Abstract
Elicited macrophages from 129sv mice with a functional deletion of the natural-resistance-associated macrophage protein 1 gene (Nramp1) were shown to be as susceptible as wild-type mice to infection with the Chlamydia trachomatis mouse pneumonitis and L3 serovars and to Chlamydia pneumoniae. Furthermore, the two groups of mice were shown to be similarly susceptible to an intranasal infection with these microorganisms. In conclusion, the Nramp1 gene does not appear to play a major role in the regulation of the susceptibility of mice to a chlamydial infection.
Some intracellular pathogens, including Chlamydia, Mycobacterium tuberculosis, Mycobacterium avium, Legionella pneumophila, Salmonella enterica serovar Typhimurium, and Toxoplasma gondii, grow in host cells inside vacuolar compartments that do not mature into phagolysosomes (3). Based on work with inbred strains of mice, natural resistance to infection with S. enterica serovar Typhimurium, Mycobacterium bovis (strain bacillus Calmette-Guérin [BCG]), Mycobacterium lepraemurium, and Leishmania donovani was shown to be controlled by the Bcg locus (11, 15). Subsequently, the natural-resistance-associated macrophage protein 1 gene (Nramp1) was identified by Vidal et al. (16) as the candidate for the Bcg mutation. Nramp1 is an integral membrane phosphoglycoprotein located in the endosome/lysosome compartment of resting macrophages and is recruited to the maturing phagosomal membrane, where it can control the growth and viability of intracellular parasites by altering the intravacuolar environment (5). In humans, a gene with a high degree of homology to the mouse Nramp1 gene has been identified and found to be expressed mainly in peripheral blood leukocytes and the lungs (2).
Chlamydiae are obligate intracellular bacteria with a unique growth cycle (8). Two morphologically and metabolically distinct forms have been identified. The elementary body (EB) measures approximately 300 nm in diameter and, although metabolically inactive, is the infectious form of this organism. The EB infects host cells by forming an inclusion using the plasma membrane of the host cell and chlamydia-specific proteins, and 2 to 6 h after penetration it initiates a transformation that leads to the formation of reticulate bodies (RB) (14). RB measure 1,000 to 1,500 nm, are metabolically active, and replicate by binary fission inside the inclusion for periods ranging from 20 to 72 h, depending on the species and strain of Chlamydia. After completion of the replication stage the RB mature again into EBs that go on to infect other cells following the rupture of the inclusion and the eukaryotic host cell membranes. The mechanisms underlying the susceptibility of certain cells to a chlamydial infection are not understood. Epithelial cells appear to be the primary targets for infection by Chlamydia pneumoniae and the Chlamydia trachomatis mouse pneumonitis (MoPn) biovar, although macrophages are also infected (6). On the other hand, isolates of the C. trachomatis lymphogranuloma venereum biovar preferentially infect cells of the reticuloendothelial system, including macrophages (6). Here, we tested the role that Nramp1 may play in chlamydial infections using mice with a deletion of that gene.
C. trachomatis serovars L3 and MoPn were purchased from the American Type Culture Collection (Manassas, Va.), and C. pneumoniae strain TW-183 was purchased from the Washington Research Foundation (University of Washington, Seattle). All three chlamydiae were grown and their titers were determined as previously described (10). Two pairs each of homozygous Nramp1 deletion (Nramp1d) and wild-type 129sv mice, a generous gift from P. Gross (McGill University), were bred at the University of California, Irvine, following an approved Institutional Animal Care and Use Committee protocol. To collect macrophages, mice were inoculated intraperitoneally with 0.5 ml of thioglycolate broth, and the cells were harvested 4 days later (17). The macrophages were counted and seeded at a density of 2.5 × 105/ml in 1-dram shell vials and incubated overnight at 37°C. The next day the medium was changed and the cells again were incubated overnight. After 24 h the medium was removed and the macrophages were infected with Chlamydia by centrifugation in parallel with control HeLa-229 and McCoy cells. The yield of inclusion-forming units (IFU) from infected macrophages and HeLa-229 and McCoy cells was assessed in McCoy cells at different times postinfection (p.i.) following sonication of the macrophages (17). For in vivo susceptibility testing, mice were infected by intranasal (i.n.) inoculation, and their weight was measured daily. Mice were euthanatized, and their lungs and spleens were harvested and processed to determine the number of chlamydial IFU (18). Mice were matched for age and sex between the Nramp1d and wild-type strains, and the experiments were done in duplicate with three to five animals per group.
