Abstract
Pertactin is an outer membrane protein expressed by Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica that induces protective immunity to Bordetella infections. The immunodominant and immunoprotective epitopes of pertactin include two repeated regions, I and II. Comparison of these two repeated regions showed that B. parapertussis pertactin is invariant, whereas B. pertussis pertactin varies mostly in region I and B. bronchiseptica pertactin varies in both repeated regions I and II, but mostly in region II. These differences may result from specific characteristics of these Bordetella species.
The genus Bordetella includes seven species; the most studied species are B. pertussis, B. parapertussis, and B. bronchiseptica. B. pertussis is responsible for respiratory infections only in humans. B. parapertussis causes infections in humans and sheep, and B. bronchiseptica infects many animal species, including humans. These pathogens produce an array of virulence factors, the synthesis of which is regulated by the two-component Bvg AS (2, 22) system. These factors include toxins such as pertussis toxin, which is the only toxin specific to B. pertussis; tracheal cytotoxin; adenylate cyclase-hemolysin; and adhesins such as filamentous hemagglutinin, fimbriae, and pertactin (PRN). PRN is an outer membrane protein with an apparent molecular mass of 69 kDa in B. pertussis, 70 kDa in B. parapertussis, and 68 kDa in B. bronchiseptica (5, 15, 16). The precursors of PRN are 91.5, 93, and 92.5 kDa in size, respectively. In B. pertussis, PRN has been demonstrated to be an agglutinogen (4), promoting attachment to certain eukaryotic cells via an Arg-Gly-Asp (RGD) motif (14).
Antibodies specific for the B. bronchiseptica-PRN are detected at high titer in immunized piglets, whereas few if any of these antibodies are detected in unprotected animals (20). Synthesis of PRN by B. bronchiseptica correlates with protection (17). The immunization of mice or piglets with preparations of PRN induces protective immunity against B. bronchiseptica infection (13, 20), and passively administered monoclonal antibodies prevent the death of animals challenged with B. bronchiseptica (17). B. pertussis PRN has also been shown to induce protective immunity to intracerebral, aerosol, and intranasal challenge with B. pertussis in mice (12, 19, 21). PRN is therefore now included in some acellular pertussis vaccines (i.e., vaccines composed of purified bacterial proteins) (10). However, the PRNs of these three species, although clearly related, have different immunogenic properties. For example, preparations of B. pertussis PRN protect mice against intranasal B. pertussis challenge but not against intranasal B. parapertussis challenge (12). They also protect mice against intracerebral B. pertussis challenge, whereas the B. bronchiseptica PRN protein does not (19).
Comparison of the deduced amino acid sequences of the three PRN proteins reveals a high degree of similarity, with the B. bronchiseptica and B. parapertussis proteins being more similar to each other than to the B. pertussis PRN protein (5, 15, 16). The sequences of the three proteins differ mostly in the number of repeats in regions I and II (Fig. 1A). Using monoclonal antibodies, Charles et al. identified and characterized a protective immunodominant epitope of the B. pertussis PRN (6). This epitope spans the (Pro-Gln-Pro)5 repeat sequences located in region II. Differences in this region may account for the observation that sera from piglets that recognize B. bronchiseptica PRN do not react with B. pertussis PRN despite the high degree of similarity between these proteins (13) and the lack of cross-protection provided by the three proteins (12, 19, 21). It has recently been shown that the PRN produced by clinical isolates of B. pertussis varies (3, 18).
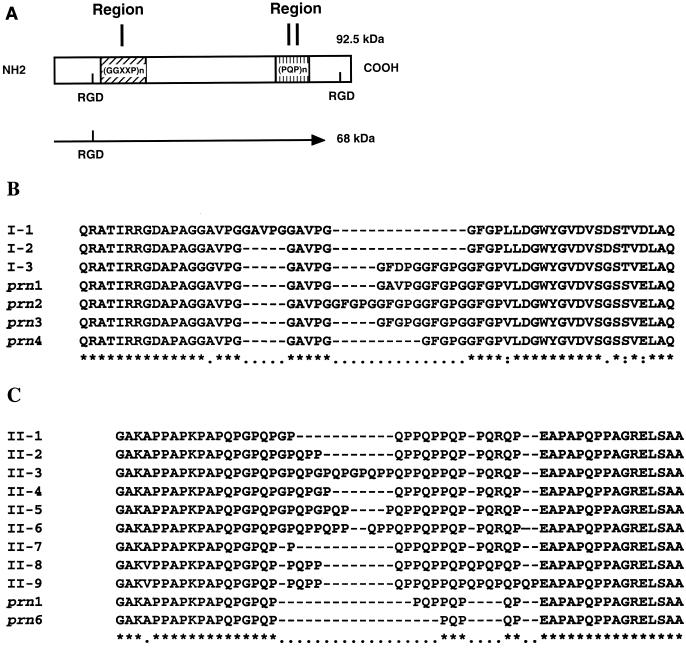
FIG. 1.
