Dick, 2017; Mariën and Borgatti, 2018], along with the prefrontal cortex region, which is a core region for speech and language [Jenkins et al., 2016]. Deficits in the frontal cortex are known to impact on executive function, and problems with executive function have been hypothesized to contribute to diverse neurodevelopmental disorders, including ASD, ADHD, and SZ [Ozonoff and Jensen, 1999; Martos-Pérez and Paula-Pérez, 2011]. As we have repeatedly pointed out, our proband exhibits ASD- and ADHD-like features. Additionally, her paternal uncle, who was also a carrier of the NRXN1 deletion inherited by our proband, presented with SZ. Furthermore, executive dysfunction has been claimed to account for the generativity deficit observed in ASD (i.e., the lack of new ideas, behaviors, and utterances tailored to new contexts) and ultimately, for the stereotypies, echolalic utterances and restrained interests typical of this condition [Turner, 2000; Gilotty et al., 2002]. Additionally, some studies [e.g., Perner and Lang, 2000; Russell, 2000; Ibáñez-Barassi, 2005] have associated executive dysfunctions with an impaired theory of mind. Overall, one could hypothesize that the NRXN1 deletion found in our proband may impact on her executive function, thus affecting a wide range of cognitive abilities, such as attention (which plays a central role in learning and communication), theory of mind (necessary for the development of adequate communication skills) and, of course, speech and language.
Brignell et al. [2018] support the view that there is no specific linguistic profile for deletions of NRXN1. We agree that the speech, language, and communication deficits found in our proband can be expected to result to a great extent from the impairment of more basic cognitive functions, which are also affected in other high-prevalent, co-morbid neurodevelopmental conditions in children with NRXN1 deletions. Nonetheless, we think that it is possible to identify and characterize a specific molecular pathway accounting for the speech and language problems observed in deletions of NRXN1. Accordingly, our hypothesis is that changes in NRXN1dosage might impact on strong candidates for language development and evolution. In our proband, NRXN1 is not significantly dysregulated compared to her healthy parents. We did not find either in silico evidence of a functional connection between the protein NRXN1 and the products of candidates for language development, impairment, and/or evolution. Nonetheless, we have found that several candidates for language disorders and/or language evolution are indeed dysregulated in the proband's blood. While as noted we have no evidence of a direct effect of the loss of NRXN1 on the dysregulation of these genes, these changes can be expected to contribute to the language deficits observed in the proband. Among the genes found downregulated, we wish to highlight the DD candidates ROBO1 and SETBP1. The former encodes a protein involved in thalamocortical axon development and plays a critical role in the establishment of vocal learning pathways in many species [Pfenning et al., 2014]. Mutations in the latter give rise to Schinzel-Giedion syndrome, a clinical condition entailing occasional epilepsy and severe developmental delay [Ko et al., 2013; Miyake et al., 2015], as well as behavioral and social deficits [Coe et al., 2014]. Also the DD candidate CNTNAP2 is found strongly downregulated in our proband. This gene encodes a protein associated with K+ voltage-gated channels in the pyramidal cells of the temporal cortex [Inda et al., 2006], which contributes to regulate brain connectivity and morphology [Scott-Van Zeeland et al., 2010; Tan et al., 2010; Dennis et al., 2011], dendritic arborization and spine development [Anderson et al., 2012], and axonogenesis in conjunction with ROBO factors [Banerjee et al., 2010]. CNTNAP2 has been also associated with language and speech regression [Strauss et al., 2006; Marchese et al., 2016; Smogavec et al., 2016], SLI [Newbury et al., 2011], ASD [Alarcón et al., 2008; Bakkaloglu et al., 2008], CAS [Worthey et al., 2013], and language impairment in SZ [Poot, 2015]. As noted, CAS is a core symptom in our proband, whereas one of her relatives has been diagnosed with SZ. Moreover, several candidates for DD are upregulated in the proband, including KIAA0319, KANSL1, and S100B. KIAA0319 encodes a membrane protein contributing to the interactions between neurons and radial glial cells during neuronal migration [Paracchini et al., 2006; Velayos-Baeza et al., 2008]. Mutations in KANSL1, which encodes a component of the NSL1 complex, important for chromatin organization and gene transcription regulation, result in Koolen-de Vries syndrome (OMIM #610443). This clinical condition features epilepsy, developmental delay, and moderate ID, which impacts mostly on expressive language development [Koolen et al., 2016]. Finally, S100B encodes a calcium-binding protein involved in neurite extension, axonal proliferation, and synaptic plasticity and learning [Sorci et al., 2013].
Among the candidates for language evolution (many of them resulting in cognitive disorders like ASD or SZ when mutated) that are found dysregulated in the proband, we wish to highlight SLIT1, SLIT2, and HES1. The SLIT/ROBO pathway is thought to play a crucial role in the externalization of language (i.e., speech) [for details, see Boeckx and Benítez-Burraco, 2014b]. Several other genes related to (the evolution of) speech are upregulated in the girl, including DUSP1, involved in vocal learning in songbirds [Horita et al., 2010, 2012]; PLAUR, part of FOXP2's interactome [Royer-Zemmour et al., 2008; Roll et al., 2010] and a candidate for speech dyspraxia [Roll et al., 2006]; TLE3, a target of FOXP2 in the inferior prefrontal cortex [Spiteri et al., 2007]; and FLNA, which encodes an actin-binding protein needed for cytoskeleton remodeling and neuronal migration [Fox et al., 1998], and that interacts with CDC42, which is also found upregulated in our proband. CDC42 regulates cortical interneuron migration [Katayama et al., 2013] and dendritic spine growth and function [Datta et al., 2015]. In mice, SLIT2 reduces CDC42 activity in living growth cones [Myers et al., 2012], whereas depletion of ROBO1 prevents SLIT2 inhibition of CDC42 activity [Yiin et al., 2009]. The abnormal expression pattern of these candidates for DD and speech-related genes might account for the speech and phonological problems exhibited by our proband.
