Abstract
Salmonella enterica serovars Gallinarum and Pullorum are S. enterica biotypes that exhibit host specificity for poultry and aquatic birds and are not normally capable of causing disease in mammalian hosts. During their evolution toward host restriction serovars Gallinarum and Pullorum lost their ability to mediate mannose-sensitive hemagglutination (MSHA), a phenotype correlated with adherence to certain cell types. Because adherence is an essential requirement for invasion of cells by bacterial pathogens, we examined whether MHSA type 1 fimbriae would increase the ability of serovars Pullorum and Gallinarum to invade normally restrictive cells. Serovars Gallinarum and Pullorum expressing S. enterica serovar Typhimurium strain LT2 type 1 fimbriae exhibited a 10- to 20-fold increased ability to adhere to and a 20- to 60-fold increased invasion efficiency of the human epithelial HEp-2 cell line. Invasion was accompanied by extensive ruffling of the membranes of the HEp-2 cells. In a murine ligated ileal loop model, a 32% increase in the number of M-cell ruffles was seen when serovar Gallinarum expressed serovar Typhimurium type 1 fimbriae.
The gram-negative bacteria of the genus Salmonella are the causative agents of numerous diseases, such as bacteremia, enteric fever, and enterocolitis in a broad range of organisms. Although some serovars of S. enterica, such as serovars Typhimurium and Enteritidis, can cause disease in a wide range of hosts, other strains have a greatly reduced range of host specificity. Two such serotypes are Gallinarum and Pullorum, which show specificity for poultry and aquatic birds (3). Several factors are most likely involved in determining the host specificity of a certain Salmonella biotype. One set of factors, for example, are the spv (for Salmonella plasmid virulence) genes which are required for the systemic phase of disease in numerous hosts (reviewed in reference 14). In addition to the virulence plasmid, it is presumed that the acquisition of several chromosomal genes has expanded the host repertoire of certain biotypes (4). Another property likely to be involved in host adaptation is the ability of the bacteria to adhere to mucosal surfaces. Most members of the Salmonella species display numerous appendages, termed fimbriae or pili, upon their cell surface that are thought to enhance the ability of the bacteria to attach to various substrates.
Type 1 fimbriae are expressed by most Salmonella serovars and are characterized by their ability to bind to mannose derivatives found on eukaryotic cells. Such binding can be inhibited by the addition of exogenous α-d-mannose, a phenotype termed mannose-sensitive hemagglutination (MSHA). It has been shown by immunoelectron microscopy and DNA hybridization techniques that serovars Pullorum and Gallinarum encode fimbriae that are closely related to the type 1 fimbriae of serovar Typhimurium (9, 22). Although closely related, the fimbriae of serovars Gallinarum and Pullorum do not exhibit MSHA (12). However, the introduction of the serovar Typhimurium LT2 fimbrial adhesin into serovar Pullorum enables this strain to exhibit MSHA and adherence to HEp-2 cells (15). To examine the relationship between MSHA, adherence, and invasion, serovar Pullorum 297 (obtained from James Duguid) and serovar Gallinarum 2933 (obtained from Bruce Stocker) were transformed with either plasmid pISF156, which confers expression of the serovar Typhimurium LT2 type 1 fimbriae (22), or, as a control, plasmid pACYC184. The ability of these transformants to agglutinate guinea pig erythrocytes and to adhere to HEp-2 cells was tested as described previously (16, 22). As shown in Table 1, the introduction of plasmid pISF156 into either serovar Gallinarum or Pullorum allowed the transformed strains to agglutinate guinea pig erythrocytes. In addition, adherence to HEp-2 cells was increased at least 10- to 20-fold when serovar Gallinarum or Pullorum contained the plasmid that confers expression of the LT2 type 1 fimbriae. The presence of 0.3% mannose abolished the abilities of these strains to hemagglutinate and to adhere to HEp-2 cells (data not shown).
TABLE 1.
