Abstract
In toxigenic Vibrio cholerae, the cholera enterotoxin (CT) is encoded by CTXΦ, a lysogenic bacteriophage. The propagation of this filamentous phage can result in the origination of new toxigenic strains. To understand the nature of possible environmental factors associated with the propagation of CTXΦ, we examined the effects of temperature, pH, salinity, and exposure to direct sunlight on the induction of the CTX prophage and studied the transmission of the phage to potential recipient strains. Exposure of cultures of CTXΦ lysogens to direct sunlight resulted in ∼10,000-fold increases in phage titers. Variation in temperature, pH, or salinity of the culture did not have a substantial effect on the induction of the prophage, but these factors influenced the stability of CTXΦ particles. Exposure of mixed cultures of CTXΦ lysogens and potential recipient strains to sunlight significantly increased both the in vitro and in vivo (in rabbit ileal loops) transduction of the recipient strains by CTXΦ. Included in these transduction experiments were two environmental nontoxigenic (CTXΦ−) strains of V. cholerae O139. These two O139 strains were transduced at high efficiency by CTXΦ, and the phage genome integrated into the O139 host chromosome. The resulting CTXΦ lysogens produced biologically active CT both in vitro and in rabbit ileal loops. This finding suggests a possible mechanism explaining the origination of toxigenic V. cholerae O139 strains from nontoxigenic progenitors. This study indicates that sunlight is a significant inducer of the CTX prophage and suggests that sunlight-induced transmission of CTXΦ may constitute part of a natural mechanism for the origination of new toxigenic strains of V. cholerae.
Seasonal outbreaks of cholera caused by toxigenic Vibrio cholerae strains are a major public health problem in many developing countries. Toxigenic V. cholerae O1 and O139 strains cause disease by colonizing the human small intestine, where they produce a potent enterotoxin, the cholera toxin (CT), which is principally responsible for the severe watery diarrhea characteristic of cholera (5, 23). The ctxAB operon, which encodes the A and B subunits of CT, is part of a larger genetic element originally termed the CTX genetic element (22). Recent studies have shown that the CTX genetic element corresponds to the genome of CTXΦ, a lysogenic filamentous bacteriophage (28). It has been previously demonstrated that naturally occurring strains of toxigenic V. cholerae produce high titers of the phage following exposure to the DNA-damaging agent mitomycin C, and cell-free phage particles can infect and convert susceptible nontoxigenic V. cholerae strains into toxigenic strains (6, 7). Similar to other temperate phages, the CTX prophage can also be induced by exposure to UV irradiation under laboratory conditions. However, no studies have been conducted so far to identify possible natural factors associated with the induction and propagation of CTXΦ.
Although toxigenic V. cholerae bacteria are human pathogens, the species can persist in the aquatic environment in the absence of human hosts. Therefore, the ecology of V. cholerae appears to involve both environmental and human host components (2, 5). During survival in the aquatic environment, the physiological state of V. cholerae is not well understood but is presumably influenced by various parameters, including sunlight, temperature, pH, and salinity (19, 25). It is not clear whether these factors can also result in the induction of CTX prophages in toxigenic V. cholerae and hence promote the transmission of CTXΦ to nontoxigenic environmental strains, leading to the origination of new toxigenic strains. In the present study, we examined the effects of temperature, pH, salinity, and exposure to direct sunlight on the induction, stability, and propagation of CTXΦ.
Naturally occurring toxigenic V. cholerae strains used in the present study included 32 strains belonging to the O1, O139, and non-O1 non-O139 serogroups, and the strains were isolated from cholera patients or environmental surface water in Bangladesh (see Table 3). The nontoxigenic V. cholerae strains used in the transduction studies or as controls included two O139 strains and three El Tor strains isolated in Bangladesh or India. The genetically marked phage IG-KmΦ was a derivative of CTXΦ into which a kanamycin resistance (Kmr) determinant was introduced into an intergenic NotI site (16). Strain ASF-1 carried a chromosomally integrated copy of the IG-KmΦ genome and was constructed by lysogenic conversion of CT-negative V. cholerae O1 El Tor strain SA-317 with IG-KmΦ. Strain SM44 was a derivative of toxigenic El Tor strain P27459 in which the CTX element was marked with a Kmr determinant (10). The genetically marked phage CTX-KmΦ was derived from strain SM44 as described previously (6, 7, 28). Properties of the control bacterial strains and phages used in this study are presented in Table 1.
TABLE 3.
