Abstract
Micropost-based microfluidic devices are widely used for microvascular network (MVN) formation in diverse research fields. However, consistently generating perfusable MVNs of physiological morphology and dimension has proven to be challenging. Here, how initial seeding parameters determine key characteristics of MVN formation is investigated and a robust two-step seeding strategy to generate perfusable physiological MVNs in microfluidic devices is established.
Keywords: cancer, microfluidics, microvasculatures, perfusable, physiological geometry
1. Introduction
Engineered microvascular networks (MVNs) have become ubiquitous tools in studies of tissue and organ function in vitro.[1,2] We and others have designed microfluidic platforms that support MVN formation within a central channel that is flanked by supporting media channels (Figure 1a).[3–5] A key design feature is the delineation of adjoining channels by a row of closely spaced microposts. Surface tension pinning at the microposts maintains infused fluids within their channels. Thus, a suspension of endothelial cells (ECs) and stromal cells within a liquid hydrogel precursor remain compartmentalized in the central channel. Within days, ECs undergo a process of spontaneous self-assembly resulting in interconnected MVNs composed of capillary-like segments. The value of MVN formed in vitro is their ability to recapitulate major functions of microvascular physiology in vivo and to elucidate the mechanisms underlying key physiological and pathological processes, such as selective permeability,[6] angiogenesis and vasculogenesis,[3,5,7] blood–brain barrier,[8,9] vascular and neuronal interactions,[10,11] transendothelial adiponectin flux,[12] vascular lung-on-chip,[13] pancreatic islet-on-chip,[14] functions of senescent stromal cells or subpopulations of stromal cells for vascularization,[15,16] nor-moxia and hypoxia,[17] tumoral vasculature regulation,[18] cancer extravasations,[19,20] glioblastoma heterogeneity,[21] procoagulant effects of ambient fine particulate matter,[22] reactions of microvascular tissues to compressive stress,[23] radiobiological effects,[24] hyperglycemia effects,[25] and others.
Figure 1.
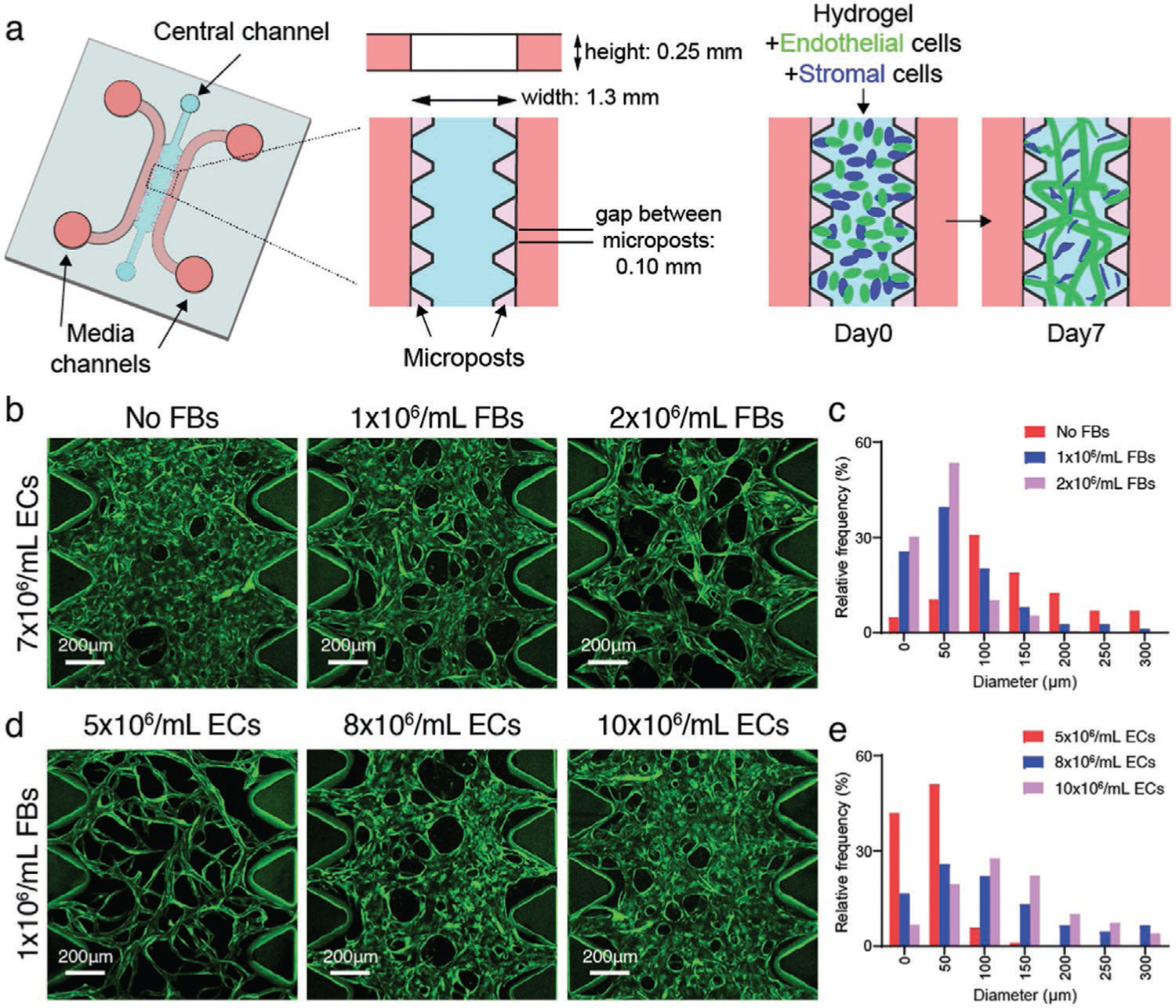
Seeding densities determine MVN diameters in microfludic devices. a) Schematic diagram of a microfluidic device with microposts for MVN formation. b) Representative images showing immortalized human umbilical vein EC (ImHUVEC, 7 × 106 mL−1) MVNs made with lung FBs at 0, 1, or 2 × 106 mL−1 concentration. ImHUVECs expressing blue fluorescent protein (BFP) were used. Green, ImHUVECs. c) Distribution of microvessel diameter of ImHUVEC MVNs made with or without FBs. Mean and SD of each group are: No FBs 164.3 ± 113.1 μm; 1 × 106 mL−1 FBs 69.45 ± 60.78 μm; 2 × 106 mL−1 FBs 47.97 ± 35.87 μm (n = 2 devices, 3 regions of interest (ROIs) each). Representative images (d) and diameter distribution (e) of MVNs made of ImHUVECs expressing BFP at 5, 8, or 10 × 106 mL−1 density with 1 × 106 mL−1 lung FBs. Mean and S.D. of each group are: 5 × 106 mL−1 ECs 36.28 ± 25.27 μm; 8 × 106 mL−1 ECs 118.9 ± 93.68 μm; 10 × 106 mL−1 ECs 128.9 ± 79.19 μm (n = 2 devices, 3 ROIs each). Green, ImHUVECs.
MVNs must be perfusable to replicate in vivo-like function. Intravascular flow is provided by the flanking media channels, but the vessels between the microposts must open at the media channel interface. However, the conditions that favor the formation of open vessels between microposts often result in MVN with diameters larger than typical microvasculature. Ensuring perfusability may explain that many published reports show perfusable MVNs with both capillary-like vessels and much larger vessels (Figure S1, Supporting Information).[3,5,8,9,13–30] To reliably form perfusable MVNs with the desired capillary-like geometry, we investigated the effect of seeding parameters on MVN geometry and, in doing so, established a robust two-step seeding strategy that allows us to provide favorable conditions for open vessels between microposts and capillary-like MVNs throughout the central region.
2. Results
2.1. Initial Seeding Parameters Control MVN Geometry In Vitro
Capillary diameters in vivo vary among the different organs, but tend to be less than ≈10 μm.[31] MVN geometry is sensitive to initial seeding parameters including stromal cell concentration, EC concentration, and cell distribution. We and others have found that co-seeding ECs with stromal cells, such as fibroblasts (FBs), produces MVNs with smaller diameters and better long-term stability[3] compared to MVNs formed without the support and interactions from stromal cells (Figure 1b,c). Lower EC seeding concentration shifts the vessel diameter distribution close to the physiological range (Figure 1d,e).
