Abstract
SNAP-25 protein is a key protein of the SNARE complex that is involved in synaptic vesicles fusion with plasma membranes and neurotransmitter release, playing a fundamental role in neural plasticity. Recently the concentration of three specific miRNAs–miR-27b-3p, miR-181a-5p and miR-23a-3p –was found to be associated with a specific SNAP-25 polymorphism (rs363050). in silico analysis showed that all the three miRNAs target SNAP-25, but the effect of the interaction between these miRNAs and the 3’UTR of SNAP-25 mRNA is currently unknown. For this reason, we verified in vitro whether miR-27b-3p, miR-181a-5p and miR-23a-3p modulate SNAP-25 gene and protein expression. Initial experiments using miRNAs-co-transfected Vero cells and SNAP-25 3’UTR luciferase reporter plasmids showed that miR-181a-5p (p≤0.01) and miR-23a-3p (p<0.05), but not miR-27b-3p, modulate the luciferase signal, indicating that these two miRNAs bind the SNAP-25 3’UTR. Results obtained using human oligodendroglial cell line (MO3.13) transfected with miR-181a-5p or miR-27b-3p confirmed that miR-181a-5p and miR-23a-3p regulate SNAP-25 gene and protein expression. Interestingly, the two miRNAs modulate in an opposite way SNAP-25, as miR-181a-5p significantly increases (p<0.0005), whereas miR-23a-3p decreases (p<0.0005) its expression. These results for the first time describe the ability of miR-181a-5p and miR-23a-3p to modulate SNAP-25 expression, suggesting their possible use as biomarkers or as therapeutical targets for diseases in which SNAP-25 expression is altered.
Introduction
Soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) is a complex that mediates pore formation and vesicle fusion and plays a key role in neural plasticity [1, 2]. Three specific proteins assemble this complex: Syntaxin 1 (STX1), synaptosomal-associated protein of 25 KDa (SNAP-25) and vesicle associated membrane protein 2 (VAMP2); these three proteins, together with others e.g. synapsins, synaptophysin and proteins of SH3 And Multiple Ankryn Repeat Domain families (SHANK), are vitally important in the mediation of synaptic communication [3–5].
Dysfunction of the SNARE complex and de-regulation of its constitutive proteins are associated to several diseases [6–8]. In particular, SNAP-25 polymorphisms, gene expression, and protein concentration, have been associated with different neurological and non-neurological diseases, including Alzheimer’s disease (AD) [9–12], Parkinson’s disease (PD) [13], neurodevelopmental disorders such as Autism and Borderline Intellectual Functioning [14–16], Attention Deficit and Hyperactivity Disorder (ADHD) [17], type 2 diabetes [18, 19] and sarcopenia [20]. Notably, SNAP-25 protein expression was shown to be reduced in brains of AD [21, 22], whereas it was observed to be increased in cerebrospinal fluids of AD [23] and PD patients [24].
The SNAP-25 gene (88,591 nucleotides) is located on chromosome 20 (20p12.2), and codes for a 206 amino acids (aa) protein, formed by three different domains: a N-terminal α-helix domain, a palmitoylation cysteine-rich domain, and a C-terminal α-helix domain1. Molecular mechanisms for SNAP-25 expression are not fully understood; microRNAs, by binding the 3’ untranslated region (3’UTR) of mRNA target [25, 26], can contribute to post-transcriptional modulation of the expression of SNAP-25, resulting in disease associated misregulation of this protein. Several works have shed light on the role played by miRNAs on the regulating key synaptic components; results [27] convincingly showed that the expression of these proteins is modulated by the interactions, between, e.g. STX1 and miR-29a [28], SNAP-25, miR-153 and miR-27 [29, 30], TXN2 (thioredoxin) and miR-27 [30] and, finally, SHANK3 (SH3 And Multiple Ankyrin Repeat Domain 3) and miR-34a [31].
We have previously shown that 3 miRNAs (miR-27b-3p, miR-23a-3p and miR-181a-5p) targeting SNAP-25 3’UTR are differentially expressed in serum of AD patients in relation with SNAP-25 rs363050 polymorphism. Thus, the concentration of these proteins was significantly reduced in AD patients carrying the SNAP-25 rs363050 GG genotype compared with those carrying the AA or AG genotypes, and was also greatly diminished when the AD patients were compared to subjects with Mild Cognitive Impairment (MCI) or healthy controls [32]. The effects of these miRNAs on SNAP-25 gene and protein expression have nevertheless never been examined; we decided to focus our efforts on clarifying this topic.
Material and methods
Cell lines
The MO3.13 immature (non-differentiated) human oligodendrocyte cell line (a 2005 kind gift from Dr. Neil Cashman, University of Toronto, Canada) [33, 34] and the Vero African green monkey kidney cell line (ATCC CCL-81, purchased from ATCC) were used in this study. The MO3.13 cell line was derived from humans with no history of clinical signs of infectious diseases, and the cell line has shown no signs of contamination, including cytopathic effects.
All cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM, PAN-Biotech GmbH, Aidenbach, Germany) supplemented with 10% fetal bovine serum (FBS, PAN-Biotech), 50 U/ml penicillin and 50 μg/ml streptomycin (Euroclone, Rho, Milan, Italy), and maintained in an incubator at 37°C with 5% CO2. Trypan blue exclusion test was used to evaluate cell viability, with an automatic cell counter (TC20 Automatic Cell Counter, BioRad, Hercules, CA, US). To note, as previously described in a recent paper, the MO3.13 remained undifferentiated throughout the transfection experiments (at least 72 hours) [35].
