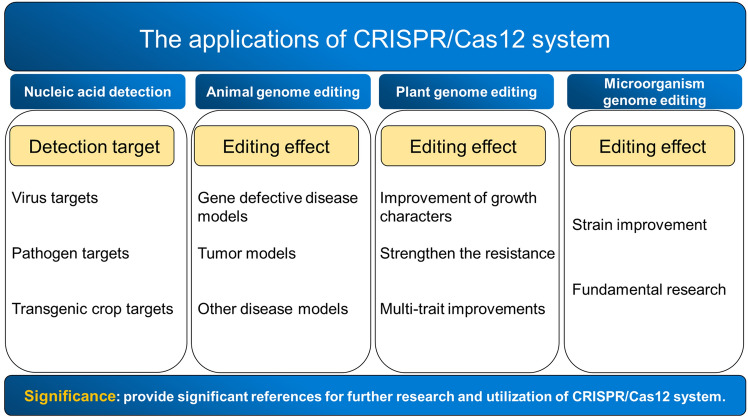
Abstract
Purpose
This work characterizes the applications of CRISPR/Cas12 system, including nucleic acid detection, animal, plant and microbial genome editing.
Methods
The literature on CRISPR/Cas12 system was collected and reviewed.
Results
CRISPR/Cas system is an acquired immune system derived from bacteria and archaea, which has become the most popular technology around the world because of its outstanding contribution in genome editing. Type V CRISPR/Cas systems are distinguished by a single RNA-guided RuvC nuclease domain with single effector molecule. Cas12a, the first reported type V CRISPR/Cas system, targets double-stranded DNA (dsDNA) adjacent to PAM sequences and trans-cleaves single-stranded DNA (ssDNA). We present the applications of CRISPR/Cas12 system for nucleic acid detection and genome editing in animals, plants and microorganisms. Furthermore, this review also summarizes the applications of other Cas12 proteins, such as Cas12b, Cas12c, Cas12d, and so on, which further widen the application prospects of CRISPR/Cas12 system.
Conclusions
Knowledge of the applications of CRISPR/Cas12 system is necessary for improving the understanding of the functional diversity of CRISPR/Cas12 system and also provides significant references for further research and utilization of CRISPR/Cas12 in other new fields.
Graphical abstract
Keywords: CRISPR/Cas12, Nucleic acid detection, Genome editing, Cas12 proteins
Introduction
The CRISPR (Clustered regularly interspaced short palindromic repeat) system is a natural acquired immune system that prevents the invasion from viruses and phage through the combination with CRISPR associated (Cas) proteins [1]. According to the 2020 classification, CRISPR/Cas systems include 2 Classes, 6 types and 33 subtypes [2]. In Class 1 CRISPR/Cas systems (including type I, type III, and type IV), the effector module composes of multisubunit effector complexes (such as Cas3, Cas5-Cas8, Cas10 and Cas11). By contrast, the effector module of Class 2 CRISPR/Cas systems (including type II, type V, and type VI) is represented by single, large multidomain protein effectors (such as Cas9, Cas12 and Cas13). The CRISPR/Cas systems, consisting of Cas gene sequences, protospacer, direct repeat sequences and the spacer sequences, can specifically recognize foreign nucleic acid molecules by the aid of CRISPR RNAs (crRNAs), and then cleave them by Cas proteins. As the spacer sequences can be easily redesigned to introduce a double-strand break within a given sequence, the CRISPR-based technology has been widely employed in genome editing, gene transcription regulation, epigenetic engineering and many other fields [3]. Currently, three Class 2 effectors, Cas9, Cas12 and Cas13, have been extensively applied in many fields because of their high sensitivity, high specificity and easy operation. The genome editing systems represented by the CRISPR/Cas9 have been successfully applied in the fields of agriculture, medicine, biology, such as the establishment of new breeding methods, the development of new gene drugs, and the exploration of biological pathways. With the in-depth study, the other types of Cas proteins, such as Cas13a and Cas12a, have been successively developed and applied in genome engineering, nucleic acid detection and molecular diagnostics. Cas12a, also known as Cpf1, is a single crRNA-guided endonuclease with a single RuvC and Nuc domain, which is responsible for the cleavage of target DNAs adjacent to PAM sequences and generates DNA ends with a 5′ overhang [4]. Type V CRISPR/Cas systems are distinguished by a single RNA-guided RuvC nuclease domain with single effector molecule. Cas12a, the first reported type V CRISPR/Cas system, has been demonstrated to be a dual-nuclease with endoribonuclease and endonuclease activities that is specific to crRNA biogenesis and target DNA interference [5]. Meanwhile, CRISPR/Cas12a has both cis- and trans-cleavage activities on single-stranded DNA (ssDNA), which further expands its potential applications [6]. Subsequently, other subtypes of type V Cas proteins, such as Cas12b (C2c1) [7, 8], Cas12c (C2c3) [8], Cas12f (Cas14) [9] and so on, have been identified one after another. Recently, CRISPR/Cas12-based methods with high sensitivity and specificity have been employed for nucleic acid detection and CRISPR-based diagnostics (CRISPR-Dx) [3]. Compared with Cas13, Cas12-based methods are more popular because of its fewer steps for target nucleic acid amplification and trans-cleavage of ssDNA probes. Furthermore, genome editing using CRISPR/Cas12 system has been widely applied in of humans, animals and plants. Although Cas12-based CRISPR-Dx methods and their applications have already been reviewed [3], the latest detection methods and their applications in genome editing still need further illustrating. In this paper, the applications of CRISPR/Cas12 system, including the applications in nucleic acid detection and molecular diagnostics, genome editing of animals, plants and microorganisms, are summarized, which can provide significant references for further research and utilization of CRISPR/Cas12 in other new fields.
CRISPR/Cas12-based nucleic acid detection
Acting as the carrier of genetic information, nucleic acids have been becoming one of the most important research fields for the development of DNA and RNA detection technologies. Previous detection and imaging technologies are mainly based on traditional methods, such as in vitro amplification, protein hybridization and microarray. Although these methods can meet the requirements for detection and imaging of targets, they still have some defects due to time-consuming, high cost, low precision and sensitivity. The Cas12-based detection methods, such as DETECTR (DNA endonuclease targeted CRISPR trans-reporter) [10], HOLMES (one-hour low cost multipurpose highly efficient system) [11], HOLMESv2 (the improved version of HOLMES) [12], CDetection (Cas12b-mediated DNA detection) [13], and Cas12aVDet (Cas12a-based visual detection) [14], have been successfully carried out to detect various pathogens with quickness, high sensitivity and specificity [3]. More recently (2021–2022), the latest detection methods with time-saving, low-cost, high sensitivity and specificity, such as LAMP-Cas12a, G-CRISPR-Cas [15], LACD [16], CLAP [17] and vis-NEAA [18] and so on, have been performed to detect Hepatitis B virus (HBV), Salmonella, Mycobacterium, COVID-19 and other pathogens.
