Abstract
Protein S-glutathionylation serves a regulatory role in proteins and modulates distinct biological processes implicated in health and diseases. Despite challenges in analyzing the dynamic and reversible nature of S-glutathionylation, recent chemical and biological methods have significantly advanced the field of S-glutathionylation, culminating in selective identification and detection, structural motif analysis, and functional studies of S-glutathionylation. This review will highlight emerging studies of protein glutathionylation, beginning by introducing biochemical tools that enable mass spectrometric identification and live-cell imaging of S-glutathionylation. Next, it will spotlight recent examples of S-glutathionylation regulating physiology and inflammation. Lastly, we will feature two emerging lines of glutathionylation research in cryptic cysteine glutathionylation and protein C-glutathionylation.
Keywords: Cysteine, S-glutathionylation, proteomics, redox signaling, cryptic cysteines, C-glutathionylation
Graphical Abstract
Introduction
Cysteine is one of the unique amino acids in proteins that retain high nucleophilicity and oxidation susceptibility in biological systems [1,2]. Due to these unique properties, cysteines play important functional roles in proteins. For example, many conserved cysteines serve as catalytic nucleophiles in enzymes (e.g., proteases), essential residues for structural folding (e.g., disulfide bond, -SS-), and metal binding [2]. Alternatively, cysteine also plays important regulatory functions in proteins via its reversible oxidations, including S-sulfenylation (SOH), S-nitrosylation (SNO), S-persulfidation (SSH), and S-glutathionylation (SSG) [3,4]. Evidence supports that individual cysteine oxoforms occur on functionally distinct protein networks and regulate different biological processes [5]. Therefore, identification and functional analyses of specific cysteine oxoforms and their complex interplay continue to be important to portray a global map of redox signaling.
Among oxoforms, protein SSG is a cysteine post-translational modification (cys-PTM) that adds bulky glutathione (GSH) to protein cysteine residues via disulfide formation, which regulates protein functions and biological processes in response to oxidants such as reactive oxygen species (ROS) [6]. As significant as ROS in physiology and pathology, the essential functions of SSG have emerged in all areas of human health, including cardiovascular regulation [6,7], inflammation and infection [8–10], apoptosis [11], and cancer [12].
At a molecular level, SSG occurs via various reaction mechanisms. SSG occurs via nucleophilic sulfur chemistry where thiolate anion (S−) reacts with oxidized glutathione (GSSG) or reactions of GSH with electrophilic sulfur intermediates, such as sulfenic acid, S-nitrosothiol, or thiyl radical [13]. SSG formation is further balanced by the activities of several enzymes, including glutaredoxin (Grx) and glutathione transferase pi and omega (GSTP and GSTO) [14]. The complexity of SSG that forms concurrently with other cysteine oxoforms (e.g., -SS-, SOH, SNO, SSH) challenges the identification of SSG sites, which are essential to uncovering new regulatory roles of SSG. Nevertheless, recent chemical and biological methods, especially combined with chemical proteomics and mass spectrometry, have advanced the field of SSG, culminating in the identification, detection, structural motif analysis, and functional studies of SSG. This review will focus on the recent studies of SSG, highlighting 1) biochemical tools that enable the discovery and detection of SSG and 2) functional analyses of SSG in biological models. Lastly, we will feature 3) newly emerging perspectives of SSG by appraising cryptic cysteine SSG and protein C-glutathionylation (C-SG).
Emerging chemical tools to study protein S-glutathionylation
SSG can be commonly detected by using a glutathione antibody. However, recent biochemical tools enabled global and individual analyses of SSG for site identifications [15], site occupancy (i.e., SSG percentage on Cys) [16], global concurrence with other cys-PTM [17], and cellular imaging [18]. The following section will showcase recent methods that detect and identify SSG.
Glutaredoxin (Grx)-mediated identification.
E. coli Grxs (Grx 1-3) are ubiquitous thiol-disulfide oxidoreductases with -CXXC- motifs that primarily reverse SSG (and disulfide reduction of ribonucleotide reductase) [19,20]. Mutating the resolving cysteine (the second Cys in -CXXC- motif) in E. coli Grx1 and Grx3 (i.e., C14S in Grx1/Grx3) confers higher specificity to deglutathionylation over disulfide reduction [21,22]. Such higher specificity of Grx1 C14S mutant (or Grx3 mutant) has been a foundation to reverse SSG selectively, which allowed selective identification of SSG among other cysteine oxidations, especially combined with the biotin-switch method (BSM) [23]. As opposed to the original BSM-based SSG detection [23], the Qian group has advanced this approach by removing a biotinylation step, thus simplifying the enrichment with the 2-thiopyridyl disulfide-based resin (Thiopropyl Sepharose), which was named as the resin-assisted capture (RAC) (Fig. 1A) [16,24,25]. In the RAC, after SSG formation, all remaining cysteines were blocked by N-ethylmaleimide (NEM). Grx1 C14S (or Grx3 C14S/C65Y in which the additional reactive cysteine, C65, was mutated to tyrosine found in other Grx isoforms [22]) was then used to reduce SSG selectively. The resulting cysteines were directly captured on the resin by forming disulfide bonds, which could reduce concerns resulting from non-specific binders of streptavidin-based enrichment. Following on-bead trypsin digestion, SSG peptides on resins were labelled by the isobaric tandem mass tag (TMT) (Fig. 1A, top). In parallel, the approach was used to capture and label all cysteine-containing peptides (SH) by TMT (Fig. 1A, bottom). TMT-based quantification of SSG versus SH samples determined levels of SSG (i.e., site occupancy) in individual cysteines (Fig. 1A) [16].
Figure 1. Biochemical methods for identification and detection of SSG.