Vero cells: Transfection and luciferase assay
Vero cells were seeded at 104 cells/well into a 96-well plate and co-transfected with a luciferase reporter plasmid containing the 3’UTR of SNAP-25 mRNA (Prod. ID: S808485, Active Motif, Carlsbad, CA, US) (genomic coordinates: chromosome 20; 10,226,822–10,228,152), together with mimic, inhibitor miRNAs (miR23a-3p, miR27b-3p and miR-181a-5p), or scramble: C. el miR-39-3p, Qiagen GmbH, Hilden, Germany) (2 μM), using the DharmaFECT Duo Transfection (SwitchGear Genomics, Carlsbad, CA, US), according to manufacturers’ instruction. The same experiments were also performed using an empty 3’UTR vector (negative control). The concentration of the plasmids was 20 μg/ml. After the co-transfection, cells were incubated in a humidified 5% CO2 incubator at 37°C for 24 hours.
A luciferase activity was assessed using the LightSwitch Luciferase Reagent (SwitchGear Genomics), as previously reported [35].
MO3.13 cells: Transfection
Plated in 6-well plates at a density of 6 x 105 cells/well, MO3.13 cells were transfected, separately, with scramble miRNA and the selected mimic or inhibitor miRNAs (Qiagen GmbH, Hilden, Germany) (50 nM). The transfection was performed by Lipofectamine RNAiMAX reagent (Life Technologies, Foster City, CA, US), according the manufacturer’s instruction. After transfection, the cells were incubated at 37°C for 6, 24, 48 and 72 h in a humidified 5% CO2 incubator. Each experiment was repeated at least 3 times and conducted in triplicate.
MO3.13 cells: RNA extraction and mRNA SNAP-25 quantitation by ddPCR and qPCR
Total RNA, extracted from the transfected MO3.13 cells using a column-based kit (QIAamp RNA Blood, Qiagen, Hilden, Germany), as previously reported [35], was retrotranscribed with High-capacity cDNA Reverse Transcription kit (Life Technologies), as specified by the manufactures.
One ng of cDNA was used for the quantification of SNAP-25 gene expression (specific assay by Life Technologies) by digital droplet PCR (ddPCR QX200, Bio-Rad, Hercules, CA, US). Absolute SNAP-25 gene expression was reported as copies/ng RNA. For the methodologies of ddPCR, see our previous article [20]. SNAP-25 gene expression was also evaluated by quantitative PCR (qPCR, CFX Touch real-time PCR, Bio-Rad, Hercules, CA, US). Specific TaqMan gene expression assays (Life Technologies, Foster City, CA, US) were utilized to detect the expression of SNAP-25 (ID: Hs00938957_m1) and of the YWHAZ reference gene (ID: Hs03044281_g1). Each cDNA template was tested in triplicate, with non-template control for each session.
MO3.13 cells: SNAP-25 protein quantitation
MO3.13 cells were mechanically disrupted by 3 repeated cycles of freezing and thawing. SNAP-25 protein was measured in MO3.13 cellular lysate (diluted 1:10) by ELISA, according to the manufacturer’s instruction (LSBio, LifeSpan BioSciences, Inc, Seattle, WA, US). The optical densities (OD) for each well were determined at 450 nm by plate reader (Sunrise, Tecan, Mannedorf, Switzerland), and optical densities (OD) were determined at 450 nm. SNAP-25 concentration was expressed as ng/mL (sensitivity: 0.78 ng/mL).
MO3.13 cells: SNAP-25 immunofluorescent staining by flow cytometry
To quantify the transmembrane SNAP-25 protein by flow cytometry, MO3.13 cells were treated with trypsin 10% for 5 minutes, collected and washed in phosphate-buffered saline (PBS) for 10 minutes at 1500 g. Cells were resuspended in 100 μl of PBS, and stained with mouse IgG1 monoclonal anti-human antibody SNAP-25 FITC (clone SP12, ThermoFisher Scientific, Rockford, IL, US). Cells were finally incubated for 30 minutes at room temperature and washed for 10 minutes at 1500 g. Analyses were performed using a Beckman-Coulter GALLIOS flow cytometer equipped with a 22 mW Blue Solid State Diode laser operating at 488 nm and with a 25 mW Red Solid State Diode laser operating at 638 nm and interfaced with Kaluza analysis software. Two-hundred-thousand events were acquired and gated on Forward and Side scatter properties. Samples were first run using mouse IgG1 isotype control (labeled FITC, code X0927, Dako, Agilent, Santa Clara, CA, US), and the mean fluorescence intensity (MFI) was calculated on MFI-positive cells alone.
Statistical analyses
Data were normally distributed and are presented as mean ± standard deviation (SD). Two-way ANOVA and, when appropriate, the Student’s t-test were used to analyze data. QuantaSoft software version 1.7.4.0917 (Bio-Rad) was used to quantify the results of ddPCR for the gene expression of SNAP-25. Threshold were determined manually for each experiment, according to the negative controls, which included a no template control. Droplet positivity was determined by fluorescence intensity; only droplets above a minimum amplitude threshold were counted as positive. For qPCR analysis, gene expression was presented as the normalization ratio calculated as follows: fold = 2-[ΔCq(sample) - ΔCq(scramble)] [36].
Results
Luciferase assay
To verify the binding of the three analyzed miRNAs–miR-23a-3p, miR-27b-3p and miR-181a-5p –to SNAP-25 mRNA 3’UTR, luciferase activity in cells lysate of Vero cells that had been co-transfected with these miRNAs was measured. Transfected Vero cells vitality was over 95%, without any toxic effect of the miRNAs and plasmid. The relative luciferase activity (RLA) was measured 24 hours after co-transfection.