The presence of plasmid pISF156 in serovars Gallinarum and Pullorum increases hemagglutination (HA) and adherence to HEp-2 cells
| Bacterium | HA | No. of adherent bacteria/HEp-2 cell |
|---|---|---|
| Serovar Typhimurium LT2 | + | 16 |
| Serovar Pullorum(pACYC184) | − | <1 |
| Serovar Pullorum(pISF156) | + | 19 |
| Serovar Gallinarum(pACYC184) | − | <1 |
| Serovar Gallinarum(pISF156) | + | 10 |
Although serovar Typhimurium SL1344 and LT2 can both mediate MSHA, the LT2 strain adheres at much higher levels (15-fold more) to HEp-2 cells than does SL1344 (not shown). Similarly, Baumler et al. observed differences in fim-mediated adherence to HeLa cells by the serovar Typhimurium isolates SR-11 and ATCC 14028 (5). Interestingly, the deduced amino acid sequence of the serovar Typhimurium LT2 fimbrial adhesin protein, FimH, (15) is slightly different from the deduced serovar Typhimurium SL1344 FimH amino acid sequence (J. Boddicker, unpublished data). It is possible that small differences in the adhesin molecules among the serovar Typhimurium strains may lead to differences in the ability of these strains to attach to the mannosyl derivatives of a certain cell type. Alternatively, as was shown by Thankavel et al. (23), the composition of the fimbrial shaft of the type 1 fimbriae can influence the adhesion of serovar Typhimurium to different cell types, perhaps by constraints imposed by the fimbrial shaft on the adhesin. In these studies we have not determined whether the serovar Typhimurium LT2 fimbriae conferred by pISF156 are solely responsible for the MSHA phenotype or whether hybrid fimbriae are produced by expression of the serovar Gallinarum and Pullorum chromosomally encoded type 1 fimbriae. Further studies are necessary to elucidate the molecular mechanisms by which the type 1 fimbriae of different serovar Typhimurium strains mediate attachment to different cell types.
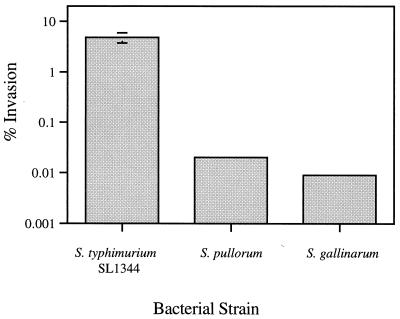
Compared to the virulent serovar Typhimurium SL1344 strain, serovar Pullorum is approximately 80-fold less efficient, and serovar Gallinarum is about 200-fold less efficient at invading HEp-2 cells in an in vitro invasion assay (Fig. 1). To determine whether the increased adherence of the pISF156-transformed serovar Gallinarum and Pullorum strains also led to an increased ability of these strains to invade HEp-2 cells, invasion assays were performed as described previously (19). Serovar Gallinarum and Pullorum strains transformed with pACYC184 showed very low levels of invasion, while the pISF156 transformants exhibited a 20- to 60-fold increase in invasion (Table 2). The addition of 0.3% mannose reduced invasion levels of the pISF156 transformants to levels similar to those of the strains transformed with the vector alone. The presence of 0.3% glucose had no effect on invasion levels (not shown), indicating that the presence of sugars other than mannose did not inhibit invasion. These results underscore the importance of cellular adherence in the infective process and suggest a role for fimbriae in the expansion or restriction of host specificity of a Salmonella biotype.
FIG. 1.
Invasion of HEp-2 cells by serovar Typhimurium SL1344, serovar Pullorum, and serovar Gallinarum. Bacteria were grown statically at 37°C in Luria broth for 48 h before infecting 5 × 104 HEp-2 cells at a multiplicity of infection of 400 in a 24-well culture dish. After the addition of bacteria, the cell culture plates were centrifuged at 750 × g for 15 min at 25°C. Data are CFU presented as the mean ± standard deviation (error bar) of triplicate wells and are presented as the percentage of the inoculum surviving gentamicin treatment. Data are representative of at least three independent experiments.
TABLE 2.