Analysis of naturally occurring toxigenic V. cholerae strains of environmental or clinical origin for sunlight-induced production of extracellular CTXΦ
| Serogroup and/or biotype | Yr of isolation | Source | No. of strains analyzed | No. of strains producing extracellular CTXΦ
|
|
|---|---|---|---|---|---|
| Induced by sunlight | Without induction | ||||
| O1, El Tor | 1995–1998 | Patient | 9 | 8 | 1 |
| 1998–1999 | Surface water | 3 | 3 | 0 | |
| O139 | 1995–1998 | Patient | 11 | 9 | 1 |
| 1995–1999 | Surface water | 4 | 4 | 0 | |
| Non-O1, non-O139 | 1995–1998 | Surface water | 5 | 1 | 0 |
TABLE 1.
Characteristics of control strains and phages used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| SM44 | Derivative of El Tor strain P27459, in which the CTX genetic element was marked with a Kmr determinant by marker exchange disrupting the ctxAB operon | 10 |
| RV508 | Derivative of classical biotype strain 569B, which is known to constitutively express CT, TCP, and other toxR-regulated gene products | 28 |
| 569B | V. cholerae O1 classical Inaba strain | Laboratory collection |
| P27457 | V. cholerae O1 El Tor Inaba strain | Laboratory collection |
| SA-317, SA-406 | Nontoxigenic (CTXΦ−) V. cholerae O1 El Tor biotype strains isolated in India | 7 |
| O395 | Classical Ogawa streptomycin-resistant strain | Laboratory collection |
| AE-2883 | Tetracycline-resistant V. cholerae O1 classical biotype strain isolated in Bangladesh | Laboratory collection |
| O395(pCTX-Km) | Strain O395 carrying the RF of CTX-KmΦ derived from strain SM44 | 28 |
| pIG-Km | Derivative of the RF DNA of wild-type CTXΦ carrying an intergenic Kmr marker | 16 |
| ASF-1 | Strain SA-317 lysogenized with IG-KmΦ | This study |
| AOE12-39, Env-99 | Environmental nontoxigenic (CTXΦ−) V. cholerae O139 strains isolated in Bangladesh | Laboratory collection |
The gene probe used in this study to detect the CTXΦ genome was a 0.5-kb EcoRI fragment of pCVD27 (6, 12). Strand-specific oligonucleotide probes with the sequences 5′TCTATCTCTGTAGCCCCTATTACG and 5′CTCAGACGGGATTTGTTAGGCACG, for probing the plus and minus strands, respectively, were also used to detect the presence of single-stranded DNA of CTXΦ and to distinguish between the replicative-form (RF) double-stranded DNA and single-stranded phage DNA in crude preparations. The O139-specific probe was a 1.3-kb EcoRI fragment of pCRII-A3 (21, 29). The nontoxigenic O139 strains were also tested for the presence of genes encoding toxin-coregulated pilus (TCP), which is the receptor for CTXΦ, the virulence regulatory gene toxR, and the CTXΦ attachment sequence attRS. The presence of the TCP pathogenicity island was determined by using PCR assays specific for the tcpA, tcpI, and acfB genes, as described previously (7, 14). The toxR gene probe was a 2.4-kb BamHI fragment of pVM7 (20), and the 18-bp attRS sequence was identified using a synthetic oligonucleotide corresponding to the attRS sequence (10). Colony blots or Southern blots were prepared using nylon filters (Hybond; Amersham International plc, Aylesbury, United Kingdom) and processed by standard methods (18). The polynucleotide probes were labeled by random priming (9) using a random primer DNA labeling kit (Bethesda Research Laboratories, Gaithersburg, Md.) and [α-32P]dCTP (3,000 Ci/mmol; Amersham). Oligonucleotide probes were labeled by 3′ tailing using terminal deoxynucleotide transferase and [α-32P]dCTP (Amersham). Southern blots and colony blots were hybridized with the labeled probes and autoradiographed as described previously (6, 7).
All strains tested for induction by sunlight were grown in Luria broth (LB) at 37°C to an absorbance at 540 nm (A540) of 0.2. The cells were collected by centrifugation, washed, and resuspended in fresh LB. Five milliliters of the suspension, containing approximately 5 × 104 bacterial cells, was spread on a sterile petri dish and exposed to direct sunlight for a specified time (30 to 45 min). The intensity of the light was measured using a digital light meter (VWR Scientific, South Plainfield, N.J.). Control plates were kept under normal light in a shaded area during the same time period. The treated cells were inoculated into test flasks containing 50 ml of fresh LB. All flasks were incubated at 37°C with shaking, and aliquots of culture were periodically removed and analyzed for the presence of extracellular CTXΦ using previously described methods (6, 7). Briefly, to detect Kmr determinant-labeled CTXΦ particles derived from strains SM44 and ASF-1, the culture supernatant was sterilized by filtration through 0.22-μm-pore-size filters (Millipore Corp., Bedford, Mass.), and the filtrate was titrated for infectious phage particles by incubating aliquots of the supernatant with the classical strain RV508 and then selecting for colonies resistant to kanamycin (50 μg/ml). To detect the induction of the CTX prophage in wild-type strains, preparations of the culture supernatants containing CTXΦ DNA were identified by Southern blot hybridization using specific probes, as described previously (6). Aliquots of LB mixed with serial dilutions of IG-KmΦ (10 to 104 particles/ml) isolated from supernatants of O395(pIG-Km) were used as positive controls to test the detection limit of the hybridization assay. An aliquot of the culture obtained for the phage assay was also diluted and plated to determine the number of viable cells.
