Abstract
Background/Aim: CIC-sarcomas are characterized by rearrangements of the capicua transcriptional repressor (CIC) gene on chromosome subband 19q13.2, generating chimeras in which CIC is the 5’-end partner. Most reported CIC-sarcomas have been detected using PCR amplifications together with Sanger sequencing, high throughput sequencing, and fluorescence in situ hybridization (FISH). Only a few CIC-rearranged tumors have been characterized cytogenetically. Here, we describe the cytogenetic and molecular genetic features of a CIC-sarcoma carrying a t(10;19)(q26;q13), a chromosomal rearrangement not previously detected in such neoplasms.
Materials and Methods: A round cell sarcoma removed from the right thigh of a 57-year-old man was investigated by G-banding cytogenetics, FISH, PCR and Sanger sequencing.
Results: The tumor cells had three cytogenetically related clones with the translocations t(9;18)(q22;q21) and t(10;19)(q26;q13) common to all of them. FISH with a BAC probe containing the CIC gene hybridized to the normal chromosome 19, to der(10)t(10;19), and to der(19)t(10;19). PCR using tumor cDNA as template together with Sanger sequencing detected two CIC::DUX4 fusion transcripts which both had a stop TAG codon immediately after the fusion point. Both transcripts are predicted to encode truncated CIC polypeptides lacking the carboxy terminal part of the native protein. This missing part is crucial for CIC’s DNA binding capacity and interaction with other proteins.
Conclusion: In addition to demonstrating that CIC rearrangement in sarcomas can occur via the microscopically visible translocation t(10;19)(q26;q13), the findings in the present case provide evidence that the missing part in CIC-truncated proteins has important functions whose loss may be important in tumorigenesis.
Keywords: CIC-sarcoma, chromosome translocation, t(10;19) (q26;q13), CIC rearrangement, CIC::DUX4 fusion, truncated CIC protein, proline rich region
According to the fifth edition of the World Health Organization classification of soft tissue and bone tumors, published in 2020, CIC-sarcoma is a high-grade, undifferentiated, round cell sarcoma characterized genomically by rearrangements of the capicua transcriptional repressor (CIC) gene on chromosome subband 19q13.2 with generation of fusions in which CIC is the 5’-end partner (1-4). The most common chimera, found in 95 % of the cases, stems from fusion of CIC with the double homeobox 4 gene (DUX4) (5,6). Rare tumors have also been reported in which CIC fused with forkhead box O4 (FOXO4 on Xq13.1), leucine twenty homeobox (LEUTX on 19q13.2), NUT midline carcinoma family member 1 (NUTM1 on 15q14), and NUT family member 2A (NUTM2A on 10q23.2) (7-15). Recently, a fusion between ataxin 1 (ATXN1 on 6p22.3) and DUX4 was found in two central nervous system sarcomas (16,17). Because the tumors with ATXN1::DUX4 had the same histology, immunohistochemical staining profile and DNA methylation pattern as CIC-rearranged sarcoma, it was concluded that the term ‘CIC-rearranged sarcoma’ may include some tumors with non-CIC alterations (16,17).
The DUX4 gene maps on the q35 band of chromosome 4 and is located within a D4Z4 repeat array associated with the autosomal dominant hereditary disease facioscapulohumeral muscular dystrophy (accession number AF117653) (18). A similar D4Z4 repeat array which also contains DUX4 maps on chromosome band 10q26 (accession number AY028079) (19). DUX4 is expressed and regulates genes during the early stages of embryogenesis (20-22). It is barely expressed in most adult tissues with the exception of testis and thymus (23,24). In the testes, the DUX4 genes from both 4q35 and 10q26 were in the germ-line lineage (23) shown to be expressed approximately equally. In vitro expression experiments showed that DUX4 protein is localized to the nucleus, induces cell death, is a transcriptional activator of paired-like homeodomain transcription factor 1 (PITX1), and also functions as a co-repressor of nuclear receptors of progesterone and glucocorticoids (25-27). In CIC-rearranged tumors, fusion of CIC with both 4q35/DUX4 and 10q26/DUX4 have been reported (5,6,28-38).
The majority of reported CIC-rearranged tumors were detected using various types of PCR amplifications together with Sanger sequencing, high throughput sequencing, and fluorescence in situ hybridization techniques (5,6,29-31,33-35,37-41). The detection of CIC::DUX4 chimera has become crucial in establishing a correct diagnosis for tumors that are otherwise difficult to classify (42,43).
Only very few CIC-rearranged tumors have been cytogenetically examined in spite of the fact that a chromosomal corollary to the gene fusion should be readily visible in suitably stained preparations intended for karyotyping. The translocation t(4;19)(q35;q13) was first described in 1992, as part of a complex karyotype, in an embryonal rhabdomyosarcoma (RMS) cell line (44) and as part of a three-way translocation, t(4;19;12)(q35;q13.1;q13), in a tumor diagnosed as undifferentiated/embryonal RMS (45). In 1996, t(4;19)(q35;q13) was found as a sole cytogenetic abnormality in a poorly differentiated extraskeletal mesenchymal sarcoma (46). That finding, together with the other two above-mentioned cases, led the authors to suggest that that t(4;19)(q35;q13) represents a recurrent chromosomal aberration typical of mesenchymal stem cells (46). It was subsequently found also in an intrabdominal teratoma (47), a subcutaneous primitive neuroectodermal tumor/Ewing sarcoma without EWSR1 rearrangements (48), and in two cases of Ewing-like sarcoma (5). Molecular investigation of the latter two tumors led to cloning and identification of the CIC::DUX4 chimera (5). Later, a t(4;19)(q35;q13) was, in some cases together with a CIC::DUX4 chimera, reported in seven more tumors diagnosed as undifferentiated round cell sarcomas (28,31,33,36). A chromosome translocation t(4;22)(q35;q12), shown by FISH to fuse EWSR1 with DUX4, was found in an embryonal rhabdomyosarcoma (49). Finally, a t(X;19) generating a CIC::FOXO4 chimera was reported in both a desmoplastic small round cell tumor and an undifferentiated round cell sarcoma (8,50).
Here, we update information about the genetics of CIC sarcomas and describe the genomic and pathological features of a diagnostically challenging tumor found to carry a t(10;19)(q26;q13) chromosome translocation leading to a CIC::DUX4 chimera.
Materials and Methods
Ethics statement. The study was approved by the regional ethics committee (Regional komité for medisinsk forskningsetikk Sør-Øst, Norge, http://helseforskning.etikkom.no). Written informed consent was obtained from the patient to publication of the case details. The ethics committee’s approval included a review of the consent procedure. All patient information has been de-identified.
Case report. The patient was a 57-year-old, previously healthy man who had experienced pain in his left inguinal region for one month. Radiological imaging showed a well demarcated tumour measuring 11.0×11.5×8.0 cm in the left adductor magnus muscle. The surgical specimen had a heterogeneous cut surface, brown and fleshy with areas of necrosis (<50% of tumour volume). Representative areas were selected for pathology analyses.
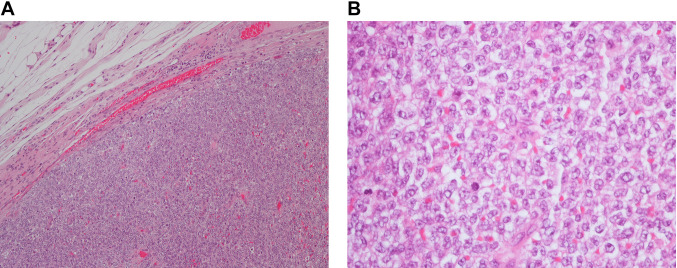
On microscopic examination (Figure 1), the tumour showed a small round cell neoplasm growing in sheets and lobules. The background was partly fibrotic, partly oedematous. The tumour cells had irregular vesicular chromatin, often with small nucleoli and a thin brim of light eosinophilic or clear cytoplasm. The mitotic rate was 23/10 HPF (1,734 mm2). Immunohistochemical staining was positive for CD99, TLE1, cylinD1, and BCOR (weak). MDM2, AE1/AE3, Cam5.2, and CD117 were focally positive. Myogenin, desmin, S100, CD34, TdT, CD3, CD20, MUM1, SOX10, Chromogranin A, and Synapthophysin were negative. Molecular analysis with the Oncomine childhood cancer panel (ThermoFisher Scientific, Waltham, MA, USA) showed no fusion transcripts.