Also of interest for explaining the language deficits observed in the proband is the upregulation of NCAM1, a gene related to working memory performance [Bisaz et al., 2013], but also to neuropsychiatric conditions like SZ, bipolar disorder, and Alzheimer disease [Atz et al., 2007]. NCAM1 is a target of FOXP2 [Konopka et al., 2009], but also of RUNX2 [Kuhlwilm et al., 2013]. RUNX2 is strongly upregulated in the proband. This is a key gene hypothesized to account for some of the changes in the human brain morphology and configuration resulting in our cognitive uniqueness, particularly, our cross-modal thinking [for a detailed discussion, see Boeckx and Benítez-Burraco, 2014a]. Also ASPM is found upregulated in our proband. This is a strong candidate for microcephaly, but also for the increase in brain size in the human lineage [Bond et al., 2002; Zhang, 2003]. Finally, the finding that DYRK1A is upregulated in the girl is of interest too. Mutations in this gene result in microcephaly, as well as in ID and absence of speech [Van Bon et al., 2011; Courcet et al., 2012]. The gene seems to play a role in learning and memory [Hämmerle et al., 2003] via its effects on synaptic plasticity and on the balance between excitation and inhibition [Souchet et al., 2014]. Overall, the abnormal expression patterns of these genes can also contribute to the speech problems of the proband through their effects on the FOXP2 interactome, but also to her problems with grammar through their effect on the RUNX2 network.
Lastly, it is worth considering the biological roles played by the genes that are strongly dysregulated in the blood of the girl compared to her healthy parents. The most promising genes for explaining her language deficits are LIN7A (found strongly downregulated) and RAP1GAP (found strongly upregulated). Both genes are expressed in the brain, particularly in the cerebellum (LIN7A) and the striatum (RAP1GAP), which are 2 regions that are crucially involved in language processing [Silveri, 2020], particularly, in speech production [Konopka and Roberts, 2016]. Deletions in LIN7A, a gene encoding a scaffold protein important for synaptic function, impact on the development of the cerebral cortex and result in ID [Matsumoto et al., 2014]. Regarding RAP1GAP, it encodes a GTPase-activating-protein acting as a negative regulator of RAP1, which is involved in cell proliferation, adhesion, differentiation, and embryogenesis and, specifically, in neuronal migration [Yang et al., 2019] and the regulation of dendritic spine morphology in the striatum [McAvoy et al., 2009] and the cortex [Chen et al., 2005]. RAP1GAP mediates aversive behaviors in the nucleus accumbens [Lin et al., 2020]. Interestingly, we have found that the products of these 2 genes interact with the products of several candidates for language impairment and/or evolution, particularly, GRIN2A, GRIN2B, HRAS, and ERBB4. GRIN2A and GRIN2B are two of the components of the NR2 subunit of a receptor for N-methyl-D-aspartate (NMDA), involved in long-term potentiation and ultimately in memory and learning. Mutations in GRIN2A give rise to speech impairment and language regression, as part of epileptic syndromes like Landau-Kleffner syndrome, continuous spike and waves during slow-wave sleep (CSWSS) syndrome, or rolandic epilepsies [Carvill et al., 2013; Lesca et al., 2013; Dimassi et al., 2014]. The speech problems observed in patients include syllable repetitions, imprecise articulation, and problems with pitch and prosody, which are usually labelled as dyspraxia or dysarthria [Turner et al., 2015], and which parallel the speech deficits observed in our proband. By contrast, mutations in GRIN2B are mostly related to behavioral problems, but also to motor and cognitive problems [Freunscht et al., 2013; Hu et al., 2016; Smigiel et al., 2016]. Concerning HRAS, it is a candidate for DD and ASD, and encodes a GTPase that contributes to regulate neural growth and differentiation, long-term potentiation, and synaptic plasticity [Comings et al., 1996]. Lastly, ERBB4is involved in the regulation of interneuron migration and in synchronizing neural oscillations in the cortex [Li et al., 2012; Hou et al., 2014]. Mutations in ERBB4 result in ID and speech delay [Kasnauskiene et al., 2013]. In our view, the strong dysregulation of these genes could contribute as well to explain the speech, language, cognitive, and behavioral problems exhibited by our proband.
Conclusions
Although the exact molecular causes of the speech and language problems observed in our proband remain to be fully elucidated, we think that they might result from the dysregulation of several known candidates for language impairment and/or evolution with a known function on brain development and with a role in cognitive diseases impacting on speech and language, particularly, ASD, ID, and DD. This hypothesis needs to be properly tested. Particularly, the link between the loss of NRXN1 and the dysregulation of these genes needs to be probed in vitro and in vivo. Still, we hope that our findings contribute to a better understanding of the speech, language, cognitive, and behavioral phenotype resulting from mutations and CNVs of this key neurexin. More detailed accounts of the neurobiological basis of the deficits (and strengths) exhibited by patients are necessary for improving (psycho)pedagogical strategies aimed to ameliorate their problems.
