Graphical abstract

Keywords: Chalcone derivatives, Biological activities, Antitumor activity, Molecular docking, CDK1
Abstract
Chalcone, a common chemical scaffold of many naturally occurring compounds, has been widely used as an effective template for drug discovery due to its broad biological activities. In this study, a series of chalcone derivatives were designed and synthesized based on the hybridization of 1-(2,4,6-trimethoxyphenyl)butan-1-one with chalcone. Interestingly, most of the target compounds exhibited inhibitory effect of tumor cells in vitro. Especially, (E)-3-(5-bromopyridin-2-yl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (B3) revealed over 10-fold potency than 5-fluorocrail against the Hela and MCF-7 cells with IC50 values of 3.204 and 3.849 μM respectively. Moreover, B3 displayed low toxicity on normal cells. Further experiments indicated that B3 effectively inhibited the proliferation and migration of tumor cells, and promoted their apoptosis. The calculation and prediction of ADME showed that the target compounds may have good pharmacokinetic properties and oral bioavailability. Reverse molecular docking suggested that the possible target of B3 is CDK1. Taken together, these results suggested that B3 appears to be a promising candidate that merits further attention in the development of anticancer drugs.
1. Introduction
The malignant tumor is one of the leading causes of death around the world. (Sung et al., 2021) Although abundant antineoplastic drugs have been studied, some of them have defects such as drug resistance and serious side effects. (Zhu et al., 2017, Pfeiffer et al., 2020) Therefore, it is still an urgent need to search for novel anticancer drugs to improve the efficacy of cancer treatment.
Chalcones are important intermediates in the biosynthesis of flavonoids with simple backbone structures and are widely distributed in sorts of natural plants such as spices, vegetables, fruits and tea. (Zhou and Xing, 2015, Zhuang et al., 2017, Zhou et al., 2021) They have a wide range of biological activities, for instance, anti-cancer, anti-inflammatory, anti-diabetic, antioxidant, anti-bacterial, anti-malarial, etc. (Mahapatra et al., 2017, Cheng et al., 2020, Gao et al., 2020, Henry et al., 2020, Rocha et al., 2020) Several chalcones derivatives have been approved for clinical applications, such as the cholagogue metochalcone, the anti-gastric ulcer drug sofalcone, and the vasoprotective agent hesperidin methylchalcone (Fig. 1). (Cheng et al., 2020) In addition, there are also plentiful chalcones in preclinical research or the initial stages of drug discovery, such as xanthohumol, curcumin, and isoliquiritigenin (Fig. 1). (Traboulsi et al., 2015, Luzak et al., 2017, Shin et al., 2020) The structural derivization of chalcones by medicinal chemists also leads to the discovery of promising drug candidates. As reported in the literature, a kind of chalcone derivative with antituberculosis activity have been discovered, and a novel chalcone derivative have been found to have the potential to treat Alzheimer's disease. (Sang et al., 2019, Gomes et al., 2017) In addition, there are also many new chalcone derivatives was found, which have anti-cancer activity (Chinthala et al., 2015, Peerzada et al., 2018, Rahimzadeh Oskuei et al., 2021, Yan et al., 2016).
Fig. 1.
Design strategy of target compounds based on clinical and preclinical representative chalcone analogues.
In our previous studies, we conducted extensive derivatization studies around phloroglucinol and finally discovered that acylphloroglucinol has better antitumor effect. According to research reports, the phenolic hydroxyl groups on the benzene ring are likely removed by metabolism in vivo, and methylation protection of the hydroxyl groups can slow down the rate of metabolism in vivo and enhance the activity.(Valdameri et al., 2012, Zhang et al., 2015). Consistent with the above findings, it has been reported that xanthohumol analogues containing 2,4,6-trimethoxybenzene have good anticancer ability, especially compound (E)-3-(3-nitrophenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (Fig. 1). (Zhang et al., 2015) In this study, we designed and synthesized a series of chalcone derivatives based on the xanthohumol analogues (E)-3-phenyl-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (A7) and evaluated the antitumor activities of these target compounds in vitro. In addition, the possible target of the target compounds were predicted by the reverse docking technique. Different from traditional molecular docking, reverse molecular docking refers to the combination of one or several small molecule drugs with a series of macromolecular targets and the targets are ranked by detailed analysis and tight binding. The highest ranked targets may be potential targets of drugs. This technology provides a reference for drug development, thereby improving the rate of success of drug discovery.
The skeleton of the lead compound A7 was structurally analyzed and divided into three parts: 2,4,6-trimethoxybenzene ring (Part A), benzene ring (Part B), and connecting bridge. Then, three series of new chalcone derivatives were designed and synthesized based on the principles of conformational analysis, scaffold hopping, and space exploration. Firstly, compounds A1-A7 were designed and synthesized by introducing different substituents into the B benzene ring to investigate the effects of different substituents on the biological activity. Secondly, the benzene ring B was replaced by aromatic rings such as monocyclic pyridine, thiophene, furan, bicyclic naphthalene ring, etc to obtain B1-B13. And it was intended to discover the effects of different structural skeletons on the biological activity. Finally, based on the introduction of phenoxy group into benzene ring B, compounds C1-C10 were designed and synthesized by introducing alicyclic or aromatic rings, such as benzyl, cyclopropylmethyl, cyclobutylmethyl, 4-methylpyridine and other structures to the phenoxy structure, to explore the effect of different chemical space structures on the biological activity. (Fig. 1).
2. Results and discussion
2.1. Chemistry
The synthetic protocols for the newly designed derivatives as described in Schemes 1-4. First, 2,4,6-trimethoxy benzene (2) was synthesized by methylation of phenolic hydroxyl groups with dimethyl sulfate. Then through the introduction of acetyl groups under Friedel-Crafts acylation, key intermediate1-(2,4,6-trimethoxyphenyl) ethyl ketone (3) was obtained. The synthesis route of compounds A1-A7 as shown in Scheme 1, treatment of 4a-4e with potassium iodide to obtain 5a-5f. Then, an aldehyde group on the benzene ring was introduced to 5a-5f via the Vilsmeier-Haack reaction catalyzed by POCl3to afford 6a-6 g. A1-A7 were obtained by the aldol condensation between compounds 6a-6 g and 3a in a basic condition.
Scheme 1.
Syntheses of A1-A7.
The synthetic route of compounds B1-B13 as shown in Scheme 2, compound 4 and 7 l-7 m underwent a Suzuki coupling reaction catalyzed by palladium diacetate to form biphenyl compounds, and then, 8a-8 m were condensated with intermediate 3 to afford compounds B1-B13.
Scheme 2.
Syntheses of B1-B13.
The synthetic route of compounds B14-B16 as shown in Scheme 3, the starting material 9a-9c were converted to 10a-10c catalyzed by acetic acid and acetic anhydride, and then10a-10c were dehydrated to form a ring catalyzed by POCl3 and DMF to produce 11a-11c. Then, 11a-11c were hydrolyzed under acidic conditions to afford intermediates 12a-12c. Finally, 12a-12c were treated with key intermediate 3 and condensated to give compounds B14-B16.
Scheme 3.
Syntheses of B14-B16.
The synthesis route of compounds C1-C10 as shown in Scheme 4. 13f-13j were treated with sodium borohydride to produce 14f-14j, followed by a chlorinative reaction catalyzed by dichlorosulfoxide to give intermediates 15f-15j, and 16a-16j were obtained by the Williamson reaction. Finally, 16a-16j were treated with key intermediate 3 to give compounds C1-C10.
Scheme 4.
Syntheses of C1-C10.
2.2. Biological evaluation
2.2.1. Anticancer activity in vitro
The anticancer activities of the synthesized compounds A1-A7, B1-B16, and C1-C10 against the Hela, A549, HepG2, and MCF-7 cancer cell lines were screened by the MTT method. 5-fluorocrail (5-Fu) was selected as a positive control because of its broad-spectrum anti-tumor activity in clinical. As the result exhibited in Table 1. Some of these compounds showed moderate to excellent anticancer activities against four cell lines. Especially compound B3 showed an IC50 value decuple higher than that of the positive control on both Hela cells and MCF-7 cells.
Table 1.
Cytotoxicity of synthetic compounds to four tumor cells.
| compound | IC50(μM) |
|||
|---|---|---|---|---|
| A549 | Hela | HepG2 | MCF-7 | |
| A1 | >100 | 72.22 ± 1.63 | 87.56 ± 1.96 | >100 |
| A2 | >100 | 54.18 ± 1.44 | 24.59 ± 1.05 | 50.79 ± 1.23 |
| A3 | >100 | 71.48 ± 1.54 | 35.41 ± 1.37 | >100 |
| A4 | 71.16 ± 1.56 | 32.31 ± 1.31 | 33.38 ± 1.37 | 32.36 ± 1.29 |
| A5 | 70.94 ± 1.82 | 38.84 ± 1.21 | 40.69 ± 1.29 | 24.67 ± 0.94 |
| A6 | 63.27 ± 1.41 | >100 | >100 | >100 |
| A7 | 59.36 ± 1.72 | 46.48 ± 1.33 | 47.12 ± 1.42 | 57.83 ± 1.45 |
| B1 | 10.55 ± 0.38 | 11.89 ± 0.44 | 10.76 ± 0.39 | 8.74 ± 0.63 |
| B2 | 9.36 ± 0.72 | 6.48 ± 0.33 | 7.12 ± 0.42 | 7.83 ± 0.45 |
| B3 | 5.19 ± 0.23 | 3.20 ± 0.21 | 5.81 ± 0.37 | 3.85 ± 0.28 |
| B4 | 19.79 ± 0.98 | 11.80 ± 0.86 | 13 ± 0.74 | 12.77 ± 0.64 |
| B5 | 46.88 ± 1.15 | 37.19 ± 1.22 | 19.92 ± 1.31 | 35.83 ± 1.35 |
| B6 | 31.33 ± 0.63 | 18.90 ± 0.73 | 19.49 ± 0.59 | 26.33 ± 0.94 |
| B7 | 47.39 ± 2.12 | 81.29 ± 2.44 | 61.44 ± 1.34 | 82.89 ± 2.65 |
| B8 | >100 | 49.80 ± 1.43 | 20.40 ± 1.52 | 48.78 ± 1.45 |
| B9 | 87.36 ± 1.83 | 21.62 ± 0.94 | 50.96 ± 0.85 | 33.32 ± 0.89 |
| B10 | 15.53 ± 0.74 | 13.57 ± 0.83 | 8.924 ± 0.68 | 10.73 ± 0.62 |
| B11 | >100 | >100 | >100 | >100 |
| B12 | 44.53 ± 1.05 | 46.34 ± 1.07 | 38.14 ± 1.45 | 51.58 ± 1.23 |
| B13 | 40.65 ± 1.51 | 30.51 ± 0.89 | 25.61 ± 0.77 | 35.32 ± 1.14 |
| B14 | 10.68 ± 0.74 | 34.97 ± 0.83 | 17.64 ± 0.74 | 7.916 ± 0.59 |
| B15 | 20.42 ± 0.67 | 4.91 ± 0.30 | 18.04 ± 0.62 | 16.04 ± 0.56 |
| B16 | 18.39 ± 0.65 | 18.09 ± 1.04 | 15.11 ± 0.62 | 12.63 ± 0.73 |
| C1 | >100 | 45.54 ± 1.59 | 39.34 ± 1.55 | >100 |
| C2 | 73.58 ± 1.17 | 22.59 ± 1.11 | >100 | >100 |
| C3 | >100 | >100 | >100 | >100 |
| C4 | >100 | >100 | >100 | >100 |
| C5 | >100 | >100 | >100 | >100 |
| C6 | 74.16 ± 1.84 | >100 | >100 | >100 |
| C7 | >100 | 34.97 ± 1.17 | 51.07 ± 2.1 | 87.76 ± 2.7 |
| C8 | >100 | >100 | >100 | >100 |
| C9 | >100 | >100 | >100 | >100 |
| C10 | >100 | >100 | >100 | >100 |
| 3 | 70.3 ± 3.17 | 26.28 ± 1.79 | 40.87 ± 1.82 | 76.3 ± 1.48 |
| 5-Fu | 43.5 ± 1.22 | 43.75 ± 1.53 | 45.38 ± 1.35 | 55.24 ± 1.92 |
In A series, compounds A1-A7 exhibited different degrees of antitumor effect in a certain pattern. Antitumor activity of compound A6 decreased significantly by introducing the methyl into the benzene ring. Compound A4 which has two methoxy groups in adjacent positions on the B-ring and compound A5 which has one methoxy group and the other one hydroxyl in adjacent positions on the B-ring, were exhibited the optimal activity against MCF-7 cell lines (IC50 = 32.36, 24.67 μM).