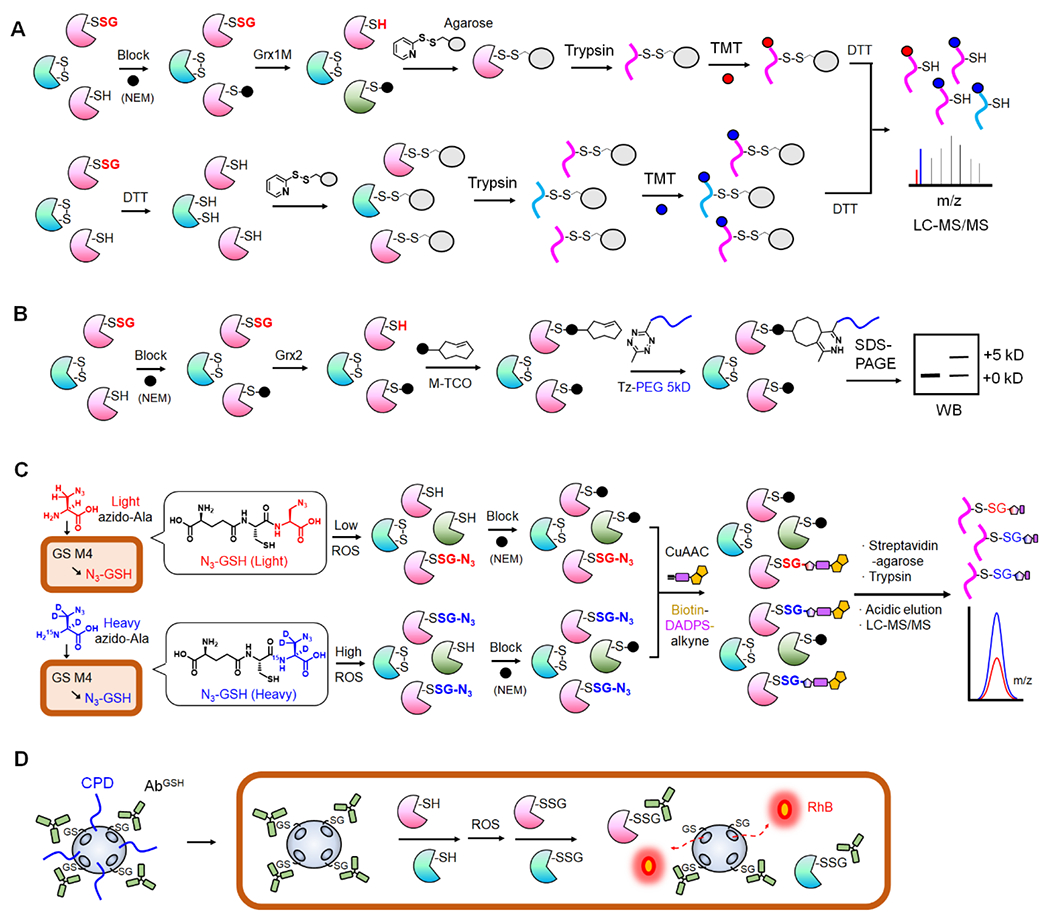
A. Grx-mediated detection of SSG or RAC-TMT. SSG is selectively reduced by Grx1 mutant (Grx1M) after blocking Cys with N-ethylmaleimide (NEM, black circle). Disulfide-based agarose captures reduced Cys in proteins (top). In parallel, all Cys-containing peptides from the same sample are captured on agarose after reducing with dithiothreitol (DTT, bottom). After trypsin digestion and TMT labeling, the bound peptides are eluted and combined for LC-MS/MS analysis to determine SSG levels. B. Grx-mediated SSG analysis with click chemistry. After selective reduction of SSG by Grx, the revealed Cys is reacted with maleimide-conjugated trans-cyclooctene (M-TCO). The subsequent reaction with 5-kD PEG-conjugated tetrazine (Tz-PEG) enables analysis of SSG levels via western blot. C. Glutathione synthetase mutant (GS M4)-based clickable glutathione. GS M4 uses light or heavy azido-Ala to synthesize isotopically labelled azido-GSH (N3-GSH) in cells. Isotopically labelled glutathionylated proteins (P-SSG) are combined, modified with biotin-DADPS-alkyne by click chemistry, captured on streptavidin-agarose, and digested by trypsin. The acidic cleavage of the DADPS linker enables elution of SSG-modified peptides for relative quantitation of SSG. D. Nanoparticle to image SSG in live cells. Mesoporous nanoparticle doped with black hole quenchers (qMSN) encapsulates and quenches rhodamine B (RhB). qMSN is modified with GSH on the surface, which binds to the GSH antibody (AbGSH) that blocks the release of RhB. The cell-penetrating disulfides (CPD) of qMSN enable its cell entry where SSG proteins (P-SSG) cause dissociation of AbGSH and releases of RhB, restoring fluorescence.
Alternatively, Grx-mediated detection of SSG has coupled with click chemistry-based polyethyleneglycol (PEG) conjugation (PEGylation) [26]. In this approach, SSG in lysates was reversed by Grx2 (mitochondrial Grx2 was used in this study, likely due to examining the mitochondrial protein). The resulting cysteines were modified by the maleimide-conjugated to trans-cyclooctene (M-TCO). The subsequent reaction with 5 kD PEG-modified methyltetrazine (Tz-PEG) enabled the detection of SSG levels in a protein via western blot analyses (Fig. 1 B). This approach was modified from a previous method that uses Cu(I)-catalyzed azide-alkyne coupling [27], which was noted to suffer from incomplete click reactions when coupling with a large size of PEG [26]. In addition, Grx-mediated detection of SSG has coupled with other detection tools, including mercury (Hg)-based enrichment or eosin-derived glutathione derivative [28,29].
The primary advantage of Grx-mediated identification is that the approach is readily applicable to biological samples from animals and patients beyond in vitro or cellular studies. For example, RAC-TMT was used to identify and quantify SSG sites upon fatiguing contraction in adult mouse skeletal muscle, finding SSG percent occupancy in 2,200 sites with an average of 4.5%, which was elevated to 5.2% upon fatiguing contraction [30]. However, potential concerns come from indirect detection of SSG in which incomplete reactions (e.g., blocking and reduction) and Grx selectivity issues during the sample processing may compromise the outcomes. Despite such concerns, Grx-mediated SSG detection stands out as a primary tool for SSG analysis.
Chemically tagged glutathione-based identification.
Metabolic labeling of biomolecules with click chemistry reporters has been crucial for PTM analysis [31]. Our group has developed an approach to label GSH with clickable reporters (e.g., azide) (Fig. 1C) [32,33]. Glutathione synthetase (GS), a non-rate limiting enzyme in GSH biosynthesis, was engineered to produce its mutants (GS M4 or M7) that use clickable Gly derivatives (e.g., azido-Ala, allyl-Gly, allyl-Ser) in place of Gly in GSH tripeptide (γGlu-Cys-Gly) [32,34]. GS M4 enabled biosynthesis of clickable GSH (e.g., azido- or allyl-GSH) in cells, which afforded SSG analysis after click chemistry (azide-alkyne or tetrazine-alkene coupling) (Fig. 1C). The approach was also coupled with mass spectrometric proteomic analysis of SSG sites using a cleavable linker (i.e., dialkoxydiphenylsilane, DADPS), finding 1,736 SSG sites in response to hydrogen peroxide (H2O2) in an HL-1 cardiomyocyte cell line [35]. Recently, isotopically labelled azido-Ala (heavy or light azido-Ala with +4 or 0 Da, respectively) was developed for quantification analysis of SSG (Fig. 1C) [15] and applied to quantify levels of 1,398 SSG induced by H2O2 and 249 SSG in response to palmitate in a cellular model of ischemic stress, discovering SSG sites associated with muscular disorders [36]. Ma and Deng et al. recently used the same approach to profile the SSG proteome, including 15-hydroxyprostaglandin dehydrogenase (15-PGDH), in cluster of differentiation 38 (CD38)-mediated epithelial-mesenchymal transition [37]. The principal merit of this approach is the direct identification of SSG with glutathionyl modification on individual peptides (Fig. 1C), unlike Grx-mediated detection, thus removing the ambiguity resulting from the labile cys-PTM, which is important when considering the complexity and reversibility of cys-PTM. Also, the approach could detect SSG occurring at a basal level, as endogenous SSG formation can be detected upon adding azido-Ala without oxidative stimulus. However, the approach may not be easily adaptable to biological samples from animal or human patients, as it needs the GS M4 mutant expressed in the system. Nevertheless, GS M4-based clickable glutathione could be used in genetically engineered animals.
Mass spectrometric identification without enrichment.