LAMP-Cas12a method (LAMP, loop-mediated isothermal amplification, combined with the CRISPR/Cas12a detection system) was developed to detect HBV, which innovatively solved the problems of point-of-care test and the nucleic acid extraction of samples. Based on LAMP-Cas12a, visualization of the assay results and a limit of detection (LOD) of 1 copy/µL within 13 min were achieved for HBV detection. Meanwhile, the sensitivity and specificity in the evaluation of 73 clinical samples were 100%. The LAMP-Cas12a-based HBV assay provides rapid, accurate test results and low costs without specialized equipment, which has important practical implications for point-of-care HBV detection [19]. By contrast, G-CRISPR-Cas (G-quadruplex-probing CRISPR/Cas12 system), the label-free assay, was used to detect foodborne pathogen, Salmonella enterica (S. enterica). The introduction of G-quadruplex probe as Cas12a substrate enabled label-free analysis of foodborne pathogens. With the help of the amplification process induced by LAMP, G-CRISPR-Cas achieved highly sensitive detection for S. enterica as low as 20 CFU (Colony-forming unit). At the same time, the double recognition of LAMP primers and Cas12a-guided RNA ensured the specificity of pathogenic gene detection. Herein, G-CRISPR-Cas assay is useful for on-site detection of the infection or pollution of foodborne pathogens, which provides a guarantee for food safety [15]. Similar to G-CRISPR-Cas, vis-NEAA (CRISPR/Cas12a combined with nicking enzyme-assisted amplification) has been taken advantage of detecting Salmonella in food. CRISPR/Cas12a can specifically identify NEAA amplicons and convert the signal into fluorescent visual readouts, thereby having the advantages of rapidity and high efficiency of NEEA method, and achieving 100% fidelity at the same time. More importantly, it solved the problem of nonspecific amplification of NEAA method. With vis-NEAA assay, Salmonella with 80 CFU/mL in spiked eggs can be detected on-site within 20 min [18]. Taken together, both G-CRISPR-Cas and vis-NEAA can be used for the detection of foodborne pathogens, thus ensuring food safety. LACD (LAMP coupled with CRISPR/Cas12a-mediated diagnostic) was developed to detect Mycobacterium tuberculosis complex (MTC) in human tuberculosis (TB). Because the engineered LAMP primers contained the specific PAM site for CRISPR/Cas12a recognition, it can be utilized to detect any target sequence, even targets without PAM sites, which provides a new idea for the application of Cas12a and widens its application [16]. AuNP-based colorimetric assay coupled with Cas12 and RT-LAMP, also called Cas12a-assisted RT-LAMP/AuNP (CLAP), is a rapid, sensitive, and visual assay for SARS-CoV-2 detection. Under the optimal conditions, the detection of SARS-CoV-2 RNA can be reduced to 4 copies/mµL within 40 min by naked eye. In addition, the advantages of the superior specificity, easy operation, high-throughput detection make it suitable for large-scale population screening of SARS-CoV-2 in public places, which is conducive to controlling and alleviating the pandemic of COVID-19. Therefore, the CLAP method has important practical roles in screening suspicious population of COVID-19 [17]. To sum up, these latest methods make use of the low equipment requirements of LAMP and the high cleavage activity of Cas12a, which make them have broad application prospects because of rapidity, high sensitivity, specificity and accuracy, high throughput, and easy operation. It should be noted that among Cas12-based detection methods, RPA (recombinant enzyme polymerase amplification), RCA (rolling circle amplification), and PCR (including rapid PCR, rRT PCR, and asymmetric PCR, Table 1) can also be used to exponentially amplify target DNA molecules. With the help of RPA, CDetection and DETECTR can achieve attomolar (åM) sensitivity for DNA detection (Table 1). The characteristics and applications of main CRISPR/Cas12 assays developed in recent years were listed in Table 1.
Table 1.
The characteristics and applications of CRISPR/Cas12-based assays
| Year | Methods | Detection targets | Reaction systems | Detection time | Sensitivity | Specificity | Multiple detections | Advantages | Applications | References |
|---|---|---|---|---|---|---|---|---|---|---|
| 2018 |
DETECTR DNA Endonuclease targeted CRISPR trans reporter |
Human papillomavirus | RPA + CRISPR/LbCas12a | 1 h | åM | Single base | No | Simple, rapid, accurate, portable; high sensitivity and specificity | Molecular diagnostics; DNA detection | [10] |
| 2018 |
HOLMES One hour low-cost multipurpose highly efficient system |
DNA and RNA targets | RPA/LAMP /rapid PCR + CRISPR/ LbCas12a | 1 h | 10 åM | Single base | No | Simple, rapid, portable; high sensitivity and specificity, low cost | DNA and RNA detection; SNP identification | [11] |
| 2018 | Cas14-DETECTR | DNA and RNA targets | RPA + CRISPR/Cas14a | 1 h | – | Single base | No | high-fidelity, high sensitivity, without PAM sequence | Molecular diagnostics; ssDNA pathogen detection | [9] |
| 2019 | HOLMESv2 | DNA and RNA targets | RPA/LAMP/ asymmetric PCR + CRISPR/ AacCas12b | 1 h | 10åM | Single base | No | Simple, inexpensive, accurate, portable; no cross contamination |
Molecular diagnostics; DNA methylation quantitation e.g., SNP identification; DNA and RNA detection, DNA quantification; DNA methylation |
[12] |
| 2019 |
CDetection Cas12b-mediated DNA detection |
DNA target | RPA + CRISPR/ AaCas12b | 0.5 ~ 2 h | åM | Single base | No | Rapid, economical; high sensitivity, specificity, and accuracy | Molecular diagnostics and clinical research | [13] |
| 2019 |
Cas12aVDet Cas12a-based visual detection |
Mycoplasma | RPA + CRISPR/LbCas12a | ~ 30 min | 10åM | Single base | No | Convenient, rapid, visual, portable; 100% accuracy | Rapid nucleic acid detection | [14] |
| 2020 | iSCAN | COVID-19 | RT-LAMP + CRISPR/LbCas12a | < 1 h | 10 copies/mµL | High specificity | No | Simple, rapid, portable; high sensitivity | Early detection of COVID-19 | [20] |
| 2020 |
STOP SHERLOCK testing in one pot |
COVID-19 | LAMP + CRISPR/AaCas12b | ~ 40 min | 100 copies/mµL | 100% specificity | No | Convenient, rapid, RNA extraction-free, low-cost, high sensitivity and specificity | Real-time detection of COVID-19 | [21] |
| 2020 |
SENA Specific enhancer for PCR-amplified nucleic acid |
COVID-19 | rRT-PCR + CRISPR/ LbCas12a | < 2 h |
1.2</=1.6</=2.1 copies/µL (95% confidence interval) |
High specificity | No | Safe, simple, stable, quick, sensitive, specific; low-cost | Clinical diagnostics of COVID-19 | [22] |
| 2020 |
Cas12a-PB CRISPR/Cas12a based portable biosensor |
GM soybean and maize powders | PCR/rapid PCR /LAMP + CRISPR/ LbCas12a | ~ 1 h |
0.1% transgenic contents |
High specificity | Yes | Simple, convenient, visual, portable; low-cost, high sensitivity and specificity | Multiple targets detection in clinic diagnostics, food safety, etc. | [23] |
| 2021 | LAMP-Cas12a | Hepatitis B virus (HBV) | LAMP + CRISPR/ LbCas12a | < 20 min | 1 copy/µL | Ultra-specificity | No | Rapid, accurate, visual; low-cost, high sensitivity and specificity | Point-of-care HBV detection | [19] |
| 2021 |
LACD Loop-mediated isothermal amplification coupled with CRISPR-Cas12a-mediated diagnostic |
Mycobacterium tuberculosis complex | LAMP + CRISPR/ LbCas12a | ~ 1 h | 50 fg genome DNA (~ 10 copies) | 5 ng of templates | No | Rapid, visual, ultrasensitive, and highly specific | Detection of a variety of target sequences | [16] |
| 2021 |
G-CRISPR-Cas G-quadruplex-probing CRISPR-Cas12 |
Salmonella enterica | G-quadruplex probe + LAMP + CRISPR/ LbCas12a | ~ 20 min | 20 CFU | High specificity | Yes | Rapid; label-free, high sensitivity, specificity, and accuracy | On-site diagnosis of the infection or contamination of foodborne pathogens | [15] |
| 2021 |
PPCas12 Proximal DNA probe-based CRISPR-Cas12 |
Salmonella enterica |
proximal DNA probe + CRISPR/ LbCas12a |
< 2 h | 619 CFU | High specificity | No | Simple; low-cost, and without amplification steps | On-site monitoring of foodborne pathogens | [24] |
| 2021 |
CLAP Cas12a-assisted RT-LAMP/AuNP |
COVID-19 | RT-LAMP/AuNP + CRISPR/LbCas12a | ~ 40 min | 4 copies/mµL | Superior specificity | Yes | Rapid; easy for operation, high-throughput, superior sensitivity, and specificity | On-site diagnosis of COVID-19 | [17] |
| 2021 | RPA-Cas12a-FS | DNA target | RPA + CRISPR/LbCas12a | 45 min | 10 copies/mµL | High specificity | No | Rapid; easy for operation, low-cost, high sensitivity and specificity | Detection of foodborne pathogenic bacteria, genetically modified crops, and meat adulteration | [25] |
| 2021 | Cas12c-DETECTOR |
Psa, HPV COVID-19 |
RPA + CRISPR/Cas12c1 | 120 min | 0.23pM | Single base | No | Rapid; high sensitivity and specificity | Plant pathogen detection, SNP identification, clinical diagnosis | [26] |
| 2022 |
vis-NEAA vis-Nicking enzyme-assisted amplification |
Salmonella | NEAA + CRISPR/ LbCas12a | ~ 20 min | 80 CFU/mL | 100% fidelity | No | Rapid, visual, and sequence-specific | Food quality monitoring, environmental analysis, and medical diagnostics | [18] |
| 2022 | isoCRISPR | Transgenic crops | RCA + CRISPR/LbCas12a | 30 min | LOD: 45.0 pM | High specificity | No | Label-free; specific and rapid; isothermal operation; single-tube test | Detection of transgenic corn; on-site rapid detection of various food contamination | [27] |
| 2022 |
CLEVER CRISPR-Cas integrated RT-LAMP easy, visual and extraction-free RNA |
COVID-19 | RT-LAMP + CRISPR/LbCas12a | ~ 100 min | 0.5 nM | 100% specificity | No | Fast, direct, versatile; RNA extraction-free; superior sensitivity and specificity | Visually diagnostics of COVID-19 | [28] |
| 2022 |
DIRECT2 DNA-immunoglobulin reporter endonuclease cleavage test |
Ribosomal intergenic spacer (IGS) of Dickeya solani | DNA-IgG probe + CRISPR/LbCas12a | 0.5 ~ 1.5 h | 0.5-1 nM | Low specificity | No | Rapid, first LFT-based platform | CRISPR-Cas12 activity detection | [29] |
CFU: colony-forming units; LAMP: loop-mediated isothermal amplification; LFT: lateral flow test; LOD: limit of detection; RCA: rolling circle amplification; RPA: recombinase polymerase amplification; AaCas12b: Alicyclobacillus acidiphilus Cas12b; AacCas12b: Alicyclobacillus acidoterrestris Cas12b; LbCas12a: Lachnospiraceae bacterium ND2006 Cas12a; –: no data
CRISPR/Cas12 system involves in genome editing
As RNA-guided endonuclease, CRISPR/Cas9 and Cas12a have been widely used for genome editing in animals, plants, and cultured cells, based on their programmable ability to trigger DNA repair at the desired sites, thereby accelerating the pace of basic research and enabling clinical and agricultural breakthroughs [30, 31]. Meanwhile, Cas12b is also a powerful tool for genome engineering because of the small size and high specificity [32–34].