In B series, the benzene ring was replaced by other heterocycles or introduced electron withdrawing groups. When the B-ring was replaced by the pyridine ring or quinolone ring, the antineoplastic activity was enhanced. While the introduction of electron withdrawing groups lead to the decrease of the antineoplastic activity Compound B3, in which B-ring was substituted by pyridine with bromine atom on the para position, exhibited potent inhibitory activity against both Hela and MCF-7 cells lines (IC50 = 3.204, 3.85 μM).
In C series, with the introduction of alicyclic or aromatic rings such as benzyl, cyclopropylmethyl, cyclobutylmethyl, 4-methylpyridine, and other structures to phenoxy position, the antitumor activity of C1-C10 were varied considerably with different substituents. Compound C1 showed comparable antitumor activity against cell lines Hela and HpeG-2 (IC50 = 45.54, 39.34 μM) with the positive control 5-Fu (IC50 = 43.75, 45.38 μM) by introducing with cyclobutylmethyl ring. However, with the introduction of rigid structures such as benzene rings, the anticancer activity decreased gradually.
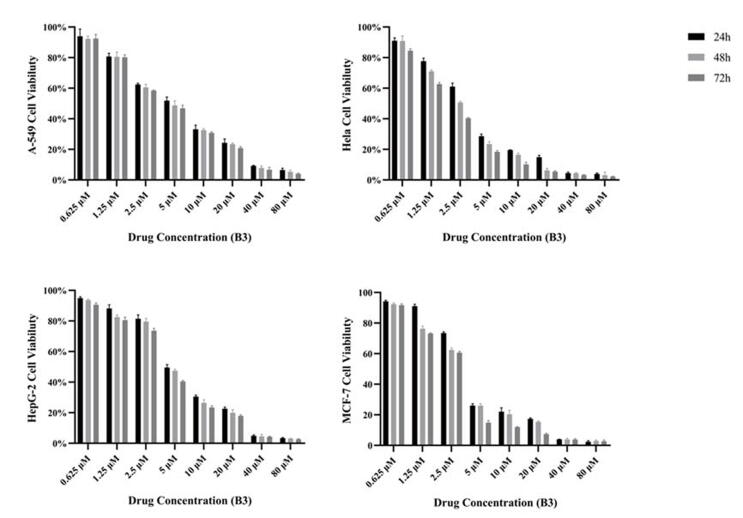
2.2.2. Drug-time curves of compound B3 against four tumor cells
Based on the results above, compound B3 was used to deal with Hela, A549, HepG2, MCF-7 cancer cell lines, to measure the relationship between time and cell viability by MTT method at 24 h, 48 h and 72 h, respectively. As shown in Fig. 2. Compound B3 showed excellent inhibitory effect on all four tumors especially Hela cells and MCF-7 cells, and had a certain concentration dependence on them. The inhibitory effect was enhanced with an increase of drug concentration, however, the time of administration had relatively little effect on tumor cell viability.
Fig. 2.
Effect of compound B3 on viability of four tumor cells at a different times of administration.
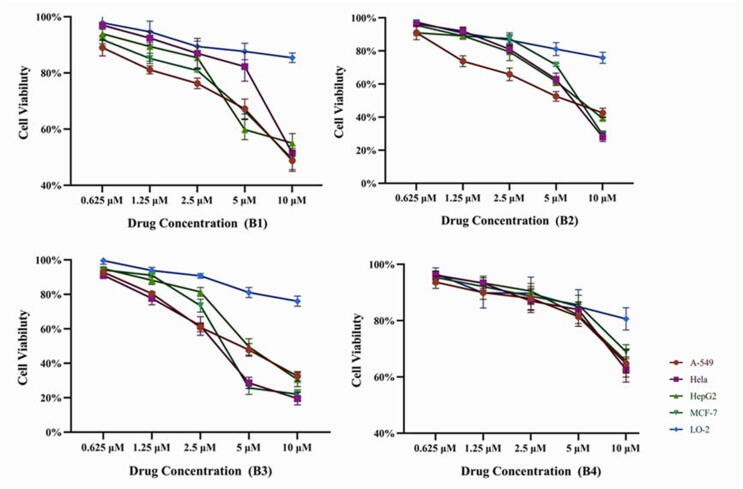
2.2.3. Cytotoxic activity on human liver cells
The toxicity of several representative compounds against normal cell lines was assessed using the human liver cell line L-O2. In a certain concentration range, the selected compounds had good inhibitory effect on tumor cells growth but were less toxic to normal cells (Fig. 3, Fig. 4).
Fig. 3.
Effect of compound B1-B4 on vability of four tumor cells and liver cell at different administration.
Fig. 4.
Effect of compound B10, B14-B16 on vability of four tumor cells and liver cell at different administration.
2.2.4. Migration ability assay
In this study, to investigate the migration inhibitory potency of compound B3 on Hela andMCF-7 cells, a scratch test was performed. Hela and MCF-7 cells were exposed to concentrations of compound B3 at 0.5 μM, 1 μM, 2 μM, 4 μM for 24 h or 48 h. The 5-Fu was used as a reference in a concentration of 5 μM. The cells treated with compound B3 at a dose of 0.5 μM revealed similar inhibition of the migration in comparison to the positive control group at a dose of 5 μM, and the effect was enhanced with the increasing of drug concentration, as shown in Fig. 5. Taken together, these results suggested that B3 showed the potential to generate an anti-metastasis effect in Hela, MCF-7 cells.
Fig. 5.
. 1 Effects of compound B3 on Hela cell migration in vitro. (A) Image of Hela migration treated with compound B3. (B) Quantitative analysis of Hela mobility. **P < 0.01, ***P < 0.001 and ****P < 0.0001. Data are presented as the mean ± SD of three separate experiments. Fig. 5-2. Effects of compound B3 on McF-7 cell migration in vitro. (C) Image of MCF-7 migration treated with compound B3. (D) Quantitative analysis of MCF-7 mobility. **P < 0.01, ***P < 0.001 and ****P < 0.0001. Data are presented as the mean ± SD of three separate experiments.
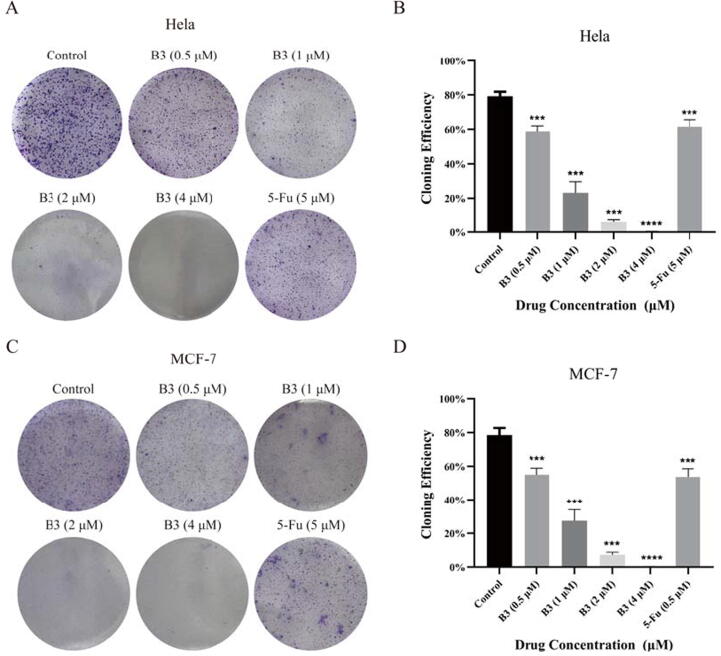
2.2.5. Colony formation assay
The cloning efficiency is used as an indicator of cell proliferation ability, invasiveness and sensitivity to killing factors. In this study, to further study the cell viability of the compound B3, the colony formation in Hela, MCF-7 cells were inspected. 5-Fu was used as the positive control. As reported in Fig. 6, the compound B3 could effectually restrain the colony formation of Hela, MCF-7 cells, especially Hela cells, with almost 100 % inhibition of tumor cells cloning at a dose of 4 μM. Moreover, compound B3 could effectively inhibit colony formation at 0.5 mM in both cell lines, which is slightly better than the positive control with 5 μM.
Fig. 6.
Cell cloning and formation experiments. (A) Image of Hela cloning treated with compound B3. (B) Quantitative analysis of clone formation rate in B3-treated Hela cells (C) Image of Hela cloning treated with compound B3. (D) Quantitative analysis of clone formation rate in B3-treated MCF-7 cells. ***P < 0.001, ****P < 0.0001. Data are presented as the mean ± SD of three separate experiments.
2.2.6. Apoptotic assay
To investigate whether the compound B3 results in cell death by inducing apoptosis, we used Hoechst 33,258 staining to observe the morphological changes of Hela cells, and Annexin V-FITC/PI assay to evaluate the apoptotic effects. Hela cells were exposed to various concentrations (0, 0.5, 1, 2 and 4 μM) of compound B3 or 5-Fu (5.0 μM) for 24 h. As shown in Fig. 7, at low doses, cell membranes blister and the cells became round and larger. As the concentration of B3 increased gradually, cells morphology changed, including cells shrinkage, nuclear division and formation of apoptotic bodies, as indicated by the red arrow. As reported in Fig. 8, the compound B3 induced apoptosis of Hela cells in a dose-dependent manner, the apoptosis index was 14.06 % (control), 16.78 % (0.5 μM), 26.41 % (1 μM), 30.95 % (2 μM), 39.8 % (4 μM), respectively. The ratio of early apoptotic cells to late apoptotic cells rose significantly with the increasing of compound concentration, suggesting that compound B3 leads to cell death by inducing apoptosis.
Fig. 7.
Hochest 33,258 staining to analyze the morphological changes of Hela apoptosis induced by compound B3.
Fig. 8.
Flow cytometry quantitative analysis of the compound B3 on apoptosis of Hela by Annexin V-FITC/PI double staining. Q1: This area contains necrotic and mechanically dead cells. Q2: This area represents late apoptotic cells. Q3: This area is an early apoptotic cell. Q3: This area is a living cell. *P < 0.05, **P < 0.01, and ***P < 0.001. Data are presented as the mean ± SD of three separate experiments.
2.3. ADMET properties assay
The calculation and prediction of important physicochemical parameters related to the absorption, distribution, metabolism and excretion (ADME) of compounds is a cost-effective strategy for filtering molecules in the early stage of drug discovery. Therefore, in this study, ADME predictions were performed for the synthesized target compounds A1-A7, B1-B16, and C1-C10. The results were presented in Table 2. First, all the 33 compounds with relative molecular masses<500, rotatable bonds no>10, hydrogen bond acceptors<10, hydrogen bond donors<5, and lipid-water partition coefficients<5, which is in accordance with Lipinski's rule of five, indicating that these compounds may have good pharmacokinetic properties and oral bioavailability. Total polar surface area (TPSA) is the major characteristic used for the optimization of the ability of drug penetration cells. From data depicted in Table 2, all synthetic compounds with TPSA<140 Å2 indicated favorable permeating cell membranes.
Table 2.
