Abstract
Background and purpose
Over the last decade, the implementation of multigene panels for hereditary tumor syndrome has increased at our institution (Inselspital, University Hospital Berne, Switzerland). The aim of this study was to determine the prevalence of variants of unknown significance (VUS) in patients with suspected Lynch syndrome and suspected hereditary breast and ovarian cancer syndrome, the latter in connection with the trend toward ordering larger gene panels.
Results
Retrospectively collected data from 1057 patients at our institution showed at least one VUS in 126 different cases (11.9%). In patients undergoing genetic testing for BRCA1/2, the prevalence of VUS was 6%. When < 10 additional genes were tested in addition to BRCA1/2, the prevalence increased to 13.8%, and 31.8% for > 10 additional genes, respectively. The gene most frequently affected with a VUS was ATM. 6% of our patients who were tested for Lynch syndrome had a VUS result in either MLH1, MSH2 or MSH6.
Conclusions
Our data demonstrate that panel testing statistically significantly increases VUS rates due to variants in non-BRCA genes. Good genetic counseling before and after obtaining results is therefore particularly important when conducting multigene panels to minimize patient uncertainty due to VUS results.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-023-01437-7.
Keywords: Hereditary breast and ovarian cancer, Lynch syndrome, VUS, BRCA1, BRCA2, MMR genes, Multigene panel
Background
Genetic testing
Genetic testing for germline sequence variants is important for assessing the risk of hereditary tumor syndromes [1]. Genetic predisposition for cancer is a complex matter because cancer risk can be increased by pathogenic variants in different genes, and a variant at a particular site may cause more than one type of cancer [2]. Researchers have identified different inherited tumor syndromes and their causative genes. According to these findings, different genes were analyzed depending on the suspected tumor syndrome [3]. Traditionally, testing for specific genes is performed after evaluating various factors such as family history, age of diagnosis and histological type of cancer [4]. This study focuses on hereditary breast and ovarian cancer (HBOC; MIM#114480) and Lynch Syndrome (LS; MIM#120435)/hereditary non-polyposis colorectal cancer (HNPCC).
Regarding HBOC the genes BRCA1 and BRCA2 have been known for more than two decades [5–7]. Myriad Genetic Laboratories introduced complete sequence analysis of these genes in 1996 as one of the first sequence-based tests for risk assessment of a common cancer [8]. Today, sequence based tests are omnipresent in genetic testing [9]. The development of next generation sequencing (NGS) technologies has greatly reduced time consumption and cost of simultaneous analysis and thereby increased the use of multigene panels in genetic testing for hereditary tumor syndromes [2, 10]. The National Comprehensive Cancer Network (NCCN) provides guidelines for genetic/familial high-risk assessment of tumor syndromes that include benefits and limitations of multigene testing using NGS [11, 12]. According to these guidelines, multigene testing can be beneficial because a family history of cancer may be explained by more than one cancer syndrome and some individuals may carry pathogenic/likely pathogenic variants in more than one gene. In addition, multigene testing may also include genes with a lower penetrance for a particular cancer syndrome, so-called moderate-risk genes. While this is an advantage of NGS, testing moderate-risk genes also comes with the limitation that some variants found will not alter patient risk management. Another limitation are higher rates of variant without clear clinical significance [11].
In addition to the NCCN guidelines, the following guidelines for genetic testing and counseling are valid in Switzerland and are used in our population: the SAKK Swiss guidelines [13, 14] for breast and ovarian cancer and the PREMM5 Model [15] for Lynch Syndrome.
Classification of variants
Genetic testing can have different results. Beside the possibilities of a variant being pathogenic (leading to a significantly elevated risk of cancer) or benign (not interfering with the function of the protein), it’s impact on the protein function can also be uncertain. Such an alteration is considered a variant of unknown significance (VUS) [9, 16, 17]. The detailed classification of the variants is shown in Table 1. To avoid misinterpretation when describing the pathogenicity of variants, the Evidence-based Network for the Interpretation of Germline Mutation Alleles (ENIGMA) has established appropriate terminology for describing genetic variants [16]. They propose to use the IARC (International Agency for Research on Cancer) five-tier variant classification system [9] to describe clinical relevance of genetic variant. This is used by ENIGMA for BRCA1/2 variant classification, and by the International Society for Gastrointestinal Hereditary Tumours (InSIGHT) group for mismatch repair (MMR) gene variant classification [17], as well as by databases used to classify genetic variants such as ClinVar [18]. A very similar probability-based method of variant classification was established by the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) [19]. The clinical implications and proposed predictive tests for relatives at risk largely overlap with the IARC and the ACMG/AMP classifications [16].
Table 1.
Classification of variants according to ENIGMA [16]
| Class | Description | Probability of being Pathogenic | Clinical implication [16] | |
|---|---|---|---|---|
| According to IARC [9] | According to ACMG/AMP [19] | |||
| 1 | Benign (B)/ of no clinical significance | < 0.001 | < 0.001 |
Treat as if ‘no mutation detected’ for this disorder No Predictive testing of at risk relatives is suggested |
| 2 | Likely benign (LB)/ of little clinical significance | 0.001–0.049 | 0.001–0.099 | |
| 3 | Variant of unknown significance (VUS) | 0.050–0.949 | 0.100–0.899 |
Surveillance based on family history and patient-based risk factors Obtaining additional data in order to reclassify, if possible Research testing of family members can be recommended |
| 4 | Likely pathogenic (LP) | 0.950–0.999 | 0.900–0.999 |
Full high-risk surveillance Predictive testing of at-risk relatives is suggested |
| 5 | Pathogenic (P) | > 0.999 | > 0.999 | |
In our study, we do not distinguish between pathogenic and likely pathogenic outcomes, but refer to them as P/LP
Variants of unknown significance
Most VUS are missense mutations, but also include intronic variants and inframe deletions and insertions [20]. There is a growing number of functional assays to test VUS on the protein level [21, 22] but in several cases the effect of a VUS remains unclear. Thus its clinical relevance remains uncertain and should not be used as a sole parameter to guide patient and relatives management [2]. Clinical management recommendations should rather be based on personal and family history of disease [16]. Therefore, VUS results in genetic testing may lead to uncertainty in patients rather than clarity. VUS are also frequently misinterpreted by physicians who order genetic testing [23, 24]. As the number of genes tested is directly related to the occurrence of VUS, the risk of inappropriate recommendations that may harm patients could increase with the use of multigene panel analysis [1, 2]. Nevertheless, studies have shown that through genetic counseling, VUS results do not lead to excessive surgery or overstress, but are only less reassuring than negative test results [25] and that surgical decisions are based heavily on the patient’s context and not just a VUS result [26].