To study the effects of pH, NaCl concentration (salinity), and temperature on the induction of CTX prophage, a series of test flasks containing LB was adjusted to different pHs (6.5, 7.5, 8.0, and 8.5), salinity levels (0.5 to 2.0%, in increments of 0.5%), or combinations of different pHs and salinities. Series of flasks containing 50 ml of LB with defined pH and salinity were inoculated with 5 × 104 cells of strain SM44 and incubated at four different temperatures, 25, 30, 37, or 40°C, with shaking. The induction of the prophage was monitored by periodically examining aliquots of culture supernatants for the presence of CTX-KmΦ during the following 12 h, as described above.
To examine the stability of CTXΦ under different conditions of temperature, pH, and salinity, CTX-KmΦ isolated from a culture of O395(pCTX-Km) was used. Approximately 5 × 106 phage particles were inoculated into a series of test tubes containing 5 ml of LB adjusted to a predefined pH and salinity. Control tubes containing normal saline or distilled water were also inoculated. The tubes were stored at different temperatures, and aliquots of the medium (100 μl) were periodically removed and analyzed for remaining infectious CTX-KmΦ particles. The Kmr colonies were counted and stability of the phage was expressed as the percentage of initial inoculum of infectious phage particles. Initially, stability was examined at room temperature at three different pHs (6.0, 7.0, and 8.0) and salinity levels (0.05, 0.5, and 1.0%), as well as at combinations of these two parameters. Later, more elaborate examinations of the effect of pH (2.0 to 12.0) at a salinity of 0.5% and that of salinity (0.05 to 1.5%) at pH 8.0 on the stability of CTXΦ were performed. The effect of temperature (4, 25, 30, 37, 40, and 45°C) on the stability of CTXΦ was examined at pH 8.0 and 0.5% salinity.
Transduction of recipient strains by CTXΦ was assayed using two different donor strains, SM44 and ASF-1, and three different recipient strains, including a tetracycline-resistant classical biotype strain, AE-2883, and two nontoxigenic O139 strains, AOE12-39 and Env-99 (Table 1). The donor and recipient strains were grown separately, washed, and resuspended in fresh LB (for the classical strain) or AKI medium (for the O139 strains). Approximately 2.5 × 104 cells of the donor or the recipient strain were mixed in 5 ml of LB or AKI medium and exposed to direct sunlight (∼42,000 lx) for 30 min. For the in vitro assay, 1 ml of the exposed cell suspension was inoculated into 50 ml of the same medium and incubated at 30°C for 16 h. Aliquots of the culture were analyzed for transduction of the recipient strain by using either appropriate antibiotics or DNA probes. In order to detect transduction of recipient strain AE-2883, dilutions of the culture were plated on Luria agar plates containing kanamycin (50 μg/ml) and tetracycline (20 μg/ml). To detect the transduction of the nontoxigenic O139 strains, dilutions of the mixed culture were grown on kanamycin plates, and colony blots prepared from the plates were hybridized with the O139-specific DNA probe to detect Kmr O139 derivatives.
For the in vivo assays, 1-ml aliquots of the sunlight-treated cells were inoculated into the ileal loops of adult New Zealand White rabbits obtained from the breeding facility of the Animal Resources Branch of the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B), and prepared as described previously (4). After 16 h, the rabbits were sacrificed and contents of the ileal loops were collected. The inside of the loops was washed out with 1 ml of 10 mM phosphate-buffered saline (pH 7.4) and collected in the same tube. Dilutions of the ileal loop fluids were analyzed by plating on appropriate antibiotic plates and using DNA probes. The ratio of Kmr-transduced colonies to total number of colonies derived from the recipient strain was calculated and expressed as the percentage of recipient cells carrying the phage genome.
Representative colonies were picked, grown in LB containing kanamycin (50 μg/ml), and further analyzed for the presence of the phage genome. Total DNA or plasmid DNA was extracted from overnight cultures by standard methods (18) and purified using microcentrifuge filter units (Ultrafree-Probind; Sigma). Integration of the phage genome into the chromosome of the recipient cells was studied by comparative Southern blot analysis of total DNA and plasmid preparations from infected and native strains (7). The ability of the CTXΦ lysogens derived from parental nontoxigenic O139 strains to produce CT was determined by the GM1 ganglioside-dependent enzyme-linked immunosorbent assay and the rabbit ileal loop assay, as described previously (4, 24).