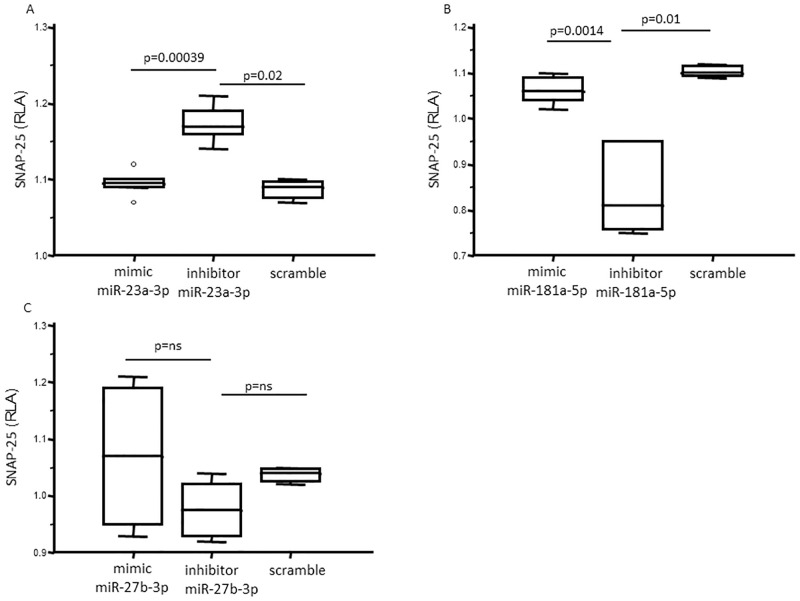
Regarding miR-23a-3p, RLA was differentially measured among Vero cells co-transfected with mimic, inhibitor and scramble (p = 0.005). In particular, the RLA of inhibitor miR23a-3p co-transfected Vero cells was significantly higher (RLA: 1.17±0.05) compared to that observed in cells co-transfected with either scramble miRNA (RLA: 1.08±0.02, p = 0.02) or mimic miR23a-3p (1.09±0.02, p = 0.004); no statistical differences were observed between Vero cells co-transfected with mimic and scramble (Fig 1(A)). Similar results were obtained with miR-181a-5p. Thus, RLA was differentially measured among Vero cells that had been co-transfected with mimic, inhibitor and scramble (p = 0.004). However, in this case the RLA of inhibitor miR-181a-5p co-transfected Vero cells was significantly lower (RLA: 0.84±0.09) than those observed in cells co-transfected with either scramble miRNA (RLA: 1.09±0.02, p = 0.01) or mimic miR-181a-5p (RLA: 1.06±0.03, p = 0.0014), whereas, again, no statistical differences were observed between Vero cells co-transfected with mimic and scramble (Fig 1(B)). Finally, no differences were observed when RLA of mimic, inhibitor and scramble miR-27b-3p-co-transfected Vero cells was analyzed (Fig 1(C)).
Fig 1. Relative luciferase activity (RLA) in Vero cells.
Different transfection experiments were performed independently with miRNA mimic, or miRNA inhibitor or scramble control. Each experiment was repeated at least 3 times and conducted in triplicate; the results presented in the figures indicate the median values obtained and 25th-75th percentile. RLA were measured in Vero cells 24 hours after transfection with (a) miR-23a-3p mimic, or inhibitor, or scramble negative control; (b) miR-181a-5p mimic, or inhibitor or scramble negative control; (c) miR-27b-3p mimic, or inhibitor or scramble negative control. RLA of cells transfected with miR-23a-3p inhibitor was significantly higher compared to that observed in cells transfected with miR-23a-3p mimic or scramble negative control; RLA of cells transfected with miR-181a-5p was significantly lower compared to that observed in cells transfected with miR-181a-5p mimic or scramble negative control. No differences were observed regarding miR-27b-3p mimic or inhibitor transfection. ns: not significative.
Taken together these results suggest that mi-23a-3p and miR-181a-5p, but not miR-27b-3p, bind the 3’UTR of SNAP-25 and, as a result, interfere with luciferase expression.
MO3.13 cells: SNAP-25 mRNA and protein quantitation
Because the luciferase experiment showed that only miR-23a-3p and miR-181a-5p modulate SNAP-25, we focused on these two miRNAs alone and examined their activity using human immature oligodendrocyte cells, MO3.13. Also in this case, cell vitality was over 95% in every transfection experiment.
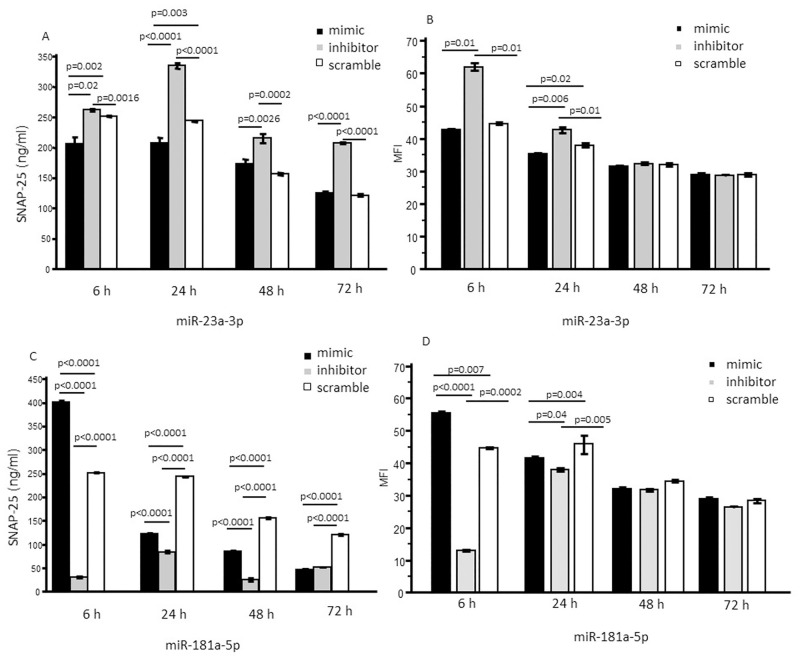
Results showed that, upon miR-23a-3p inhibitor transfection, SNAP-25 production by MO3.13 was significantly increased at each time-point (6, 24, 48 and 72 hours post transfection) (p<0.005 for each time point) compared to what was observed in scramble-transfected cells. An opposite effect was detected when SNAP-25 protein production was analyzed in miR-23a-3p mimic-transfected MO3.13 cells. In this case SNAP-25 production was significantly reduced compared to scramble-transfected MO3.13 cells, 6 and 24 hours post transfection. Notably, SNAP-25 generation returned to normal 72 hours post transfection (Fig 2(A)). Immunostaining analysis by flow cytometry showed comparable effects for mimic and inhibitory transfection experiments on SNAP-25 protein expression. In particular, the miR-23a-3p inhibitor significantly increased SNAP-25 production as compared to what was observed in cells that had been transfected by the scrambled miRNA 6 and 24 hours post transfection (p<0.05). On the converse, transfection with miR-23a-3p significantly down-regulated SNAP-25 production compared to scramble miRNA transfected cells 24 hours post transfection (p = 0.02) (Fig 2(B)).