Statement of Ethics
Ethics approval for this research was granted by the Comité Ético del Hospital “Reina Sofía”. Written informed consent was obtained from the proband's parents for conducting the psycholinguistic evaluations and the molecular analyses, and for publicizing this case report and all the accompanying tables and images in scientific journals and meetings.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
This research was conducted in absence of external financial aid.
Author Contributions
Antonio Benítez-Burraco conceived the paper, planned the molecular experiments, analyzed the genomic/genetic data, and wrote the paper. Maite Fernández-Urquiza and M. Salud Jiménez-Romero conducted the clinical evaluation of the proband, analyzed the psycholinguistic data, and wrote the paper. All authors approved the final version of the manuscript.
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article. A preprint version of this article is available on PsyArXiv [https://doi.org/10.31234/osf.io/aw25n].
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
We would like to thank the proband and her family for their participation in this research. We wish to thank also Dr. Montserrat Barcos Martínez and Dr. Isabel Espejo Portero, from the “Reina Sofía” hospital in Córdoba, for granting us access to the patient and her clinical history.
Funding Statement
This research was conducted in absence of external financial aid.
References
- 1.Alarcón M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al Shehhi M, Forman EB, Fitzgerald JE, McInerney V, Krawczyk J, Shen S, et al. NRXN1 deletion syndrome: phenotypic and penetrance data from 34 families. Eur J Med Genet. 2019;62((3)):204–209. doi: 10.1016/j.ejmg.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 3.Anderson GR, Galfin T, Xu W, Aoto J, Malenka RC, Südhof TC. Candidate autism gene screen identifies critical role for cell-adhesion molecule CASPR2 in dendritic arborization and spine development. Proc Natl Acad Sci U S A. 2012;109((44)):18120–18125. doi: 10.1073/pnas.1216398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atz ME, Rollins B, Vawter MP. NCAM1 association study of bipolar disorder and schizophrenia: polymorphisms and alternatively spliced isoforms lead to similarities and differences. Psychiatr Genet. 2007;17((2)):55. doi: 10.1097/YPG.0b013e328012d850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakkaloglu B, O'Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82((1)):165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee S, Blauth K, Peters K, Rogers SL, Fanning AS, Bhat MA. Drosophila neurexin IV interacts with roundabout and is required for repulsive midline axon guidance. J Neurosci. 2010;30((16)):5653–5667. doi: 10.1523/JNEUROSCI.6187-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Béna F, Bruno DL, Eriksson M, van Ravenswaaij-Arts C, Stark Z, Dijkhuizen T, et al. Molecular and clinical characterization of 25 individuals with exonic deletions of NRXN1 and comprehensive review of the literature. Am J Med Genet B Neuropsychiatr Genet. 2013;162B((4)):388–403. doi: 10.1002/ajmg.b.32148. [DOI] [PubMed] [Google Scholar]
- 8.Benítez-Burraco A, Boeckx C. Possible functional links among brain-and skull-related genes selected in modern humans. Front Psychol. 2015;6:794. doi: 10.3389/fpsyg.2015.00794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benítez-Burraco A, Murphy E. The oscillopathic nature of language deficits in autism: from genes to language evolution. Front Hum Neurosci. 2016;10:120. doi: 10.3389/fnhum.2016.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bisaz R, Boadas-Vaello P, Genoux D, Sandi C. Age-related cognitive impairments in mice with a conditional ablation of the neural cell adhesion molecule. Learn Mem. 2013;20((4)):183–193. doi: 10.1101/lm.030064.112. [DOI] [PubMed] [Google Scholar]
- 11.Boeckx C, Benítez-Burraco A. The shape of the human language-ready brain. Front Psychol. 2014a;5:282. doi: 10.3389/fpsyg.2014.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boeckx C, Benítez-Burraco A. Globularity and language-readiness: generating new predictions by expanding the set of genes of interest. Front Psychol. 2014b;5:1324. doi: 10.3389/fpsyg.2014.01324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32((2)):316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 14.Brignell A, St John M, Boys A, Bruce A, Dinale C, Pigdon L, et al. Characterization of speech and language phenotype in children with NRXN1 deletions. Am J Med Genet B Neuropsychiatr Genet. 2018;177:700–708. doi: 10.1002/ajmg.b.32664. [DOI] [PubMed] [Google Scholar]