CRISPR/Cas12 system involves in genome editing of human and animals
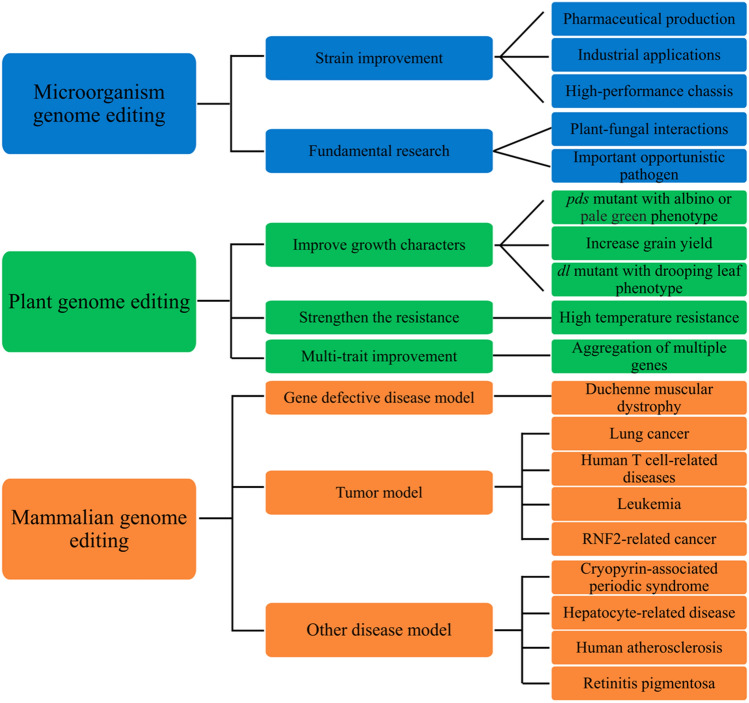
As the second CRISPR/Cas system for mammalian genome editing, Cas12a recognizes the PAM sequence of 5′-TTN, which improves the recognition range of the genome compared with the PAM sequence of Cas9 recognizing 5′-NGG. The CRISPR/Cas12a and Cas9 systems complement each other and further accelerate the development of CRISPR genome editing tools. In 2017, LbCas12a-mediated genome editing was successfully used to induce pluripotent stem cells (IPSCs) from patients with Duchenne muscular dystrophy (DMD) and correct target genes of DMD in mouse embryos, making it an important contribution to DMD correction in gene therapy [35]. In the same year, by the aid of Cas12a’s ability of processing its own crRNA, Zhang Feng’s group accomplished simultaneously editing up to four genes in mammalian cells and three genes in mouse brains [36]. Furthermore, Liu et al. validated that the engineered variants of Cas12a with two different nuclear localization sequences (NLS) at the C terminus can improve the mutagenesis efficiency of LbCas12a and FnoCas12a in mammalians, which provide a scheme for broadening the application of Cas12a in vertebrates [37]. In addition, AsCas12a and LbCas12a were used for genome editing in mouse embryos, Drosophila and zebrafish by means of their high activity, but low genome coverage and low targeting efficiency hinder their applications in biomedical fields [38].
CRISPR/Cas12b/C2c1 (type V-B) is a dual-RNA-guided DNA endonuclease, which contains RuvC-like endonuclease domain distantly related to Cas12a and relies on both crRNA and tracrRNA for DNA cleavage [7, 8]. The optimal cleavage activity of Cas12b from Alicyclobacillus acidiphilus (AaCas12b) was maintained in a wide temperature range (31 ~ 59 °C), together with a small size, high specificity, increased stability, and minimal off-target effects, making it suitable for mammalian genome editing and clinical applications, including single and multiplex genome editing, gene activation, and establishment of gene mutant mouse models, suggesting that CRISPR/Cas12b can be used as a versatile tool for mammalian genome engineering [33, 39]. Subsequently, Strecker et al. identified a promising candidate Cas12b for human genome editing from Bacillus hisashii (BhCas12b) and obtained the gain-of-function mutation of BhCas12b. The results showed that the mutant BhCas12b promoted in vitro genome editing in human cell lines and primary human T cells and displayed higher specificity. This work further confirmed that Cas12b can be used for genome editing of human cells [32]. More recently, Un1Cas12f1 (type V-I) was redesigned and optimized for genome editing in human cells. The results demonstrated that optimized Un1Cas12f1 system enabled efficient, specific genome editing in human cells with an efficiency comparable to SpCas9 and a specificity similar to AsCas12a [40]. These observations suggest that CRISPR/Cas12 can be used as a versatile tool for mammalian genome editing (Table 2).
Table 2.
