Abstract
For coronavirus disease 2019 (COVID-19), a pandemic disease characterized by strong immune dysregulation in severe patients, convenient and efficient monitoring of the host immune response is critical. Human hosts respond to viral and bacterial infections in different ways, the former is characterized by the activation of interferon stimulated genes (ISGs) such as IFI27, while the latter is characterized by the activation of anti-bacterial associated genes (ABGs) such as S100A12. This two-tiered innate immune response has not been examined in COVID-19. In this study, the activation patterns of this two-tiered innate immune response represented by IFI27 and S100A12 were explored based on 1421 samples from 17 transcriptome datasets derived from the blood of COVID-19 patients and relevant controls. It was found that IFI27 activation occurred in most of the symptomatic patients and displayed no correlation with disease severity, while S100A12 activation was more restricted to patients under severe and critical conditions with a stepwise activation pattern. In addition, most of the S100A12 activation was accompanied by IFI27 activation. Furthermore, the activation of IFI27 was most pronounced within the first week of symptom onset, but generally waned after 2–3 weeks. On the other hand, the activation of S100A12 displayed no apparent correlation with disease duration and could last for several months in certain patients. These features of the two-tiered innate immune response can further our understanding on the disease mechanism of COVID-19 and may have implications to the clinical triage. Development of a convenient two-gene protocol for the routine serial monitoring of this two-tiered immune response will be a valuable addition to the existing laboratory tests.
Introduction
The COVID-19 pandemic has relentlessly taken over 6.6 million lives in the past three years (https://covid19.who.int/). Although vaccination and drug treatment have saved millions of lives [1, 2], we have not seen the ending sight of the pandemic yet. The pandemic has presented major challenges to the global medical systems, including the overwhelmingly large volume of hospitalized patients and fast deteriorating conditions for many patients. To meet these challenges, convenient and effective ways for continuous patient monitoring need to be deployed for quick and better clinical decision making.
To achieve this goal, a great variety of clinical tests have been explored in hospitals around the globe [3, 4]. Conventional blood tests included blood cell counts (especially neutrophils and lymphocytes) and size distribution [5–8]. Common laboratory tests included procalcitonin (PCT) [9], C-reactive protein (CRP) [10], D-dimer [11], interleukin-6 (IL-6) [12], and others [13]. In addition, parameters reflecting organ dysfunction have been extensively studied [14–17]. To assist diagnosis and prognosis, simple models have been constructed based on some of these parameters and demographic information such as age, sex, and number of comorbidities [18–22]. However, these models are far from perfect, reflecting the heterogeneity and complexity of COVID-19.
For more in-depth understanding of this disease, modern technologies have also been applied to the investigation. Deep immune profiling based on single-cell technologies have revealed profound change in the adaptive immunity [23–26]. Clinically relevant novel factors have been discovered using proteomics [27, 28], metabolomics [29, 30] and lipidomics [31, 32]. Mechanistic and biomarker studied have also employed transcriptomics of mRNA [33], miRNA [34], and cell-free RNA [35]. Unfortunately, the novel factors from these discovery investigations have rarely been replicated at independent institutions. Ideally, for the purpose of daily monitoring, the biomarkers need to be easily accessible and robustly reproducible under a variety of circumstances.
Gene activation or suppression in peripheral blood can be easily accessed and evaluated. In fact, activation of interferon stimulated genes is the most prominent signature of host response to viral infection [36], while activation of antibacterial associated genes has been consistently observed in patients with bacterial infection [37]. These two groups of genes represent a two-tiered innate immune response to infection and potentially other medical conditions. In previous works, we have proposed S100A12 as the most prominent host marker for bacterial infection which was compared favorably against PCT and CRP in over a thousand clinical samples [38]. Others have extensively utilized IFI27 and other ISGs for the identification of viral infection in clinics [39–41]. Since these two genes are directly linked to disease mechanism, the simultaneous monitoring of IFI27 and S100A12 in COVID-19 will give us a clearer insight into the two-tiered innate immune response to this devastating medical condition.
The goal of this work is to find out whether IFI27 and S100A12 can be used as effective markers to monitor COVID-19 progression. The following features will be examined: How prevalent is the gene activation in the blood of COVID-19 patients? Is the gene activation correlated with disease severity? Is the gene activation correlated with disease duration? Additionally, is the activation pattern consistent in whole blood and peripheral blood mononuclear cells (PBMCs)? The robustness of the patterns will be corroborated by the consistency among independent studies from around the globe. Following the investigation, it will be clear that the two genes can be used as valuable markers for the convenient and efficient monitoring of the pandemic disease.
Materials and methods
Transcriptome datasets for COVID-19
Most of the transcriptome datasets were downloaded from gene expression omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). Only one of the datasets were downloaded from the Zenodo repository (https://zenodo.org/). All of these transcriptome studies on COVID-19 employed RNA-Seq (RNA sequencing) technology, while the only transcriptome study on seasonal flu utilized microarray technology.
The tissue source was whole blood for 12 of the 17 transcriptome datasets on COVID-19. Dataset GSE152641 contains 86 samples, including 24 samples from healthy donors and 62 samples from COVID-19 patients [42]. Dataset GSE171110 contains 54 samples, including 10 samples from healthy donors and 44 samples from severe COVID-19 patients [43]. Dataset GSE167000 contains 95 samples from hospitalized subjects, including 30 samples from SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) negative patients and 65 samples from SARS-CoV-2 positive patients [44]. Dataset GSE172114 contains 69 samples from COVID-19 patients, including 46 samples from critical patients and 23 samples from non-critical patients [45]. Dataset Zenodo 6120249 contains 143 samples, including 10 samples from healthy volunteers, 13 samples from non-hospitalized COVID-19 patients, 78 samples from hospitalized COVID-19 patients, and 42 samples from sepsis patients [46]. Dataset GSE189990 contains 24 samples, including 4 samples from healthy controls and 20 samples from COVID-19 patients [47]. Dataset GSE161777 contains 56 samples, including 14 samples from healthy controls and 42 samples from COVID-19 patients [48]. Dataset GSE161731 contains 155 samples, including 19 samples from healthy controls, 12 samples from hospitalized COVID-19 patients, 35 samples from non-hospitalized COVID-19 patients, 23 samples for patients with bacterial infection, 17 samples from patients with influenza infection, and 49 samples from patients with other coronavirus infection [49]. Dataset GSE155454 contains 58 samples, including 6 samples from healthy controls and 52 samples from COVID-19 patients (reference not available). Dataset GSE166190 contains 98 samples, including 35 samples from children and 63 samples from adults [50]. Dataset GSE178967 contains 144 samples from COVID-19 patients at day 0 and day 5 of the hospital admission [51]. Dataset GSE169687 contains 152 samples, including 14 samples from healthy controls and 138 samples from recovered COVID-19 patients at 3 time points [52].
The tissue source was leukocytes for two transcriptome datasets. Dataset GSE154998 contains 14 samples from intensive care unit (ICU) patients, including 7 samples from SARS-CoV-2 negative patients and 7 samples from SARS-CoV-2 positive patients [53]. Dataset GSE157103 contains 126 samples, including 100 samples from COVID-19 patients (50 ICU patients and 50 non-ICU patients) and 26 samples from patients with other diseases [54].