- 15.Carvill GL, Regan BM, Yendle SC, O'Roak BJ, Lozovaya N, Bruneau N, et al. GRIN2A mutations cause epilepsy-aphasia spectrum disorders. Nat Genet. 2013;45((9)):1073–1076. doi: 10.1038/ng.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castronovo P, Baccarin M, Ricciardello A, Picinelli C, Tomaiuolo P, Cucinotta F, et al. Phenotypic spectrum of NRXN1 mono- and bi-allelic deficiency: A systematic review. Clin Genet. 2020;97((1)):125–137. doi: 10.1111/cge.13537. [DOI] [PubMed] [Google Scholar]
- 17.Chen XS, Reader RH, Hoischen A, Veltman JA, Simpson NH, Francks C, et al. Next-generation DNA sequencing identifies novel gene variants and pathways involved in specific language impairment. Sci Rep. 2017;7((1)):46105–17. doi: 10.1038/srep46105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, Wang PY, Ghosh A. Regulation of cortical dendrite development by Rap1 signaling. Mol Cell Neurosci. 2005;28((2)):215–228. doi: 10.1016/j.mcn.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Ching MS, Shen Y, Tan WH, Jeste SS, Morrow EM, Chen X, et al. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am J Med Genet B Neuropsychiatr Genet. 2010;153b((4)):937–947. doi: 10.1002/ajmg.b.31063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coe BP, Witherspoon K, Rosenfeld JA, Van Bon BW, Vulto-van Silfhout AT, Bosco P, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46((10)):1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comings DE, Wu S, Chiu C, Muhleman D, Sverd J. Studies of the c-Harvey-Ras gene in psychiatric disorders. Psychiatry Res. 1996;63((1)):25–32. doi: 10.1016/0165-1781(96)02829-6. [DOI] [PubMed] [Google Scholar]
- 22.Courcet JB, Faivre L, Malzac P, Masurel-Paulet A, Lopez E, Callier P, et al. The DYRK1A gene is a cause of syndromic intellectual disability with severe microcephaly and epilepsy. J Med Genet. 2012;49((12)):731–736. doi: 10.1136/jmedgenet-2012-101251. [DOI] [PubMed] [Google Scholar]
- 23.Curran S, Ahn JW, Grayton H, Collier DA, Ogilvie CM. NRXN1 deletions identified by array comparative genome hybridisation in a clinical case series - further understanding of the relevance of NRXN1 to neurodevelopmental disorders. J Mol Psychiatry. 2013;1((1)):4. doi: 10.1186/2049-9256-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dabell MP, Rosenfeld JA, Bader P, Escobar LF, El-Khechen D, Vallee SE, et al. Investigation of NRXN1 deletions: Clinical and molecular characterization. Am J Med Genet A. 2013;161A((4)):717–731. doi: 10.1002/ajmg.a.35780. [DOI] [PubMed] [Google Scholar]
- 25.Datta D, Arion D, Corradi JP, Lewis DA. Altered expression of CDC42 signaling pathway components in cortical layer 3 pyramidal cells in schizophrenia. Biol Psychiatry. 2015;78((11)):775–785. doi: 10.1016/j.biopsych.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De La Cruz MV, González M. Madrid: TEA; 2011. Adaptación Española del Inventario de Desarrollo Battelle. [Google Scholar]
- 27.Dennis EL, Jahanshad N, Rudie JD, Brown JA, Johnson K, McMahon KL, et al. Altered structural brain connectivity in healthy carriers of the autism risk gene, CNTNAP2. Brain Connect. 2011;1((6)):447–459. doi: 10.1089/brain.2011.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diez-Itza E, Martínez V, Pérez V, Fernández-Urquiza M. Explicit oral narrative intervention for students with Williams syndrome. Front Psychol. 2017;8:2337. doi: 10.3389/fpsyg.2017.02337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diez-Itza E, Viejo A, Fernández-Urquiza M. Pragmatic Profiles of Adults with Fragile X Syndrome and Williams Syndrome. Brain Sci. 2022;12((3)):385. doi: 10.3390/brainsci12030385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dimassi S, Labalme A, Lesca G, Rudolf G, Bruneau N, Hirsch E, et al. A subset of genomic alterations detected in rolandic epilepsies contains candidate or known epilepsy genes including GRIN2A and PRRT2. Epilepsia. 2014;55((2)):370–378. doi: 10.1111/epi.12502. [DOI] [PubMed] [Google Scholar]
- 31.Fernández-Urquiza M, Díaz F, Moreno V, Lázaro M, Simón T. Protocolo Rápido de Evaluación Pragmática Revisado. Universidad de Valencia; 2015. PREP-R. [Google Scholar]
- 32.Fernández-Urquiza M, Miranda M, Martínez V, Diez-Itza E. Pragmática textual de las narraciones en el síndrome de Down: Perfiles de coherencia y cohesión. In: Aguilar E, Adrover D, Bull L, López R, editors. Proceedings of the VIIIth International Conference of Language Acquisition. Universidad de las Islas Baleares; 2016. p. p. 30. [Google Scholar]
- 33.Fernández-Urquiza M, Diez-Itza E, Cortiñas S. PREP-CORP: Sistema de etiquetado pragmático de corpus clínicos de lengua oral. In: Fernández MC, Martí M, Ruiz AM, editors. Investigaciones actuales en Lingüística. Volumen VI: Aplicaciones de la Lingüística. Universidad de Alcalá de Henares; 2017a. pp. p. 167–183. [Google Scholar]
- 34.Fernández-Urquiza M, Shiro M, Viejo A, Miranda M, Diez-Itza E. 14th International Conference for the Study of Child Language. University of Lyon; 2017b. Pragmatic Profiles of Williams and Down Syndromes Narratives: Textual Coherence and Evaluative Language. [Google Scholar]