Hereditary breast and ovarian cancer
BRCA1 and BRCA2 are well known genes with a proven association to HBOC [5–7]. The prospective cohort study by Kuchenbaecker et al. [27] showed that the cumulative risk of breast cancer by age 80 years was approximately 72% for BRCA1 and 69% for BRCA2 mutation carriers. The corresponding ovarian cancer risks was found to be 44% for BRCA1 and 17% for BRCA2 mutation carriers. Therefore, a diagnosis with a pathogenic variant in one of those genes clearly affects the patient, its treatment and prevention from other cancers [28].
Back in 1999, Easton stated in a paper on predisposition genes for breast cancer [29] that only 20–25% of the familial risk for breast cancer is due to susceptibility genes known at that time, including BRCA1, BRCA2, TP53, PTEN and ATM. More recent studies describe other high or moderately high risk and susceptibility genes of breast [30] and ovarian [31, 32] cancer.
The expected prevalence of pathogenic mutations in BRCA1/BRCA2 genetic testing of patients who meet testing guidelines is about 9% according to previous studies [30, 33, 34]. These and other studies have also shown, that next generation sequencing multigene panels can detect pathogenic variants in susceptibility genes, that would be missed in traditional BRCA1/BRCA2 testing [30, 33–36]. This means that larger gene panels will detect more pathogenic variants than if only the high risk genes BRCA1 and BRCA2 are tested. At the same time, the prevalence of VUS is also increasing while testing more and more susceptibility genes.
Several studies attempted to determine the prevalence of VUS in BRCA1 and BRCA2 genetic testing, and the results varied widely by population, with an overall VUS-prevalence of 7–15% [37]. With further testing and reclassification of VUS, the detection rate declined to approximately 5–10% in BRCA genetic testing conducted in the United States [38]. Myriad Genetic Laboratories [39] recorded a VUS decline from 13% to only 2.1% in 2013. In less well studied genes, the number of VUS remains high [35].
Lynch syndrome
LS, also known as hereditary nonpolyposis colorectal cancer, is the most common hereditary colorectal cancer type [40]. It is responsible for about 1% of colorectal tumors [41, 42]. LS is caused by heterozygous germline inactivation of the DNA mismatch repair genes (MMR) MLH1, MSH2, MSH6 and PMS2 [40]. In addition, Ligtenberg et al. [43] described that a deletion at the 3' end of the gene EPCAM, which is located upstream of the MSH2 gene, can cause epigenetic silencing of the MSH2 gene and thus also lead to LS. Mutations in MMR genes lead to an accumulation of mutations in microsatellites in tumors, so called microsatellites instability (MSI) [44]. Patients with LS have a very high risk of developing colorectal cancer (25–70%) and endometrial cancer (30–70%), and are also at higher risk of ovarian, gastric, small bowel, pancreatic, urothelial, renal, prostate, breast cancer and central nervous system tumors than the average population [44–49].
Aims and benefit of the study
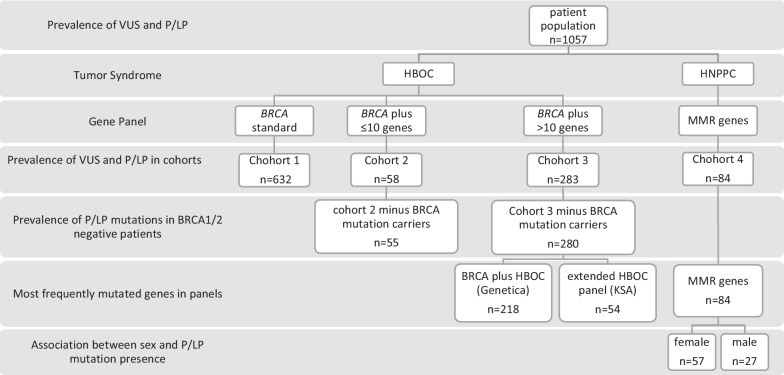
In our study, patients genetically tested for one of the hereditary tumor syndromes HBOC or LS are divided into cohorts according to the genetic test performed. In the beginning of genetic testing in the UKMO (University Hospital for Medical Oncology) at the Inselspital, mainly BRCA1 and BRCA2 genes were tested for HBOC and DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 were tested for LS. While testing for LS has remained the same over the years more multigene panels have been conducted for HBOC since the beginning of 2016.
The aim of this study is to determine the rates of pathogenic variants and VUS in genetic testing and the increase of VUS regarding broader use of multigene panels. We are investigating the hypothesis that the larger the gene panels for HBOC, the higher the incidence of VUS in our patient population. Therefore, the null hypothesis to be refuted is that there is no correlation between the number of genes tested and the occurrence of VUS. At the same time, we do not expect an increase of VUS in the Lynch Syndrome cohort, as the panel did not change over time. In addition, we evaluate in which genes included in commonly used HBOC panels [50, 51] the most variants (P/LP and VUS) occur in our study.
This study evaluates genetic panels currently used in our clinic and compares the outcome to international literature. This contributes to the quality assurance of genetic testing at the UKMO, as the actual rates of P/LP and VUS are used to assess the utility of multi-gene panels [52].
Methods
Study design and setting
In this retrospective cohort study the primary aim is to show the prevalence of VUS in our patient population and its increase regarding broader use of gene panels.
Participant characteristics
Included in the study are patients who underwent genetic testing in the UKMO at the Inselspital between 2010 and 2021. The patient population contains male and female participants over the age of 18 years, who were genetically tested for HBOC or HNPCC, due to a diagnosis with breast, ovarian, endometrial, cervical, fallopian tube, colon cancer and/or several other types of carcinomas or whose family history points to one of these hereditary tumor syndromes (n = 1057).