Prediction of “ADME” related physicochemical properties of target compounds.
| Compound | MW | Rotatable bonds | H-bond acceptors | H-bond donors | TPSA | Consensus Log P | ESOL Log S |
|---|---|---|---|---|---|---|---|
| A1 | 388.41 | 9 | 7 | 0 | 72.45 | 3.32 | −4.19 |
| A2 | 328.36 | 7 | 5 | 0 | 53.99 | 3.34 | −4.04 |
| A3 | 358.39 | 8 | 6 | 0 | 63.22 | 3.31 | −4.11 |
| A4 | 358.39 | 8 | 6 | 0 | 63.22 | 3.34 | −4.11 |
| A5 | 344.36 | 7 | 6 | 1 | 74.22 | 3.00 | −3.90 |
| A6 | 402.44 | 9 | 7 | 0 | 72.45 | 3.62 | −4.49 |
| A7 | 298.33 | 6 | 4 | 0 | 44.76 | 3.34 | −3.97 |
| B1 | 299.32 | 6 | 5 | 0 | 57.65 | 2.57 | −3.32 |
| B2 | 378.22 | 6 | 5 | 0 | 57.65 | 3.26 | −4.44 |
| B3 | 378.22 | 6 | 5 | 0 | 57.65 | 3.21 | −4.23 |
| B4 | 377.23 | 6 | 4 | 0 | 44.76 | 3.96 | −4.88 |
| B5 | 304.36 | 6 | 4 | 0 | 73.00 | 3.26 | −3.79 |
| B6 | 383.26 | 6 | 4 | 0 | 73.00 | 3.92 | −4.91 |
| B7 | 288.30 | 6 | 5 | 0 | 57.90 | 2.55 | −3.30 |
| B8 | 367.19 | 6 | 5 | 0 | 57.90 | 3.26 | −4.42 |
| B9 | 348.39 | 6 | 4 | 0 | 44.76 | 4.25 | −5.11 |
| B10 | 349.38 | 6 | 5 | 0 | 57.65 | 3.56 | −4.50 |
| B11 | 351.40 | 6 | 4 | 0 | 49.69 | 3.42 | −4.37 |
| B12 | 374.43 | 7 | 4 | 0 | 44.76 | 4.66 | −5.47 |
| B13 | 392.42 | 7 | 5 | 0 | 44.76 | 4.99 | −5.63 |
| B14 | 365.38 | 6 | 5 | 1 | 77.62 | 3.17 | −4.06 |
| B15 | 379.41 | 6 | 5 | 1 | 77.62 | 3.50 | −4.35 |
| B16 | 383.37 | 6 | 6 | 1 | 77.62 | 3.47 | −4.22 |
| C1 | 368.42 | 9 | 5 | 0 | 53.99 | 3.98 | −4.58 |
| C2 | 396.48 | 9 | 5 | 0 | 53.99 | 4.64 | −5.41 |
| C3 | 410.50 | 9 | 5 | 0 | 53.99 | 4.93 | −5.84 |
| C4 | 404.46 | 9 | 5 | 0 | 53.99 | 4.55 | −5.39 |
| C5 | 483.35 | 9 | 5 | 0 | 53.99 | 5.20 | −6.30 |
| C6 | 405.44 | 9 | 6 | 0 | 66.88 | 3.77 | −4.74 |
| C7 | 410.48 | 9 | 5 | 0 | 82.23 | 4.50 | −5.22 |
| C8 | 454.51 | 9 | 5 | 0 | 53.99 | 5.45 | −6.52 |
| C9 | 455.50 | 9 | 6 | 0 | 66.88 | 4.74 | −5.91 |
| C10 | 471.50 | 9 | 6 | 1 | 86.85 | 4.41 | −5.46 |
2.4. Molecular docking
In order to validate the docking reliability, the ligand was removed from the active site and docked back into the binding pocket. The root mean square deviation (RMSD) between the predicted conformation and the actual conformation from the crystal structure of ligand was 0.619Ǻ, which is smaller than the resolution of X-ray crystallography, 1.8Ǻ. It indicated that the parameter set for the Surflex-dock simulation was reasonable to reproduce the X-ray structure. Therefore, the simulation method and the parameter set could be extended to study the binding conformations of the other inhibitors.
In this study, we investigated pieces of literature related to the anti-tumor effects of chalcone natural products and derivatives, selected 20 receptor proteins as structural models for molecular docking studies. The putative target selection was based on the availability of the three-dimensional crystal structure of the protein and the importance of the target for tumor growth and survival (Table 3). Then, 6 representative compounds B1-B4, B10, and B16 were selected to calculate docking scores with 20 proteins mentioned above, among which binding energies with small molecules of 12 proteins can be calculated, as shown in Table 4. Among them, CDK1 (PDB:4Y72) protein showed the best binding potential with six active small molecules. Therefore, CDK1 may be the target for predicting the antitumor effect of this class of compounds.
Table 3.
The selected 20 anticancer drug targets that were used for molecular modelling studies.
| Number | Protein Target | Targeted pathway | PDB ID | References |
|---|---|---|---|---|
| 1 | Tubulin | Wnt/β-catenin | 1SA0 | (Li et al., 2018) |
| 2 | XO | Erk signal pathway | 3NVY | (Burmaoglu et al., 2019) |
| 3 | ABCG2 | ABC transporters pathway | 6FFC | (Roussel et al., 2019) |
| 4 | TrXR | MAPKs signal transduction pathway | 3EAN | (Wang et al., 2020) |
| 5 | Fli-1 | Notch1 signaling pathway | 5JVT | (Ma et al., 2020) |
| 6 | Caspase-3 | Apoptosis pathway | 2 J30 | (Dong et al., 2018) |
| 7 | Caspase-8 | Apoptosis pathway | 3KJn | (Dong et al., 2018) |
| 8 | EGFR | PI3K/AKT/mTOR Pathway | 1 M17 | (Abou-Zied et al., 2019) |
| 9 | BRAF | RAF/MEK/ERK pathway | 3OG7 | (Mohassab et al., 2021) |
| 10 | ALK | PI3K/AKT/mTOR Pathway | 2XP2 | (Kommidi et al., 2015) |
| 11 | ER-α | MAPK signaling pathway | 3ERT | (Muchtaridi et al., 2017) |
| 12 | VEGFR-2 | VEGFA/VEGFR2 pathway | 4ASD | (Ahmed et al., 2021) |
| 13 | EHMT2 | PI3K/AKT/mTOR pathway | 3K5K | (Vanaparthi et al., 2020) |
| 14 | HDAC8 | Epac2-Rap1A-Akt pathway | 3SFH | (Vanaparthi et al., 2020) |
| 15 | CDK1 | Apoptosis pathway | 4Y72 | (Farghaly et al., 2020) |
| 16 | TOPO1 | Apoptosis pathway | 1SC7 | (Kamal et al., 2014) |
| 17 | GSK-3β | PP2A-GSK3β-MCL-1 pathway | 1Q41 | (Dan et al., 2020) |
| 18 | DNA dodecamer | DNA cleavage pathway | 2DND | (El-Wakil et al., 2020) |
| 19 | PPARα | Wnt/β-Catenin | 2P54 | (Niu et al., 2017) |
| 20 | TOPOIIβ | Apoptosis pathway | 5JL9 | (Mohammed et al., 2021) |
Table 4.
Binding energy of representative compounds to the receptor.
| PDB ID | Binding energy (Kcal/mol) |
|||||
|---|---|---|---|---|---|---|
| B1 | B2 | B3 | B4 | B10 | B16 | |
| 1SA0 | −7.3 | −7.0 | −6.7 | −6.9 | −7.8 | −8.5 |
| 3NVY | ND | ND | ND | ND | ND | ND |
| 6FFC | ND | ND | ND | ND | ND | ND |
| 3EAN | ND | ND | ND | ND | ND | ND |
| 5JVT | ND | ND | ND | ND | ND | ND |
| 2 J30 | ND | ND | ND | ND | ND | ND |
| 3KJn | −5.8 | −5.9 | −5.8 | −5.6 | −6.8 | −7.0 |
| 1 M17 | −6.8 | −6.8 | −7.0 | −6.9 | −8.2 | −8.3 |
| 3OG7 | −7.5 | −7.5 | −6.9 | −7.0 | −8.4 | −8.0 |
| 2XP2 | −6.7 | −6.6 | −6.3 | −6.3 | −7.4 | −7.7 |
| 3ERT | −7.3 | −7.5 | −7.0 | −6.9 | −8.3 | −8.7 |
| 4ASD | −6.8 | −7.2 | −6.5 | −6.7 | −7.6 | −8.1 |
| 3K5K | −6.9 | −6.6 | −6.6 | −6.8 | −7.4 | −7.8 |
| 3SFH | −6.1 | −5.8 | −5.4 | −5.7 | −6.5 | −6.7 |
| 4Y72 | −8.2 | −8.1 | −8.4 | −8.1 | −9.0 | −9.3 |
| 1SC7 | ND | ND | ND | ND | ND | ND |
| 1Q41 | −7.5 | −7.9 | −7.6 | −7.4 | −8.0 | −8.5 |
| 2DND | ND | ND | ND | ND | ND | ND |
| 2P54 | −7.4 | −7.3 | −7.2 | −7.1 | −8.2 | −8.3 |
| 5JL9 | ND | ND | ND | ND | ND | ND |
ND = not determined.
The 2D and 3D visualization analysis of the interaction between small molecule ligand and CDK1 were shown in Fig. 9, Fig. 10. As shown in Fig. 9 (A), oxygen on the methyl phenolic hydroxyl and the carbonyl group of compound B1 formed a hydrogen bond with TYR15. The pyridine ring of compound B1 formed hydrophobic interactions with the residues VAL18, ALA145, VAL64, and ALA31, and the pyridine ring formed hydrophobic interactions with residues VAL18, ALA145, VAL64, and ALA31. As shown in Fig. 9 (B), compound B2 is structurally similar to compound B1 and also produced hydrogen bonds, hydrophobic interactions, van der Waals interacsmall-molecule ion forces, and acted on mostly the same amino acid residues. As reported in Fig. 10 (D), Compound B4 interacted with the protein without hydrogen bonds, mainly by forming van der Waals interaction with residues LEU83, LYS89, PHE82, ASP86, SER84, and so on, benzene and pyridine rings of compound B4 formed hyperconjugation with residues ILE10 and VAL18, respectively. As described in Fig. 10 (C), compound B3 is structurally similar to compounds B1, B2, and B4, however, exhibited stronger interaction with the protein. The benzene ring formed an anion-π interaction with ASP86, methyl group in the benzene ring of compound B3 formed C—H interaction with LEU83, pyridine ring forms hyperconjugation with residues VAL18, hydrophobic interaction with residues LYS89, LEU135, ILE10, PHE80, LYS33, and van der Waals interaction with residues ASP146, ALA145, MET85. As shown in Fig. 11 (E), the oxygen atom in the benzene ring of compound B10 produced hydrogen bond interaction with GLY11, the naphthalene ring formed hyperconjugation interaction with VAL18 and hydrophobic interaction with LYS35, and formed van der Waals interaction with LYS89, MET85, ASP146, TYR15, and so on, the benzene ring of compound B10 produced anion -π interaction with ASP86. As reported in Fig. 11 (F), the hydrogen in the lactam group of compound B16 formed a hydrogen bond with ILE10, the F atom formed a hydrogen bond with residue GLN134, quinolone produced anion -π interaction with ASP86 and π-π interaction force with PHE80, and formed van der Waals interaction forces with residues GLY11, GLU51, PHE147.
Fig. 9.
2D and 3D interaction modes of compounds with 4Y72. (A) 2D and 3D interaction modes of compound B1 with 4Y72. (B) 2D and 3D interaction modes of compound B2 with 4Y72.
Fig. 10.
2D and 3D interaction modes of compounds with 4Y72. (C) 2D and 3D interaction modes of compound B3 with 4Y72. (D) 2D and 3D interaction modes of compound B4 with 4Y72.
Fig. 11.
2D and 3D interaction modes of compounds with 4Y72. (E) 2D and 3D interaction modes of compound B10 with 4Y72. (F) 2D and 3D interaction modes of compound B16 with 4Y72.