- 35.Fox JW, Lamperti ED, Ekşioğlu YZ, Hong SE, Feng Y, Graham DA, et al. Mutations in Filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- 36.Freunscht I, Popp B, Blank R, Endele S, Moog U, Petri H, et al. Behavioral phenotype in five individuals with de novo mutations within the GRIN2B gene. Behav Brain Funct. 2013;9:20. doi: 10.1186/1744-9081-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilotty L, Kenworthy L, Sirian L, Black DO, Wagner AE. Adaptive skills and executive function in autism spectrum disorders. Child Neuropsychol. 2002;8:241–248. doi: 10.1076/chin.8.4.241.13504. [DOI] [PubMed] [Google Scholar]
- 38.Gjørlund MD, Nielsen J, Pankratova S, Li S, Korshunova I, Bock E, et al. Neuroligin-1 induces neurite outgrowth through interaction with neurexin-1β and activation of fibroblast growth factor receptor-1. FASEB J. 2012;26((10)):4174–4186. doi: 10.1096/fj.11-202242. [DOI] [PubMed] [Google Scholar]
- 39.González-Mantilla AJ, Moreno-De-Luca A, Ledbetter DH, Martin CL. A cross-disorder method to identify novel candidate genes for developmental brain disorders. JAMA Psychiatry. 2016;73((3)):275–283. doi: 10.1001/jamapsychiatry.2015.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregor A, Albrecht B, Bader I, Bijlsma EK, Ekici AB, Engels H, et al. Expanding the clinical spectrum associated with defects in CNTNAP2 and NRXN1. BMC Med Genet. 2011;12:106. doi: 10.1186/1471-2350-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grice HP. Logic and Conversation. In: Cole P, Morgan J, editors. Syntax and Semantics Volume 3: Speech Acts. Academic Press; 1975. pp. p. 41–58. [Google Scholar]
- 42.Hämmerle B, Carnicero A, Elizalde C, Ceron J, Martínez S, Tejedor FJ. Expression patterns and subcellular localization of the Down syndrome candidate protein MNB/DYRK1A suggest a role in late neuronal differentiation. Eur J Neurosci. 2003;17((11)):2277–2286. doi: 10.1046/j.1460-9568.2003.02665.x. [DOI] [PubMed] [Google Scholar]
- 43.Horita H, Kobayashi M, Oka K, Jarvis E, Wada K. Specialized motor-driven dusp1 expression in song nuclei of vocal learning birds. Neurosci Res. 2010;68((S1)):e179. doi: 10.1371/journal.pone.0042173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horita H, Kobayashi M, Liu WC, Oka K, Jarvis ED, Wada K. Specialized motor-driven dusp1 expression in the song systems of multiple lineages of vocal learning birds. PLoS One. 2012;7((8)):e42173. doi: 10.1371/journal.pone.0042173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou XJ, Ni KM, Yang JM, Li XM. Neuregulin 1/ErbB4 enhances synchronized oscillations of prefrontal cortex neurons via inhibitory synapses. Neuroscience. 2014;261:107–117. doi: 10.1016/j.neuroscience.2013.12.040. [DOI] [PubMed] [Google Scholar]
- 46.Hu C, Chen W, Myers SJ, Yuan H, Traynelis SF. Human GRIN2B variants in neurodevelopmental disorders. J Pharmacol Sci. 2016;132((2)):115–121. doi: 10.1016/j.jphs.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Z, Xiao X, Zhang Z, Li M. Genetic insights and neurobiological implications from NRXN1 in neuropsychiatric disorders. Mol Psychiatry. 2019;24((10)):1400–1414. doi: 10.1038/s41380-019-0438-9. [DOI] [PubMed] [Google Scholar]
- 48.Ibáñez-Barassi AM. Autismo, funciones ejecutivas y mentalismo: reconsiderando la heurística de descomposición modular. Revista Argentina de Neuropsicología. 2005;6:25–49. [Google Scholar]
- 49.Inda MC, DeFelipe J, Muñoz A. Voltage-gated ion channels in the axon initial segment of human cortical pyramidal cells and their relationship with chandelier cells. Proc Natl Acad Sci U S A. 2006;103((8)):2920–2925. doi: 10.1073/pnas.0511197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4((2)):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 51.Jenkins AK, Paterson C, Wang Y, Hyde TM, Kleinman JE, Law AJ. Neurexin 1 (NRXN1) splice isoform expression during human neocortical development and aging. Mol Psychiatry. 2016;21((5)):701–706. doi: 10.1038/mp.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kasnauskiene J, Ciuladaite Z, Preiksaitiene E, Utkus A, Peciulyte A, Kučinskas V. A new single gene deletion on 2q34: ERBB4 is associated with intellectual disability. Am J Med Genet A. 2013;161A((6)):1487–1490. doi: 10.1002/ajmg.a.35911. [DOI] [PubMed] [Google Scholar]
- 53.Katayama K, Imai F, Campbell K, Lang RA, Zheng Y, Yoshida Y. RhoA and Cdc42 are required in pre-migratory progenitors of the medial ganglionic eminence ventricular zone for proper cortical interneuron migration. Development. 2013;140((15)):3139–3145. doi: 10.1242/dev.092585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ko JM, Lim BC, Kim KJ, Hwang YS, Ryu HW, Lee JH, et al. Distinct neurological features in a patient with Schinzel-Giedion syndrome caused by a recurrent SETBP1 mutation. Childs Nerv Syst. 2013;29((4)):525–529. doi: 10.1007/s00381-013-2047-2. [DOI] [PubMed] [Google Scholar]