Patients tested for known familial mutations or germline analysis based on somatically detected variants were excluded from the study, as these cases were not relevant to the research question.
Tests performed for hereditary tumor syndromes other than HBOC or HNPCC (e.g., Li-Fraumeni syndrome and familial adenomatous polyposis) were also excluded.
The indication for test followed national (SAKK) [14] and international guidelines (NCCN) [11] and the costs were covered by the medical insurance and complying with the Swiss law.
Materials
The different genetic tests and multigene panels used in our study are listed in Table 2. The commissioned tests were carried out in different laboratories in Switzerland, namely Genetica [50] in Zurich, Center for Laboratory Medicine at the Cantonal Hospital Aarau (KSA) [51], Medical Genetics Department at the Valais Hospital [53], Department of Genetic Medicine at the University Hospital of Geneva (HUG) [54] and Institute of Medical Genetics and Pathology at the University Hospital Basel [55]. In most cases, gene panel diagnostics provided by the laboratory were performed. However, the laboratories also offered us the possibility to assemble personalized panels depending on the patient's medical history and pedigree. 38 cases of personalized panels are included in our analyses and assigned to the corresponding cohorts according to the number of genes investigated.
Table 2.
Panels for hereditary tumor syndromes
| Panel | Lab | List of genes | Cohort |
|---|---|---|---|
| Hereditary breast and ovarian cancer | |||
| BRCA standard | Genetica, KSA, HUG or Valais | BRCA1 and BRCA2 | BRCA1/2 = 1 |
| BRCA plus Mamma/Breast cancer panel | Genetica, KSA | BRCA1, BRCA2, ATM, CDH1, CHEK2, PALB2, PTEN, STK11, TP53 | BRCA plus ≤ 10 genes = 2 |
| BRCA plus Ovar/Ovarian cancer panel | Genetica, KSA | BRCA1, BRCA2, BRIP1, EPCAM (3’UTR), MLH1, MSH2, MSH6, PMS2, RAD51C, RAD51D | 2 |
| Customized | BRCA plus ≤ 10 | 2 | |
| BRCA plus Lynch | Genetica, KSA | BRCA1, BRCA2, EPCAM (3'UTR), MLH1, MSH2, MSH6, PMS2 | 2 |
| BRCA plus MMR genes | KSA | BRCA1, BRCA2 MLH1, MSH2, MSH6 and PMS2 | 2 |
| BRCA plus HBOC | Genetica | BRCA1, BRCA2, ATM, BARD1, BRIP1, CDH1, CHEK2, EPCAM (3'UTR), MLH1, MSH2, MSH6, PALB2, PMS2, PTEN, RAD51C, RAD51D, STK11, TP53 | BRCA plus > 10 genes = 3 |
| Extended breast- and ovarian cancer panel | KSA | BRCA1, BRCA2, ATM, BARD1, BLM, BRIP1, CDH1, CHEK2, EPCAM, FAM175A, MEN1, MLH1, MRE11A, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, PTEN, RAD50, RAD51C, RAD51D, STK11, TP53, XRCC2 | 3 |
| Customized | BRCA plus > 10 genes | 3 | |
| Lynch syndrome | |||
| MMR genes | Basel | MLH1, MSH2, MSH6 and PMS2 | 4 |
All cited laboratories underwent Swiss Accreditation (SAS) [56] based on the relevant international standards. The analyses were performed according to international standard and the results—positive and negative—were interpreted taking into account the limitation of used techniques. They provided the VUS’s scores using the most current classification, in particular according to the ACMG-Criteria [19] and using the support of Genome Aggregation Database gnomAD [57], ClinVar [18] as well as REVEL [58] and providing the newest literature citations.
Interventions, comparisons and statistical analysis
First, we determined the overall prevalence of VUS and pathogenic/likely pathogenic (P/LP) variants in all the genetic tests of our patient group. Because some patients had more than one result (for instant a pathogenic variant in the BRIP1 gene and a VUS in the PMS2 gene), we used the multiple response function in SPSS. To determine the increase in VUS and P/LP variant prevalence with broader use of gene panels, our population was divided into 4 cohorts (Table 2).
Regarding HBOC, the group that underwent standard BRCA testing, referred to as cohort 1 (n = 632), was compared with patients who underwent gene panel testing (cohort 2 and 3). The difference between cohort 2 and 3 lies in the number of additional genes tested. Cohort 2 (n = 58) contains cases tested for BRCA1/2 plus ≤ 10 other genes, whereas those cases tested for > 10 additional genes are included in cohort 3 (n = 283). A detailed list of all panels used can be found under Additional file 1.
For the descriptive statistics we divided our database according to the suspected tumor syndrome, hereby cohorts 1–3 are compared to each other and cohort 4 (n = 84) is looked at separately. The comparison of cohorts 1–3 refers to the frequencies of VUS and P/LP variants and the relevance of the association was evaluated with χ2 test. A P value of less than 0.05 indicated statistical relevance.
In a further step, the two most frequently used panels for HBOC in our clinic, BRCA plus HBOC from Genetica [50] (n = 218) and the extended breast and ovarian cancer panel from KSA [51] (n = 54) were analyzed to determine in which genes VUS and P/LP variants occur most frequently.
Since the same four genes have been tested for LS since the beginning of genetic testing at the Inselspital, no control group could be determined. Therefore, we have identified the prevalence of VUS in patients tested for Lynch Syndrome (cohort 4) and their change over the years.
In addition we have examined in which genes variants most frequently occur and whether there is a difference of P/LP rates in male and female subjects, as previous MMR gene analysis have shown, that male sex is a predictor of variant [15, 59].
Figure 1 gives an overview of the grouping and interventions carried out.
Fig. 1.
Grouping and Interventions
Results
Participant characteristics
Of 1503 patients who underwent genetic testing at the Inselspital during the given period, 1057 met the inclusion criteria for our study. The other tests are not relevant for our analysis and are therefore excluded in the statistical evaluation. The ethnic background of participants is similar to the one of the general Swiss population (German 65%, French 18%, Italian 10%, Romansch 1%, other 6%) [60]
Main findings
In 785 cases from our population (74.3%), no clinically relevant variant was found. In 147 cases (13.9%) at least one pathogenic variant and in 126 cases (11.9%) at least one VUS could be detected. In one case, we found a pathogenic variant and a VUS simultaneously and in another case two pathogenic variants were detected. 141 VUS were found in 126 different cases, as some genetic tests detected 2 or even 3 different VUS in one or several genes.