The applications, characteristics, and advantages of CRISPR/Cas12 system in genome editing of human and animals
| Disease types | Editor | Target genes | Plasmids | Transformation methods | Editing strategies | Editing effect | Advantages | References |
|---|---|---|---|---|---|---|---|---|
|
Duchenne muscular dystrophy (DMD) |
AsCas12a LbCas12a |
Exon 51 of the human DMD gene Exon 23 of the mouse Dmd gene |
pLbCpf1-2 A-GFP pAsCpf1-2 A-GFP |
Human: nucleofection Mouse: one-cell embryo injection |
Reframing and exon skipping | Correct human DMD and mouse Dmd mutations in vitro and in vivo | High efficiency and specificity; low frequency of off-targeting effects; heritable | [35] |
| Human atherosclerosis |
AsCas12a LbCas12a |
Apoe and/or Ldlr genes |
pcDNA3.1-hAsCpf1 pcDNA3.1-hLbCpf1 |
Microinjection | Knockout | Generate Apoe- and/or Ldlr-deficient rats | High efficiency and specificity; multiple gene targeting capacity; few or no off-targeting effects; heritable | [41] |
| Cryopyrin-associated periodic syndrome (CAPS) | AsCas12a | NLRP3 | NLRP3-Cpf1-sgRNA | Electroporation | Point mutation | Produce NLRP3 R259W homo-cloned pigs | No sign of off-targeting; precise NLRP3 point mutation; accurate | [42] |
| Hepatocyte-related disease |
AsCas12a LbCas12a |
AAVS1 locus |
pHM-CBh-LbCpf1 pHM-CBh-AsCpf1 pHM-U6-lbgRNA-AAVS1 pHM-U6-asgRNA-AAVS1 |
Co-transfection | InDels | Generate genome cleavages in PHHs | Larger capacity; efficiently targets human hepatocytes | [43] |
| Leukemia | AsCas12a | Exon 3 of Lif gene | AsCpf1/mRNA AsCpf1/RNP | Electroporation | Knockout | Generate LIF knock out mice | Easy, fast, and technically less demanding | [44] |
| Lung cancer | LbCas12a | EGFR | oAd/Cas12a/crEGFR | Liposome transfection | InDels | Precise genomic reprogramming | Efficient precise, and cancer-specific; no off-target; can be used as an alternative cancer therapy | [45] |
| Human T cell-related diseases | BhCas12b | Human CD4 + T cell containing TTTV PAM site | BhCas12b v4-sgRNA | Electroporation | InDels | Exhibit indel rates of 32 ~ 49% across three tested targets | Small size and high target specificity; low off-targeting activity; a promising tool for in vivo genome editing | [32] |
| RNF2 related cancer | AaCas12b | Human RNF2 gene and mouse Nrl gene | AaCas12b RNPs | Microinjection | InDels | Produce indels with the frequency at up to 66.7% | Single and multiplex genome editing, gene activation, and generation of gene mutant mouse models; high specificity and minimal off-target effects | [33] |
AaCas12b: Alicyclobacillus acidiphilus Cas12b; FnCas12a: Francisella tularensis Cas12a; LbCas12a: Lachnospiraceae bacterium ND2006 Cas12a; BhCas12b: Bacillus hisashii Cas12b; InDels: insertions and deletions
CRISPR/Cas12 system involves in genome editing of plants
CRISPR/LbCas12a is a temperature-sensitive system in genome editing of plants, which has been successfully applied in rice, Arabidopsis, maize, soya bean, and other species [46]. The temperature tolerance and precise cleavage ability of Cas12a open up new prospects for creating GMO (genetically modified organism)-free crops with valuable traits. In rice, the average mutation frequency of target sites was 47.2%, indicating that FnCas12a can effectively induce gene-targeted mutations [47]. Subsequently, Zhong et al. used rice as a model system to investigate the PAM requirement of FnCas12a in plant genome editing, suggesting that FnCas12a preferred TTTV PAMs for efficient genome editing in rice [48]. AsCas12a was successfully used to induce heritable mutations with 77.8% and 92.8% frequencies at two target sites among rice T0 lines [49]. In transgenic maize, LbCas12a-based genome editing achieved 100% high-frequency at a daytime temperature of 28 °C [49]. Nowadays, Cas12a has also been used for genome editing of woody plants. The CRISPR/AsCas12a has been verified to be the most effective at simultaneously knocking out the members of multigene families, which makes up for the deficiency of woody plant mutants and promotes the research of forest genetics [50]. Editing the coding sequences of GhPGF and GhCLA1 genes by CRISPR/LbCas12a can accurately edit tetraploid cottons (Gossypium hirsutum). The results showed that non-transgenic and gossypol-free cottons were successfully created, which provided valuable germplasm resources for molecular breeding of cottons [46].
CRISPR/Cas12b has been proved to be effective in plant genome editing. AacCas12b and AaCas12b against OsEPFL9-sgRNA02 were transferred into rice callus by Agrobacterium tumefaciens-mediated transformation. The mutation rate of AacCas12b T0 transgenic line was 36.4%, and that of AaCas12b was 54.2%, which can effectively generate stable rice mutants [34]. Moreover, the potential applications of the CRISPR/Cas12b in Arabidopsis were also explored. With BvCas12b and BhCas12b v4, a large number of deletions were produced at multiple sites in Arabidopsis, and stable mutants were successfully obtained without obvious mutations at potential off-target sites [51]. These results elucidated the potential utility of CRISPR/Cas12b system for genome editing in rice and Arabidopsis. The applications, characteristics and advantages of CRISPR/Cas12 system in plant genome editing were listed in Table 3. Notably, the CRISPR/Cas12 system mainly achieves plant-targeted mutagenesis through insertions and deletions, and its temperature dependence limits its utility in plant genome editing. In the future, it is necessary to further explore and resolve the temperature dependence of Cas12a, so as to facilitate its utilization in genome editing of other species.
Table 3.
The applications, characteristics, and advantages of CRISPR/Cas12 system in plant genome editing
| Species | Editor | Target genes | Plasmids | Transformation methods | Editing strategies | Editing efficiency or mutation rate | Advantages | References |
|---|---|---|---|---|---|---|---|---|
|
Arabidopsis Arabidopsis thaliana |
BhCas12b v4 BvCas12b |
AtPDS3 AtFLS2 AtGL2 AtTT4 |
BhCas12b-sgRNA BhCas12b v4-sgRNA | Agrobacterium-mediated transformation | InDels | 0 ~ 4.3% | Multiplex genome editing; large deletions at multiple loci; no off-target mutations at the tested loci | [51] |
|
LbCas12a ttLbCas12a |
AtALS |
ttLbCas12a-sgRNA LbCas12a-sgRNA |
Agrobacterium-mediated transformation | InDels |
LbCas12a: 0.26% (22℃), 0.79% (28℃) ttLbCas12a: 0.63% (22℃), 1.34% (28℃) |
ttLbCas12a increases gene targeting efficiencies and achieves high temperature plant gene targeting | [52] | |
| AsCas12a, LbCas12a |
AtGL2 AtTT4 |
AtGL2-crRNA1 AtTT4-crRNA1 |
Agrobacterium-mediated transformation | InDels |
T2 plants AtGL2: ~ 35% AtTT4: ~ 15% |
High-temperature regimes to achieve high editing efficiencies | [49] | |
|
Rice Oryza sativa |
AaCas12b AacCas12b BhCas12b BthCas12b |
OsEPFL9 OsGS3 |
OsEPFL9-sgRNA02 OsGS3-sgRNA02 |
Agrobacterium-mediated transformation | Deletions |
T0 lines OSEPFL9: 66.7% OsGS3: 70.85% |
Multiplexed genome editing; no off-target mutations | [34] |
| AsCas12a | OsDEP1 OsROC5 |
OsROC5-crRNA01 OsDEP1-crRNA02 |
Polyethylene glycol (PEG)-mediated transfection | Deletions |
T0 lines OsDEP1: 77.8%, OsROC5: 92.8% |
High-frequency mutation | [49] | |
| FnCas12a |
OsDL OsALS |
crOsDL-1 ~ 2 crOsALS-1 ~ 2 |
Agrobacterium-mediated transformation | Deletions | The average mutant rate: 47.2% | Biallelic mutants obtained in the T0 generation in rice | [47] | |
|
Maize Zea mays |
LbCas12a | ZmGL2 | LbCas12a-ZmGL2-crRNA1&2 | Agrobacterium-mediated transformation | InDels |
A842B-2-2-2: 51.4% A842B-5-1 :100% |
High mutagenesis frequencies | [49] |
|
Cotton Gossypium hirrntnml |
AacCas12b | GhCLA | pRGEB32-AacCas12b-GhCLA | Agrobacterium-mediated transformation | Deletions (9 ~ 14 bp) | Mutation rate: ~ 20% | Efficient precise and faithfully inheritable; no off-target mutations | [53] |
| LbCas12a | GhPGF | LbCas12a-GhPGF | Agrobacterium-mediated transformation | InDels |
GhPGF-crRNA1: 91.5% (34 °C) GhPGF-crRNA2: 67.6% (34 °C) |
Creation of nontransgenic, gossypol-free cotton; no off-target mutations | [46] | |
|
Tobacco Nicotiana tabacum |
LbCas12a |
NtPDS NtSTF1 |
crNtPDS-1, 2 crNtSTF1-1 ~ 4 |
Agrobacterium-mediated transformation | InDels (majority) | The average mutant rate: 28.2% | Chimeric mutations | [47] |
|
Citrus Citrus reticulata |
LbCas12a |
CsPDS CsLOBP |
GFPp1380N-35 S-LbCas12a-crRNA-cspds GFPp1380N-Yao-LbCas12a-crRNA-lobp |
Agrobacterium-mediated transformation | Short deletions | The average mutant rate: 28.3% | No potential off-targets | [54] |
|
Poplar Populus tomentosa |
AsCas12a LbCas12a FnCas12a |
PagPDS |
PagPDS-AsCas12a PagPDS-LbCas12a PagPDSFnCas12a |
Agrobacterium-mediated transformation | Large-fragment deletions |
AsCas12a: ~ 70% LbCas12a: ~ 33% FnCas12a: 6.7% |
Simple and high efficiency; multigene simultaneous knockout mutants obtained | [55] |
AaCas12b: Alicyclobacillus acidiphilus Cas12b; AacCas12b: Alicyclobacillus acidoterrestris Cas12b; AsCas12a: Acidaminococcus sp.BV3L6 Cas12a; BhCas12b: Bacillus hisashii Cas12b; BhCas12b v4: a variant of BhCas12b from Bacillus hisashii; BthCas12b: Bacillus thermoamylovorans Cas12b; BvCas12b: Bacillus sp. V3-13 Cas12b; FnCas12a: Francisella novicida U112 Cas12a; FnoCas12a: Francisella tularensis novicida Cas12a; LbCas12a: Lachnospiraceae bacterium ND2006 Cas12a
CRISPR/Cas12 system involves in genome editing of microorganisms
As a versatile tool for genome editing, CRISPR/Cas12 can also be used for gene editing of microorganisms, including cyanobacteria, bacteria, and fungi. Cyanobacteria are photoautotrophic microorganisms and also an important model organism for physiological and ecological studies. CRISPR/Cas12a has been verified to be suitable for precise genome editing of cyanobacteria through homologous recombination [56, 57]. Subsequently, the Standard European Vector Architecture (SEVA)-based plasmids (containing CRISPR/Cas12a system) has been shown to be effective for genome editing of different genera of cyanobacteria [57]. Similarly, Mycolicibacterium neoaurum ATCC 25,795, a classical bacterium producing valuable steroidal drugs, has achieved efficient and accurate genetic manipulation through CRISPR/Cas12a system. Liu et al. confirmed that CRISPR/Cas12a system had great potential in precise genome editing of M. neoaurum, such as integration of targeted genes into desired sites and targeted deletion of DNA sequences of different lengths [58]. Recently, CRISPR/Cas12a has been proved to be able to simultaneously edit pyrG, pksP, and kusA genes of Aspergillus aculeatus TBRC 277 (an industrially related cell factory), with an efficiency of up to 40% [59]. In conclusion, CRISPR/Cas12-mediated microbial genome editing mainly involves in strain improvement, such as the production of valuable steroidal pharmaceuticals [58] and bioproducts [59], as well as high-performance chassis [60], and fundamental research, for instance, the study of plant-fungal interactions [61, 62] and the pathogenesis of important opportunistic pathogens [63] (Table 4). For ease of understanding, based on existing reports, CRISPR/Cas12-mediated genome editing was represented in Fig. 1.
