Abstract
Purpose:
Genetic researchers’ selection of a database can have scientific, regulatory, and ethical implications. It is important to understand what is driving database selection such that database stewards can be responsive to user needs while balancing the interests of communities in equitably benefiting from advances.
Methods:
We conducted 23 semistructured interviews with US academic genetic researchers working with private, government, and collaboratory data stewards to explore factors that they consider when selecting a genetic database.
Results:
Interviewees used existing databases to avoid burdens of primary data collection, which was described as expensive and time-consuming. They highlighted ease of access as the most important selection factor, integrating concepts of familiarity and efficiency. Data features, such as size and available phenotype, were also important. Demographic diversity was not originally cited by any interviewee as a pivotal factor; when probed, most stated that the option to consider diversity in database selection was limited. Database features, including integrity, harmonization, and storage were also described as key components of efficient use.
Conclusion:
There is a growing market and competition between genetic data stewards. Data need to be accessible, harmonized, and administratively supported for their existence to be translated into use and, in turn, result in scientific advancements across diverse communities.
Introduction
To meet researcher needs and promote high quality research, the US federal government has invested intensive resources in building demographically diverse and accessible databases. These initiatives include the National Institutes of Health’s (NIH) mandated inclusion of diverse populations in research1,2 and new efforts to maximize data sharing by requiring investigators to deposit data generated from funded research into NIH-administered databases.3,4 The government also has created several major genetic databases, such as the database of Genotypes and Phenotypes (dbGaP),5 and the All of Us research program, with the specific goal of increasing representation from historically underrepresented populations.6 Despite these efforts, US academic genetic researchers are increasingly using privately-held genetic data (eg, generated by direct-to-consumer (DTC) or clinical genetic testing entities) for their work, despite almost half reporting some form of NIH funding for their research.7
The use of different kinds of genetic databases for research can have both ethical and regulatory implications. For example, a lack of demographic diversity in research databases can lead to misleading results, incorrect diagnoses, or findings that are not socially contextualized for historically excluded patients.8–11 Although the All of Us database currently supports many participants traditionally underrepresented in biomedical research,12 private genetic databases are historically populated with European participants with higher socioeconomic status who can afford recreational genetic testing or the insurance required to cover it clinically.13,14 The use of private genetic databases also gives industry a gatekeeping role to choose which research protocols they will support with data access. Commercial data use agreements can also limit the amount of information that can be shared in resulting publications or research databases, potentially undermining the goals of new NIH data sharing requirements.15
While the US government continues to invest in building accessible and demographically diverse genetic databases, it is important to understand what is driving the decisions researchers make about which databases to use. Toward this end, we conducted semistructured interviews with US academic genetic researchers, exploring the factors they consider if given the choice between genetic data sources to answer their research questions.
Materials and Methods
Recruitment
We identified prospective interviewees through a PubMed review of articles published between January 2017 and December 2019. We sampled articles with at least 1 corresponding author with a US academic affiliation that indicated the use of data from at least one of the following types of genetic data stewards (based in the United States or abroad): (1) a private steward (based on their inclusion in Research and Markets’ rank of DTC genetic testing companies, ie, 23andMe, Ambry Genetics, Ancestry.com, Color Genomics, Gene by Gene)16 or (2) an academic, government, or consortia-related steward (Table 1).
Table 1:
Interview Sampling
| Database Type | Search Results | Evaluated for Eligibility | Eligible | Contacted by Email | Interviewed | Response Rate |
|---|---|---|---|---|---|---|
| Privatea | 605 | 605 | 63 | 22 | 11 | 50% |
| Other | 8361 | 1382 | 48 | 28 | 12 | 43% |
| Total | 8966 | 1987 | 111 | 50 | 23 | 46% |
Private databases were sampled based on their inclusion in Research and Markets’ rank of DTC genetic testing companies (i.e. 23andMe, Ambry Genetics, Ancestry.com, Color Genomics, Gene by Gene)
We contacted authors from approximately half of these eligible articles, oversampling for female and Latino/Hispanic, African American or Black, or Asian researchers, via an email to the corresponding author. We sent interested authors additional information about the study and obtained their informed consent over the phone.
Interviews and analyses
We generated a semistructured interview protocol based on a literature review of different attributes of genetic databases and solicited expert input from qualitative experts and genetic researchers to identify confusing or unclear phrasing. We asked interviewees questions regarding employment, why they chose a specific data steward or stewards to answer their research question (if they had the choice to begin with), participant protections, data usage agreements, funding, data sharing, and research outcomes. In this paper, we focus on their selection of database(s) (Appendix: Interview Guide). Although interview questions focused on the database(s) used to answer the research question in the publication identified in our PubMed search, we also asked interviewees to compare their experience with other databases they previously used.