An anonymized list of all identified variants is provided in Additional file 2.
The prevalence of each test result (no variant, pathogenic variant, or VUS) in absolute numbers and percent within each cohort is shown in Table 3.
Table 3.
Prevalence of test results in absolute numbers and percent within each cohort
| Test result*cohorts Cross-tabulationsa | ||||||
|---|---|---|---|---|---|---|
| Cohorts | Total | |||||
| 1 | 2 | 3 | ||||
| Test result | No variant | n | 494 | 44 | 189 | 727 |
| % within cohorts | 78.2% | 75.9% | 66.8% | |||
| Pathogenic | n | 105 | 6 | 16 | 127 | |
| % within cohorts | 16.6% | 10.3% | 5.7% | |||
| VUS | n | 38 | 8 | 90 | 136 | |
| % within cohorts | 6.0% | 13.8% | 31.8% | |||
| Total | n | 632 | 58 | 283 | 973 | |
| Percentages and totals are based on respondents and values may exceed 100% as patients may have multiple mutations (multiple responses possible) | ||||||
| aTumor syndrome = HBOC | ||||||
| Test result*cohorts Cross-tabulationsa | ||||
|---|---|---|---|---|
| Cohorts | Total | |||
| 4 | ||||
| Test result | No variant | Count | 58 | 58 |
| % within cohorts | 69.0% | |||
| Pathogenic | Count | 22 | 22 | |
| % within cohorts | 26.2% | |||
| VUS | Count | 5 | 5 | |
| % within cohorts | 6.0% | |||
| Total | Count | 84 | 84 | |
Percentages and totals are based on respondents
aTumor syndrome = Lynch
Variants of unknown significance
In cohort 1, 38 VUS (6.0%) were found in the BRCA1 or BRCA2 gene. There were 8 VUS (13.8%) identified in cohort 2 and 90 VUS (31.8%) in cohort 3. As expected, VUS rates were directly proportional to the number of genes analyzed. Using the χ2 test, the null hypothesis can be rejected because the association between the prevalence of VUS and the increasing number of genes tested for HBOC in the cohorts is statistically significant at a level of 0.05 (P = 0.0E0).
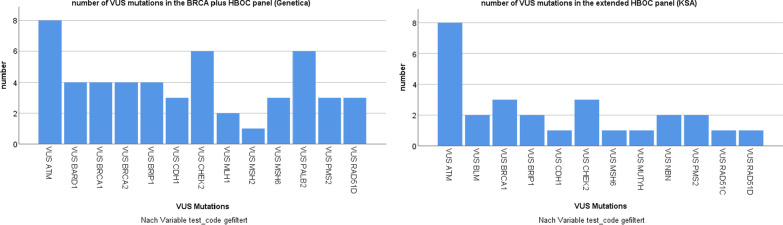
In the BRCA plus HBOC panel from Genetica containing 18 genes (Table 2), VUS were identified in 13 different genes, most frequently in ATM (17% of all VUS found in this Panel). Among 26 genes included in the KSA extended breast and ovarian cancer panel, ATM was also the gene in which the highest number of VUS was detected at 32%. In total, VUS were found in 12 different genes in this panel (Fig. 2).
Fig. 2.
Number of VUS in the HBOC panels from the laboratories of Genetica and KSA
In cohort 4 (Lynch Syndrome patients), 5 subjects (6.0%) were identified with a VUS. All of them were female subjects and none had an MSI-H tumor. 2 variants were found in MLH1, 1 in MSH2, 2 in MSH6 and none in PMS2. Since the number of VUS found in this cohort is very small, it cannot serve as a reliable indication of whether prevalence has remained constant over the years. Only 4 VUS were found in 2019 and 1 in 2021, which made it impossible to perform a valid χ2 test.
Pathogenic variants
We observed that the prevalence of P/LP variants in cohorts 1–3 relatively decrease with increasing size of the gene panel (Table 3). When BRCA1/2 variant carriers were excluded from cohort 2 and 3, 3 variants were detected in 55 individuals in cohort 2 (5.5%) and 11 P/LP variants in 280 individuals (3.9%) in cohort 3.
11 P/LP variants in 8 different genes could be detected in the BRCA plus HBOC panel from Genetica and 5 P/LP variants in 3 different genes in the extended HBOC panel from KSA.
22 (26.2%) P/LP variants were found in 21 cases of our LS population. 3 P/LP variants were found in the MLH1 gene, 11 in MSH2 and 8 in MSH6, two of them in the same subject. No deleterious variant was detected in the PMS2 gene. 10 pathogenic variants were found in female subjects (n = 57) and 11 in male subjects (n = 27), resulting in a prevalence of pathogenic variants in female subjects of 17.5% and 40.7% in male subjects, respectively. Using χ2 the association between sex and prevalence of pathogenic variants in MMR genes is statistically relevant at a level of 0.05 (P = 0.023).
Discussion
Implications on our findings concerning HBOC
Consistant to other publications [61, 62], we can see a clear trend towards ordering larger panels for HBOC at our clinic over the last decade. A strong argument for panel testing is the fact that phenotypes of different cancer syndromes often overlap [53]. Breast cancer, for example, can be caused by a pathogenic variant in BRCA1/2 genes (HBOC syndrome), but also due to a mutation in the TP53 gene (Li-Fraumeni Syndrome). Similarly, ovarian cancer can be caused by pathogenic variants in BRCA1/2 and the MMR genes (LS). By testing multiple genes, several syndromes can be examined at once [62].
