Abstract
Oligodendrocytes are central nervous system glial cells that wrap neuronal axons with their differentiated myelin membranes as biological insulators. There has recently been an emerging concept that multiple sclerosis could be triggered and promoted by various risk genes that appear likely to contribute to the degeneration of oligodendrocytes. Despite the known involvement of vitamin D, immunity, and inflammatory cytokines in disease progression, the common causes and key genetic mechanisms remain unknown. Herein, we focus on recently identified risk factors and risk genes in the background of multiple sclerosis and discuss their relationships.
Keywords: multiple sclerosis, oligodendrocyte, risk gene
1. Introduction
The myelin sheath is formed as a multilamellar membrane structure through the spiral wrapping of neuronal axons that act as insulators [1,2,3,4]. The transmission of each action potential on a limited membrane region is significantly promoted by the resulting saltatory conduction. Electrical signals are quickly derived to adjacent or distant neuronal cells and neuronal networks. If the myelin is damaged, however, fast signal transmission is not achieved, which causes defective neuronal function. This phenomenon is typically observed in demyelinating states. One well-known demyelinating disease is multiple sclerosis (MS) of the central nervous system (CNS). It is thought that MS is often caused by an abnormal autoimmune reaction in the CNS.
First defined by the National Multiple Sclerosis Society (in the United States) in 1996, MS is a chronic inflammatory disease that is characterized by demyelination mainly in the brain and, in turn, axonal degeneration [5]. The prevalence of MS is higher in Caucasians in the United States and Europe. The incidence rate is more than 100 patients per 100,000 population members in some areas of Northern Europe [6]. In Japan, the prevalence was estimated to be 1 to 5 patients per 100,000 population members, but this number has reportedly increased to 14 to 18 over a single decade [7]. The incidence of MS is increasing in both developed and developing countries [8]. The average age of onset of MS is middle-age and the disease is approximately twice as common in women than in men [9,10].
It is unclear why the number of MS patients has increased recently across countries and regions. There are various risks and possible reasons for the development of the disease, including smoking, vitamin D deficiency, obesity, and Epstein-Barr virus, which is a type of herpes virus [11]. MS also has genetic factors, as first-degree relatives and identical twins have a 25% chance of being affected [12]. The major histocompatibility complex (MHC) HLA-DRB1*15:01 allele was the first factor identified as a risk factor for MS [13]. Subsequent studies have shown that interleukin (IL) 2Rα and IL7R are also genetic factors [14].
MS symptoms likely depend on the tissues and regions where demyelination occurs. Some of the most common symptoms are optic neuritis and brainstem and spinal cord syndromes. Early clinical symptoms usually recover, but relapses are often followed by sequelae [15]. The onset is related to the location and size of the lesion, and even small lesions in the symptomatic zone are likely to cause symptoms, with magnetic resonance imaging showing typical “Dawson’s fingers” with periventricular lesions [16].
This narrative review will focus on the previously reported major risk factors of MS. We describe the disease and the possible therapeutic signaling pathways related to the risk factors as well as risk gene products in MS. We have selected the references using inclusion criteria that focus on reviews of MS and original papers. We have also included large-scale meta-analyses of original genetic studies.
2. MS and Environmental Factors (Vitamin D)
Epidemiological studies have shown that there are racial differences in developing MS, with a minimum prevalence at the equator and an increase with northern or southern latitudes [17]. Vitamin D is produced primarily by the action of ultraviolet B rays on the skin. This is supported by circumstantial evidence suggesting environmental factors of vitamin D deficiency due to a lack of sunlight as a predisposing factor for MS [18]. Indeed, vitamin D deficiency has been suggested as a possible cause of MS and/or contributor to the progression of MS, but it likely has limited pathological effects [19]. It has been reported that people with blood 25-OH-D levels of 40 ng/mL or higher have a 62% lower risk of MS than those with levels below 25 ng/mL [20], suggesting that having normal vitamin D levels reduces the risk of MS [9]. Serum 25-OH-D is a metabolite of vitamin D used to assess vitamin D levels in vivo (Figure 1) [20]. Adil et al. reported levels of vitamin D bioavailability and adipose tissue–secreted hormones such as adiponectin and leptin [21]. MS risk correlated with a genetic predisposition to the body mass index (BMI) but anti-correlated with the 25-OH-D level. Leptin and adiponectin have no effect on the increased risk of MS due to lowered vitamin D levels. Vitamin D supplementation modestly reverses the effect of obesity on MS [22]. In support of this study, Michaela et al. examined the association between 52 risk variants identified through genome-wide association studies (GWASs) and disease severity in MS and found that they were not associated with MS severity in terms of cohort, gender, age of onset, and HLA-DRB1*15:01 allele [21].
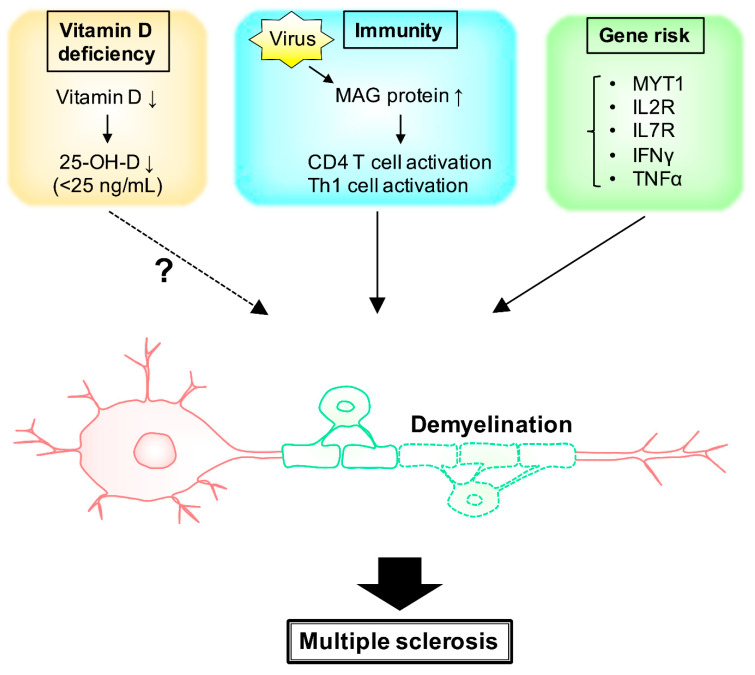
Figure 1.
Schematic diagram of some putative major factors associated with multiple sclerosis. Deficiency of vitamin D results in demyelination. The levels of 25-OH-D, a metabolite from vitamin D, is one of the risks of MS. MAG proteins, as well as signaling molecules around immune cells, are also related to MS demyelination.
Scazzone et al. investigated the effects of vitamin D–related genes on MS susceptibility. Of the 12 vitamin D gene product pathways investigated, the most studied was the vitamin D receptor and the least studied were other vitamin D–related gene products. Scazzone et al. reported that it is not clear whether these mutations directly affect the risk of MS [23]. When vitamin D supplementation is used as a treatment, no statistically significant differences were found and its effectiveness could not be demonstrated [19,24]. Despite the lack of significant difference, vitamin D alters the transcriptome profile of macrophages and microglia. In addition, vitamin D activates T cells (Figure 1) [25]. MS risk and genetic abnormalities in vitamin D metabolism have been reported in several cases, with genetic abnormalities for CYP27B1 in the cytochrome P450 family gene products, which is a regulator of calcitriol synthesis, influencing MS risk [26].
Genetic polymorphisms in the gene-encoding molecules involved in vitamin D homeostasis are associated with vitamin D deficiency. However, Klotho, which is coded as a protein with vitamin D metabolism, has no genotypic frequencies that differ between MS patients and controls [27]. This finding means that the role of Klotho does not involve genetic susceptibility to MS.
Several studies have strengthened the candidacy of the environmental factors between vitamin D and the most major risk gene HLA-DRB1*15:01 allele. Vitamin D deficiency is also reported to be an important MS disease pathogenesis.