Elicited peritoneal macrophages were infected in vitro at multiplicities of infection (MOI) of 1 and 10 with the C. trachomatis L3 or MoPn serovar or C. pneumoniae, and the yield of IFU was determined at different times p.i. As shown in Table 1, the number of C. trachomatis MoPn IFU detected at 5, 24, 48 and 72 h p.i. from the macrophages of the Nramp1d mice that were infected at an MOI of 1 was not significantly different from the yield of IFU from the chlamydia-infected macrophages of the wild-type animals. The number of IFU was low at 5 h p.i., most likely reflecting the transformation of EB to RB, and subsequently the number of IFU increased. Similar results were obtained when macrophages were infected at an MOI of 10 with MoPn or when the L3 serovar or C. pneumoniae was tested (data not shown). As expected, the yields of chlamydial IFU from macrophages were lower than those from the infected control HeLa-229 and McCoy cells.
TABLE 1.
Yield of chlamydial IFU from macrophages infected at an MOI of 1
| Time p.i. (h) | Yield of chlamydiae (mean log 10 IFU ± 1 SD)
|
|||||
|---|---|---|---|---|---|---|
|
C. trachomatis L3
|
C. trachomatis MoPn
|
|||||
| Nramp1d mice | Wild-type mice | HeLa cells | Nramp1d mice | Wild-type mice | McCoy cells | |
| 0a | 4.69 ± 0.10 | 4.69 ± 0.10 | 4.69 ± 0.10 | 5.28 ± 0 | 5.28 ± 0 | 5.28 ± 0 |
| 5 | 3.58 ± 0.25 | 3.43 ± 0.40 | 2.80 ± 0 | 2.46 ± 0.09 | 2.60 ± 0.2 | 2.93 ± 0 |
| 24 | 4.01 ± 0.08 | 4.02 ± 0.26 | 7.68 ± 0.5 | 5.87 ± 0.02 | 6.36 ± 0.08 | 6.14 ± 0 |
| 48 | 5.73 ± 0.14 | 5.64 ± 0.23 | 8.71 ± 0.05 | 6.34 ± 0.06 | 6.28 ± 0.09 | 8.12 ± 0 |
| 72 | 5.69 ± 0.17 | 5.06 ± 0.05 | 8.55 ± 0.07 | 6.71 ± 0.29 | 6.49 ± 0.09 | 6.63 ± 0 |
For 0 h, values are numbers of C. trachomatis IFU used to infect the macrophages.
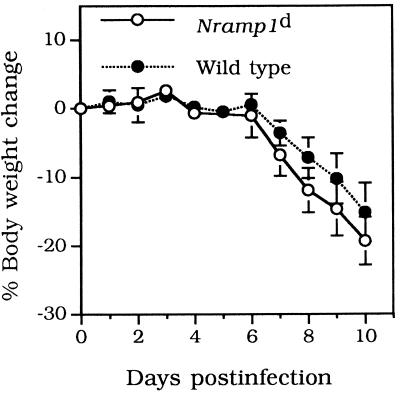
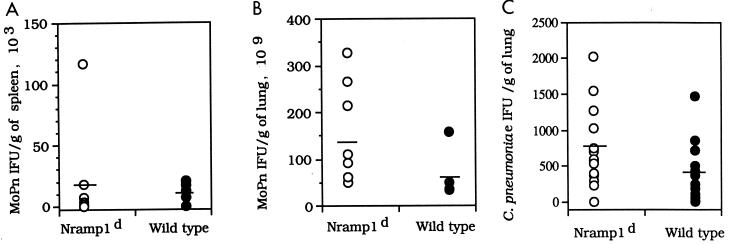
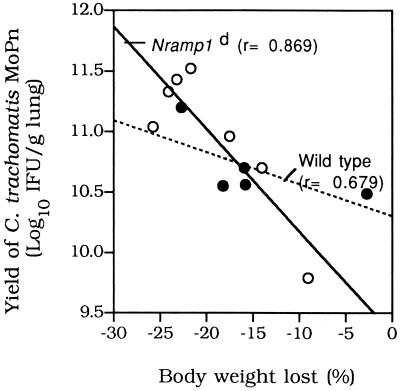
The weights of the 7- to 9-week-old mice infected i.n. with 500 IFU of C. trachomatis MoPn are shown in Fig. 1A. Overall, the decreases in body weight were similar in the Nramp1d and wild-type mice. By day 10 p.i., both groups had lost approximately 15 to 20% of their initial body weight. Lungs and spleens harvested from both groups of animals were cultured, and the number of IFU per gram of tissue was calculated. Figure 2A and B show that there were no statistically significant differences between the numbers of IFU recovered from the lungs or spleens of Nramp1d and wild-type mice, although overall fewer chlamydiae were recovered from the wild-type animals. Similar results were obtained when mice were challenged i.n. with 104 IFU of C. trachomatis MoPn. Groups of Nramp1d and wild-type mice were also infected i.n. with 105 IFU of C. pneumoniae. The mice were euthanatized at 5 days p.i. and the number of IFU per gram of lung tissue was determined (Fig. 2C). Overall, although the number of IFU recovered from the control group was less than the number recovered from the Nramp1d group, the difference was not statistically significant (Fig. 2C). As shown in Fig. 3, there was a positive correlation between weight loss and yield of C. trachomatis IFU from the lungs in both groups of mice. Animals that had lost more weight at the end of the 10 days of observation had a more severe infection, as shown by the higher number of C. trachomatis IFU recovered from their lungs.