Table 4.
The applications, characteristics, and advantages of CRISPR/Cas12 system in microorganism genome editing
| Species | Editor | Target genes | Plasmids | Transformation methods | Editing strategies | Editing efficiency or mutation rate | Advantages | References |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli | FnCas12a |
ahpC tyrR |
pTF pTF-lacZ pSIMcpf1 |
Electroporation |
Knock-outs knock-ins genome integration of large DNA fragment |
Markerless deletion: > 80% genome integration: 50% |
Simple; markerless deletion or genome integration; high efficiency | [64] |
| Cyanobacteria | FnCas12a | nblA |
pSEVA351-Cpf1 pSEVA451-Cpf1 pSEVACpf1nblA |
Natural transformation or conjugation | Deletions | 40% | High efficiency; free cost; simplicity | [57] |
| FnCas12a |
nblA psbA |
pSL2680 | Conjugation |
Point mutations knock-outs knock-ins direct gene replacement |
~ 20% |
Multiplex targeting; markerless editing; nontoxic engineering; low cost |
[65] | |
| Mycolicibacterium neoaurum | FnCas12a |
hsd4A KshA2 |
p261-Cas12a pCM-cr_hsd4A pCM-cr_L2 pCM-cr_KshA2 pCM-cr_L1-L2 pCM-cr_T1-cr_T2 pCM-cr_T1-cr_T3 pCM-cr_T1-cr_T4 pCM-cr_T1-cr_T5 pCM-cr_T1-cr_T6 |
Electroporation |
1 kb, 5 kb, 10 kb, 15 kb, 20 kb and 24 kb deletions knock-ins |
Deletion efficiencies: 70%, 30%, 30%, 20%, 20% and 10% Integration efficiencies: single crossover: 100% double crossovers: 9% |
Targeted deletion of DNA sequences and precise genomic integration | [58] |
| Magnaporthe oryzae | LbCas12a | BUF1 |
pFGL821 pFGL921 |
PEG-mediated transformation | Large-scale insertions and deletions | 50%~100% | Variable DNA repair | [61, 62] |
| Enterococcus faecium | AsCas12a |
lacL acpH treA unaG |
pJC005.em.XlacL pJC005.em.XacpH pJC005.em.XtreA pJC005.em.XtreA::unaG etc. |
Electro- transformation | InDels | > 80% | Rapid, efficient, and cost-effective | [63] |
| Shewanella oneidensis |
AsCas12a BhCas12b |
ampC nagK gfp |
pAsCpf1-ampC pBhCas12b-ampC etc. |
Conjugation |
knock-outs knock-ins gene replacement |
Insertion 94.4%, 83.9% Deletion: 41.67% Replacement: 25% |
Ideally suitable to target T-rich DNA sequences | [60] |
| Aspergillus aculeatus |
FnCas12a AsCas12a LbCas12a |
pyrG pksP kusA |
pCRISPR01-FnCpf1 pCRISPR01-AsCpf1 pCRISPR01-LbCpf1 pCRISPR01-FnCpf1-crRNA-kusA1-pksP2-pyrG3 etc. |
Protoplast-mediated transformation | InDels |
FnCas12a: 93% AsCas12a: 43% LbCas12a: 13% FnCas12a: three genes combined editing efficiency: 40% |
Versatile, flexible, precise and highly efficient; multiplex gene-editing |
[59] |
AsCas12a: Acidaminococcus sp.BV3L6 Cas12a; BhCas12b: Bacillus hisashii Cas12b; FnCas12a: Francisella novicida U112 Cas12a; LbCas12a: Lachnospiraceae bacterium ND2006 Cas12a
Fig. 1.
CRISPR/Cas12-mediated genome editing of animals, plants and microorganisms
The other Cas12 proteins and their applications
As shown in Table 5, Cas proteins of type V can be divided into 11 subtypes, including subtype V-A (Cas12a), subtype V-B (Cas12b), subtype V-C (Cas12c), subtype V-D (Cas12d), subtype V-E (Cas12e), subtype V-F (Cas12f), subtype V-G (Cas12g), subtype V-H (Cas12h), subtype V-I (Cas12i), subtype V-K (Cas12k), and subtype V-U [2]. CRISPR/Cas12c (C2c3) and Cas12d (CasY) proteins represent compact CRISPR/Cas systems, which have limited sequence homology with the crRNAs of Cas12a and Cas9 [66]. The study demonstrated that Cas12d-catalyzed DNA cleavage required a short complementary untranslated RNA (scoutRNA) and crRNA, and the scoutRNA was an important cofactor for Cas12c-catalyzed pre-crRNA maturation. Cas12c can be applied for plant pathogen detection and single nucleotide polymorphism (SNP) identification. In addition, Cas12d can boost RNA-guided DNA interference in bacteria [66]. Cas12e (CasX) is an RNA-guided DNA endonuclease that modifies the genomes of humans, mouse, Drosophila, yeast, Escherichia coli, and other model organisms, thus having the potential as a universal genome editor [67]. The miniature Cas12f1 is an RNA-guided endonuclease, which has been demonstrated to be an efficient genome editing tool in bacteria and human cells [68]. Cas12g is an RNA-guided ribonuclease that cleaves single-stranded RNA (ssRNA) and ssDNA by targeting ssRNA substrate. CRISPR/Cas12g system provides a promising platform for RNA editing and nucleic acid detection by virtue of the small molecular weight and high thermal stability of Cas12g protein [69]. Cas12i, the subtype V-I CRISPR/Cas effector, is an endonuclease that predominantly recognizes and cleaves non-target strand of 28 bp double-stranded DNA (dsDNA) substrate. Compared with 20 bp dsDNA substrate recognized by Cas9, Cas12a, Cas12b, and Cas12e, Cas12i has the potential to be exploited into a high-fidelity genome editor [70]. Cas12k, encoded by cyanobacterial Scytonema hofmannisystem Tn7-like transposon, has no endonuclease activity and mediates guide RNA-dependent transposition [2, 71]. By combining the transposase with the CRISPR effector Cas12k, DNA fragments can be directly integrated into the target sites with 80% frequency, which laid the foundation for precise insertion of DNA [71]. Querques et al. demonstrated the feasibility of using CRISPR-associated transposons as a tool of programmable site-specific gene insertion [72]. The functions of Cas12 proteins from subtype V-H and V-U need to be further explored. The classifications and applications of CRISPR/Cas12 system were listed in Table 5.