Plasmid pISF156 increases invasion of HEp-2 cells by serovars Gallinarum and Pullorum
| Bacterial strain | % Invasiona with:
|
|
|---|---|---|
| No mannose | Mannose | |
| Serovar Pullorum pACYC184 | 0.011 ± 0.008 | 0.006 ± 0.001 |
| Serovar Pullorum pISF156 | 0.651 ± 0.202 | 0.003 ± 0.000 |
| Serovar Gallinarum pACYC184 | 0.013 ± 0.005 | 0.009 ± 0.001 |
| Serovar Gallinarum pISF156 | 0.306 ± 0.059 | 0.009 ± 0.005 |
| Serovar Typhimurium hilA(pACYC184) | 0.038 ± 0.002 | 0.038 ± 0.012 |
| Serovar Typhimurium hilA(pISF156) | 0.057 ± 0.007 | 0.015 ± 0.005 |
Bacteria were grown and invasion assays were performed as described in the legend to Fig. 1. Data are CFU presented as the means ± standard deviations of triplicate wells and are presented as the percentage of the inoculum surviving gentamicin treatment.
To examine whether this enhanced invasion of HEp-2 cells could be due to a result of host cell responses triggered by increased adherence of the bacteria, the serovar Typhimurium SL1344 hilA mutant BJ70 was transformed with plasmid pISF156. HilA is a positive regulator of Salmonella pathogenicity island 1 (SPI-1) virulence gene expression, and serovar Typhimurium strains carrying mutations in hilA are 100-fold less invasive for HEp-2 cells than wild-type serovar Typhimurium (2). As is the case with SL1344, BJ70 is not very adherent to HEp-2 cells (fewer than two adherent bacteria per cell) (not shown). If BJ70(pISF156) were to exhibit increased invasion of HEp-2 cells, it would suggest that the invasion exhibited by serovar Gallinarum(pISF156) and serovar Pullorum(pISF156) is a result of nonspecific responses, such as pinocytosis, by the host cell. Invasion assays were performed using wild-type BJ70 transformed with either plasmid pISF156 or pACYC184. BJ70(pISF156) exhibited extremely high levels of MSHA and adherence to HEp-2 cells (>50 adherent bacteria per cell) (not shown). However, BJ70 expressing the serovar Typhimurium LT2 type 1 fimbriae exhibits less than a twofold increase in invasion of HEp-2 cells (Table 2). This slight increase in invasion is significantly less than the 20- to 60-fold increase shown by the pISF156-transformed serovars Pullorum and Gallinarum. This suggests that the presence of LT2 type 1 fimbriae alone is insufficient to increase invasion levels; rather, an active, bacterium-dependent uptake mechanism is also required. The efficiency of such a mechanism may be increased by fimbria-mediated adherence that facilitates a close proximity between the bacterium and the host cell.
Several of the genes on SPI-1 (i.e., prgH, prgK, orgA, and invH) encode components of a type III secretion system that directs the export of bacterial proteins across the inner and outer membranes and are involved in delivering bacterial effector proteins (SipA, SipC, SptP, and AvrA) into eukaryotic cells. Delivery of these proteins leads to the induction of cytoskeletal rearrangements in the host cell, ruffling of the cell membrane, and macropinocytosis of the invading bacterium (reviewed in reference 10). DNA hybridization studies using probes specific for invH (1) and PCR analysis using invA-specific primers (20) have indicated that serovars Gallinarum and Pullorum possess at least some SPI-1 genes. To determine whether additional genes spanning the length of the island are present in these strains, PCR was performed on DNA isolated from serovars Pullorum and Gallinarum using primers specific for the serovar Typhimurium hilA, invH, orgA, and sipC genes. The presence of the hilA, invH, orgA, and sipC SPI-1 genes in serovars Pullorum and Gallinarum was detected by the generation of DNA products the same size as those from invasive serovar Typhimurium (data not shown). In contrast, PCR performed on DNA isolated from serovar Litchfield (obtained from Kris Rahn), which does not possess the SPI-1 genes (13), did not generate any DNA products.
