The kidney is a complex organ and a key regulator of homeostasis. It comprises various functional units (nephrons, vasculature, and interstitium) that are organized spatially, so that location and physiological roles are synchronized (1). Hence, renal microenvironments harbor diverse and specialized cells. Defining protein expression at the cellular level in the various renal regions is essential for making breakthroughs in understanding the physiology of health and pathogenesis of disease. In addition, the successful development of therapeutic strategies depends on precisely targeting the desired molecule in the correct cells.
Advances in microscopy techniques allow large-scale imaging of entire kidney sections at high resolution (2). Fluorescence microscopy, used in this setting, can define the expression of a protein across the entire kidney with cellular resolution (Fig. 1, A–H). Multiplexed immunofluorescence (IF) strategies allow the detection of several protein targets simultaneously in the same tissue section (3). The use of the appropriate analytic strategies can then offer unique opportunities to explore the spatial distribution of cells or proteins of interest and link to biological function. However, several important steps are needed to ensure that high-quality data are obtained to support a rigorous analytic pipeline. A key aspect of such quality assurance is antibody selection and validation.
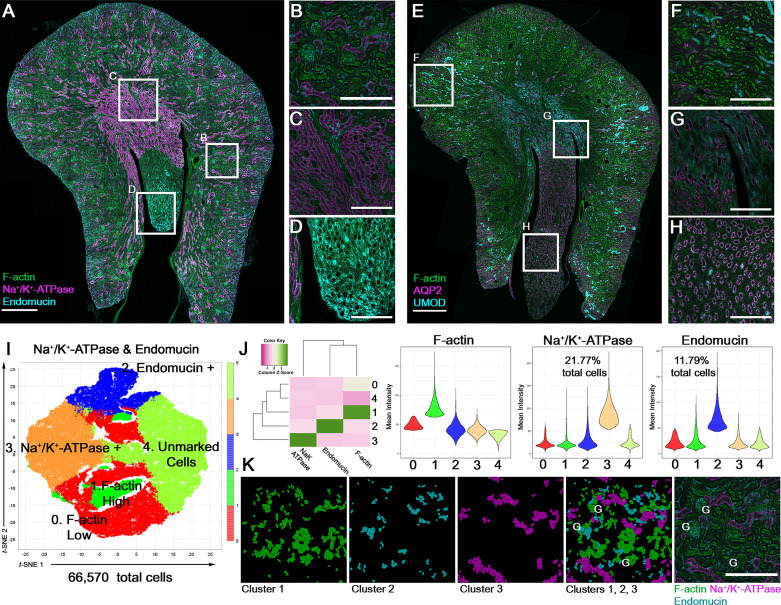
Figure 1.
Confocal fluorescence large-scale imaging of kidney sections. A: image of a mouse kidney section (produced by digital stitching after tile scanning using a ×20 objective) stained with phalloidin (F-actin), Na+-K+-ATPase (sc-28800, Santa Cruz Biotechnology), endomucin (ab106100, Abcam), and DAPI (not shown). B–D: enlarged regions from the boxed areas in A showing the cortex (B), outer medulla (C), and inner medulla/papilla (D). E: image of a kidney section stained with phalloidin, aquaporin-2 (AQP2; PA5-22865, Thermo Fisher), uromodulin (UMOD; AF5175, R&D), and DAPI (not shown). F–H: enlarged regions from E similar to what was shown for A. I: t-distributed stochastic neighbor embedding (t-SNE) plots generated with volumetric tissue exploration and analysis from the image shown in A, following cell segmentation, and using cell-associated mean fluorescence intensity for each channel. G-means unsupervised based clusters are labeled with the putative major cell types. The heatmap (z-scaled) and violin plots in J show the mean fluorescence intensity of each target in the five clusters shown. Nuclear overlays mapping back the three main clusters to the images are shown in K, displayed in the same color as the corresponding label for each target protein (compare merged overlays with the staining in the two rightmost panels). G, glomeruli. Scale bars in A and E = 500 µm; scale bars for all other images = 200 µm.
In this feature, we present large-scale imaging data with examples of four antibodies [Na+-K+-ATPase, endomucin, uromodulin (UMOD), and aquaporin 2 (AQP2)] commonly used in our laboratory to differentially label distinct cell populations. We also discuss antibody validation and demonstrate quantitative analysis using a software tool [volumetric tissue exploration and analysis (VTEA)] developed by our group (3, 4) and publicly available with a tutorial (https://vtea.wiki/) as a plugin for the ImageJ (https://imagej.net) platform (Fig. 1, I–K).