Fig 2. SNAP-25 protein concentration in MO3.13.
Different transfection experiments were performed independently with miRNA mimic, or miRNA inhibitor or scramble control. Each experiment was repeated at least 3 times and conducted in triplicate; the results presented in the figures indicate the mean values obtained. SNAP-25 protein concentration (ng/ml) were measured in MO3.13 cells in different time points: 6, 24, 48 and 72 hours after transfection with (a) miR-23a-3p mimic, or inhibitor or scramble negative control by ELISA (a) and by flow cytometry (b); miR-181a-5p mimic, or inhibitor or scramble negative control by ELISA (c) and by flow cytometry (d). SNAP-25 concentration in cells transfected with miR-23a-3p inhibitor was significantly higher compared to that observed in cells transfected with miR-23a-3p mimic or scramble negative control in each time point; SNAP-25 concentration in cells transfected with miR-181a-5p inhibitor was significantly lower compared to that observed in cells transfected with miR-181a-5p mimic or scramble negative control in each time point.
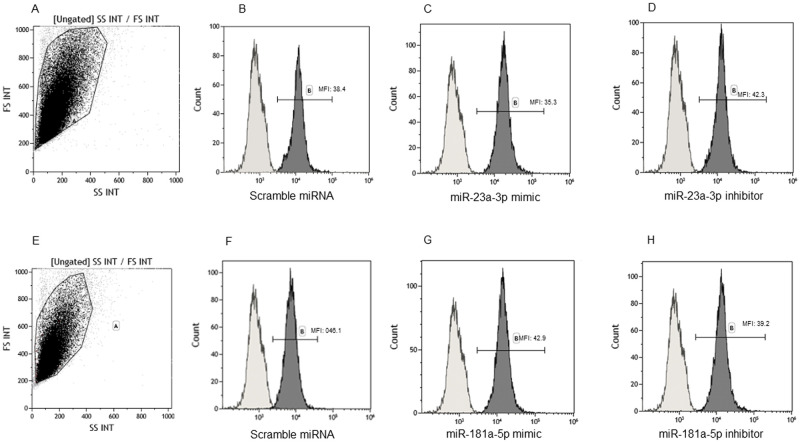
On the converse, SNAP-25 protein production was significantly lower at each time-point (6, 24, 48 and 72 hours post transfection) (p<0.0001 for each time point) in MO3.13 cells transfected with miR-181a-5p inhibitor compared to those transfected with scramble miRNA. Finally, the concentration of this protein was significantly augmented in MO3.13 cells transfected with the mimic compared to those transfected with scramble at 6 hours post transfection (p<0.05) (Fig 2(C)). Flow cytometry analysis showed comparable effects for mimic and inhibitory transfection experiments. In particular, SNAP-25 expression was significantly reduced at 6 and 24 hours post transfection in MO3.13 cells by the miR-23a-3p inhibitor, whereas MO3.13 cells transfected by the mimic miRNA were characterized by a significantly increased production of SNAP-25 compared to those transfected by scramble miRNA (p<0.05) (Fig 2(D)). Representative dot plot and histograms are shown in Fig 3.
Fig 3. SNAP-25 immunostaining in MO3.13.
Representative results obtained by staining MO3.13 with mAbs specific for SNAP-25. Gate on forward and side scatter plot (a and e) and (in dark grey diagrams) SNAP-25 signal intensity of MO3.13 cells transfected with scramble miRNA (b and f), miR-23a-3p mimic (c), or inhibitor (d), and miR-181a-5p mimic (g), or inhibitor (h). Light grey diagrams in b, c, d, f, g and h represent the isotype control. MFI: mean fluorescence intensity.
No statistical differences were observed when SNAP-25 gene expression was investigated (Table 1) by ddPCR; notably, at 6 hours post transfection miR-181a-5p mimic transfection SNAP-25 mRNA concentration was much higher compared to all the other conditions, analogously to what was observed when protein production was analyzed. Comparable results were obtained with qPCR analysis (S1 Table). Importantly, data obtained using ddPCR and qPCR significantly and positively correlated with each other (p = 0.02).
Table 1. SNAP-25 gene expression (copies/ng) in MO3.13 cells transfected with miR-23a-3p mimic, miR23a-3p inhibitor, miR-181a-5p mimic, miR-181a-5p inhibitor, and scramble negative miRNA.