Figure 1. Microscopic examination of the CIC-sarcoma. (A) Hematoxylin and eosin (HE)-stained section showing the small round cell tumor, welldemarcated from the surrounding tissue, magnification ×100. (B) HE-stained section showing the round cell morphology. Irregular vesicular nuclei, some with distinct nucleoli, magnification ×400.
Chromosome banding analysis. A representative tumor area was investigated cytogenetically as previously described (51). The material was mechanically and enzymatically disaggregated, and the resulting cells were short-term cultured, harvested and processed for cytogenetic examination. To obtain G-banding of chromosomes, Wright’s stain was used (Sigma Aldrich; St Louis, MO, USA). The cytogenetic analysis and karyotype description followed the recommendations of the International System for Human Cytogenomic Nomenclature (ISCN) 2020 guidelines (52).
Fluorescence in situ hybridization (FISH). The BAC clone RP11-556K23, which maps to 19q13.2 and contains the CIC gene, was used (33). The FISH probe was prepared from bacteriophage Phi29 DNA polymerase amplified BAC DNAs using previously described methodology and labelled with fluorescein-12-dCTP (PerkinElmer, Boston, MA, USA) to obtain a green signal (53,54). Fluorescent signals were captured and analyzed using the CytoVision system (Leica Biosystems, Newcastle, UK). BAC DNA was also sequenced with T7 (5’-TAATACGACTCACTATAGGG-3’) and SP6 (5’- ATTTAGGTGACACTATAG-3’) primers using the BigDye terminator v1.1 cycle sequencing kit (ThermoFisher Scientific) in order to obtain BAC-end sequences and verify the map position of BAC clone RP11-556K23.
Reverse transcription (RT) PCR and Sanger sequencing analyses. Total RNA was extracted using the miRNeasy Mini Kit (Qiagen, Hilden, Germany) from a frozen (–80˚C) part of the tumor specimen adjacent to where material had been taken for cytogenetic analysis and histologic examination. cDNA was synthesized from one μg of total RNA in a 20 μl reaction volume using iScript Advanced cDNA Synthesis Kit for RT-qPCR according to the manufacturer’s instructions (Bio-Rad, Hercules, CA, USA). cDNA corresponding to 20 ng total RNA was used as template in a 25 μl reaction volume PCR assay containing 12.5 μl Premix Ex Taq™ DNA Polymerase Hot Start Version (Takara Bio Europe/SAS, Saint-Germain-en-Laye, France) and 0.4 μM of each of the forward (CIC-4377F) and reverse (DUXL4-1553R1) primers. The sequence of the CIC-4377F primer was 5’-CCG AGG ACG TGC TTG GGG AGC TA-3’ corresponding to position 4573-4595 of the NCBI Reference Sequence NM_015125.5 [Homo sapiens capicua transcriptional repressor (CIC), mRNA, reported 12-JUN-2022]. The sequence of the DUXL4-1553R1 primer was 5’-CCA GGA AAG AAT GGC AGT TCT CCG C-3’ corresponding to position 1577-1553 of the reference sequence XM_047445716.1 (reported 05-April-2022) which represents a predicted Homo sapiens double homeobox protein 4-like protein 4 and maps on chromosome 10 (https://www.ncbi.nlm.nih.gov/nuccore/XM_047445716.1). The DUXL4-1553R1 primer corresponds also to position 1472-1448 (with a nucleotide substitution) of the reference sequence NM_001306068.3 (reported 24-July-2022) which represents the Homo sapiens double homeobox 4 (DUX4), transcript variant 1, mRNA and maps on 4q35.2.
PCR amplification was conducted on a C-1000 Thermal cycler (Bio-Rad) using the following thermal cycling profile: An initial denaturation step of 30 sec at 94˚C followed by 35 cycles of 7 s at 98˚C, 30 s at 60˚C, 30 s at 72˚C and a final extension step for 5 min at 72˚C. Three μl of the PCR products were stained with GelRed (Biotium, Fremont, CA, USA), analyzed by electrophoresis through 1.0 % agarose gel, and photographed. DNA gel electrophoresis was performed using lithium borate buffer (55). The remaining PCR products were purified with the MinElute PCR Purification Kit (Qiagen) and cloned to pCR4-TOPO TA vector using the TOPO TA cloning kit for sequencing (ThermoFischer Scientific). Twelve colonies were sequenced with the dideoxy procedure using the BigDye terminator v1.1 cycle sequencing kit following the company’s recommendations (ThermoFisher Scientific).
Bioinformatics. The sequences obtained by Sanger sequencing were compared to the NCBI Reference Sequences NM_015125.3 (CIC) and NM_001306068.3 (DUX4) using the Basic Local Alignment Search Tool (BLAST) (56). They were aligned to the sequences on the Human GRCh37/hg19 assembly and the recently described T2T-CHM13 v2.0 human genome (57) using the BLAST-like alignment tool (BLAT) and the human genome browser at UCSC (58,59). They were also compared with the sequences reported in the articles by Italiano et al. (6), Machado et al. (32), Gambarotti et al. (34), Kao et al. (60), Tsukamoto et al. (37), Yoshida et al. (41), and Cocchi et al. (38). For multiple sequence alignment, the MultAlin software (61) was used (http://multalin.toulouse.inra.fr/multalin/).
Results
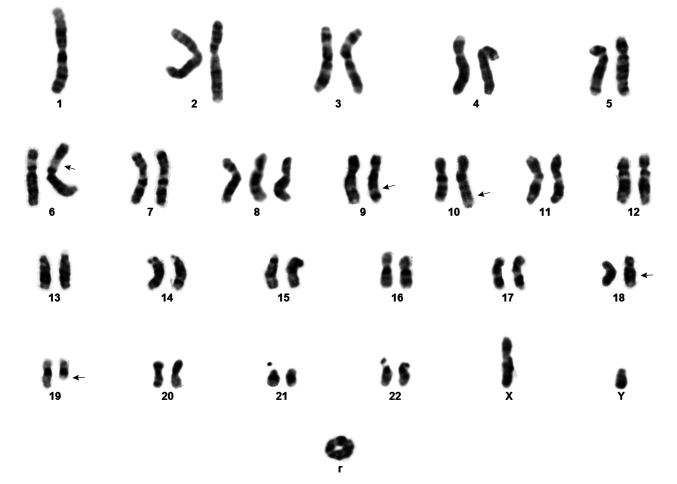
G-banding analysis of tumor cells detected three related clones which had the translocations t(9;18)(q22;q21) and t(10;19)(q26;q13) in common. The karyotype describing clonal evolution was: 46,XY,del(1)(p34p35),t(9;18)(q22;q21), t(10;19)(q26;q13)[5]/46,idem,-del(1),add(6)(p21), +r[3]/47,idem,-del(1),add(6),+8,+r[5]. A karyogram of the third clone is shown in Figure 2.
Figure 2. G-Banding analysis of the CIC-sarcoma represented by a karyogram of the clone 47,XY,-1,add(6)(p21),+8,t(9;18)(q22;q21),t(10;19)(q26;q13),+r. Arrows indicate breakpoints.