FIG. 1.
Change in body weight of 7- to 9-week-old Nramp1d and wild-type 129sv mice infected i.n. with 500 IFU of C. trachomatis MoPn.
FIG. 2.
(A and B) Number of IFU recovered at 10 days p.i. from the spleens and lungs of 7- to 9-week-old Nramp1d and wild-type 129sv mice infected i.n. with 500 IFU of C. trachomatis MoPn. (C) Number of IFU recovered at 5 days p.i. from the lungs of mice infected with 105 IFU of C. pneumoniae.
FIG. 3.
Correlation between weight loss and yield of C. trachomatis IFU from the lungs at 10 days p.i.
In this group of experiments we have shown an apparent lack of significant effect of the Nramp1 deletion on the susceptibility of 129sv mice to a chlamydial respiratory infection. These results were supported by the finding that peritoneal macrophages elicited from Nramp1d and wild-type mice yielded similar numbers of chlamydial IFU following infection. The fact that the Nramp1d mice with a 129sv background are not significantly more susceptible to chlamydial infection than control mice does not mean that in other strains of mice the results will be the same. It is possible that another gene(s) may mask the effect of the Nramp1 gene in 129sv mice but not in other strains. However, the fact that C3H/He, a strain of mice that has the resistant Nramp gene, is generally more susceptible to chlamydial infection than the BALB/c and C57BL/6 strains of mice, which carry the susceptible gene, appears to substantiate our results (7). We tested several MOI in vitro and challenged the mice with different quantities of chlamydial IFU since in other experimental models variations in the infectious dose have uncovered different susceptibilities to infection in Nramp1d and wild-type strains of mice (4, 13). However, within the limited range that we tested we could not find differences. Also, North et al. (9) recently have reported that the Nramp1 gene is of limited importance in susceptibility to M. tuberculosis in 129sv mice. On the other hand, differences in susceptibility to the BCG strain of M. bovis are well established (4). It is then possible, for example, that the infectivity of Chlamydia psittaci could be more affected by the Nramp1 gene than the infectivities of isolates of C. trachomatis or C. pneumoniae.
The role that the Nramp1 gene plays in susceptibility or resistance to certain infections in the human population is still controversial. For instance, in a study performed in West Africa, individuals with a polymorphism in the Nramp1 gene were overrepresented in the group with M. tuberculosis infection (1). On the other hand, a study carried out in Brazil failed to confirm this association (12). Thus, more data are needed to determine the role that the Nramp1 gene may play in intracellular infections in humans. In the case of chlamydial infections we are not aware of any studies that have addressed this issue in the human population. Although our investigation using a mouse model did not show a significant association between the Nramp1 gene and susceptibility to chlamydial infection, that should not deter studies with humans.
Acknowledgments
This work was supported by Public Health Service grants AI-32248 and AI-30499 from the National Institute of Allergy and Infectious Diseases.
We thank P. Gros for generously providing breeding pairs of 129sv mice with the null allele at the Nramp1 locus and wild-type mice.