3. MS and Environmental Factors (Immunity)
CNS fibers are covered with myelin sheaths whose composition contains abundant lipids and proteins. In MS, demyelinating plaques are involved in an immune response triggered by T cells. Proteolipid protein (PLP), myelin basic protein (MBP), and myelin oligodendrocyte glycoprotein (MOG) proteins have been well-studied as self-antigens involved in demyelination in MS [28]. The autoimmune disease against MOG is called MOG antibody (MOG-IgG)-associated disease [29]. The clinical features are considered to reflect a unique disease with a different etiology from MS and optic neuritis, related to aquaporin-4 (AQP-4)-IgG [30,31].
Immune responses to myelin-associated glycoprotein (MAG) have been primarily implicated in the development of MS. Increased MAG-recognizing T and B cells in MS patients have been observed [32]. However, the MAG peptide itself did not elicit disease-specific T and B cell responses, suggesting that this is secondary to demyelination rather than an attack on MAG by immune responses [33]. In addition to MAG proteins, environmental factors such as viral infection trigger demyelination somewhat. Then, differentiation into CD4-positive T cells and Th1-type cells results in one of the key events in the early stages of MS (Figure 1) [34].
Most of the more than 100 mutations in MS reported to date are related to the human leukocyte antigen (HLA) and the immune system, supporting the idea that MS is an immune disease. However, these mutations account for only 25% of heritability, leading to the new concept of “phantom heritability.” Sawcer et al. proposed insufficient non-redundant unnecessary sufficient (INUS), which describes the plurality of causation when a mutation cannot be found [35].
Experimental autoimmune encephalomyelitis (EAE) is a typical mouse model of MS and has also been the basis for its etiology and therapeutic development with regard to induced CNS inflammation. A few researchers have put forth that the debate should not be focused on EAE, arguing that the phenotype is weak [36]. Microglia, macrophages, and dendritic cells, which are potent antigen-presenting cells, have been reported to be increased in EAE mice [37]. Microglia and macrophages are present in MS lesions, myelin proteins MBP, and PLP as well as the minor myelin proteins [38]. Faber et al. compared gene expression in opticospinal EAE (OSE) and MOG EAE models. They demonstrated a more extensive enrichment of human MS risk genes among transcripts differentially expressed in OSE than in MOG EAE [39].
When Li et al. analyzed the transcriptional profiling data in the human brain in MS, 133 known and unknown genes were identified [40]. They included genes encoding a number of extracellular matrixes, such as collagen, signal-triggering receptor, and molecules involved in immune-related pathways and phosphatidylinositol-3 kinase (PI3K)-Akt pathways. Among them, four major extracellular and transmembrane proteins, IL17A, IL2, CD44, and IGF1, and 16 extracellular proteins interacting with IL17A have been associated with MS pathogenesis. Additionally, Del-1, which is an interacting protein with IL17A that may be associated with MS progression and relapse, has been identified as a probable biomarker.
Regulatory T cell (Treg) alteration has also been implicated in the pathogenesis of MS. X-linked forkhead box P3 (FoxP3) plays a crucial role in the development and stability of Tregs. However, FoxP3 and vitamin D3 did not have any association with MS [41].
The humoral immune response to Epstein-Barr virus nuclear antigen 1 (EBNA-1)-specific immunoglobulin γ (IgG) titers in families with MS was determined as a result of investigating the role of specific genetic loci on the antiviral IgG titers. The EBNA-1 IgG gradient being the highest in MS patients and the lowest in biologically unrelated spouses indicates a genetic contribution to EBNA-1 IgG levels that is only partially explained by HLA-DRB1*15:01 carriership [42].
Although it was previously known that non-coding RNAs (ncRNAs) create transcription noise, they are also now believed to be regulators of immune responses. Dysregulation of ncRNAs is one of the underlying mechanisms of immune disorders such as MS. Several studies have reported the aberrant expression of ncRNAs in the sera or blood cells of MS patients [43,44,45]. The results of these studies propose different classes of ncRNAs (long non-coding RNAs, microRNAs, and circular RNAs) as diagnostic or predictive markers in MS [46].
Demyelinating plaques are related to the autoimmune response in MS. The production of inflammatory cytokines caused by the immune response, such as PLP, MBP, and MOG, revealed MS as a chronic inflammatory disease.
4. Gene Risk and Signaling Pathway
Environmental cues associated with the increased risk of developing MS have been established, and over 200 risk loci with moderate to subtle effects have been described. To dissect the influence of genetic predisposition and environmental factors, Florian et al. investigated the peripheral immune signatures of 61 monozygotic twin pairs discordant for MS. They revealed an inflammatory shift in a monocyte cluster of twins with MS, coupled with the emergence of a population of naive helper T cells that have a transient response IL2 as MS-related immune alterations [47]. The research on genetically identical (monozygotic) twins shows that the concordance rate for MS is approximately 30%. This indicates that genetic and environmental factors interact with MS. Baranzini et al. examined DNA methylation and gene expression across the genome in three monozygotic twins discordant for MS; however, there were no consistent differences in DNA sequence [48]. It is surprising that the environment strongly indicated epigenetic modifications to germline susceptibility based on studies of adoptees, half-siblings, and avuncular pairs. The fact that complete explanations for disease heritability were unachieved after whole-genome association studies warrants consideration of all the factors contributing to disease risk, such as genetic, epigenetic, and environmental factors [49].
As many as 200 single-nucleotide polymorphisms (SNPs) are associated with MS risk (Table 1) [50,51,52,53]. Gresle et al. analyzed MS risk expression quantitative trait loci associations for 129 distinct genes in MS patients [54]. They identified the MS risk SNPs, rs2256814 Myelin transcription factor 1 (MYT1) in CD4 cells and rs12087340 RF00136 in monocyte cells. IL7 receptor (IL7R) is a member of the type I cytokine receptor family and is a primary pleiotropic receptor in immune cells (Figure 1). Two GWASs of MS reported that three SNPs outside of the MHC region were associated with MS: rs6897032 within the IL7R gene and two SNPs (rs2104286 and rs12722489) in the IL2R gene [14,55]. Omraninava et al. revealed that the IL7RA gene rs6897932 SNP decreases MS susceptibility (Figure 1) [56]. Infection with the herpes virus and Mycoplasma pneumonia create grounds for MS. The T allele in the IFNγ gene (+874) and the genotypes of AA and AG at the TNFα gene (-308) at position−308 were considered potential risk factors for MS (Figure 1) [57]. Despite GWASs explaining that there are common SNPs associated with various diseases, known common variants only account for part of the estimated heritability of common complex diseases. Nadia et al. identified the rare functional variants analyzed within a large Italian MS multiplex family with five affected members [58]. Another recent study showed that up to 5% of MS inheritability may be accounted for by rare variations in the gene coding sequence, with four novel low genes driving MS risk independently of common variant signals [59]. Based on the research of a large cohort of Italian individuals, researchers identified three SNPs (rs4267364, rs8070463, rs67919208) that were involved in the regulation of TBK1 Binding Protein 1 (TBKBP1) and prioritized them as functionally relevant in MS [60]. Recent GWAS research in MS that has analyzed up to 47,000 MS patients and 68,000 healthy controls has determined more than 200 non-MHC genome-wide associations. The results show that immune cells, such as T cells, B cells, and monocytes, have susceptible gene specificity [61]. The International Multiple Sclerosis Consortium analyzed the large-scale GWAS data of 47,000 MS patients and 68,000 healthy controls and established a reference genetic map of MS. Their findings demonstrate the enrichment of MS genes in these brain-resident immune cells, suggesting that they may have a role in targeting an autoimmune process to the CNS, although MS is most likely initially triggered by a perturbation in peripheral immune responses [52].
Table 1.