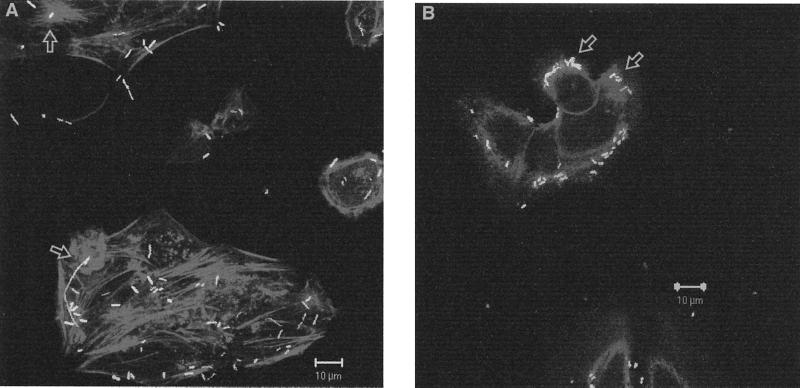
Because serovars Gallinarum and Pullorum appear to possess several of the SPI-1 genes, we wanted to determine whether the invasion of HEp-2 cells by the pISF156-transformed serovar Gallinarum and Pullorum strains might occur in an actin-dependent manner similar to that of serovar Typhimurium. In order to visualize the bacteria using fluorescent microscopy, these strains were also transformed with a plasmid carrying the gene for green fluorescent protein (8) in vector pKSV7 (21). After allowing the doubly transformed strains to invade HEp-2 tissue culture cells for 1 h, actin was visualized using rhodamine phalloidin. Ruffling events were seen in both the serovar Pullorum- and serovar Gallinarum-invaded HEp-2 cells, and green fluorescent bacteria were visualized near the ruffling (Fig. 2). Because actin rearrangement could occasionally be visualized after invasion of HEp-2 cells with the pACYC184-transformed parent strains but at a much lower frequency (not shown), we interpret this result to mean that all of the components necessary for the serovar Gallinarum- or serovar Pullorum-induced ruffling are intact in the wild-type strains and that plasmid pISF156 simply promotes the contact of the bacteria with the HEp-2 cells to allow the exchange of invasion signals to occur at a higher frequency. Studies using SPI-1 mutants of serovars Gallinarum and Pullorum are needed to confirm this interpretation.
FIG. 2.
Serovars Gallinarum and Pullorum can induce actin rearrangement upon invasion of HEp-2 cells. HEp-2 cells (5 × 104) were infected at a multiplicity of infection of 400 with either serovar Gallinarum (A) or serovar Pullorum (B) containing a green fluorescent protein-expressing plasmid and pISF156. The plates were centrifuged at 1,400 rpm for 15 min. After 1 h of incubation actin was visualized using rhodamine phalloidin (Molecular Probes, Eugene, Oreg.) and coverslips viewed with a Zeiss LSM 510 confocal microscope. The arrows denote the actin rearrangement of HEp-2 cells and associated bacteria.
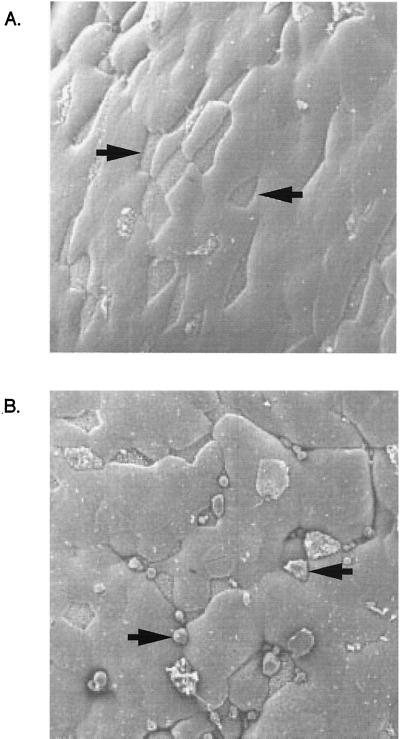
A previous study examining the ability of serovar Gallinarum to invade murine M cells showed no evidence of entry into or destruction of M cells (18). However, the results of our in vitro invasion assay led us to examine whether the presence of the serovar Typhimurium LT2 type 1 fimbriae might also lead to an ability of serovar Gallinarum to invade murine M cells in a ligated ileal loop model. Approximately 4 × 109 serovar Gallinarum bacteria transformed with either pISF156 or vector alone were grown under fimbria-inducing conditions and were used to inoculate ligated ileal loops according to a previously published protocol (17) using 8- to 12-week-old BALB/c mice. The number of M-cell ruffles seen by electron microscopy increased from 0% (0 of 50) to 32% (16 of 50) when serovar Gallinarum expressed the serovar Typhimurium type 1 fimbrial genes as compared to serovar Gallinarum transformed with vector alone (Fig. 3). Wild-type serovar Typhimurium SL1344 induces ruffles in about 50 to 60% of the M cells in ligated ileal loops (unpublished data). These results could explain why Pascopella et al. did not see entry of serovar Gallinarum into murine M cells (18). It is possible that expression of serovar Typhimurium LT2 type 1 fimbriae by serovar Gallinarum facilitated contact of the bacteria with the M cells in order for the invasion process to occur. This would mean that, in addition to lpf (6), under certain conditions the serovar Typhimurium type 1 fimbriae may be important for the attachment of Salmonella to the small intestine.