(A) Structure of the B. bronchiseptica PRN, which harbors two repeated regions (I and II) sequenced in this study. (B and C) Multiple alignments of region I (B) and region II (C).
The sequences of the prn gene of various clinical isolates revealed three major types of PRN variant. It has been suggested that epidemics result from changes in the sequences of the genes encoding PRN and pertussis toxin because the proteins present in the clinical isolates currently in circulation differ in sequence from those expressed by the vaccine strains used in the Netherlands (18). For PRN, all the observed deduced amino acid differences are located in region I. The deduced amino acid sequences of B. pertussis prn types A=1 and C=3 are very similar, differing by only two amino acids, whereas type B=2 is quite different, having a five-amino-acid insertion in the same region (18). Only one type was found to differ in region II. This type, A*=6, is produced by the B. pertussis World Health Organization reference strain 18323 and one French clinical isolate (3). It does not, however, seem to be common because it has been detected in only one clinical isolate (3). The production by this B. pertussis strain of this unusual PRN reflects the many common properties shared with the B. parapertussis and B. bronchiseptica species. No differences were found in the phenotype and behavior in the animal model of B. pertussis clinical isolates expressing different PRNs (3).
In this study, we sequenced and compared the deduced amino acid sequences of the repeated regions of the prn genes of 10 B. parapertussis isolates of human origin and 40 B. bronchiseptica isolates of animal and human origin (Table 1). The aim of this study was to analyze whether the PRN polymorphism observed in B. pertussis species also occurs in B. parapertussis and B. bronchiseptica. Sequencing was limited to regions I and II mostly because variation in the B. pertussis prn gene was observed in these regions and because these regions are immunodominant (Fig. 1A).
TABLE 1.
Isolates used in this study and corresponding PRN types
| Bordetella speciesa | Isolate | PRN types (regions I and II)/no. of isolates | Accession no. (region I, region II) |
|---|---|---|---|
| BB | CN7531 | I-2, II-4 | Q03035 |
| BB | 9.73H+ | I-1, II-3/3 | AJ250076, AJ250077 |
| BB | LAPR | I-2, II-3/8 | AJ250078, AJ250079 |
| BB | 5 | I-2, II-4/8 | AJ250080, AJ250081 |
| BB | 335 | I-2, II-1/3 | AJ250082, AJ250083 |
| BB | CVGEO | I-2, II-5/6 | AJ250084, AJ250085 |
| BB | BBCH | I-2, II-6/4 | AJ250086, AJ250087 |
| BB | DEL | I-1, II-2/5 | AJ250088, AJ250089 |
| BB | CAT1 | I-1, II-7/1 | AJ250090, AJ250091 |
| BB | 286 | I-3, II-8/1 | AJ250093, AJ250092 |
| BB | SEI | I-3, II-9/1 | AJ250094, AJ250095 |
| BPP | CN2591 | I-1, II-2 | P24328 |
| BPP | 63.2 | I-1, II-2/10 | Identical to P24328 |
| BP | Tohama | prn1 | AJ006158 |
| BP | 18323 | prn6 | AJ006152 |
| BP | Hav | prn2 | AJ007361 |
| BP | Fr287 | prn3 | AJ006156 |
BB, B. bronchiseptica; BP, B. pertussis; BPP, B. parapertussis.
DNA was extracted, amplified by PCR, and sequenced as previously described (3). Amplified PCR products were purified and sequenced by ESGS (Cybergene Group, Evry, France). Deduced amino acid sequences were analyzed with GCG software (Wisconsin package version 9.1; Genetics Computer Group, Madison, Wis.) (11). The deduced amino acid sequences of regions I and II were compared, and multiple alignments of the amino acid sequences were created with the CLUSTAL W program of GCG for each region (Fig. 1B and C).
No difference was found between the sequences of regions I and II of the PRNs produced by the 10 B. parapertussis isolates and the published sequence (16). However, three different types were found among the 40 B. bronchiseptica prn genes analyzed, with differences in the number of repeats in region I (Fig. 1B). The largest group corresponded to sequences with three copies of the repeated sequence, identical to the sequence reported previously (15). A higher degree of variability was observed in the second repeated region of the B. bronchiseptica PRN (Fig. 1C). Nine variants were observed. Again, the differences concerned the number of repeats (6 to 9). Some B. bronchiseptica variants are of the B. parapertussis type, but no B. bronchiseptica variant presented the same pattern as the B. pertussis isolates. Furthermore, no unique association between one type of region I and one type of region II was observed. We did not observe in any of the three species a pattern similar to those of the 18323 strain and the CZ isolate (3), which are considered intermediate between B. pertussis, B. bronchiseptica, and B. parapertussis. These data are consistent with the B. parapertussis and B. bronchiseptica prn genes' being more similar to each other than to the B. pertussis prn gene (1). The lack of variation in the PRN expressed by the 10 human B. parapertussis isolates is consistent with other studies which showed that these isolates constitute a highly clonal group (23, 25).