We conducted each 30 to 60-minute interview via phone or Zoom between March and July 2020 (K.S.-B., C.D.K.,M.K.).We mailed interviewees a $100 gift card after the completion of the interview. We audio recorded and transcribed the interviews, reviewed the resulting transcripts for accuracy, and cleaned and de-identified them (C.D.K.). For the thematic analysis, we employed a method of iterative description using grounded theory.17–19 We characterized themes common across interviews as well as captured individual variation (K.S.-B., K.A.R., M.G.T., C.D.K., R.D.V.).20 Our original codebook was developed via the structure of the interview guide, but we iteratively added additional variation and complexity concurrently with conducting interviews. All analysts concurred that thematic saturation was reached after 23 interviews. We then double coded all transcripts (K.A.R., M.G.T., C.D.K.) and reconciled discrepancies through discussion (K.A.R., M.G.T., C.D.K., K.S.-B.). We read through various subsets of coded excerpts to identify relevant themes, which were then discussed with the entire group and consolidated into the final thematic analysis. This study was approved by the University of Michigan Institutional Review Board
Results
We interviewed 23 US academic genetic researchers (Table 2). In total, 11 were sampled for their use of a private database and 12 for their use of an academic, government, or consortia database (Table 3). Most interviewees were female (n = 13), non-Hispanic White researchers (n = 14), with an average of 8.5 years at their institution (Table 2). Almost all compared different kinds of databases beyond the one for which they were sampled, for a total of 70 distinct databases discussed (30 academic data stewards, 13 government, 11 private, 8 nongovernmental organizations, and the remaining 8 via collaborations) (Figure 1).
Table 2:
Interviewee Characteristics (n = 23)
| n (% or SD) | |
|---|---|
| Gender | |
| Female | 13 (57%) |
| Male | 10 (43%) |
|
| |
| Years at Institution (SD) | |
| 8.5 (5.3) | |
|
| |
| Race/Ethnicity | |
| Non-Hispanic White/Caucasian | 14 (61%) |
| Asian American/Asian | 7 (30%) |
| Hispanic/Latino | 2 (9%) |
|
| |
| Job Title | |
| Full Professor/Researcher | 5 (22%) |
| Associate Professor/Researcher | 10 (43%) |
| Assistant Professor/Researcher | 5 (22%) |
| Trainee | 2 (8%) |
| Other title | 1 (4%) |
|
| |
| School | |
| School of Public Health | 7 (30%) |
| School of Medicine | 13 (52%) |
| School of Dentistry | 1 (4%) |
| School of Literature, Science, and the Arts | 1 (4%) |
| School of Pharmacology | 1 (4%) |
This journal no longer allows the term “Caucasian” except as it refers to the people from the Caucasus region. As respondents in this study self identified using this term in response to an open-ended question, we have reported it here.
Table 3:
Primary Dataset Characteristics (n = 23)a
| n (%) | |
|---|---|
| Type(s) of Data Source(s) Discussed | |
| Governmental | 7 (30%) |
| Private (DTC) | 6 (26%) |
| Academic | 5 (22%) |
| Consortium | 5 (22%) |
| Private: (not DTC) | 5 (22%) |
|
| |
| Analysis Type(s) | |
| Genome-Wide Association Study | 15 (65%) |
| Whole Exome Sequencing | 3 (13%) |
| Targeted Sequencing | 2 (9%) |
| Multiplex Genetic Panel Testing | 1 (4%) |
| Case reports | 1 (4%) |
| Clinical germline testing | 1 (4%) |
| Genome-wide Copy Number Variant | 1 (4%) |
| Mendelian Randomization | 1 (4%) |
|
| |
| Data Type | |
| Individual | 16 (70%) |
| Aggregate | 7 (30%) |
|
| |
| Paid for access? | |
| No | 16 (70%) |
| Private dataset | 8 (50%) |
| Other dataset | 8 (50%) |
| Yes | 7 (30%) |
| Private dataset | 3 (43%) |
| Other dataset | 4 (57%) |
DTC, direct-to-consumer.
Primary dataset = The dataset used in the PubMed article from which the researcher was first identified. This dataset may contain data from multiple different sources; data source and data type may sum to more than 100%.
Figure 1: Databases referenced by interviewees, by category of primary management and funding (n=70).