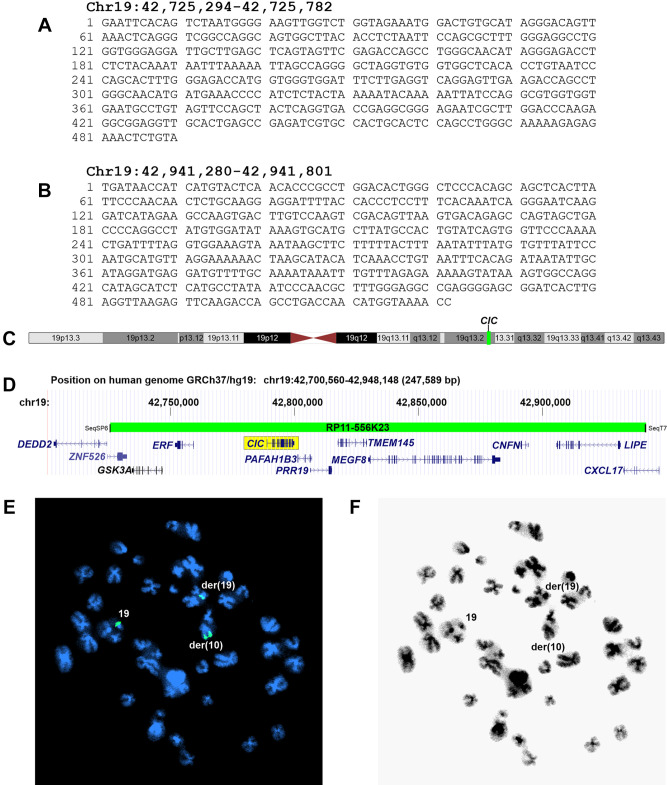
Sequencing of the BAC-probe RP11-556K23-end with the primer SP6 showed that this part of the clone corresponds to chr19:42,725,294-42,725,782 (GRCh37/hg19) (Figure 3A). Sequencing of the other end of the probe with the T7 primer showed it to correspond to chr19:42,941,280-42,941,801 (Figure 3B). The sequencing data verified that the BAC probe RP11-556K23 maps to subband 19q13.2 (Figure 3C) and contains many genes, among them CIC (Figure 3D). FISH with the BAC probe RP11-556K23 on metaphase spreads showed the probe hybridizing to three chromosomes resulting in green signals on the normal chromosome 19, on der(10)t(10;19), and on der(19)t(10;19) (Figures 3E and 3F).
Figure 3. Fluorescence in situ hybridization (FISH) examination of the CIC-sarcoma. (A) Partial sequence of the BAC clone RP11-556K23 using the SP6 primer. (B) Partial sequence of the BAC clone RP11-556K23 using the T7 primer. (C) Ideogram of the chromosome 19 showing the mapping positions of the CIC gene (vertical line) and the BAC clone RP11-556K23 probe (green box). (D) Diagram showing the mapping pattern of the FISH probe RP11-556K23 for the CIC gene. Some neighboring genes in the region are also shown. (E) FISH results on a metaphase spread. Green signals are seen on chromosomes 19, der(10)t(10;19) and der(19)t(10;19). (F) Inverted DAPI staining of the metaphase spread used for FISH experiments.
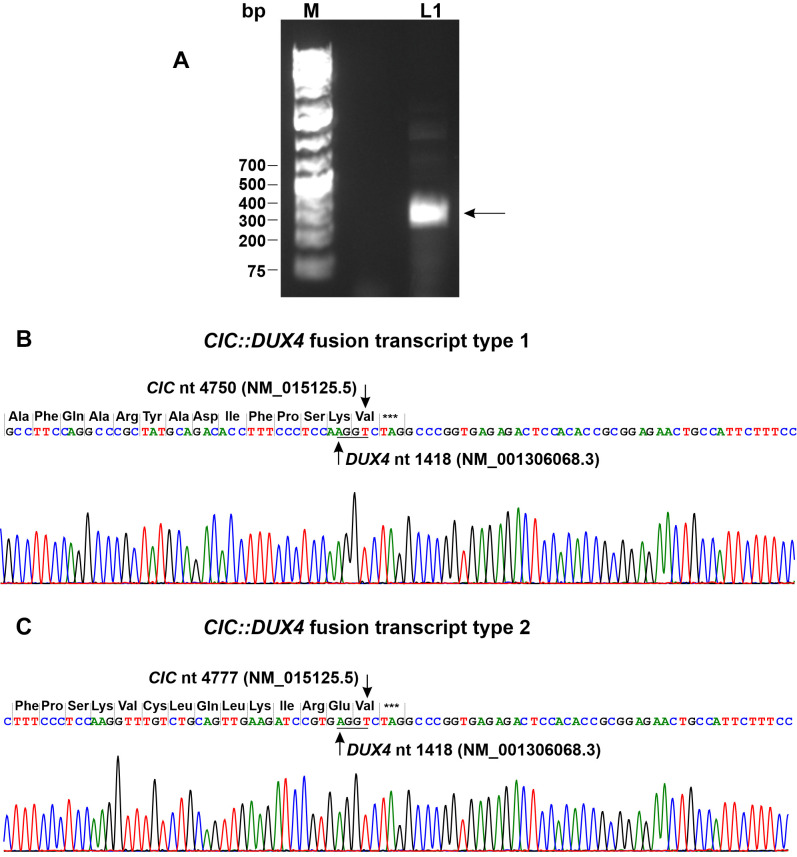
RT-PCR with primers CIC-4377F and DUXL4-1553R1 amplified an approximately 300 bp cDNA fragment (Figure 4A). Cloning to pCR4-TOPO TA vector and sequencing of the cloned amplified PCR product showed that it actually consisted of two chimeric CIC::DUX4 cDNA fragments. In the first fragment, named type 1 fusion, nucleotide 4750 of the CIC sequence with accession number NM_015125.5 fused to nucleotide 1418 of DUX4 sequence with accession number NM_001306068.3 (Figure 4B). In the second fragment, the type 2 CIC::DUX4 fusion transcript, nucleotide 4777 of the CIC/NM_015125.3 sequence had fused to nucleotide 1418 of DUX4/NM_001306068 (Figure 4C). At the junction, both transcripts had four nucleotides, AGGT, which were thus common to both CIC and DUX4 sequences (Figure 4B and C). Comparison of the type 1 and 2 CIC::DUX4 fusion transcripts with other published sequences showed that they are recurrent (Figure 5).
Figure 4. Molecular genetic analysis of the CIC-sarcoma. (A) Gel electrophoresis of reverse transcription (RT) PCR amplification products: lane 1, amplification of a 300 bp cDNA fragment using the forward primer CIC-4377F and the reverse primer DUXL4-1553R1. M, GeneRuler 1 kb Plus DNA ladder (ThermoFisher Scientific). (B) Partial sequence chromatogram showing the junction in the CIC::DUX4 fusion transcript type 1. (C) Partial sequence chromatogram showing the junction in the CIC::DUX4 fusion transcript type 2. The triplets and the corresponding coding amino acids are shown. Stop codon is noted with ***. The common four nucleotides AGGT at the junctions are underlined.
Figure 5. Multiple sequence alignment of the CIC::DUX4 fusion transcripts type 1 and type 2 with previously reported CIC::DUX4 fusion transcripts by Cocchi et al. (38), Gambarotti et al. (34), Kao et al. (60), Machado et al. (32), Tsukamoto et al. (37), Yoshida et al. (41) and Italiano et al. (6). CIC sequence is written in red and DUX4 sequence in black letters. The tetranucleotide AGGT which was found at the junction and was common to both CIC and DUX4 is written in blue letters. The stop codon TAG or TGA is underlined.
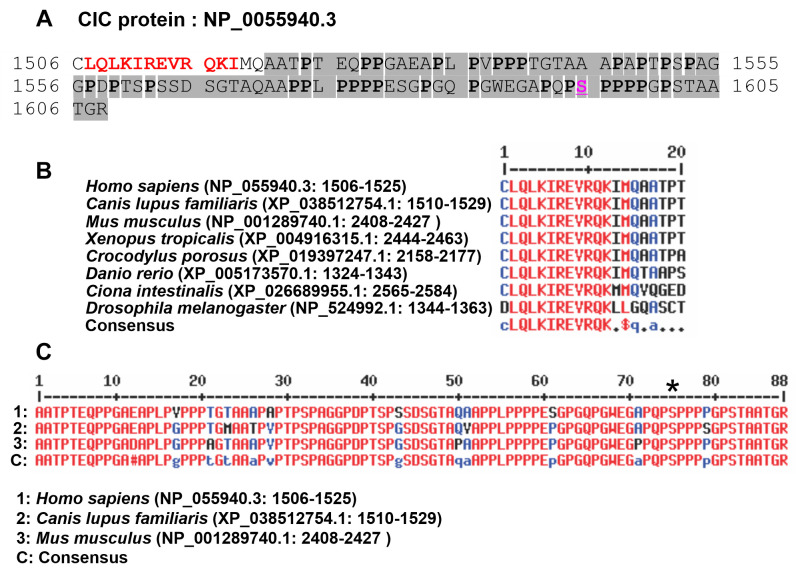
Because both CIC::DUX4 cDNA fragments had a stop TAG codon immediately after the fusion point (Figure 4B and C), they would code for a truncated CIC protein. Thus, CIC::DUX4 fusion transcript type 1 would code for a truncated protein lacking the last 103 amino acids corresponding to amino acids 1506 to 1608 of the CIC protein with reference number NP_055940.3 (isoform CIC-S), whereas the type 2 CIC::DUX4 transcript would code for a truncated protein that lacks the last 94 aa of CIC corresponding to amino acids 1515 to 1608 of the CIC protein/NP_055940.3 (isoform CIC-S) (Figure 6).