The SNAP-25 expression was measured in different time points: 6 hours after transfection, 24 hours after transfection, 48 hours after transfection, and 72 hours after transfection. Data are expressed as mean ± standard deviation.
| Timing | Transfection | SNAP-25 mRNA (copies/ng) |
|---|---|---|
| 6 hours after transfection | scramble miRNA | 14.31±2.88 |
| miRNA-23a-3p mimic | 21.65±10.34 | |
| miRNA-23a-3p inhibitor | 19.36±7.35 | |
| miRNA-181a-5p mimic | 57.42±22.60 | |
| miRNA-181a-5p inhibitor | 21.45±16.32 | |
| 24 hours after transfection | scramble miRNA | 13.75±7.15 |
| miRNA-23a-3p mimic | 23.36±10.26 | |
| miRNA-23a-3p inhibitor | 22.55±5.42 | |
| miRNA-181a-5p mimic | 9.26±4.83 | |
| miRNA-181a-5p inhibitor | 7.24±4.21 | |
| 48 hours after transfection | scramble miRNA | 3.24±1.14 |
| miRNA-23a-3p mimic | 3.56±0.12 | |
| miRNA-23a-3p inhibitor | 0.65±0.02 | |
| miRNA-181a-5p mimic | 3.13±0.22 | |
| miRNA-181a-5p inhibitor | 4.22±1.36 | |
| 72 hours after transfection | scramble miRNA | 2.12±0.13 |
| miRNA-23a-3p mimic | 3.76±0.01 | |
| miRNA-23a-3p inhibitor | 0.72±0.32 | |
| miRNA-181a-5p mimic | 2.24±0.15 | |
| miRNA-181a-5p inhibitor | 2.25±0.91 |
These results confirm that miR-23a-3p and miR-181a-5p interact with 3’UTR SNAP-25, and demonstrate that the effect of such interaction is diametrically different, as miR-23a-3p down-regulates whereas miR-181a-5p up-regulates SNAP-25 protein and gene activity.
Discussion
The aim of the present study was to verify whether miR-23a-3p, miR-181a-5p and miR-27b-3p can modulate the expression of SNAP-25, as in silico they are able to bind its 3’UTR. It is very important to verify this interaction: it is known that SNAP-25 is involved in Alzheimer’s disease [5], and also these three miRNAs seem to be associated with the disease [32, 37–39]. Moreover, miR-23a-3p and miR181b-5p were shown to be associated as well with Autism Syndrome Disorder (ASD) [40–43], a disease in which even SNAP-25 is also suggested to play a role. In particular, in ASD children SNAP-25 polymorphisms were shown to be associated with hyperactivity and cognitive impairment [14, 15], leading to the hypothesis that the functioning of synapses related with cognitive activity are influenced by different expression of SNAP-25 protein [15]. The real nature of the interaction between these SNPs and SNAP-25, as well as the possible functional consequences of such interaction, though, have not yet been analyzed.
To address this question, we initially used a non-human cell line–Vero cells–that as per definition does not express the human SNAP-25. These cells were co-transfected independently with the three selected miRNAs and with a plasmid containing the 3’UTR of human SNAP-25 linked with luciferase, so that the luciferase activity was subordinated by the potential binding between the 3’UTR and miRNA. Results showed that miR-23a-3p and miR-181a-5p modulate the luciferase activity, and have an opposite effect on such activity, thus miR-181a-5p enhances and miR-23a-3p reduces the expression of luciferase. In contrast with the previous results, miR-27b-3p, had no effect on luciferase activity, indicating that that this miRNA, which in silico is theoretically able to interact with the SNAP-25 3’UTR, actually cannot bind this genetic region. To note, in a previous work, Machitani and coworkers [30], found that pre-miR-27b is able to suppress the SNAP-25 expression: as they performed transfection experiments with a pre-miRNA, and on the base of our results, it is plausible that their results are due to miR-27b-5p and not to miR-27b-3p. Next, on the basis of these results, we focused our attention of miR-23a-3p and miR-181a-5p, to verify whether these two miRNAs can modulate the SNAP-25 expression in human oligodendrocyte cells. To this end we used the MO3.13 cell line, a cell line that expresses SNAP-25. Transfection of MO3.13 cells with the appropriate mimic and inhibitor miRNAs were performed, and SNAP-25 mRNA and protein were quantified. Results confirmed that miR-181a-5p increases whereas miR-23a-3p decreases SNAP-25 protein expression. This effect was very clear and significant for protein concentration, whereas results for mRNA expression were not as compelling. However, this is not an unexpected result, as miRNA-mediated gene silencing is a complex mechanism in which several and redundant mechanisms are involved [44]. Moreover, the predominant mechanism of direct miRNAs action is translational inhibition [45], as several studies found that miRNAs can inhibit protein synthesis without affecting mRNA levels [46, 47], possibly because of the presence of alternate isoforms, post-translational modification or other factors involved in the silencing. The results regarding the miR-181a-5p capacity to increase the SNAP-25 expression is quite unexpected, as usually miRNAs act as suppressor of gene expression: future studies are required to address this issue.
miR-23a-3p is localized on chromosome 19 (19p13.12), and was identified for the first time in 2001 [48]. miR-23a-3p is a little-known miRNA, mainly found to be deregulated in cancers [49, 50]. However, it is important to underline that this miRNA was found to be expressed in synapses in animal models [51]. Additional results indicated that miR-23a-3p expression increases in isolated neuropiles after long-term potentiation, a synaptic plasticity form whereby a long-lasting enhancement of synaptic transmission is due to high-frequency stimulation [52]. Because SNAP-25 is a synaptic protein, these findings support our results, and it is plausible that the results obtained in rats could be the consequence of the binding between SNAP-25 and miR-23a-3p.
miR-181a-5p is localized on chromosome 1 (1q32.1), and was identified in human cells for the first time in 2003 [53]. Also this miRNA is barely known, although it seems to be involved in cell proliferation and differentiation in cancer [54], and as regulator of inflammation in macrophages and dendritic cells [55]. Importantly, recent studies showed that this miRNA is expressed in neurons, where it could have a role in synaptic development, function and plasticity [56, 57], possibly via SNAP-25 interaction.