Induction of CTX prophage by sunlight.
Two strains, SM44 and ASF-1, harboring Kmr determinant-marked CTX prophages were initially used to study parameters influencing prophage induction. CTXΦ does not form plaques, and genetically marked prophages allow quantification of the number of phage particles secreted in supernatants via transduction assays (6, 7, 28). Initially, a large number of individual colonies of strains SM44 and ASF-1 were picked and tested separately for the production of Kmr-transducing particles. Most colonies tested either were negative or produced a very small number of phage particles (between 5 and 127 particles per ml) when grown without artificial induction. The number of spontaneous CTXΦ-transducing particles produced by SM44 or ASF-1 was significantly lower than the titers previously reported for two other CTXΦ lysogens (15). Presumably, this reflects strain differences.
To examine the effect of sunlight on the induction of CTX prophages in SM44 and ASF-1, we selected colonies that spontaneously produced a small number of phage particles (<50/ml). No significant difference in the CTX-KmΦ transducing activity of the culture supernatants was noted when the pH, salinity, or temperature of the cultures was varied. However, when the cultures were exposed to direct sunlight (mean intensity of 42,000 lx) for 30 min prior to incubation, the number of phage particles increased more than 10,000-fold (Table 2). No increase was noted in control cultures which were stored in a well-lighted area (average of 3,180 lx) but away from direct sunshine. This showed that direct sunlight is a significant natural inducer of the CTX prophage. Examination of the production of phage particles in sunlight-induced cultures at different stages of cell growth showed a steady increase of phage particles for nearly 6 h. This was followed by a sharp decline which corresponded to the stationary phase of the culture (Fig. 1). These findings are in agreement with a previous report (15) that infectious CTXΦ particles are rapidly inactivated during the stationary phase of a culture by a secreted CTXΦ-destroying factor identified as the hemagglutinin protease of V. cholerae. We confirmed that the rapid decline in CTXΦ titers in stationary-phase cultures of SM44 and ASF-1 was secondary to CTXΦ-destroying factor activity (data not shown).
TABLE 2.
Sunlight-induced production of extracellular CTXΦ particles by genetically marked strains of V. cholerae carrying a kanamycin-resistant determinant in the CTXΦ genome
| Strain | Culture conditionsa | Titer of CTXΦ particles/mlb |
|---|---|---|
| SM44 | Without exposure to direct sunlight | 0.3 × 102 |
| After exposure to direct sunlight | 4.2 × 105 | |
| ASF-1 | Without exposure to direct sunlight | 0.1 × 102 |
| After exposure to direct sunlight | 3.7 × 105 | |
| SA-317 | Without exposure to direct sunlight | 0 |
| After exposure to direct sunlight | 0 | |
| SA-406(pCTX-Km) | Without exposure to direct sunlight | 8.1 × 104 |
| After exposure to direct sunlight | 9.7 × 104 |
Cells were exposed to direct sunlight for 30 min prior to incubation, and supernatants of cultures were analyzed after incubation for 5 h at 30°C.
Median value of five observations.
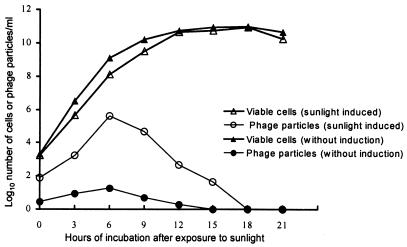
FIG. 1.
Kinetics of cell growth and production of extracellular CTXΦ particles by strain SM44 induced with exposure to direct sunlight (42,000 lx) for 30 min. Values are the averages of five different observations.
CTXΦ was detectable in culture supernatants from 25 of 32 wild-type (nonmarked) strains induced with sunlight (Table 3) by Southern blot hybridization using a plus strand-specific oligonucleotide probe (Fig. 2). The bands corresponding to the single-stranded phage DNA were absent in 30 of 32 preparations derived from supernatants of wild-type strains grown without sunlight induction. In control assays, approximately 2 × 102 transducing particles per ml produced a visible band. Thus, some of these wild-type strains may have produced a small number of phage particles during normal growth that was below this level of detection.
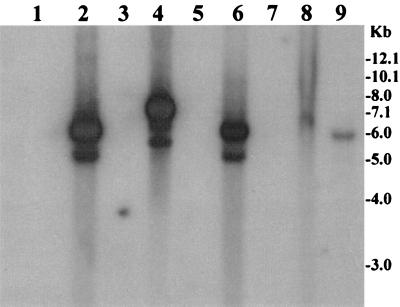
FIG. 2.