Figure 6. The C-terminal part of the CIC protein is absent from the truncated protein encoded by CIC::DUX4 fusion transcript 1. (A) The amino acid sequence of the missing part corresponds to amino acids 1506-1608 in CIC/NP_055940.3. The intrinsically disordered region containing 30 P (in bold) and 15 A amino acid residues is in gray background. The serine in the PSPP motif, which can be phosphorylated, is in red and underlined. (B). The phylogenetically conserved peptide CLQLKIREVRQKIMQ in various species. (C) Conservation of the intrinsically disordered region in mammals. *above serine in the PSPP motif.
Discussion
In the present study, we used chromosome banding, FISH and RT-PCR/Sanger sequencing methodologies to reach the diagnosis CIC sarcoma for a tumor whose morphologic features alone were insufficient to arrive at this conclusion. Cytogenetic analysis showed, among several aberrations, a t(10;19)(q26;q13) chromosome translocation, while FISH showed splitting of a probe which contained the CIC gene and RT-PCR/Sanger sequencing confirmed the presence of a CIC::DUX4 chimeric gene (Figure 2 and Figure 3).
To the best of our knowledge, this is the second solid tumor (of altogether four malignant neoplasms) in which a t(10;19)(q24;q13) translocation was detected. The translocation was previously reported in two acute lymphoblastic leukemias as the sole cytogenetic abnormality (62,63), and in a primary malignant neuroepithelial tumor of the kidney which had the karyotype 45,XX,der dic(1)t(1;13)(p1?3;p13),del(9)(p13),t(10;19)(q26;q13),-13 (64). At the DNA level, t(10;19)(q24;q13) has been shown to target different genes. It resulted in truncation of the FAM53B gene on subband 10q26.1 (63) in acute lymphoblastic leukemia, whereas in sarcomas it generates CIC::DUX4 chimeric genes as shown by the present case and the tumors of references (6) and (37).
We detected two types, 1 and 2, of CIC::DUX4 fusion transcript (Figure 4 and Figure 5), both of which were also reported in previous articles (Figure 5) (6,32,34,37,38,41,60). The common features of both transcripts are: 1) that the CIC gene breakpoint occurs in the coding region of the last exon (exon 20 in reference sequence NM_015125.5); 2) that the breakpoint in the DUX4 gene occurs within the 3’-end untranslated region; 3) that at the junction there is a four nucleotide sequence, AGGT, common to both CIC and DUX4, and 4) that a stop TGA codon is introduced after the fusion point resulting in a truncated CIC protein instead of the chimeric CIC::DUX4 protein.
The truncated protein encoded by CIC::DUX4 fusion transcript 1 lacks the last 103 amino acids of the normal CIC protein (amino acid 1506-1608 in CIC/NP_055940.3), whereas the truncated protein coded for by CIC::DUX4 fusion transcript 2 lacks the last 94 amino acids of the CIC protein (Figure 6A) (amino acids 1515-1608 in CIC/NP_055940.3). The N-terminal of the missing part contains the phylogenetically conserved peptide CLQLKIREVRQKIMQ (or RQKIMQ for the protein coded by CIC::DUX4 fusion transcript 2) (Figure 6A and B) that is part of the C1 region of the CIC protein important for the repressor activity of CIC (65-67). Electrophoretic mobility shift assays with CIC protein from Drosophila melanogaster showed that deletion of the LKIREV or RQKL was enough for the CIC protein to lose its ability to bind at optimal CIC binding sites T(C/G)AATGAA (66,67).
The carboxyl-terminal part of the missing sequence (amino acids 1521-1608 in CIC/NP_055940.3) is conserved in placental mammals and is 34% rich in the hydrophobic amino acid proline (P) and 17% rich in alanine (A) (Figure 6A and C). Proline rich regions are found in intrinsically disordered regions of proteins and are involved in protein-protein interactions by binding SRC homology 3 (SH3), WW, GYF and EVH1 domains (68-76). Intrinsically disordered regions play important roles in a plethora of cellular functions (76-83). According to the database of protein disorder and mobility annotations (MobiDB), the missing part of the CIC protein (amino acids 1521-1608 in CIC/NP_055940.3) is an intrinsically disordered region (https://mobidb.org/Q96RK0) (84-86). It contains short motifs which may interact with the above-mentioned domains (Figure 6C). For example, LPVPP, APPLP and LPPPP may bind to the SH3 domain of a number of proteins (87,88). The PPLP short motif may bind to Group II WW domains, such as the WW domains of the amyloid beta precursor protein binding family B member 1 (APBB1, also known as FE65) and pre-mRNA processing factor 40 homolog A (PRPF40A, also known as FBP11) (89,90). The motifs PPPP, LPPP and PSPP may bind to EVH1 domains from various proteins (69,74). In addition, the serine (S) in PSPP (position 1595 in reference sequence NP_055940.3) can be phosphorylated (pS). Motif pSP was shown to bind to group IV WW domains, such as those from Pin1 (49), PDX-1 C-terminus-interacting factor, and NEDD4 proteins (91-93).
Proline rich regions are also found in repression domains of various transcription factors. The TP53 transcription factor has a 41.4% proline-rich and a 34.5% alanine-rich region between amino acids 64-92 in sequence with accession number NP_000537.3. This transcription repression domain is essential for the induction of apoptosis, for the activation of TP53 DNA binding capacity to tumor protein p53 inducible protein 3 (TP53I3, also known as PIG3), and for activation of TP53 following ionizing radiation (94-96). The HHEX (officially full name is haematopoietically expressed homeobox, also known as PRH) transcription factor has an N-terminal transcription repression domain between amino acids 1-143 in sequence with accession number NP_002720.1. It is 20% rich in proline and 11.4% rich in alanine (97-100). Finally, the transcription repression domain of the WT1 protein, found between amino acids 71-180 in sequence with accession number NP_000369.4, is 20%, 14%, and 13.6% rich in amino acids proline, glycine, and alanine, respectively (101-103).
Conclusion
Although functional studies are still lacking, current knowledge suggests that the missing part of CIC in the truncated proteins translated from CIC::DUX4 fusion transcripts 1 and 2 influences the CIC protein’s DNA binding capacity, the transcription repression function, and interaction with other proteins, possibly in particular interactions with proteins carrying an SH3, WW, GYF and EVH1 domain. Absence of this part of CIC seems to be crucial in CIC-mediated tumorigenesis.
Conflicts of Interest
The Authors declare that they have no potential conflicts of interest.
Authors’ Contributions
IP designed and supervised the research, performed molecular genetic experiments and bioinformatics analysis, and wrote the manuscript. KA performed molecular genetic experiments and interpreted the data. LG performed cytogenetic analysis. HRH, TDP, and IL performed the pathological examination. FM evaluated the data. SH assisted with experimental design and writing of the manuscript. All Authors read and approved of the final manuscript.
Acknowledgements
This study was supported by grants from Radiumhospitalets Legater.