- 55.Konopka G, Roberts TF. Insights into the Neural and Genetic Basis of Vocal Communication. Cell. 2016;10164((6)):1269–1276. doi: 10.1016/j.cell.2016.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, et al. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462((7270)):213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koolen DA, Pfundt R, Linda K, Beunders G, Veenstra-Knol HE, Conta JH, et al. The Koolen-de Vries syndrome: a phenotypic comparison of patients with a 17q21.31 microdeletion versus a KANSL1 sequence variant. Eur J Hum Genet. 2016;24((5)):652–659. doi: 10.1038/ejhg.2015.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuhlwilm M, Davierwala A, Pääbo S. Identification of putative target genes of the transcription factor RUNX2. PLoS One. 2013;8((12)):e83218. doi: 10.1371/journal.pone.0083218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lesca G, Rudolf G, Bruneau N, Lozovaya N, Labalme A, Boutry-Kryza N, et al. GRIN2A mutations in acquired epileptic aphasia and related childhood focal epilepsies and encephalopathies with speech and language dysfunction. Nat Genet. 2013;45((9)):1061–1066. doi: 10.1038/ng.2726. [DOI] [PubMed] [Google Scholar]
- 60.Levy SE, Giarelli E, Lee LC, Schieve LA, Kirby RS, Cunniff C, et al. Autism spectrum disorder and co-occurring developmental, psychiatric, and medical conditions among children in multiple populations of the United States. J Dev Behav Pediatr. 2010;31((4)):267–275. doi: 10.1097/DBP.0b013e3181d5d03b. [DOI] [PubMed] [Google Scholar]
- 61.Li H, Chou SJ, Hamasaki T, Perez-Garcia CG, O'Leary DD. Neuregulin repellent signaling via ErbB4 restricts GABAergic interneurons to migratory paths from ganglionic eminence to cortical destinations. Neural Dev. 2012;7:10. doi: 10.1186/1749-8104-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao SF, Liu JC, Hsu CL, Chang MY, Chang TM, Cheng H. Cognitive development in children with language impairment, and correlation between language and intelligence development in kindergarten children with developmental delay. J Child Neurol. 2015;30((1)):42–47. doi: 10.1177/0883073814535486. [DOI] [PubMed] [Google Scholar]
- 63.Lin YH, Yamahashi Y, Kuroda K, Faruk MO, Zhang X, Yamada K, et al. Accumbal D2R-medium spiny neurons regulate aversive behaviors through PKA-Rap1 pathway. Neurochem Int. 2020;143:104935. doi: 10.1016/j.neuint.2020.104935. [DOI] [PubMed] [Google Scholar]
- 64.Lowther C, Speevak M, Armour CM, Goh ES, Graham GE, Li C, et al. Molecular characterization of NRXN1 deletions from 19,263 clinical microarray cases identifies exons important for neurodevelopmental disease expression. Genet Med. 2017;19((1)):53–61. doi: 10.1038/gim.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacWhinney B. Transcription format and programs. Lawrence Erlbaum; 2000. The CHILDES Project: Tools for Analyzing Talk Volume I. [Google Scholar]
- 66.Marchese M, Valvo G, Moro F, Sicca F, Santorelli FM. Targeted Gene Resequencing (Astrochip) to Explore the Tripartite Synapse in Autism–Epilepsy Phenotype with Macrocephaly. Neuromolecular Med. 2016;18((1)):69–80. doi: 10.1007/s12017-015-8378-2. [DOI] [PubMed] [Google Scholar]
- 67.Mariën P, Borgatti R. Language and the cerebellum. Handb Clin Neurol. 2018;154:181–202. doi: 10.1016/B978-0-444-63956-1.00011-4. [DOI] [PubMed] [Google Scholar]
- 68.Martos-Pérez J, Paula-Pérez I. [An approach to the executive functions in autism spectrum disorder] Rev Neurol. 2011;52((Suppl 1)):S147–53. [PubMed] [Google Scholar]
- 69.Matsumoto A, Mizuno M, Hamada N, Nozaki Y, Jimbo EF, Momoi MY, et al. LIN7A depletion disrupts cerebral cortex development, contributing to intellectual disability in 12q21-deletion syndrome. PLoS One. 2014;9((3)):e92695. doi: 10.1371/journal.pone.0092695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McAvoy T, Zhou MM, Greengard P, Nairn AC. Phosphorylation of Rap1GAP, a striatally enriched protein, by protein kinase A controls Rap1 activity and dendritic spine morphology. Proc Natl Acad Sci U S A. 2009;106((9)):3531–3536. doi: 10.1073/pnas.0813263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Missler M, Südhof TC. Neurexins: three genes and 1001 products. Trends Genet. 1998;14((1)):20–26. doi: 10.1016/S0168-9525(97)01324-3. [DOI] [PubMed] [Google Scholar]
- 72.Miyake F, Kuroda Y, Naruto T, Ohashi I, Takano K, Kurosawa K. West syndrome in a patient with Schinzel-Giedion syndrome. J Child Neurol. 2015;30((7)):932–936. doi: 10.1177/0883073814541468. [DOI] [PubMed] [Google Scholar]
- 73.Murphy E, Benítez-Burraco A. Bridging the gap between genes and language deficits in schizophrenia: an oscillopathic approach. Front Hum Neurosci. 2016;10:422. doi: 10.3389/fnhum.2016.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murphy E, Benítez-Burraco A. Language deficits in schizophrenia and autism as related oscillatory connectomopathies: an evolutionary account. Neurosci Biobehav Rev. 2017;83:742–764. doi: 10.1016/j.neubiorev.2016.07.029. [DOI] [PubMed] [Google Scholar]