Typically, the PTM-modified proteome needs enrichment before mass spectrometric analysis, which maximizes PTM detection within the resolution and sensitivity power of mass spectrometry. However, the advance in mass spectrometry has brought PTM proteomic analyses without PTM enrichment, including SSG identification [17,38,39]. One example is the top-down proteomics that uses a Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer with electron-capture dissociation (ECD) [40]. The Ge group used this approach to examine sarcomeric protein PTM, including SSG, that occurs upon sarcopenia (age-dependent loss of skeletal muscle mass and function). The study found SSG of troponin I (TnI) (2-3% SSG abundance) increased in soleus skeletal muscle with older versus younger ages [39]. The approach is robust such that it enables the detection of labile PTMs and the comprehensive analyses of PTMs (i.e., different proteoforms and PTM sites and abundance), thus detecting direct glutathione conjugation (+305.03 Da) to proteoforms [39]. However, top-down proteomics has more challenges in preparing and analyzing the protein samples (e.g., difficulty in protein-level fractionation, solubilization, and data analysis) than the peptides in bottom-up proteomics [41,42], which hampers its use. However, it is notable that the advancement in instrumentation (e.g., high-resolution Orbitrap) and the analytic technique over the years is rapidly broadening the field of top-down proteomics [43].
Alternatively, the bottom-up proteomics using the Orbitrap has advanced to direct detection of SSG and cys-PTM without enrichment [17]. Mouse pancreatic β-cell lysates were collected and analyzed after blocking nucleophilic reduced cysteines, quantifying about 231 cys-PTM sites (sulfinic acid, sulfonic acid, -SS-, SSH, and SSG; about 10 SSG) increased or decreased upon endoplasmic reticulum (ER) stress out of 9342 proteins detected in lysates [17]. The approach is significant, enabling a global view of complex cys-PTM simultaneously and estimating the relative abundance of each cys-PTM on individual cysteines. However, the approach illustrates a challenge for direct SSG detection, identifying a small number of SSG sites [17].
Imaging protein S-glutathionylation.
Despite high interest in proteome-wide identification of SSG, the approach to detect SSG in live cells has been limited. Recently, the Yao group developed nanoquencher-based probes that sense SSG formation in live cells (Fig. 1D) [18]. Mesoporous silica-based nanoparticle covalently doped with black hole quenchers (qMSN) was loaded with a fluorescent dye (rhodamine B, RhB), which emits no fluorescence due to quenchers. qMSN surface was covalently immobilized with GSH, followed by non-covalently binding to GSH antibody (AbSSG) (Fig. 1D). The large size of AbSSG acts as a gatekeeper to the pores of qMSN, blocking the release of encapsulated RhB. However, exogenous GSH or glutathionylated proteins (P-SSG) displaces the gatekeeper AbSSG from the qMSN surface, thus releasing RhB with the restoration of fluorescence (Fig. 1D). The qMSN surface was also conjugated with cell-penetrating poly(disulfide) (CPD), which mediates delivery of the qMSN-based probe into cells via a thiol-mediated uptake that enables its delivery to the cytoplasm, unlike endocytosis (Fig. 1D) [44]. Interestingly, AbSSG retains higher selectivity to P-SSG over GSH (5,000:1 selectivity), thus enabling the probe to sense a low level of P-SSG that occurs inside cells, even in the presence of a high concentration of GSH [18]. The probe is distinct from GSH probes [45,46], thus promising to contribute to SSG biology, such as high-throughput screening of small molecules in live cells that modulate levels of SSG.
Alternatively, protein A conjugated with CPD was developed for simple delivery of antibodies, including fluorescein isothiocyanate (FITC)-labelled AbSSG, namely the “Mix-and Go” approach, which enabled fluorescence imaging of SSG in live cells [47]. These approaches may progress to image SSG of a specific protein in future studies.
Emerging biological targets of protein S-glutathionylation
The identification of protein targets and sites of SSG enables understanding the functional and regulatory roles of SSG. Previous studies have found that SSG regulates diverse cellular pathways, depending on their targets. For example, individual target analysis along with proteomic analysis support that SSG regulates metabolism (e.g., glycolysis, tricarboxylic acid cycle, and the electron transport chain [48]), translation (e.g., initiation and elongation factors [36]), signal transduction (e.g., G-protein, kinase, and phosphatase, including Ras [49], Src [50], and low molecular weight protein tyrosine phosphatase [51]), transcription factors (e.g., p53 [52]), cytoskeletal structure (e.g., actin [53,54] and vimentin [55]), inflammation (e.g., inhibitor of nuclear factor kappa-B kinase subunit β, IKKβ [56,57]), apoptosis (e.g., Fas and caspase 3 [58,59]), and calcium release (e.g., sarcoendoplasmic reticulum calcium ATPase, SERCA [60]) among others, showing a broad role of SSG in regulating biological processes. Therefore, functional analyses of SSG continue to expand our understanding of SSG in all areas of human health. This section will highlight the significance of SSG with recent examples of target proteins regulating cardiac contraction, inflammation, and coronavirus.
Titin.
Titin is an elastic myofilament protein essential for producing passive force during muscle contraction (Fig. 2A) [61]. Many studies have demonstrated that titin truncations, mutations, and PTM result in several forms of cardiomyopathy [62], supporting its critical role in heart muscle elasticity. In the structure, titin is a modular protein with immunoglobulin (Ig)-like domains and fibronectin-domains, especially with unique sequences (N2A and N2B domains) and a segment rich in proline, glutamate, valine, and lysine (PEVK domain) at the elastic I-band (Fig. 2A) [63]. The titin elasticity is governed by the unfolding and refolding of Ig-domains in addition to stretches of PEVK and unstructured inter-domain sequences [63]. Importantly, many cysteines in I-band Ig domains were found to form SSG in the unfolded state at the cryptic cysteine (Fig. 2A), which reduces dynamics of re-folding and weakens Ig-domain stability, thus decreasing titin-based passive tension or stiffness [64]. Alternatively, intramolecular disulfide (-SS-) within Ig domains hinders titin stretches, thus increasing its stiffness [65]. Recent studies support that increased sarcomere strain induces unfolded domain oxidation (UnDox) more prevalently on elastic I-band Ig-domains of titin [65]. Notably, titin SSG was found elevated, especially at I-band, in the mouse heart during ischemia, which correlated with titin phosphorylation that also reduces titin stiffness [65]. However, UnDOx, including SSG, was suggested to increase the aggregation of titin domains [65] that can cause myocardial stiffness commonly seen in heart diseases.
Figure 2. Examples of SSG in biology.