In our study, we find that the number of VUS increases while P/LP variants relatively decrease when we test a broader variety of susceptibility genes for HBOC in our population. This can be explained by the fact that in some patients screened with multigene panels, a standard BRCA test was performed first. Only if the result was negative (no P/LP mutation) the whole panel was analyzed. This leads to a biased low result in the number of pathogenic variants in multigene panels, as 0% had a pathogenic variant in BRCA1/2 in the pre-tested cases, instead of about 9–10% as in other studies [30, 33, 34]. The similar bias can be observed in the prevalence of pathogenic variants in BRCA1/2 tests. Pathogenic variants turn out to be higher than expected, possibly because many standard BRCA tests with negative results have been expanded with a multigene panel and these cases therefore show up in cohorts 2 and 3. In particular, patients who are tested with BRCA plus HBOC panel from Genetica (cases of cohort 3) are often first screened for BRCA1/2 and the panel is directly expanded by the laboratory in case of a negative result. To avoid this bias, we specified the prevalence of pathogenic variants when BRCA1/2 variant carriers were excluded from cohort 2 and 3. In this case, 16 out of 347 subjects were found to have a pathogenic variant. In a similar study from Tung et al. [33] over 1700 individuals were tested with a gene panel of 11 breast cancer susceptibility genes other than BRCA1/2. Our result is consistent with their finding that the frequency of P/LP variants in genes susceptible to breast cancer is 4.7% in BRCA1/2-negative candidates. Our data support previous studies showing that multigene panel testing improves the identification of hereditary cancer risk in patients who meet guidelines for genetic testing for HBOC [30, 33–36].
To describe the utility of multigene panels for clinical practice, not only the increase in pathogenic variants needs to be looked at, but it is also important whether the genes are considered high or moderate risk [30] and whether they are clinically accessible or not [63]—factors we did not focus on in this study. However, another important element for the evaluation of multigene panels, which is specifically addressed in our study, is the increase in VUS rates.
The VUS rate increased from 6% when only BRCA1/2 was tested to 13.8% when BRCA1/2 plus ≤ 10 genes was tested and to 31.8% when BRCA1/2 plus > 10 genes was tested, respectively. In previous studies, VUS rates have been reported to range from 6.7 to 41.7%, depending on the number of genes tested [33, 35, 64, 65]. Our results confirm this dependence between the prevalence of VUS and the number of genes tested. However, it is expected and already shown in some studies that VUS rates in less known genes will decrease over time (as they did in BRCA1/2 [39]). The reason for this development is the ever-improving variant classification, which is progressing, among other things due to the rapid accumulation of data from family tests [35, 66] and the standardization of classification in working groups like the ClinGen Expert Panels (Variant Curation Expert Panels) [67]. The increase in the rate of VUS also correlates with the ethnicity of patients undergoing genetic testing, as a recent Stanford University study of over 1400 patients clearly showed [68]. VUS rates increased in non-white subjects from 4.4% (BRCA1/2 only) to 25–38% when 13 and 28 genes were tested, respectively. In white subjects, the prevalence of VUS increased from 3.3% only to 17% and 25%, respectively. Our study did not examine the VUS rates depending on the ethnic background of the patients. Since our patient population includes predominantly white subjects, it is reasonable to assume that the VUS rate would likely be higher in a similar study with a more diverse patient population.
The increased likelihood of an inconclusive result is an important factor in deciding between targeted testing for only one gene and the use of multigene panels, as this is obviously a challenge for both patients and physicians. Although a VUS does not change the medical course of action in a patient, growing VUS rates must not only be considered as a disadvantage of increased use of NGS gene panels. Particularly in pedigrees with an extensive cancer history, such a result may contribute to increased follow-up, taking into account the medical history, and furthermore, patients can be directly informed if a VUS result later proves to be benign or pathogenic. Of course, the usefulness of multigene panels must also take into account that with larger panels not only the number of VUS increases, but also the number of pathogenic variants found, which definitely can lead to clinical consequences.
In terms of patient management of a VUS result, Culver et al. [25] evaluated differences in cancer destress and surgical decision making between patients with BRCA negative results and patients with a VUS in BRCA 1 or 2, using a two year follow-up questionnaire in US patients who underwent genetic counseling due to personal or family history of breast or ovarian cancer. They concluded, that even though genetic counseling was less reassuring for participants in the VUS cohort, the result did not cause excessive surgery or exaggerated cancer distress compared to negative results.
Implications on our findings concerning Lynch syndrome
Our results regarding LS are not very conclusive due to the small number. Nevertheless, we can give a descriptive analysis of our results and try to compare them with the existing literature. Our data show a much higher prevalence than expected, with 26.2% P/LP variants, compared with 5% in the publication by Kastrinos et al. [15]—the study on which our test criteria are based. Other than the bias due to the small number, the difference could be explained by the fact that in most cases of our study molecular testing was performed only when loss of MMR gene expression was detected in tumor tissue (exceptions were made for very young patients with a personal history strongly suggestive of LS). The number of cases tested for loss of expression was not assessed. Pathogenic variants were more common in males (40.7% of male subjects) than in females (17.5% of female subjects), suggesting that male sex is likely a predictor of variants in MMR gene analysis [15, 59]. The most frequent variants were found in the MSH2 gene (n = 11), followed by MSH6 (n = 8) and MLH1 (n = 3). Other reports tend to show higher MLH1 variant rates [69] and also databases for MMR variants list most known variants in the MLH1 gene [17, 70]. Because our results are based on only a few pathogenic variants (n = 22), it is difficult to assess whether the low MLH1 variant rate in our cohort is significant. None of our patients had a P/LP variant in the PMS2 gene—consistent with the literature that variants in this gene are the least likely of all MMR genes to cause LS [17, 71].
Our VUS rate of 6% for MLH1/MSH2/MSH6 and PMS2 matches the prevalence of VUS in MMR genes by Eggington et al. [72]. In their poster presented at the American College of Medical Genetics and Genomics Annual Meeting, they reported the observation that VUS rates have decreased over time due to reclassification of VUS, similar to observations made for BRCA1/2 genes. Evaluation of our annual percentage of VUS in the LS cohort should control for our assumption that the prevalence of VUS would not increase over the years because the gene panel was not expanded. However, this control proved to be inconclusive due to the very few VUS cases in this cohort.
Limitations/strengths
Since this is a retrospective study and the cases are from multiple departments in our clinic, it is possible that individual cases were lost and not included in the study. However, a large amount of data was available to us, which increases the significance of our results. Our clinic has the largest department for oncology in the canton of Berne and thus a large access area for patients. Nevertheless, the study is not representative for the whole canton of Berne or even Switzerland, as many other (private) clinics also perform genetic testing for cancer, and we did not perform a randomized selection of patients.