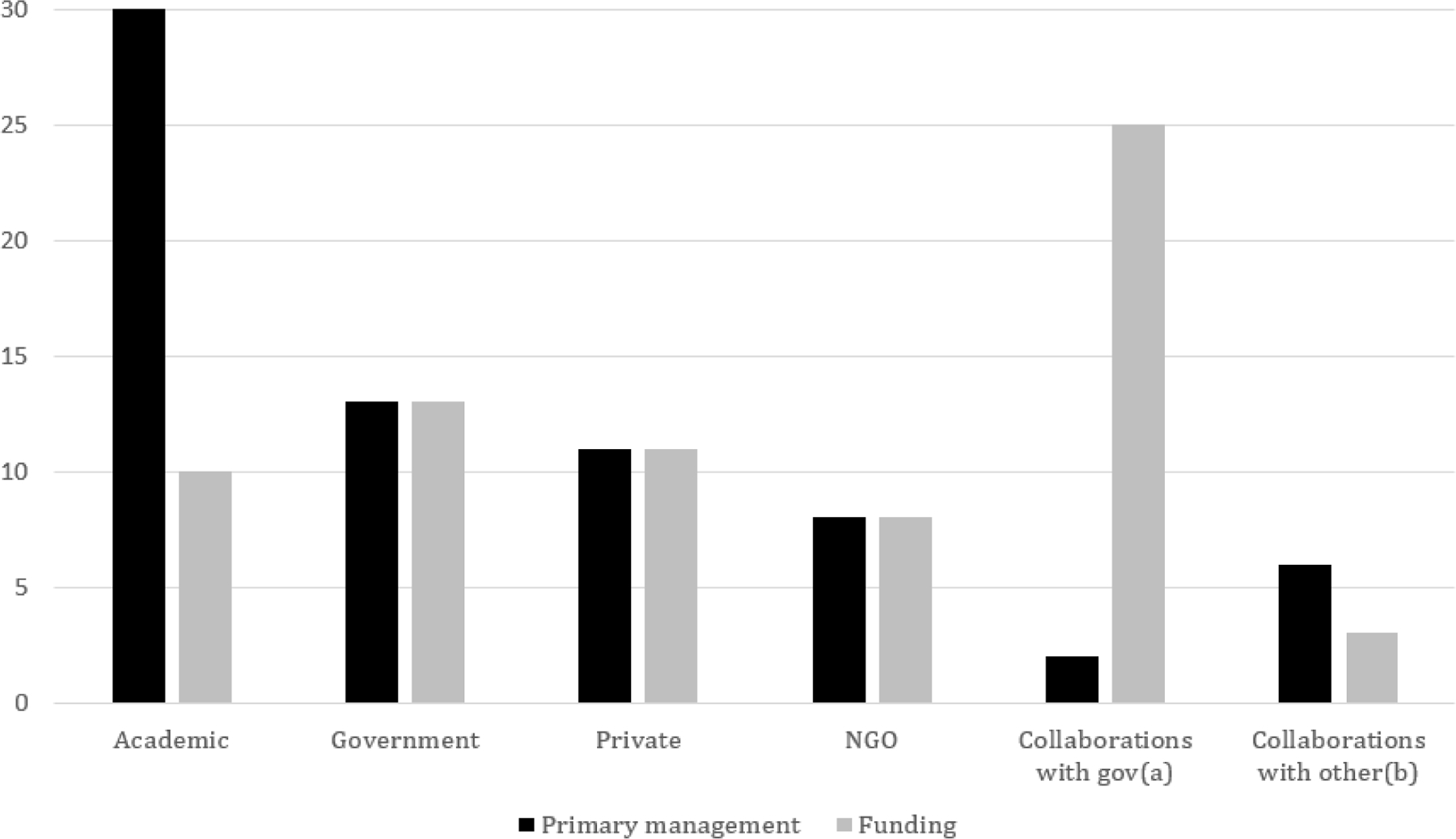
(a) Includes: 1 Acad/Govt and 1 Acad/NGO/Govt/Priv collaborations in primary management; and 11 Acad/Govt, 4 Acad/Govt/NGO, 3 Acad/Govt/NGO/Priv, and 7 Govt/NGO collaborations
(b) Includes: 5 Acad/NGO and 1 Acad/NGO/Priv collaborations in primary management; and 3 Acad/NGO collaborations in funding
Theme 1: Motivation to use existing databases
First, we found many interviewees were motivated to use existing databases for their research to avoid burdens of primary data collection, including time and financial cost. Interviewees who struggled to accumulate data on their own found themselves underpowered:“Collecting data…is one of the most horrific experiences known to man. Takes a massive amount of time… But other peoples’ old data is really much, much better because there’s more of it around.” Interviewees also sought to avoid the data harmonization necessary to bring smaller existing databases together: “It took two years off my life when I was [analyzing these data] because I literally had 35 different [genome-wide association studies (GWAS)] data sets, and every sample had a different [clinical condition] battery. […] almost universally, every sample used a different micro-array for their genotyping… That was a massive undertaking, trying to get all those individual, independent samples aligned on the same genotype reference panels…it was brutal.” Another pointed out the advantage of existing databases managed by professional teams, which can allow the researcher to “actually focus on running the studies instead of doing annoying data cleaning tasks.”
Many pointed out that analyzing already existing data was less expensive or was the only option for unfunded research. In total, 70% of interviewees reported that they did not pay for data access (split evenly between those who used private databases vs other) (Table 3). One interviewee described the 2-pronged cost structure of 23andMe, including an annual competition in which it selects those “that align with whatever their priorities are for that year” to support for free as well as a fee-for-access service.The influence of cost as a factor also seemed to vary by seniority, with more junior interviewees less able to pay for data access. As 1 research fellow explained, “I don’t have much money. Getting papers out quickly is important.” Therefore, “cost and ease of access were two huge factorsthat were amplified even greater for somebody who’s in a more junior position...”
If interviewees decided that generating their own data was not the right approach, they also identified multiple factors they considered when choosing which database to use, categorized into the additional themes of ease of access, database features, and data management/downstream effects of database selection.