During the hydrogel curing process, cells sink to the bottom of the device due to gravity, which leads to flatter MVNs with systematically larger diameters near the bottom of the device (Figure 2a,b). Counteracting the pooling of ECs by flipping the devices several times during hydrogel polymerization improves the cell distribution and results in a more 3D distributed MVN with diameters closer to those of capillary beds in vivo (Figure 2c,d).
Figure 2.
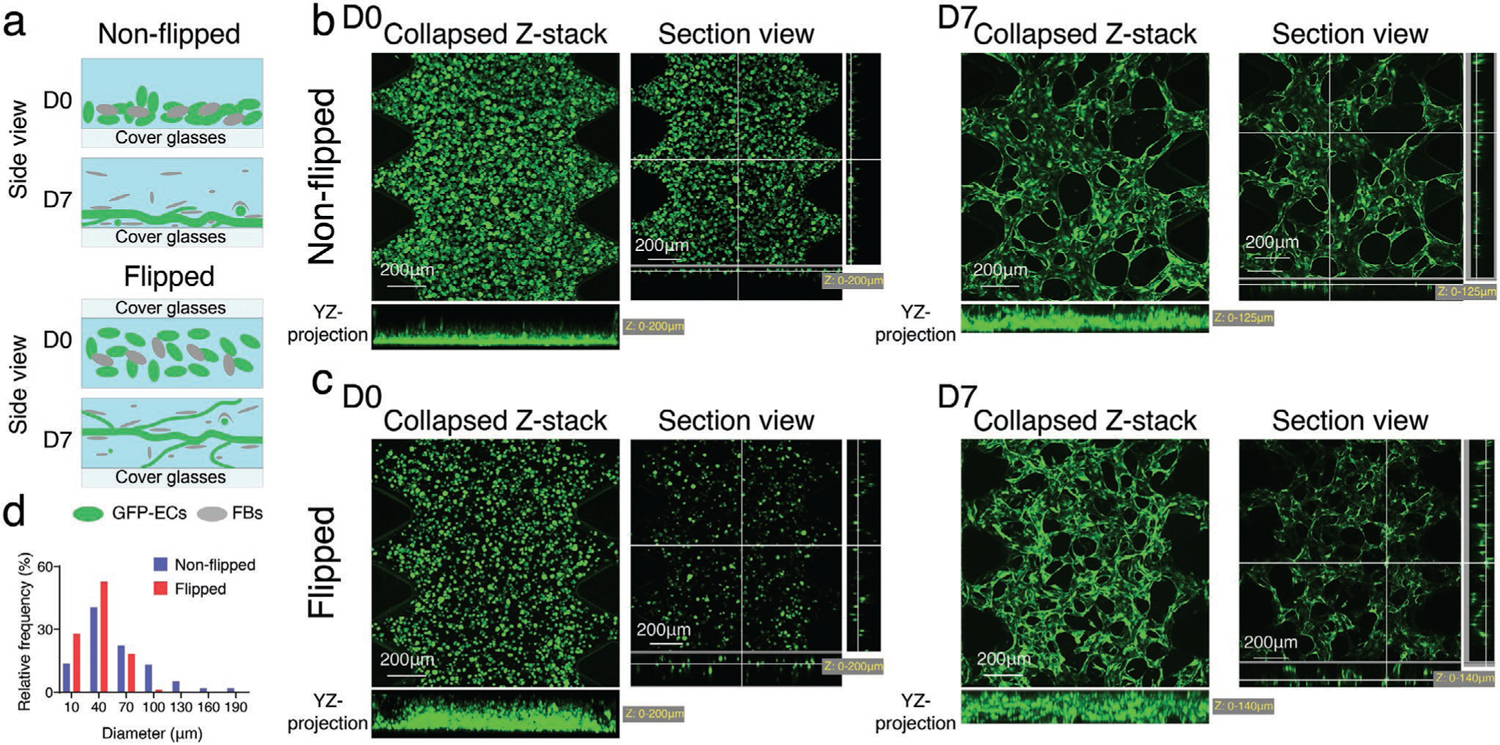
Initial cell spatial distribution determines MVN morphology. a) Schematic illustration of non-flipped and flipped seeding method. EC-FB mixture was seeded in the microfluidic device, the device either stayed right side up (no flipping), or was flipped —three to five times. The cell mixture is more evenly distributed in all three dimensions in flipped devices, leading to narrower and more 3D distributed MVNs at day 7, compared to non-flipped method. Representative confocal images of ImHUVECs expressing GFP and MVNs seeding in non-flipped b) or flipped c) method at day 0 (left) and day 7 (right). d) Diameter distribution of microvessels made in non-flipped or flipped method. Mean and SD of each group are: non-flipped 63.27 ± 45.29 μm; flipped 37.17 ± 19.00 μm (n = 3 devices, 2 ROIs each).
However, a drawback of manipulating initial seeding parameters to produce physiological MVNs with capillary-like vessels is impeded perfusability, generating non-perfusable or partially perfusable MVNs. We quantified vessel opening percentage and central perfusable MVN percentage of MVNs formed with ImHUVECs and lung FBs at different densities (Figure 3). We observed that the MVNs with larger diameters are more likely to form open interfaces with the neighboring media channel (Figure 3). On the other hand, the smaller diameter (more capillary-like) MVNs often lacked open microvessels, causing the vessels to be either non-perfusable or only partially perfusable (Figure 3). Thus, higher EC density with lower FB density at the regions between microposts promotes open microvessel and perfusable MVN formation.
Figure 3.
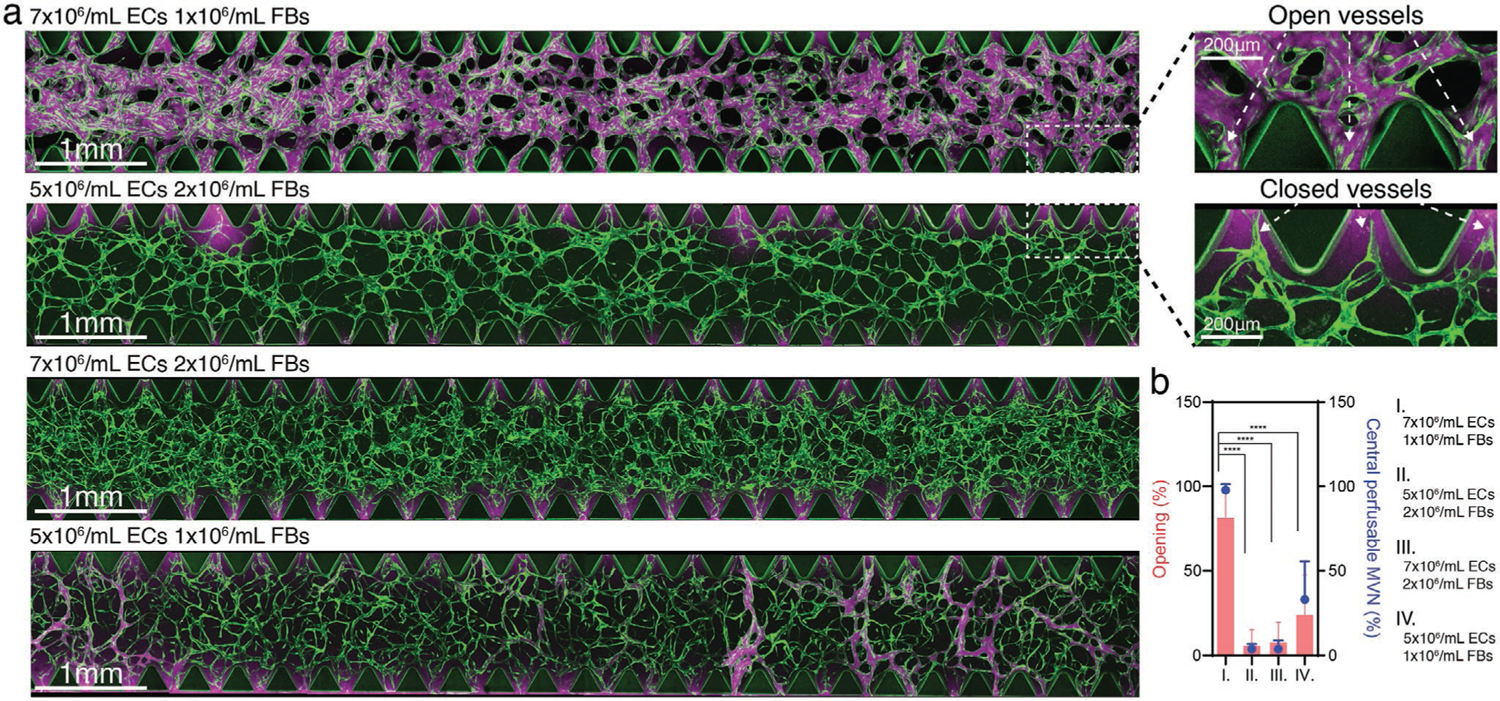
Seeding density determines MVN perfusability. a) Representative confocal images of MVNs formed with ImHUVECs expressing BFP and lung FBs at various concentrations (left). Zoomed in images of microvessels in the regions between microposts showing vessel openings and closed vessels (right). Lower EC and higher FB densities led to lower opening percentation and less or non-perfusable MVN formation. Green, ImHUVECs. Magenta, dextran (70 kDa). b) Statistical analysis of microvessel opening percentage (bars) and central perfusable MVN percentage (dots) in each ROI of MVNs made at various EC and FB concentrations. Bars represent mean ± SD. Two-tailed t tests were performed for the statistical comparisons (n = 3 devices, 5 ROIs each).