Supernatant fluids of toxigenic V. cholerae strains, exposed to direct sunlight (42,000 lx) for 30 min and subsequently grown for 5 h, were sterilized by filtration through a 0.22-μm-pore-size filter and were used to precipitate possible bacteriophage particles. The precipitates were dissolved in appropriate buffer and treated with DNase I and RNase A to remove contaminating exogenous DNA or RNA. Total phage nucleic acids were isolated as described in the text and analyzed using the ctxA probe. Lanes 1, 3, 5, and 7 contain phage DNA preparations from four toxigenic strains grown without exposure to sunlight, whereas lanes 2, 4, 6, and 8 contain phage DNA preparations derived from the corresponding sunlight-induced cultures. Lane 9 contains phage DNA preparations from approximately 3 × 102 IG-KmΦ particles added to LB and used as a control. Numbers indicating molecular sizes of bands correspond to a supercoiled DNA ladder (Bethesda Research Laboratories).
The duration of sunlight exposure as well as the intensity of sunlight had a significant effect on both phage titer and viable cell number. The effect of sunlight was roughly proportional to the product of light intensity and duration of exposure. Exposure to sunlight resulted in cell death, but phage production by the surviving cells was increased. For example, immediately after exposure to sunlight at an average intensity of 42,000 lx for 30 min, the viable cell count was reduced by approximately 6%, and the total phage production by the surviving cells increased more than 20-fold (Fig. 1).
The CTXΦ genome is comprised of a core region and an RS2 region (30). With the exception of ctxAB, the genes of the core region are thought to be essential for morphogenesis of CTXΦ particles and hence for its propagation as an infectious phage. The open reading frames in RS2, rstR, rstA, and rstB, encode a repressor (RstR) as well as products required for the replication and integration of CTXΦ (30). In lysogens, RstR is thought to repress transcription of rstA, a gene required for CTXΦ replication. Our data suggest that RstR repressor function is probably inactivated by direct sunlight. Inactivation of RstR presumably leads to the expression of rstA and thereby allows the CTX prophage to enter into a replicative pathway. To begin to address this possibility, we examined the sunlight-exposed cells for the presence of CTXΦ RF DNA. This was accomplished by hybridizing plasmid preparations with a minus strand-specific oligonucleotide probe which would not hybridize with the plus strand found in the CTXΦ genome. The CTXΦ RF DNA was detectable in Southern blots of plasmids prepared from sunlight-exposed cells and not from unexposed cells (data not shown). This suggests that the sunlight-induced increases in phage production in strains carrying the CTX prophage were preceded by and mediated by production of the RF DNA of CTXΦ. This is further supported by the observation that phage titers in the culture supernatants of strain SA-406(pCTX-Km), which carried the RF of the phage genome, did not increase substantially after the culture was exposed to sunlight (Table 2). High titers of the phage were detected in the supernatants from cultures of this strain, irrespective of whether the cells were exposed to sunlight or kept in the shade. However, further studies are required to investigate the detailed molecular mechanisms associated with sunlight-mediated induction of the CTX prophage.
Stability of CTXΦ.
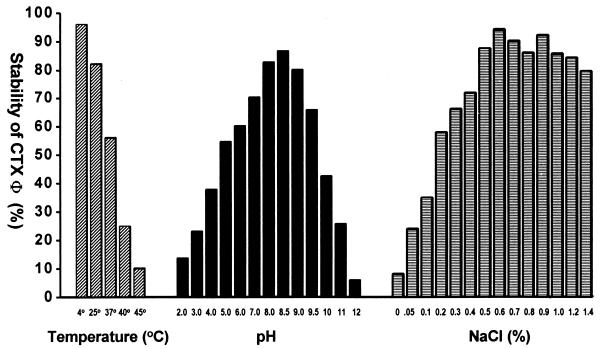
The effects of temperature, pH, and salinity on the stability of CTX-KmΦ were assessed by adding a defined number of phage particles into medium adjusted to different conditions of salinity and pH or into normal saline. The titers of CTX-KmΦ transducing particles remaining after 12 h, expressed as a percentage of the original titer, are shown in Fig. 3. Phage particles remained infectious for more than 4 weeks when stored at room temperature in normal saline or in LB containing a minimum of 0.5% NaCl. The transducing phages were fairly stable over a pH range between 4.0 and 10.0 (Fig. 3). At temperatures above 37°C or at a salinity below 0.1%, the majority of the transducing particles were inactivated. These findings suggest that phage particles may persist in the aquatic environment as infectious agents, depending on these and possibly other parameters.
FIG. 3.
Effects of temperature, pH, and salinity on the stability of CTXΦ, assayed after 12 h of incubation in LB adjusted to different pHs or salinity levels, using the genetically marked phage CTX-KmΦ (see text for details). The effect of salinity was assayed at pH 8.0 and that of pH was assayed at a salinity of 0.5% at room temperature. The effect of temperature was measured at pH 8.0 and a salinity of 0.5%. Values are the averages of three independent observations.