An important aspect of producing reproducible data is quality assurance and ongoing quality control measures (5). In our pipeline, this is a multistep process, divided into microscope performance monitoring, reagents validation, tissue quality, and image analysis. A well-developed analytic pipeline could also provide another layer of validation and identify potential blind spots in the entire process. For consistent confocal microscope performance, we monitor laser power, axial and lateral resolution, and the signal-to-noise ratio using an Argo-POWER system (https://argolight.com). Tissue quality is usually assured by proper handling, processing, and sectioning. We routinely perform cross sections across the larger part of the mouse kidney to capture all regions of the kidney (cortex, outer medulla, inner medulla, and papilla; Fig. 1, A and E). We have applied these methods for frozen sections (in optimum cutting temperature compound) or for formalin-fixed specimens (sectioned on a vibratome) (6).
Antibody validation is a major part of assuring high-quality IF data. Choosing antibodies with a track record of publication or available data is essential not only for increased chances of success but also to ensure reproducibility. The use of curated databases for antibodies such as www.antibodypedia.com or data repositories for validated antibodies (e.g., the GUDMAP project, www.gudmap.org) can play a key role in promoting best practices in this domain. A primary fluorophore-conjugated antibody could be used if the target antigen is expressed at a high level. A primary unconjugated antibody with a secondary fluorophore conjugated is commonly used to amplify the signal for low-abundance antigens. Staining protocols have been previously published (2, 4, 6). A negative control will assure that the signal obtained is specific to the antibody staining. The ideal negative control will also demonstrate specificity of the lost signal to the target antigen, by showing the absence of staining in tissues from a target gene knockout animal. Using UMOD−/− mice, we performed such validation for the antibody against UMOD (7) used in Fig. 1. However, such an ideal setting is not always available. When staining with unconjugated antibodies, it is important to verify the absence of nonspecific binding of secondary antibodies by performing control experiments without the primary antibodies. This was performed for all antibodies used in this study. Antibody-antigen-specific blocking peptides, when available, can also be used to test the specificity of the staining. Furthermore, to accurately assign signal to antibody staining, it is important to understand the autofluorescence of the tissue, which requires scanning tissue sections without or before staining. For example, endogenous fluorescence in the green wavelengths is detected in tubules and vessels, thereby potentially obfuscating low signals from common green fluorophores. In such cases, it is imperative to show that a true signal is several folds above the level of endogenous fluorescence, and the availability of appropriate controls will facilitate such a key task.
Positive controls are more challenging, particularly if the expression pattern of a protein is limited or unknown. Availability of a knockout animal can be of tremendous benefit since the wild-type animal becomes a de facto positive control. Ideally, the expression pattern of a protein should be consistent with published literature. In our example (Fig. 1, A–D), Na+-K+-ATPase is highly expressed at the basolateral domain of thick ascending limbs and distal convoluted tubules, more so than other nephron segments such as proximal tubules (8). Endomucin is expressed in (venous/capillary) endothelial cells (9). As shown in Fig. 1, E–H, we observed the expression of UMOD in thick ascending limb cells and AQP2 in collecting ducts (evident in the medulla and papilla; Fig. 1, G and H) (8). After validation of staining, large-scale imaging allows a comprehensive survey of protein expression across all regions of the kidney and may provide intriguing observations that could form a basis for discovery. For example, a survey of endomucin expression (Fig. 1A) reveals an underappreciated endothelial cell population in the papillary area (Fig. 1D).
A consistent and automated (or semiautomated) analytic pipeline is essential to perform quantitative analysis (abundance, distribution, and fluorescence intensity associated with cell types), as shown in Fig. 1, I–K, using VTEA software. The latest version of VTEA incorporates unsupervised clustering algorithms and machine learning classifiers that can be used for semiautomated detection and graphing of various cell types, based on features such as fluorescence intensity (10). Displaying cell census and cell-associated fluorescence intensity for each cluster can confirm the identity of cell types and allow quantitative comparisons when used for various replicates and/or experimental groups. Consistent results between samples also provide an additional layer of validation for the entire process. Furthermore, mapping back the analysis to the images (Fig. 1K) also validates the clusters and allows for interactive exploration of the data and hypothesis generation. As an example, the data shown in Fig. 1K suggest that endomucin is expressed uniquely in a subpopulation of glomerular endothelial cells, which raises novel inquiries for future studies.