2.2. Two-Step Method to Generate Perfusable MVNs with Physiological Geometry
Our novel 2-step method decouples the region between the microposts from the bulk of the central channel to independently provide conditions favorable for generating open, large-diameter vessels between microposts and capillary-like MVNs in the central region (Figure 4a). First, we infused ECs alone at a high concentration into the central channel, then immediately aspirated the uncured solution. Due to surface tension, a small volume of ECs suspended in gel solution remained between microposts (termed “outer layer ECs”). Second, we infused EC and stromal cell mixture to the central region to form the main MVNs. Within 7 days, the ECs in the central region self-organized into MVNs with small diameters, while the higher concentration of outer layer ECs formed wider and open lumens connecting the MVNs to the flanking media channels (Figure 4a,c). The two-step seeding strategy produced MVNs with smaller diameter microvessels in the central regions compared to the one-step method producing perfusable but wider MVNs; and a higher perfusability (more open outer layer microvessels) than the one-step narrower but not perfusable MVNs (Figure 4b–e, also refer to Figures S2 and S3, Supporting Information, for detailed characterization).
Figure 4.
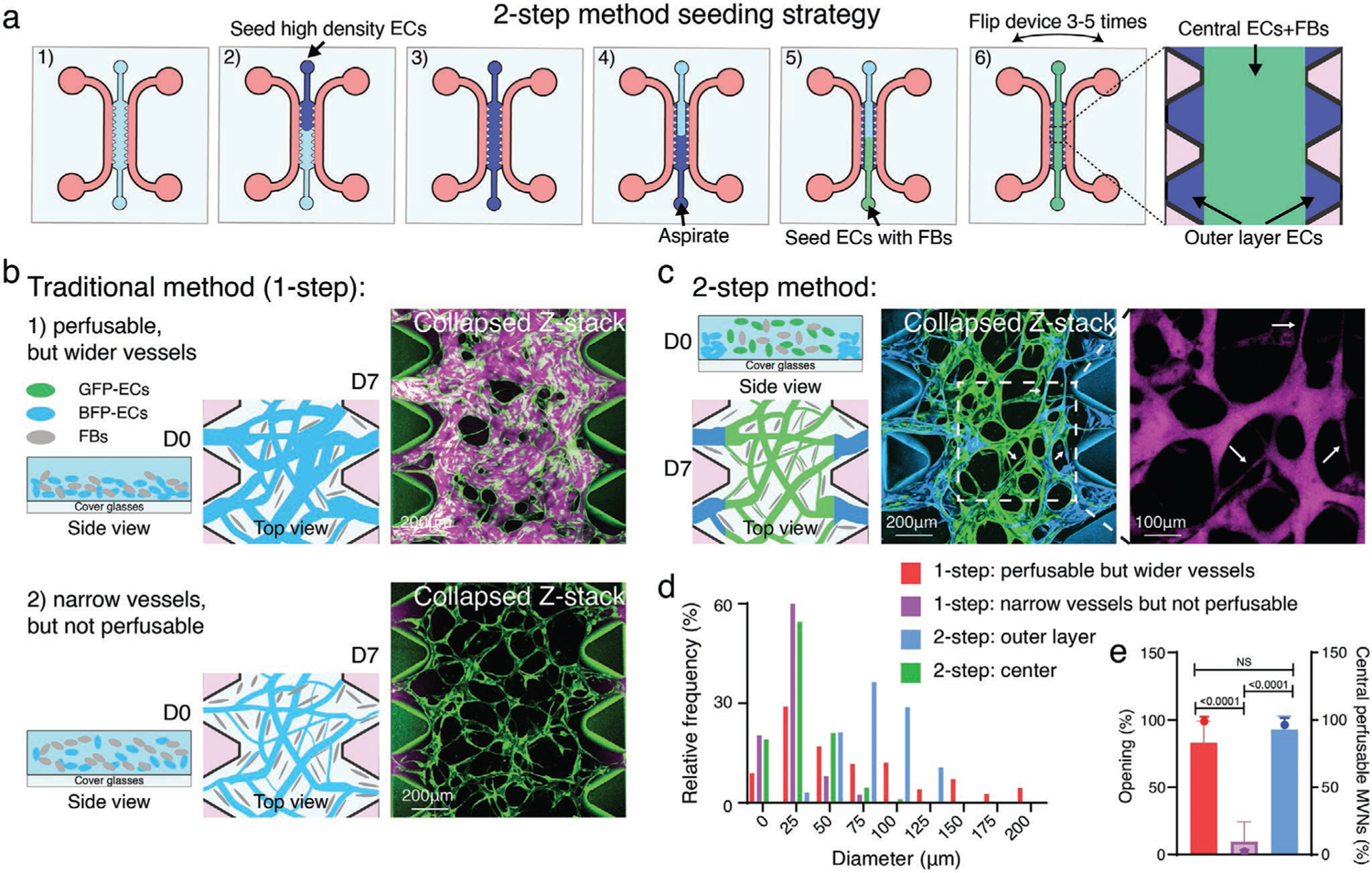
Two-step strategy for robustly generating perfusable MVNs of physiological geometry. a) Schematic diagram of two-step seeding method in the microfluidic device with microposts. All cells were suspended in hydrogel precursor (fibrinogen/thrombin). First, high density of ECs are seeded in the device. Second, the uncured gel is carefully aspirated, and the ECs remaining in the regions between microposts are referred to as outer layer ECs. Third, the ECs/stromal cells, such as FBs, are seeded into the central gel region. Microfluidic devices are flipped several times to improve vascular cell 3D distribution in the center. MVNs will form within 7 days. b) Schematic illustration and representative images of the traditional 1-step method of forming MVNs. 1) Non-flipped, higher EC (8 × 106 mL−1) with lower FB (1 × 106 mL−1) cell density tend to form perfusable but wider diameter MVNs. 2) Evenly distributed (flipped), lower EC (5 × 106 mL−1) with higher FB (1.5 × 106 mL−1) cell density form non-perfusable, narrower diameter MVNs. ImHUVECs expressing BFP and lung FBs were used. Green, ImHUVECs. Magenta, dextran (70 kDa). c) Schematic cartoon and representative images of the two-step method of generating perfusable MVNs with narrower diameters in the center. In this experiment, BFP-ImHUVECs (cyan, 10 × 106 mL−1) were used as outer layer ECs; GFP-ImHUVECs (green, 5 × 106 mL−1) with FBs (1.5 × 106 mL−1) were seeded in the center. Devices were flipped several times during hydrogel polymerization. The dotted square indicates an enlarged dextran image that demonstrates perfusibility of the narrow vessels. White arrows point to narrow microvessels. d) Diameter distribution of microvessels from MVNs made by one- or two-step seeding method. Mean and SD of each group are one-step: perfusable but wider vessels 74.17 ± 62.27 μm; one-step: narrow vessels but not perfusable 21.16 ± 12.95 μm; two-step: outer layer 80.60 ± 25.36 μm; two-step: center 27.46 ± 18.63 μm (n = 3 devices, 3 ROIs each). e) Statistical analysis of microvessel opening percentage (bars) and central perfusable MVN percentage (dots) in each ROI of MVNs made by one- or two-step seeding method. Bars represent mean ± SD. Two-tailed t-tests were performed for the statistical comparisons (n = 3 devices, 5 ROIs each).