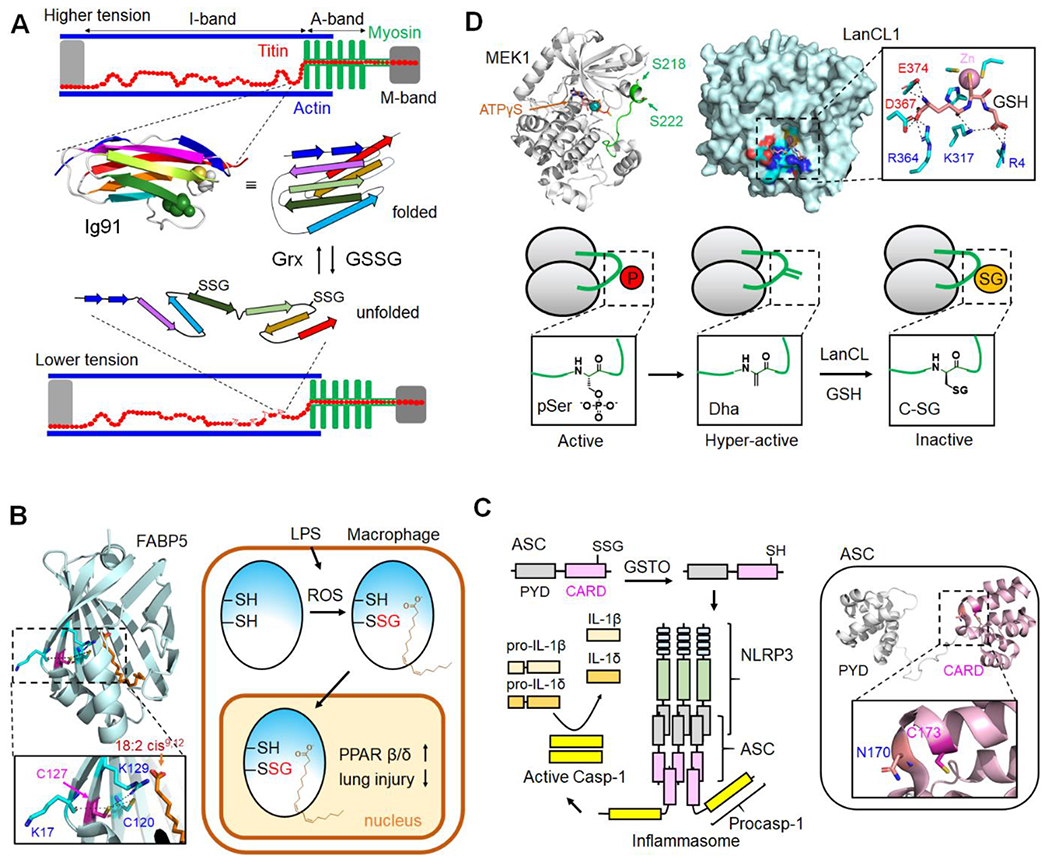
A. Titin SSG in muscle contraction. Titin has Ig-domains, including Ig91 (two cysteines in a space-filling model, PDB: 1TIT), at the elastic I-band that can unfold during sarcomere stretches. Ig-domain unfolding exposes cryptic cysteines susceptible to SSG. SSG in Ig-domain retards refolding and promotes its relaxed state, which reduces the titin-based passive tension. B. FABP5 SSG in macrophage. FABP5 C127 (pink in β-strand, PDB: 4LKT) is susceptible to SSG in response to LPS-induced ROS, promoting fatty acid binding and translocation to the nucleus. FABP5 in the nucleus activates PPAR β/δ genes for pro-survival, suppressing macrophage inflammation. C. ASC SSG in inflammasome formation. LPS-ATP or mitochondrial ROS promotes GSTO interactions with ASC, causing its deglutathionylation. ASC deglutathionylation enables its oligomerization, which bridges and forms an inflammasome with NLRP3 and pro-caspases. Inflammasome complex activates caspase-1, which processes and activates cytokines for pyroptosis. ASC has C173 in the human CARD domain (PDB: 2KN6, equivalent to mouse C171). D. LanCL catalyzed C-glutathionylation (C-SG) in kinases. The phosphate sites (S218 and S222) in the activation loop (green) of MEK1 (PDB: 3EQD) and other kinases are susceptible to forming Dha/Dhb at physiological conditions, which cause hyperactivation of kinases. Dha/Dhb at the activation loop reacts with GSH to form C-SG catalyzed by LanCL, which inactivates the hyperactivity of kinases, thus preventing kinase dysregulation. The LanCL active site has a binding site for GSH (PDB: 3E73).
Fatty acid-binding protein 5 (FABP5).
FABPs are small lipid chaperons that bind hydrophobic ligands such as long-chain fatty acids and transport them to cellular compartments [66]. Guo et al. found that macrophage-specific Grx1 knockout alleviates inflammatory acute lung injury [67]. The subsequent Grx1-mediated SSG proteomics found FABP5 is regulated via SSG at Cys127, which can form a disulfide bond with Cys120 (see Fig. 2B). Biochemical studies support that FABP5 SSG promotes its fatty acid binding and nuclear translocation in response to ROS, activating peroxisome proliferator-activated receptor β/δ (PPAR β/δ) pro-survival target genes (Fig. 2B). Therefore, FABP5 SSG was found to suppress inflammation in macrophages and alleviate acute lung injury induced by lipopolysaccharide (LPS). It is worth noting that Cys127 is buried and positioned in the inner structure (Fig. 2B), which could disfavor SSG formation. However, Cys127 pKa is predicted to be low (7.8 in the PropKa program [68]; in contrast, Cys120 pKa is 12.9 using Protein Data Bank, PDB, 4LKT), supporting its potential reactivity. Notably, sulfenylation proteomics found FABP5 C127 sulfenylation elevated by H2O2 [69]. In addition, chemical proteomics in C. elegans identified that lipid binding protein-3 (LBP-3) Cys154 (high homology to FABP5 C127) is susceptible to cysteine oxidations, modulating C. elegans lifespan [70].
Apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC).
Inflammasomes, including NACHT, LRR, and PYD domains-containing protein 3 (NLRP3), are crucial regulators of the innate immune system in response to pathogens [71]. Inflammasome activation leads to caspase-1 activation that cleaves and activates proinflammatory cytokines (e.g., interleukin 1β and δ) (Fig. 1C), eventually inducing pyroptosis [72]. Li et al. found that GSTO1 knockout mice were resistant to NLRP3 inflammasome-dependent arthritis, which correlated with reduced oligomerization of ASC [73]. ASC oligomerization is necessary to bridge NLRP3 and caspase-1 to form an inflammasome complex in macrophages (Fig. 2C). ASC has only one cysteine at 171 in the caspase recruitment domain (CARD). The PropKa analysis predicts that C171 is buried but has relatively low pKa (7.4-9.0 with PDB 6N1H) (Fig. 2C). Biochemical studies found that ASC SSG inhibits its oligomerization, resulting in the deactivation of NLRP3 inflammasome in macrophages [73]. Interestingly, mitochondrial ROS promoted GSTO1-ASC interaction, which induced GSTO1-mediated ASC deglutathionylation and NLRP3 inflammasome activation (Fig. 2C). Lastly, ASC C171A mice induced NLRP3-dependent hyper-inflammation, suggesting that ASC SSG serves as a brake to prevent NLRP3 inflammasome-induced hyper-inflammation [73].
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) main protease (Mpro).
Recent studies have found the potential role of GSH and SSG in coronavirus disease 19 (COVID-19) [74,75]. The causative agent, SARS-CoV-2, encodes two proteases for replication. One protease, Mpro, is only active as a homodimer [76]. In contrast, the monomeric active site pocket disintegrates, thus decreasing its substrate binding [76,77]. Mpro was found susceptible to SSG at Cys300, which is located at the dimer interface [75]. Importantly, Mpro Cys300 SSG inhibits its dimerization, leading to deactivation [75]. This finding highlights that Mpro activity is regulated by SSG, suggesting a potential therapeutic approach that targets Mpro Cys300.
New research direction of protein glutathionylation
The research on GSH and redox biology has brought new perspectives on SSG in recent years. This section will highlight emerging lines of SSG research, such as cryptic cysteine SSG and protein C-SG.