The risk allele and its possible role for the 200 autosomal non-MHC genome-wide effects. This list shows the 200 SNP regions and the possible roles of probable genes associated with MS risks, as identified by SNP analyses [50,51,52,53].
| SNP Region | Gene | Protein | Possible Role of Nearest Gene |
|---|---|---|---|
| rs6742 | rtel1 | RTEL1 | DNA helicase |
| rs32658 | fam170a | FAM170A | DNA binding activator |
| rs137955 | rpl3 | RPL3 | ribosomal protein |
| rs140522 | hdac10 | HDAC10 | deacetylase |
| rs198398 | mtor | MTOR | rapamycin kinase |
| rs244656 | ppp2ca | PPP2CA | catalytic subunit of protein phosphatase |
| rs249677 | arhgap26 | ARHGAP26 | GTPase activating protein |
| rs354033 | znf862 | ZNF862 | zinc finger protein |
| rs405343 | axin1 | AXIN1 | cytoplasmic protein |
| rs438613 | eomes | EOMES | DNA binding domain |
| rs483180 | notch2 | NOTCH2 | notch receptor |
| rs531612 | rela | RELA | proto-oncogene transcription factor |
| rs631204 | tnfaip3 | TNFAIP3 | cytokine |
| rs701006 | arhgap9 | ARHGAP9 | GTPase activating protein |
| rs719316 | atxn1 | ATXN1 | DNA binding protein |
| rs735542 | myc | MYC | proto-oncogene transcription factor |
| rs760517 | lgals1 | LGALS1 | galactoside binding protein |
| rs802730 | ptprk | PTPRK | protein tyrosine phosphatase receptor |
| rs883871 | rara | RARA | retinoic acid receptor |
| rs962052 | rnd3 | RND3 | Rho family GTPase |
| rs983494 | cd48 | CD48 | immune response regulator |
| rs1014486 | il12a | IL12A | cytokine |
| rs1026916 | hoxa13 | HOXA13 | homeobox |
| rs1076928 | pim1 | PIM1 | proto-oncogene kinase |
| rs1077667 | c3 | C3 | complement component |
| rs1087056 | znf438 | ZNF438 | zinc finger protein |
| rs1112718 | ide | IDE | insulin enzyme |
| rs1177228 | commd1 | COMMD1 | copper metabolism |
| rs1250551 | zmiz1 | ZMIZ1 | zinc finger protein |
| rs1323292 | rgs1 | RGS1 | G Protein Signaling |
| rs1365120 | traf6 | TRAF6 | adaptor protein |
| rs1399180 | gata3 | GATA3 | transcription factor |
| rs1415069 | bcar3 | BCAR3 | anti-estrogen resistance protein |
| rs1465697 | atf5 | ATF5 | transcription factor |
| rs1738074 | synj2 | SYNJ2 | inositol polyphosphate 5-phosphatase |
| rs1800693 | cd9 | CD9 | immune response regulator |
| rs2084007 | ppp2ca | PPP2CA | catalytic subunit of protein phosphatase |
| rs2150879 | rps6kb1 | RPS6KB1 | ribosomal protein |
| rs2248137 | znf217 | ZNF217 | zinc finger protein |
| rs2269434 | celf1 | CELF1 | alternative splicing |
| rs2286974 | litaf | LITAF | cytokine |
| rs2289746 | alcam | ALCAM | immunoglobulin receptor |
| rs2317231 | cd1e | CD1E | immune response regulator |
| rs2327586 | sgk1 | SGK1 | serine/threonine kinase |
| rs2331964 | cd86 | CD86 | immune response regulator |
| rs2364485 | cd9 | CD9 | immune response regulator |
| rs2469434 | cd226 | CD226 | immune response regulator |
| rs2546890 | il12b | IL12B | cytokine |
| rs2585447 | znf217 | ZNF217 | zinc finger protein |
| rs2590438 | bcl6 | BCL6 | immune signaling receptor |
| rs2705616 | mapk10 | MAPK10 | MAPK |
| rs2726479 | cxxc4 | CXXC4 | zinc finger protein |
| rs2836438 | ets2 | ETS2 | transcription factor |
| rs2986736 | camta1 | CAMTA1 | transcription activator |
| rs3184504 | arpc3 | ARPC3 | cell polymerization |
| rs3737798 | cd48 | CD48 | immune response regulator |
| rs3809627 | mapk3 | MAPK3 | MAPK |
| rs3923387 | plec | PLEC | cytoskeleton |
| rs4262739 | ets1 | ETS1 | proto-oncogene transcription factor |
| rs4325907 | rpl24 | RPL24 | ribosomal protein |
| rs4409785 | maml2 | MAML2 | cytoplasmic protein |
| rs4728142 | smo | SMO | G protein-coupled receptor |
| rs4796224 | acaca | ACACA | acetyl-CoA carboxylase |
| rs4808760 | ifi30 | IFI30 | lysosomal thiol reductase |
| rs4812772 | mybl2 | MYBL2 | proto-oncogene transcription factor |
| rs4820955 | lif | LIF | cytokine |
| rs4896153 | bclaf1 | BCLAF1 | BCL transcription factor |
| rs4939490 | fads1 | FADS1 | fatty acid desaturase |
| rs4940730 | malt1 | MALT1 | caspase-like protease |
| rs5756405 | rac2 | RAC2 | GTP binding protein |
| rs6020055 | cebpb | CEBPB | transcriptional activator protein |
| rs6032662 | cd40 | CD40 | immune response regulator |
| rs6072343 | plcg1 | PLCG1 | phospholipase |
| rs6427540 | cd48 | CD48 | immune response regulator |
| rs6496663 | iqgap1 | IQGAP1 | GTPase activating protein |
| rs6533052 | nfkb1 | NFKB1 | cytokine |
| rs6564681 | maf | MAF | proto-oncogene kinase |
| rs6589706 | kmt2a | KMT2A | Lysine Methyltransferase |
| rs6589939 | clmp | CLMP | transmembrane protein |
| rs6670198 | prdm16 | PRDM16 | zinc finger protein |
| rs6672420 | runx3 | RUNX3 | transcription factor |
| rs6738544 | stat1 | STAT1 | transcription activator |
| rs6789653 | zbtb38 | ZBTB38 | zinc finger protein |
| rs6837324 | tec | TEC | tyrosine kinase |
| rs6911131 | hivep2 | HIVEP2 | zinc finger protein |
| rs6990534 | myc | MYC | proto-oncogene transcription factor |
| rs7222450 | crhr1 | CRHR1 | G-protein coupled receptor |
| rs7260482 | apoe | APOE | apoprotein |
| rs7731626 | map3k1 | MAP3K1 | MAPK kinase |
| rs7855251 | anp32b | ANP32B | RNA polymerase binding protein |
| rs7975763 | mphosph9 | MPHOSPH9 | M phase phosphoprotein |
| rs7977720 | olr1 | OLR1 | low density lipoprotein receptor |
| rs8062446 | nlrc5 | NLRC5 | cytokine receptor |
| rs9308424 | batf3 | BATF3 | basic leucine zipper protein |
| rs9568402 | rnaseh2b | RNASEH2B | ribonuclease |
| rs9591325 | rnaseh2b | RNASEH2B | ribonuclease |
| rs9610458 | ube2l3 | UBE2L3 | ubiquitin conjugating enzyme |
| rs9808753 | ifnar2 | IFNAR2 | interferon receptor |
| rs9843355 | cd80 | CD80 | immune response regulator |
| rs9863496 | satb1 | SATB1 | matrix protein |
| rs9878602 | rybp | RYBP | DNA binding protein |
| rs9900529 | grb2 | GRB2 | growth factor receptor |
| rs9909593 | rara | RARA | retinoic acid receptor |
| rs9955954 | malt1 | MALT1 | caspase-like protein |