2.3. Applications of MVNs Made with Two-Step Method
The two-step seeding method produced MVNs that are suitable for immunofluorescent staining (Figure 5a) and cell perfusion experiments. As a demonstration, we perfused tumor cells or tumor clusters (Figure 5b–d) into MVNs formed from the traditional one-step seeding method and our two-step seeding strategy. More tumor cells were arrested by physical trapping in MVNs formed using the two-step than the one-step method (Figure 5b,c). A similar approach allows immune cells, such as monocytes, to be perfused in the MVNs made by the two-step method (Figure 5e).
Figure 5.
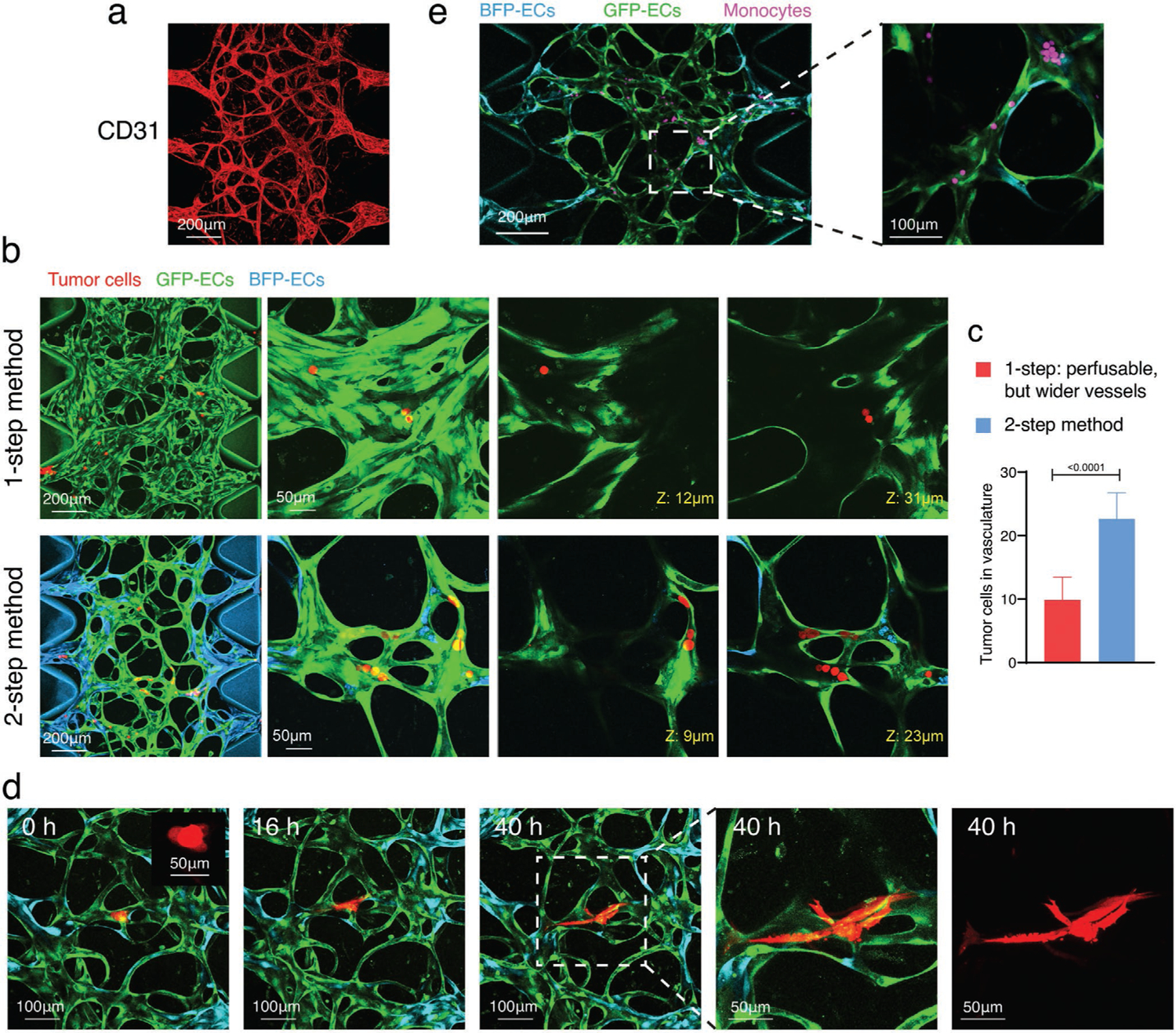
Applications of two-step seeding method produced MVNs. a) Two-step MVNs formed with ImHUVECs and lung FBs were stained with CD31 antibody to demonstrate the immunostaining applications of this method. b) Representative confocal images of perfused tumor cells (red) in MVNs made by one- or two-step seeding method with ImHUVECs and lung FBs. Zoomed in images on the right show H69M tumor cells that are arrested in these two MVNs. Red, tumor cells. Cyan, ImHUVEC expressing BFP. Green, ImHUVECs expressing GFP. c) Quantification of tumor cell number arrested in the MVNs made by one- or two-step seeding method. Bars represent mean ± SD. Two-tailed t-tests were performed for the statistical comparisons (n = 3 devices, 6 ROIs each). d) Tumor clusters (2–4 cells, mimicking circulating tumor clusters) made from the H69M cell line were perfused in two-step MVNs and imaged for 40 h. Red, tumor cells. Cyan, ImHUVEC expressing BFP. Green, ImHUVECs expressing GFP. Inserted image shows tumor cell cluster in a higher magnification. e) Monocytes were perfused in two-step MVNs formed with ImHUVECs and lung FBs as an example of immune cell perfusion studies.
The two-step seeding strategy also resulted in the formation of physiologically relevant MVNs using ImHUVECs with FBs derived from patients with thyroid cancer, brain microvascular ECs with astrocytes and brain pericytes, dermal microvascular ECs with dermal FBs, and iPSC-derived ECs with lung FBs (Figure 6). These MVNs are also suitable for immunofluorescent staining and permeability tests (Figure 6; Figure S4a,b, Supporting Information). For example, immunostaining shows that astrocytes are in the stroma and bridging nearby ECs (Figure 6b). More studies demonstrating interactions of brain EC, brain pericytes, and astrocytes in our blood–brain barrier model system can be found in our recent publication.[32]
Figure 6.