FIG. 3.
Serovar Typhimurium type 1 fimbriae increase M-cell disruption by serovar Gallinarum. Approximately 4 × 109 serovar Gallinarum bacteria transformed with either vector alone (A) or pISF156 (B) were grown under fimbria-inducing conditions and were used to inoculate BALB/C ligated ileal loops. Scanning electron microscopy was used to examine M cells (denoted by arrows) in the Peyer's patch tissue. Magnification is 1,000-fold.
Because it appeared that the pISF156-transformed serovar Gallinarum was able to invade murine M cells, we were interested in determining whether the transformants would also be lethal in mice. BALB/c mice were orally inoculated with 2 × 109 pACY184- or pISF156-transformed serovar Gallinarum organisms. There were no obvious signs of disease, nor did any deaths occur, in either group of mice (not shown). Some isolates of serovar Gallinarum 2933 lack the spv virulence plasmid (4), and most have been found to be nonflagellate (7, 11). Therefore, decreased virulence in the mouse model of typhoidal infection by avian-adapted serovars is undoubtedly due to the lack of numerous factors. Additional studies are needed to define the components required for the expansion or restriction of the host repertoire by a specific Salmonella biotype.
Acknowledgments
We thank Matt Browning for technical assistance and Julie Tinker for careful review of the manuscript.
This work was supported by NIH grant AI38268 to B. D. Jones and USDA grant 97-35204-4616 to S. Clegg. R.L.W. was supported by NIH postdoctoral training grant HL07638.
REFERENCES
- 1.Altmeyer R M, McNern J K, Bossio J C, Rosenshine I, Finlay B B, Galan J E. Cloning and molecular characterization of a gene involved in Salmonella adherence and invasion of cultured epithelial cells. Mol Microbiol. 1993;7:89–98. doi: 10.1111/j.1365-2958.1993.tb01100.x. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj V, Hwang C, Lee C A. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 3.Barrow P A, Huggins M B, Lovell M A. Host specificity of Salmonella infection in chickens and mice is expressed in vivo primarily at the level of the reticuloendothelial system. Infect Immun. 1994;62:4602–4610. doi: 10.1128/iai.62.10.4602-4610.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumler A J, Tsolis R M, Ficht T A, Adams L G. Evolution of host adaptation in Salmonella enterica. Infect Immun. 1998;66:4579–4587. doi: 10.1128/iai.66.10.4579-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumler A J, Tsolis R M, Heffron F. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect Immun. 1996;64:1862–1865. doi: 10.1128/iai.64.5.1862-1865.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumler A J, Tsolis R M, Heffron F. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer's patches. Proc Natl Acad Sci USA. 1996;93:279–283. doi: 10.1073/pnas.93.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chart H, Conway D, Rowe B. Outer membrane characteristics of Salmonella enteritidis phage type-4 growing in chickens. Epidemiol Infect. 1993;111:449–454. doi: 10.1017/s0950268800057174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 9.Crichton P B, Yakubu D E, Old D C, Clegg S. Immunological and genetical relatedness of type-1 and type-2 fimbriae in salmonellas of serotypes Gallinarum, Pullorum and Typhimurium. J Appl Bacteriol. 1989;67:283–291. doi: 10.1111/j.1365-2672.1989.tb02497.x. [DOI] [PubMed] [Google Scholar]