Theme 2: Importance of ease of access
When asked about the single most important factor in selecting a genetic database for their work, interviewees consistently discussed topics related to the theme of ease of access. As one interviewee summarized“… by and large the [type of] data wasn’t the issue. It was getting access to the data…” Familiarity, or the presence of a preexisting relationship,with the data steward was described as one of the most powerful determinants of access: “I know these people and I know their quality, so I just go with the people I know.” When interviewees had previously worked with a data steward, they described of having to do less work to assess quality or gain access. These relationships were often maintained as interviewees moved to other institutions.
Efficiency in access was also an important consideration and was often described as based on whether legal agreements needed to be established. One interviewee described the benefits of working with their collaboratory: “We didn’t have to go through a lot of data access agreements and data use agreements... In some sense, one might even say that the project was really defined to some extent by the fact that we had access to those data, rather than the other way around.” By contrast, databases with inefficient access were described as those that “require writing a proposal to apply, and wait for permission, and wait for material transfer agreements between institutions.” Another described the access process for dbGaP as “extremely slow and capricious.” Other examples of legal agreements causing inefficiencies included compliance with the European General Data Protection Regulation (GDPR), intellectual property, nondisclosure, or coauthorship agreements—which could add months to negotiations. One interviewee also pointed out inequity in the consistency of what agreements their university would support: “The sign off on it for some people…[and], candidly, how senior you are and how much pull you have matters.”
Others discussed efficiency in terms of a customerservice orientation and the scientific value added from such “collaborations.” In these cases, private databases were generally described more favorably than government ones. One interviewee compared a private data steward “which is very responsive and cares a lot about giving us the highest quality results” to UK Biobank, which they described as a “bureaucracy dealing with lots and lots of researchers. They’re very slow, in general, to reply about potential issues, and it’s not really a personal interaction at all.” Interviewees were conflicted when comparing the efficiency of government databases with each other.
Finally, several interviewees spoke about the potential for publicly available, open access data to ease access burdens. As the interviewee who was frustrated with their “horrific” data collection experience lauded, publicly available GWAS data are “just fantastic. It’s amazing. It’s brilliant. And it’s an enormous benefit…to the whole research community...”
Theme 3: Importance of specific database features
Interviewees also described selecting databases based on specific features they valued, such as size and phenotypes. Several pointed out that the size of the database matters most when conducting GWAS. Interviewees described having to share their data with other laboratories because “everybody was doing very small sample research for genetics, and nothing was being replicated, nothing was being discovered. So, we realized that we needed to start to collaborate and pool our genetic data to increase our sample sizes.” Several interviewees extolled the “astronomical number” of cases they were able to access via private databases: “people were just really excited about the enormous size of the [Private Company] data set and the value of the [statistical] power that that entails...” The large size of private databases, as compared to many government ones, led several interviewees to speculate that research using DTCgenerated genetic data would outpace others: “…if they haven’t already, [DTC Companies] are going to pull very far ahead of traditional NIH-funded research.
Because [DTC Company] collaborated with us on this…paper, and they contributed like 600,000 subjects, just at the drop of a hat. Whereas it took us like four or five years to get 35,000 people!” Another agreed that DTC companies will “leapfrog over everybody” because they actually get paid to collect data and “they don’t have to get all their grants rejected all the time and keep reapplying. They’re not begging for money, the way people in academia are, to do this kind of research.”
Access to relevant phenotypes was described as another important aspect of database selection: “A lot of the genetics cohorts out there have really just information on case status and genotype information… [But] we wanted to be able to review people’s charts, confirm the diagnoses that we thought the individuals had.” Interviewees valued the phenotypic data availability at UK Biobank as well as from private companies. Limited sequencing methods (eg, chromatinimmunoprecipitation sequencing) or only allowing access to summary statistics (as some private companies did) were described as constraining analyses or quality.
Interestingly, valuing the demographic diversity of communities represented in the database was not a theme that emerged organically from the interviews. When specifically prompted how diversity might affect their database selection, many interviewees explained that they did not consider data diversity because they already knew that most databases are homogenous, therefore, it “didn’t really play a role…” Another described homogeneity as a strength in GWAS protocols because “you’d better have a single ethnicity sample to avoid spurious associations.” Another pointed out that some publicly available genomic data do not include self-identified racial information at all.
Several interviewees were reflective of the fact that this homogeny would limit the generalizability of their research. This frustrated some: “there’s just not enough [participants with] African ancestry, so we’re going to ignore them? …that’s actually not acceptable anymore, right?” But others emphasized the self-reenforcing nature of the exclusion cycle21: “Europeans are the most prevalent [participants] that we have, so if we’re studying another ancestry, which we very much want to, but if no one else is doing it, it just makes your work harder, especially when you do secondary analysis that can only be done across the same ancestry...we were quite limited by what are the others doing.” Some interviewees hypothesized that most private databases were demographically homogenous owing to selfselection biases: “people are choosing to become customers; they tend to be of especially high socioeconomic status.” Another, who had generated their own data, stated that they had not actively prohibited non-White participants from enrolling, and therefore, could not have controlled their 90% non-Hispanic White enrollment. But others described how they were able to enable work on diverse populations by combining strengths of different databases or using government databases as references. One interviewee, with a majority African American patient population, described how “finding ancestrally matched samples can be a challenge,” but highlighted the strengths of dbGaP in so doing. Another suggested taking advantage of “the large sample sizes of [Private Company] and the UK Biobank, but then show that things like the predictive power of the polygenic score hold up and representative samples...”Several interviewees cited the All of Us program as an exemplar for collecting data from diverse populations as well as use as a reference data set.