Conformation-dependent cryptic cysteine glutathionylation.
pKa and residue-surface accessibility (RSA) are two determining factors for cysteine oxidations [3]. Low cysteine pKa enables thiolate formation, increasing susceptibility to oxidation. Higher RSA increases the probability of reactions. Despite their importance, recent experimental data support low or minimal differences in average pKa and RSA values in groups of oxidized versus non-oxidized cysteines [5,16]. Such data could arise from the limited availability of structural information. However, the discrepancy is also supported by an emerging idea of cryptic cysteines or conformational-dependent cysteine SSG [64], defined as SSG of cysteines only accessible to oxidants upon conformational changes or potentially protein-protein interaction changes. The term cryptic cysteine was used with an example of titin [64] (Fig. 2A). In this gigantic protein, 89 out of 93 Ig-domains in the elastic I-band have cysteines buried in their β-sandwich structures [64] (e.g., two cysteines buried in the folded structure, Fig. 2A). However, these cysteines can be exposed upon unfolding of Ig-domain induced by sarcomere stretch, thus susceptible to SSG (e.g., cysteine SSG in the unfolded structure, Fig. 2A). In addition, the Held group recently reported redox regulation of cryptic cysteines that are only exposed and oxidized upon epidermal growth factor (EGF)-induced conformational changes of proteins [78], suggesting that cysteine-mediated redox signaling depends on its stimulus that alters protein structures and interactions.
Protein C-glutathionylation.
For many years, it has been observed that β-elimination of water (H2O) and hydrogen sulfide (H2S) from serine, threonine, and cysteine in proteins or β-elimination of phosphate from phosphoserine and phosphothreonine give rise to dehydro-amino acids, such as 2,3-dehydroalanine (Dha) and 2,3-didehydrobutyrine (Dhb) [79]. The Dha/Dhb are reactive, thus forming a thioether linkage with GSH via a Michael addition, namely C-SG [80]. The Dha/Dhb are often found in the long-lived proteins in eyes, neurons, and cardiac myocytes, thus observing C-SG in aged and cataractous lens proteins [81]. C-SG presumably protects proteins by preventing protein crosslinking and aggregation during aging and cataractogenesis [81,82]. The proteomic approach has led to C-SG identification in lens proteins [81,83].
Although C-SG is thought to occur via non-enzymatic reactions, Van Der Donk and Davis groups recently reported an example of enzyme-catalyzed C-SG formation in kinases [84]. Kinases are activated by phosphorylation at the activation loop (left box, Figure 2D). The phosphate groups at the activation loop were found to undergo β-elimination under physiological conditions, generating Dha/Dhb (middle box, Fig. 2D) [84]. Eukaryotic LanC-like proteins (LanCL) are homologous to bacterial LanC proteins that catalyze lanthionine formation between cysteine and Dha/Dhb [85]. Notably, consistent with their roles in bacteria, LanCL catalyzed C-SG formation on the activation loop of mitogen-activated protein kinase (MAPK), including mitogen-activated protein kinase kinase (MEK1) and extracellular signal-regulated kinase 1/2 (ERK1/2) (right box, Fig. 2D) [84]. LanCL-induced C-SG was specific to Dha/Dhb formed at phosphorylation sites. Therefore, the study discovers evidence of C-SG formation modulated by LanCL in the eukaryotic system.
Dha/Dhb formation at the activation loop of MEK1 was found to confer hyperactivity, while C-SG causes almost inactive MEK1 (Fig. 2D) [84]. This implied that Dha/Dhb formation in kinases causes aberrantly activated kinases, which could be rescued by C-SG formation, suggesting the protective role of C-SG. The study also identified the LanCL interactome, implying that LanCL quenches deleterious effects of Dha/Dhb in many damaged proteins (namely eliminylome, proteins bearing β-elimination) [84]. Pre-mature death of LanCL knockout mice further demonstrates the protective role of C-SG [84]. Therefore, discovering C-SG mediated by LanCLs in kinases unfolds a new research area in irreversible glutathionylation.
Conclusion
SSG is a major form of reversible cysteine oxidation responsive to the cellular redox environment modulated by oxidases, redox enzymes, and metabolic states [36,86]. The continuing research has significantly advanced the field of SSG in recent years. Proteome-wide analyses found about 4-5% basal levels of SSG (and ca. 10% basal cysteine oxidation) in proteins [16], suggesting their responsive capacity upon stimulus. Chemical proteomics has discovered many SSG sites (>2,000 in database) in the proteome [15,30,35,87]. Functional analyses of individual proteins unveiled SSG in regulating protein structure and functions. Although not included here, many examples of animal models unraveled the biological roles of enzymes regulating SSG [88–92]. Lastly, examples of conformation-dependent cryptic cysteines and C-SG provide a new venue to appraise SSG biology. Future studies will continue developing new approaches and uncovering SSG biology, culminating in discovering therapeutic opportunities.
Highlights.
Cysteine glutathionylation serves distinct regulatory roles in proteins
Various biochemical tools have been developed for the identification, detection, and imaging of glutathionylation.
Proteomic strategies for site specific identification and site occupancy analyses of glutathionylation are available.
Functional analysis of glutathionylation spans all areas of human health, including cardiac regulation, inflammation, and infection.
Evidence for cryptic cysteine glutathionylation and irreversible C-glutathionylation is emerging.
Acknowledgments
This work was supported by the National Institute of Health (NIH) [R01 HL131740 (Y.-H.A) and R01 GM143214 (Y.-H.A)].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
Authors declare no competing interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Fomenko DE, Marino SM, Gladyshev VN: Functional diversity of cysteine residues in proteins and unique features of catalytic redox-active cysteines in thiol oxidoreductases. Mol Cells (2008) 26(3):228–235. [PMC free article] [PubMed] [Google Scholar]
- 2.Pace NJ, Weerapana E: Diverse functional roles of reactive cysteines. ACS Chem Biol (2013) 8(2):283–296. [DOI] [PubMed] [Google Scholar]
- 3.Paulsen CE, Carroll KS: Cysteine-mediated redox signaling: Chemistry, biology, and tools for discovery. Chem Rev (2013) 113(7):4633–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng J, Fu L, Liu K, Tian C, Wu Z, Jung Y, Ferreira RB, Carroll KS, Blackwell TK, Yang J: Global profiling of distinct cysteine redox forms reveals wide-ranging redox regulation in c. Elegans. Nat Commun (2021) 12(1):1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gould NS, Evans P, Martinez-Acedo P, Marino SM, Gladyshev VN, Carroll KS, Ischiropoulos H: Site-specific proteomic mapping identifies selectively modified regulatory cysteine residues in functionally distinct protein networks. Chem Biol (2015) 22(7):965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper reports the proteome-wide analyses of four different cysteine modifications, including SSG, and compares their distinct biochemical properties and protein networks.