In conclusion, these six compounds exhibited potent interaction with CDK1 protein and completely occupied the protein pocket groove. However, the docking fraction of compounds B10 and B16 with protein CDK1 was higher than that of the optimal active molecule B3. We predicted the permeability and hydrophobic effect of all synthetic compounds, compounds B3, B10, and B16 had TPSA with 57.65, 57.65, and 77.62 Å2, respectively. Simultaneously, consensus lipophilicity (LogPO/W) of compounds B3, B10, and B16 were 3.21, 3.56, and 3.47, respectively. These results suggested that B3 may have better solubility and membrane permeability than B10 and B16. Therefore, we speculated that compound B3 is more likely to access the cell through the lipid bilayer and has superior antitumor activity than other molecules.
3. Conclusion
In summary, 33 novel chalcone derivatives were designed and synthesized according to the structure–activity relationship, skeleton transition and space exploration. It was found that these compounds have good antitumor activity. The dominant compound B3 was selected for normal cytotoxicity test, scratch test, cell cloning test and apoptosis test in vitro. These tests revealed that B3 inhibited the migration and proliferation of tumor cells and promoted the apoptosis of tumor cells. Besides, it had a low toxicity to normal cells. Reverse molecular docking predicted that the possible target of B3 was a cyclin-dependent kinase CDK1. The above studies show that B3 has the potential to become a candidate compound for cancer treatment.
4. Experimental section
4.1. General methods for chemistry
All reagents and solvents were purchased from commercial vendors and were dried and used in this study. The purities of all synthesized compounds were verified to be > 95 % by area normalization analysis in high performance liquid chromatography. Thin-Layer Chromatography (TLC) analysis were executed on silica gel 60 F254 silica plates with Merck KGaA. The 1H and 13C NMR spectral data of targeted compounds were recorded on c Avance 300 and 400 MHz spectrometers with TMS as the internal standard (Bruker, Rheinstatten, Germany). Mass spectrometry data were recorded on an Agilent 1260 mass spectrometer. Synthesized compounds were analyzed by the Chromaster HPLC instrument (5 μm, 4.6 mm × 250 mm Supersil ODS2 C18 column). Melting points (m.p.) of all final compounds were recorded on a SGW X-4 apparatus and uncorrected.
1,3,5-trimethoxybenzene (5a). To a solution of phloroglucinol (1 g, 0.008 mol) in methanol was added anhydrous potassium carbonate (4.38 g, 0.032 mol). The potassium iodide (4 mL, 0.032 mol) was added dropwise without light and the mixture was stirred at room temperature for 3 h monitored by TLC. Then the reactant was filtered, and washed with methanol. The filtrate was collected, concentrated and purified by flash column chromatography to afford 5a.
Preparations of compounds 5b-5e were similar to 5a.
1,3,5-trimethoxy-2-methylbenzene (5f). The compound 4f (1 g, 0.005 mol) was dissolved in tetrahydrofuran by stirring under ice bath, followed by the addition of sodium cyanoborohydride (0.6 g, 0.01 mol). The mixture was stirred at room temperature for 3 h and monitored by TLC. Dilute hydrochloric acid was added to quench the reaction, and the reactant was extracted with ethyl acetate and condensed and dried to obtain 5f.
1-(2,4,6-trimethoxyphenyl) ethyl ketone (3). To a solution of 5a (2 g, 0.012 mol) in dichloromethane was added aluminum chloride catalyst (3.17 g, 0.024 mol) with stirring under ice bath. Acetyl chloride (1 mL, 0.013 mol) was slowly dropped into the mixture with accelerated stirring. The reaction was conducted at 0℃ and monitored by TLC. Then, the solution was quenched with water, filtered, and washed with ethyl acetate, the filtrate was collected, concentrated, extracted with ethyl acetate and the organic layer was collected, concentrated. The residue was purified by flash column chromatography to afford 3 as a white solid, 80 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 6.25 (s, 2H), 3.80 (s, 3H), 3.74 (s, 6H), 2.31 (s, 3H). ESI-MS m/z: 210.09 [M−H]−, C11H14O4.
2,4,6-trimethoxy benzaldehyde (6a). To a solution of 5a (0.5 g, 0.003 mol) in DMF was added phosphorus oxychloride (3 mL, 0.032 mol) slowly under ice bath. The mixture was stirred at 0 °C for 3 h and monitored by TLC. Then, the potassium carbonate solution prepared in advance was added dropwise to quench the reaction. The reactant was adjusted pH = 9–10, filtered and dried to afford 6a as a white solid, 90 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 10.20 (s, 1H), 6.27 (s, 2H), 3.88 (s, 3H), 3.83 (s, 6H). ESI-MS m/z: 196.07 [M−H]−, C10H12O4.
Preparations of compounds 6d-6f were similar to 6a.
3,4-dimethoxybenzaldehyde (6d). A pale pink solid, 75 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 9.85 (s, 1H), 7.60 – 7.54 (m, 1H), 7.39 (d, J = 1.9 Hz, 1H), 7.18 (d, J = 8.2 Hz, 1H), 3.85 (d, J = 16.0 Hz, 6H). ESI-MS m/z: 166.23[M-H]−, C9H10O3.
2,4,6-trimethoxy-3-methylbenzaldehyde (6f). A gray solid, 81 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 10.19 (s, 1H), 6.54 (s, 1H), 3.92 (s, 3H), 3.89 (s, 3H), 3.68 (s, 3H), 1.98 (s, 3H). ESI-MS m/z: 210.27 [M−H]−, C11H14O4.
[1,1′-biphenyl]-4-formaldehyde (8 l). To a solution of 4-bromobenzaldehyde (0.2 g, 0.001 mol) and phenylboronic acid (0.2 g, 0.002 mol) in anhydrous tetrahydrofuran was added potassium carbonate (0.39 g, 0.002 mol). The palladium acetate (0.05 g, 0.02 mol) was added under nitrogen protection. The reaction system was heated to 80 °C after stirring. When reaction was finished, the mixture was filtered and washed with ethyl acetate. The filtrate was collected, concentrated and purified by flash column chromatography to afford 8 l as a white solid, 72 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 10.06 (s, 1H), 8.09 – 7.95 (m, 2H), 7.97 – 7.89 (m, 2H), 7.84 – 7.73 (m, 2H), 7.58 – 7.40 (m, 3H). ESI-MS m/z: 182.37 [M−H]−, C13H10O.
Preparation of compound 8 m was similar to 8 l.
4′-fluoro-[1,1′-biphenyl]-4-carbaldehyde (8 m). A white solid, 68 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 10.11 (s, 1H), 8.07 – 8.03 (m, 2H), 7.98 – 7.94 (m, 2H), 7.91 – 7.86 (m, 2H), 7.44 – 7.37 (m, 2H). ESI-MS m/z: 200.25 [M−H]−, C13H9FO.
1-methyl-1H-indole (8 k). 1H-indole (0.3 g, 0.002 mol), sodium hydride (0.1 g, 0.004 mol) and DMF (20 mL) were added to a round bottom flask with a magnetic stirring bar under ice bath, followed by the addition of potassium iodide (1 mL, 0.004 mol). The mixture was stirred at 0 °C for 3 h without light and monitored by TLC. After that, the reaction was quenched with water, the solid was precipitated, extracted and filtrated, washed with methanol. The filtrate was collected, concentrated to afford compound 8 k.
N-Phenylacetamide (10a). Aniline (2 g, 0.022 mol) was placed in a 50 mL round-bottom flask, and acetic anhydride (2 mL, 0.043 mol) was added followed by the addition of acetic acid (1 mL, 0.043 mol). The mixture was stirred at 60 °C for 3 h without light and monitored by TLC. The reaction was quenched by the addition of water, and the solid was precipitated. The reactant was filtered and the filtrate was collected, concentrated and dried under vacuum to afford 10a as a white solid, 93 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 9.93 (s, 1H), 7.65 – 7.51 (m, 2H), 7.38 – 7.18 (m, 2H), 7.16 – 6.89 (m, 1H), 2.04 (s, 3H). ESI-MS m/z: 135.07[M-H]−, C8H9NO.
Preparations of compounds 10b-10c were similar to 10a.
N-(p-tolyl)acetamide (10b). A pale-yellow solid, 92 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 9.82 (s, 1H), 7.48 – 7.44 (m, 2H), 7.08 (d, J = 8.2 Hz, 2H), 2.24 (s, 3H), 2.02 (s, 3H). ESI-MS m/z: 149.08[M-H]−, C9H11NO.
N-(4-fluorophenyl)acetamide (10c). A white solid, 94 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 9.97 (s, 1H), 7.61 – 7.56 (m, 2H), 7.16 – 7.10 (m, 2H), 2.03 (s, 3H). ESI-MS m/z: 153.06[M-H]−, C8H8FNO.
2-chloroquinoline-3-carbaldehyde (11a). To a solution of 10a (1 g, 0.007 mol) in DMF was added phosphorus oxychloride (10 mL, 0.06 mol) under ice bath. The mixture was stirred at 90 °C for 5 h without light and monitored by TLC. Then, the reaction was cooled to room temperature and quenched with water. The solid was separated out with continuously stirring, filtered, washed, and the filter cake was collected and dried in a vacuum drying oven to afford compound 11a.
Preparations of compounds 11b-11c were similar to 11a.
2-Oxo-1,2-dihydroquinoline-3-carbaldehyde (12a). Compound 11a (1 g, 0.006 mol) was placed in a 100 mL round-bottom flask, and 70 % acetic acid solution prepared in advance was added in it. The mixture was refluxed and stirred at 90 °C for 5 h and monitored by TLC. Then, the reaction was cooled to room temperature and was quenched by the addition of water. Until a large amount of solid was precipitated with stirring, the reactant was filtered and dried under vacuum to afford 12a as a white solid, 99 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 10.39 (s, 1H), 9.02 (s, 1H), 8.30 (d, J = 8.2 Hz, 1H), 8.17 – 7.94 (m, 2H), 7.78 (t, J = 7.5 Hz, 1H). ESI-MS m/z: 173.34[M-H]−, C10H7NO2.
Preparations of compounds 12b-12c were similar to 12a.
7-methyl-2-oxo-1,2-dihydroquinoline-3-carbaldehyde (12b). A pale yellow solid, 97 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 12.18 (s, 1H), 10.24 (s, 1H), 8.43 (s, 1H), 7.71 (s, 1H), 7.50 (s, 1H), 7.28 (d, J = 8.4 Hz, 1H), 2.35 (s, 3H). ESI-MS m/z: 187.14[M-H]−, C11H9NO2.
7-fluoro-2-oxo-1,2-dihydroquinoline-3-carbaldehyde (12c). A pale yellow solid, 96 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 12.33 (s, 1H), 10.25 (s, 1H), 8.50 (s, 1H), 7.95 – 7.79 (m, 1H), 7.71 – 7.50 (m, 1H), 7.50 – 7.37 (m, 1H). ESI-MS m/z: 191.04[M-H]−, C10H6FNO2.
(4-Bromophenyl)methanol (14f). To a solution of p-bromobenzaldehyde (0.2 g, 0.001 mol) in methanol was added sodium borohydride (0.06 g, 0.002 mol) dropwise. The mixture was refluxed and stirred at 0 °C for 2 h and monitored by TLC. Then, the reaction was quenched by the addition of dilute hydrochloric acid. The reactant was extracted with ethyl acetate and the combined organic phase was concentrated and dried under vacuum to afford 14f as a white solid, 90 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 7.64 – 7.50 (m, 2H), 7.33 (d, J = 8.3 Hz, 2H), 5.34 (t, J = 5.7 Hz, 1H), 4.52 (d, J = 5.7 Hz, 2H). ESI-MS m/z: 185.97[M-H]−, C7H7BrO.
1-bromo-4-(chloromethyl)benzene (15f). To a solution of 14f (0.2 g, 0.001 mol) in dichloromethane was added thionyl chloride (1 mL) slowly with stirring under ice bath. The mixture was refluxed and stirred at 0 °C for 2 h and monitored by TLC. Then, the reaction was quenched by the addition of water and the reactant was extracted with ethyl acetate. The combined organic phase was concentrated and dried under vacuum to afford 15f.