Theme 4: Importance of data management support and downstream effects of selection
A last thematic area was considerations associated with data management or downstream effects of data selection. Whereas we had hypothesized that data integrity would play a large role in selection, and this was true for many interviewees, others who used large, established, databases stated that they relied on the data depositors or stewards to ensure quality. Although 2 interviewees spoke positively about a specific private company’s quality control, another dismissed the need to independently verify its data because “we were just trying to get to big numbers, hoping that with big numbers we will achieve more or less accuracy...” Several mentioned specific concerns regarding the accuracy of self-reported phenotypic data from DTC databases. Many interviewees put an emphasis on “well-respected and regarded” databases to reassure colleagues and reviewers of quality.
One interviewee pointed out the additional, often hidden, costs of data storage and the computational infrastructure necessary to extract and analyze large amounts of data. Although they were able to access individual-level data from UK Biobank, “that is a mega hassle because you have to have terabytes of data to store it on. And then you have to have someone who has the skills to extract the bits you want.” Therefore, although the UK Biobank data access was inexpensive, the cost of use was higher: “we needed to have the IT support to help us download all the data and arrange it and show us how to access it,” otherwise “the answers turned out rubbish.”
Discussion
In contrast to the laborious and expensive process of generating genetic data de novo, the availability of large and inexpensive databases has created a genetic data market in which academic researchers sometimes have the option to choose between different databases or select more than one. But, beyond convenience, uses of different kinds of databases can have important implications for what science is advanced—and to which communities it will generalize. Our findings elicit important considerations for database stewards focusing on not only building large and comprehensive databases but also ensuring that they are used by genetic researchers for the benefit of diverse communities (Table 4).
Table 4.
Major findings and considerations for database stewards
| Sub-theme | Finding(s) from interviews | Recommendations |
|---|---|---|
| Theme 1: Use of existing databases for genetics research | ||
| Time spent | Primary data collection was described as more time-consuming and costly than existing database use | Efforts focused on building accessible databases allow researchers to focus time and funding on analysis, as opposed to primary data collection |
| Financial cost | ||
| Theme 2: Importance of ease of access | ||
| Familiarity | Familiarity was described in terms of a relationship with a specific person, rather than the database itself | Investing resources in long-term employees who support researchers by facilitating access, acclimating them with the data, and answering questions is valuable |
| Efficiency | Efficiency was described as the ability to spend time on research, rather than administrative matters such as legal agreements | Limit the burdensome nature of legal negotiations for the researcher and ensure that all “rules” are equitably applied across researchers |
| Theme 3: Importance of specific data features | ||
| Size | The number of contributors represented, and the phenotypic information included, were critical factors and sometimes even drove research questions | Enabling efforts to encourage and facilitate datasharing in widely accessible databases should be a key component of balancing access for researchers |
| Phenotypes | ||
| Demographic diversity | Interviewees described not having the choice to select databases based on demographic diversity, due to the homogeny of existing cohorts, or incomplete demographic information. While this was seen as a limitation, it was a commonly accepted one. | Efforts to increase the diversity of participants represented in databases should focus on the enrollment of historically excluded communities at the point of data collection; support may be needed for researchers attempting to do research with diverse databases who might have additional data harmonization, computational, and analysis needs |
| Theme 4: Importance of data management support and downstream effects of selection | ||
| Integrity | Data stewards were relied upon to ensure integrity of databases | Ensuring data quality at the point of deposit, rather than assuming secondary users will or can do so, as well as supporting interoperability across data resources, are critical |
| Harmonization | Data harmonization was seen as a time-consuming chore that distracted from valuable research. | |
| Storage | Some databases were hard to use due to vast data storage and computational infrastructure necessary to extract and analyze data | Supporting researchers with limited storage and computing infrastructure may help enable less well-resourced researchers |
First, many interviewees enthusiastically highlighted existing databases (including those held by academia, government, private industry, nongovernmental organizations, and collaborations) as an excellent alternative to trying to collect data alone or harmonizing small databases—an experience described as expensive, time-consuming, and generally agonizing. Many were strongly supportive of government and collaboratory efforts to do so, such that the burden did not fall on individual researchers.
Second, interviewees generally stated that the ease of access was the most important factor in their selection, describing the interrelated components of familiarity with the data steward and efficiency. Interviewees indicated that a previous, positive experience with the data steward strongly influenced their decision to work with them again and encourage others to do the same. But interviewees most often discussed valuing a familiar relationship with the people who supported them, rather than with the database brand per se. Long-term employees of data stewards who support researchers by facilitating access, acclimating researchers to the data, and answering questions were seen as key attributes.