The tissue source was PBMCs for three transcriptome datasets. Dataset GSE152418 contains 34 samples, including 17 samples from healthy controls and 17 samples from COVID-19 patients [55]. Dataset GSE184401 contains 43 samples, including 21 samples from patients with mild COVID-19 and 22 samples from patients with severe COVID-19 (reference not available). Dataset GSE179627 contains 70 samples, including 22 samples from healthy controls and 48 samples from COVID-19 patients [56].
All of the COVID-19 patients were tested positive for SARS-CoV-2 infection and had symptoms compatible with COVID-19 infection. In total, 1421 samples were included in these 17 RNA-Seq studies on COVID-19, including 1134 whole blood samples, 140 leukocyte samples, and 147 PBMC samples.
Additionally, dataset GSE68310 contains 281 samples from non-hospitalized patients with mild influenza infection sampled day 2, day 4, day 6 and day 21 from symptom onset [57]. The tissue source was whole blood for this dataset.
Data analysis and presentation
For the RNA-Seq datasets, expression values of IFI27 and S100A12 were extracted from the original expression matrix file and normalized based on the total number of reads for each sample and then log transformed. The normalization step was skipped if the normalized data was already provided by the original data submitters. For the microarray dataset, the normalized data were downloaded from GEO and the expression values of IFI27 and S100A12 were extracted from the gene expression matrix. The gene expression and sample information were merged for further analysis. Empirical cut-off values for gene activation were usually based on the highest values in the healthy control groups. The correlation analysis was done in Excel. The group comparison (p-value calculation) for each dataset was done in R (https://www.r-project.org/) using t-test. The figures were drawn with ggplot2 package in R.
Results
Activation of IFI27 and S100A12 in COVID-19 patients
As host markers for infection, activation of both IFI27 and S100A12 was observed in a significant portion of patients with COVID-19 compared to healthy controls. In the dataset GSE152641 (Fig 1A), sampling was conducted within 24 hours of hospital admission with a median of 6 days from symptom onset (4–8 days). In this dataset, S100A12 expression was significantly higher in the patient group (p = 1.019e-07). It was below 11.20 in all of the 24 healthy controls (median value 9.66), while it was above 11.20 in 25 of the 62 patients (40.3%) with COVID-19 (median value 11.03). In the meantime, IFI27 expression was also significantly higher in the patient group (p = 1.09e-10). It was below 9.0 in 23 of the 24 healthy controls (median value 5.65, the other one being an outlier), while it was above 9.0 in 47 of these 62 patients (75.8%) with COVID-19 (median value 10.65). It showed that activation of both genes can be observed in the first week of the disease course, and activation of IFI27 was more common and at larger scale than that of S100A12 in hospitalized patients with COVID-19. Furthermore, activation of both genes was observed in 22 of the 62 patients (35.5%), suggesting that activation of S100A12 was generally accompanied by the activation of IFI27 (22 out of 25, 88%). In addition, non-activation of these two genes was observed in 12 of the 62 hospitalized patients (19.4%).
Fig 1. Activation of S100A12 and IFI27 in a significant portion of hospitalized COVID-19 patients compared to healthy controls (whole blood).
A) upper panel (dataset GSE152641), comparison of S100A12 and IFI27 expression in healthy controls and hospitalized COVID-19 patients. B) lower panel (dataset GSE171110), comparison of S100A12 and IFI27 expression in healthy controls and severe COVID-19 patients.
Since patients with severe disease are more concerned, the comparison between severe COVID-19 patients and healthy controls are of high interest. In the dataset GSE171110 (Fig 1B), 36 of the 44 severe patients were ICU patients, and sampling was conducted within 3 days of hospital admission with a median of 11 days from symptom onset (7–14 days). In this dataset, S100A12 expression was significantly higher in the patient group (p = 1.076e-08). It was below 12.0 in all of the 10 healthy controls (median value 11.16), while it was above 12.0 in 35 of the 44 patients (79.5%) with severe COVID-19 (median value 13.33). In the meantime, IFI27 expression was significantly higher in the patient group (p = 3.465e-05). It was below 11.0 in 9 of the 10 healthy controls (median value 7.57), while it was above 11.0 in 37 of these 44 patients (84.1%) with severed COVID-19 (median value 13.99). It showed that activation of these two genes were commonly observed in the second week of the disease course for severe patients. Compared to the dataset GSE152641, activation of S100A12 was more common in patients with severe COVID-19. In addition, activation of both genes was observed in 29 of the 44 severe patients (65.9%), while non-activation of these two gene was only observed in one severe patient. Again, activation of S100A12 was generally accompanied by the activation of IFI27 (29 out of 35, 82.9%).
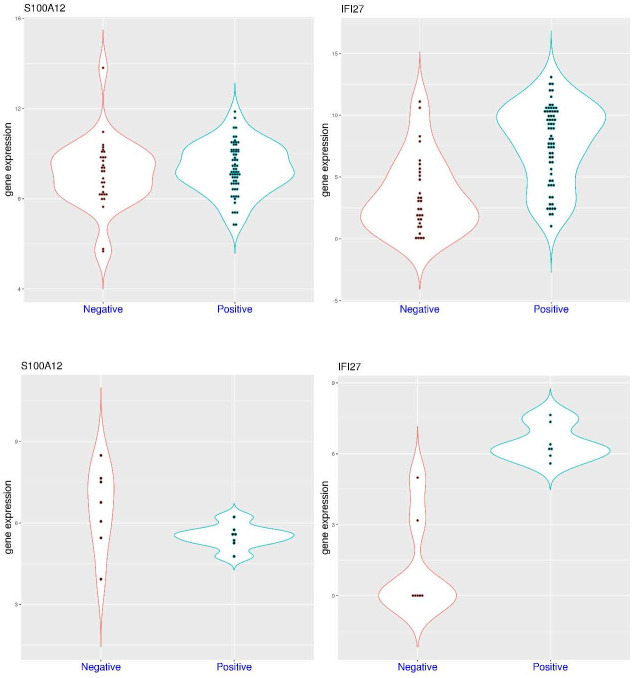
To have a broader understanding of gene activation, IFI27 and S100A12 expression can be compared between patients admitted to the same hospital with and without COVID-19. In the dataset GSE167000 (Fig 2A), all of the recruited non-ICU patients had relatively short hospital stay (2–9.5 days), some of which were tested positive for COVID-19. In this dataset, S100A12 expression was similarly distributed in both groups (p = 0.402) with median values of 9.28 and 9.22 in the patient groups with and without COVID-19, respectively. For example, S100A12 expression was above 11.5 in only 1 of the 30 negative patients and 2 of the 65 positive patients. Interestingly, the highest value (13.8) was observed in the negative group, and the highest value in the positive group was below 12. In the meantime, IFI27 expression was significantly higher in the positive group (median value 8.32) compared to the negative group (median value 2.74) (p = 6.413e-08). For example, IFI27 expression was above 6.5 in 4 of the 30 negative patients (13.3%) and 45 of the 65 positive patients (69.2%). The top six IFI27 expression values were all within the positive group. Overall, among hospitalized non-ICU patients, S100A12 expression might not be distinguishable between the positive and negative groups, while IFI27 activation was more restricted to the positive group.
Fig 2. Expression level of S100A12 and IFI27 in hospitalized patients with and without COVID-19 (whole blood).
A) upper panel (dataset GSE167000), comparison of S100A12 and IFI27 expression in non-critical patients with and without COVID-19. B) lower panel (dataset GSE154998), comparison of S100A12 and IFI27 expression in intensive care unit (ICU) patients with and without COVID-19.