- 75.Myers JP, Robles E, Ducharme-Smith A, Gomez TM. Focal adhesion kinase modulates Cdc42 activity downstream of positive and negative axon guidance cues. J Cell Sci. 2012;125((12)):2918–2929. doi: 10.1242/jcs.100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Newbury DF, Paracchini S, Scerri TS, Winchester L, Addis L, Richardson AJ, et al. Investigation of dyslexia and SLI risk variants in reading- and language-impaired subjects. Behav Genet. 2011;41((1)):90–104. doi: 10.1007/s10519-010-9424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ozonoff S, Jensen J. Brief report: specific executive function profiles in three neurodevelopmental disorders. J Autism Dev Disord. 1999;29:171–177. doi: 10.1023/a:1023052913110. [DOI] [PubMed] [Google Scholar]
- 78.Paracchini S, Thomas A, Castro S, Lai C, Paramasivam M, Wang Y, et al. The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum Mol Genet. 2006;15((10)):1659–1666. doi: 10.1093/hmg/ddl089. [DOI] [PubMed] [Google Scholar]
- 79.Paracchini S, Diaz R, Stein J. Advances in Dyslexia Genetics-New Insights Into the Role of Brain Asymmetries. Adv Genet. 2016;96:53–97. doi: 10.1016/bs.adgen.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 80.Perner J, Lang B. Theory of mind and executive function: is there a developmental relationship? In: Baron-Cohen S, Tager-Flugsber H, Cohen D, editors. Understanding others minds: perspectives from developmental cognitive neuroscience. Oxford University Press; 2000. pp. p. 150–181. [Google Scholar]
- 81.Pettigrew KA, Frinton E, Nudel R, Chan MTM, Thompson P, Hayiou-Thomas ME, et al. Further evidence for a parent-of-origin effect at the NOP9 locus on language-related phenotypes. J Neurodev Disord. 2016;8((1)):24–28. doi: 10.1186/s11689-016-9157-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pfenning AR, Hara E, Whitney O, Rivas MV, Wang R, Roulhac PL, et al. Convergent transcriptional specializations in the brains of humans and song-learning birds. Science. 2014;346:1256846. doi: 10.1126/science.1256846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Poot M. Connecting the CNTNAP2 networks with neurodevelopmental disorders. Mol Syndromol. 2015;6((1)):7–22. doi: 10.1159/000371594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Roll P, Rudolf G, Pereira S, Royer B, Scheffer IE, Massacrier A, et al. SRPX2 mutations in disorders of language cortex and cognition. Hum Mol Genet. 2006;15((7)):1195–1207. doi: 10.1093/hmg/ddl035. [DOI] [PubMed] [Google Scholar]
- 85.Roll P, Vernes SC, Bruneau N, Cillario J, Ponsole-Lenfant M, Massacrier A, et al. Molecular networks implicated in speech-related disorders: FOXP2 regulates the SRPX2/uPAR complex. Hum Mol Genet. 2010;19((24)):4848–4860. doi: 10.1093/hmg/ddq415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rowen L, Young J, Birditt B, Kaur A, Madan A, Philipps DL, et al. Analysis of the human neurexin genes: alternative splicing and the generation of protein diversity. Genomics. 2002;79((4)):587–597. doi: 10.1006/geno.2002.6734. [DOI] [PubMed] [Google Scholar]
- 87.Royer-Zemmour B, Ponsole-Lenfant M, Gara H, Roll P, Lévêque C, Massacrier A, et al. Epileptic and developmental disorders of the speech cortex: ligand/receptor interaction of wild-type and mutant SRPX2 with the plasminogen activator receptor uPAR. Hum Mol Genet. 2008;17((23)):3617–3630. doi: 10.1093/hmg/ddn256. [DOI] [PubMed] [Google Scholar]
- 88.Russell J. Editorial Médica Panamericana; 2000. El autismo como trastorno de la función ejecutiva. [Google Scholar]
- 89.Sacks H, Schegloff EA, Jefferson G. A simplest systematics for the organization of turn-taking for conversation. Language. 1974;50((4)):696–735. [Google Scholar]
- 90.Schaaf CP, Boone PM, Sampath S, Williams C, Bader PI, Mueller JM, et al. Phenotypic spectrum and genotype-phenotype correlations of NRXN1 exon deletions. Eur J Hum Genet. 2012;20((12)):1240–1247. doi: 10.1038/ejhg.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Scott-Van Zeeland AA, Abrahams BS, Alvarez-Retuerto AI, Sonnenblick LI, Rudie JD, Ghahremani D, et al. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci Transl Med. 2010;2((56)):56ra80. doi: 10.1126/scitranslmed.3001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shiro M, Diez-Itza E, Viejo A, Fernández-Urquiza M. Pragmática evaluativa de las narraciones en el síndrome de Williams. In: Aguilar-Mediavilla E, Adrover-Roig D, Buil-Legaz L, López-Penadés R, editors. Proceedings of the VIIIth International Conference of Language Acquisition. Universidad de las Islas Baleares; 2016. pp. p. 31–2. [Google Scholar]
- 93.Shiro M, Diez-Itza E, Fernández-Urquiza M. Evaluative language and component structure of oral narratives in Williams Syndrome. In: Aguilar-Mediavilla E, Buil-Legaz L, López-Penadés R, Adrover-Roig D, Sanchez-Azanza VA, editors. Atypical Language Development in Romance Languages. John Benjamins; 2019. pp. p. 235–252. [Google Scholar]