- 6.Rashdan NA, Shrestha B, Pattillo CB: S-glutathionylation, friend or foe in cardiovascular health and disease. Redox Biol (2020) 37:101693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns M, Rizvi SHM, Tsukahara Y, Pimentel DR, Luptak I, Hamburg NM, Matsui R, Bachschmid MM: Role of glutaredoxin-1 and glutathionylation in cardiovascular diseases. Int J Mol Sci (2020) 21(18):6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chia SB, Elko EA, Aboushousha R, Manuel AM, van de Wetering C, Druso JE, van der Velden J, Seward DJ, Anathy V, Irvin CG, Lam YW et al. Dysregulation of the glutaredoxin/s-glutathionylation redox axis in lung diseases. Am J Physiol Cell Physiol (2020) 318(2):C304–C327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullen L, Mengozzi M, Hanchmann EM, Alberts B, Gjezzo P: How the redox state regulates immunity. Free Radical Bio Med (2020) 157:3–14. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez-Dosal R, Horan KA, Rahbek SH, Ichijo H, Chen ZJ, Mieyal JJ, Hartmann R, Paludan SR: HSV infection induces production of ROS, which potentiate signaling from pattern recognition receptors: Role for S-glutathionylation of TRAF3 and 6. Plos Pathog (2011) 7(9):e1002250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anathy V, Roberson EC, Guala AS, Godburn KE, Budd RC, Janssen-Heininger YMW: Redox-based regulation of apoptosis: S-glutathionylation as a regulatory mechanism to control cell death. Antioxid Redox Sign (2012) 16(6):496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayes JD, Dinkova-Kostova AT, Tew KD: Oxidative stress in cancer. Cancer Cell (2020) 38(2):167–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong Y, Uys JD, Tew KD, Townsend DM: S-glutathionylation: From molecular mechanisms to health outcomes. Antioxid Redox Sign (2011) 15(1):233–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsui R, Ferran B, Oh A, Croteau D, Shao D, Han J, Pimentel DR, Bachschmid MM: Redox regulation via glutaredoxin-1 and protein S-glutathionylation. Antioxid Redox Signal (2020) 32(10):677–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.VanHecke GC, Yapa Abeywardana M, Huang B, Ahn YH: Isotopically labeled clickable glutathione to quantify protein s-glutathionylation. Chembiochem (2020) 21(6):853–859. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper introduces a proteomic quantification strategy with isotopically labelled clickable glutathione and quantitative analyses of SSG in response to hydrogen peroxide.
- 16.Duan J, Zhang T, Gaffrey MJ, Weitz KK, Moore RJ, Li X, Xian M, Thrall BD, Qian WJ: Stochiometric quantification of the thiol redox proteome of macrophages reveals subcellular compartmentalization and susceptibility to oxidative perturbations. Redox Biol (2020) 36:101649. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This paper describes the global profiling of glutathionylation with the RAC-TMT approach and analyzes site occupancy of glutathionylation and cysteine oxidations at a basal level in macrophages.
- 17.Li X, Day NJ, Feng S, Gaffrey MJ, Lin TD, Paurus VL, Monroe ME, Moore RJ, Yang B, Xian M, Qian WJ: Mass spectrometry-based direct detection of multiple types of protein thiol modifications in pancreatic beta cells under endoplasmic reticulum stress. Redox Biol (2021) 46:102111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao X, Yuan P, Yu C, Li L, Yao SQ: Nanoquencher-based selective imaging of protein glutathionylation in live mammalian cells. Angew Chem Int Ed Engl (2018) 57(32):10257–10262. [DOI] [PubMed] [Google Scholar]; • This paper reports the nanoparticle-based sensor selectively responsive to SSG over GSH in live cells. This paper is the first example of selectively imaging SSG in live cells.
- 19.Gallogly MM, Starke DW, Mieyal JJ: Mechanistic and kinetic details of catalysis of thioldisulfide exchange by glutaredoxins and potential mechanisms of regulation. Antioxid Redox Sign (2009) 11(5):1059–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Begas P, Liedgens L, Moseler A, Meyer AJ, Deponte M: Glutaredoxin catalysis requires two distinct glutathione interaction sites. Nat Commun (2017) 8:14835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elgan TH, Berndt KD: Quantifying escherichia coli glutaredoxin-3 substrate specificity using ligand-induced stability. J Biol Chem (2008) 283(47):32839–32847. [DOI] [PubMed] [Google Scholar]
- 22.Nordstrand K, slund F, Holmgren A, Otting G, Berndt KD: NMR structure of escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: Implications for the enzymatic mechanism. J Mol Biol (1999) 286(2):541–552. [DOI] [PubMed] [Google Scholar]
- 23.Lind C, Gerdes R, Hamnell Y, Schuppe-Koistinen I, von Lowenhielm HB, Holmgren A, Cotgreave IA: Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch Biochem Biophys (2002) 406(2):229–240. [DOI] [PubMed] [Google Scholar]
- 24.Su D, Gaffrey MJ, Guo J, Hatchell KE, Chu RK, Clauss TR, Aldrich JT, Wu S, Purvine S, Camp DG, Smith RD et al. Proteomic identification and quantification of S-glutathionylation in mouse macrophages using resin-assisted enrichment and isobaric labeling. Free Radic Biol Med (2014) 67:460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duan J, Kodali VK, Gaffrey MJ, Guo J, Chu RK, Camp DG, Smith RD, Thrall BD, Qian WJ: Quantitative profiling of protein S-glutathionylation reveals redox-dependent regulation of macrophage function during nanoparticle-induced oxidative stress. ACS Nano (2016) 10(1):524–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cobley JN, Noble A, Jimenez-Fernandez E, Valdivia Moya MT, Guille M, Husi H: Catalyst-free click PEGylation reveals substantial mitochondrial ATP synthase sub-unit alpha oxidation before and after fertilisation. Redox Biol (2019) 26:101258. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper describes a click reaction-based PEGylation approach to quantify levels of SSG on individual proteins and its applications.
- 27.van Leeuwen LAG, Hinchy EC, Murphy MP, Robb EL, Cocheme HM: Click-PEGylation - a mobility shift approach to assess the redox state of cysteines in candidate proteins. Free Radic Biol Med (2017) 108:374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doulias PT, Gould NS: Analysis of cysteine post-translational modifications using organic mercury resin. Curr Protoc Protein Sci (2018) 94(1):e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coppo L, Ogata FT, Santhosh SM, Sventelius T, Holmgren A: Enzymatic glutaredoxin-dependent method to determine glutathione and protein S-glutathionylation using fluorescent eosin-glutathione. Anal Biochem (2019) 568:24–30. [DOI] [PubMed] [Google Scholar]
- 30.Kramer PA, Duan J, Gaffrey MJ, Shukla AK, Wang L, Bammler TK, Qian WJ, Marcinek DJ: Fatiguing contractions increase protein S-glutathionylation occupancy in mouse skeletal muscle. Redox Biol (2018) 17:367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper describes the identification of over 2,000 cysteines forming glutathionylation in skeletal muscle upon fatiguing contractions.
- 31.Parker CG, Pratt MR: Click chemistry in proteomic investigations. Cell (2020) 180(4):605–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samarasinghe KTG, Godage DNPM, VanHecke GC, Ahn YH: Metabolic synthesis of clickable glutathione for chemoselective detection of glutathionylation. J Am Chem Soc (2014) 136(33):11566–11569. [DOI] [PubMed] [Google Scholar]; •• This paper describes engineering glutathione synthetase that enables biosynthesis of clickable glutathione in cells for selective detection of glutathionylation.