GRANTS
This work was supported by Veterans Affairs Merit Award I01BX003935 and National Institute of Diabetes and Digestive and Kidney Diseases Grants R01DK111651 and P30DK079312 (to T.M.E-A.).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
A.R.S., S.W., and T.M.E.-A. conceived and designed research; A.R.S. and S.W. performed experiments; A.R.S., S.W., and T.M.E.-A. analyzed data; A.R.S., S.W., and T.M.E.-A. interpreted results of experiments; A.R.S., S.W., and T.M.E.-A. prepared figures; A.R.S., S.W., and T.M.E.-A. drafted manuscript; A.R.S., S.W., and T.M.E.-A. edited and revised manuscript; A.R.S., S.W., and T.M.E.-A. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors acknowledge Shehnaz Kahn, Daria Barwinska, Michael Ferkowicz, Ken Dunn, and Pierre Dagher for support of various aspects of this research. The authors also thank the Indiana Center for Biological Microscopy.
REFERENCES
- 1. Stewart BJ, Ferdinand JR, Young MD, Mitchell TJ, Loudon KW, Riding AM, et al. Spatiotemporal immune zonation of the human kidney. Science 365: 1461–1466, 2019. doi: 10.1126/science.aat5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ferkowicz MJ, Winfree S, Sabo AR, Kamocka MM, Khochare S, Barwinska D, Eadon MT, Cheng YH, Phillips CL, Sutton TA, Kelly KJ, Dagher PC, El-Achkar TM, Dunn KW; Kidney Precision Medicine Project. Large-scale, three-dimensional tissue cytometry of the human kidney: a complete and accessible pipeline. Lab Invest 101: 661–676, 2021. doi: 10.1038/s41374-020-00518-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dunn KW, Molitoris BA, Dagher PC. The Indiana O'Brien Center for Advanced Renal Microscopic Analysis. Am J Physiol Renal Physiol 320: F671–F682, 2021. doi: 10.1152/ajprenal.00007.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Winfree S, Khan S, Micanovic R, Eadon MT, Kelly KJ, Sutton TA, Phillips CL, Dunn KW, El-Achkar TM. Quantitative three-dimensional tissue cytometry to study kidney tissue and resident immune cells. J Am Soc Nephrol 28: 2108–2118, 2017. doi: 10.1681/ASN.2016091027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. El-Achkar TM, Eadon MT, Menon R, Lake BB, Sigdel TK, Alexandrov T, et al. A multimodal and integrated approach to interrogate human kidney biopsies with rigor and reproducibility: guidelines from the Kidney Precision Medicine Project. Physiol Genomics 53: 1–11, 2021. doi: 10.1152/physiolgenomics.00104.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Black LM, Farrell ER, Barwinska D, Osis G, Zmijewska AA, Traylor AM, Esman SK, Bolisetty S, Whipple G, Kamocka MM, Winfree S, Spangler DR, Khan S, Zarjou A, El-Achkar TM, Agarwal A. VEGFR3 tyrosine kinase inhibition aggravates cisplatin nephrotoxicity. Am J Physiol Renal Physiol 321: F675–F688, 2021. doi: 10.1152/ajprenal.00186.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. El-Achkar TM, Wu XR, Rauchman M, McCracken R, Kiefer S, Dagher PC. Tamm-Horsfall protein protects the kidney from ischemic injury by decreasing inflammation and altering TLR4 expression. Am J Physiol Renal Physiol 295: F534–544, 2008. doi: 10.1152/ajprenal.00083.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee JW, Chou CL, Knepper MA. Deep sequencing in microdissected renal tubules identifies nephron segment-specific transcriptomes. J Am Soc Nephrol 26: 2669–2677, 2015. doi: 10.1681/ASN.2014111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Samulowitz U, Kuhn A, Brachtendorf G, Nawroth R, Braun A, Bankfalvi A, Bocker W, Vestweber D. Human endomucin: distribution pattern, expression on high endothelial venules, and decoration with the MECA-79 epitope. Am J Pathol 160: 1669–1681, 2002. doi: 10.1016/S0002-9440(10)61114-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Winfree S, McNutt AT, Khochare S, Borgard TJ, Barwinska D, Sabo AR, Ferkowicz MJ, Williams JC, Lingeman JE, Gulbronson CJ, Kelly KJ, Sutton TA, Dagher PC, Eadon MT, Dunn KW, El-Achkar TM. Integrated cytometry with machine learning applied to high-content imaging of human kidney tissue for in-situ cell classification and neighborhood analysis. bioRxiv, 2022. doi: 10.1101/2021.12.27.474025. [DOI] [PMC free article] [PubMed]