Our study is also limited because the outcomes (VUS and P/LP) were compared over the years that data were collected with the most recent versions of the databases (ClinVar [18] and others) at the time of collection. Thus, for the early cases in our study, it is possible that VUS would no longer be classified as such based on current knowledge. In a future study, it would be interesting to see whether the classification of each VUS remains the same over the years or is reclassified as pathogenic/likely pathogenic or benign/likely benign and how this affects the results on the prevalence of VUS depending on the panel used.
Another limitation is that although it was investigated which genes of the HBOC panels were most frequently mutated, no distinction was made according to the indication of the test (breast/ovarian or other cancer or familial predisposition). Thus, although it is possible to estimate which genes are frequently involved in HBOC, it is not possible to say how strongly the genes are associated with the occurrence of each tumor.
Conclusions
We found a statistically significant correlation between the rate of VUS and the increasing use of multigene panels. Our results support the use of NGS gene panels in routine diagnostics, as there is also an increase in clinically relevant variants with greater use of larger panels. With good genetic counseling before and after obtaining results, patients at risk for HBOC may benefit from larger gen panels despite increased rates of VUS.
Supplementary Information
Additional file 2: Anonymized list of identified variants.
Acknowledgements
Not applicable.
Abbreviations
- ACMG
American College of Medical Genetics and Genomics
- AMP
Association for Molecular Pathology
- ENIGMA
Evidence-based Network for the Interpretation of Germline Mutation Alleles
- HBOC
Hereditary breast and ovarian cancer
- HNPPC
Hereditary non-polyposis colorectal cancer
- IARC
International Agency for Research on Cancer
- InSiGHT
International Society for Gastrointestinal Hereditary Tumours
- LS
Lynch syndrome
- MMR
Mismatch repair
- NCCN
National Comprehensive Cancer Network
- NGS
Next generation sequencing
- P/LP
Pathogenic/likely pathogenic mutation
- SAKK
Schweizerische Arbeitsgemeinschaft für Klinische Krebsforschung
- UKMO
Universitätsklinik für Medizinische Onkologie
- VUS
Variant of unknown significance
Author contributions
FA was involved in the data analysis, performed the literature research and statistical analysis and wrote the first draft of the manuscript. MF participated in data collection and analysis. AS was involved in the editing of the manuscript. MR designed the concept and supervised the study and edited the manuscript. All authors read and approved the final manuscript.
Funding
No funding was received for this study.
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
This study was approved by the Swiss Ethics Commission: Kantonale Ethikkommission des Kanton Bern, Switzerland. Patients/Probands gave informed consent to tests as well as to use of data/material for further anonymous use including publication according to local and national guidelines. All methods were carried out in accordance with relevant guidelines and regulation (Declaration of Helsinki).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mahon S. Management of patients with a genetic variant of unknown significance. Oncol Nurs Forum. 2015;42(3):316–318. doi: 10.1188/15.ONF.316-318. [DOI] [PubMed] [Google Scholar]
- 2.Domchek SM, Bradbury A, Garber JE, Offit K, Robson ME. Multiplex genetic testing for cancer susceptibility: Out on the high wire without a net? J Clin Oncol. 2013;31(10):1267–1270. doi: 10.1200/JCO.2012.46.9403. [DOI] [PubMed] [Google Scholar]
- 3.Lindor NM, McMaster ML, Lindor CJ, Greene MH. Concise handbook of familial cancer susceptibility syndromes. JNCI Monogr. 2008;2008(38):3–93. doi: 10.1093/jncimonographs/lgn001. [DOI] [PubMed] [Google Scholar]
- 4.Genetics, genomics, and cancer risk assessment—Weitzel—2011—CA: A Cancer Journal for Clinicians - Wiley Online Library. Accessed March 21, 2022. https://acsjournals.onlinelibrary.wiley.com/doi/full/10.3322/caac.20128.
- 5.Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266(5182):66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 6.Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378(6559):789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 7.Futreal PA, Liu Q, Shattuck-Eidens D, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266(5182):120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 8.Collins FS. BRCA1—lots of mutations, lots of dilemmas. N Engl J Med. 1996;334(3):186–188. doi: 10.1056/NEJM199601183340311. [DOI] [PubMed] [Google Scholar]
- 9.Plon SE, Eccles DM, Easton D, et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29(11):1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh T, Lee MK, Casadei S, et al. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci. 2010;107(28):12629–12633. doi: 10.1073/pnas.1007983107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines). Genetic/familial high risk assessment: Breast, Ovarian, and Pancreatic. Version 2.2022. NCCN. Published March 9, 2022. Accessed April 4, 2022. https://www.nccn.org/guidelines/guidelines-detail.
- 12.NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines). Genetic/familial high risk assessment: Colorectal. Version 1.2021. NCCN. Published May 1, 2021. Accessed April 4, 2022. https://www.nccn.org/guidelines/guidelines-detail
- 13.Chappuis PO, Bolliger B, Bürki N, Buser K, Heini K. Genetic predisposition to breast and ovarian cancer. :6.
- 14.Stoll S, Unger S, Azzarello-Burri S, et al. Update Swiss guideline for counselling and testing for predisposition to breast, ovarian, pancreatic and prostate cancer: Swiss Group for Clinical Cancer Research (SAKK) N for CPT and C (CPTC), ed. Swiss Med Wkly. 2021 doi: 10.4414/smw.2021.w30038. [DOI] [PubMed] [Google Scholar]
- 15.Kastrinos F, Uno H, Ukaegbu C, et al. Development and validation of the PREMM5 model for comprehensive risk assessment of lynch syndrome. J Clin Oncol. 2017;35(19):2165–2172. doi: 10.1200/JCO.2016.69.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spurdle AB, Greville-Heygate S, Antoniou AC, et al. Towards controlled terminology for reporting germline cancer susceptibility variants: an ENIGMA report. J Med Genet. 2019;56(6):347–357. doi: 10.1136/jmedgenet-2018-105872. [DOI] [PubMed] [Google Scholar]
- 17.Thompson BA, Spurdle AB, Plazzer JP, et al. Application of a five-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants lodged on the InSiGHT locus-specific database. Nat Genet. 2014;46(2):107–115. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ClinVar - ClinGen | Clinical Genome Resource. Accessed October 30, 2022. https://www.clinicalgenome.org/data-sharing/clinvar/.