Different types of MVNs made with two-step method. Representative images of two-step seeding method produced MVNs formed by a) ImHUVECs expressing BFP with patient-derived thyroid FBs. Opening percentage (%) is 67.46 ± 15.95%; central perfusable MVN percentage is 77.67 ± 16.81% (n = 3 devices, 5 ROIs each). Green, ECs. Magenta, dextran. b) Two-step brain MVNs formed with brain ECs expressing GFP with brain pericytes and astrocytes. Opening percentage is 80.80 ± 14.02%; central perfusable MVN percentage is 87.20 ± 12.22% (n = 3 devices, 5 ROIs each). Green, ECs. Magenta, dextran. Yellow, GFAP immunofluorescent staining for activated astrocytes. c) Two-step dermal MVNs formed with human dermal blood microvascular ECs (HDBMECs) expressing BFP and dermal FBs. Opening percentage is 93.20 ± 8.621%; central perfusable MVN percentage is 98.93 ± 4.652% (n = 3 devices, 5 ROIs each). Green, ECs. Magenta, dextran. d) Permeability analysis of two-step dermal MVNs treated with or without TNF-α. Bars represent mean ± SD. Two-tailed t-tests were performed for the statistical comparisons (n = 4 devices, 4 ROIs each). e) Two-step dermal MVNs stained with CD31 antibody (red). f) Two-step method MVNs formed with iPSC-derived ECs and lung FBs. Opening percentage is 77.37 ± 15.03%; central perfusable MVN percentage is 76.47 ± 13.43% (n = 3 devices, 5 ROIs each). Green, iPSC-ECs stained with UEA-I. Magenta, dextran. g) Statistical analysis of permeability of two-step iPSC-EC MVNs treated with or without TNF-α. Bars represent mean ± SD. Two-tailed t-tests were performed for the statistical comparisons (n = 4 devices, 4 ROIs each). h) CD31 immunostaining (red) of two-step MVNs made with iPSC-ECs and lung FBs. In all MVN images, white arrows point to the perfusable narrow microvessels. Dashed rectangles identify regions shown in inserts at higher magnification. 10 kDa dextran was used for brain MVNs perfusion. The 70 kDa dextran was used for all other perfusion and permeability experiments.
2.4. Hybrid MVN Formation using Two-Step Method
Beyond improving MVN formation, the two-step seeding method opens the door for the formation of hybrid MVNs that allow parsimonious use of vascular cells that may be difficult to obtain in large quantities, such as brain ECs, brain pericytes, astrocytes, and patient-derived ECs. The ECs of interest can be used exclusively for MVN formation. We formed MVNs consisting of an outer layer of readily available ECs that connect to central MVNs comprised of the ECs of interest. For example, we seeded readily available immortalized HUVECs as the outer layer ECs with much scarcer brain ECs along with brain pericytes and astrocytes in the central region (Figure 7a). These two different EC types readily anastomosed into a perfusable MVN (Figure 7b). Functionally, the permeability of the hybrid brain MVNs in the central region was similar to pure brain MVN controls (Figure 7c, Figure S4c, Supporting Information). Similarly, we formed hybrid MVNs with immortalized HUVEC and iPSC-ECs (Figure 7d).
Figure 7.
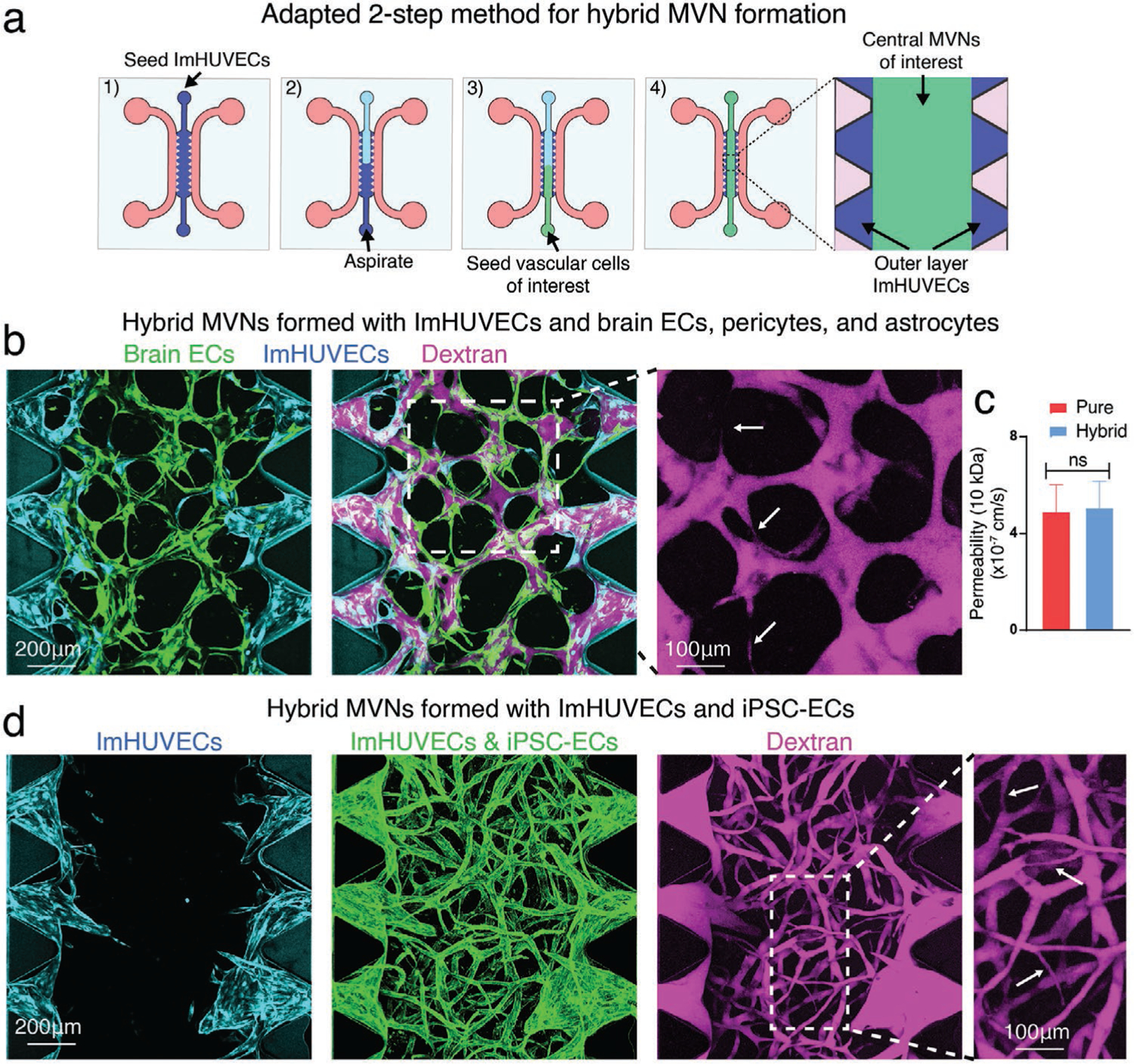
Adapted two-step seeding method for hybrid MVN formation. a) Schematics of adapted two-step seeding method in the microfluidic device. To reduce the usage of vascular cells of interest for MVN formation, ImHUVECs were used for outer layer EC seeding. After the uncured ImHUVECs/gel precursor liquids were carefully aspirated, the vascular cells of interest, such as brain primary ECs along with astrocytes, and brain pericytes, were seeded to the central gel region. b) Representative images of hybrid MVNs formed by ImHUVECs (outer layer) and brain specific MVNs (central region). Opening percentage is 90.93 ± 8.78%; central perfusable MVN percentage is 96.22 ± 5.91% (n = 3 devices, 5 ROIs each). Cyan, ImHUVECs expressing BFP. Green, brain ECs expressing GFP. Magenta, dextran (10 kDa). White arrows point to the perfusable narrow microvessels. c) Permeability measurements made in the central region of brain MVNs formed with two-step pure or hybrid MVN strategy using 10 kDa dextran. Bars represent mean ± SD. Two-tailed t-tests were performed for the statistical comparisons (n = 3 devices, 4 ROIs each). d) Representative images of hybrid MVNs formed by ImHUVECs expressing BFP (outer layer, cyan) and iPSC-ECs along with lung FBs (central region). Opening percentage is 97.73 ± 5.982%; central perfusable MVN percentage is 93.89 ± 6.166% (n = 3 devices, 5 ROIs each). ECs were stained with UEA-I (green). Magenta, dextran (70 kDa). White arrows point to the perfusable narrow microvessels.
Finally, the two-step seeding strategy can also be adapted for hybrid MVNs formed in two different extracellular matrices (ECMs). We formed MVNs within ECMs with different densities and ECMs derived from different species between the microposts and throughout the central channel (Figure 8).
Figure 8.