The situation is somewhat different for ICU patients. In the dataset GSE154998 (Fig 2B), sampling was conducted at ICU admission for all of the recruited patients, some of which were tested positive for COVID-19. In this dataset, S100A12 expression was mostly higher in the negative group (median values 6.76) compared to the positive group (median value 5.57) (p-value not calculated due to small sample size). The top four S100A12 expression values were all within the negative group. On the other hand, IFI27 expression was significantly higher in the positive group (median value 6.21) compared to the negative group (median value 0, not detected in most samples due to technical reasons). The IFI27 expression values in the positive group were all higher than the top value in the negative group. Overall, among ICU patients, S100A12 activation could be even stronger in certain patients within the negative groups (such as bacterial sepsis), while IFI27 activation was more restricted to the positive group. Overall, for both ICU and non-ICU patients, IFI27 activation was more specific to COVID-19.
S100A12 activation is correlated with COVID-19 severity
A practical question is whether the activation of IFI27 or S100A12 is correlated with COVID-19 severity. In the dataset GSE172114 (Fig 3A), sampling was conducted upon ICU admission (for critical patients, a median of 7 days post symptom onset) or hospital admission (for non-critical patients, a median of 9.5 days post symptom onset). In this dataset, S100A12 expression was significantly higher in the critical patient group (p = 1.285e-12). It was above 6.0 in only 3 of the 23 patients (13.0%) with non-critical COVID-19 (median value 4.61), while it was above 6.0 in 41 of the 46 patients (89.1%) with critical COVID-19 (median value 7.63). In the meantime, IFI27 expression was similarly distributed in both groups (p>0.05). It was above 5.0 in 15 of the 23 patients (65.2%) with non-critical COVID-19 (median value 6.46), while it was above 5.0 in 38 of the 46 patients (82.6%) with critical COVID-19 (median value 7.87). It showed that activation of S100A12 was much more pronounced in critical COVID-19 patients than that of non-critical counterparts. In addition, lower expression of both genes (empirical cutoff defined above) was not observed in patients with critical COVID-19, while higher expression of both genes was observed in 33 of those 46 patients (71.7%). Again, activation of S100A12 was generally accompanied by the activation of IFI27 (33 out of 41, 80.5%).
Fig 3. Expression level of S100A12 and IFI27 in COVID-19 patients with different severity (whole blood).
A) upper panel (dataset GSE172114), comparison of S100A12 and IFI27 expression in moderate and critical COVID-19 patients. B) lower panel (dataset Zenodo 6120249), comparison of S100A12 and IFI27 expression in COVID-19 patients with different severity (non-hospitalized, moderate, severe and critical).
To have a strong correlation with disease severity, a step-wise increase of activation would be preferred. In the dataset Zenodo 6120249 (Fig 3B), sampling was conducted upon hospital or ICU admission (for hospitalized patients with varying time post symptom onset) or in the recovery stage (for community cases, at least 7 days post symptom onset). In this dataset, S100A12 expression was significantly different in the COVID-19 patient groups with different severity. The p-values were 0.00142, 5.803e-05, and 0.000363 for the comparison of community cases to mild cases, mild cases to severe cases, and severe cases to critical cases, respectively. The expression was below 8.0 in all of the 10 healthy volunteers (median value 6.22) and same for all of the 13 non-hospitalized COVID-19 patients (median value 6.33), while it was above 8.0 in 4 of the 18 patients (22.2%) with mild COVID-19 (median value 7.31), 23 of the 41 patients (56.1%) with severe COVID-19 (median value 8.11), and 17 of the 19 patients (89.5%) with critical COVID-19 (median value 9.15). In addition, the highest value was only 8.41 in the mild COVID-19 group. Therefore, S100A12 activation was a marker for severe and critical COVID-19. In the meantime, IFI27 expression was similar in the mild, severe and critical COVID-19 patient groups (p>0.05 for pairwise comparison). It was below 4.5 in all of the 10 healthy volunteers (median value 1.75), while it was above 4.5 in 6 of the 13 non-hospitalized COVID-19 patients (46.2%, median value 3.09), 14 of the 18 patients (77.8%) with mild COVID-19 (median value 7.57), 34 of the 41 patients (82.9%) with severe COVID-19 (median value 7.52), and 15 of the 19 patients (78.9%) with critical COVID-19 (median value 6.39). Overall, this clearly demonstrated a stepwise increase of S100A12 activation in COVID-19, while IFI27 activation was similarly predominant in hospitalized COVID-19 patients irrespective of disease severity. Again, activation of S100A12 was generally accompanied by the activation of IFI27 in COVID-19 patients (84.1% for this cohort). As an additional comparison, S100A12 expression was above 8.0 in 39 of the 42 sepsis patients (92.9%, median value 10.23), while IFI27 expression was above 4.5 in only 10 of these 42 sepsis patients (23.8%, median value 1.94). Therefore, activation of S100A12 could be even stronger in sepsis, but it was infrequently accompanied by the activation of IFI27 (except for viral sepsis).
This trend of step-wise S100A12 activation in COVID-19 was replicated in two more datasets. In the dataset GSE189990 (Fig 4A), sampling was conducted upon hospital admission. In this dataset, S100A12 expression showed a step-wise increase trend (p-values not calculated due to small sample size). It was below 8.0 in all of the 4 healthy controls (median value 7.27), while it was above 8.0 in the two patients (100%) with incidental COVID-19 (median value 8.31), all of the 4 patients (100%) with moderate COVID-19 (median value 9.44), 8 of the 9 patients (88.9%) with critical COVID-19 (median value 9.42), and all of the 5 patients (100%) with fatal COVID-19 (median value 11.16). In another dataset GSE161777 (Fig 4B), this trend could even be quantified by correlation analysis. In this dataset, multiple patients were sampled at multiple time points. The disease severity/stage was indexed as 2 to 7 with decreasing clinical score, representing critical, highly complicated, complicated, moderate, convalescence, and recovery. There was a moderate correlation (r = 0.54) between this severity index and S100A12 expression.
Fig 4. Correlation of S100A12 expression and the severity of COVID-19 (whole blood).

A) upper panel (dataset GSE189990), comparison of S100A12 expression in COVID-19 patients with different severity (incidental, moderate, critical and fatal). B) lower panel (dataset GSE161777), correlation between S100A12 expression and severity index (from critical to recovery).
This trend was also clear when combining the pictures from two more datasets. In the dataset GSE161731 (Fig 5A), hospitalized COVID-19 patients were compared with non-hospitalized patients and control subjects. In this dataset, S100A12 expression was significantly higher in hospitalized COVID-19 patients compared to non-hospitalized patients (p = 0.000386). It was below 10.6 in all of the 19 healthy controls (median value 9.36), while it was above 10.6 in only one of the 35 non-hospitalized patients with COVID-19 (median value 8.72), and 5 of the 12 hospitalized patients (41.7%) with COVID-19 (median value 10.02). In the meantime, IFI27 expression was below 6.2 in all of the 19 healthy controls (median value 1.0), while it was above 6.2 in 11 of the 35 non-hospitalized patients (31.4%) with COVID-19 (median value 4.3), and 11 of the 12 hospitalized patients (91.7%) with COVID-19 (median value 8.56). Overall, S100A12 activation was rarely observed in non-hospitalized patients with COVID-19, while it could be observed in a significant portion of hospitalized patients with COVID-19, depending on the distribution of severity. For comparison, S100A12 activation was not observed in the influenza group, rarely (1 out of 49) in the other coronavirus infection group (similar to the non-hospitalized COVID-19 group), but commonly (19 out of 23) in the bacterial infection group. On the other hand, IFI27 activation was not observed in the bacterial infection group, infrequently (7 out of 49) in the other coronavirus infection group, but commonly (23 out of 23) in the influenza group.