No host specificity was observed with respect to PRN type. It was thought that B. pertussis allelic prn types 2 and 3 emerged after 30 years of high-coverage vaccination with the same vaccine strains producing allelic prn type 1. If this is the case, the lack of variation in the PRN of the 10 B. parapertussis PRN isolates, 6 of which were collected recently (1990 to 1997) and 4 of which were collected some years ago (1963 to 1964), suggests that selection pressure due to 30 years of vaccination with a B. pertussis whole-cell vaccine may have affected B. pertussis PRN but not B. parapertussis PRN. However, more B. parapertussis isolates must be analyzed before any firm conclusion can be drawn.
It has been shown that region II plays a role in the induction of protective immunity (6). The lack of cross-protection between B. pertussis, B. parapertussis, and B. bronchiseptica PRN is consistent with this because the major differences between these proteins occur in this region. No variation in this region was observed for the PRN produced by B. pertussis isolates. These data suggest that 30 years of vaccination may have induced variation in one immunodominant repeat region (I) but not in the other region (II). One can make the hypothesis that region II, which was already known to be implicated in the induction of protective immunity (6), is the most involved region. In contrast, analysis of the PRN of B. bronchiseptica shows polymorphism in both regions. This may account for the inability of B. bronchiseptica vaccines to induce long-lasting protection. This polymorphism may also be linked to the ability of B. bronchiseptica to induce chronic infections (7, 9, 24). In fact, we have previously shown that B. bronchiseptica may persist in the host (9) by varying the expression of one of its toxins (adenylate cyclase hemolysin) or the structure of its lipopolysaccharide (8, 9). Variation in PRN region II may also provide a mean for this bacterium to escape host immune responses. Our observations, as well as those published previously (18), indicate that a continuous surveillance of antigens expressed by B. pertussis isolates is necessary to verify whether vaccine formulation should be adapted.
Acknowledgments
We thank the Collège de Bactériologie, Virologie et Hygiène des Hôpitaux Généraux de France for the gift of some B. bronchiseptica isolates.
REFERENCES
- 1.Arico B, Gross R, Smida J, Rappuoli R. Evolutionary relationships in the genus Bordetella. Mol Microbiol. 1987;1:301–308. doi: 10.1111/j.1365-2958.1987.tb01936.x. [DOI] [PubMed] [Google Scholar]
- 2.Arico B, Miller J F, Roy C, Stibitz S, Monack D, Falkow S, Gross R, Rappuoli R. Sequences required for expression of Bordetella pertussis virulence factors share homology with prokaryotic signal transduction proteins. Proc Natl Acad Sci USA. 1989;86:6671–6675. doi: 10.1073/pnas.86.17.6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boursaux-Eude C, Thiberge S, Carletti G, Guiso N. Intranasal murine model of Bordetella pertussis infection. II. Sequence variation and protection induced by a tricomponent acellular vaccine. Vaccine. 1999;17:2651–2660. doi: 10.1016/s0264-410x(99)00038-9. [DOI] [PubMed] [Google Scholar]
- 4.Brennan M J, Li Z M, Cowell J L, Bisher M E, Steven A C, Novotny P, Manclark C R. Identification of a 69-kilodalton nonfimbrial protein as an agglutinogen of Bordetella pertussis. Infect Immun. 1988;56:3189–3195. doi: 10.1128/iai.56.12.3189-3195.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charles I G, Dougan G, Pickard D, Chatfield S, Smith M, Novotny P, Morrissey P, Fairweather N F. Molecular cloning and characterization of protective outer membrane protein P.69 from Bordetella pertussis. Proc Natl Acad Sci USA. 1989;86:3554–3558. doi: 10.1073/pnas.86.10.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charles I G, Li J L, Roberts M, Beesley K, Romanos M, Pickard D J, Francis M, Campbell D, Dougan G, Brennan M J, Manclarck C R, Jensen M A, Heron I, Chubb A, Novotny P, Fairweather N F. Identification and characterization of a protective immunodominant B cell epitope of pertactin (P.69) from Bordetella pertussis. Eur J Immunol. 1991;21:1147–1153. doi: 10.1002/eji.1830210509. [DOI] [PubMed] [Google Scholar]
- 7.Goodnow R A. Biology of Bordetella bronchiseptica. Microbiol Rev. 1980;44:722–738. doi: 10.1128/mr.44.4.722-738.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gueirard P, Le Blay K, Le Coustumier A, Chaby R, Guiso N. Variation in Bordetella bronchiseptica lipopolysaccharide during human infection. FEMS Microbiol Lett. 1998;162:331–337. doi: 10.1111/j.1574-6968.1998.tb13017.x. [DOI] [PubMed] [Google Scholar]