Adapted two-step seeding method for MVN formation in hybrid matrices. a) Schematics of adapted two-step seeding method for two different types of ECMs (ECM-A and ECM-B). To reduce the usage of the scarce ECMs (such as patient-derived or tissue specific ECM), common ECMs (such as bovine fibrin), can be used for outer layer EC seeding. After the uncured ECs/ECM-A liquids were carefully aspirated, ECs with FBs in scarce ECM-B were seeded to the central gel region to form MVNs in hybrid ECMs. b) Representative images of wide perfusable MVNs formed in fibrin at 3 mg mL−1 (ECM-A) or 5 mg mL−1 (ECM-B) using one-step seeding method (ImHUVECs expressing BFP, 8 × 106 mL−1; lung FBs, 1 × 106 mL−1). Green, ImHUVECs. Magenta, dextran (70 kDa). c) Confocal images of perfusable MVNs formed in hybrid fibrin (outer layer 3 mg mL−1, central 5 mg mL−1) using two-step seeding strategy (outer layer ImHUVECs expressing BFP, 10 × 106 mL−1; central ImHUVECs expressing BFP, 5 × 106 mL−1; and lung FBs 1.5 × 106 mL−1). Opening percentage is 94.33 ± 8.295%; central perfusable MVN percentage is 97.89 ± 3.63% (n = 3 devices, 5 ROIs each). Green, ImHUVECs. Magenta, dextran (70 kDa). White arrows point to the perfusable narrow microvessels. d) Experimental design of seeding MVNs in bovine or human fibrin. To discriminate between the two types of ECMs, Alexa Fluor 488 labeled human fibrin was added to non-labeled human fibrin. Left, one-step seeding method in bovine fibrin gel; middle, one-step seeding in human fibrin gel (containing 10% Alexa Fluor 488 labeled human fibrin); right, adapted two-step seeding for hybrid fibrin (outer layer, bovine fibrin; central, human fibrin mixed with 10% of Alexa Fluor 488 labeled human fibrin). e) Representative images of wide perfusable MVNs formed in fibrin derived from bovine (ECM-A) or from human (ECM-B) using one-step seeding method (ImHUVECs expressing BFP, 8 × 106 mL−1; lung FBs, 1 × 106 mL−1). f) Representative images of perfusable MVNs formed in hybrid ECMs (outer layer bovine fibrin, central human fibrin) using two-step seeding method (outer layer ImHUVECs 10 × 106 mL−1, central ImHUVECs 5 × 106 mL−1, and lung FBs 1.5 × 106 mL−1). Opening percentage is 92.07 ± 10.88%; central perfusable MVN percentage is 98.25 ± 3.99% (n = 3 devices, 5 ROIs each). Cyan, ImHUVECs expressing BFP. Green, human fibrin. Magenta, dextran (70 kDa). White arrows point to the perfusable narrow microvessels.
3. Discussion
Blood cells, including immune cells, travel through the capillaries that have diameters as small as 5–10 μm. Circulating tumor cells and clusters are arrested in capillaries by mechanical trapping and endothelial adhesion during metastasis.[33] Thus, perfusable MVNs with capillary-like geometry and dimensions are critical to interrogate both normal and pathological processes in vitro. Microfluidic-based approaches to capillary network formation have become ubiquitous tools to study physiology at the level of individual capillaries.[1] But despite the desire for capillary morphology and size in many applications, a survey of published reports reveals that many studies fail to recapitulate the small diameters found in typical capillaries. The bias toward larger vessel diameters is often by necessity. Many studies represent a vessel by seeding an endothelial monolayer on the walls of one or more of the channels in the microfluidic device.[34–37] In others, in which the microcirculation self assembles from cells (often ECs and FBs), small-diameter vessels are possible, but even in this case, it is rare to find capillary-sized structures.[3,5,8,9,13–30] A common reason for this is the need to generate perfusable vessels, and MVNs with larger diameters are more likely to be perfusable. In contrast, narrow, capillary-like vessels are less likely to be perfusable because they fail to open at the interface to the media channel. This suggests that the immediate vicinity of the interface between the bulk vascular network and abutting media channel is a critical determinant of microvascular network function. To address this limitation, we have developed a simple 2-step procedure that increases the likelihood of MVNs opening to the media channels while maintaining capillary-like morphology.
Microvascular network seeding parameters are cell- and device-specific. Thus, it is essential to titrate seeding parameters for different types of vascular cells. However, we believe the trends observed in our devices with our cells are generally consistent with results obtained in other labs, and with other devices and cells. We generally find that the ratio of EC-to-FB concentrations is one of the primary determinants of vascular dimension, with lower values favoring narrower, more physiological microvessels. This ratio, however, is dynamic, due to different cell types having distinct proliferation and/or migration tendencies that result in changes in local cell density and MVN geometry both initially and during the course of an experiment. With the seeding parameters we used in this paper (Table S1, Supporting Information), at ≈7 days the MVNs are able to form and become relatively stable, making it a good time point for various types of study, such as morphology, perfusability, and permeability. But, we want to stress that, if using different primary cells at different seeding conditions, the optimal time window for functional test might vary.
We chose to address these problems and produce vascular beds with smaller vessel diameters, by manipulating the spatial distribution of the EC-to-FB ratio in such a way as to generate large, highly perfusable vessels adjacent to the media channels and smaller, more physiological ones in the central gel region. To do this, we devised a two-step seeding method in which FBs are seeded in the central gel region only. However, even using the two-step method, we observed that FBs tend to appear in the regions between microposts by approximately day 7, indicating that FBs migrate from the central region to the outer layer region.
We found that two-step MVNs formed with various types of vascular cells show different geometry (Figure 6). For example, the diameters of MVNs made from iPSC-derived ECs are more homogenous than MVNs made with other types of ECs. Such differences emphasize the importance of establishing tissue specific MVNs for particular studies. Future comparisons among these vascular cells will promote our understanding of tissue specific vascularization.
A drawback of using the two-step method to form MVNs is that cells forming the outer EC layer are wasted, especially if the cell sources are scarce, such as human brain ECs or patient-derived primary cells. An alternative approach is to use immortalized HUVEC or other readily available ECs in the outer layer ECs. We demonstrated that the ImHUVECs could anastomose with brain ECs or iPSC-EC MVNs with similar functionality to pure MVNs.
A major challenge is to generate organoids with internal vasculature that functionally connects with an external vasculature.[2] Based on the present findings, one approach is to induce anastomoses between vasculatures developed independently inside and outside the organoid. Our two-step seeding protocol leads to perfusable networks composed of differing EC types within microvascular devices. Future studies could leverage the ability to visualize vascular networks with high spatial and temporal resolution to better understand the mechanism underlying the vascular interconnections formed in microfluidic devices.
The two-step seeding method can also be adapted to form MVNs in hybrid ECMs. It has been shown that the ECM density and stiffness within tumors is higher than in normal tissues.[38] However, replicating this condition within a microfluidic device is problematic, because ECMs with higher density produce narrower microvessels.[3] Thus, it is unlikely that MVNs formed by traditional seeding protocols with dense ECM would be perfusable and our two-step method could potentially solve this problem by using different ECM densities in the outer layer and central regions.
4. Conclusion
In summary, we developed a novel two-step method for seeding vascular cells that facilitates the formation of functional and perfusable MVNs with physiologically relevant network geometries. This method is amenable to many types of ECs, stromal cells, and ECMs, including those derived from patients. The two-step seeding strategy produces more physiologically relevant MVNs to study microvascular diseases, immune cell trafficking, tumor cell extravasation, drug transport and kinetics, and vascularized tumor spheroids and organoids.