4-(Cyclopropylmethoxy)benzaldehyde (16a). To a solution of 7f (0.15 g, 0.001 mol) in DMF was added potassium carbonate (0.3 g, 0.002 mol) with stirring. The mixture was refluxed and stirred at 70–80 °C for 5 h and monitored by TLC and then the reaction was cooled to room temperature. The reactant was filtered and the filtrate was collected. The solid was precipitated by addition of a small amount of water and was filtered and dried to afford 16a as a white solid, 84 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 9.78 (s, 1H), 7.89 – 7.71 (m, 2H), 7.02 (d, J = 8.7 Hz, 2H), 3.85 (d, J = 7.1 Hz, 2H), 1.22 – 1.11 (m, 1H), 0.57 – 0.47 (m, 2H), 0.34 – 0.19 (m, 2H). ESI-MS m/z: 176.08[M-H]−, C11H12O2.
Preparations of compounds 16b-16j were similar to 16a.
4-(cyclohexylmethoxy) benzaldehyde (16c). A white solid, 77 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 9.86 (s, 1H), 7.85 (d, J = 8.4 Hz, 2H), 7.12 (d, J = 8.4 Hz, 2H), 3.90 (d, J = 6.1 Hz, 2H), 1.91 – 1.63 (m, 6H), 1.35 – 1.01 (m, 5H). ESI-MS m/z: 218.13[M-H]−, C14H18O2.
4-(benzyloxy)benzaldehyde (16d). A white solid, 80 % yield, 1H NMR (400 MHz, DMSO‑d6) δ 9.88 (s, 1H), 7.93 – 7.85 (m, 2H), 7.51 – 7.33 (m, 5H), 7.22 (d, J = 8.7 Hz, 2H), 5.24 (s, 2H). ESI-MS m/z: 212.08[M-H]−, C14H12O2.
4-(pyridin-3-ylmethoxy)benzaldehyde (16e). A white solid, 86 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 9.87 (s, 1H), 8.68 – 8.52 (m, 1H), 7.95 – 7.81 (m, 3H), 7.54 (d, J = 7.8 Hz, 1H), 7.50 – 7.33 (m, 1H), 7.28 – 7.18 (m, 2H), 5.31 (s, 2H). ESI-MS m/z: 213.08[M-H]−, C13H11NO2.
4-(thiophen-3-ylmethoxy)benzaldehyde (16 g). A brown solid, 73 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 9.88 (s, 1H), 7.96 (s, 2H), 7.90 – 7.86 (m, 2H), 7.24 – 7.19 (m, 3H), 5.23 (s, 2H). ESI-MS m/z: 218.04[M-H]−, C12H10O2S.
4-(quinolin-3-ylmethoxy)benzaldehyde (16i). 86 % yield. 1H NMR (400 MHz, DMSO‑d6) δ 9.93 (s, 1H), 8.03 – 7.92 (m, 5H), 7.70 – 7.58 (m, 3H), 7.33 (d, J = 8.5 Hz, 2H), 5.47 (s, 2H). ESI-MS m/z: 263.09[M-H]−, C17H13NO2.
1,3-bis(2,4,6-trimethoxyphenyl)2-propenone (A1). To a solution of 3a (0.154 g, 0.0008 mol) in methanol was added 50 % sodium hydroxide solution which was prepared in advance, and the mixture was stirred at room temperature for 5 h and monitored by TLC. Then, the reaction was quenched by the addition of water. And then the reactant was extracted with ethyl acetate. The organic layer was collected, concentrated and dried to obtain crude product. Then the crude product was purified by flash column chromatography to afford A1 as a pale yellow solid, 72 % yield, m.p.150-152℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.61 – 7.50 (m, 1H), 7.08 – 6.96 (m, 1H), 6.28 (d, J = 6.7 Hz, 4H), 3.85 – 3.80 (m, 12H), 3.70 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 194.82, 163.50, 161.98, 161.31, 158.32, 135.47, 128.85, 112.30, 105.14, 91.49, 56.47, 56.19, 55.98, 55.87. ESI-MS m/z: 389.0 [M−H]−. Anal. calcd. For C21H24O7: C, 64.94; H, 6.23; Found: C, 64.95; H, 6.21.
Preparations of compounds A2-A7, B1-B16 and C1-C16 were similar to A1.
(E)-3-(4-methoxyphenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (A2). A pale yellow solid, 70 % yield, m.p.116-117℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.62 (d, J = 8.6 Hz, 2H), 7.15 (d, J = 16.1 Hz, 1H), 6.96 (d, J = 8.5 Hz, 2H), 6.82 (d, J = 16.0 Hz, 1H), 6.31 (s, 2H), 3.82 (d, J = 15.6 Hz, 6H), 3.71 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.81, 162.23, 161.65, 158.40, 144.25, 130.79, 127.33, 127.31, 114.91, 111.61, 91.48, 56.23, 55.90, 55.80. ESI-MS m/z: 329.0 [M−H]−. Anal. calcd. For C19H20O5: C, 69.50; H, 6.14; Found: C, 69.55; H, 6.11.
(E)-3-(3,4-dimethoxyphenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (A3). A pale yellow solid, 73 % yield, m.p.123-124℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.34 (s, 1H), 7.27 – 7.13 (m, 2H), 7.05 – 6.90 (m, 2H), 6.36 (s, 2H), 3.89 (s, 3H), 3.85 (d, J = 2.5 Hz, 6H), 3.76 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 194.05, 162.16, 158.33, 151.46, 149.42, 144.91, 127.56, 123.47, 111.98, 111.64, 111.00, 91.48, 56.23, 56.05, 56.00, 55.90. ESI-MS m/z: 358.9 [M−H]−. Anal. calcd. For C20H22O6: C, 67.03; H, 6.19; Found: C, 67.05; H, 6.13.
(E)-3-(3,5-dimethoxyphenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (A4). A pale yellow solid, 71 % yield, m.p.114-118℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.62 (d, J = 8.5 Hz, 1H), 7.42 (d, J = 16.1 Hz, 1H), 6.82 (d, J = 16.1 Hz, 1H), 6.64 – 6.53 (m, 2H), 6.30 (s, 2H), 3.87 – 3.78 (m, 9H), 3.70 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.94, 163.29, 162.13, 159.90, 158.38, 139.22, 130.46, 127.29, 115.98, 111.86, 106.73, 98.83, 91.50, 56.25, 56.22, 55.95, 55.89. ESI-MS m/z: 359.0 [M−H]−. Anal. calcd. For C20H22O6: C, 67.03; H, 6.19; Found: C, 67.07; H, 6.15.
(E)-3-(3-hydroxy-4-methoxyphenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (A5). A yellow solid, 69 % yield, m.p.145-146℃. 1H NMR (400 MHz, DMSO‑d6) δ 9.16 (s, 1H), 7.14 – 6.96 (m, 3H), 6.87 (d, J = 8.8 Hz, 1H), 6.62 (d, J = 16.0 Hz, 1H), 6.24 (s, 2H), 3.75 (d, J = 11.9 Hz, 6H), 3.64 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.63, 162.21, 158.39, 150.61, 147.17, 144.69, 127.57, 127.08, 122.11, 114.50, 112.42, 111.61, 91.47, 56.23, 56.06, 55.90. ESI-MS m/z: 345.0 [M−H]−. Anal. calcd. For C19H20O6: C, 66.27; H, 5.85; Found: C, 66.25; H, 5.82.
(E)-3-(2,4,6-trimethoxy-3-methylphenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (A6). A pale yellow solid, 67 % yield, m.p.137-138℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.40 (d, J = 16.3 Hz, 1H), 7.04 (d, J = 16.3 Hz, 1H), 6.52 (s, 1H), 6.30 (s, 2H), 3.93 – 3.83 (m, 9H), 3.70 (s, 6H), 3.48 (s, 3H), 1.96 (s, 3H). 13C NMR (101 MHz, DMSO‑d6) δ 195.49, 162.08, 161.21, 159.57, 159.39, 158.25, 137.02, 129.86, 111.55, 111.43, 109.07, 92.63, 91.29, 61.19, 56.39, 56.30, 56.15, 55.89, 8.77. ESI-MS m/z: 403.0 [M−H]−. Anal. calcd. For C22H26O7: C, 65.66; H, 6.51; Found: C, 65.61; H, 6.55.
(E)-3-phenyl-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (A7). A yellow solid, 60 % yield, m.p.126-128℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.70 – 7.64 (m, 2H), 7.41 (d, J = 5.1 Hz, 3H), 7.22 (d, J = 16.1 Hz, 1H), 6.97 (d, J = 16.1 Hz, 1H), 6.32 (s, 2H), 3.84 (s, 3H), 3.72 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.75, 162.43, 158.55, 144.02, 134.83, 130.94, 129.44, 128.94, 111.44, 91.54, 56.28, 55.94. ESI-MS m/z: 299.0 [M−H]−. Anal. calcd. For C18H18O4: C, 72.47; H, 6.08; Found: C, 72.41; H, 6.05.
(E)-3-(pyridin-2-yl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (B1). A yellow solid, 69 % yield, m.p.93-94℃. 1H NMR (400 MHz, DMSO‑d6) δ 8.63 (d, J = 4.3 Hz, 1H), 7.89 – 7.71 (m, 2H), 7.44 – 7.35 (m, 1H), 7.23 (d, J = 2.2 Hz, 2H), 6.33 (s, 2H), 3.85 (s, 3H), 3.72 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.91, 162.61, 158.61, 153.15, 150.51, 143.10, 137.65, 132.21, 125.14, 125.10, 111.18, 91.52, 56.31, 55.96. ESI-MS m/z: 299.9 [M−H]−. Anal. calcd. For C17H17NO4: C, 68.22; H, 5.72; N, 4.68; Found: C, 68.21; H, 5.75; N, 4.61.
(E)-3-(6-bromopyridin-2-yl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (B2). A yellow solid, 71 % yield, m.p.130-132℃. 1H NMR (400 MHz, DMSO‑d6) δ 8.66 (d, J = 2.5 Hz, 1H), 8.17 – 8.11 (m, 1H), 7.69 (d, J = 8.4 Hz, 1H), 7.29 – 7.10 (m, 2H), 6.32 (s, 2H), 3.84 (d, J = 1.2 Hz, 3H), 3.72 (d, J = 3.2 Hz, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.26, 162.67, 158.73, 151.19, 143.12, 138.79, 138.25, 131.85, 130.61, 128.74, 111.14, 91.55, 56.31, 56.26, 55.97. ESI-MS m/z: 378.0 [M−H]−. Anal. calcd. For C17H16BrNO4: C, 53.99; H, 4.26; N, 3.70; Found: C, 53.94; H, 4.21; N, 3.74.
(E)-3-(5-bromopyridin-2-yl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (B3). A pale yellow solid, 75 % yield, m.p.123-124℃. 1H NMR (400 MHz, DMSO‑d6) δ 8.80 (d, J = 2.3 Hz, 1H), 8.23 – 8.11 (m, 1H), 7.81 (d, J = 8.4 Hz, 1H), 7.38 – 7.18 (m, 2H), 6.37 (s, 2H), 3.89 (s, 3H), 3.78 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.59, 162.70, 158.68, 152.03, 151.27, 141.55, 140.20, 132.77, 126.62, 121.61, 111.10, 91.52, 56.32, 55.97. ESI-MS m/z: 378.8 [M−H]−. Anal. calcd. For C17H16BrNO4: C, 53.99; H, 4.26; N, 3.70; Found: C, 53.96; H, 4.29; N, 3.72.
(E)-3-(4-bromophenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (B4). A pale yellow solid, 75 % yield, m.p.121-122℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.72 – 7.55 (m, 4H), 7.21 (d, J = 16.1 Hz, 1H), 7.00 (d, J = 16.1 Hz, 1H), 6.31 (s, 2H), 3.84 (s, 3H), 3.72 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.53, 162.51, 158.60, 142.45, 134.17, 132.37, 130.88, 130.14, 124.23, 111.38, 91.53, 56.28, 55.94. ESI-MS m/z: 377.0 [M−H]−. Anal. calcd. For C18H17BrO4: C, 57.31; H, 4.54; Found: C, 57.34; H, 4.59.