- 9.Gueirard P, Weber C, Le Coustumier A, Guiso N. Human Bordetella bronchiseptica infection related to contact with infected animals: persistence of bacteria in host. J Clin Microbiol. 1995;33:2002–2006. doi: 10.1128/jcm.33.8.2002-2006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hewlett E L, Cherry J D. New and improved vaccines against pertussis: new generation vaccines. New York, N.Y: Marcel Dekker; 1997. [Google Scholar]
- 11.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignments on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 12.Khelef N, Danve B, Quentin-Millet M J, Guiso N. Bordetella pertussis and Bordetella parapertussis: two immunologically distinct species. Infect Immun. 1993;61:486–490. doi: 10.1128/iai.61.2.486-490.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobisch M, Novotny P. Identification of a 68-kilodalton outer membrane protein as the major protective antigen of Bordetella bronchiseptica by using specific-pathogen-free piglets. Infect Immun. 1990;58:352–357. doi: 10.1128/iai.58.2.352-357.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leininger E, Roberts M, Kenimer J G, Charles I G, Fairweather N, Novotny P, Brennan M J. Pertactin, an Arg-Gly-Asp-containing Bordetella pertussis surface protein that promotes adherence of mammalian cells. Proc Natl Acad Sci USA. 1991;88:345–349. doi: 10.1073/pnas.88.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Fairweather N F, Novotny P, Dougan G, Charles I G. Cloning, nucleotide sequence and heterologous expression of the protective outer-membrane protein P.68 pertactin from Bordetella bronchiseptica. J Gen Microbiol. 1992;138:1697–1705. doi: 10.1099/00221287-138-8-1697. [DOI] [PubMed] [Google Scholar]
- 16.Li L J, Dougan G, Novotny P, Charles I G. P.70 pertactin, an outer-membrane protein from Bordetella parapertussis: cloning, nucleotide sequence and surface expression in Escherichia coli. Mol Microbiol. 1991;5:409–417. doi: 10.1111/j.1365-2958.1991.tb02123.x. [DOI] [PubMed] [Google Scholar]
- 17.Montaraz J A, Novotny P, Ivanyi J. Identification of a 68-kilodalton protective protein antigen from Bordetella bronchiseptica. Infect Immun. 1985;47:744–751. doi: 10.1128/iai.47.3.744-751.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mooi F R, van Oirschot H, Heuvelman K, van der Heide H G J, Gaastra W, Willems R J L. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in the Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect Immun. 1998;66:670–675. doi: 10.1128/iai.66.2.670-675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novotny P, Chubb A P, Cownley K, Montaraz J A, Beesley J E. Bordetella adenylate cyclase: a genus specific protective antigen and virulence factor. Dev Biol Stand. 1985;61:27–41. [PubMed] [Google Scholar]
- 20.Novotny P, Kobisch M, Cownley K, Chubb A P, Montaraz J A. Evaluation of Bordetella bronchiseptica vaccines in specific-pathogen-free piglets with bacterial cell surface antigens in enzyme-linked immunosorbent assay. Infect Immun. 1985;50:190–198. doi: 10.1128/iai.50.1.190-198.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shahin R D, Brennan M J, Li Z M, Meade B D, Manclark C R. Characterization of the protective capacity and immunogenicity of the 69-kD outer membrane protein of Bordetella pertussis. J Exp Med. 1990;171:63–73. doi: 10.1084/jem.171.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stibitz S, Aaronson W, Monack D, Falkow S. Phase variation in Bordetella pertussis by frameshift mutation in a gene for a novel two-component system. Nature. 1989;338:266–269. doi: 10.1038/338266a0. [DOI] [PubMed] [Google Scholar]
- 23.van der Zee A, Groenendijk H, Peeters M, Mooi F R. The differentiation of Bordetella parapertussis and Bordetella bronchiseptica from humans and animals as determined by DNA polymorphism mediated by two different insertion sequence elements suggests their phylogenetic relationship. Int J Syst Bacteriol. 1996;46:270–273. doi: 10.1099/00207713-46-3-640. [DOI] [PubMed] [Google Scholar]
- 24.Woolfrey B F, Moody J A. Human infections associated with Bordetella bronchiseptica. Clin Microbiol Rev. 1991;4:243–255. doi: 10.1128/cmr.4.3.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuk M H, Heininger U, Martinez de Tejada G, Miller J F. Human but not ovine isolates of Bordetella parapertussis are highly clonal as determined by PCR-based RAPD fingerprinting. Infection. 1998;26:270–273. doi: 10.1007/BF02962245. [DOI] [PubMed] [Google Scholar]