Table 5.
The classifications, characteristics and functions of Cas12 proteins [2]
| Subtype | Variant | tracrRNAa | Gene nomenclature | Cleavage activity | Amino acids | Functions | References |
|---|---|---|---|---|---|---|---|
| V-A | No |
Cas12ab (Cpf1)c |
dsDNA, ssDNA | 1310 | Gene editing; nucleic acid detection; molecular diagnosis | [8, 11, 47] | |
| V-B | V-B1 | Yes |
Cas12b1 (C2c1) |
dsDNA, ssDNA | ~ 1100 | Gene editing; nucleic acid detection | [12, 32, 39] |
| V-B2 | Yes | Cas12b2 | dsDNA, ssDNA | Gene editing | |||
| V-C | No |
Cas12c (C2c3) |
dsDNA | 1253 | Nucleic acid detection | [26] | |
| V-D | No |
Cas12d (CasY) |
dsDNA | ~ 1200 | DNA interference | [73] | |
| V-E | Yes |
Cas12e (CasX) |
dsDNA | ~ 980 | Genome editor | [67] | |
| V-F | V-F1 | Yes |
Cas12f1 (Cas14a) |
ssDNA | 400 ~ 700 | Gene editing | [73] |
| High-fidelity SNP genotypes | |||||||
| V-F2 | No |
Cas12f2 (Cas14b) |
– | ||||
| V-F3 | No |
Cas12f3 (Cas14c) |
– | ||||
| V-G | Yes | Cas12g | RNA, ssDNA | 768 | Nucleic acid detection | [69] | |
| V-H | No | Cas12h | dsDNA | 871 | – | ||
| V-I | No | Cas12i | dsDNA | 1055 | Gene editing | [70] | |
| V-K | V-U5 | Yes |
Cas12k (C2c5) |
Transposases | –d | Help Tn-7 jump gene find specific sequence | [71, 72] |
| V-U | V-U1 | No | C2c4 | Transposon-encoded nucleases | – | – | [74] |
| V-U2 | No | C2c8 | |||||
|
V-U3 (V-F1) |
No | C2c10 | Originate independently from different TnpB families | ||||
| V-U4 | No | C2c9 |
dsDNA double-stranded DNA, ssDNA single-stranded DNA
aTransactivating CRISPR RNA (tracrRNA)
bSystematic name
cLegacy name
d–No data
It should be pointed out that in addition to the functions mentioned above, Cas12 also participates in gene regulation through CRISPR interference (CRISPRi) [64, 75]. Recent study has shown that CRISPRi can efficiently (> 90%) achieve transcriptional inhibition of target genes [64]. Taken together, these findings can improve our understanding of the functional diversity of CRISPR/Cas12 system. With further research, Cas12 will exhibit an increasingly extensive application prospect.
Conclusion
CRISPR/Cas technology, as the most commonly used genome editing tool, is known as the “magic scissors” for genome editing because of its simplicity, cheapness and high efficiency, which allows researchers to efficiently and precisely alter, edit or replace genes in plants, animals and even humans. Nowadays, the improved CRISPR/Cas technology is widely employed in the breeding of new varieties of animals and plants and biomedical fields, such as searching for genes related to signal pathways, screening drug targets, and gene therapy. Undoubtedly, CRISPR/Cas-mediated genome editing in vivo has certain off-target effects, which may lead to the loss of large DNA fragments at the distal end of the cleavage site, and even other more complex gene mutations [76, 77]. However, with the further development, modification, and improvement of CRISPR/Cas technology, the problems mentioned above will be solved gradually. Meanwhile, based on the CRISPR/Cas system, combined with isothermal amplification, electrochemical sensor, optical imaging and other technologies, the new biosensors with high specificity and sensitivity, as well as rapidity, simplicity and convenience have been established by researchers, which are extensively applied in the fields of disease-related detection and imaging, detection of various disease markers, thereby enabling high accuracy and reliability of disease diagnosis at the molecular level. Recently, Ivanov et al. developed a platform for detecting Cas12 trans-cleavage activity based on lateral flow test (LFT), and its assay result only depends on composite DNA-IgG probe [29]. In the future, the development of a CRISPR/Cas12-based probe detection platform holds great promise. Additionally, the novel RNA editing tool Cas7-11 can be fused together for genome editing, which may be used in tissue regeneration and antiviral drugs [78]. Cas12k has no nuclease activity and only mediates guide RNA-dependent transposition [2, 79]. The development of transposase component engineering may expand its application in eukaryotic cells in the future. A latest study reported a surprising finding that the designed CRISPR/Cas12 DNA device can degrade the native chromosome and convert bacterial cells into biosynthetic chassis for high-efficiency molecular biomanufacturing [80]. As a consequence, with the unremitting efforts of researchers from all over the world, the new functions and new types of Cas proteins will be further discovered and deciphered, which will further broaden their diverse biotechnological applications, so as to better utilize the CRISPR/Cas system to serve humans.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31872425), National Natural Science Foundation of China (31861143051). We are thankful to the anonymous reviewers for reviewing our manuscript and providing helpful comments and suggestions.
Author contributions
YY: conceptualization, methodology, validation, supervision, writing—review and editing. DW: conceptualization, methodology, investigation, formal analysis, writing—original draft. PL: methodology, validation. SM: validation, visualization. KC: funding acquisition, resources.
Funding
Funding was provided by National Natural Science Foundation of China (Grant No. 31872425, 31861143051).
Declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent was obtained from all the individual participants included in this study.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18(2):67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leung RK, Cheng QX, Wu ZL, Khan G, Liu Y, Xia HY, et al. Review: CRISPR/Cas12-based nucleic acids detection systems. Methods. 2021;203:276–281. doi: 10.1016/j.ymeth.2021.02.018. [DOI] [PubMed] [Google Scholar]
- 4.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532(7600):517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- 6.Li SY, Cheng QX, Liu JK, Nie XQ, Zhao GP, Wang J. CRISPR/Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018;28(4):491–493. doi: 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H, Gao P, Rajashankar KR, Patel DJ. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell. 2016;167(7):1814–1828e1812. doi: 10.1016/j.cell.2016.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60(3):385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrington LB, Burstein D, Chen JS, Paez-Espino D, Ma E, Witte IP, et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362(6416):839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, et al. CRISPR/Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li SY, Cheng QX, Wang JM, Li XY, Zhang ZL, Gao S, et al. CRISPR/Cas12a-assisted nucleic acid detection. Cell Discov. 2018;4:20. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, Li S, Wu N, Wu J, Wang G, Zhao G, et al. HOLMESv2: a CRISPR/Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth Biol. 2019;8(10):2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- 13.Teng F, Guo L, Cui T, Wang XG, Xu K, Gao Q, et al. CRISPR/Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol. 2019;20(1):132. doi: 10.1186/s13059-019-1742-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang B, Wang R, Wang D, Wu J, Li J, Wang J, et al. A CRISPR/Cas12a-based platform for rapid and visual nucleic acid detection. Anal Chem. 2019;91(19):12156–12161. doi: 10.1021/acs.analchem.9b01526. [DOI] [PubMed] [Google Scholar]