References
- 1.WHO Classification of Tumours Editorial Board . Lyon, France, International agency for research on cancer. 2020. WHO classification of Tumours of Soft Tissue and Bone. Fifth edn. [Google Scholar]
- 2.Anderson WJ, Doyle LA. Updates from the 2020 World Health Organization Classification of soft tissue and bone tumours. Histopathology. 2021;78(5):644–657. doi: 10.1111/his.14265. [DOI] [PubMed] [Google Scholar]
- 3.Choi JH, Ro JY. The 2020 WHO Classification of tumors of bone: an updated review. Adv Anat Pathol. 2021;28(3):119–138. doi: 10.1097/PAP.0000000000000293. [DOI] [PubMed] [Google Scholar]
- 4.Kallen ME, Hornick JL. The 2020 WHO Classification: What’s new in soft tissue tumor pathology. Am J Surg Pathol. 2021;45(1):e1–e23. doi: 10.1097/PAS.0000000000001552. [DOI] [PubMed] [Google Scholar]
- 5.Kawamura-Saito M, Yamazaki Y, Kaneko K, Kawaguchi N, Kanda H, Mukai H, Gotoh T, Motoi T, Fukayama M, Aburatani H, Takizawa T, Nakamura T. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15(13):2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- 6.Italiano A, Sung YS, Zhang L, Singer S, Maki RG, Coindre JM, Antonescu CR. High prevalence of CIC fusion with double-homeobox (DUX4) transcription factors in EWSR1-negative undifferentiated small blue round cell sarcomas. Genes Chromosomes Cancer. 2012;51(3):207–218. doi: 10.1002/gcc.20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, Patidar R, Hurd L, Chen L, Shern JF, Liao H, Wen X, Gerard J, Kim JS, Lopez Guerrero JA, Machado I, Wai DH, Picci P, Triche T, Horvai AE, Miettinen M, Wei JS, Catchpool D, Llombart-Bosch A, Waldman T, Khan J. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet. 2014;10(7):e1004475. doi: 10.1371/journal.pgen.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugita S, Arai Y, Tonooka A, Hama N, Totoki Y, Fujii T, Aoyama T, Asanuma H, Tsukahara T, Kaya M, Shibata T, Hasegawa T. A novel CIC-FOXO4 gene fusion in undifferentiated small round cell sarcoma: a genetically distinct variant of Ewing-like sarcoma. Am J Surg Pathol. 2014;38(11):1571–1576. doi: 10.1097/PAS.0000000000000286. [DOI] [PubMed] [Google Scholar]
- 9.Huang SC, Zhang L, Sung YS, Chen CL, Kao YC, Agaram NP, Singer S, Tap WD, D’Angelo S, Antonescu CR. Recurrent CIC gene abnormalities in angiosarcomas: a molecular study of 120 cases with concurrent investigation of PLCG1, KDR, MYC, and FLT4 gene alterations. Am J Surg Pathol. 2016;40(5):645–655. doi: 10.1097/PAS.0000000000000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu W, Wang J, Yuan L, Zhang X, Ji Y, Song C, Zeng J, Sun X. Case report: a unique case of pediatric central nervous system embryonal tumor harboring the CIC-LEUTX fusion, germline NBN variant and somatic TSC2 mutation: expanding the spectrum of CIC-rearranged neoplasia. Front Oncol. 2020;10:598970. doi: 10.3389/fonc.2020.598970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangray S, Kelly DR, LeGuellec S, Fridman E, Aggarwal S, Shago M, Matoso A, Madison R, Pramanik S, Zhong S, Li R, Lombardo KA, Cramer S, Pressey J, Ross JS, Corona RJ, Bratslavsky G, Argani P, Coindre JM, Somers GR, Ali SM, Yakirevich E. Clinicopathologic features of a series of primary renal CIC-rearranged sarcomas with comprehensive molecular analysis. Am J Surg Pathol. 2018;42(10):1360–1369. doi: 10.1097/PAS.0000000000001098. [DOI] [PubMed] [Google Scholar]
- 12.Schaefer IM, Dal Cin P, Landry LM, Fletcher CDM, Hanna GJ, French CA. CIC-NUTM1 fusion: A case which expands the spectrum of NUT-rearranged epithelioid malignancies. Genes Chromosomes Cancer. 2018;57(9):446–451. doi: 10.1002/gcc.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watson S, Perrin V, Guillemot D, Reynaud S, Coindre JM, Karanian M, Guinebretière JM, Freneaux P, Le Loarer F, Bouvet M, Galmiche-Rolland L, Larousserie F, Longchampt E, Ranchere-Vince D, Pierron G, Delattre O, Tirode F. Transcriptomic definition of molecular subgroups of small round cell sarcomas. J Pathol. 2018;245(1):29–40. doi: 10.1002/path.5053. [DOI] [PubMed] [Google Scholar]
- 14.Le Loarer F, Pissaloux D, Watson S, Godfraind C, Galmiche-Rolland L, Silva K, Mayeur L, Italiano A, Michot A, Pierron G, Vasiljevic A, Ranchère-Vince D, Coindre JM, Tirode F. Clinicopathologic features of CIC-NUTM1 sarcomas, a new molecular variant of the family of CIC-fused sarcomas. Am J Surg Pathol. 2019;43(2):268–276. doi: 10.1097/PAS.0000000000001187. [DOI] [PubMed] [Google Scholar]
- 15.Mantilla JG, Ricciotti RW, Chen E, Hoch BL, Liu YJ. Detecting disease-defining gene fusions in unclassified round cell sarcomas using anchored multiplex PCR/targeted RNA next-generation sequencing-Molecular and clinicopathological characterization of 16 cases. Genes Chromosomes Cancer. 2019;58(10):713–722. doi: 10.1002/gcc.22763. [DOI] [PubMed] [Google Scholar]
- 16.Pratt D, Kumar-Sinha C, Cieślik M, Mehra R, Xiao H, Shao L, Franson A, Cantor E, Chinnaiyan AM, Mody R, Abdullaev Z, Aldape K, Quezado M, Camelo-Piragua S. A novel ATXN1-DUX4 fusion expands the spectrum of ‘CIC-rearranged sarcoma’ of the CNS to include non-CIC alterations. Acta Neuropathol. 2021;141(4):619–622. doi: 10.1007/s00401-021-02278-3. [DOI] [PubMed] [Google Scholar]
- 17.Satomi K, Ohno M, Kubo T, Honda-Kitahara M, Matsushita Y, Ichimura K, Narita Y, Ichikawa H, Yoshida A. Central nervous system sarcoma with ATXN1::DUX4 fusion expands the concept of CIC-rearranged sarcoma. Genes Chromosomes Cancer. 2022;61(11):683–688. doi: 10.1002/gcc.23080. [DOI] [PubMed] [Google Scholar]
- 18.Gabriëls J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, Belayew A. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236(1):25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 19.van Geel M, Dickson MC, Beck AF, Bolland DJ, Frants RR, van der Maarel SM, de Jong PJ, Hewitt JE. Genomic analysis of human chromosome 10q and 4q telomeres suggests a common origin. Genomics. 2002;79(2):210–217. doi: 10.1006/geno.2002.6690. [DOI] [PubMed] [Google Scholar]
- 20.Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, Nix DA, Peterson CM, Tapscott SJ, Carrell DT, Cairns BR. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet. 2017;49(6):925–934. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whiddon JL, Langford AT, Wong CJ, Zhong JW, Tapscott SJ. Conservation and innovation in the DUX4-family gene network. Nat Genet. 2017;49(6):935–940. doi: 10.1038/ng.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mocciaro E, Runfola V, Ghezzi P, Pannese M, Gabellini D. DUX4 role in normal physiology and in FSHD muscular dystrophy. Cells. 2021;10(12):3322. doi: 10.3390/cells10123322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, Miller DG. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6(10):e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das S, Chadwick BP. Influence of repressive histone and DNA methylation upon D4Z4 transcription in non-myogenic cells. PLoS One. 2016;11(7):e0160022. doi: 10.1371/journal.pone.0160022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dixit M, Ansseau E, Tassin A, Winokur S, Shi R, Qian H, Sauvage S, Mattéotti C, van Acker AM, Leo O, Figlewicz D, Barro M, Laoudj-Chenivesse D, Belayew A, Coppée F, Chen YW. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci U S A. 2007;104(46):18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kowaljow V, Marcowycz A, Ansseau E, Conde CB, Sauvage S, Mattéotti C, Arias C, Corona ED, Nuñez NG, Leo O, Wattiez R, Figlewicz D, Laoudj-Chenivesse D, Belayew A, Coppée F, Rosa AL. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord. 2007;17(8):611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 27.Quintero J, Saad NY, Pagnoni SM, Jacquelin DK, Gatica LV, Harper SQ, Rosa AL. The DUX4 protein is a co-repressor of the progesterone and glucocorticoid nuclear receptors. FEBS Lett. 2022;596(20):2644–2658. doi: 10.1002/1873-3468.14416. [DOI] [PubMed] [Google Scholar]