Efficiency of database access was another central component of “ease of access,” and was often described in terms a lack of legal red tape or a high customer-service focus (which seemed to favor private stewards). Although legal protections surrounding genetic data use are a critical component of protecting the autonomy rights of individual contributors—in addition to intellectual property and other related rights of researchers and their institutions22,23—they were roundly described as burdensome, unrelated to the science, and sometimes applied by institutions in a discriminatory fashion. These findings are consistent with previous assessments of data use agreements being viewed as overly burdensome23 as well as existing “bottlenecks” to data access.24
Third, interviewees also prioritized data features such as size of the database and access to related phenotypes. Several interviewees described the “astronomical” size of DTC databases and speculated that without increasing the size of current government data resources, use of DTC databanks might outpace government ones—with attendant limitations.15 Enabling efforts to encourage and facilitate data sharing in widely accessible databases should be a key component of balancing access for researchers without DTC collaborations. Importantly, demographic diversity was not described de novo as a factor that interviewees considered when selecting a database. When probed, almost all interviewees pointed out that genetic databases are predominantly made up of participants of European ancestry; they described not having the choice to consider the demographic diversity of communities included in databases to begin with. Most interviewees presented this as a limitation of their research in terms of generalizability but one that was widely accepted as unavoidable. Private databases may also be particularly limited in terms of socioeconomic diversity because, as one interviewee noted, their consumers are self-selecting and tend to be of a higher socioeconomic status (which can also be associated with a lack of racial and ethnic diversity). Several interviewees discussed the importance of access to smaller demographically diverse databases to act as a reference tool to further validate results explored in a larger Euro-centric database. The salience of familiarity with the data steward, paired with this Euro-centric focus of existing databases and the high socioeconomic status of participants in private ones, limits the broad applicability of research findings. These limitations are cyclical; if researchers must rely on pooled resources to access the necessary statistical power to do their work, their work can only be as diverse as the overall pool. And recent assessments have shown that global GWAS catalogs are overwhelmingly populated by those of European ancestry25 (eg, only 2.4% of participants in the National Human Genome Research Institute-European Bioinformatics Institute GWAS Catalog are of African ancestry26,27). Those independently recruiting diverse community participants to provide research samples may also face a more expensive and time-consuming process owing to the many disparities and differences already built into recruitment and consent methods from the ground up. Without external forces compelling more researchers to prioritize diverse sample use, other priorities—like publishing results quickly in high-impact journals with strict requirements regarding statistical significance—might drive continued use of predominantly European populations. Efforts to increase the demographic diversity of communities represented in databases need to focus on the enrollment of historically excluded communities at the point of data collection, eg, the NIH’s policy for the inclusion of women and minorities as participants in human subjects research28 (including ensuring the validity of results via evolving best practices for the collection of demographic information29–31). Additional support, or education regarding such support, may also be needed for researchers who are attempting to do research with historically excluded communities who might have additional data harmonization, computational, and analysis needs.
Fourth, interviewees took into consideration potential challenges regarding data management, including issues related to data integrity, harmonization, and storage. These limitations could lead to significant downstream work if the limitations were not, or could not be, accounted for at the data access or feature assessment stage. Many interviewees relied on data stewards to ensure data integrity and spoke of frustrations with data that lacked interoperability. Those using individual-level data from some government databases complained that storing data required large, expensive, technical resources and trained professionals to extract relevant information.
These findings highlight the need for quality assurance at the repository level, rather than assuming database users will or can do so, as well as supporting interoperability and external computing opportunities across data resources. These data management challenges are intersectional with challenges to increasing the demographic diversity of communities represented in databases. Our interviewees described a world in which researchers who do not have the resources, infrastructure, or an academic network of experienced data users are at a disadvantage. Those who lack sufficient resources or institutional support to access large databases, navigate the attendant contractual considerations, and secure the necessary computing infrastructure to store and analyze large databases, must resort to the use of smaller databases with lower statistical power for their research, limit analyses to summary statistics, or undergo the onerous process of data harmonization and cleaning themselves.
Given the increased use of private genetic databases for academic research,7 we hypothesized that private genetic databases might offer some advantages over other existing resources and that researchers would value data features— such as size, available phenotype, demographic diversity, and quality—in that choice. What we found was that database features such as familiarity, efficiency, analytical support, and necessary data management tools appear to have had an even greater effect on database selection and that data availability might define the research question pursued—rather than the other way around. Overall, this work highlights an inherent tension between the government goal of alleviating “health data poverty”32 for historically excluded communities and the structural factors and institutional values that drive researcher choice and use of those databanks. The type of prolonged relationship with a community, foundational to establishing trust and promoting research engagement,33–37 can be in direct conflict with these institutional pressures. If academia continues to reward the publication of large numbers of pieces in high-impact journals over the production of generalizable research, it will continue to limit the effect of government efforts. Decentralized approaches, such as journals prioritizing the publication of research with historically underrepresented populations,38 may help alleviate this tension. In the meantime, lack of demographic diversity in those large, accessible databases can and will continue to have tangible consequences for populations excluded from research.8–10
It is important to note that the findings reported in this research represent the views of a specific group of academic genetic researchers and are only a snapshot of the varied considerations researchers make when selecting a database for their work. Further research is necessary to generalize their experiences, such as surveys across a wider population, and an assessment of the relationship (if any) between researcher demographics, professional status, type of work, values, and choice of database. These interviews are a valuable first step in this process.