- 33.Samarasinghe KTG, Ahn Y-H: Synthesizing clickable glutathione by glutathione synthetase mutant for detecting protein glutathionylation. Synlett (2015) 26(03):285–293. [Google Scholar]
- 34.Kekulandara DN, Samarasinghe KTG, Munkanatta Godage DNP, Ahn YH: Clickable glutathione using tetrazine-alkene bioorthogonal chemistry for detecting protein glutathionylation. Org Biomol Chem (2016) 14(46):10886–10893. [DOI] [PubMed] [Google Scholar]
- 35.VanHecke GC, Abeywardana MY, Ahn YH: Proteomic identification of protein glutathionylation in cardiomyocytes. J Proteome Res (2019) 18(4):1806–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yapa Abeywardana M, Samarasinghe KTG, Munkanatta Godage DNP, Ahn YH: Identification and quantification of glutathionylated cysteines under ischemic stress. J Proteome Res (2021) 20(9):4529–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Y, Zhu S, Yi M, Zhang W, Xue Y, Liu X, Deng H: Profiling glutathionylome in CD38-mediated epithelial-mesenchymal transition. J Proteome Res (2022) 21(5):1240–1250. [DOI] [PubMed] [Google Scholar]
- 38.Jin Y, Diffee GM, Colman RJ, Anderson RM, Ge Y: Top-down mass spectrometry of sarcomeric protein post-translational modifications from non-human primate skeletal muscle. J Am Soc Mass Spectrom (2019) 30(12):2460–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei L, Gregorich ZR, Lin Z, Cai W, Jin Y, McKiernan SH, McIlwain S, Aiken JM, Moss RL, Diffee GM, Ge Y: Novel sarcopenia-related alterations in sarcomeric protein post-translational modifications (PTMs) in skeletal muscles identified by top-down proteomics. Mol Cell Proteomics (2018) 17(1): 134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui W, Rohrs HW, Gross ML: Top-down mass spectrometry: Recent developments, applications and perspectives. Analyst (2011) 136(19):3854–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown KA, Melby JA, Roberts DS, Ge Y: Top-down proteomics: Challenges, innovations, and applications in basic and clinical research. Expert Rev Proteomics (2020) 17(10):719–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melby JA, Roberts DS, Larson EJ, Brown KA, Bayne EF, Jin S, Ge Y: Novel strategies to address the challenges in top-down proteomics. J Am Soc Mass Spectrom (2021) 32(6):1278–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen B, Brown KA, Lin Z, Ge Y: Top-down proteomics: Ready for prime time? Anal Chem (2018) 90(1):110–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laurent Q, Martinent R, Lim B, Pham AT, Kato T, Lopez-Andarias J, Sakai N, Matile S: Thiol-mediated uptake. JACS Au (2021) 1(6):710–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gutscher M, Pauleau AL, Marty L, Brach T, Wabnitz GH, Samstag Y, Meyer AJ, Dick TP: Real-time imaging of the intracellular glutathione redox potential. Nat Methods (2008) 5(6):553–559. [DOI] [PubMed] [Google Scholar]
- 46.Jiang X, Chen J, Bajic A, Zhang C, Song X, Carroll SL, Cai ZL, Tang M, Xue M, Cheng N, Schaaf CP et al. Quantitative real-time imaging of glutathione. Nat Commun (2017) 8:16087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du S, Liew SS, Zhang CW, Du W, Lang W, Yao CCY, Li L, Ge J, Yao SQ: Cell-permeant bioadaptors for cytosolic delivery of native antibodies: A “mix-and-go” approach. ACS Cent Sci (2020) 6(12):2362–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This paper describes a simple and modular approach for intracellular delivery of antibodies and its applications, including SSG detection, in cells.
- 48.Mailloux RJ: Protein S-glutathionylation reactions as a global inhibitor of cell metabolism for the desensitization of hydrogen peroxide signals. Redox Biol (2020) 32:101472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adachi T, Pimentel DR, Heibeck T, Hou X, Lee YJ, Jiang B, Ido Y, Cohen RA: S-glutathiolation of RAS mediates redox-sensitive signaling by angiotensin II in vascular smooth muscle cells. J Biol Chem (2004) 279(28):29857–29862. [DOI] [PubMed] [Google Scholar]
- 50.Yang Y, Dong X, Zheng S, Sun J, Ye J, Chen J, Fang Y, Zhao B, Yin Z, Cao P, Luo L: GSTpi regulates VE-cadherin stabilization through promoting S-glutathionylation of Src. Redox Biol (2020) 30:101416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abdelsaid MA, El-Remessy AB: S-glutathionylation of LMW-PTP regulates VEGF-mediated FAK activation and endothelial cell migration. J Cell Sci (2012) 125(Pt 20):4751–4760. [DOI] [PubMed] [Google Scholar]
- 52.Velu CS, Niture SK, Doneanu CE, Pattabiraman N, Srivenugopal KS: Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry (2007) 46(26):7765–7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sakai J, Li J, Subramanian KK, Mondal S, Bajrami B, Hattori H, Jia Y, Dickinson BC, Zhong J, Ye K, Chang CJ et al. Reactive oxygen species-induced actin glutathionylation controls actin dynamics in neutrophils. Immunity (2012) 37(6):1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J, Boja ES, Tan W, Tekle E, Fales HM, English S, Mieyal JJ, Chock PB: Reversible glutathionylation regulates actin polymerization in A431 cells. J Biol Chem (2001) 276(51):47763–47766. [DOI] [PubMed] [Google Scholar]
- 55.Kaus-Drobek M, Mucke N, Szczepanowski RH, Wedig T, Czarnocki-Cieciura M, Polakowska M, Herrmann H, Wyslouch-Cieszynska A, Dadlez M: Vimentin S-glutathionylation at cys328 inhibits filament elongation and induces severing of mature filaments in vitro. FEBS J (2020) 287(24) :5304–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reynaert NL, van der Vliet A, Guala AS, McGovern T, Hristova M, Pantano C, Heintz NH, Heim J, Ho YS, Matthews DE, Wouters EF et al. Dynamic redox control of NF-kappaB through glutaredoxin-regulated S-glutathionylation of inhibitory kappaB kinase beta. Proc Natl Acad Sci U S A (2006) 103(35):13086–13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Checconi P, Limongi D, Baldelli S, Ciriolo MR, Nencioni L, Palamara AT: Role of glutathionylation in infection and inflammation. Nutrients (2019) 11(8):1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anathy V, Aesif SW, Guala AS, Havermans M, Reynaert NL, Ho YS, Budd RC, Janssen-Heininger YM: Redox amplification of apoptosis by caspase-dependent cleavage of glutaredoxin 1 and S-glutathionylation of fas. J Cell Biol (2009) 184(2):241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anathy V, Roberson EC, Guala AS, Godburn KE, Budd RC, Janssen-Heininger YM: Redox-based regulation of apoptosis: S-glutathionylation as a regulatory mechanism to control cell death. Antioxid Redox Signal (2012) 16(6):496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adachi T, Weisbrod RM, Pimentel DR, Ying J, Sharov VS, Schoneich C, Cohen RA: S-glutathiolation by peroxynitrite activates serca during arterial relaxation by nitric oxide. Nat Med (2004) 10(11):1200–1207. [DOI] [PubMed] [Google Scholar]
- 61.Granzier HL, Labeit S: The giant protein titin: A major player in myocardial mechanics, signaling, and disease. Circ Res (2004) 94(3):284–295. [DOI] [PubMed] [Google Scholar]
- 62.Tharp CA, Haywood ME, Sbaizero O, Taylor MRG, Mestroni L: The giant protein titin’s role in cardiomyopathy: Genetic, transcriptional, and post-translational modifications of TTN and their contribution to cardiac disease. Front Physiol (2019) 10:1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Freundt JK, Linke WA: Titin as a force-generating muscle protein under regulatory control. J Appl Physiol (1985) (2019) 126(5):1474–1482. [DOI] [PubMed] [Google Scholar]
- 64.Alegre-Cebollada J, Kosuri P, Giganti D, Eckels E, Rivas-Pardo JA, Hamdani N, Warren CM, Solaro RJ, Linke WA, Fernandez JM: S-glutathionylation of cryptic cysteines enhances titin elasticity by blocking protein folding. Cell (2014) 156(6):1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This paper describes glutathionylation in titin immunoglobulin domains and its implications in titin-mediated force generation during contraction.