- 15.Xia X, Ma B, Zhang T, Lu Y, Khan MR, Hu Y, et al. G-quadruplex-probing CRISPR/Cas12 assay for label-free analysis of foodborne pathogens and their colonization in vivo. ACS Sens. 2021;6(9):3295–3302. doi: 10.1021/acssensors.1c01061. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Li J, Li S, Zhu X, Wang X, Huang J, et al. LAMP-CRISPR/Cas12-based diagnostic platform for detection of Mycobacterium tuberculosis complex using real-time fluorescence or lateral flow test. Mikrochim Acta. 2021;188(10):347. doi: 10.1007/s00604-021-04985-w. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Chen M, Liu C, Chen J, Luo X, Xue Y, et al. Sensitive and rapid on-site detection of SARS-CoV-2 using a gold nanoparticle-based high-throughput platform coupled with CRISPR/Cas12-assisted RT-LAMP. Sens Actuators B Chem. 2021;345:130411. doi: 10.1016/j.snb.2021.130411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai L, Wang L, Huang S, Bai R, Lv X, Sun L, et al. Rapid, visual, and sequence-specific detection of Salmonella in egg liquid with vis-NEAA, a CRISPR/Cas12 empowered new strategy. J Agric Food Chem. 2022;70(7):2401–2409. doi: 10.1021/acs.jafc.1c06715. [DOI] [PubMed] [Google Scholar]
- 19.Ding R, Long J, Yuan M, Zheng X, Shen Y, Jin Y, et al. CRISPR/Cas12-based ultra-sensitive and specific point-of-care detection of HBV. Int J Mol Sci. 2021;22(9):4842. doi: 10.3390/ijms22094842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali Z, Aman R, Mahas A, Rao GS, Tehseen M, Marsic T, et al. iSCAN: an RT-LAMP-coupled CRISPR/Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 2020;288:198129. doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joung J, Ladha A, Saito M, Segel M, Bruneau R, Huang MW, et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. MedRxiv. 2020 doi: 10.1101/2020.05.04.20091231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W, Yu L, Wen D, Wei D, Sun Y, Zhao H, et al. A CRISPR/Cas12a-based specific enhancer for more sensitive detection of SARS-CoV-2 infection. EBioMedicine. 2020;61:103036. doi: 10.1016/j.ebiom.2020.103036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, Qian C, Wu C, Wang Z, Wang D, Ye Z, et al. End-point dual specific detection of nucleic acids using CRISPR/Cas12a based portable biosensor. Biosens Bioelectron. 2020;157:112153. doi: 10.1016/j.bios.2020.112153. [DOI] [PubMed] [Google Scholar]
- 24.Zhang T, Li HT, Xia X, Liu J, Lu Y, Khan MR, et al. Direct detection of foodborne pathogens via a proximal DNA probe-based CRISPR/Cas12 assay. J Agric Food Chem. 2021;69(43):12828–12836. doi: 10.1021/acs.jafc.1c04663. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Wang J, Zeng H, Liu X, Jiang W, Wang Y, et al. RPA-Cas12a-FS: a frontline nucleic acid rapid detection system for food safety based on CRISPR/Cas12a combined with recombinase polymerase amplification. Food Chem. 2021;334:127608. doi: 10.1016/j.foodchem.2020.127608. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Zhong C. Cas12c-DETECTOR: a specific and sensitive Cas12c-based DNA detection platform. Int J Biol Macromol. 2021;193(PtA):441–449. doi: 10.1016/j.ijbiomac.2021.10.167. [DOI] [PubMed] [Google Scholar]
- 27.Zhu X, Yang H, Wang M, Wu M, Khan MR, Luo A, et al. Label-free detection of transgenic crops using an isothermal amplification reporting CRISPR/Cas12 assay. ACS Synth Biol. 2022;11(1):317–324. doi: 10.1021/acssynbio.1c00428. [DOI] [PubMed] [Google Scholar]
- 28.Bhatt A, Fatima Z, Ruwali M, Misra CS, Rangu SS, Rath D, et al. CLEVER assay:a visual and rapid RNA extraction-free detection of SARS-CoV-2 based on CRISPR-Cas integrated RT-LAMP technology. J Appl Microbiol. 2022 doi: 10.1111/jam.15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanov AV, Safenkova IV, Zherdev AV, Dzantiev BB. DIRECT(2): a novel platform for a CRISPR/Cas12-based assay comprising universal DNA-IgG probe and a direct lateral flow test. Biosens Bioelectron. 2022;208:114227. doi: 10.1016/j.bios.2022.114227. [DOI] [PubMed] [Google Scholar]
- 30.Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361(6405):866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu WY, Lebbink JHG, Kanaar R, Geijsen N, van der Oost J. Genome editing by natural and engineered CRISPR-associated nucleases. Nat Chem Biol. 2018;14(7):642–651. doi: 10.1038/s41589-018-0080-x. [DOI] [PubMed] [Google Scholar]
- 32.Strecker J, Jones S, Koopal B, Schmid-Burgk J, Zetsche B, Gao L, et al. Engineering of CRISPR/Cas12b for human genome editing. Nat Commun. 2019;10(1):212. doi: 10.1038/s41467-018-08224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teng F, Cui T, Feng G, Guo L, Xu K, Gao Q, et al. Repurposing CRISPR/Cas12b for mammalian genome engineering. Cell Discov. 2018;4:63. doi: 10.1038/s41421-018-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ming M, Ren Q, Pan C, He Y, Zhang Y, Liu S, et al. CRISPR/Cas12b enables efficient plant genome engineering. Nat Plants. 2020;6(3):202–208. doi: 10.1038/s41477-020-0614-6. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Long C, Li H, McAnally JR, Baskin KK, Shelton JM, et al. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci Adv. 2017;3(4):e1602814. doi: 10.1126/sciadv.1602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol. 2017;35(1):31–34. doi: 10.1038/nbt0217-178b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu P, Luk K, Shin M, Idrizi F, Kwok S, Roscoe B, et al. Enhanced Cas12a editing in mammalian cells and zebrafish. Nucleic Acids Res. 2019;47(8):4169–4180. doi: 10.1093/nar/gkz184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teng F, Li J, Cui T, Xu K, Guo L, Gao Q, et al. Enhanced mammalian genome editing by new Cas12a orthologs with optimized crRNA scaffolds. Genome Biol. 2019;20(1):15. doi: 10.1186/s13059-019-1620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L, Chen P, Wang M, Li X, Wang J, Yin M, et al. C2c1-sgRNA complex structure reveals RNA-Guided DNA cleavage mechanism. Mol Cell. 2017;65(2):310–322. doi: 10.1016/j.molcel.2016.11.040. [DOI] [PubMed] [Google Scholar]
- 40.Kim DY, Lee JM, Moon SB, Chin HJ, Park S, Lim Y, et al. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat Biotechnol. 2022;40(1):94–102. doi: 10.1038/s41587-021-01009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JG, Ha CH, Yoon B, Cheong SA, Kim G, Lee DJ, et al. Knockout rat models mimicking human atherosclerosis created by Cpf1-mediated gene targeting. Sci Rep. 2019;9(1):2628. doi: 10.1038/s41598-019-38732-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W, Shi L, Zhuang Z, Wu H, Lian M, Chen Y, et al. Engineered pigs carrying a gain-of-function NLRP3 homozygous mutation can survive to adulthood and accurately recapitulate human systemic spontaneous inflammatory responses. J Immunol. 2020;205(9):2532–2544. doi: 10.4049/jimmunol.1901468. [DOI] [PubMed] [Google Scholar]
- 43.Tsukamoto T, Sakai E, Iizuka S, Taracena-Gandara M, Sakurai F, Mizuguchi H. Generation of the adenovirus vector-mediated CRISPR/Cpf1 system and the application for primary human hepatocytes prepared from humanized mice with chimeric liver. Biol Pharm Bull. 2018;41(7):1089–1095. doi: 10.1248/bpb.b18-00222. [DOI] [PubMed] [Google Scholar]