| rs9992763 | rpl34 | RPL34 | ribosomal protein |
| rs10063294 | slc1a3 | EAA1 | transporter |
| rs10191360 | cxcr4 | CXCR4 | chemokine receptor |
| rs10230723 | ikzf1 | IKAROS | DNA binding protein |
| rs10245867 | hoxa13 | HOXA13 | homeobox |
| rs10271373 | tbxas1 | TBXAS1 | lipid synthase |
| rs10801908 | atp1a1 | ATP1A1 | transporting subunit |
| rs10936182 | il12a | IL12A | cytokine |
| rs10936602 | mecom | MDS1 And EVI1 Complex Locus | zinc finger protein |
| rs10951042 | mad1l1 | MAD1 | cell cycle controller |
| rs10951154 | hoxa4 | HOXA4 | homeobox |
| rs11079784 | npepps | NPEPPS | peptidase |
| rs11083862 | c5ar1 | C5AR1 | complement component receptor |
| rs11125803 | adcy3 | ADCY3 | adenylate cyclase |
| rs11161550 | bcl10 | BCL10 | immune signaling receptor |
| rs11231749 | esrra | ESRRA | estrogen related receptor |
| rs11256593 | pfkfb3 | PFKFB3 | phosphofructo kinase |
| rs11578655 | extl2 | EXTL2 | glycosyltransferase |
| rs11749040 | dab2 | DAB2 | adaptor protein |
| rs11809700 | rpl5 | RPL5 | ribosomal protein |
| rs11852059 | ptger2 | PTGER2 | prostaglandin receptor |
| rs11899404 | lpin1 | LPIN1 | lipid phosphohydrolase |
| rs11919880 | cnot10 | CNOT10 | transcription complex |
| rs12133753 | cdc7 | CDC7 | cell cycle kinase |
| rs12147246 | rcor1 | RCOR1 | transcription factor |
| rs12211604 | rreb1 | RREB1 | binding protein |
| rs12365699 | kmt2a | KMT2A | methyltransferase |
| rs12434551 | zfp36l1 | ZFP36L1 | zinc finger protein |
| rs12478539 | zfp36l2 | ZFP36L2 | zinc finger protein |
| rs12588969 | rcor1 | RCOR1 | chromatin binding |
| rs12609500 | tyk2 | TYK2 | tyrosine kinase |
| rs12614091 | cd28 | CD28 | immune response regulator |
| rs12622670 | aplf | APLF | component of the cellular response |
| rs12722559 | pfkfb3 | PFKFB3 | glycolysis-related biphosphatase |
| rs12832171 | cd9 | CD9 | immune response regulator |
| rs12925972 | maf | MAF | proto-oncogene kinase |
| rs12971909 | map2k2 | MAP2K2 | MAPK kinase |
| rs13066789 | bcl6 | BCL6 | immune signaling receptor |
| rs13136820 | uchl1 | UCHL1 | ubiquitin hydrolase |
| rs13327021 | eomes | EOMES | DNA binding domain |
| rs13385171 | sertad2 | SERTAD2 | transcription activator |
| rs13414105 | alk | ALK | tyrosine kinase |
| rs17051321 | qrfpr | QRFPR | pyroglutamylated receptor |
| rs17724508 | maf | MAF | proto-oncogene kinase |
| rs17741873 | camk2g | CAMK2G | CAM kinase |
| rs17780048 | tnfaip3 | TNFAIP3 | cytokine |
| rs28703878 | pkia | PKIA | protein kinase inhibitor |
| rs28834106 | dnm2 | DNM2 | GTP binding protein |
| rs34026809 | kmt2a | KMT2A | methyltransferase |
| rs34536443 | tyk2 | TYK2 | tyrosine kinase |
| rs34681760 | adcy2 | ADCY2 | adenylate cyclase |
| rs34695601 | fos | FOS | proto-oncogene transcription factor |
| rs34723276 | extl2 | EXTL2 | glycosyltransferase |
| rs34947566 | litaf | LITAF | cytokine |
| rs35218683 | deaf1 | DEAF1 | zinc finger protein |
| rs35486093 | bcl10 | BCL10 | adaptor protein |
| rs35540610 | sp110 | SP110 | nuclear body protein |
| rs35703946 | irf8 | IRF8 | cytokine |
| rs55858457 | mad1l1 | MAD1L1 | cell cycle controller |
| rs56095240 | maml2 | MAML2 | transcriptional activator |
| rs57116599 | il1b | IL1B | cytokine |
| rs58166386 | rasal3 | RASAL3 | Ras GTPase activating protein |
| rs58394161 | rpl5 | RPL5 | ribosomal protein |
| rs59655222 | znf281 | ZNF281 | zinc finger protein |
| rs60600003 | elmo1 | ELMO1 | adaptor protein |
| rs61708525 | plxnc1 | PLXNC1 | transmembrane receptor |
| rs61863928 | egr2 | EGR2 | transcription factor |
| rs61884005 | arntl | ARNTL | transcriptional activator |
| rs62013236 | acsbg1 | ACSBG1 | acyl-CoA synthetase |
| rs62420820 | tnfaip3 | TNFAIP3 | cytokine |
| rs67111717 | nsd1 | NSD1 | transcriptional regulator |
| rs67934705 | rpl11 | RPL11 | ribosomal protein |
| rs71329256 | cd86 | CD86 | immune response regulator |
| rs72922276 | pde4b | PDE4B | phosphodiesterase |
| rs72928038 | rragd | RRAGD | Ras related GTPase binding protein |
| rs72989863 | march1 | MARCH1 | ubiquitin protein ligase |
| rs73414214 | pik3cg | PIK3CG | Phosphoinositide 3-kinase |
| chr1:154983036 | arhgef2 | ARHGEF2 | Rho/Rac guanine nucleotide exchanger |
| chr1:32738415 | hdac1 | HDAC1 | histone deacetylase |
| chr2:112492986 | anapc1 | ANAPC1 | anaphase-promoting complex |
| chr3:100848597 | rpl24 | RPL24 | ribosomal protein |
| chr3:112693983 | cd200 | CD200 | immune response regulator |
| chr3:121783015 | cd86 | CD86 | immune response regulator |
| chr5:40429250 | dab2 | DAB2 | DAB adaptor protein |
| chr6:119215402 | mcm9 | MCM9 | ATP hydrolysis activity |
| chr6:130348257 | arhgap18 | ARHGAP18 | Ras GTPase activating protein |
| chr6:14691215 | jarid2 | JARID2 | transcriptional repressor |
| chr7:50328339 | ikzf1 | IKZF1 | zinc finger protein |
| chr8:129177769 | myc | MYC | proto-oncogene transcription factor |
| chr8:95851818 | rad54b | RAD54B | DEAD-like helicase |
| chr11:118783424 | kmt2a | KMT2A | lysine methyltransferase |
| chr11:14868316 | pde3b | PDE3B | phosphodiesterase |
| chr13:100026952 | dock9 | DOCK9 | Cdc42 guanine nucleotide exchanger |
| chr14:88523488 | kcnk10 | KCNK10 | potassium channel protein |
| chr16:11213951 | litaf | LITAF | cytokine |
| chr16:11353879 | litaf | LITAF | cytokine |
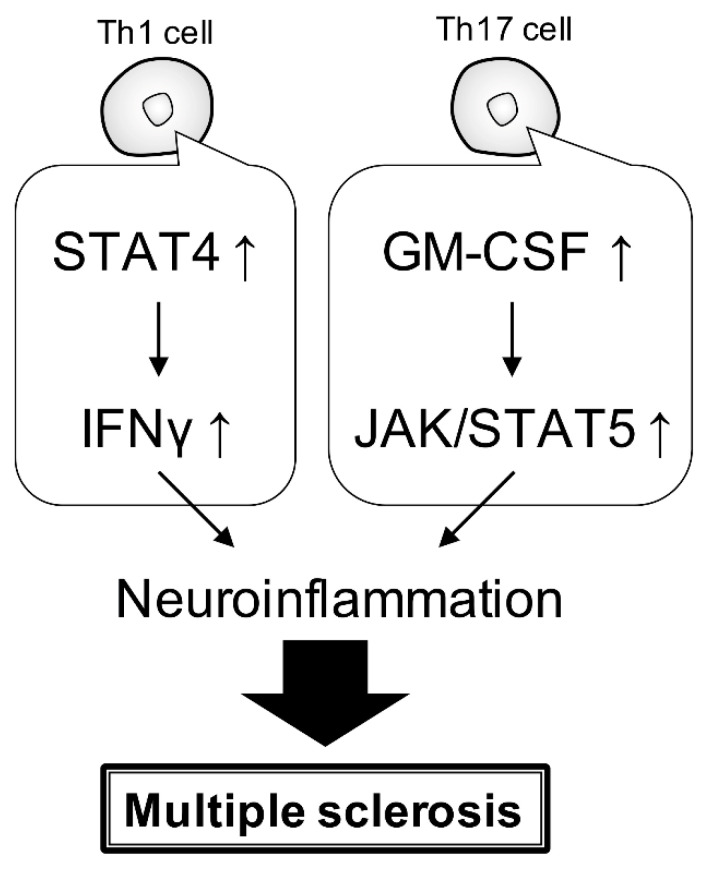
The Janus kinase and signal transducer and activator of the transcription (JAK/STAT) pathway is essential for both innate and acquired immunity. It has also been reported to be associated with several neuroinflammatory diseases (Figure 2) [62]. In EAE mice, Th1 cells produce interferon-gamma (IFNγ) via STAT4 and inflammatory macrophages, which promote macrophage activation. Similarly, Th17 produces granulocyte-macrophage colony-stimulating factor (GM-CSF) in the CNS and promotes macrophage polarization to inflammation via JAK/STAT5 (Figure 2) [63].