REFERENCES
- 1.Bellamy R, Ruwende C, Corra T, McAdam K P W J, Whittle H C, Hill A V S. Variation in the NRAMP1 susceptibility gene and susceptibility to tuberculosis in West Africa. N Engl J Med. 1998;338:640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 2.Cellier M, Govoni G, Vidal S, Kwan T, Groulx N, Liu J, Sanchez E, Skamene E, Schurr E, Gros P. Human natural resistance-associated macrophage protein: cDNA cloning, chromosomal mapping, genomic organization, and tissue-specific expression. J Exp Med. 1994;180:1741–1752. doi: 10.1084/jem.180.5.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia del Portillo F, Finlay B B. The varied lifestyles of intracellular pathogens within eukaryotic vacuolar compartments. Trends Microbiol. 1995;3:373–380. doi: 10.1016/s0966-842x(00)88982-9. [DOI] [PubMed] [Google Scholar]
- 4.Gros P, Skamene E, Forget A. Genetic control of natural resistance to Mycobacterium bovis (BCG) in mice. J Immunol. 1981;127:2417–2421. [PubMed] [Google Scholar]
- 5.Gruenheid S, Pinner E, Desjardins M, Gros P. Natural resistance to infection with intracellular pathogens: the Nramp1 protein is recruited to the membrane of the phagosome. J Exp Med. 1997;185:717–730. doi: 10.1084/jem.185.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.la Verda D, Byrne G I. Interactions between macrophages and Chlamydiae. In: Zwilling B S, Eisenstein T K, editors. Macrophage-pathogen interactions. New York, N.Y: Marcel Dekker, Inc.; 1993. pp. 381–399. [Google Scholar]
- 7.Malo D, Vogan K, Vidal S, Hu J, Cellier M, Schurr E, Fuks A, Bumsstead N, Morgan K, Gros P. Haplotype mapping and sequence analysis of the mouse Nramp gene predict susceptibility to infection with intracellular parasites. Genomics. 1994;23:51–61. doi: 10.1006/geno.1994.1458. [DOI] [PubMed] [Google Scholar]
- 8.Moulder J W. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.North R J, LaCourse R, Ryan L, Gros P. Consequence of Nramp1 deletion to Mycobacterium tuberculosis infection in mice. Infect Immun. 1999;67:5811–5814. doi: 10.1128/iai.67.11.5811-5814.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pal S, Fielder T J, Peterson E M, de la Maza L M. Protection against infertility in a BALB/c mouse salpingitis model by intranasal immunization with the mouse pneumonitis biovar of Chlamydia trachomatis. Infect Immun. 1994;62:3354–3362. doi: 10.1128/iai.62.8.3354-3362.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plant J E, Glynn A. Genetics of resistance to infection with Salmonella typhimurium in mice. J Infect Dis. 1976;133:72–78. doi: 10.1093/infdis/133.1.72. [DOI] [PubMed] [Google Scholar]
- 12.Shaw M A, Collins A, Peakock C S, Miller E N, Black G F, Sibthorpe D, Lins-Lainson Z, Shaw J J, Ramos F, Silveira F, Blackwell J M. Evidence that genetic susceptibility to Mycobacterium tuberculosis in a Brazilian population is under oligogenic control: linkage study of the candidate genes NRAMP1 and TNFA. Tuber Lung Dis. 1997;78:35–45. doi: 10.1016/s0962-8479(97)90014-9. [DOI] [PubMed] [Google Scholar]
- 13.Skamene E, Gros P, Forget A, Kongshavn P A L, St. Charles C, Taylor B A. Genetic regulation of resistance to intracellular pathogens. Nature (London) 1982;297:506–509. doi: 10.1038/297506a0. [DOI] [PubMed] [Google Scholar]
- 14.Taraska T, Ward D M, Ajioka R S, Wyrick P B, Davis-Kaplan S R, Davis C H, Kaplan J. The late chlamydial inclusion membrane is not derived from the endocytic pathway and is relatively deficient in host proteins. Infect Immun. 1996;64:3713–3727. doi: 10.1128/iai.64.9.3713-3727.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vidal S, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate gene for Bcg. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 16.Vidal S, Tremblay M, Govoni G, Gauthier S, Sebastiani G, Malo D, Skamene E, Olivier M, Jothy S, Gros P. The Ity/Lsh/Bcg locus; natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1 gene. J Exp Med. 1995;182:655–666. doi: 10.1084/jem.182.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong G, de la Maza L M. Activation of mouse peritoneal macrophages in vitro or in vivo by recombinant murine gamma interferon inhibits the growth of Chlamydia trachomatis serovar L1. Infect Immun. 1988;56:3322–3325. doi: 10.1128/iai.56.12.3322-3325.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong G, Peterson E M, Czarniecki C W, de la Maza L M. Recombinant murine gamma interferon inhibits Chlamydia trachomatis serovar L1 in vivo. Infect Immun. 1988;56:283–286. doi: 10.1128/iai.56.1.283-286.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]