- 28.Rakheja D, Goldman S, Wilson KS, Lenarsky C, Weinthal J, Schultz RA. Translocation (4;19)(q35;q13.1)-associated primitive round cell sarcoma: report of a case and review of the literature. Pediatr Dev Pathol. 2008;11(3):239–244. doi: 10.2350/07-06-0296.1. [DOI] [PubMed] [Google Scholar]
- 29.Yoshimoto M, Graham C, Chilton-MacNeill S, Lee E, Shago M, Squire J, Zielenska M, Somers GR. Detailed cytogenetic and array analysis of pediatric primitive sarcomas reveals a recurrent CIC-DUX4 fusion gene event. Cancer Genet Cytogenet. 2009;195(1):1–11. doi: 10.1016/j.cancergencyto.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 30.Graham C, Chilton-MacNeill S, Zielenska M, Somers GR. The CIC-DUX4 fusion transcript is present in a subgroup of pediatric primitive round cell sarcomas. Hum Pathol. 2012;43(2):180–189. doi: 10.1016/j.humpath.2011.04.023. [DOI] [PubMed] [Google Scholar]
- 31.Choi EY, Thomas DG, McHugh JB, Patel RM, Roulston D, Schuetze SM, Chugh R, Biermann JS, Lucas DR. Undifferentiated small round cell sarcoma with t(4;19)(q35;q13.1) CIC-DUX4 fusion: a novel highly aggressive soft tissue tumor with distinctive histopathology. Am J Surg Pathol. 2013;37(9):1379–1386. doi: 10.1097/PAS.0b013e318297a57d. [DOI] [PubMed] [Google Scholar]
- 32.Machado I, Cruz J, Lavernia J, Rubio L, Campos J, Barrios M, Grison C, Chene V, Pierron G, Delattre O, Llombart-Bosch A. Superficial EWSR1-negative undifferentiated small round cell sarcoma with CIC/DUX4 gene fusion: a new variant of Ewing-like tumors with locoregional lymph node metastasis. Virchows Arch. 2013;463(6):837–842. doi: 10.1007/s00428-013-1499-9. [DOI] [PubMed] [Google Scholar]
- 33.Panagopoulos I, Gorunova L, Bjerkehagen B, Heim S. The “grep” command but not FusionMap, FusionFinder or ChimeraScan captures the CIC-DUX4 fusion gene from whole transcriptome sequencing data on a small round cell tumor with t(4;19)(q35;q13) PLoS One. 2014;9(6):e99439. doi: 10.1371/journal.pone.0099439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gambarotti M, Benini S, Gamberi G, Cocchi S, Palmerini E, Sbaraglia M, Donati D, Picci P, Vanel D, Ferrari S, Righi A, Dei Tos AP. CIC-DUX4 fusion-positive round-cell sarcomas of soft tissue and bone: a single-institution morphological and molecular analysis of seven cases. Histopathology. 2016;69(4):624–634. doi: 10.1111/his.12985. [DOI] [PubMed] [Google Scholar]
- 35.Antonescu CR, Owosho AA, Zhang L, Chen S, Deniz K, Huryn JM, Kao YC, Huang SC, Singer S, Tap W, Schaefer IM, Fletcher CD. Sarcomas with CIC-rearrangements are a distinct pathologic entity with aggressive outcome: a clinicopathologic and molecular study of 115 cases. Am J Surg Pathol. 2017;41(7):941–949. doi: 10.1097/PAS.0000000000000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krskova L, Stejskalova E, Kabickova E, Mrhalova M, Kodet R. A t(4;19) pediatric undifferentiated sarcoma with a novel variant of the CIC-DUX4 fusion transcript. Pathol Res Pract. 2017;213(3):281–285. doi: 10.1016/j.prp.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 37.Tsukamoto Y, Futani H, Yoshiya S, Watanabe T, Kihara T, Matsuo S, Hirota S. Primary undifferentiated small round cell sarcoma of the deep abdominal wall with a novel variant of t(10;19) CIC-DUX4 gene fusion. Pathol Res Pract. 2017;213(10):1315–1321. doi: 10.1016/j.prp.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 38.Cocchi S, Gamberi G, Magagnoli G, Maioli M, Righi A, Frisoni T, Gambarotti M, Benini S. CIC rearranged sarcomas: A single institution experience of the potential pitfalls in interpreting CIC FISH results. Pathol Res Pract. 2022;231:153773. doi: 10.1016/j.prp.2022.153773. [DOI] [PubMed] [Google Scholar]
- 39.Kajtár B, Tornóczky T, Kálmán E, Kuzsner J, Hogendoorn PC, Szuhai K. CD99-positive undifferentiated round cell sarcoma diagnosed on fine needle aspiration cytology, later found to harbour a CIC-DUX4 translocation: a recently described entity. Cytopathology. 2014;25(2):129–132. doi: 10.1111/cyt.12079. [DOI] [PubMed] [Google Scholar]
- 40.Loke BN, Lee VKM, Sudhanshi J, Wong MK, Kuick CH, Puhaindran M, Chang KTE. Novel exon-exon breakpoint in CIC-DUX4 fusion sarcoma identified by anchored multiplex PCR (Archer FusionPlex Sarcoma Panel) J Clin Pathol. 2017;70(8):697–701. doi: 10.1136/jclinpath-2016-204247. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida A, Arai Y, Kobayashi E, Yonemori K, Ogura K, Hama N, Mukai W, Motoi T, Kawai A, Shibata T, Hiraoka N. CIC break-apart fluorescence in-situ hybridization misses a subset of CIC-DUX4 sarcomas: a clinicopathological and molecular study. Histopathology. 2017;71(3):461–469. doi: 10.1111/his.13252. [DOI] [PubMed] [Google Scholar]
- 42.Donthi D, Malik P, Prenshaw KL, Hong H. A rare case of round cell sarcoma with CIC-DUX4 mutation mimicking a phlegmon: review of literature. Am J Case Rep. 2020;21:e925683. doi: 10.12659/AJCR.925683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aranza S, Roydhouse C, Kho J. A rare diagnostically challenging case of CIC-DUX4 sarcoma arising in the neck. Pathology. 2022 doi: 10.1016/j.pathol.2022.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Urumov IJ, Manolova Y. Cytogenetic analysis of an embryonal rhabdomyosarcoma cell line. Cancer Genet Cytogenet. 1992;61(2):214–215. doi: 10.1016/0165-4608(92)90092-m. [DOI] [PubMed] [Google Scholar]
- 45.Roberts P, Browne CF, Lewis IJ, Bailey CC, Spicer RD, Williams J, Batcup G. 12q13 abnormality in rhabdomyosarcoma. A nonrandom occurrence. Cancer Genet Cytogenet. 1992;60(2):135–140. doi: 10.1016/0165-4608(92)90005-s. [DOI] [PubMed] [Google Scholar]
- 46.Richkind KE, Romansky SG, Finklestein JZ. t(4;19)(q35;q13.1): a recurrent change in primitive mesenchymal tumors. Cancer Genet Cytogenet. 1996;87(1):71–74. doi: 10.1016/0165-4608(95)00240-5. [DOI] [PubMed] [Google Scholar]
- 47.Bussey KJ, Lawce HJ, Olson SB, Arthur DC, Kalousek DK, Krailo M, Giller R, Heifetz S, Womer R, Magenis RE. Chromosome abnormalities of eighty-one pediatric germ cell tumors: sex-, age-, site-, and histopathology-related differences—a Children’s Cancer Group study. Genes Chromosomes Cancer. 1999;25(2):134–146. [PubMed] [Google Scholar]