Taken together, although they will need to be further confirmed by other analyses, results herein for the first time strongly suggest that miR-23a-3p and miR-181a-5p interact with the SNAP-25 gene acting as modulators of protein expression and having an opposite role on such expression. A limitation of our study is the lacking of immunoblotting and immunostaining experiments to confirm the exact relation between miRNAs and SNAP-25.
It remains to be clarified whether these miRNAs bind SNAP-25 mRNA directly or by interacting with other molecular factors, why they have an opposite effect on gene expression and if they have an effect also on other SNARE complex proteins. Because both SNAP-25 and miR-23a-3p and miR-181a-5p were found associated with Alzheimer’s disease, Mild Cognitive Impairment and ASD, the combined study of these biological parameters could yeld new important information regarding the mechanisms underlying these conditions. Finally, results also suggest that these miRNAs could be considered as therapeutical target in diseases in which SNAP-25 is involved, and that the analysis of their concentration in biological samples could offer a novel diagnostic and prognostic tool.
Supporting information
The SNAP-25 expression was measured in different time points by qPCR: 6 hours after transfection, 24 hours after transfection, 48 hours after transfection and 72 hours after transfection. Data are expressed as mean ± standard deviation.
(DOCX)
Acknowledgments
The authors kindly acknowledge the SA.M.B.A. project members, as well Dr. Claudia Vanetti, and all the Laboratory of Immunology of Department of Biomedical and Clinical Sciences, Sacco, University of Milan, for the support on cytoflow analysis. Fondazione Alessandro e Vincenzo Negroni Prati Morosini and Fondazione Romeo ed Enrica Invernizzi.
Data Availability
All relevant data are within the paper.
Funding Statement
This study was financial funded by Ministero della Salute (Award number: Ricerca Corrente 2020-2022) and by Fondazione Cariplo (Award number: 2017-0622). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Antonucci F, Corradini I, Fossati G, Tomasoni R, Menna E, Matteoli M. SNAP-25, a known presynaptic protein with emerging postsynaptic functions. Front Synaptic Neurosci. 2016; 8: 7. doi: 10.3389/fnsyn.2016.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holz RW, Zimmerberg J. Dynamic relationship of the SNARE complex with a membrane. Biophys J. 2019; 117: 627–630. doi: 10.1016/j.bpj.2019.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monteiro P, Feng G. SHANK proteins: roles at the synapse and in autism spectrum disorder. Nat Rev Neurosci 2017; 18: 147–157. doi: 10.1038/nrn.2016.183 [DOI] [PubMed] [Google Scholar]
- 4.Jahn R, Sudhof TC. Membrane fusion and exocytosis. Annu Rev Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863 [DOI] [PubMed] [Google Scholar]
- 5.Longhena F, Faustini G, Brembati V, Pizzi M, Benfenati F, Bellucci A. An updated reappraisal of synapsins: structure, function and role in neurological and psychiatric disorders. Neurosci Biobehav Rev 2021; 130: 33–60. doi: 10.1016/j.neubiorev.2021.08.011 [DOI] [PubMed] [Google Scholar]
- 6.Margiotta A. Role of SNAREs in neurodegenerative diseases. Cells 2021; 10: 991. doi: 10.3390/cells10050991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen F, Chen H, Chen Y, Wei W, Sun Y, Zhang L, et al. Dysfunction of the SNARE complex in neurological and psychiatric disorders. Pharmacol Res. 2021; 165: 105469. doi: 10.1016/j.phrs.2021.105469 [DOI] [PubMed] [Google Scholar]
- 8.Tang BL. SNAREs and developmental disorders. J Cell Physiol. 2021; 236: 2482–2504. doi: 10.1002/jcp.30067 [DOI] [PubMed] [Google Scholar]
- 9.Guerini FR, Farina E, Costa AS, Baglio F, Saibene FL, Margaritella N, et al. ApoE and SNAP-25 polymorphisms predict of multidimensional stimulation therapy rehabilitation in Alzheimer’s disease. Neurorehabil Neural Repair. 2016; 30: 883–893. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Therriault J, Kang MS, Ng KP, Pascoal TA, Rosa-Neto P, et al. Cerebrospinal fluid synaptosomal-associated protein 25 is a key player in synaptic degeneration in mild cognitive impairment and Alzheimer’s disease. Alzheimers Res Ther. 2018; 10: 80. doi: 10.1186/s13195-018-0407-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agliardi C, Guerini FR, Zanzottera M, Bianchi A, Nemni R, Clerici M. SNAP-25 in serum is carried by exosome of neuronal origin and is a potential biomarker of Alzheimer’s disease. Mol Neurobiol. 2019; 56: 5792–5798. [DOI] [PubMed] [Google Scholar]
- 12.Costa AS, Guerini FR, Arosio B, Galimberti D, Zanzottera M, Bianchi A, et al. SNARE complex polymorphisms associate with alterations of visual selective attention in Alzheimer’s disease. J Alzheimers Dis. 2019; 69: 179–188. doi: 10.3233/JAD-190147 [DOI] [PubMed] [Google Scholar]
- 13.Agliardi C, Guerini FR, Zanzottera M, Riboldazzi G, Zangaglia R, Sturchio A, et al. SNAP25 gene polymorphisms protect against Parkinson’s disease and modulate disease severity in patients. Mol Neurobiol. 2019; 56: 4455–4463. doi: 10.1007/s12035-018-1386-0 [DOI] [PubMed] [Google Scholar]
- 14.Guerini FR, Bolognesi E, Chiappedi M, Manca S, Ghezzo A, Agliardi C, et al. SNAP-25 single nucleotide polymorphisms are associated with hyperactivity in autism spectrum disorders. Pharmacol Res. 2011; 64: 283–288. doi: 10.1016/j.phrs.2011.03.015 [DOI] [PubMed] [Google Scholar]