(E)-3-(thiophen-3-yl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (B5). A yellow solid, 71 % yield, m.p.71-73℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.99 – 7.93 (m, 1H), 7.65 – 7.52 (m, 2H), 7.19 (d, J = 16.0 Hz, 1H), 6.79 (d, J = 16.0 Hz, 1H), 6.31 (s, 2H), 3.83 (s, 3H), 3.71 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 194.43, 162.27, 158.39, 138.19, 130.45, 129.26, 128.27, 126.28, 111.32, 91.49, 56.31, 56.25, 55.91. ESI-MS m/z: 305.0 [M−H]−. Anal. calcd. For C16H16O4S: C, 63.14; H, 5.30; Found: C, 63.16; H, 5.35.
(E)-3-(6-bromo-4H-thiopyran-3-yl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (B6). A yellow solid, 71 % yield, m.p.80-81℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.50 – 7.28 (m, 3H), 6.63 (d, J = 15.8 Hz, 1H), 6.36 (s, 2H), 3.89 (s, 3H), 3.77 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 192.92, 162.52, 158.58, 141.62, 135.88, 133.74, 132.56, 128.53, 116.16, 110.99, 91.53, 56.28, 55.94. ESI-MS m/z: 382.9 [M−H]−. Anal. calcd. For C16H15BrO4S: C, 50.14; H, 3.95; Found: C, 50.16; H, 3.92.
(E)-3-(furan-3-yl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (B7). A yellow solid, 71 % yield, m.p.104-106℃. 1H NMR (400 MHz, DMSO‑d6) δ 8.03 (s, 1H), 7.68 (s, 1H), 7.04 (d, J = 16.0 Hz, 1H), 6.90 (d, J = 2.0 Hz, 1H), 6.61 (d, J = 15.9 Hz, 1H), 6.24 (s, 2H), 3.76 (s, 3H), 3.64 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 194.14, 162.24, 158.33, 146.64, 145.52, 135.38, 129.48, 123.19, 111.21, 108.36, 91.46, 56.23, 55.90. ESI-MS m/z: 289. 0 [M−H]−. Anal. calcd. For C16H16O5: C, 66.66; H, 5.59; Found: C, 66.61; H, 5.56.
(E)-3-(5-bromofuran-3-yl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (B8). A yellow solid, 71 % yield, m.p.107-109℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.03 – 6.81 (m, 2H), 6.70 (d, J = 3.5 Hz, 1H), 6.50 (d, J = 15.9 Hz, 1H), 6.23 (s, 2H), 3.76 (s, 3H), 3.64 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 192.90, 162.51, 158.53, 153.04, 129.53, 126.64, 126.18, 119.33, 115.61, 110.94, 91.49, 56.29, 55.94. ESI-MS m/z: 367.0 [M−H]−. Anal. calcd. For C16H15BrO5: C, 52.34; H, 4.12; Found: C, 52.32; H, 4.17.
(E)-3-(naphthalen-2-yl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (B9). A pale yellow solid, 76 % yield, m.p.142-144℃. 1H NMR (400 MHz, DMSO‑d6) δ 8.15 (s, 1H), 7.99 – 7.85 (m, 4H), 7.61 – 7.52 (m, 2H), 7.38 (d, J = 16.1 Hz, 1H), 7.09 (d, J = 16.0 Hz, 1H), 6.35 (s, 2H), 3.86 (s, 3H), 3.74 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.96, 162.44, 158.53, 144.34, 134.25, 133.38, 132.43, 130.87, 129.76, 129.04, 128.98, 128.14, 127.88, 127.24, 124.36, 111.40, 91.52, 56.29, 55.95. ESI-MS m/z: 349.0 [M−H]−. Anal. calcd. For C22H20O4: C, 75.84; H, 5.79; Found: C, 75.88; H, 5.77.
(E)-3-(quinolin-3-yl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (B10). A yellow solid, 79 % yield, m.p.160-161℃. 1H NMR (400 MHz, DMSO‑d6) δ 9.23 (d, J = 2.2 Hz, 1H), 8.69 (d, J = 2.1 Hz, 1H), 8.08 – 7.95 (m, 2H), 7.84 – 7.78 (m, 1H), 7.69 – 7.62 (m, 1H), 7.44 (d, J = 16.2 Hz, 1H), 7.28 (d, J = 16.2 Hz, 1H), 6.35 (s, 2H), 3.86 (s, 3H), 3.74 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.57, 162.61, 158.68, 150.34, 148.35, 140.75, 136.49, 131.17, 130.99, 129.25, 129.18, 128.09, 127.81, 111.24, 91.55, 56.32, 55.97. ESI-MS m/z: 349.9 [M−H]−. Anal. calcd. For C21H19NO4: C, 72.19; H, 5.48; N, 4.01; Found: C, 72.16; H, 5.46; N, 4.06.
(E)-3-(1-methyl-1H-indol-3-yl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (B11). A yellow solid, 72 % yield, m.p.181-182℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.91 (s, 1H), 7.83 (d, J = 7.9 Hz, 1H), 7.54 (d, J = 8.2 Hz, 1H), 7.38 (d, J = 16.0 Hz, 1H), 7.26 (dt, J = 27.1, 7.4 Hz, 2H), 6.76 (d, J = 16.0 Hz, 1H), 6.32 (s, 2H), 3.82 (d, J = 15.7 Hz, 6H), 3.71 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.64, 161.95, 158.31, 139.00, 138.45, 136.36, 125.79, 124.07, 123.15, 121.82, 120.46, 111.89, 111.35, 91.46, 56.19, 55.88, 33.39. ESI-MS m/z: 352.0 [M−H]−. Anal. calcd. For C21H21NO4: C, 71.78; H, 6.02; N, 3.99; Found: C, 71.76; H, 6.06; N, 3.96.
(E)-3-([1,1′-biphenyl]-4-yl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (B12). A yellow solid, 78 % yield, m.p.109-110℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.83 – 7.68 (m, 6H), 7.54 – 7.38 (m, 3H), 7.27 (d, J = 16.1 Hz, 1H), 7.01 (d, J = 16.1 Hz, 1H), 6.33 (s, 2H), 3.85 (s, 3H), 3.73 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.69, 162.45, 158.58, 143.49, 142.37, 139.60, 133.97, 130.43, 129.63, 129.51, 129.43, 129.40, 128.47, 127.58, 127.14, 111.48, 91.53, 56.29, 55.94. ESI-MS m/z: 375.0 [M−H]−. Anal. calcd. For C24H22O4: C, 76.99; H, 5.92; Found: C, 76.96; H, 5.96.
(E)-3-(4′-fluoro-[1,1′-biphenyl]-4-yl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (B13). A yellow solid. 74 % yield, m.p.137-138℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.86 – 7.68 (m, 6H), 7.39 – 7.21 (m, 3H), 7.02 (d, J = 16.1 Hz, 1H), 6.33 (s, 2H), 3.85 (s, 3H), 3.73 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.68, 162.45, 161.40, 158.58, 143.41, 141.29, 136.10, 133.94, 129.64, 129.44, 129.26, 129.17, 127.52, 116.42, 116.21, 111.48, 91.54, 56.28, 55.94. ESI-MS m/z: 393.0 [M−H]−. Anal. calcd. For C24H21FO4: C, 73.46; H, 5.39; Found: C, 73.49; H, 5.34.
(E)-3-(3-oxo-3-(2,4,6-trimethoxyphenyl)prop-1-en-1-yl)quinolin-2(1H)-one (B14). A pale yellow solid, 71 % yield, m.p.170-172℃. 1H NMR (400 MHz, DMSO‑d6) δ 12.06 (s, 1H), 8.44 (s, 1H), 7.69 (d, J = 7.9 Hz, 1H), 7.55 (t, J = 7.7 Hz, 1H), 7.44 – 7.26 (m, 3H), 7.21 (t, J = 7.5 Hz, 1H), 6.33 (s, 2H), 3.85 (s, 3H), 3.72 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 194.56, 162.37, 161.18, 158.43, 141.24, 139.43, 139.34, 132.17, 131.44, 129.22, 126.12, 122.73, 119.54, 115.53, 111.30, 91.50, 56.28, 55.94. ESI-MS m/z: 365.9 [M−H]−. Anal. calcd. For C21H19NO5: C, 69.03; H, 5.24; N, 3.83; Found: C, 69.06; H, 5.26; N, 3.81.
(E)-6-methyl-3-(3-oxo-3-(2,4,6-trimethoxyphenyl)prop-1-en-1-yl)quinolin-2(1H)-one (B15). A yellow solid, 72 % yield, m.p.198-200℃. 1H NMR (400 MHz, DMSO‑d6) δ 12.06 (s, 1H), 8.35 (s, 1H), 7.48 (s, 1H), 7.45 – 7.29 (m, 3H), 7.29 – 7.18 (m, 1H), 6.32 (s, 2H), 3.84 (s, 3H), 3.71 (s, 6H), 2.34 (s, 3H). 13C NMR (101 MHz, DMSO‑d6) δ 194.60, 162.37, 161.06, 158.44, 141.02, 139.43, 137.43, 133.62, 131.81, 131.27, 128.53, 126.00, 119.50, 115.44, 111.30, 91.48, 56.27, 55.93, 20.83. ESI-MS m/z: 380.0 [M−H]−. Anal. calcd. For C22H21NO5: C, 69.03; H, 5.24; N, 3.83; Found: C, 69.06; H, 5.26; N, 3.81.
(E)-6-fluoro-3-(3-oxo-3-(2,4,6-trimethoxyphenyl)prop-1-en-1-yl)quinolin-2(1H)-one(B16). An Orange solid, 70 % yield, m.p.171-172℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.91 (s, 1H), 7.25 (d, J = 4.0 Hz, 2H), 7.18 – 7.02 (m, 3H), 6.22 (s, 2H), 3.75 (s, 3H), 3.61 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 195.03, 166.60, 162.03, 158.60, 158.25, 143.72, 137.68, 129.89, 125.42, 120.79, 120.29, 120.06, 117.30, 111.79, 111.61, 91.45, 56.20, 55.89. ESI-MS m/z: 384.0 [M−H]−. Anal. calcd. For C22H21NO5: C, 65.79; H, 4.73; N, 3.65; Found: C, 65.76; H, 4.76; N, 3.61.
(E)-3-(4-(cyclopropylmethoxy)phenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (C1). A yellow solid, 70 % yield, m.p.92-93℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.59 (d, J = 8.5 Hz, 2H), 7.15 (d, J = 16.0 Hz, 1H), 6.94 (d, J = 8.5 Hz, 2H), 6.81 (d, J = 16.0 Hz, 1H), 6.30 (s, 2H), 3.88 – 3.82 (m, 5H), 3.71 (s, 6H), 1.31 – 1.15 (m, 1H), 0.68 – 0.49 (m, 2H), 0.41 – 0.28 (m, 2H). 13C NMR (101 MHz, DMSO‑d6) δ 193.76, 162.21, 161.07, 158.39, 144.26, 130.77, 127.21, 127.16, 115.36, 111.64, 91.48, 72.70, 56.23, 55.90, 10.50, 3.58. ESI-MS m/z: 368.9 [M−H]−. Anal. calcd. For C22H24O5: C, 71.72; H, 6.57; Found: C, 71.76; H, 6.53.
(E)-3-(4-(cyclopentylmethoxy)phenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (C2). A yellow solid, 74 % yield, m.p.78-80℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.59 (d, J = 8.5 Hz, 2H), 7.15 (d, J = 16.0 Hz, 1H), 6.94 (d, J = 8.4 Hz, 2H), 6.81 (d, J = 16.0 Hz, 1H), 6.31 (s, 2H), 3.95 – 3.75 (m, 6H), 3.71 (s, 3H), 3.36 (s, 2H), 2.36 – 2.24 (m, 1H), 1.81 – 1.72 (m, 2H), 1.64 – 1.50 (m, 4H), 1.36 – 1.26 (m, 2H). 13C NMR (101 MHz, DMSO‑d6) δ 193.74, 162.22, 161.25, 158.40, 144.24, 130.76, 127.20, 115.37, 111.66, 91.48, 72.22, 56.22, 55.89, 38.90, 29.41, 25.39. ESI-MS m/z: 397.0 [M−H]−. Anal. calcd. For C24H28O5: C, 72.71; H, 7.12; Found: C, 72.76; H, 7.13.