Conclusion
As the federal government continues to invest in building genetic databases that represent demographically diverse communities, it should be aware of the growing market and competition between data stewards. As our interviews show, the existence of a database is only one step on the journey toward enabling generalizable science. Data need to be accessible, harmonized, and administratively supported to be translated into use and, in turn, result in scientific advancements across diverse communities.
Supplementary Material
Acknowledgments
This work was funded in part by the National Human Genome Research Institute (K01HG010496 and T32- HG010030), the National Center for Advancing Translational Sciences (UL1TR002240), the National Institute of Mental Health (R01MH126937), the National Cancer Institute (R01CA237118), and the National Science Foundation (IIS1553146).
Footnotes
Ethics Declaration
This study was approved by the University of Michigan Institutional Review Board (HUM00175088). The study data were de-identified. This study was performed in accordance with relevant guidelines and regulations, including those set forth in the Declaration of Helsinki. Informed consent was obtained from all subjects, and deidentified data were used for analysis and reporting.
Conflict of Interest
The authors declare no conflicts of interest.
Additional Information
The online version of this article (https://doi.org/10.1016/j.gim.2022.09.003) contains supplementary material, which is available to authorized users.
Data Availability
De-identified qualitative quotes, organized by theme rather than in full transcript presentation to further protect the identity of the interviewee, are available upon request.
References
- 1.Saulsberry L, Olopade OI. Precision oncology: directing genomics and pharmacogenomics toward reducing cancer inequities. Cancer Cell 2021;39(6):730–733. 10.1016/j.ccell.2021.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins FS, Adams AB, Aklin C, et al. Affirming NIH’s commitment to addressing structural racism in the biomedical research enterprise. Cell 2021;184(12):3075–3079. 10.1016/j.cell.2021.05.014 [DOI] [PubMed] [Google Scholar]
- 3.National Institutes of Health Office of Extramural Research. Final NIH policy for data management and sharing National Institutes of Health. Published October 29, 2020. Accessed September 29, 2022. https://grants.nih.gov/grants/guide/notice-files/NOT-OD-21-013.html [Google Scholar]
- 4.Jorgenson LA, WolinetzCD,Collins FS. Incentivizing a newculture of data stewardship: the NIH policy for data management and sharing. JAMA 2021;326(22):2259–2260. 10.1001/jama.2021.20489 [DOI] [PubMed] [Google Scholar]
- 5.National Center for Biotechnology Information. dbGaP. National Center for Biotechnology Information Accessed April 7, 2022. https://www.ncbi.nlm.nih.gov/gap/ [Google Scholar]
- 6.National Institutes of Health. All of Us Research Program. National Institutes of Health Accessed April 7, 2022. https://allofus.nih.gov/ [Google Scholar]
- 7.Spector-Bagdady K, Fakih A, Krenz C, Marsh EE, Roberts JS. Genetic data partnerships: academic publications with privately owned or generated genetic data. Genet Med 2019;21(12):2827–2829. 10.1038/s41436-019-0569-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonham VL, Callier SL, Royal CD. Will precision medicine move us beyond race? N Engl J Med 2016;374(21):2003–2005. 10.1056/NEJMp1511294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caswell-Jin JL, Gupta T, Hall E, et al. Racial/ethnic differences in multiple-gene sequencing results for hereditary cancer risk. Genet Med 2018;20(2):234–239. 10.1038/gim.2017.96 [DOI] [PubMed] [Google Scholar]
- 10.Manrai AK, Funke BH, Rehm HL, et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med 2016;375(7):655–665. 10.1056/NEJMsa1507092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SS, Appelbaum PS, Chung WK. Challenges and potential solutions to health disparities in genomic medicine. Cell 2022;185(12):2007–2010. 10.1016/j.cell.2022.05.010 [DOI] [PubMed] [Google Scholar]
- 12.Blizinsky K. Precision medicine research, “All of Us”, and Inclusion. The Hastings Center & Center for ELSI Resources & Analysis (CERA) Published November 16, 2021. Accessed April 7, 2022. https://www.thehastingscenter.org/precision-medicine-research-all-of-us-and-inclusion/ [Google Scholar]
- 13.Carere DA, Couper MP, Crawford SD, et al. Design, methods, and participant characteristics of the Impact of Personal Genomics (PGen) Study, a prospective cohort study of direct-to-consumer personal genomic testing customers. Genome Med 2014;6(12):96. 10.1186/s13073-014-0096-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tung JY, Eriksson N, Kiefer AK, et al. Characteristics of an online consumer genetic research cohort, 23andMe 2011. Accessed April 7, 2022. https://blog.23andme.com/wp-content/uploads/2011/10/ASHG2011poster-JYT.pdf [Google Scholar]