5. Experimental Section
Cells, Antibodies, and Reagents:
ImHUVECs expressing BFP or GFP (P20-P30),[16] HDBMECs (Lifeline Cell Technology, P5–P8), and iPSC-ECs (Alstem, P4–P5) were cultured in VascuLife VEGF Endothelial Medium (Lifeline Cell Technology). Lung FBs (Lonza, P7), immortalized lung FBs expressing mCherry (P10),[16] dermal FBs (Lifeline Cell Technology, P5), and patient-derived thyroid FBs (P4) were cultured in FibroLife S2 Fibroblast Medium (Lifeline Cell Technology). Human brain microvascular endothelial cells (Angioproteomie, P4–P5) were cultured on fibronectin (30 μg mL−1) coated flasks in Vasculife VEGF endothelial medium with 5% FBS. Human astrocytes (ScienCell, P5) and brain pericytes (ScienCell, P5) were cultured on poly-l-lysine coated flasks (0.15 μg μL−1) in suggested commercial media from ScienCell. The medium was replenished every 2 days. Cells were detached and seeded into microfluidic devices after reaching 80% confluency. H69M cells expressing mCherry were cultured in RPMI1640 (Thermo Fisher) with 10% FBS and 1% penicillin-streptomycin (Sigma). CD31 and GFAP monoclonal antibody (Abcam and Invitrogen, respectively). Fibrinogen from bovine plasma, fibrinogen from human plasma, and thrombin from bovine plasma were used to form gels (Sigma), and Alexa Fluor 488-conjugate human fibrinogen (Thermo Fisher) was mixed with human fibrinogen (1:10) to discriminate from bovine fibrin. Ulex Europaeus Agglutinin I (UEA I), Fluorescein (1:100) was used for human EC labeling (Vector laboratories).
Isolation of Primary FBs:
FBs were isolated from patient thyroid cancer surgical resections in accordance with a protocol approved by the Institutional Review Board of Dana Farber Cancer Institute (09-472). Briefly, specimens were minced with sterile forceps and scalpels until pieces were smaller than 1 mm. Next, the tissue was digested in a warm solution of collagenase P, DNAse, and dispase in RPMI medium supplemented with 2% FBS. Every 15 min, cells released from the minced pieces were collected with a pipette and transferred to a tube of cold FACS buffer containing 2% FBS and 2 × 10−3 m EDTA in PBS. After 75 min, all cells were pelleted via centrifugation and passed through a 30 μm cell filter. The single cell fraction was plated at low density on tissue culture dishes pre-coated with 1% gelatin. FibroLife S2 medium was added and refreshed every 2–3 days until FBs reached ≈90% confluency in ≈7–12 days.
Microvascular Network Formation:
3D cell culture chips (AIM Biotech) were utilized to generate in vitro MVNs. AIM chip body was made of cyclic olefin polymer (COP) with a type of gas-permeable plastic serving as the bottom film. AIM Biotech chips contain three parallel channels: a central gel channel flanked by two media channels. Microposts separate fluidic channels and serve to confine the liquid gelling solution in the central channel by surface tension before polymerization. The gel channel was 1.3 mm wide and 0.25 mm tall, the gap between microposts was 0.1 mm, and the width of media channels was 0.5 mm. For the one-step seeding method, ECs and FBs were co-seeded into the chip as previously described.[16] Briefly, ECs and FBs were concentrated in VascuLife containing thrombin (4 U mL−1). Cell mixture solution was then further mixed with fibrinogen (3 mg mL−1 final concentration, unless otherwise specified) at a 1:1 ratio and quickly pipetted into the chip through the gel inlet with a final concentration of 7 × 106 mL−1 for ECs and 1 × 106 mL−1 for stromal cells, unless otherwise specified. For the two-step seeding strategy, the outer layer EC solution was made with a final concentration of 10 × 106 mL−1 for ECs only, then pipetted into the gel inlet, immediately followed by aspirating from the gel outlet, leaving only residual solution around the microposts. Another solution with final concentrations of 5 × 106 mL−1 ECs and 1.5 × 106 mL−1 stromal cells (unless otherwise specified) was then pipetted into the same chip through the gel outlet. The EC/stromal cell mixture was seeded through a gel outlet port to avoid the outer layer ECs being pushed out to media channel, if there was any liquid left in the central gel channel after aspiration. For flipped devices, the chip was inverted roughly every 30 s —three to five times right after seeding. The device was placed upside down to polymerize in a humidified enclosure and allowed to polymerize at 37 °C for 15 min in a 5% CO2 incubator, before VascuLife was introduced to the media channels. After seeding, culture medium was added and changed daily in the device. After 7 days, the MVNs were ready for further experiments. A detailed troubleshooting table for generating MVNs can be found in previous protocols.[32,39] All experimental parameters of MVN formation for each figure are summarized in Table S1 (Supporting Information).
MVN Perfusion, Permeability Measurement, Immunofluorescent Staining, Imaging, and Analysis:
To confirm the perfusability of MVNs, the culture medium in one media channel was aspirated, followed by the injection of 20 μL of 10 μg mL−1 70 kDa MW Texas red dextran solution or Alexa Fluor 647-conjugated 10 kDa MW dextran (Invitrogen). The process was then repeated for the other media channel, before imaging under a confocal microscope. To quantify perfusability, microvessel opening percentage was calculated by dividing the number of open vessels between microposts by the total numbers of vessels between microposts in each ROI; central perfusable MVN percentage was calculated by dividing the area of perfused dextran in the central region by the area of fluorescent MVNs in the same region (images were maximum intensity projections). The permeability of the brain MVN was measured as previously described,[6] by quantifying the increase in fluorescence intensity of Alexa Fluor 647-conjugated 10 kDa MW dextran in the matrix over 10 min. For permeability tests of dermal MVNs and iPSC-EC MVNs, MVNs were treated with 5 ng mL−1 TNF-α (Sino Biological) overnight prior to being perfused with 70 kDa MW Texas red dextran. The detailed permeability measurement protocol and ImageJ Marco for permeability analysis can be found in previous protocol.[32] For immunofluorescent staining, MVNs were fixed by 4% paraformaldehyde. After thorough washing with PBS, MVNs were permeabilized with 2% Triton X-100 and then blocked with 10% goat serum (Invitrogen). Subsequently, MVNs were stained with CD31 (5 μg mL−1) or GFAP antibody (Invitrogen, 1:500) on a rocker at 4 °C overnight. Next, the microfluidic devices were washed five times with PBS on a rocker at room temperature, the MVNs were stained with secondary antibody Alexa Fluor 633-conjugated goat anti-rabbit IgG (H+L) or Alexa Fluor 594-conjugated goat anti-rat IgG (H+L) (Invitrogen, 1:500) on a rocker at 4 °C overnight. After extensive washing, the MVNs were ready for confocal imaging using an Olympus FLUOVIEW FV1200 confocal laser scanning microscope with a 10× objective. Z-stack images were acquired with a 5 μm step size. All images shown are collapsed Z-stacks, displayed using range-adjusted Imaris software, unless otherwise specified. Vessel diameters were measured manually using ImageJ (NIH, U.S., demonstrated in Figure S5, Supporting Information). FB accumulation at the outer layer or central regions was quantified using ImageJ (NIH, U.S.) by dividing the total fluorescence intensity of FBs located at the outer layer or central regions by the area of the regions between microposts. Vessel area percentage, joint density, and average vessel length were analyzed and quantified using AngioTool.[40] Images of the central region of MVNs were used for analysis.
Tumor Cell, Tumor Cluster, and Monocyte Perfusion in MVNs:
Tumor cells were resuspended at a concentration of 1 × 106 mL−1 in culture medium. Similar to dextran perfusion as described above, a 20 μL tumor cell suspension was perfused into the MVNs and incubated for 15 min. Tumor cell medium was aspirated from the media channels to remove the unattached cells, and Vasculife was replenished.The devices were then ready for imaging. To form tumor clusters to mimic circulating tumor clusters, tumor cell suspension was seeded in a 96-well ultra-low attachment plate (Wako Chemicals USA) at 2–4 cells per well. 24 h later, the tumor clusters were perfused. Monocytes were isolated from healthy donor’s blood by the monocyte core at MIT, and followed by CellTracker Deep Red (Invitrogen) staining. The experiments with primary blood cells were approved by the Committee on the Use of Humans as Experimental Subjects. After washing, monocytes were suspended at a concentration of 1 × 106 mL−1 in Vasculife and perfused.