- 19.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Easton DF, Deffenbaugh AM, Pruss D, et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007;81(5):873–883. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boonen RACM, Wiegant WW, Celosse N, et al. Functional analysis identifies damaging CHEK2 missense variants associated with increased cancer risk. Cancer Res. 2022;82(4):615–631. doi: 10.1158/0008-5472.CAN-21-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shuen AY, Lanni S, Panigrahi GB, et al. Functional repair assay for the diagnosis of constitutional mismatch repair deficiency from non-neoplastic tissue. J Clin Oncol. 2019;37(6):461–470. doi: 10.1200/JCO.18.00474. [DOI] [PubMed] [Google Scholar]
- 23.Plon SE, Cooper HP, Parks B, et al. Genetic testing and cancer risk management recommendations by physicians for at-risk relatives. Genet Med. 2011;13(2):148–154. doi: 10.1097/GIM.0b013e318207f564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richter S, Haroun I, Graham TC, Eisen A, Kiss A, Warner E. Variants of unknown significance in BRCA testing: impact on risk perception, worry, prevention and counseling. Ann Oncol. 2013;24:viii69–viii74. doi: 10.1093/annonc/mdt312. [DOI] [PubMed] [Google Scholar]
- 25.Culver J, Brinkerhoff C, Clague J, et al. Variants of uncertain significance in BRCA testing: evaluation of surgical decisions, risk perception, and cancer distress. Clin Genet. 2013;84(5):464–472. doi: 10.1111/cge.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray ML, Cerrato F, Bennett RL, Jarvik GP. Follow-up of carriers of BRCA1 and BRCA2 variants of unknown significance: variant reclassification and surgical decisions. Genet Med. 2011;13(12):998–1005. doi: 10.1097/GIM.0b013e318226fc15. [DOI] [PubMed] [Google Scholar]
- 27.Kuchenbaecker KB, Hopper JL, Barnes DR, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317(23):2402. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 28.Kotsopoulos J. BRCA mutations and breast cancer prevention. Cancers. 2018;10(12):524. doi: 10.3390/cancers10120524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Easton DF. How many more breast cancer predisposition genes are there? Breast Cancer Res. 1999;1(1):14. doi: 10.1186/bcr6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurian AW, Hare EE, Mills MA, et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol. 2014;32(19):2001–2009. doi: 10.1200/JCO.2013.53.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh T, Casadei S, Lee MK, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci. 2011;108(44):18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norquist BM, Harrell MI, Brady MF, et al. Inherited mutations in women with ovarian carcinoma. JAMA Oncol. 2016;2(4):482–490. doi: 10.1001/jamaoncol.2015.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tung N, Battelli C, Allen B, et al. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer. 2015;121(1):25–33. doi: 10.1002/cncr.29010. [DOI] [PubMed] [Google Scholar]
- 34.Lincoln SE, Kobayashi Y, Anderson MJ, et al. A systematic comparison of traditional and multigene panel testing for hereditary breast and ovarian cancer genes in more than 1000 patients. J Mol Diagn. 2015;17(5):533–544. doi: 10.1016/j.jmoldx.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 35.LaDuca H, Stuenkel AJ, Dolinsky JS, et al. Utilization of multigene panels in hereditary cancer predisposition testing: analysis of more than 2,000 patients. Genet Med. 2014;16(11):830–837. doi: 10.1038/gim.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maxwell KN, Wubbenhorst B, D’Andrea K, et al. Prevalence of mutations in a panel of breast cancer susceptibility genes in BRCA1/2-negative patients with early-onset breast cancer. Genet Med. 2015;17(8):630–638. doi: 10.1038/gim.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frank TS, Deffenbaugh AM, Reid JE, et al. Clinical Characteristics of Individuals With Germline Mutations in BRCA1 and BRCA2: Analysis of 10,000 Individuals. 11. [DOI] [PubMed]
- 38.Saam J, Burbidge L, Bowles K. Decline in rate of BRCA1/2 variants of uncertain significance: 2002–2008. Poster presented at: 27th Annual Education Conference of the National Society of Genetic Counselors; 2008; Los Angeles.
- 39.Eggington J, Bowles K, Moyes K, et al. A comprehensive laboratory-based program for classification of variants of uncertain significance in hereditary cancer genes. Clin Genet. 2014;86(3):229–237. doi: 10.1111/cge.12315. [DOI] [PubMed] [Google Scholar]
- 40.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348(10):919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 41.Aaltonen LA, Salovaara R, Kristo P, et al. Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338(21):1481–1487. doi: 10.1056/NEJM199805213382101. [DOI] [PubMed] [Google Scholar]
- 42.Samowitz WS, Curtin K, Lin HH, et al. The colon cancer burden of genetically defined hereditary nonpolyposis colon cancer. Gastroenterology. 2001;121(4):830–838. doi: 10.1053/gast.2001.27996. [DOI] [PubMed] [Google Scholar]
- 43.Ligtenberg MJL, Kuiper RP, Chan TL, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet. 2009;41(1):112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 44.Vasen HFA, Blanco I, Aktan-Collan K, et al. Revised guidelines for the clinical management of Lynch syndrome (HNPCC): recommendations by a group of European experts. Gut. 2013;62(6):812–823. doi: 10.1136/gutjnl-2012-304356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Win AK, Young JP, Lindor NM, et al. Colorectal and other cancer risks for carriers and noncarriers from families with a DNA mismatch repair gene mutation: a prospective cohort study. J Clin Oncol. 2012;30(9):958–964. doi: 10.1200/JCO.2011.39.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watson P, Lynch HT. The tumor spectrum in HNPCC. Anticancer Res. 1994;14(4B):1635–1639. [PubMed] [Google Scholar]
- 47.Aarnio M, Sankila R, Pukkala E, et al. Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int J Cancer. 1999;81(2):214–218. doi: 10.1002/(sici)1097-0215(19990412)81:2<214::aid-ijc8>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 48.Dominguez-Valentin M, Sampson JR, Seppälä TT, et al. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: findings from the Prospective Lynch Syndrome Database. Genet Med. 2020;22(1):15–25. doi: 10.1038/s41436-019-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monahan KJ, Bradshaw N, Dolwani S, et al. Guidelines for the management of hereditary colorectal cancer from the British Society of Gastroenterology (BSG)/Association of Coloproctology of Great Britain and Ireland (ACPGBI)/United Kingdom Cancer Genetics Group (UKCGG) Gut. 2020;69(3):411–444. doi: 10.1136/gutjnl-2019-319915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Genpanel Diagnostik – Genetica AG. Accessed March 14, 2022. https://genetica-ag.ch/genpanel-diagnostik/.