- 65.Loescher CM, Breitkreuz M, Li Y, Nickel A, Unger A, Dietl A, Schmidt A, Mohamed BA, Kotter S, Schmitt JP, Kruger M et al. Regulation of titin-based cardiac stiffness by unfolded domain oxidation (UNDOX). Proc Natl Acad Sci U S A (2020) 117(39):24545–24556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Furuhashi M, Hotamisligil GS: Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov (2008) 7(6):489–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Y, Liu Y, Zhao S, Xu W, Li Y, Zhao P, Wang D, Cheng H, Ke Y, Zhang X: Oxidative stress-induced FABP5 S-glutathionylation protects against acute lung injury by suppressing inflammation in macrophages. Nat Commun (2021) 12(1):7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rostkowski M, Olsson MH, Sondergaard CR, Jensen JH: Graphical analysis of pH-dependent properties of proteins predicted using propka. BMC Struct Biol (2011) 11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang J, Gupta V, Carroll KS, Liebler DC: Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat Commun (2014) 5:4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martell J, Seo Y, Bak DW, Kingsley SF, Tissenbaum HA, Weerapana E: Global cysteine-reactivity profiling during impaired insulin/IGF-1 signaling in C. elegans identifies uncharacterized mediators of longevity. Cell Chem Biol (2016) 23(8):955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng D, Liwinski T, Elinav E: Inflammasome activation and regulation: Toward a better understanding of complex mechanisms. Cell Discov (2020) 6:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Angosto-Bazarra D, Molina-Lopez C, Penin-Franch A, Hurtado-Navarro L, Pelegrin P: Techniques to study inflammasome activation and inhibition by small molecules. Molecules (2021) 26(6):1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li S, Wang L, Xu Z, Huang Y, Xue R, Yue T, Xu L, Gong F, Bai S, Wu Q, Liu J et al. ASC deglutathionylation is a checkpoint for NLRP3 inflammasome activation. J Exp Med (2021) 218(9):e20202637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Polonikov A: Endogenous deficiency of glutathione as the most likely cause of serious manifestations and death in COVID-19 patients. ACS Infect Dis (2020) 6(7):1558–1562. [DOI] [PubMed] [Google Scholar]
- 75.Davis DA, Bulut H, Shrestha P, Yaparla A, Jaeger HK, Hattori SI, Wingfield PT, Mieyal JJ, Mitsuya H, Yarchoan R: Regulation of the dimerization and activity of SARS-CoV-2 main protease through reversible glutathionylation of cysteine 300. Mbio (2021) 12(4):e0209421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi J, Sivaraman J, Song J: Mechanism for controlling the dimer-monomer switch and coupling dimerization to catalysis of the severe acute respiratory syndrome coronavirus 3c-like protease. J Virol (2008) 82(9):4620–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R: Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science (2020) 368(6489):409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Behring JB, van der Post S, Mooradian AD, Egan MJ, Zimmerman MI, Clements JL, Bowman GR, Held JM: Spatial and temporal alterations in protein structure by EGF regulate cryptic cysteine oxidation. Sci Signal (2020) 13(615):eaay7315. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This paper describes the global analysis of conformation-dependent redox signaling in kinases with examples of cysteine oxidations induced by EGF.
- 79.Siodlak D: Alpha,beta-dehydroamino acids in naturally occurring peptides. Amino Acids (2015) 47(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Townsend DM, Lushchak VI, Cooper AJ: A comparison of reversible versus irreversible protein glutathionylation. Adv Cancer Res (2014) 122:177–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Z, Lyons B, Truscott RJ, Schey KL: Human protein aging: Modification and crosslinking through dehydroalanine and dehydrobutyrine intermediates. Aging Cell (2014) 13(2):226–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Linetsky M, LeGrand RD: Glutathionylation of lens proteins through the formation of thioether bond. Mol Cell Biochem (2005) 272(1-2):133–144. [DOI] [PubMed] [Google Scholar]
- 83.Wang Z, Schey KL: Quantification of thioether-linked glutathione modifications in human lens proteins. Exp Eye Res (2018) 175:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lai KY, Galan SRG, Zeng Y, Zhou TH, He C, Raj R, Riedl J, Liu S, Chooi KP, Garg N, Zeng M et al. LanCLs add glutathione to dehydroamino acids generated at phosphorylated sites in the proteome. Cell (2021) 184(10):2680–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This paper reports LanCL catalyzing the formation of irreversible C-glutathionylation between dehydroalanine and GSH at the activation loop of kinases and the biological significance of C-glutathionylation.
- 85.Repka LM, Chekan JR, Nair SK, van der Donk WA: Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem Rev (2017) 117(8):5457–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Samarasinghe KTG, Godage DNPM, Zhou YN, Ndombera FT, Weerapana E, Ahn YH: A clickable glutathione approach for identification of protein glutathionylation in response to glucose metabolism. Mol Biosyst (2016) 12(8):2471–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang P, Zhang Q, Li S, Cheng B, Xue H, Wei Z, Shao T, Liu ZX, Cheng H, Wang Z: ICysMod: An integrative database for protein cysteine modifications in eukaryotes. Brief Bioinform (2021) 22(5):bbaa400. [DOI] [PubMed] [Google Scholar]
- 88.Hanschmann EM, Berndt C, Hecker C, Garn H, Bertrams W, Lillig CH, Hudemann C: Glutaredoxin 2 reduces asthma-like acute airway inflammation in mice. Front Immunol (2020) 11:561724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van de Wetering C, Manuel AM, Sharafi M, Aboushousha R, Qian X, Erickson C, MacPherson M, Chan G, Adcock IM, ZounematKermani N, Schleich F et al. Glutathione-S-transferase P promotes glycolysis in asthma in association with oxidation of pyruvate kinase m2. Redox Biol (2021) 47:102160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scalcon V, Folda A, Lupo MG, Tonolo F, Pei N, Battisti I, Ferri N, Arrigoni G, Bindoli A, Holmgren A, Coppo L et al. Mitochondrial depletion of glutaredoxin 2 induces metabolic dysfunction-associated fatty liver disease in mice. Redox Biol (2022) 51:102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chia SB, Nolin JD, Aboushousha R, Erikson C, Irvin CG, Poynter ME, van der Velden J, Taatjes DJ, van der Vliet A, Anathy V, Janssen-Heininger YMW: Glutaredoxin deficiency promotes activation of the transforming growth factor beta pathway in airway epithelial cells, in association with fibrotic airway remodeling. Redox Biol (2020) 37:101720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cha SJ, Lee S, Choi HJ, Han YJ, Jeon YM, Jo M, Lee S, Nahm M, Lim SM, Kim SH, Kim HJ et al. Therapeutic modulation of GSTO activity rescues Fus-associated neurotoxicity via deglutathionylation in als disease models. Dev Cell (2022) 57(6):783–798. [DOI] [PubMed] [Google Scholar]