Fig 5. Expression level of S100A12 and IFI27 in COVID-19 patients and patients with other diseases (whole blood).
A) upper panel (dataset GSE161731), comparison of S100A12 and IFI27 expression in healthy controls and patients with COVID-19 (hospitalized and non-hospitalized) or other types of infection. B) lower panel (dataset GSE157103), comparison of S100A12 and IFI27 expression in intensive care unit (ICU) and non-ICU patients with or without COVID-19.
In another dataset GSE157103 (Fig 5B), hospitalized patients were further divided into ICU patients and non-ICU patients. In this dataset, S100A12 expression was significantly higher in the ICU group with COVID-19 compared to the non-ICU group (p = 7.815e-07). It was above 12.2 in 14 of the 50 non-ICU patients (28%) with COVID-19 (median value 11.74), while it was above 12.2 in 36 of the 50 ICU patients (72%) with COVID-19 (median value 12.62). In the meantime, IFI27 expression was above 6.0 in 38 of the 50 non-ICU patients (76%) with COVID-19 (median value 8.11), and it was above 6.0 in 24 of the 50 ICU patients (48%) with COVID-19 (median value 5.74). In comparison, for patients without COVID-19, S100A12 expression was above 12.2 in one of the 10 non-ICU patients, and 12 of the 16 ICU patients (75%), while IFI27 expression was above 6.0 in none of the 10 non-ICU patients, and only 2 of the 16 ICU patients. Overall, S100A12 activation was correlated with disease severity (with or without COVID-19), while IFI27 activation was more specific to COVID-19. The combined observations from these two datasets (GSE161731 and GSE157103) showed the stepwise increase of S100A12 activation in COVID-19 patients from non-hospitalized patients to hospitalized patients, and from hospitalized non-ICU patients to ICU patients.
Fast waning of IFI27 activation during COVID-19 disease course
As markers of host immune response, the dynamic properties of IFI27 and S100A12 expression can also be examined. In the dataset Zenodo 6120249 (Fig 6A), IFI27 expression was moderately correlated with the time since symptom onset (r = 0.54). In another dataset GSE155454 (Fig 6B), IFI27 expression displayed even stronger correlation with the time since symptom onset (r = 0.70). In both datasets, IFI27 expression was generally back to the control level after day 15. On the other hand, S100A12 expression displayed no correlation with the time since symptom onset in both datasets (r close to 0).
Fig 6. Correlation of IFI27 expression with the duration of COVID-19 infection.
A) upper left (dataset Zenodo 6120249), correlation between IFI27 expression and time since symptom onset in COVID-19 patients. B) upper right (dataset GSE155454), correlation between IFI27 expression and time since symptom onset in COVID-19 patients. C) lower panel (dataset GSE166190), correlation between IFI27 expression and time interval index in COVID-19 patients (both adults and children).
This fast waning of IFI27 activation was similarly observed in both adults and children. In the dataset GSE166190 (Fig 6C), time intervals were defined according to the days post symptom onset, including interval 1 (0–5 days), interval 2 (6–14 days), interval 3 (15–22 days), interval 4 (23–35 days), and interval 5 (36–81 days). In this dataset, IFI27 expression displayed strong correlation with the time interval in both the adult group (r = 0.69) and the children group (r = 0.82). In both groups, IFI27 expression was activated in the first time-interval (day 0–5), and mostly came down to the control level at time interval 3 (day 15 and after). This feature was consistent with the previous two datasets.
Not surprisingly, this fast waning of IFI27 activation was also observed in seasonal influenza infection. In the dataset GSE68310 (Fig 7A), community monitoring of seasonal flu was conducted. Again, IFI27 expression was already activated at day 2 after symptom onset and gradually came back to normal level before day 21. The correlation was also very strong between IFI27 expression and the time since symptom onset (r = 0.87). Since the same group of people were sampled at multiple time points, this trend was even more convincing.
Fig 7. Longitudinal tracking of the expression level of IFI27 and S100A12 during the course of infection.
A) upper left (dataset GSE68310), tracking of IFI27 expression in a community cohort with seasonal influenza. B) upper right (dataset GSE161731), tracking of IFI27 expression in non-hospitalized patients with COVID-19. C) lower left (dataset GSE178967), change of IFI27 expression in COVID-19 patients 5 days after study enrollment. D) lower right (dataset GSE169687), long-term tracking of S100A12 expression in COVID-19 patients.
Serial sampling was also conducted for patients with COVID-19. In the dataset GSE161731 (Fig 7B), some of the non-hospitalized COVID-19 patients were sampled at multiple time points and grouped into three time intervals (1-early, <11 days, 2-middle, 11–21 days, and 3-late, >21 days). Moderate correlation (r = 0.60) was observed between IFI27 expression and the time interval. Activation of IFI27 was observed in 7 of the 9 samples within time interval 1 (please refer to Fig 5A for the cutoff value), while only in 2 of 18 samples within time interval 2, and only in 1 of 16 samples within time interval 3. In another dataset GSE178967 (Fig 7C), COVID-19 outpatients were enrolled and examined at day 0 and 5 for gene expression (median days from symptom onset was 5 at enrollment). Compared to the expression level at the baseline, IFI27 expression was down for most of the patients (52 out of 72, 72.2%) after only 5 days since enrollment. To be more specific, significant increase of IFI27 expression (>1) only occurred to patients with low baseline expression (<4.5), while most of the patients (25 out of 28, 89.3%) with higher baseline expression (>6) displayed decrease of IFI27 expression. Overall, the features from both datasets were consistent with the feature of fast waning of IFI27 activation within the first two weeks.
The same does not apply to S100A12 activation which may sustain for much longer time. In the dataset GSE169687 (Fig 7D), recovering COVID-19 patients were followed up to 6 months from the first positive polymerase chain reaction (PCR) test. In this cohort, S100A12 expression was below 6.0 in all of the 14 healthy controls (median value 4.41), but it was above 6.0 in 9 of the 24 patients at week 12 (median value 5.51), 12 of 58 patients at week 16 (median value 5.32), and 11 of the 56 patients at week 24 (median value 5.17). Although higher activation of S100A12 (>7.0) was rarely observed (0 at week 12, and only one at week 16 and 24 each), this moderate activation in a significant portion of patients may still be related to the long-term health, since long COVID was observed later for some of these patients.
Activation pattern of IFI27 and S100A12 in PBMC
The observations above were found in studies with whole blood (or leukocytes). A few other studies were conducted using PBMC. In the dataset GSE152418, sampling was mostly conducted within 3 weeks post symptom onset. In this dataset (Fig 8A), S100A12 expression was below 11.6 in all of the 17 healthy volunteers (median value 10.01) and same for all of the 4 patients with moderate COVID-19 (median value 9.75), while it was above 11.6 in 4 of the 8 patients (50%) with severe COVID-19 (median value 11.62), and all of the 4 patients (100%) with critical COVID-19 (median value 12.96). This clearly demonstrated a step-wise increase of S100A12 activation in the COVID-19 patients with increasing severity (p-value not calculated due to small sample size). In the meantime, IFI27 expression was below 7.0 in all of the 17 healthy volunteers (median value 4.0), while it was above 7.0 in 15 of the 16 COVID-19 patients (median value 13.3, 10.33, and 9.98 for the moderate, severe and critical group, respectively). In addition (Fig 8B), IFI27 expression was highly correlated with disease duration (r = 0.82), while S100A12 expression displayed no correlation with disease duration (r = 0.1). Overall, the stepwise increase of S100A12 activation and fast waning of IFI27 activation were consistently observed in whole blood and PBMC of patients with COVID-19.