- 94.Silveri MC. Contribution of the cerebellum and the basal ganglia to language production: speech, word fluency, and sentence construction-Evidence from pathology. Cerebellum. 2021;20((2)):282–294. doi: 10.1007/s12311-020-01207-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smigiel R, Kostrzewa G, Kosinska J, Pollak A, Stawinski P, Szmida E, et al. Further evidence for GRIN2B mutation as the cause of severe epileptic encephalopathy. Am J Med Genet A. 2016;170((12)):3265–3270. doi: 10.1002/ajmg.a.37887. [DOI] [PubMed] [Google Scholar]
- 96.Smogavec M, Cleall A, Hoyer J, Lederer D, Nassogne MC, Palmer EE, et al. Eight further individuals with intellectual disability and epilepsy carrying bi-allelic CNTNAP2 aberrations allow delineation of the mutational and phenotypic spectrum. J Med Genet. 2016;53((12)):820–827. doi: 10.1136/jmedgenet-2016-103880. [DOI] [PubMed] [Google Scholar]
- 97.Sorci G, Riuzzi F, Arcuri C, Tubaro C, Bianchi R, Giambanco I, et al. S100B protein in tissue development, repair and regeneration. World J Biol Chem. 2013;4((1)):1–12. doi: 10.4331/wjbc.v4.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Souchet B, Guedj F, Sahún I, Duchon A, Daubigney F, Badel A, et al. Excitation/inhibition balance and learning are modified by Dyrk1a gene dosage. Neurobiol Dis. 2014;69:65–75. doi: 10.1016/j.nbd.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 99.Spiteri E, Konopka G, Coppola G, Bomar J, Oldham M, Ou J, et al. Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am J Hum Genet. 2007;81((6)):1144–1157. doi: 10.1086/522237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354((13)):1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 101.Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455((7215)):903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43((Database issue)):D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tan GC, Doke TF, Ashburner J, Wood NW, Frackowiak RS. Normal variation in fronto-occipital circuitry and cerebellar structure with an autism-associated polymorphism of CNTNAP2. Neuroimage. 2010;53((3)):1030–1042. doi: 10.1016/j.neuroimage.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tromp A, Mowry B, Giacomotto J. Neurexins in autism and schizophrenia-a review of patient mutations, mouse models and potential future directions. Mol Psychiatry. 2021;26((3)):747–760. doi: 10.1038/s41380-020-00944-8. [DOI] [PubMed] [Google Scholar]
- 105.Turner M. Hacia una explicación de la conducta repetitiva en el autismo basada en la disfunción ejecutiva. In: Russell J, editor. El autismo como trastorno de la función ejecutiva. Editorial Médica Panamericana; 2000. pp. p. 55–98. [Google Scholar]
- 106.Turner SJ, Mayes AK, Verhoeven A, Mandelstam SA, Morgan AT, Scheffer IE. GRIN2A: an aptly named gene for speech dysfunction. Neurology. 2015;84((6)):586–593. doi: 10.1212/WNL.0000000000001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Van Bon BWM, Hoischen A, Hehir‐Kwa J, De Brouwer APM, Ruivenkamp C, Gijsbers ACJ, et al. Intragenic deletion in DYRK1A leads to mental retardation and primary microcephaly. Clin Genet. 2011;79((3)):296–299. doi: 10.1111/j.1399-0004.2010.01544.x. [DOI] [PubMed] [Google Scholar]
- 108.Velayos-Baeza A, Toma C, Paracchini S, Monaco AP. The dyslexia-associated gene KIAA0319 encodes highly N- and O-glycosylated plasma membrane and secreted isoforms. Hum Mol Genet. 2008;17((6)):859–871. doi: 10.1093/hmg/ddm358. [DOI] [PubMed] [Google Scholar]
- 109.Vias C, Dick AS. Cerebellar contributions to language in typical and atypical development: a review. Dev Neuropsychol. 2017;42((6)):404–421. doi: 10.1080/87565641.2017.1334783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Worthey EA, Raca G, Laffin JJ, Wilk BM, Harris JM, Jakielski KJ, et al. Whole-exome sequencing supports genetic heterogeneity in childhood apraxia of speech. J Neurodev Disord. 2013;5((1)):29. doi: 10.1186/1866-1955-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang C, Li X, Zhang B, Fu S, Li S, Shen J, et al. The mechanism of Rap1 regulates N-cadherin to control neuronal migration. J Mol Neurosci. 2019;68:539–548. doi: 10.1007/s12031-019-01316-w. [DOI] [PubMed] [Google Scholar]
- 112.Yiin JJ, Hu B, Jarzynka MJ, Feng H, Liu KW, Wu JY, et al. Slit2 inhibits glioma cell invasion in the brain by suppression of Cdc42 activity. Neuro Oncol. 2009;11((6)):779–789. doi: 10.1215/15228517-2008-017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang J. Evolution of the human ASPM gene, a major determinant of brain size. Genetics. 2003;165((4)):2063–2070. doi: 10.1093/genetics/165.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article. A preprint version of this article is available on PsyArXiv [https://doi.org/10.31234/osf.io/aw25n].