Transmission of CTXΦ.
We examined the transfer of CTXΦ between strains when mixed cultures of potential donor and recipient strains were exposed to sunlight. Transduction of the Kmr determinant-marked CTX prophages carried by strains SM44 and ASF-1 was monitored by transfer of kanamycin resistance to the recipient strains. One recipient, AE-2883, could be differentiated from the donor strains by its resistance to tetracycline, whereas the other two strains were of the O139 serogroup and could be differentiated from the donor strains by using an O139-specific probe. Transduction was assayed both under in vitro laboratory conditions and in vivo inside the ileal loops of adult rabbits. Previous investigators have used infant mice to study in vivo transfer of CTXΦ, but we found that the rabbit ileal loop model is also useful for in vivo CTXΦ transduction assays. Sunlight exposure significantly increased in vitro transduction of the recipient strains. When the mixed cultures were exposed to sunlight, the number of transductants was more than 20-fold higher than the number obtained in the absence of sunlight (Table 4). The overall transduction efficiency was higher in the rabbit ileal loops than in vitro, and the in vivo efficiency increased further when the mixed cultures were exposed to sunlight prior to inoculation in rabbits (Table 4). TCP, the receptor for CTXΦ, is known to be expressed more adequately in vivo (17). Since the induction studies clearly showed a marked increase in extracellular CTXΦ, it is most likely that in addition to adequate expression of TCP, the increased efficiency of CTXΦ transduction in vivo was due to the availability of high titers of transducing particles in the inoculum which was preexposed to sunlight.
TABLE 4.
Transduction of V. cholerae strains by CTXΦ derived from sunlight-treated lysogens carrying genetically marked CTX prophages
| Donor strain | Recipient strain | % of recipient cells transduced
|
|||
|---|---|---|---|---|---|
| In vitro
|
In rabbit ileal loops
|
||||
| Without induction | After exposure to sunlight | Without induction | After exposure to sunlight | ||
| SM44 | AE-2883 | 0.23 | 5.36 | 15.23 | 45.62 |
| ASF-1 | AE-2883 | 0.16 | 5.25 | 17.51 | 39.51 |
| SM44 | AOE12-39 | 0.06 | 3.45 | 11.82 | 25.21 |
| ASF-1 | AOE12-39 | 0.12 | 3.94 | 7.49 | 15.12 |
| SM44 | Env-99 | 0.06 | 3.45 | 9.52 | 25.21 |
| ASF-1 | Env-99 | 0.12 | 3.94 | 4.49 | 17.16 |
Transduction of AE-2883 was detected by plating on Luria agar plates containing kanamycin and tetracycline, which counterselected the donor strains. Transductants of the O139 strains were enumerated by screening kanamycin-resistant colonies using an O139-specific DNA probe (see text for details). Values are medians of five different observations made in different rabbits. Results are expressed as percentage of total number of recipient cells recovered from rabbit ileal loops.
The origin of strains belonging to the newly emerged epidemic serogroup O139 has been investigated in several previous studies (1, 3, 8, 26, 29). These studies suggested that the O139 serogroup probably originated from a toxigenic strain of the El Tor biotype by changes in the genes determining the serogroup antigen. In the present study, two nontoxigenic O139 strains were transduced by a genetically marked phage, IG-KmΦ, which carried a functional ctxAB operon. Subsequent analysis of the transductants showed that the phage genome integrated into the O139 chromosome, forming stable lysogens. These lysogens of the O139 strains were tested for production of CT and were found to produce high levels of biologically active CT (Table 5). Thus, we demonstrated the lysogenic conversion of two nontoxigenic O139 strains into toxigenic derivatives, and this process was significantly enhanced by sunlight (Table 4). Although we have previously demonstrated lysogenic conversion of nontoxigenic O1 El Tor strains by CTXΦ, this is the first demonstration of the origination of toxigenic O139 strains from nontoxigenic progenitors. Thus, lysogenic conversion may constitute an alternative mechanism for the emergence of epidemic O139 strains from nontoxigenic progenitors in the environment.
TABLE 5.