Fig 8. Expression level of S100A12 and IFI27 in peripheral blood mononuclear cells (PBMCs) as correlated with COVID-19 severity or disease duration (dataset GSE152418).
A) upper panel, comparison of S100A12 and IFI27 expression in healthy controls and COVID-19 patients with different severity (moderate, severe and critical). B) lower panel, correlation between IFI27 expression and time since symptom onset in COVID-19 patients.
This feature was further replicated in two more datasets for PBMC. In the dataset GSE184401 (Fig 9A), severe patients were further divided into two groups, with or without secondary infection. In this dataset, S100A12 expression was significantly higher in the severe patient group compared to the mild patient group (p = 8.989e-08). It was below 8.15 in all of the 21 patients with mild COVID-19 (median value 6.43), while it was above 8.15 in 11 of the 17 severe COVID-19 patients without secondary infection (median value 9.23), and all of the 5 severe COVID-19 patients with secondary infection (median value 8.72). On the other hand, IFI27 expression was not significantly higher in the severe group compared to the mild group (p>0.05). The median value of IFI27 expression was 3.51, 3.3, and 1.44 for the mild group, severe without secondary infection group, and severe with secondary infection group, respectively. For example, IFI27 expression was above 5.0 in 6 of the 21 patients with mild COVID-19, 4 of the 17 severe COVID-19 patients without secondary infection, and one of the 5 severe COVID-19 patients with secondary infection.
Fig 9. Expression levels of S100A12 and IFI27 in peripheral blood mononuclear cells (PBMCs) as markers for the infection in COVID-19 patients.
A) upper panel (dataset GSE184401), comparison of S100A12 and IFI27 expression in COVID-19 patients with different severity (mild or severe) and secondary infection (with or without). B) lower panel (dataset GSE179627), comparison of S100A12 and IFI27 expression in healthy controls and COVID-19 patients with different severity (asymptomatic, mild and moderate) and different stages (acute, recovering and re-positive).
In another dataset GSE179627 (Fig 9B), in addition to patients undergoing treatment (asymptomatic, mild or moderate), the study also included recovering patients at the time of hospital discharge and those who were tested positive again weeks after being discharged. In this dataset, S100A12 expression was below 7.0 in all of the 22 control subjects, as well as all of the 21 hospitalized patients, while it was above 7.0 in 4 of the 15 recovering patients, 3 of the 12 re-positive patients. This showed that S100A12 activation may last for a long time in certain COVID-19 patients. On the other hand, IFI27 activation was mostly restricted to the acute phase of the disease. For example, IFI27 expression was below 5.0 in all of the 22 control subjects as well as all of the 8 asymptomatic patients, while it was above 5.0 in 2 of the 3 mild patients, 7 of the 10 moderate patients, 2 of the 15 recovering patients, and none of the 12 re-positive patients. Overall, the fast waning of IFI27 activation and the long tail of S100A12 activation were consistently observed in whole blood and PBMC.
Discussion
Based on the data presented above, a two-tiered innate immune response was clearly observed in patients with COVID-19, the first tier being the activation of ISGs, while the second tier being the activation of ABGs. Because interferon signaling is likely a universal response to respiratory viral infection, the activation of ISGs was highly prevalent in patients with symptomatic COVID-19. However, unlike seasonal flu, the activation of ABGs was observed in a significant portion of patients with COVID-19, along with much higher death rate in this pandemic disease [58]. This is likely because the first-tier response is far from sufficient for some patients with COVID-19, leading to the ensuing deployment of the second-tier response. The second-tier response is more restricted to severe or even critical patients, reflecting the overwhelming infection status including the high load of viral antigen and RNA in the blood [59]. Clearly, this response is not a mere reflection of neutrophil expansion [5], because similar pattern was observed in PBMCs.
This study included 1421 samples from 17 independent studies around the globe, including Europe, North America, Asia and Australia. The consistent features from independent studies suggest that geographic locations do not have a significant impact on the two-tiered innate immune response in COVID-19 patients. Our previous works also demonstrated that age and sex do not affect the expression of the marker genes for the two-tiered innate immune response [38]. Thus, the observations presented above are robust and not confounded by these common factors.
An advantage of this two-gene marker panel is the direct link to the two-tiered innate immune response which makes it different from other markers or marker panels. Importantly, the first-tier response is strongly correlated with disease duration, while the second-tier response is strongly correlated with disease severity. Thus, rich clinically relevant information can be obtained through simultaneous monitoring of the two-tiered innate immune response. This has a clear advantage over existing markers such as PCT and CRP.
The characteristic features of this two-tiered response in COVID-19 can have implications in clinical decisions. Activation of several inflammatory markers has been observed in patients with severe COVID-19 and has been proposed as prognostic markers [19, 60, 61]. Since S100A12 expression displayed a stepwise activation pattern and demonstrated strong association with severe COVID-19, it may serve as a good prognostic marker as well. In fact, S100A12 expression showed superior prognostic power compared to PCT, CRP, D-dimer and other parameters in our previous reanalysis of data from a prospective study [62]. This is likely because the second-tier innate immune response is highly connected to the disease mechanism in severe COVID-19 including hyperinflammation. Several studies showed that better prognostic power can be achieved by serial sampling of clinical parameters [63, 64]. Thus, serial sampling of S100A12 expression is recommended, and sustained or even increased activation shall be taken as an alarming sign. Since S100A12 activation may last for several months in certain patients with COVID-19, it warrants further investigation whether this sustained activation is associated with some of the lingering symptoms in long COVID [65]. In addition, another application is to rule out secondary bacterial co-infection when S100A12 activation is not detected, similar to the recently proposed usage of PCT [66]. This is because S100A12 activation is the most prominent signature of bacterial infection (with or without viral co-infection).
On the other hand, the fast waning of IFI27 activation suggests that IFI27 expression may serve as a surrogate of disease duration in patients with COVID-19. In the meantime, patients with sustained IFI27 activation over 2–3 weeks may deserve special attention. Another application is to alert nosocomial SARS-CoV-2 infection for patients admitted for other reasons when IFI27 activation is newly detected (compared to baseline value at admission), because IFI27 activation is rarely observed in hospitalized patients with other diseases. Additionally, since most of the S100A12 activation is accompanied by IFI27 activation in patients with severe COVID-19, it is unlikely that the inability of ISG activation is to blame for most of the severe symptoms, which has implications in the usage of interferon as a therapeutic option.
This study is limited by the availability of more high-quality transcriptome data with enriched clinical information of individual patients, likely due to privacy concern and other practical reasons. For example, laboratory test results and time from symptom onset for each sample are not provided in most of the datasets. In addition, sample size is not ideal in many studies. It’s also desirable to have high quality data with the inclusion of symptomatic patients, asymptomatic patients and healthy controls in the same study. Sampling at multiple time points is critical for disease monitoring but is rarely conducted in these transcriptome studies. In the case of long COVID, follow-up well over six months may be needed. It’s also of strong interest whether and how the activation pattern may change with different variants of the virus.