- 15.Spector-Bagdady K. Governing secondary research use of health data and specimens: the inequitable distribution of regulatory burden between federally funded and industry research. J Law Biosci 2021;8(1):lsab008. 10.1093/jlb/lsab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Research and Markets. Global consumer DNA (genetic) testing market—forecasts from 2018–2023. Research and Markets Accessed April 7, 2022. https://www.researchandmarkets.com/research/w4fsmm/global_928?w=5 [Google Scholar]
- 17.Thorne S, Kirkham SR, O’Flynn-Magee K. The analytic challenge in interpretive description. Int J Qual Methods 2004;3(1):1–11. 10.1177/160940690400300101 [DOI] [Google Scholar]
- 18.Thorne S, Kirkham SR, MacDonald-Emes J. Interpretive description: a noncategorical qualitative alternative for developing nursing knowledge. Res Nurs Health 1997;20(2):169–177. [DOI] [PubMed] [Google Scholar]
- 19.Thorne S. Interpretive Description: Qualitative Research for Applied Practice Routledge; 2016. [Google Scholar]
- 20.Vaismoradi M, Turunen H, Bondas T. Content analysis and thematic analysis: implications for conducting a qualitative descriptive study. Nurs Health Sci 2013;15(3):398–405. 10.1111/nhs.12048 [DOI] [PubMed] [Google Scholar]
- 21.Bracic A, Callier SL, Price WN 2nd. Exclusion cycles: Reinforcing disparities in medicine. Science 2022;377(6611):1158–1160. [DOI] [PubMed] [Google Scholar]
- 22.Heeney C, Kerr SM. Balancing the local and the universal in maintaining ethical access to a genomics biobank. BMC Med Ethics 2017;18(1):80. 10.1186/s12910-017-0240-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mello MM, Triantis G, Stanton R, Blumenkranz E, Studdert DM. Waiting for data: barriers to executing data use agreements. Science 2020;367(6474):150–152. 10.1126/science.aaz7028 [DOI] [PubMed] [Google Scholar]
- 24.van Schaik TA, Kovalevskaya NV, Protopapas E, Wahid H, Nielsen FGG. The need to redefine genomic data sharing: a focus on data accessibility. Appl Transl Genom 2014;3(4):100–104. 10.1016/j.atg.2014.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature 2016;538(7624):161–164. 10.1038/538161a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morales J, Welter D, Bowler EH, et al. A standardized framework for representation of ancestry data in genomics studies, with application to the NHGRI-EBI GWAS Catalog. Genome Biol 2018;19(1):21. 10.1186/s13059-018-1396-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bentley AR, Callier SL, Rotimi CN. Evaluating the promise of inclusion of African ancestry populations in genomics. NPJ Genom Med 2020;5:5. 10.1038/s41525-019-0111-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Institutes of Health. Inclusion of women and minorities as participants in research involving human subjects National Institutes of Health. Accessed April 7, 2022. https://grants.nih.gov/policy/inclusion/women-and-minorities.htm [Google Scholar]
- 29.Bonham VL, Green ED, P´ erez-Stable EJ. Examining how race, ethnicity, and ancestry data are used in biomedical research. JAMA 2018;320(15):1533–1534. 10.1001/jama.2018.13609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis ACF, Molina SJ, Appelbaum PS, et al. Getting genetic ancestry right for science and society. Science 2022;376(6590):250–252. 10.1126/science.abm7530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callier SL. The use of racial categories in precision medicine research. Ethn Dis 2019;29(Suppl 3):651–658. 10.18865/ed.29.S3.651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibrahim H, Liu X, Zariffa N, Morris AD, Denniston AK. Health data poverty: an assailable barrier to equitable digital health care. Lancet Digit Health 2021;3(4):e260–e265. 10.1016/S2589-7500(20)30317-4 [DOI] [PubMed] [Google Scholar]
- 33.Yu JH, Crouch J, Jamal SM, Tabor HK, Bamshad MJ. Attitudes of African Americans toward return of results from exome and whole genome sequencing. Am J Med Genet A 2013;161A(5):1064–1072. 10.1002/ajmg.a.35914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Millon Underwood S, Buseh AG, Kelber ST, Stevens PE, Townsend L. Enhancing the participation of African Americans in health-related genetic research: findings of a collaborative academic and community-based research study. Nurs Res Pract 2013;2013:749563. 10.1155/2013/749563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dang JHT, Rodriguez EM, Luque JS, Erwin DO, Meade CD, Chen MS Jr. Engaging diverse populations about biospecimen donation for cancer research. J Community Genet 2014;5(4):313–327. 10.1007/s12687-014-0186-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamin R. Race for cures: rethinking the racial logics of ‘trust’ in biomedicine. Sociol Compass 2014;8(6):755–769. 10.1111/soc4.12167 [DOI] [Google Scholar]
- 37.Isler MR, Sutton K, Cadigan RJ, Corbie-Smith G. Community perceptions of genomic research: implications for addressing health disparities. N C Med J 2013;74(6):470–476. 10.18043/ncm.74.6.470 [DOI] [PubMed] [Google Scholar]
- 38.Brothers KB, Bennett RL, Cho MK. Taking an antiracist posture in scientific publications in human genetics and genomics. Genet Med 2021;23(6):1004–1007. 10.1038/s41436-021-01109-w [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
De-identified qualitative quotes, organized by theme rather than in full transcript presentation to further protect the identity of the interviewee, are available upon request.