Figure 2.
JAK/STAT signaling pathway associated with MS. Cytokines, through JAK/STAT signaling, especially in Th1 and Th17 cells, are putatively considered responsible for the progression of MS.
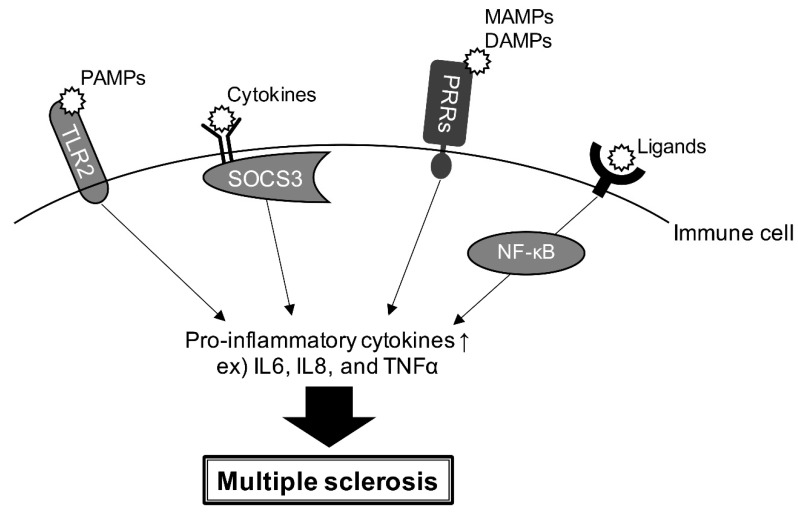
A comprehensive analysis of genes in the brain of MS patients has shown increased levels of immune cell populations and decreased ones of endothelial cells, Th1 cells, and Treg cells in MS lesions [64]. Toll-like receptors (TLRs) have a variety of roles, including axonal pathway formation and dorsoventral patterning in the CNS. TLR ligands, such as pathogen-associated molecular patterns (PAMPs), have been identified as T cell promoters in MS. In particular, TLR2 expression is high in MS lesions and TLR2 activation induces the expression of pro-inflammatory cytokines such as IL-6, IL-8, and TNF-α, which are implicated in exacerbated inflammation (Figure 3) [65]. The HLA signal in the Italian population maps to a glycoprotein involved in dendritic cell (DC) maturation, such as TNFSF14 gene encoding LIGHT. Miriam et al. reported that the TNFSF14 intronic SNP rs1077667 was the main MS-associated variant in the region. That means that the intronic variant rs1077667 alters the expression of TNFSF14 in DCs, which may play a role in MS pathogenesis [66]. A variant in TNHSH13B, encoding the cytokine and drug target B-cell activating factor (BAFF), was associated with upregulated humoral immunity through increased levels of soluble BAFF, B lymphocytes, and immunoglobulins in MS [67]. Leptin (LEP) and leptin receptor (LEPR) overexpression are related to MS activity and progression, and peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC1A) is able to affect the reactive oxygen species production in the pathogenesis of MS. LEP rs7799039 and LEPR rs1137101 genetic variants modify the serum LEP levels and PGC1A rs8192678 alters the PGC1A activity. Ivana et al. revealed that the PGC1A rs8192678 minor allele had an increased risk for the occurrence of MS, and LEP rs7799039 affected the LEP gene expression in relapsing-remitting patients [68].
Figure 3.
Interaction of some receptors with their cognate ligands induces the expression of pro-inflammatory cytokines in immune cells. Pathogen-associated molecular patterns interaction with TLRs, SOCS3 activation by cytokine receptors, microbe-associated molecular patterns or damage-associated molecular patterns binding to pattern recognition receptors, and/or activation through NF-κB are involved in the regulation of the expression of inflammatory cytokines, which are responsible for MS, in immune cells.
Furthermore, in relapsing MS, reduced suppression of cytokine signaling-3 (SOCS3) expression in the CNS and immune cells may induce LEP-mediated overexpression of pro-inflammatory cytokines (Figure 3) [69]. Pattern recognition receptors, which are triggered by both microbe-associated molecular patterns and damage-associated molecular patterns, have been reported to regulate innate immune responses in MS and an EAE model. Pattern recognition receptor signaling promotes inflammatory-producing cytokine production in CNS autoimmune diseases (Figure 3) [70]. NF-κB is involved in a wide range of vital processes, including inflammation, cell proliferation, and differentiation. Abnormal NF-κB activation has been reported to be closely associated with the development of MS and EAE [71].
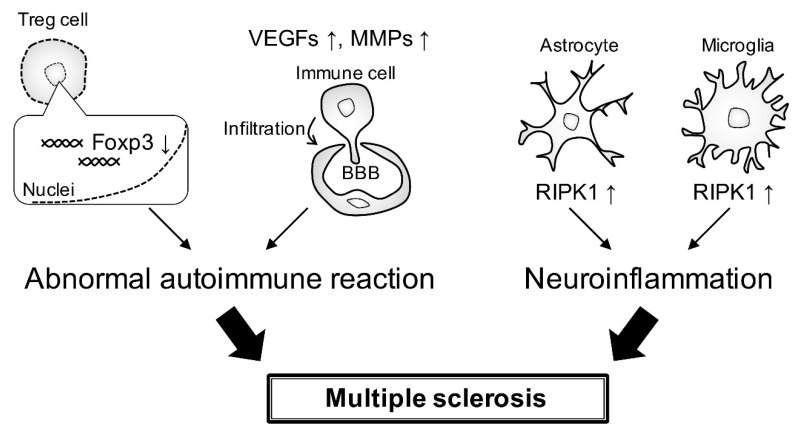
In MS, the altered Foxp3-E2 variant-associated inhibitory activity of Treg cells is associated with defective signaling via IL-2 and glycolysis, which modulates Treg cell induction and function in autoimmunity [72]. The expression of vascular endothelial growth factors and matrix metallopeptidases involved in angiogenesis is increased in MS. These genes are also involved in basement membrane degradation and blood–brain barrier disruption, which allows immune cells to infiltrate the CNS in EAE and MS (Figure 4) [73]. Programmed cell death 1 (PD-1) is known as an immune checkpoint that is associated with several autoimmune diseases. Research on the frequency of PD-1 genotypes and alleles in MS patients shows that PD-1 gene polymorphisms may be associated with MS [74]. Phosphorylation of receptor-interacting protein kinase 1 (RIPK1) in astrocytes and microglia triggers a detrimental neuroinflammatory program that contributes to the neurodegenerative environment in MS (Figure 4) [75].
Figure 4.
Abnormal autoimmune reaction and neuroinflammation in MS. Altered Foxp3 expression in Treg cells induces an abnormal autoimmune reaction. Expression levels of vascular endothelial growth factors and matrix metallopeptidases are increased, probably disrupting the blood–brain barrier. This disruption allows immune cells to infiltrate. Phosphorylation of RIPK1 in astrocytes and microglia is involved in the promotion of the neuroinflammatory program.
Risk genes have been well studied by meta-analyses and many SNPs have been identified. MYT1, IL2R, IL7R, IFNγ, and TNFα, among others, are considered to be the major risk genes in MS. The related major signaling in MS is the JAK/STAT pathway.