- 48.Somers GR, Shago M, Zielenska M, Chan HS, Ngan BY. Primary subcutaneous primitive neuroectodermal tumor with aggressive behavior and an unusual karyotype: case report. Pediatr Dev Pathol. 2004;7(5):538–545. doi: 10.1007/s10024-004-2024-6. [DOI] [PubMed] [Google Scholar]
- 49.Sirvent N, Trassard M, Ebran N, Attias R, Pedeutour F. Fusion of EWSR1 with the DUX4 facioscapulohumeral muscular dystrophy region resulting from t(4;22)(q35;q12) in a case of embryonal rhabdomyosarcoma. Cancer Genet Cytogenet. 2009;195(1):12–18. doi: 10.1016/j.cancergencyto.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 50.Yustein JT, Rednam S, Bertuch AA, Goss JA, Brandt ML, Eldin K, Lu X, Hicks J. Abdominal undifferentiated small round cell tumor with unique translocation (X;19)(q13;q13.3) Pediatr Blood Cancer. 2010;54(7):1041–1044. doi: 10.1002/pbc.22437. [DOI] [PubMed] [Google Scholar]
- 51.Panagopoulos I, Gorunova L, Lund-Iversen M, Andersen K, Andersen HK, Lobmaier I, Bjerkehagen B, Heim S. Cytogenetics of spindle cell/pleomorphic lipomas: Karyotyping and FISH analysis of 31 tumors. Cancer Genomics Proteomics. 2018;15(3):193–200. doi: 10.21873/cgp.20077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McGowan-Jordan J, Hastings RJ, Moore S. Basel, Karger. 2020. ISCN 2020: An International system for human cytogenomic nomenclature; p. pp. 164. [DOI] [PubMed] [Google Scholar]
- 53.Roohi J, Cammer M, Montagna C, Hatchwell E. An improved method for generating BAC DNA suitable for FISH. Cytogenet Genome Res. 2008;121(1):7–9. doi: 10.1159/000124374. [DOI] [PubMed] [Google Scholar]
- 54.Panagopoulos I, Gorunova L, Andersen K, Lund-Iversen M, Hognestad HR, Lobmaier I, Micci F, Heim S. Chromosomal translocation t(5;12)(p13;q14) leading to fusion of high-mobility group AT-hook 2 gene with intergenic sequences from chromosome sub-band 5p13.2 in benign myoid neoplasms of the breast: a second case. Cancer Genomics Proteomics. 2022;19(4):445–455. doi: 10.21873/cgp.20331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singhal H, Ren YR, Kern SE. Improved DNA electrophoresis in conditions favoring polyborates and lewis acid complexation. PLoS One. 2010;5(6):e11318. doi: 10.1371/journal.pone.0011318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 57.Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, Aganezov S, Hoyt SJ, Diekhans M, Logsdon GA, Alonge M, Antonarakis SE, Borchers M, Bouffard GG, Brooks SY, Caldas GV, Chen NC, Cheng H, Chin CS, Chow W, de Lima LG, Dishuck PC, Durbin R, Dvorkina T, Fiddes IT, Formenti G, Fulton RS, Fungtammasan A, Garrison E, Grady PGS, Graves-Lindsay TA, Hall IM, Hansen NF, Hartley GA, Haukness M, Howe K, Hunkapiller MW, Jain C, Jain M, Jarvis ED, Kerpedjiev P, Kirsche M, Kolmogorov M, Korlach J, Kremitzki M, Li H, Maduro VV, Marschall T, McCartney AM, McDaniel J, Miller DE, Mullikin JC, Myers EW, Olson ND, Paten B, Peluso P, Pevzner PA, Porubsky D, Potapova T, Rogaev EI, Rosenfeld JA, Salzberg SL, Schneider VA, Sedlazeck FJ, Shafin K, Shew CJ, Shumate A, Sims Y, Smit AFA, Soto DC, Sović I, Storer JM, Streets A, Sullivan BA, Thibaud-Nissen F, Torrance J, Wagner J, Walenz BP, Wenger A, Wood JMD, Xiao C, Yan SM, Young AC, Zarate S, Surti U, McCoy RC, Dennis MY, Alexandrov IA, Gerton JL, O’Neill RJ, Timp W, Zook JM, Schatz MC, Eichler EE, Miga KH, Phillippy AM. The complete sequence of a human genome. Science. 2022;376(6588):44–53. doi: 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kao YC, Sung YS, Chen CL, Zhang L, Dickson BC, Swanson D, Vaiyapuri S, Latif F, Alholle A, Huang SC, Hornick JL, Antonescu CR. ETV transcriptional upregulation is more reliable than RNA sequencing algorithms and FISH in diagnosing round cell sarcomas with CIC gene rearrangements. Genes Chromosomes Cancer. 2017;56(6):501–510. doi: 10.1002/gcc.22454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Braekeleer M, Poon MC, Russell J, Lin CC. A case of acute lymphoblastic leukemia with t(10;19)(q26;q13) Cancer Genet Cytogenet. 1985;16(4):369–372. doi: 10.1016/0165-4608(85)90247-x. [DOI] [PubMed] [Google Scholar]
- 63.Panagopoulos I, Gorunova L, Torkildsen S, Tierens A, Heim S, Micci F. FAM53B truncation caused by t(10;19)(q26;q13) chromosome translocation in acute lymphoblastic leukemia. Oncol Lett. 2017;13(4):2216–2220. doi: 10.3892/ol.2017.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parham DM, Roloson GJ, Feely M, Green DM, Bridge JA, Beckwith JB. Primary malignant neuroepithelial tumors of the kidney: a clinicopathologic analysis of 146 adult and pediatric cases from the National Wilms’ Tumor Study Group Pathology Center. Am J Surg Pathol. 2001;25(2):133–146. doi: 10.1097/00000478-200102000-00001. [DOI] [PubMed] [Google Scholar]
- 65.Astigarraga S, Grossman R, Díaz-Delfín J, Caelles C, Paroush Z, Jiménez G. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J. 2007;26(3):668–677. doi: 10.1038/sj.emboj.7601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Forés M, Simón-Carrasco L, Ajuria L, Samper N, González-Crespo S, Drosten M, Barbacid M, Jiménez G. A new mode of DNA binding distinguishes Capicua from other HMG-box factors and explains its mutation patterns in cancer. PLoS Genet. 2017;13(3):e1006622. doi: 10.1371/journal.pgen.1006622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Papagianni A, Forés M, Shao W, He S, Koenecke N, Andreu MJ, Samper N, Paroush Z, González-Crespo S, Zeitlinger J, Jiménez G. Capicua controls Toll/IL-1 signaling targets independently of RTK regulation. Proc Natl Acad Sci U.S.A. 2018;115(8):1807–1812. doi: 10.1073/pnas.1713930115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14(2):231–241. [PubMed] [Google Scholar]
- 69.Renfranz PJ, Beckerle MC. Doing (F/L)PPPPs: EVH1 domains and their proline-rich partners in cell polarity and migration. Curr Opin Cell Biol. 2002;14(1):88–103. doi: 10.1016/s0955-0674(01)00299-x. [DOI] [PubMed] [Google Scholar]
- 70.Ball LJ, Kühne R, Schneider-Mergener J, Oschkinat H. Recognition of proline-rich motifs by protein-protein-interaction domains. Angew Chem Int Ed Engl. 2005;44(19):2852–2869. doi: 10.1002/anie.200400618. [DOI] [PubMed] [Google Scholar]
- 71.Kofler MM, Freund C. The GYF domain. FEBS J. 2006;273(2):245–256. doi: 10.1111/j.1742-4658.2005.05078.x. [DOI] [PubMed] [Google Scholar]
- 72.Freund C, Schmalz HG, Sticht J, Kühne R. Proline-rich sequence recognition domains (PRD): ligands, function and inhibition. Handb Exp Pharmacol. 2008;(186):407–429. doi: 10.1007/978-3-540-72843-6_17. [DOI] [PubMed] [Google Scholar]