(E)-3-(4-(cyclohexylmethoxy)phenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (C3). A pale yellow solid, 74 % yield, m.p.100-101℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.59 (d, J = 8.4 Hz, 2H), 7.15 (d, J = 16.0 Hz, 1H), 6.94 (d, J = 8.4 Hz, 2H), 6.81 (d, J = 16.0 Hz, 1H), 6.31 (s, 2H), 3.82 (d, J = 12.3 Hz, 5H), 3.71 (s, 6H), 1.85 – 1.58 (m, 6H), 1.32 – 0.98 (m, 5H). 13C NMR (101 MHz, DMSO‑d6) δ 193.74, 162.21, 161.24, 158.39, 144.25, 130.77, 127.20, 127.17, 115.36, 111.64, 91.47, 73.29, 56.22, 55.89, 37.44, 29.63, 26.48, 25.70. ESI-MS m/z: 410.9 [M−H]−. Anal. calcd. For C25H30O5: C, 73.15; H, 7.37; Found: C, 73.16; H, 7.33.
(E)-3-(4-(benzyloxy)phenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (C4). A yellow solid, 72 % yield, m.p.132-134℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.62 (d, J = 8.5 Hz, 2H), 7.49 – 7.30 (m, 5H), 7.15 (d, J = 16.1 Hz, 1H), 7.04 (d, J = 8.6 Hz, 2H), 6.82 (d, J = 16.1 Hz, 1H), 6.31 (s, 2H), 5.16 (s, 2H), 3.84 (s, 3H), 3.71 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.80, 162.24, 160.70, 158.41, 144.16, 137.13, 130.78, 128.93, 128.41, 128.20, 127.54, 127.41, 115.74, 111.59, 91.47, 69.78, 56.23, 55.90. ESI-MS m/z: 404.8 [M−H]−. Anal. calcd. For C25H24O5: C, 74.24; H, 5.98; Found: C, 74.26; H, 5.93.
(E)-3-(4-((4-bromobenzyl)oxy)phenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (C5). A yellow solid, 77 % yield, m.p.148-149℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.65 – 7.58 (m, 4H), 7.41 (d, J = 8.4 Hz, 2H), 7.15 (d, J = 16.0 Hz, 1H), 7.03 (d, J = 8.5 Hz, 2H), 6.83 (d, J = 16.0 Hz, 1H), 6.30 (s, 2H), 5.15 (s, 2H), 3.83 (s, 3H), 3.71 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.75, 162.24, 160.47, 158.42, 136.66, 131.86, 130.78, 130.29, 127.51, 121.50, 115.77, 91.51, 68.96, 56.25, 55.91. ESI-MS m/z: 482.9 [M−H]−. Anal. calcd. For C25H23BrO5: C, 62.12; H, 4.80; Found: C, 62.16; H, 4.83.
(E)-3-(4-(pyridin-2-ylmethoxy)phenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (C6). A yellow solid, 70 % yield, m.p.132-133℃. 1H NMR (400 MHz, DMSO‑d6) δ 8.59 (d, J = 4.8 Hz, 1H), 7.84 (t, J = 7.6 Hz, 1H), 7.63 (d, J = 8.4 Hz, 2H), 7.51 (d, J = 7.8 Hz, 1H), 7.40 – 7.30 (m, 1H), 7.15 (d, J = 16.0 Hz, 1H), 7.05 (d, J = 8.3 Hz, 2H), 6.83 (d, J = 16.0 Hz, 1H), 6.31 (s, 2H), 5.24 (s, 2H), 3.83 (s, 3H), 3.71 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.77, 162.24, 160.47, 158.42, 156.75, 149.63, 144.05, 137.50, 130.81, 127.75, 127.54, 123.54, 122.23, 115.73, 111.60, 91.48, 70.81, 56.23, 55.90. ESI-MS m/z: 406.0 [M−H]−. Anal. calcd. For C24H23NO5: C, 71.10; H, 5.72; N, 3.45; Found: C, 71.16; H, 5.73; N, 3.41.
(E)-3-(4-(thiophen-3-ylmethoxy)phenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (C7). A yellow solid, 76 % yield, m.p.117-118℃. 1H NMR (400 MHz, DMSO‑d6) δ 7.65 – 7.53 (m, 4H), 7.22 – 7.12 (m, 2H), 7.03 (d, J = 8.5 Hz, 2H), 6.83 (d, J = 16.0 Hz, 1H), 6.31 (s, 2H), 5.15 (s, 2H), 3.84 (s, 3H), 3.71 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.76, 162.24, 160.64, 158.42, 144.13, 137.99, 130.76, 128.06, 127.52, 127.41, 127.20, 124.70, 115.68, 111.64, 91.50, 65.39, 56.24, 55.91. ESI-MS m/z: 411.0 [M−H]−. Anal. calcd. For C23H22O5S: C, 67.30; H, 5.40; Found: C, 67.34; H, 5.43.
(E)-3-(4-(naphthalen-2-ylmethoxy)phenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (C8). A yellow solid, 74 % yield, m.p.124-126℃. 1H NMR (400 MHz, DMSO‑d6) δ 8.03 – 7.88 (m, 4H), 7.69 – 7.46 (m, 5H), 7.20 – 7.05 (m, 3H), 6.82 (d, J = 16.0 Hz, 1H), 6.30 (s, 2H), 5.34 (s, 2H), 3.83 (s, 3H), 3.70 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.81, 162.24, 160.71, 158.41, 144.15, 134.77, 133.21, 133.02, 130.81, 128.59, 128.25, 128.08, 127.60, 127.44, 126.84, 126.81, 126.67, 126.12, 115.83, 111.59, 91.47, 69.90, 56.23, 55.90. ESI-MS m/z: 455.0 [M−H]−. Anal. calcd. For C29H26O5: C, 76.63; H, 5.77; Found: C, 76.66; H, 5.73.
(E)-3-(4-(quinolin-3-ylmethoxy)phenyl)-1-(2,4,6-trimethoxyphenyl)prop-2-en-1-one (C9). A yellow solid, 71 % yield, m.p.191-192℃. 1H NMR (400 MHz, DMSO‑d6) δ 11.97 (s, 1H), 8.00 (s, 1H), 7.75 – 7.60 (m, 3H), 7.50 (t, J = 7.7 Hz, 1H), 7.33 (d, J = 8.3 Hz, 1H), 7.22 – 7.13 (m, 2H), 7.07 (d, J = 8.4 Hz, 2H), 6.83 (d, J = 16.1 Hz, 1H), 6.31 (s, 2H), 5.06 (s, 2H), 3.83 (s, 3H), 3.71 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 194.14, 162.23, 160.65, 158.41, 152.15, 152.14, 144.12, 138.49, 137.28, 135.40, 130.87, 128.42, 127.48, 124.55, 122.90, 122.55, 122.37, 119.36, 115.66, 91.48, 65.34, 56.24, 55.91. ESI-MS m/z: 456.0 [M−H]−. Anal. calcd. For C28H25NO5: C, 73.83; H, 5.53; N, 3.08; Found: C, 73.86; H, 5.57; N, 3.03.
(E)-3-((4-(3-oxo-3-(2,4,6-trimethoxyphenyl)prop-1-en-1-yl)phenoxy)methyl)quinolin-2(1H)-one (C10). A yellow solid, 71 % yield, m.p.196-197℃. 1H NMR (400 MHz, DMSO‑d6) δ 12.02 (s, 1H), 8.01 (s, 1H), 7.76 – 7.59 (m, 3H), 7.51 (t, J = 7.7 Hz, 1H), 7.34 (d, J = 8.2 Hz, 1H), 7.23 – 7.01 (m, 4H), 6.83 (d, J = 16.0 Hz, 1H), 6.31 (s, 2H), 5.06 (s, 2H), 3.83 (s, 3H), 3.71 (s, 6H). 13C NMR (101 MHz, DMSO‑d6) δ 193.78, 162.24, 161.38, 160.58, 158.41, 144.11, 138.71, 137.41, 130.87, 128.64, 128.45, 127.70, 127.49, 122.48, 119.32, 115.65, 115.48, 111.60, 91.48, 65.29, 56.24, 55.91. ESI-MS m/z: 471.9 [M−H]−. Anal. calcd. For C28H25NO6: C, 71.33; H, 5.34; N, 2.97; Found: C, 71.36; H, 5.37; N, 2.92.
4.2. Biological activities evaluation
4.2.1. Reagents and materials
The Hep2, A549, MCF-7, Hela, and L-02 cells were purchased from Shanghai Institute of Biochemistry, Chinese Academy of Sciences. The target compounds and positive control 5-fluorouracil were dissolved with DMSO at a concentration of 20 mM, then, stored at −20 °C. Dulbecco's Modified Eagle Medium (DMEM), trypsin, Penicillin-Streptomycin solution and fetal bovine serum (FBS) were purchased from GIBCO, phosphate buffer (PBS) was purchased from Hyclone. Hoechst 33,258 staining solution and dimethyl sulfoxide were purchased from Beyotime. Annexin V-FITC/PI apoptosis detection reagent was purchased from CWBIO.
4.2.2. MTT cell viability assay
The anti-proliferation activities of 2,4, 6-trimethoxychalcone derivatives A1-A7, B1-B16, C1-C10 against four tumor cells (Hela, A549, HepG2, McF-7) and one normal cell (L-02) were evaluated by MTT assay. All the cells were cultured in DMEM medium and 10 % fetal bovine serum and 1 % penicillin–streptomycin solution were added at 37 °C under 5 % CO2. The cells were inoculated into 96-well plates with 2 × 104 cells per well, then, treated with the target compounds and positive control at various concentrations for 48 h. Then, 10 μL of re-configured MTT (5 mg/mL in PBS) was added to each well under dark conditions, and the cells were cultivated at 37 °C for 4 h. The old culture medium was discarded, and 100 μL DMSO was added. The absorbance was measured with a microplate reader (490 nm). Each experiment was repeated three times. GraphPad Prism (8.0.2 version) was used to process experimental data.
4.2.3. Migration ability assay
Hela and MCF-7 cells were inoculated into six-well plates with a density of 1 × 105-2 × 105 cells per well and cultivated until 90 % confluence, the cell monolayer was scratched vertically with the tip of a 200 μL pipetting spear. Then, cells were washed three times with PBS and incubated with different concentrations of compound B3 and positive control for 24 or 48 h. The cells were photographed at 0 h, 24 h and 48 h along the scrape line under an inverted microscope (Olympus, Tokyo, Japan). The area of cell migration was measured using Image J software (version 1.80), and calculated the cell migration rate.
4.2.4. Colony formation assay
Hela and MCF-7 cells were inoculated into six-well plates with a density of 1500 cells per well and cultivated in a regular culture medium for 48 h. Cells were exposed to compound B3 or 5-Fu at indicated concentrations for 7 days. Then, they were washed twice with PBS, fixed with paraformaldehyde for 30 min, and dyed with crystal violet staining solution for 30 min. Finally, cells were washed with PBS. The colony formation was photographed by microscope.
4.2.5. Hoechst 33,258 staining assay
Hela cells were inoculated into twelve-well plates with a density of 1 × 104 per well and cultivated in a regular culture medium for 24 h. Cells were exposed to compound B3 or 5-Fu at indicated concentrations for 24 h. Then, these cells were washed twice with PBS and 180 μL Hoechst 33,258 staining solution was added in each well. The plate was placed in an incubator for 30 min. Photographs were taken with the fluorescence microscope after the cells were washed by PBS.