5. Conclusions and Perspective
We have examined and discussed the genetic risks in the background of MS. The major risks include (1) the genes related to vitamin D deficiency, (2) the genes involved in the immune response, and (3) the genes responsible for inflammatory cytokines and the related signaling molecules. Nucleotide sequence analyses with advancing technologies have clarified that there are an increasing number of other possible categories of risk genes besides these in MS. In the future, molecules related to these risk gene products may be promising therapeutic target candidates.
Acknowledgments
We thank Takako Morimoto and Yoichi Seki (Tokyo University of Pharmacy and Life Sciences) for their insightful comments.
Author Contributions
Conceptualization, R.S. and J.Y.; methodology, R.S.; software, R.S.; validation, R.S. and J.Y.; formal analysis, R.S.; investigation, R.S.; resources, R.S.; data curation, R.S. and J.Y.; writing—original draft preparation, R.S.; writing—review and editing, R.S. and J.Y.; visualization, R.S. and J.Y.; supervision, J.Y.; project administration, J.Y. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors have declared that no competing interest exist.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Simons M., Nave K.A. Oligodendrocytes: Myelination and axonal xsupport. Cold Spring Harb. Perspect. Biol. 2015;8:a020479. doi: 10.1101/cshperspect.a020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barateiro A., Brites D., Fernandes A. Oligodendrocyte development and myelination in neurodevelopment: Molecular mechanisms in health and disease. Curr. Pharm. Des. 2016;22:656–679. doi: 10.2174/1381612822666151204000636. [DOI] [PubMed] [Google Scholar]
- 3.Elbaz B., Popko B. Molecular control of oligodendrocyte development. Trends Neurosci. 2019;42:263–277. doi: 10.1016/j.tins.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams K.L., Dahl K.D., Gallo V., Macklin W.B. Intrinsic and extrinsic regulators of oligodendrocyte progenitor proliferation and differentiation. Semin. Cell. Dev. Biol. 2021;116:16–24. doi: 10.1016/j.semcdb.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lublin F.D., Reingold S.C. Defining the clinical course of multiple sclerosis: Results of an international survey. National Multiple Sclerosis Society (USA) Advisory Committee on Clinical Trials of New Agents in Multiple Sclerosis. Neurology. 1996;46:907–911. doi: 10.1212/WNL.46.4.907. [DOI] [PubMed] [Google Scholar]
- 6.Glaser A., Stahmann A., Meissner T., Flachenecker P., Horakova D., Zaratin P., Brichetto G., Pugliatti M., Rienhoff O., Vukusic S., et al. Multiple sclerosis registries in Europe—An updated mapping survey. Mult. Scler. Relat. Disord. 2019;27:171–178. doi: 10.1016/j.msard.2018.09.032. [DOI] [PubMed] [Google Scholar]
- 7.Houzen H., Kondo K., Horiuchi K., Niino M. Consistent increase in the prevalence and female ratio of multiple sclerosis over 15 years in northern Japan. Eur. J. Neurol. 2018;25:334–339. doi: 10.1111/ene.13506. [DOI] [PubMed] [Google Scholar]
- 8.Browne P., Chandraratna D., Angood C., Tremlett H., Baker C., Taylor B.V., Thompson A.J. Atlas of Multiple Sclerosis 2013: A growing global problem with widespread inequity. Neurology. 2014;83:1022–1024. doi: 10.1212/WNL.0000000000000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pierrot-Deseilligny C., Souberbielle J.C. Vitamin D and multiple sclerosis: An update. Mult. Scler. Relat. Disord. 2017;14:35–45. doi: 10.1016/j.msard.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Nohara C., Hase M., Liebert R., Wu N. The burden of multiple sclerosis in Japan. J. Med. Econ. 2017;20:1290–1298. doi: 10.1080/13696998.2017.1373653. [DOI] [PubMed] [Google Scholar]
- 11.Alfredsson L., Olsson T. Lifestyle and Environmental Factors in Multiple Sclerosis. Cold Spring Harb. Perspect. Med. 2019;9:a028944. doi: 10.1101/cshperspect.a028944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baecher-Allan C., Kaskow B.J., Weiner H.L. Multiple sclerosis: Mechanisms and immunotherapy. Neuron. 2018;97:742–768. doi: 10.1016/j.neuron.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 13.Sawcer S., Ban M., Maranian M., Yeo T.W., Compston A., Kirby A., Daly M.J., De Jager P.L., Walsh E., Lander E.S., et al. A high-density screen for linkage in multiple sclerosis. Am. J. Hum. Genet. 2005;77:454–467. doi: 10.1086/444547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.International Multiple Sclerosis Genetics C., Hafler D.A., Compston A., Sawcer S., Lander E.S., Daly M.J., De Jager P.L., de Bakker P.I., Gabriel S.B., Mirel D.B., et al. Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 15.Dobson R., Giovannoni G. Multiple sclerosis—A review. Eur. J. Neurol. 2019;26:27–40. doi: 10.1111/ene.13819. [DOI] [PubMed] [Google Scholar]
- 16.Hemond C.C., Bakshi R. Magnetic resonance imaging in multiple sclerosis. Cold Spring Harb. Perspect. Med. 2018;8:a028969. doi: 10.1101/cshperspect.a028969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simpson S., Jr., Blizzard L., Otahal P., Van der Mei I., Taylor B. Latitude is significantly associated with the prevalence of multiple sclerosis: A meta-analysis. J. Neurol. Neurosurg. Psychiatry. 2011;82:1132–1141. doi: 10.1136/jnnp.2011.240432. [DOI] [PubMed] [Google Scholar]
- 18.Jagannath V.A., Filippini G., Di Pietrantonj C., Asokan G.V., Robak E.W., Whamond L., Robinson S.A. Vitamin D for the management of multiple sclerosis. Cochrane Database Syst. Rev. 2018;9:CD008422. doi: 10.1002/14651858.CD008422.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lincoln M.R., Schneider R., Oh J. Vitamin D as disease-modifying therapy for multiple sclerosis? Expert Rev. Clin. Immunol. 2021;17:691–693. doi: 10.1080/1744666X.2021.1915772. [DOI] [PubMed] [Google Scholar]
- 20.Munger K.L., Levin L.I., Hollis B.W., Howard N.S., Ascherio A. Serum 25-hydroxyvitamin D levels and risk of multiple sclerosis. JAMA. 2006;296:2832–2838. doi: 10.1001/jama.296.23.2832. [DOI] [PubMed] [Google Scholar]
- 21.George M.F., Briggs F.B., Shao X., Gianfrancesco M.A., Kockum I., Harbo H.F., Celius E.G., Bos S.D., Hedstrom A., Shen L., et al. Multiple sclerosis risk loci and disease severity in 7,125 individuals from 10 studies. Neurol. Genet. 2016;2:e87. doi: 10.1212/NXG.0000000000000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harroud A., Manousaki D., Butler-Laporte G., Mitchell R.E., Davey Smith G., Richards J.B., Baranzini S.E. The relative contributions of obesity, vitamin D, leptin, and adiponectin to multiple sclerosis risk: A Mendelian randomization mediation analysis. Mult. Scler. 2021;27:1994–2000. doi: 10.1177/1352458521995484. [DOI] [PubMed] [Google Scholar]
- 23.Scazzone C., Agnello L., Bivona G., Lo Sasso B., Ciaccio M. Vitamin D and genetic susceptibility to multiple sclerosis. Biochem. Genet. 2021;59:1–30. doi: 10.1007/s10528-020-10010-1. [DOI] [PubMed] [Google Scholar]
- 24.McLaughlin L., Clarke L., Khalilidehkordi E., Butzkueven H., Taylor B., Broadley S.A. Vitamin D for the treatment of multiple sclerosis: A meta-analysis. J. Neurol. 2018;265:2893–2905. doi: 10.1007/s00415-018-9074-6. [DOI] [PubMed] [Google Scholar]