Production of cholera toxin by CTXΦ-infected derivatives of nontoxigenic V. cholerae O139 strains
| Strain | Characteristic(s) | CT productiona | Fluid accumulationb in rabbit ileal loops (ml/cm of ileal loop) |
|---|---|---|---|
| P27457 | Toxigenic El Tor biotype strain of V. cholerae O1 | 2.65 ± 0.56 | 2.27 ± 0.42 |
| AOE12-39 | Wild-type, CTX-negative V. cholerae O139 strain | UD (<0.01)c | 0 |
| AOE12-39(IG-Km) | Strain AOE12-39 lysogenized with IG-KmΦ | 3.29 ± 0.71 | 2.53 ± 0.73 |
| AOE12-39(CTX-Km) | Strain AOE12-39 lysogenized with CTX-KmΦ | UD (<0.01) | 0 |
| Env-99 | Wild-type, CTX-negative V. cholerae O139 strain | UD (<0.01) | 0 |
| Env-99(IG-Km) | Strain Env-99 lysogenized with IG-KmΦ | 2.79 ± 0.62 | 2.45 ± 0.65 |
| Env-99(CTX-Km) | Strain Env-99 lysogenized with CTX-KmΦ | UD (<0.01) | 0 |
Determined by GM1 ganglioside-dependent enzyme-linked immunosorbent assay. Toxin amounts are expressed in micrograms per unit of optical density of the culture at 600 nm. Values are the averages of five independent observations.
Values are the averages of five independent observations made in different rabbits.
UD, undetectable. The toxin amounts were less than 0.01 μg/ml, which was the lowest concentration of purified toxin used as a control (see text for details).
Environmental factors in the emergence of epidemic strains.
Species of the genus Vibrio have been regarded as a group of organisms whose major habitats are aquatic ecosystems (2). It has been suggested that acquisition of virulence-associated genes, particularly those encoding intestinal colonization factors and enterotoxins, has allowed specific vibrios to adapt to the human intestinal environment (5, 13). The genes for the major enterotoxin, ctxAB, reside in the genome of CTXΦ, and the propagation of this phage is assumed to be associated with the horizontal transmission of genes encoding CT. In the present study, we demonstrated the conversion of nontoxigenic O139 strains into toxigenic derivatives. These strains were also positive for genes encoding the intestinal colonization factor and CTXΦ receptor, TCP, and the virulence regulatory gene toxR, involved in the expression of both TCP and CT, which are the major virulence factors found in epidemic strains (11).
Since it does not appear that CTXΦ lysogens often spontaneously produce high titers of CTXΦ particles, efficient propagation of this phage under natural conditions may be mediated by environmental factors which result in the induction of CTX prophages and the persistence of free CTXΦ particles in aquatic environments. In the present study, we showed that the CTX prophage is induced by sunlight. The stability of the phage is favored at a salinity above 0.5%, temperatures below 37°C, and a pH range of 5.0 to 10.0. In most countries in which cholera is endemic, substantial fluctuations in water temperature and salinity may occur between the dry and wet seasons, or between winter and summer. It would be interesting to study whether these fluctuating environmental conditions induce CTX prophages and influence the stability of CTXΦ particles. Furthermore, since cholera outbreaks in these areas also occur in a seasonal pattern, it is possible that these changes in environmental conditions play a role in initiation of cholera epidemics caused by new strains of toxigenic V. cholerae. The full array of factors associated with the propagation of CTXΦ in natural environments where V. cholerae can survive have yet to be elucidated. Our efforts are at present directed towards understanding the transmission of the CTX phage in the aquatic environment and its possible relationship with the emergence of seasonal epidemics in areas where cholera is endemic.
Acknowledgments
We thank V. I. Mathan for helpful discussion.
This research was funded in part by the United States Agency for International Development (USAID) under grant HRN-5986-A-00-6005-00 with the ICDDR,B, and by the National Institutes of Health under grant no. RO1 AI39129-01A1 with the Department of International Health, Johns Hopkins University, and the ICDDR,B. The ICDDR,B is supported by countries and agencies which share its concern for the health problems of developing countries.