This ongoing pandemic has made reverse transcriptase PCR (RT-PCR) platforms widely accessible and cost effective (~4$/test in mainland China), which can facilitate the realization of routine monitoring of the two-tiered innate immune response. In our previous works, we have conducted RT-PCR experiments on S100A12, IFI27 and other relevant genes in well over a thousand clinical blood samples [38, 67, 68]. We have also designed a single-tube assay for three genes including S100A12, IFI27 and an internal reference gene (unpublished work). Hopefully, this kind of assays can be applied to the routine monitoring of COVID-19 and other infectious diseases in the future.
Conclusions
In this study, a two-tiered innate immune response was demonstrated in COVID-19 patients, and a two-gene marker (IFI27/S100A12) for this two-tiered innate immune response was proposed. For most of the symptomatic COVID-19 patients, IFI27 activation was observable within the first 2–3 weeks of symptom onset and gradually came back to the baseline level after this acute phase. On the other hand, S100A12 activation displayed no correlation with disease duration but was highly correlated with disease severity. Simultaneous serial monitoring of these two genes can examine the two-tiered innate immune response in real time and may better guide clinical triage of COVID-19.
Data Availability
All of the datasets analyzed in the current study are publicly available. Most of the transcriptome datasets are available at GEO (gene expression omnibus, https://www.ncbi.nlm.nih.gov/geo/). The accession numbers are: GSE152641, GSE171110, GSE167000, GSE172114, GSE189990, GSE161777, GSE161731, GSE155454, GSE166190, GSE178967, GSE169687, GSE154998, GSE157103, GSE152418, GSE184401, GSE179627 and GSE68310. One of the datasets is available at the Zenodo repository (https://zenodo.org/). The accession number is 6120249.
Funding Statement
This work was supported by grants from the Strategic Priority Research Program of Chinese Academy of Sciences (Grant No. XDB38030200), the National Key Research and Development Program of China (grant no. 2016YFC0901700), and the National High Technology Program of China (863 Program; Grant No. 2015AA020108) awarded to HL by the Ministry of Science and Technology of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dye C. The benefits of large scale covid-19 vaccination. Bmj. 2022;377:o867. doi: 10.1136/bmj.o867 [DOI] [PubMed] [Google Scholar]
- 2.A living WHO guideline on drugs for covid-19. Bmj. 2022;377:o1045. doi: 10.1136/bmj.o1045 [DOI] [PubMed] [Google Scholar]
- 3.Battaglini D, Lopes-Pacheco M, Castro-Faria-Neto HC, Pelosi P, Rocco PRM. Laboratory Biomarkers for Diagnosis and Prognosis in COVID-19. Frontiers in immunology. 2022;13:857573. doi: 10.3389/fimmu.2022.857573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rovito R, Augello M, Ben-Haim A, Bono V, d’Arminio Monforte A, Marchetti G. Hallmarks of Severe COVID-19 Pathogenesis: A Pas de Deux Between Viral and Host Factors. Frontiers in immunology. 2022;13:912336. doi: 10.3389/fimmu.2022.912336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Zhao J, Yang L, Hu J, Yao Y. Value of the Neutrophil-Lymphocyte Ratio in Predicting COVID-19 Severity: A Meta-analysis. Dis Markers. 2021;2021:2571912. doi: 10.1155/2021/2571912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett TJ, Bilaloglu S, Cornwell M, Burgess HM, Virginio VW, Drenkova K, et al. Platelets contribute to disease severity in COVID-19. Journal of thrombosis and haemostasis: JTH. 2021;19(12):3139–53. doi: 10.1111/jth.15534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alsuwaidi L, Al Heialy S, Shaikh N, Al Najjar F, Seliem R, Han A, et al. Monocyte distribution width as a novel sepsis indicator in COVID-19 patients. BMC infectious diseases. 2022;22(1):27. doi: 10.1186/s12879-021-07016-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banon T, Wortsman J, Ben Moshe S, Gazit S, Peretz A, Ben Tov A, et al. Evaluating red blood cell distribution width from community blood tests as a predictor of hospitalization and mortality in adults with SARS-CoV-2: a cohort study. Annals of medicine. 2021;53(1):1410–8. doi: 10.1080/07853890.2021.1968484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrido P, Cueto P, Rovira C, Garcia E, Parra A, Enriquez R, et al. Clinical value of procalcitonin in critically ill patients infected by SARS-CoV-2. The American journal of emergency medicine. 2021;46:525–31. doi: 10.1016/j.ajem.2020.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goncalves FAR, Besen B, Lima CA, Cora AP, Pereira AJR, Perazzio SF, et al. Use and misuse of biomarkers and the role of D-dimer and C-reactive protein in the management of COVID-19: A post-hoc analysis of a prospective cohort study. Clinics. 2021;76:e3547. doi: 10.6061/clinics/2021/e3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poudel A, Poudel Y, Adhikari A, Aryal BB, Dangol D, Bajracharya T, et al. D-dimer as a biomarker for assessment of COVID-19 prognosis: D-dimer levels on admission and its role in predicting disease outcome in hospitalized patients with COVID-19. PloS one. 2021;16(8):e0256744. doi: 10.1371/journal.pone.0256744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lampart M, Zellweger N, Bassetti S, Tschudin-Sutter S, Rentsch KM, Siegemund M, et al. Clinical utility of inflammatory biomarkers in COVID-19 in direct comparison to other respiratory infections-A prospective cohort study. PloS one. 2022;17(5):e0269005. doi: 10.1371/journal.pone.0269005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stauning MA, Altintas I, Kallemose T, Eugen-Olsen J, Lindstrom MB, Rasmussen LJH, et al. Soluble Urokinase Plasminogen Activator Receptor as a Decision Marker for Early Discharge of Patients with COVID-19 Symptoms in the Emergency Department. The Journal of emergency medicine. 2021;61(3):298–313. doi: 10.1016/j.jemermed.2021.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su C, Xu Z, Hoffman K, Goyal P, Safford MM, Lee J, et al. Identifying organ dysfunction trajectory-based subphenotypes in critically ill patients with COVID-19. Sci Rep. 2021;11(1):15872. doi: 10.1038/s41598-021-95431-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Araya S, Tsegay YG, Atlaw A, Aragaw M, Tadlo G, Tsegaye N, et al. Organ function biomarker abnormalities, associated factors and disease outcome among hospitalized patients with COVID-19. Biomarkers in medicine. 2022;16(6):417–26. doi: 10.2217/bmm-2021-0681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng QL, Yu ZJ, Ji F, Li GM, Zhang GF, Xu JH, et al. Dynamic changes in liver function parameters in patients with coronavirus disease 2019: a multicentre, retrospective study. BMC infectious diseases. 2021;21(1):818. doi: 10.1186/s12879-021-06572-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrow AJ, Sykes R, McIntosh A, Kamdar A, Bagot C, Bayes HK, et al. A multisystem, cardio-renal investigation of post-COVID-19 illness. Nature medicine. 