- 15.Braida D, Guerini FR, Ponzoni L, Corradini I, De Astis S, Pattini L, et al. Association between SNAP-25 gene polymorphisms and cognition in autism: functional consequences and potential therapeutic strategies. Transl Psychiatry 2015; 5: e500. doi: 10.1038/tp.2014.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blasi V, Bolognesi E, Ricci C, Baglio G, Zanzottera M, Canevini MP, et al. SNAP-25 Single Nucleotide Polymorphisms, Brain Morphology and Intelligence in Children with Borderline Intellectual Functioning: A Mediation Analysis. Front Neurosci. 2021; 15: 715048. doi: 10.3389/fnins.2021.715048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu YS, Dai X, Wu W, Yuan FF, Gu X, Chen JG, et al. The association of SNAP25 gene polymorphisms in attention deficit/hyperactivity disorder: a systematic review and meta-analysis. Mol Neurbiol. 2017; 54: 2189–2200. doi: 10.1007/s12035-016-9810-9 [DOI] [PubMed] [Google Scholar]
- 18.Chen YL, Pei D, Hung YL, Lee CH, Hsiao FC, Wu CZ, et al. Associations between genetic variants and the severity of metabolic syndrome in subjects with type 2 diabetes. Genet Mol Res. 2015; 14: 2518–2526. doi: 10.4238/2015.March.30.10 [DOI] [PubMed] [Google Scholar]
- 19.Al-Dagri NM, Costa AS, Alokail MS, Zanzottera M, Alenad AM, Mohammed AK, et al. Synaptosomal protein of 25 kDa (Snap25) polymorphisms associated with glycemic parameters in type 2 diabetes patients. J Diabetes Res. 2016; 2016: 8943092. doi: 10.1155/2016/8943092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agostini S, Mancuso R, Costa AS, Guerini FR, Trecate F, Miglioli R, et al. Sarcopenia associates with SNAP-25 SNPs and a miRNAs profile which is modulated by structured rehabilitation treatment. J Transl Med. 2021; 19: 315. doi: 10.1186/s12967-021-02989-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greber S, Lubec G, Cairns N, Fountoulakis M. Decreased levels of synaptosomal associated protein 25 in the brain of patients with Down Syndrome and Alzheimer’s disease. Electrophoresis 1999; 20: 928–934. doi: [DOI] [PubMed] [Google Scholar]
- 22.Musunuri S, Khoonsari PE, Mikus M, Wetterhall M, Haggmark-Manberg A, Lannfelt L, et al. Increased levels of extracellular microvesicle markers and decreased levels of endocytic/exocytis proteins in the Alzheimer’ disease brain. J Alzheimers Dis. 2016; 54: 1671–1686. [DOI] [PubMed] [Google Scholar]
- 23.Brinkmalm A, Brinkmalm G, Honer WG, Frolich L, Hausner L, Minthon L, et al. SNAP-25 is a promising nolev cerebrospinal fluid biomarker for synapse degeneration in Alzheimer’s disease. Mol Neurodegener. 2014; 9: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bereczki E, Borgstedt A, Hoglund K, Tsitsi P, Brodin L, Ballard C, et al. Synaptic proteins in CSF relate to Parkinson’s disease stage markers. NPJ Parkinsons Dis. 2017; 3: 7. doi: 10.1038/s41531-017-0008-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–97. doi: 10.1016/s0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 26.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018; 141: 1202–7. doi: 10.1016/j.jaci.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukiw WJ. NF-kB-regulated micro RNAs (miRNAs) in primary human brain cells. Exp Neurol 2012; 235: 484–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bagge A, Dahmcke DM, Dalgaard LT. Syntaxin-1a is a direct target of miRNA-29a in insulin-producing β-cells. Horm Metab Res. 2013; 45: 463–6. [DOI] [PubMed] [Google Scholar]
- 29.Wei C, Thatcher EJ, Olena AF, Cha DJ, Perdigoto AL, Marshall AF, et al. miRNA-153 regulates SNAP-25, synaptic transmission, and neuronal development. PLoS One 2013; 8: e57080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Machitani M, Sakurai F, Wakabayashi K, Nakatani K, Tachibana M, Mizuguchi H. microRNA miR-27 inhibits adenovirus infection by suppressing the expression of SNAP25 and TXN2. J Virol 2017; 91: e00159–17. doi: 10.1128/JVI.00159-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y, Jaber VR, LeBeauf A, Sharfman NM, Lukiw WJ. microRNA-34a (miRNA-34a) mediated down-regulation of the post-synaptic cytoskeletal element SHANK3 in sporadic Alzheimer’s disease (AD). Front Neurol. 2019; 10: 28. doi: 10.3389/fneur.2019.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agostini S, Mancuso R, Liuzzo G, Bolognesi E, Costa AS, Bianchi A, et al. Serum miRNAs expression and SNAP-25 genotype in Alzheimer’s disease. Front Aging Neurosci. 2019; 11: 52. doi: 10.3389/fnagi.2019.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLaurin J, Trudel GC, Shaw IT, Antel JP, Cashman NR. A human glial hybrid cell line differentially expressing genes subserving oligodendrocyte and astrocytes phenotype. J Neurobiol. 1995; 26: 283–293. [DOI] [PubMed] [Google Scholar]
- 34.Buntix M, Vanderlocht J, Hellings N, Vandenabeele F, Lambrichts I, Raus J, et al. Characterization of three human oligodendroglial cell lines as a model to study oligodendrocyte injury: morphology and oligodendrocyte-specific gene expression. J Neurocytol. 2003; 32: 25–38. doi: 10.1023/a:1027324230923 [DOI] [PubMed] [Google Scholar]
- 35.Mancuso R, Agostini S, Marventano I, Hernis A, Saresella M, Clerici M. NCAM1 is the target of miRNA-572: validation in the human oligodendroglial cell line. Cell Mol Neurobiol. 2018; 38: 431–440. doi: 10.1007/s10571-017-0486-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fleige S, Walf V, Huch S, Prgomet C, Sehm J, Pfaffl MW. Comparison of relative mRNA quantification modles and the impact of RNA integrity in quantitative real-time PCR RT-PCR. Biotechnol Lett 2006; 28: 1601–1613. [DOI] [PubMed] [Google Scholar]