- 44.Kim YS, Kim GR, Park M, Yang SC, Park SH, Won JE, et al. Electroporation of AsCpf1/RNP at the zygote stage is an efficient genome editing method to generate knock-out mice deficient in leukemia inhibitory factor. Tissue Eng Regen Med. 2020;17(1):45–53. doi: 10.1007/s13770-019-00225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoon AR, Jung BK, Choi E, Chung E, Hong J, Kim JS, et al. CRISPR/Cas12a with an oAd induces precise and cancer-specific genomic reprogramming of EGFR and efficient tumor regression. Mol Ther. 2020;28(10):2286–2296. doi: 10.1016/j.ymthe.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li B, Liang S, Alariqi M, Wang F, Wang G, Wang Q, et al. The application of temperature sensitivity CRISPR/LbCpf1 (LbCas12a) mediated genome editing in allotetraploid cotton (G. hirsutum) and creation of nontransgenic, gossypol-free cotton. Plant Biotechnol J. 2021;19(2):221–223. doi: 10.1111/pbi.13470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Endo A, Masafumi M, Kaya H, Toki S. Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci Rep. 2016;6:38169. doi: 10.1038/srep38169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong ZH, Zhang YX, You Q, Tang X, Ren QR, Liu SS, et al. Plant genome editing using FnCpf1 and LbCpf1 nucleases at redefined and altered PAM sites. Mol Plant. 2018;11(7):999–1002. doi: 10.1016/j.molp.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 49.Malzahn AA, Tang X, Lee K, Ren Q, Sretenovic S, Zhang Y, et al. Application of CRISPR/Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol. 2019;17(1):9. doi: 10.1186/s12915-019-0629-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arizti-Sanz J, Freije CA, Stanton AC, Petros BA, Boehm CK, Siddiqui S, et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat Commun. 2020;11(1):5921. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu F, Qiao X, Zhao Y, Zhang Z, Gao Y, Shi L, et al. Targeted mutagenesis in Arabidopsis thaliana using CRISPR/Cas12b/C2c1. J Integr Plant Biol. 2020;62(11):1653–1658. doi: 10.1111/jipb.12944. [DOI] [PubMed] [Google Scholar]
- 52.Merker L, Schindele P, Huang TK, Wolter F, Puchta H. Enhancing in planta gene targeting efficiencies in Arabidopsis using temperature-tolerant CRISPR/LbCas12a. Plant Biotechnol J. 2020;18(12):2382–2384. doi: 10.1111/pbi.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Q, Alariqi M, Wang F, Li B, Ding X, Rui H, et al. The application of a heat-inducible CRISPR/Cas12b (C2c1) genome editing system in tetraploid cotton (G. hirsutum) plants. Plant Biotechnol J. 2020;18(12):2436–2443. doi: 10.1111/pbi.13417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jia H, Orbovic V, Wang N. CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol J. 2019;17(10):1928–1937. doi: 10.1111/pbi.13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.An Y, Geng Y, Yao J, Fu C, Lu M, Wang C, et al. Efficient genome editing in populus using CRISPR/Cas12a. Front Plant Sci. 2020;11:593938. doi: 10.3389/fpls.2020.593938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ng IS, Keskin BB, Tan SI. A critical review of genome editing and synthetic biology applications in metabolic engineering of microalgae and cyanobacteria. Biotechnol J. 2020;15(8):e1900228. doi: 10.1002/biot.201900228. [DOI] [PubMed] [Google Scholar]
- 57.Baldanta S, Guevara G, Navarro-Llorens JM. SEVA-Cpf1, a CRISPR/Cas12a vector for genome editing in cyanobacteria. Microb Cell Fact. 2022;21(1):103. doi: 10.1186/s12934-022-01830-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu K, Gao Y, Li ZH, Liu M, Wang FQ, Wei DZ. CRISPR/Cas12a assisted precise genome editing of Mycolicibacterium neoaurum. New Biotechnol. 2022;66:61–69. doi: 10.1016/j.nbt.2021.10.003. [DOI] [PubMed] [Google Scholar]
- 59.Abdulrachman D, Champreda V, Eurwilaichitr L, Chantasingh D, Pootanakit K. Efficient multiplex CRISPR/Cpf1 (Cas12a) genome editing system in Aspergillus aculeatus TBRC 277. J Biotechnol. 2022;355:53–64. doi: 10.1016/j.jbiotec.2022.06.011. [DOI] [PubMed] [Google Scholar]
- 60.Chen Y, Cheng M, Feng X, Niu X, Song H, Cao Y. Genome editing by CRISPR/Cas12 recognizing AT-Rich PAMs in Shewanella oneidensis MR-1. ACS Synth Biol. 2022;11(9):2947–2955. doi: 10.1021/acssynbio.2c00208. [DOI] [PubMed] [Google Scholar]
- 61.Huang J, Cook DE. CRISPR/Cas12a ribonucleoprotein-mediated gene editing in the plant pathogenic fungus Magnaporthe oryzae. STAR Protoc. 2022;3(1):101072. doi: 10.1016/j.xpro.2021.101072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang J, Rowe D, Zhang W, Suelter T, Valent B, Cook DE. CRISPR/Cas12a induced DNA double-strand breaks are repaired by locus-dependent and error-prone pathways in a fungal pathogen. BioRxiv. 2021 doi: 10.1101/2021.09.08.459484. [DOI] [Google Scholar]
- 63.Chua MJ, Collins J. Rapid, eficient, and cost-effective gene editing of Enterococcus faecium with CRISPR/Cas12a. Microbiol Spectr. 2022;10(1):e0242721. doi: 10.1128/spectrum.02427-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jervis AJ, Hanko EKR, Dunstan MS, Robinson CJ, Takano E, Scrutton NS. A plasmid toolset for CRISPR-mediated genome editing and CRISPRi gene regulation in Escherichia coli. Microb Biotechnol. 2021;14(3):1120–1129. doi: 10.1111/1751-7915.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ungerer J, Pakrasi HB. Cpf1 is a versatile tool for CRISPR genome editing across diverse species of cyanobacteria. Sci Rep. 2016;6:39681. doi: 10.1038/srep39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harrington LB, Ma E, Chen JS, Witte IP, Gertz D, Paez-Espino D, et al. A scoutRNA is required for some type V CRISPR-Cas systems. Mol Cell. 2020;79(3):416–424e415. doi: 10.1016/j.molcel.2020.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roberson EDO. A catalog of CasX genome editing sites in common model organisms. BMC Genom. 2019;20(1):528. doi: 10.1186/s12864-019-5924-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu Z, Zhang Y, Yu H, Pan D, Wang Y, Wang Y, et al. Programmed genome editing by a miniature CRISPR/Cas12f nuclease. Nat Chem Biol. 2021;17(11):1132–1138. doi: 10.1038/s41589-021-00868-6. [DOI] [PubMed] [Google Scholar]
- 69.Li Z, Zhang H, Xiao R, Han R, Chang L. Cryo-EM structure of the RNA-guided ribonuclease Cas12g. Nat Chem Biol. 2021;17(4):387–393. doi: 10.1038/s41589-020-00721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang H, Li Z, Xiao R, Chang L. Mechanisms for target recognition and cleavage by the Cas12i RNA-guided endonuclease. Nat Struct Mol Biol. 2020;27(11):1069–1076. doi: 10.1038/s41594-020-0499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Strecker J, Ladha A, Gardner Z, Schmid-Burgk JL, Makarova KS, Koonin EV, et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science. 2019;365(6448):48–53. doi: 10.1126/science.aax9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Querques I, Schmitz M, Oberli S, Chanez C, Jinek M. Target site selection and remodelling by type V CRISPR-transposon systems. Nature. 2021;599(7885):497–502. doi: 10.1038/s41586-021-04030-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen LX, Al-Shayeb B, Meheust R, Li WJ, Doudna JA, Banfield JF. Candidate phyla radiation roizmanbacteria from hot springs have novel and unexpectedly abundant CRISPR-Cas systems. Front Microbiol. 2019;10:928. doi: 10.3389/fmicb.2019.00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, et al. New CRISPR–Cas systems from uncultivated microbes. Nature. 2017;542(7640):237–241. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fontana J, Dong C, Ham JY, Zalatan JG, Carothers JM. Regulated expression of sgRNAs tunes CRISPRi in E. coli. Biotechnol J. 2018;13(9):e1800069. doi: 10.1002/biot.201800069. [DOI] [PubMed] [Google Scholar]
- 76.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36(8):765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shin HY, Wang C, Lee HK, Yoo KH, Zeng X, Kuhns T, et al. CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat Commun. 2017;8:15464. doi: 10.1038/ncomms15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ozcan A, Krajeski R, Ioannidi E, Lee B, Gardner A, Makarova KS, et al. Programmable RNA targeting with the single-protein CRISPR effector Cas7-11. Nature. 2021;597(7878):720–725. doi: 10.1038/s41586-021-03886-5. [DOI] [PubMed] [Google Scholar]
- 79.Xiao R, Wang S, Han R, Li Z, Gabel C, Mukherjee IA, et al. Structural basis of target DNA recognition by CRISPR/Cas12k for RNA-guided DNA transposition. Mol Cell. 2021;81(21):4457–4466e4455. doi: 10.1016/j.molcel.2021.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pantoja Angles A, Ali Z, Mahfouz M. CS-Cells: a CRISPR/Cas12 DNA device to generate chromosome-shredded cells for efficient and safe molecular biomanufacturing. ACS Synth Biol. 2022;11(1):430–440. doi: 10.1021/acssynbio.1c00516. [DOI] [PubMed] [Google Scholar]