2022;28(6):1303–13. doi: 10.1038/s41591-022-01837-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jong VMT, Rousset RZ, Antonio-Villa NE, Buenen AG, Van Calster B, Bello-Chavolla OY, et al. Clinical prediction models for mortality in patients with covid-19: external validation and individual participant data meta-analysis. Bmj. 2022;378:e069881. doi: 10.1136/bmj-2021-069881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubio-Rivas M, Mora-Lujan JM, Formiga F, Corrales Gonzalez MA, Garcia Andreu MDM, Moreno-Torres V, et al. Clusters of inflammation in COVID-19: descriptive analysis and prognosis on more than 15,000 patients from the Spanish SEMI-COVID-19 Registry. Internal and emergency medicine. 2022;17(4):1115–27. doi: 10.1007/s11739-021-02924-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varikasuvu SR, Varshney S, Dutt N, Munikumar M, Asfahan S, Kulkarni PP, et al. D-dimer, disease severity, and deaths (3D-study) in patients with COVID-19: a systematic review and meta-analysis of 100 studies. Sci Rep. 2021;11(1):21888. doi: 10.1038/s41598-021-01462-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chomistek AK, Liang C, Doherty MC, Clifford CR, Ogilvie RP, Gately RV, et al. Predictors of critical care, mechanical ventilation, and mortality among hospitalized patients with COVID-19 in an electronic health record database. BMC infectious diseases. 2022;22(1):413. doi: 10.1186/s12879-022-07383-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashktorab H, Pizuorno A, Adeleye F, Laiyemo A, Dalivand MM, Aduli F, et al. Symptomatic, clinical and biomarker associations for mortality in hospitalized COVID-19 patients enriched for African Americans. BMC infectious diseases. 2022;22(1):552. doi: 10.1186/s12879-022-07520-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adamo S, Michler J, Zurbuchen Y, Cervia C, Taeschler P, Raeber ME, et al. Signature of long-lived memory CD8(+) T cells in acute SARS-CoV-2 infection. Nature. 2022;602(7895):148–55. doi: 10.1038/s41586-021-04280-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unterman A, Sumida TS, Nouri N, Yan X, Zhao AY, Gasque V, et al. Single-cell multi-omics reveals dyssynchrony of the innate and adaptive immune system in progressive COVID-19. Nature communications. 2022;13(1):440. doi: 10.1038/s41467-021-27716-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Notarbartolo S, Ranzani V, Bandera A, Gruarin P, Bevilacqua V, Putignano AR, et al. Integrated longitudinal immunophenotypic, transcriptional and repertoire analyses delineate immune responses in COVID-19 patients. Science immunology. 2021;6(62). doi: 10.1126/sciimmunol.abg5021 [DOI] [PubMed] [Google Scholar]
- 26.Kreutmair S, Unger S, Nunez NG, Ingelfinger F, Alberti C, De Feo D, et al. Distinct immunological signatures discriminate severe COVID-19 from non-SARS-CoV-2-driven critical pneumonia. Immunity. 2021;54(7):1578–93 e5. doi: 10.1016/j.immuni.2021.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura Y, Nakai Y, Shin J, Hara M, Takeda Y, Kubo S, et al. Identification of serum prognostic biomarkers of severe COVID-19 using a quantitative proteomic approach. Sci Rep. 2021;11(1):20638. doi: 10.1038/s41598-021-98253-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McArdle A, Washington KE, Chazarin Orgel B, Binek A, Manalo DM, Rivas A, et al. Discovery Proteomics for COVID-19: Where We Are Now. Journal of proteome research. 2021;20(10):4627–39. doi: 10.1021/acs.jproteome.1c00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li ZB, Liu J, Zhang SQ, Yu Y, Liang HF, Lu QQ, et al. Novel potential metabolic biomarker panel for early detection of severe COVID-19 using full-spectrum metabolome and whole-transcriptome analyses. Signal transduction and targeted therapy. 2022;7(1):129. doi: 10.1038/s41392-022-00976-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sindelar M, Stancliffe E, Schwaiger-Haber M, Anbukumar DS, Adkins-Travis K, Goss CW, et al. Longitudinal metabolomics of human plasma reveals prognostic markers of COVID-19 disease severity. Cell reports Medicine. 2021;2(8):100369. doi: 10.1016/j.xcrm.2021.100369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda R, Lodge S, Nitschke P, Spraul M, Schaefer H, Bong SH, et al. Integrative Modeling of Plasma Metabolic and Lipoprotein Biomarkers of SARS-CoV-2 Infection in Spanish and Australian COVID-19 Patient Cohorts. Journal of proteome research. 2021;20(8):4139–52. doi: 10.1021/acs.jproteome.1c00458 [DOI] [PubMed] [Google Scholar]
- 32.He X, Liu C, Peng J, Li Z, Li F, Wang J, et al. COVID-19 induces new-onset insulin resistance and lipid metabolic dysregulation via regulation of secreted metabolic factors. Signal transduction and targeted therapy. 2021;6(1):427. doi: 10.1038/s41392-021-00822-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masood KI, Yameen M, Ashraf J, Shahid S, Mahmood SF, Nasir A, et al. Upregulated type I interferon responses in asymptomatic COVID-19 infection are associated with improved clinical outcome. Sci Rep. 2021;11(1):22958. doi: 10.1038/s41598-021-02489-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez-Pato A, Virseda-Berdices A, Resino S, Ryan P, Martinez-Gonzalez O, Perez-Garcia F, et al. Plasma miRNA profile at COVID-19 onset predicts severity status and mortality. Emerging microbes & infections. 2022;11(1):676–88. doi: 10.1080/22221751.2022.2038021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Li J, Zhang L, Sun HX, Zhang Z, Xu J, et al. Plasma cell-free RNA characteristics in COVID-19 patients. Genome research. 2022;32(2):228–41. doi: 10.1101/gr.276175.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annual review of immunology. 2014;32:513–45. doi: 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song F, Qian Y, Peng X, Li X, Xing P, Ye D, et al. The frontline of immune response in peripheral blood. PloS one. 2017;12(8):e0182294. doi: 10.1371/journal.pone.0182294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lei H, Xu X, Wang C, Xue D, Wang C, Chen J. A host-based two-gene model for the identification of bacterial infection in general clinical settings. International journal of infectious diseases: IJID: official publication of the International Society for Infectious Diseases. 2021. doi: 10.1016/j.ijid.2021.02.112 [DOI] [PubMed] [Google Scholar]
- 39.Tang BM, Shojaei M, Parnell GP, Huang S, Nalos M, Teoh S, et al. A novel immune biomarker IFI27 discriminates between influenza and bacteria in patients with suspected respiratory infection. The European respiratory journal. 2017;49(6). doi: 10.1183/13993003.02098-2016 [DOI] [PubMed] [Google Scholar]
- 40.Xu N, Hao F, Dong X, Yao Y, Guan Y, Yang L, et al. A two-transcript biomarker of host classifier genes for discrimination of bacterial from viral infection in acute febrile illness: a multicentre discovery and validation study. The Lancet Digital health. 2021;3(8):e507–e16. doi: 10.1016/S2589-7500(21)00102-3 [DOI] [PubMed] [Google Scholar]