- 37.Gamez-Valero A, Campdelacreu J, Vilas D, Isperto L, Rene R, Alvarez R, et al. Exploratory study on microRNA profiles from plasma-derived extracellular vesicles in Alzheimer’s disease and dementia with Lewy bodies. Transl Neurodegener. 2019; 8: 31. doi: 10.1186/s40035-019-0169-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serpente M, Fenoglio C, D’Anca M, Arcaro M, Sorrentino F, Visconte C, et al. MiRNA profiling in plasma neural-derived samall extracellular vesicles from patients with Alzheimer’s disease. Cells 2020; 9: 1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin P, Xue Y, Wang T, Zhong D, Li G. The therapeutic targets of Fingolimod (FTY720) are involved in pathological processes in the frontal cortex of Alzheimer’s disease patients: a network pharmacology study. Front Aging Neurosci. 2021; 13: 609679. doi: 10.3389/fnagi.2021.609679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mundalil Vasu M, Anitha A, Thanseem I, Suzuki K, Yamada K, Takahashi T, et al. Serum microRNA profiles in children with autism. Mol Autism 2014; 5: 40. doi: 10.1186/2040-2392-5-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hicks SD, Ignacio C, Gentile K, Middleton FA. Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatr. 2016; 16: 52. doi: 10.1186/s12887-016-0586-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sehovic E, Spahic L, Smajlovic-Skenderagic L, Pistoljevic N, Dzanko E, Hajdarpasic A. Identification of developmental disorders including autism spectrum disorder using salivary miRNAs in children from Bosnia and Herzegovina. PLoS One 2020; 15: e0232351. doi: 10.1371/journal.pone.0232351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frye RE, Rose S, McCullough S, Bennuri SC, Porter-Gill PA, Dweep H. MicroRNA Expression Profiles in Autism Spectrum Disorder: Role for miR-181 in Immunomodulation. J Pers Med. 2021; 11: 922. doi: 10.3390/jpm11090922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015; 16: 421–433. doi: 10.1038/nrg3965 [DOI] [PubMed] [Google Scholar]
- 45.Cloonan N. Re-thinking miRNA-mRNA interactions: intertwining issues cofound target discovery. Bioessay 2015; 37: 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in c. elegans. Cell 1993; 75: 855–862. doi: 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- 47.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabbditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999; 216: 671–680. [DOI] [PubMed] [Google Scholar]
- 48.Lagos-Quintana M, Rahut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Sciences 2001; 294: 853–858. doi: 10.1126/science.1064921 [DOI] [PubMed] [Google Scholar]
- 49.Quan J, Pan X, Li Y, Hu Y, Tao L, Li Z, et al. miR-23a-3p acts as an oncogene and potential prognostic biomarker by targeting PNRC2 in RCC. Biomed Pharmacother. 2019; 110: 656–666. doi: 10.1016/j.biopha.2018.11.065 [DOI] [PubMed] [Google Scholar]
- 50.Lee Y, Kim SJ, Choo J, Heo G, Yoo JW, Jung Y, et al. miR-23a-3p is a key regulator of IL-17C-induced tumor angiogenesis in colorectal cancer. Cells 2020; 9: 1363. doi: 10.3390/cells9061363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pichardo-Casas I, Goff LA, Swerdel MR, Athie A, Davila J, Ramos-Brossier M. Expression profiling of synaptic microRNAs from the adult rat brain identifies regional differences and seizure-induced dynamic modulation. Brain Research 2012; 1436: 20–33. doi: 10.1016/j.brainres.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryan B, Logan BJ, Abraham WC, Williams JM. MicroRNAs, miR-23a-3p and miR-151-3p, are regulated in dentate gyrus neuropil following induction of long-term potentiation in vivo. PLoS One 2017; 12: e0170407. doi: 10.1371/journal.pone.0170407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dostie J, Mourelatos Z, Yang M, Sharma A, Dreyfuss G. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 2003; 9: 180–186. doi: 10.1261/rna.2141503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yishioka H, Ramakrishnan SS, Suzuki A, Iwata J. Phenytoin inhibits cell proliferation through microRNA-196a-5p in mouse lip mesenchymal cells. Int J Mol Sci. 2021; 22: 1746. doi: 10.3390/ijms22041746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie W, Li M, Xu N, Lv Q, Huang N, He J, et al. miR-181a regulates inflammation responses in monocytes and macrophages. PLoS One 2013; 8: e58639. doi: 10.1371/journal.pone.0058639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saba R, Storchel PH, Aksoy-Aksel A, Kepura F, Lippi G, Plant TD, et al. Dopamine-regulated microRNA miR-181a controls GluA2 surface expression in hippocampal neurons. Mol Cell Biol. 2012; 32: 619–632. doi: 10.1128/MCB.05896-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu Z, Li Z. miRNAs in synapse development and synaptic plasticity. Curr Opin Neurobiol 2017; 45: 24–31. doi: 10.1016/j.conb.2017.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The SNAP-25 expression was measured in different time points by qPCR: 6 hours after transfection, 24 hours after transfection, 48 hours after transfection and 72 hours after transfection. Data are expressed as mean ± standard deviation.
(DOCX)
Data Availability Statement
All relevant data are within the paper.