- 10.Darwin K H, Miller V L. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin Microbiol Rev. 1999;12:405–428. doi: 10.1128/cmr.12.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duguid J P, Anderson E S, Alfredsson G A, Barker R, Old D C. A new biotyping scheme for Salmonella typhimurium and its phylogenetic significance. J Med Microbiol. 1975;8:149–166. doi: 10.1099/00222615-8-1-149. [DOI] [PubMed] [Google Scholar]
- 12.Duguid J P, Anderson E S, Campbell I. Fimbriae and adhesive properties in Salmonellae. J Pathol Bacteriol. 1966;92:107–138. doi: 10.1002/path.1700920113. [DOI] [PubMed] [Google Scholar]
- 13.Ginocchio C C, Rahn K, Clarke R C, Galan J E. Naturally occurring deletions in the centisome 63 pathogenicity island of environmental isolates of Salmonella spp. Infect Immun. 1997;65:1267–1272. doi: 10.1128/iai.65.4.1267-1272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guiney D G, Fang F C, Krause M, Libby S, Buchmeier N A, Fierer J. Biology and clinical significance of virulence plasmids in Salmonella serovars. Clin Infect Dis. 1995;21(Suppl. 2):S146–S151. doi: 10.1093/clinids/21.supplement_2.s146. [DOI] [PubMed] [Google Scholar]
- 15.Hancox L S, Yeh K S, Clegg S. Construction and characterization of type 1 non-fimbriate and non-adhesive mutants of Salmonella typhimurium. FEMS Immunol Med Microbiol. 1997;19:289–296. doi: 10.1111/j.1574-695X.1997.tb01099.x. [DOI] [PubMed] [Google Scholar]
- 16.Hornick D B, Allen B L, Horn M A, Clegg S. Adherence to respiratory epithelia by recombinant Escherichia coli expressing Klebsiella pneumoniae type 3 fimbrial gene products. Infect Immun. 1992;60:1577–1588. doi: 10.1128/iai.60.4.1577-1588.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones B D, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pascopella L, Raupach B, Ghori N, Monack D, Falkow S, Small P L. Host restriction phenotypes of Salmonella typhi and Salmonella gallinarum. Infect Immun. 1995;63:4329–4335. doi: 10.1128/iai.63.11.4329-4335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Penheiter K L, Mathur N, Giles D, Fahlen T, Jones B D. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer's patches. Mol Microbiol. 1997;24:697–709. doi: 10.1046/j.1365-2958.1997.3741745.x. [DOI] [PubMed] [Google Scholar]
- 20.Rahn K, De Grandis S A, Clarke R C, McEwen S A, Galan J E, Ginocchio C, Curtiss III R, Gyles C L. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes. 1992;6:271–279. doi: 10.1016/0890-8508(92)90002-f. [DOI] [PubMed] [Google Scholar]
- 21.Smith K, Youngman P. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie. 1992;74:705–711. doi: 10.1016/0300-9084(92)90143-3. [DOI] [PubMed] [Google Scholar]
- 22.Swenson D L, Clegg S, Old D C. The frequency of fim genes among Salmonella serovars. Microb Pathog. 1991;10:487–492. doi: 10.1016/0882-4010(91)90115-q. [DOI] [PubMed] [Google Scholar]
- 23.Thankavel K, Shah A H, Cohen M S, Ikeda T, Lorenz R G, Curtiss III R, Abraham S N. Molecular basis for the enterocyte tropism exhibited by Salmonella typhimurium type 1 fimbriae. J Biol Chem. 1999;274:5797–5809. doi: 10.1074/jbc.274.9.5797. [DOI] [PubMed] [Google Scholar]