- 41.Herberg JA, Kaforou M, Wright VJ, Shailes H, Eleftherohorinou H, Hoggart CJ, et al. Diagnostic Test Accuracy of a 2-Transcript Host RNA Signature for Discriminating Bacterial vs Viral Infection in Febrile Children. JAMA. 2016;316(8):835–45. doi: 10.1001/jama.2016.11236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thair SA, He YD, Hasin-Brumshtein Y, Sakaram S, Pandya R, Toh J, et al. Transcriptomic similarities and differences in host response between SARS-CoV-2 and other viral infections. iScience. 2021;24(1):101947. doi: 10.1016/j.isci.2020.101947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levy Y, Wiedemann A, Hejblum BP, Durand M, Lefebvre C, Surenaud M, et al. CD177, a specific marker of neutrophil activation, is associated with coronavirus disease 2019 severity and death. iScience. 2021;24(7):102711. doi: 10.1016/j.isci.2021.102711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galbraith MD, Kinning KT, Sullivan KD, Baxter R, Araya P, Jordan KR, et al. Seroconversion stages COVID19 into distinct pathophysiological states. eLife. 2021;10. doi: 10.7554/eLife.65508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carapito R, Li R, Helms J, Carapito C, Gujja S, Rolli V, et al. Identification of driver genes for critical forms of COVID-19 in a deeply phenotyped young patient cohort. Science translational medicine. 2022;14(628):eabj7521. doi: 10.1126/scitranslmed.abj7521 [DOI] [PubMed] [Google Scholar]
- 46.Consortium CO-M-oBA. A blood atlas of COVID-19 defines hallmarks of disease severity and specificity. Cell. 2022;185(5):916–38 e58. doi: 10.1016/j.cell.2022.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wargodsky R, Dela Cruz P, LaFleur J, Yamane D, Kim JS, Benjenk I, et al. RNA Sequencing in COVID-19 patients identifies neutrophil activation biomarkers as a promising diagnostic platform for infections. PloS one. 2022;17(1):e0261679. doi: 10.1371/journal.pone.0261679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernardes JP, Mishra N, Tran F, Bahmer T, Best L, Blase JI, et al. Longitudinal Multi-omics Analyses Identify Responses of Megakaryocytes, Erythroid Cells, and Plasmablasts as Hallmarks of Severe COVID-19. Immunity. 2020;53(6):1296–314 e9. doi: 10.1016/j.immuni.2020.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McClain MT, Constantine FJ, Henao R, Liu Y, Tsalik EL, Burke TW, et al. Dysregulated transcriptional responses to SARS-CoV-2 in the periphery. Nature communications. 2021;12(1):1079. doi: 10.1038/s41467-021-21289-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vono M, Huttner A, Lemeille S, Martinez-Murillo P, Meyer B, Baggio S, et al. Robust innate responses to SARS-CoV-2 in children resolve faster than in adults without compromising adaptive immunity. Cell reports. 2021;37(1):109773. doi: 10.1016/j.celrep.2021.109773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu Z, van der Ploeg K, Chakraborty S, Arunachalam P, Mori D, Jacobson K, et al. Early immune responses have long-term associations with clinical, virologic, and immunologic outcomes in patients with COVID-19. Research square. 2022. doi: 10.21203/rs.3.rs-847082/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryan FJ, Hope CM, Masavuli MG, Lynn MA, Mekonnen ZA, Yeow AEL, et al. Long-term perturbation of the peripheral immune system months after SARS-CoV-2 infection. BMC Med. 2022;20(1):26. doi: 10.1186/s12916-021-02228-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gill SE, Dos Santos CC, O’Gorman DB, Carter DE, Patterson EK, Slessarev M, et al. Transcriptional profiling of leukocytes in critically ill COVID19 patients: implications for interferon response and coagulation. Intensive care medicine experimental. 2020;8(1):75. doi: 10.1186/s40635-020-00361-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Overmyer KA, Shishkova E, Miller IJ, Balnis J, Bernstein MN, Peters-Clarke TM, et al. Large-Scale Multi-omic Analysis of COVID-19 Severity. Cell systems. 2021;12(1):23–40 e7. doi: 10.1016/j.cels.2020.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arunachalam PS, Wimmers F, Mok CKP, Perera R, Scott M, Hagan T, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369(6508):1210–20. doi: 10.1126/science.abc6261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang J, Lin D, Li K, Ding X, Li L, Liu Y, et al. Transcriptome Analysis of Peripheral Blood Mononuclear Cells Reveals Distinct Immune Response in Asymptomatic and Re-Detectable Positive COVID-19 Patients. Frontiers in immunology. 2021;12:716075. doi: 10.3389/fimmu.2021.716075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhai Y, Franco LM, Atmar RL, Quarles JM, Arden N, Bucasas KL, et al. Host Transcriptional Response to Influenza and Other Acute Respiratory Viral Infections—A Prospective Cohort Study. PLoS pathogens. 2015;11(6):e1004869. doi: 10.1371/journal.ppat.1004869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murphy SL, Kochanek KD, Xu J, Arias E. Mortality in the United States, 2020. NCHS data brief. 2021(427):1–8. [PubMed] [Google Scholar]
- 59.Brasen CL, Christensen H, Olsen DA, Kahns S, Andersen RF, Madsen JB, et al. Daily monitoring of viral load measured as SARS-CoV-2 antigen and RNA in blood, IL-6, CRP and complement C3d predicts outcome in patients hospitalized with COVID-19. Clinical chemistry and laboratory medicine. 2021;59(12):1988–97. doi: 10.1515/cclm-2021-0694 [DOI] [PubMed] [Google Scholar]
- 60.Tong-Minh K, van der Does Y, Engelen S, de Jong E, Ramakers C, Gommers D, et al. High procalcitonin levels associated with increased intensive care unit admission and mortality in patients with a COVID-19 infection in the emergency department. BMC infectious diseases. 2022;22(1):165. doi: 10.1186/s12879-022-07144-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masotti L, Grifoni E, Pelagalli G, Cioni E, Mattaliano C, Cioffi E, et al. Prognostic role of Interleukin-6/lymphocytes ratio in SARS-CoV2 related pneumonia. Int Immunopharmacol. 2022;103:108435. doi: 10.1016/j.intimp.2021.108435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lei H. A single transcript for the prognosis of disease severity in COVID-19 patients. Sci Rep. 2021;11(1):12174. doi: 10.1038/s41598-021-91754-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Velavan TP, Kuk S, Linh LTK, Lamsfus Calle C, Lalremruata A, Pallerla SR, et al. Longitudinal monitoring of laboratory markers characterizes hospitalized and ambulatory COVID-19 patients. Sci Rep. 2021;11(1):14471. doi: 10.1038/s41598-021-93950-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li T, Wang X, Zhuang X, Wang H, Li A, Huang L, et al. Baseline characteristics and changes of biomarkers in disease course predict prognosis of patients with COVID-19. Internal and emergency medicine. 2021;16(5):1165–72. doi: 10.1007/s11739-020-02560-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crook H, Raza S, Nowell J, Young M, Edison P. Long covid-mechanisms, risk factors, and management. Bmj. 2021;374:n1648. doi: 10.1136/bmj.n1648 [DOI] [PubMed] [Google Scholar]
- 66.Carbonell R, Urgeles S, Salgado M, Rodriguez A, Reyes LF, Fuentes YV, et al. Negative predictive value of procalcitonin to rule out bacterial respiratory co-infection in critical covid-19 patients. The Journal of infection. 2022. doi: 10.1016/j.jinf.2022.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Z, Peng X, Li HY, Wang Y, Qian Y, Wang Z, et al. Evaluation of Peripheral Immune Dysregulation in Alzheimer’s Disease and Vascular Dementia. J Alzheimers Dis. 2019;71(4):1175–86. doi: 10.3233/JAD-190666 [DOI] [PubMed] [Google Scholar]
- 68.Lei H, Wang C, Wang Y, Wang C. Single-cell RNA-Seq revealed profound immune alteration in the peripheral blood of patients with bacterial infection. International journal of infectious diseases: IJID: official publication of the International Society for Infectious Diseases. 2021;103:527–35. doi: 10.1016/j.ijid.2020.11.205 [DOI] [PubMed] [Google Scholar]