- 73.Kofler M, Schuemann M, Merz C, Kosslick D, Schlundt A, Tannert A, Schaefer M, Lührmann R, Krause E, Freund C. Proline-rich sequence recognition: I. Marking GYF and WW domain assembly sites in early spliceosomal complexes. Mol Cell Proteomics. 2009;8(11):2461–2473. doi: 10.1074/mcp.M900191-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peterson FC, Volkman BF. Diversity of polyproline recognition by EVH1 domains. Front Biosci (Landmark Ed) 2009;14(3):833–846. doi: 10.2741/3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morgan AA, Rubenstein E. Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS One. 2013;8(1):e53785. doi: 10.1371/journal.pone.0053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Theillet FX, Kalmar L, Tompa P, Han KH, Selenko P, Dunker AK, Daughdrill GW, Uversky VN. The alphabet of intrinsic disorder: I. Act like a Pro: On the abundance and roles of proline residues in intrinsically disordered proteins. Intrinsically Disord Proteins. 2013;1(1):e24360. doi: 10.4161/idp.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 78.Uversky VN. The alphabet of intrinsic disorder: II. Various roles of glutamic acid in ordered and intrinsically disordered proteins. Intrinsically Disord Proteins. 2013;1(1):e24684. doi: 10.4161/idp.24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uversky VN. The intrinsic disorder alphabet. III. Dual personality of serine. Intrinsically Disord Proteins. 2015;3(1):e1027032. doi: 10.1080/21690707.2015.1027032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Babu MM. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem Soc Trans. 2016;44(5):1185–1200. doi: 10.1042/BST20160172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bondos SE, Dunker AK, Uversky VN. On the roles of intrinsically disordered proteins and regions in cell communication and signaling. Cell Commun Signal. 2021;19(1):88. doi: 10.1186/s12964-021-00774-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bondos SE, Dunker AK, Uversky VN. Intrinsically disordered proteins play diverse roles in cell signaling. Cell Commun Signal. 2022;20(1):20. doi: 10.1186/s12964-022-00821-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Malagrinò F, Pennacchietti V, Santorelli D, Pagano L, Nardella C, Diop A, Toto A, Gianni S. On the effects of disordered tails, supertertiary structure and quinary interactions on the folding and function of protein domains. Biomolecules. 2022;12(2):209. doi: 10.3390/biom12020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Potenza E, Di Domenico T, Walsh I, Tosatto SC. MobiDB 2.0: an improved database of intrinsically disordered and mobile proteins. Nucleic Acids Res. 2015;43(Database issue):D315–D320. doi: 10.1093/nar/gku982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Piovesan D, Tabaro F, Paladin L, Necci M, Micetic I, Camilloni C, Davey N, Dosztányi Z, Mészáros B, Monzon AM, Parisi G, Schad E, Sormanni P, Tompa P, Vendruscolo M, Vranken WF, Tosatto SCE. MobiDB 3.0: more annotations for intrinsic disorder, conformational diversity and interactions in proteins. Nucleic Acids Res. 2018;46(D1):D471–D476. doi: 10.1093/nar/gkx1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Piovesan D, Necci M, Escobedo N, Monzon AM, Hatos A, Mičetić I, Quaglia F, Paladin L, Ramasamy P, Dosztányi Z, Vranken WF, Davey NE, Parisi G, Fuxreiter M, Tosatto SCE. MobiDB: intrinsically disordered proteins in 2021. Nucleic Acids Res. 2021;49(D1):D361–D367. doi: 10.1093/nar/gkaa1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390(Pt 3):641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saksela K, Permi P. SH3 domain ligand binding: What’s the consensus and where’s the specificity. FEBS Lett. 2012;586(17):2609–2614. doi: 10.1016/j.febslet.2012.04.042. [DOI] [PubMed] [Google Scholar]
- 89.Ermekova KS, Zambrano N, Linn H, Minopoli G, Gertler F, Russo T, Sudol M. The WW domain of neural protein FE65 interacts with proline-rich motifs in Mena, the mammalian homolog of Drosophila enabled. J Biol Chem. 1997;272(52):32869–32877. doi: 10.1074/jbc.272.52.32869. [DOI] [PubMed] [Google Scholar]
- 90.Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CS, Yu J, Hersi K, Raaijmakers J, Gish G, Mbamalu G, Taylor L, Yeung B, Vassilovski G, Amin M, Chen F, Matskova L, Winberg G, Ernberg I, Linding R, O’donnell P, Starostine A, Keller W, Metalnikov P, Stark C, Pawson T. WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol. 2005;25(16):7092–7106. doi: 10.1128/MCB.25.16.7092-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283(5406):1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 92.Yaffe MB, Elia AE. Phosphoserine/threonine-binding domains. Curr Opin Cell Biol. 2001;13(2):131–138. doi: 10.1016/s0955-0674(00)00189-7. [DOI] [PubMed] [Google Scholar]
- 93.Yaffe MB, Smerdon SJ. PhosphoSerine/threonine binding domains: you can’t pSERious. Structure. 2001;9(3):R33–R38. doi: 10.1016/s0969-2126(01)00580-9. [DOI] [PubMed] [Google Scholar]
- 94.Campbell HG, Mehta R, Neumann AA, Rubio C, Baird M, Slatter TL, Braithwaite AW. Activation of p53 following ionizing radiation, but not other stressors, is dependent on the proline-rich domain (PRD) Oncogene. 2013;32(7):827–836. doi: 10.1038/onc.2012.102. [DOI] [PubMed] [Google Scholar]
- 95.Hoyos D, Greenbaum B, Levine AJ. The genotypes and phenotypes of missense mutations in the proline domain of the p53 protein. Cell Death Differ. 2022;29(5):938–945. doi: 10.1038/s41418-022-00980-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 1998;17(16):4668–4679. doi: 10.1093/emboj/17.16.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guiral M, Bess K, Goodwin G, Jayaraman PS. PRH represses transcription in hematopoietic cells by at least two independent mechanisms. J Biol Chem. 2001;276(4):2961–2970. doi: 10.1074/jbc.M004948200. [DOI] [PubMed] [Google Scholar]
- 98.Swingler TE, Bess KL, Yao J, Stifani S, Jayaraman PS. The proline-rich homeodomain protein recruits members of the Groucho/Transducin-like enhancer of split protein family to co-repress transcription in hematopoietic cells. J Biol Chem. 2004;279(33):34938–34947. doi: 10.1074/jbc.M404488200. [DOI] [PubMed] [Google Scholar]
- 99.Soufi A, Gaston K, Jayaraman PS. Purification and characterisation of the PRH homeodomain: Removal of the N-terminal domain of PRH increases the PRH homeodomain-DNA interaction. Int J Biol Macromol. 2006;39(1-3):45–50. doi: 10.1016/j.ijbiomac.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 100.Gaston K, Tsitsilianos MA, Wadey K, Jayaraman PS. Misregulation of the proline rich homeodomain (PRH/HHEX) protein in cancer cells and its consequences for tumour growth and invasion. Cell Biosci. 2016;6:12. doi: 10.1186/s13578-016-0077-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Madden SL, Cook DM, Morris JF, Gashler A, Sukhatme VP, Rauscher FJ 3rd. Transcriptional repression mediated by the WT1 Wilms tumor gene product. Science. 1991;253(5027):1550–1553. doi: 10.1126/science.1654597. [DOI] [PubMed] [Google Scholar]
- 102.Madden SL, Cook DM, Rauscher FJ 3rd. A structure-function analysis of transcriptional repression mediated by the WT1, Wilms’ tumor suppressor protein. Oncogene. 1993;8(7):1713–1720. [PubMed] [Google Scholar]
- 103.Toska E, Roberts SG. Mechanisms of transcriptional regulation by WT1 (Wilms’ tumour 1) Biochem J. 2014;461(1):15–32. doi: 10.1042/BJ20131587. [DOI] [PMC free article] [PubMed] [Google Scholar]