- 25.Clarke J., Yaqubi M., Futhey N.C., Sedaghat S., Baufeld C., Blain M., Baranzini S., Butovsky O., Antel J., White J.H., et al. Vitamin D regulates MerTK-dependent phagocytosis in human myeloid cells. J. Immunol. 2020;205:398–406. doi: 10.4049/jimmunol.2000129. [DOI] [PubMed] [Google Scholar]
- 26.Ramasamy A., Trabzuni D., Forabosco P., Smith C., Walker R., Dillman A., Sveinbjornsdottir S., North American Brain Expression Consortium U.K.B.E.C., Hardy J., Weale M.E., et al. Genetic evidence for a pathogenic role for the vitamin D3 metabolizing enzyme CYP24A1 in multiple sclerosis. Mult. Scler. Relat. Disord. 2014;3:211–219. doi: 10.1016/j.msard.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scazzone C., Agnello L., Sasso B.L., Ragonese P., Bivona G., Realmuto S., Iacolino G., Gambino C.M., Bellia C., Salemi G., et al. Klotho and vitamin D in multiple sclerosis: An Italian study. Arch. Med. Sci. 2020;16:842–847. doi: 10.5114/aoms.2019.86969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohammadi-Milasi F., Mahnam K., Shakhsi-Niaei M. In silico study of the association of the HLA-A*31:01 allele (human leucocyte antigen allele 31:01) with neuroantigenic epitopes of PLP (proteolipid protein), MBP (myelin basic protein) and MOG proteins (myelin oligodendrocyte glycoprotein) for studying the multiple sclerosis disease pathogenesis. J. Biomol. Struct. Dyn. 2021;39:2526–2542. doi: 10.1080/07391102.2020.1751291. [DOI] [PubMed] [Google Scholar]
- 29.Dolbec K., Chalkley J., Sudhakar P. Atypical MOG antibody disease presenting with typical multiple sclerosis lesions. Mult. Scler. Relat. Disord. 2020;44:102342. doi: 10.1016/j.msard.2020.102342. [DOI] [PubMed] [Google Scholar]
- 30.Jarius S., Ruprecht K., Stellmann J.P., Huss A., Ayzenberg I., Willing A., Trebst C., Pawlitzki M., Abdelhak A., Gruter T., et al. MOG-IgG in primary and secondary chronic progressive multiple sclerosis: A multicenter study of 200 patients and review of the literature. J. Neuroinflam. 2018;15:88. doi: 10.1186/s12974-018-1108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mariano R., Messina S., Roca-Fernandez A., Leite M.I., Kong Y., Palace J.A. Quantitative spinal cord MRI in MOG-antibody disease, neuromyelitis optica and multiple sclerosis. Brain. 2021;144:198–212. doi: 10.1093/brain/awaa347. [DOI] [PubMed] [Google Scholar]
- 32.Andersson M., Yu M., Soderstrom M., Weerth S., Baig S., Solders G., Link H. Multiple MAG peptides are recognized by circulating T and B lymphocytes in polyneuropathy and multiple sclerosis. Eur. J. Neurol. 2002;9:243–251. doi: 10.1046/j.1468-1331.2002.00391.x. [DOI] [PubMed] [Google Scholar]
- 33.Moller J.R., Johnson D., Brady R.O., Tourtellotte W.W., Quarles R.H. Antibodies to myelin-associated glycoprotein (MAG) in the cerebrospinal fluid of multiple sclerosis patients. J. Neuroimmunol. 1989;22:55–61. doi: 10.1016/0165-5728(89)90009-X. [DOI] [PubMed] [Google Scholar]
- 34.Sospedra M., Martin R. Immunology of multiple sclerosis. Annu. Rev. Immunol. 2005;23:683–747. doi: 10.1146/annurev.immunol.23.021704.115707. [DOI] [PubMed] [Google Scholar]
- 35.Sawcer S., Franklin R.J.M., Ban M. Multiple sclerosis genetics. Lancet Neurol. 2014;13:700–709. doi: 10.1016/S1474-4422(14)70041-9. [DOI] [PubMed] [Google Scholar]
- 36.Banwell B., Giovannoni G., Hawkes C., Lublin F. Multiple sclerosis is a multifaceted disease. Mult. Scler. Relat. Disord. 2014;3:553–554. doi: 10.1016/j.msard.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 37.McMahon E.J., Bailey S.L., Castenada C.V., Waldner H., Miller S.D. Epitope spreading initiates in the CNS in two mouse models of multiple sclerosis. Nat. Med. 2005;11:335–339. doi: 10.1038/nm1202. [DOI] [PubMed] [Google Scholar]
- 38.Guerrero B.L., Sicotte N.L. Microglia in multiple sclerosis: Friend or foe? Front. Immunol. 2020;11:374. doi: 10.3389/fimmu.2020.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faber H., Kurtoic D., Krishnamoorthy G., Weber P., Putz B., Muller-Myhsok B., Weber F., Andlauer T.F.M. Gene expression in spontaneous experimental autoimmune encephalomyelitis is linked to human multiple sclerosis risk genes. Front. Immunol. 2020;11:2165. doi: 10.3389/fimmu.2020.02165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li M., Chen H., Yin P., Song J., Jiang F., Tang Z., Fan X., Xu C., Wang Y., Xue Y., et al. Identification and clinical validation of key extracellular proteins as the potential biomarkers in relapsing-remitting multiple sclerosis. Front. Immunol. 2021;12:753929. doi: 10.3389/fimmu.2021.753929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scazzone C., Agnello L., Lo Sasso B., Salemi G., Gambino C.M., Ragonese P., Candore G., Ciaccio A.M., Giglio R.V., Bivona G., et al. FOXP3 and GATA3 polymorphisms, vitamin D3 and multiple sclerosis. Brain Sci. 2021;11:415. doi: 10.3390/brainsci11040415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mescheriakova J.Y., van Nierop G.P., van der Eijk A.A., Kreft K.L., Hintzen R.Q. EBNA-1 titer gradient in families with multiple sclerosis indicates a genetic contribution. Neurol. Neuroimmunol. Neuroinflamm. 2020;7:e872. doi: 10.1212/NXI.0000000000000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eftekharian M.M., Komaki A., Mazdeh M., Arsang-Jang S., Taheri M., Ghafouri-Fard S. Expression profile of selected MicroRNAs in the peripheral blood of multiple sclerosis patients: A multivariate statistical analysis with ROC Curve to find new biomarkers for fingolimod. J. Mol. Neurosci. 2019;68:153–161. doi: 10.1007/s12031-019-01294-z. [DOI] [PubMed] [Google Scholar]
- 44.Ridolfi E., Fenoglio C., Cantoni C., Calvi A., De Riz M., Pietroboni A., Villa C., Serpente M., Bonsi R., Vercellino M., et al. Expression and genetic analysis of MicroRNAs involved in multiple sclerosis. Int. J. Mol. Sci. 2013;14:4375–4384. doi: 10.3390/ijms14034375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore C.S., Rao V.T.S., Durafourt B.A., Bedell B.J., Ludwin S.K., Bar-Or A., Antel J.P. miR-155 as a multiple sclerosis-relevant regulator of myeloid cell polarization. Ann. Neurol. 2013;74:709–720. doi: 10.1002/ana.23967. [DOI] [PubMed] [Google Scholar]
- 46.Ghafouri-Fard S., Taheri M. A comprehensive review of non-coding RNAs functions in multiple sclerosis. Eur. J. Pharmacol. 2020;879:173127. doi: 10.1016/j.ejphar.2020.173127. [DOI] [PubMed] [Google Scholar]
- 47.Ingelfinger F., Gerdes L.A., Kavaka V., Krishnarajah S., Friebel E., Galli E., Zwicky P., Furrer R., Peukert C., Dutertre C.A., et al. Twin study reveals non-heritable immune perturbations in multiple sclerosis. Nature. 2022;603:152–158. doi: 10.1038/s41586-022-04419-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Handunnetthi L., Handel A.E., Ramagopalan S.V. Contribution of genetic, epigenetic and transcriptomic differences to twin discordance in multiple sclerosis. Expert Rev. Neurother. 2010;10:1379–1381. doi: 10.1586/ern.10.116. [DOI] [PubMed] [Google Scholar]