REFERENCES
- 1.Calia K E, Murtagh M, Ferraro M J, Calderwood S B. Comparison of Vibrio cholerae O139 with V. cholerae O1 classical and El Tor biotypes. Infect Immun. 1994;62:1504–1506. doi: 10.1128/iai.62.4.1504-1506.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colwell R R, Spira W M. The ecology of Vibrio cholerae. In: Barua D, Greenough III W B, editors. Cholera. New York, N.Y: Plenum Medical Book Co.; 1992. pp. 107–127. [Google Scholar]
- 3.Comstock L E, Johnson J A, Michshalski J M, Morris J G, Jr, Kaper J B. Cloning and sequencing of a region encoding a surface polysaccharide of Vibrio cholerae O139 and characterization of the insertion site in the chromosome of V. cholerae O1. Mol Microbiol. 1996;19:815–826. doi: 10.1046/j.1365-2958.1996.407928.x. [DOI] [PubMed] [Google Scholar]
- 4.De S N, Chatterje D N. An experimental study of the mechanisms of action of Vibrio cholerae on the intestinal mucous membrane. J Pathol Bacteriol. 1953;46:559–562. doi: 10.1002/path.1700660228. [DOI] [PubMed] [Google Scholar]
- 5.Faruque S M, Albert M J, Mekalanos J J. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faruque S M, Asadulghani, Alim A R M A, Albert M J, Islam K M N, Mekalanos J J. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic Vibrio cholerae O1 and O139. Infect Immun. 1998;66:3752–3757. doi: 10.1128/iai.66.8.3752-3757.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faruque S M, Asadulghani, Saha M N, Alim A R M A, Albert M J, Islam K M N, Mekalanos J J. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXΦ: molecular basis for origination of new strains with epidemic potential. Infect Immun. 1998;66:5819–5825. doi: 10.1128/iai.66.12.5819-5825.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faruque S M, Alim A R M A, Roy S K, Khan F, Nair G B, Sack R B, Albert M J. Molecular analysis of rRNA and cholera toxin genes carried by the new epidemic strain of toxigenic Vibrio cholerae O139 synonym Bengal. J Clin Microbiol. 1994;32:1050–1053. doi: 10.1128/jcm.32.4.1050-1053.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feinberg A, Volgelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg I, Mekalanos J J. Effect of a recA mutation on cholera toxin gene amplification and deletion events. J Bacteriol. 1986;165:723–731. doi: 10.1128/jb.165.3.723-731.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herrington D A, Hall R H, Losonsky G, Mekalanos J J, Taylor R K, Levine M M. Toxin, toxin-coregulated pili and ToxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaper J B, Morris J G, Jr, Nishibuchi M. DNA probes for pathogenic Vibrio species. In: Tenover F C, editor. DNA probes for infectious diseases. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 65–67. [Google Scholar]
- 13.Karaolis D K R, Lan R, Reeves P R. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J Bacteriol. 1995;177:3191–3198. doi: 10.1128/jb.177.11.3191-3198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keasler S P, Hall R H. Detection and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet. 1993;341:1661. doi: 10.1016/0140-6736(93)90792-f. [DOI] [PubMed] [Google Scholar]
- 15.Kimsey H H, Waldor M K. Vibrio cholerae hemagglutinin/protease inactivates CTXφ. Infect Immun. 1998;66:4025–4029. doi: 10.1128/iai.66.9.4025-4029.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazar S, Waldor M K. ToxR-independent expression of cholera toxin from the replicative form of CTXΦ. Infect Immun. 1998;66:394–397. doi: 10.1128/iai.66.1.394-397.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S H, Hava D L, Waldor M K, Camilli A. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell. 1999;99:625–634. doi: 10.1016/s0092-8674(00)81551-2. [DOI] [PubMed] [Google Scholar]
- 18.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 19.Miller C J, Drasar B S, Feachem R G. Response of toxigenic Vibrio cholerae O1 to physicochemical stresses in aquatic environments. J Hyg. 1984;93:475–495. doi: 10.1017/s0022172400065074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair G B, Bag P K, Shimada T, Ramamurthy T, Takeda T, Yamamoto S, Kurazono H, Takeda Y. Evaluation of DNA probes for specific detection of Vibrio cholerae O139 Bengal. J Clin Microbiol. 1995;33:2186–2187. doi: 10.1128/jcm.33.8.2186-2187.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson G D N, Woods A, Chiang S L, Mekalanos J J. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc Natl Acad Sci USA. 1993;90:3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabbani G H, Greenough W B. Cholera. In: Lebenthal E, Duffy M, editors. Textbook of secretory diarrhea. New York, N.Y: Raven Press Ltd.; 1990. pp. 233–253. [Google Scholar]
- 24.Sack D A, Huda S, Neogi P K B, Daniel R R, Spira W M. Microtiter ganglioside enzyme-linked immunosorbent assay for vibrio and Escherichia coli heat-labile enterotoxins and antitoxin. J Clin Microbiol. 1980;11:35–40. doi: 10.1128/jcm.11.1.35-40.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singleton F L, Attwell R, Jangi S, Colwell R R. Effects of temperature and salinity on Vibrio cholerae growth. Appl Environ Microbiol. 1982;44:1047–1058. doi: 10.1128/aem.44.5.1047-1058.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stroeher U H, Jedani K E, Dredge B K, Morona R, Brown M H, Karageorgos L E, Albert M J, Manning P A. Genetic rearrangements in the rfb regions of Vibrio cholerae O1 and O139. Proc Natl Acad Sci USA. 1995;92:10374–10378. doi: 10.1073/pnas.92.22.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 29.Waldor M K, Mekalanos J J. Vibrio cholerae O139 specific gene sequence. Lancet. 1994;343:1366. doi: 10.1016/s0140-6736(94)92504-6. [DOI] [PubMed] [Google Scholar]
- 30.Waldor M K, Rubin E J, Pearson G D, Kimsey H, Mekalanos J J. Regulation, replication, and integration functions of the Vibrio cholerae CTXΦ are encoded by region RS2. Mol Microbiol. 1997;24:917–926. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]