- 51.DGVademecum. Accessed March 14, 2022. https://dgvademecum.ksa.ch/.
- 52.Robson ME, Bradbury AR, Arun B, et al. American society of clinical oncology policy statement update: genetic and genomic testing for cancer susceptibility. J Clin Oncol. 2015;33(31):3660–3667. doi: 10.1200/JCO.2015.63.0996. [DOI] [PubMed] [Google Scholar]
- 53.Liste der Analysen - Spital Wallis. Accessed April 6, 2022. https://www.spitalwallis.ch/gesundheitsfachpersonal/zentralinstitut-der-spitaeler/laboratorien/fachbereiche/genetik/liste-der-analysen.
- 54.Liste des panels de gènes. Accessed April 6, 2022. https://www.hug.ch/medecine-genetique/liste-panels-genes.
- 55.Universitätsspital Basel. Auftrag Lynch-Syndrom. Accessed March 14, 2022. https://www.unispital-basel.ch/fileadmin/unispitalbaselch/Lehre_Forschung/Dep_Biomedizin/Medizinische_Genetik/Labordiagnostik/Auftragsformulare/Auftrag_Lynch-Syndrom_HNPCC_18_11_2015.pdf.
- 56.SAS SAS. Swiss Accreditation Service SAS. Accessed October 30, 2022. https://www.sas.admin.ch/sas/en/home.html.
- 57.Daly MJ. gnomAD Genome Aggregation Database. Accessed October 30, 2022. https://gnomad.broadinstitute.org/.
- 58.Ioannidis NM, Rothstein JH, Pejaver V, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99(4):877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kastrinos F, Steyerberg EW, Mercado R, et al. The PREMM(1,2,6) model predicts risk of MLH1, MSH2, and MSH6 germline mutations based on cancer history. Gastroenterology. 2011;140(1):73–81. doi: 10.1053/j.gastro.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Switzerland Population and Demographics from Switzerland | - CountryReports. Accessed October 30, 2022. https://www.countryreports.org/country/Switzerland/population.htm.
- 61.O’Leary E, Iacoboni D, Holle J, et al. Expanded gene panel use for women with breast cancer: identification and intervention beyond breast cancer risk. Ann Surg Oncol. 2017;24(10):3060–3066. doi: 10.1245/s10434-017-5963-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hall MJ, Forman AD, Pilarski R, Wiesner G, Giri VN. Gene panel testing for inherited cancer risk. J Natl Compr Canc Netw. 2014;12(9):1339–1346. doi: 10.6004/jnccn.2014.0128. [DOI] [PubMed] [Google Scholar]
- 63.Desmond A, Kurian AW, Gabree M, et al. Clinical actionability of multigene panel testing for hereditary breast and ovarian cancer risk assessment. JAMA Oncol. 2015;1(7):943–951. doi: 10.1001/jamaoncol.2015.2690. [DOI] [PubMed] [Google Scholar]
- 64.Kapoor NS, Curcio LD, Blakemore CA, et al. Multigene panel testing detects equal rates of pathogenic BRCA1/2 mutations and has a higher diagnostic yield compared to limited BRCA1/2 analysis alone in patients at risk for hereditary breast cancer. Ann Surg Oncol. 2015;22(10):3282–3288. doi: 10.1245/s10434-015-4754-2. [DOI] [PubMed] [Google Scholar]
- 65.Doherty J, Bonadies DC, Matloff ET. Testing for hereditary breast cancer: panel or targeted testing? Experience from a clinical cancer genetics practice. J Genet Couns. 2015;24(4):683–687. doi: 10.1007/s10897-014-9796-2. [DOI] [PubMed] [Google Scholar]
- 66.McVean GA, Altshuler (Co-Chair) DM, Durbin (Co-Chair) RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed]
- 67.Kanavy DM, McNulty SM, Jairath MK, et al. Comparative analysis of functional assay evidence use by ClinGen Variant Curation Expert Panels. Genome Med. 2019;11(1):77. doi: 10.1186/s13073-019-0683-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caswell-Jin JL, Gupta T, Hall E, et al. Racial/ethnic differences in multiple-gene sequencing results for hereditary cancer risk. Genet Med. 2018;20(2):234–239. doi: 10.1038/gim.2017.96. [DOI] [PubMed] [Google Scholar]
- 69.Niessen RC, Berends MJW, Wu Y, et al. Identification of mismatch repair gene mutations in young patients with colorectal cancer and in patients with multiple tumours associated with hereditary non-polyposis colorectal cancer. Gut. 2006;55(12):1781–1788. doi: 10.1136/gut.2005.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Woods MO, Williams P, Careen A, et al. A new variant database for mismatch repair genes associated with Lynch syndrome. Hum Mutat. 2007;28(7):669–673. doi: 10.1002/humu.20502. [DOI] [PubMed] [Google Scholar]
- 71.Niessen RC, Kleibeuker JH, Westers H, et al. PMS2 involvement in patients suspected of Lynch syndrome. Genes Chromosomes Cancer. 2009;48(4):322–329. doi: 10.1002/gcc.20642. [DOI] [PubMed] [Google Scholar]
- 72.Eggington JM, Burbidge LA, Roa B, et al. Current Variant of Uncertain Significance Rates in BRCA1/2 and Lynch Syndrome Testing. Poster presented at: American College of Medical Genetics and Genomics Annual Meeting; March 2012; Myriad Genetic Laboratories, Inc., 320 Wakara Way, Salt Lake City, Utah 84108, USA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 2: Anonymized list of identified variants.
Data Availability Statement
The datasets used during the current study are available from the corresponding author on reasonable request.