- 49.Ramagopalan S.V., Dyment D.A., Ebers G.C. Genetic epidemiology: The use of old and new tools for multiple sclerosis. Trends Neurosci. 2008;31:645–652. doi: 10.1016/j.tins.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 50.International Multiple Sclerosis Genetics C., Beecham A.H., Patsopoulos N.A., Xifara D.K., Davis M.F., Kemppinen A., Cotsapas C., Shah T.S., Spencer C., Booth D., et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat. Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.International Multiple Sclerosis Genetics C., Wellcome Trust Case Control C., Sawcer S., Hellenthal G., Pirinen M., Spencer C.C., Patsopoulos N.A., Moutsianas L., Dilthey A., Su Z., et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.International Multiple Sclerosis Genetics C. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science. 2019;365:eaav7188. doi: 10.1126/science.aav7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu A., Manuel A.M., Dai Y., Zhao Z. Prioritization of risk genes in multiple sclerosis by a refined Bayesian framework followed by tissue-specificity and cell type feature assessment. BMC Genom. 2022;23:362. doi: 10.1186/s12864-022-08580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gresle M.M., Jordan M.A., Stankovich J., Spelman T., Johnson L.J., Laverick L., Hamlett A., Smith L.D., Jokubaitis V.G., Baker J., et al. Multiple sclerosis risk variants regulate gene expression in innate and adaptive immune cells. Life Sci. Alliance. 2020;3 doi: 10.26508/lsa.202000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jonasdottir A., Thorlacius T., Fossdal R., Jonasdottir A., Benediktsson K., Benedikz J., Jonsson H.H., Sainz J., Einarsdottir H., Sigurdardottir S., et al. A whole genome association study in Icelandic multiple sclerosis patients with 4804 markers. J. Neuroimmunol. 2003;143:88–92. doi: 10.1016/j.jneuroim.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 56.Omraninava M., Mehranfar S., Vahedi P., Razi B., Imani D., Aslani S., Feyzinia S. Association between IL7 Receptor Alpha (Il7ra) gene rs6897932 polymorphism and the risk of multiple sclerosis: A meta-regression and meta-analysis. Mult. Scler. Relat. Disord. 2021;48:102687. doi: 10.1016/j.msard.2020.102687. [DOI] [PubMed] [Google Scholar]
- 57.Asgharzadeh M., Najafi-Ghalehlou N., Poor B.M., Asgharzadeh V., Pourostadi M., Vegari A., Kafil H.S., Fadaee M., Farhoudi M., Rashedi J. IFN-gamma and TNF-alpha gene polymorphisms in multiple sclerosis patients in Northwest Iran. Endocr. Metab. Immune Disord. Drug Targets. 2021;21:520–525. doi: 10.2174/1871530320666200505123443. [DOI] [PubMed] [Google Scholar]
- 58.Barizzone N., Cagliani R., Basagni C., Clarelli F., Mendozzi L., Agliardi C., Forni D., Tosi M., Mascia E., Favero F., et al. An investigation of the role of common and rare variants in a large italian multiplex family of multiple sclerosis patients. Genes. 2021;12:1607. doi: 10.3390/genes12101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.International Multiple Sclerosis Genetics Consortium Electronic address, c.c.y.e.; International Multiple Sclerosis Genetics, C. Low-frequency and rare-coding variation contributes to multiple sclerosis risk. Cell. 2018;175:1679–1687. doi: 10.1016/j.cell.2018.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sorosina M., Barizzone N., Clarelli F., Anand S., Lupoli S., Salvi E., Mangano E., Bordoni R., Roostaei T., Mascia E., et al. A multi-step genomic approach prioritized TBKBP1 gene as relevant for multiple sclerosis susceptibility. J. Neurol. 2022;269:4510–4522. doi: 10.1007/s00415-022-11109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.International Multiple Sclerosis Genetics C. A systems biology approach uncovers cell-specific gene regulatory effects of genetic associations in multiple sclerosis. Nat. Commun. 2019;10:2236. doi: 10.1038/s41467-019-09773-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan Z., Gibson S.A., Buckley J.A., Qin H., Benveniste E.N. Role of the JAK/STAT signaling pathway in regulation of innate immunity in neuroinflammatory diseases. Clin. Immunol. 2018;189:4–13. doi: 10.1016/j.clim.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamilton J.A. Colony-stimulating factors in inflammation and autoimmunity. Nat. Rev. Immunol. 2008;8:533–544. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 64.Dou M., Zhou X., Li L., Zhang M., Wang W., Wang M., Jing Y., Ma R., Zhao J., Zhu L. Illumination of molecular pathways in multiple sclerosis lesions and the immune mechanism of matrine treatment in EAE, a mouse model of MS. Front. Immunol. 2021;12:640778. doi: 10.3389/fimmu.2021.640778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen J.Q., Szodoray P., Zeher M. Toll-like receptor pathways in autoimmune diseases. Clin. Rev. Allergy Immunol. 2016;50:1–17. doi: 10.1007/s12016-015-8473-z. [DOI] [PubMed] [Google Scholar]
- 66.Zuccala M., Barizzone N., Boggio E., Gigliotti L., Sorosina M., Basagni C., Bordoni R., Clarelli F., Anand S., Mangano E., et al. Genomic and functional evaluation of TNFSF14 in multiple sclerosis susceptibility. J. Genet. Genom. 2021;48:497–507. doi: 10.1016/j.jgg.2021.03.017. [DOI] [PubMed] [Google Scholar]
- 67.Steri M., Orru V., Idda M.L., Pitzalis M., Pala M., Zara I., Sidore C., Faa V., Floris M., Deiana M., et al. Overexpression of the Cytokine BAFF and autoimmunity risk. N. Engl. J. Med. 2017;376:1615–1626. doi: 10.1056/NEJMoa1610528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kolic I., Stojkovic L., Stankovic A., Stefanovic M., Dincic E., Zivkovic M. Association study of rs7799039, rs1137101 and rs8192678 gene variants with disease susceptibility/severity and corresponding LEP, LEPR and PGC1A gene expression in multiple sclerosis. Gene. 2021;774:145422. doi: 10.1016/j.gene.2021.145422. [DOI] [PubMed] [Google Scholar]
- 69.Bjørbaek C., Elmquist J.K., Frantz J.D., Shoelson S.E., Flier J.S. Identification of SOCS-3 as a potential mediator of central leptin resistance. Mol. Cell. 1998;1:619–625. doi: 10.1016/S1097-2765(00)80062-3. [DOI] [PubMed] [Google Scholar]
- 70.Deerhake M.E., Biswas D.D., Barclay W.E., Shinohara M.L. Pattern recognition receptors in multiple sclerosis and its animal models. Front. Immunol. 2019;10:2644. doi: 10.3389/fimmu.2019.02644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou Y., Cui C., Ma X., Luo W., Zheng S.G., Qiu W. Nuclear factor kappaB (NF-kappaB)-mediated inflammation in multiple sclerosis. Front. Immunol. 2020;11:391. doi: 10.3389/fimmu.2020.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Rosa V., Galgani M., Porcellini A., Colamatteo A., Santopaolo M., Zuchegna C., Romano A., De Simone S., Procaccini C., La Rocca C., et al. Glycolysis controls the induction of human regulatory T cells by modulating the expression of FOXP3 exon 2 splicing variants. Nat. Immunol. 2015;16:1174–1184. doi: 10.1038/ni.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hamid K.M., Mirshafiey A. Role of proangiogenic factors in immunopathogenesis of multiple sclerosis. Iran J. Allergy Asthma Immunol. 2016;15:1–12. [PubMed] [Google Scholar]
- 74.Hassani N., Salmaninejad A., Aslani S., Kamali-Sarvestani E., Vessal M. The association between PD-1 gene polymorphisms and susceptibility to multiple sclerosis. Immunol. Med. 2022:1–8. doi: 10.1080/25785826.2022.2137967. [DOI] [PubMed] [Google Scholar]
- 75.Zelic M., Pontarelli F., Woodworth L., Zhu C., Mahan A., Ren Y., LaMorte M., Gruber R., Keane A., Loring P., et al. RIPK1 activation mediates neuroinflammation and disease progression in multiple sclerosis. Cell. Rep. 2021;35:109112. doi: 10.1016/j.celrep.2021.109112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.