Statistical Analysis:
All error bars are shown as mean ± SD. Statistical analysis was detected via by Student’s t-test with GraphPad Prism. Sample number (n) and p-value are provided in the figures or figure legends.
Supplementary Material
Acknowledgements
Z.W. was supported by a Ludwig Center Fund Post-Doctoral Research Fellowship from the Koch Institute for Integrative Cancer Research and by National Cancer Institute (U01 CA214381). S.E.S. was supported by a fellowship from the National Cancer Institute (K00CA212227). This work was supported by a National Cancer Institute Physical Sciences-Oncology Network supplement to award U01 CA214381. The monocytes were kindly provided by Prof. Byran Bryson.
Conflict of Interest
R.D.K. is the co-founder of and holds a significant financial interest in AIM Biotech, a company that produces microfluidic devices. R.D.K. also receives research support from Amgen, Novartis, Boehringer Ingelheim, GSK, AbbVie and Roche. D.A.B. is a co-founder of and holds a significant financial interest in Xsphera Biosciences, and is a consultant for Qiagen/N of One. D.A.B. has also received research funding from Novartis, Gilead Sciences, BMS, and Lilly Oncology.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Zhengpeng Wan, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02215, USA.
Amy X. Zhong, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Shun Zhang, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Georgios Pavlou, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Mark F. Coughlin, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
Sarah E. Shelton, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02215, USA.
Huu Tuan Nguyen, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Jochen H. Lorch, Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02215, USA
David A. Barbie, Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02215, USA
Roger D. Kamm, Department of Mechanical Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- [1].Ewald ML, Chen YH, Lee AP, Hughes CCW, Lab Chip 2021, 3244, 10.1039/d1lc00530h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zhang S, Wan Z, Kamm RD, Lab Chip 2021, 21, 473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Whisler JA, Chen MB, Kamm RD, Tissue Eng Part C Methods 2014, 20, 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Moya ML, Hsu YH, Lee AP, Hughes CC, George SC, Tissue Eng Part C Methods 2013, 19, 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kim S, Lee H, Chung M, Jeon NL, Lab Chip 2013, 13, 1489. [DOI] [PubMed] [Google Scholar]
- [6].Offeddu GS, Haase K, Gillrie MR, Li R, Morozova O, Hickman D, Knutson CG, Kamm RD, Biomaterials 2019, 212, 115. [DOI] [PubMed] [Google Scholar]
- [7].Kim S, Chung M, Ahn J, Lee S, Jeon NL, Lab Chip 2016, 16, 4189. [DOI] [PubMed] [Google Scholar]
- [8].Bang S, Lee SR, Ko J, Son K, Tahk D, Ahn J, Im C, Jeon NL, Sci. Rep 2017, 7, 8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Campisi M, Shin Y, Osaki T, Hajal C, Chiono V, Kamm RD, Biomaterials 2018, 180, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Uwamori H, Higuchi T, Arai K, Sudo R, Sci. Rep 2017, 7, 17349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Osaki T, Sivathanu V, Kamm RD, Sci. Rep 2018, 8, 5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yoon N, Kim S, Sung HK, Dang TQ, Jeon JS, Sweeney G, Biochim Biophys Acta Gen Subj 2021, 1865, 129796. [DOI] [PubMed] [Google Scholar]
- [13].Mejías JC, Nelson MR, Liseth O, Roy K, Lab Chip 2020, 20, 3601. [DOI] [PubMed] [Google Scholar]
- [14].Rambol MH, Han E, Niklason LE, Tissue Eng Part A 2020, 26, 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Xiao Y, Liu C, Chen Z, Blatchley MR, Kim D, Zhou J, Xu M, Gerecht S, Fan R, Adv. Biosyst 2019, 3, 1900089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wan Z, Zhang S, Zhong AX, Shelton SE, Campisi M, Sundararaman SK, Offeddu GS, Ko E, Ibrahim L, Coughlin MF, Liu T, Bai J, Barbie DA, Kamm RD, Biomaterials 2021, 276, 121032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Song J, Miermont A, Lim CT, Kamm RD, Sci. Rep 2018, 8, 17949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Campisi M, Sundararaman SK, Shelton SE, Knelson EH, Mahadevan NR, Yoshida R, Tani T, Ivanova E, Canadas I, Osaki T, Lee SWL, Thai T, Han S, Piel BP, Gilhooley S, Paweletz CP, Chiono V, Kamm RD, Kitajima S, Barbie DA, Front Immunol 2020, 11, 2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen MB, Whisler JA, Jeon JS, Kamm RD, Integr. Biol 2013, 5, 1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M, Kamm RD, Proc Natl Acad Sci USA 2015, 112, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Xiao Y, Kim D, Dura B, Zhang K, Yan R, Li H, Han E, Ip J, Zou P, Liu J, Chen AT, Vortmeyer AO, Zhou J, Fan R, Adv. Sci. (Weinh) 2019, 6, 1801531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Li Y, Hu C, Wang P, Liu Y, Wang L, Pi Q, Gong Z, Yang X, Mak M, Wu Y, J Nanobiotechnology 2019, 17, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ahn J, Lee H, Kang H, Choi H, Son K, Yu J, Lee J, Lim J, Park D, Cho M, Appl. Sci 2020, 10, 2027. [Google Scholar]
- [24].Guo Z, Yang CT, Maritz MF, Wu H, Wilson P, Warkiani ME, Chien CC, Kempson I, Aref AR, Thierry B, Adv. Mater. Technol 2019, 4, 1800726. [Google Scholar]
- [25].Yoon N, Dadson K, Dang T, Chu T, Noskovicova N, Hinz B, Raignault A, Thorin E, Kim S, Jeon JS, Jonkman J, McKee TD, Grant J, Peterson JD, Kelly SP, Sweeney G, Am. J. Physiol. Endocrinol. Metab 2019, 317, E760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Akasaka R, Ozawa M, Nashimoto Y, Ino K, Shiku H, Micromachines (Basel) 2021, 12, 1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Haase K, Piatti F, Marcano M, Shin Y, Visone R, Redaelli A, Rasponi M, Kamm RD, Biomaterials 2022, 280, 121248. [DOI] [PubMed] [Google Scholar]
- [28].Nelson MR, Ghoshal D, Mejias JC, Rubio DF, Keith E, Roy K, Biomaterials 2021, 270, 120683. [DOI] [PubMed] [Google Scholar]
- [29].Kim S, Park J, Kim J, Jeon JS, ACS Biomater. Sci. Eng 2021, 7, 1230. [DOI] [PubMed] [Google Scholar]
- [30].Akinbote A, Beltran-Sastre V, Cherubini M, Visone R, Hajal C, Cobanoglu D, Haase K, Front Physiol 2021, 12, 735915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Osaki T, Sivathanu V, Kamm RD, Curr. Opin. Biotechnol 2018, 52, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hajal C, Offeddu GS, Shin Y, Zhang S, Morozova O, Hickman D, Knutson CG, Kamm RD, Nat. Protoc 2022, 17, 95. [DOI] [PubMed] [Google Scholar]
- [33].Chaffer CL, Weinberg RA, Science 2011, 331, 1559. [DOI] [PubMed] [Google Scholar]
- [34].Jimenez-Torres JA, Peery SL, Sung KE, Beebe DJ, Adv. Healthcare Mater 2016, 5, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sato M, Sasaki N, Ato M, Hirakawa S, Sato K, Sato K, PLoS One 2015, 10, e0137301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tsai M, Kita A, Leach J, Rounsevell R, Huang JN, Moake J, Ware RE, Fletcher DA, Lam WA, J. Clin. Invest 2012, 122, 408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhang Q, Liu T, Qin J, Lab Chip 2012, 12, 2837. [DOI] [PubMed] [Google Scholar]
- [38].Wullkopf L, West AV, Leijnse N, Cox TR, Madsen CD, Oddershede LB, Erler JT, Mol. Biol. Cell 2018, 29, 2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chen MB, Whisler JA, Frose J, Yu C, Shin Y, Kamm RD, Nat. Protoc 2017, 12, 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zudaire E, Gambardella L, Kurcz C, Vermeren S, PLoS One 2011, 6, e27385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