4.2.6. Annexin V-FITC/PI apoptosis assay
Hela cells were inoculated into six-well plates with a density of 2 × 105 per well and cultivated in a regular culture medium for 24 h. Cells were exposed to compound B3 or 5-Fu at specified concentrations for 24 h and then, they were collected and washed with PBS. 5 μL Annexin V-FITC and 10 μL PI were added to each well in a dark place for 15 min. The processes were conducted according to the kit manual.
4.3. ADME prediction
The physicochemical properties, ADME parameters and the prediction of drug properties of the target compounds were obtained through the SwissADME website (https://www.swissadme.ch) provided free of charge by the Swiss Institute of Bioinformatics (SIB).
4.4. Molecular docking
The Chem3D software (19.0.0.22 Version) was used to process and construct the 3D structure of small molecules, which were saved into mol2 format and then converted into PDBQT format with openbable for stand-by. The X-ray crystal structures of proteins were derived from the PDB Database. After the target protein molecules were treated in AutoDock software (1.5.6 Version) by dehydration, hydrogenation, extracting the ligand and retaining the single chain, they were stored in PDBQT format. The position and size of the docking box were determined by the docking site of the original ligand in the crystal complex. Then docking binding energy was calculated, the lower the energy was, the more stable the binding was. Finally, Pymol (2.3.1 Version) software was used for visual analysis of the docking results.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The work was supported by Innovation and strengthening project of Guangdong Pharmaceutical University-Special Innovation Project of Guangdong Education Department (Natural Science) (2020KZDZX1131, 2022ZDZX2030) and Special Projects in Key Fields of General Colleges and Universities in Guangdong Provinceand ‘Guangdong Province Graduate Education Innovation Program in 2020 and 2021 (2021JGXM071)’, Scientific Research Project of Guangdong Provincial Bureau of traditional Chinese Medicine (20231205).
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsps.2022.11.006.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Abou-Zied H.A., Youssif B.G.M., Mohamed M.F.A., Hayallah A.M., Abdel-Aziz M. EGFR inhibitors and apoptotic inducers: Design, synthesis, anticancer activity and docking studies of novel xanthine derivatives carrying chalcone moiety as hybrid molecules. Bioorg. Chem. 2019;89 doi: 10.1016/j.bioorg.2019.102997. [DOI] [PubMed] [Google Scholar]
- Ahmed M.F., Santali E.Y., El-Haggar R. Novel piperazine-chalcone hybrids and related pyrazoline analogues targeting VEGFR-2 kinase; design, synthesis, molecular docking studies, and anticancer evaluation. J. Enzyme Inhib. Med. Chem. 2021;36:307–318. doi: 10.1080/14756366.2020.1861606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmaoglu S., Ozcan S., Balcioglu S., Gencel M., Noma S.A.A., Essiz S., Ates B., Algul O. Synthesis, biological evaluation and molecular docking studies of bis-chalcone derivatives as xanthine oxidase inhibitors and anticancer agents. Bioorg. Chem. 2019;91:103149. doi: 10.1016/j.bioorg.2019.103149. [DOI] [PubMed] [Google Scholar]
- Cheng P., Yang L., Huang X., Wang X., Gong M. Chalcone hybrids and their antimalarial activity. Arch. Pharm. 2020;353(4):1900350. doi: 10.1002/ardp.201900350. [DOI] [PubMed] [Google Scholar]
- Chinthala Y., Thakur S., Tirunagari S., Chinde S., Domatti A.K., Arigari N.K., Srinivas K.V.N.S., Alam S., Jonnala K.K., Khan F. Synthesis, docking and ADMET studies of novel chalcone triazoles for anti-cancer and anti-diabetic activity. Eur. J. Med. Chem. 2015;93:564–573. doi: 10.1016/j.ejmech.2015.02.027. [DOI] [PubMed] [Google Scholar]
- Dan N.T., Quang H.D., Van Truong V., Huu Nghi D., Cuong N.M., Cuong T.D., Toan T.Q., Bach L.G., Anh N.H.T., Mai N.T. Design, synthesis, structure, in vitro cytotoxic activity evaluation and docking studies on target enzyme GSK-3β of new indirubin-3'-oxime derivatives. Sci. Rep. 2020;10(1):11429. doi: 10.1038/s41598-020-68134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Liu X., Zhao T., Wang L., Li H., Zhang S., Li X., Bai X., Zhang Y., Yang B. Apoptosis-inducing effects and growth inhibitory of a novel chalcone, in human hepatic cancer cells and lung cancer cells. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;105:195–203. doi: 10.1016/j.biopha.2018.05.126. [DOI] [PubMed] [Google Scholar]
- El-Wakil M.H., Khattab S.N., El-Yazbi A.F., El-Nikhely N., Soffar A., Khalil H.H. New chalcone-tethered 1,3,5-triazines potentiate the anticancer effect of cisplatin against human lung adenocarcinoma A549 cells by enhancing DNA damage and cell apoptosis. Bioorg. Chem. 2020;105 doi: 10.1016/j.bioorg.2020.104393. [DOI] [PubMed] [Google Scholar]
- Farghaly T.A., Masaret G.S., Muhammad Z.A., Harras M.F. Discovery of thiazole-based-chalcones and 4-hetarylthiazoles as potent anticancer agents: Synthesis, docking study and anticancer activity. Bioorg. Chem. 2020;98 doi: 10.1016/j.bioorg.2020.103761. [DOI] [PubMed] [Google Scholar]
- Gao F., Huang G., Xiao J. Chalcone hybrids as potential anticancer agents: Current development, mechanism of action, and structure-activity relationship. Med. Res. Rev. 2020;40(5):2049–2084. doi: 10.1002/med.21698. [DOI] [PubMed] [Google Scholar]
- Gomes M.N., Braga R.C., Grzelak E.M., Neves B.J., Muratov E., Ma R., Klein L.L., Cho S., Oliveira G.R., Franzblau S.G., Andrade C.H. QSAR-driven design, synthesis and discovery of potent chalcone derivatives with antitubercular activity. Eur. J. Med. Chem. 2017;137:126–138. doi: 10.1016/j.ejmech.2017.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry E.J., Bird S.J., Gowland P., Collins M., Cassella J.P. Ferrocenyl chalcone derivatives as possible antimicrobial agents. J. Antibiot. 2020;73(5):299–308. doi: 10.1038/s41429-020-0280-y. [DOI] [PubMed] [Google Scholar]
- Kamal A., Srinivasulu V., Nayak V.L., Sathish M., Shankaraiah N., Bagul C., Reddy N.V.S., Rangaraj N., Nagesh N. Design and synthesis of C3-pyrazole/chalcone-linked beta-carboline hybrids: antitopoisomerase I, DNA-interactive, and apoptosis-inducing anticancer agents. ChemMedChem. 2014;9(9):2084–2098. doi: 10.1002/cmdc.201300406. [DOI] [PubMed] [Google Scholar]
- Kommidi D.R., Pagadala R., Rana S., Singh P., Shintre S.A., Koorbanally N.A., Jonnalagadda S.B., Moodley B. Novel carbapenem chalcone derivatives: synthesis, cytotoxicity and molecular docking studies. Org. Biomol. Chem. 2015;13(14):4344–4350. doi: 10.1039/c5ob00197h. [DOI] [PubMed] [Google Scholar]
- Li L., Jiang S., Li X., Liu Y., Su J., Chen J. Recent advances in trimethoxyphenyl (TMP) based tubulin inhibitors targeting the colchicine binding site. Eur. J. Med. Chem. 2018;151:482–494. doi: 10.1016/j.ejmech.2018.04.011. [DOI] [PubMed] [Google Scholar]
- Luzak B., Kassassir H., Rój E., Stanczyk L., Watala C., Golanski J. Xanthohumol from hop cones (Humulus lupulus L.) prevents ADP-induced platelet reactivity. Arch. Physiol. Biochem. 2017;123(1):54–60. doi: 10.1080/13813455.2016.1247284. [DOI] [PubMed] [Google Scholar]
- Ma Y., Xu B., Yu J., Huang L., Zeng X., Shen X., Ren C., Ben-David Y., Luo H. Fli-1 Activation through Targeted Promoter Activity Regulation Using a Novel 3', 5'-diprenylated Chalcone Inhibits Growth and Metastasis of Prostate Cancer Cells. Int. J. Mol. Sci. 2020;21(6):2216. doi: 10.3390/ijms21062216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra D.K., Bharti S.K., Asati V. Chalcone Derivatives: Anti-inflammatory Potential and Molecular Targets Perspectives. Curr. Top. Med. Chem. 2017;17(28):3146–3169. doi: 10.2174/1568026617666170914160446. [DOI] [PubMed] [Google Scholar]
- Mohammed H.H.H., Abbas S.H., Hayallah A.M., Abuo-Rahma G.-D., Mostafa Y.A. Novel urea linked ciprofloxacin-chalcone hybrids having antiproliferative topoisomerases I/II inhibitory activities and caspases-mediated apoptosis. Bioorg. Chem. 2021;106:104422. doi: 10.1016/j.bioorg.2020.104422. [DOI] [PubMed] [Google Scholar]
- Mohassab A.M., Hassan H.A., Abdelhamid D., Gouda A.M., Youssif B.G.M., Tateishi H., Fujita M., Otsuka M., Abdel-Aziz M. Design and synthesis of novel quinoline/chalcone/1,2,4-triazole hybrids as potent antiproliferative agent targeting EGFR and BRAF(V600E) kinases. Bioorg. Chem. 2021;106 doi: 10.1016/j.bioorg.2020.104510. [DOI] [PubMed] [Google Scholar]
- Muchtaridi M., Syahidah H., Subarnas A., Yusuf M., Bryant S., Langer T. Molecular Docking and 3D-Pharmacophore Modeling to Study the Interactions of Chalcone Derivatives with Estrogen Receptor Alpha. Pharmaceuticals (Basel, Switzerland). 2017;10(4):81. doi: 10.3390/ph10040081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Wang W., Li J., Lei Y.u., Zhao Y., Yang W., Zhao C., Lin B., Song S., Wang S. A novel structural class of coumarin-chalcone fibrates as PPARα/γ agonists with potent antioxidant activities: Design, synthesis, biological evaluation and molecular docking studies. Eur. J. Med. Chem. 2017;138:212–220. doi: 10.1016/j.ejmech.2017.06.033. [DOI] [PubMed] [Google Scholar]
- Peerzada M.N., Khan P., Ahmad K., Hassan M.I., Azam A. Synthesis, characterization and biological evaluation of tertiary sulfonamide derivatives of pyridyl-indole based heteroaryl chalcone as potential carbonic anhydrase IX inhibitors and anticancer agents. Eur. J. Med. Chem. 2018;155:13–23. doi: 10.1016/j.ejmech.2018.05.034. [DOI] [PubMed] [Google Scholar]
- Pfeiffer P., Yilmaz M., Möller S., Zitnjak D., Krogh M., Petersen L.N., Poulsen L.Ø., Winther S.B., Thomsen K.G., Qvortrup C. TAS-102 with or without bevacizumab in patients with chemorefractory metastatic colorectal cancer: an investigator-initiated, open-label, randomised, phase 2 trial. Lancet Oncol. 2020;21(3):412–420. doi: 10.1016/s1470-2045(19)30827-7. [DOI] [PubMed] [Google Scholar]
- Rahimzadeh Oskuei S., Mirzaei S., Reza Jafari-Nik M., Hadizadeh F., Eisvand F., Mosaffa F., Ghodsi R. Design, synthesis and biological evaluation of novel imidazole-chalcone derivatives as potential anticancer agents and tubulin polymerization inhibitors. Bioorg. Chem. 2021;112:104904. doi: 10.1016/j.bioorg.2021.104904. [DOI] [PubMed] [Google Scholar]
- Rocha S., Ribeiro D., Fernandes E., Freitas M. A Systematic Review on Anti-diabetic Properties of Chalcones. Curr. Med. Chem. 2020;27(14):2257–2321. doi: 10.2174/0929867325666181001112226. [DOI] [PubMed] [Google Scholar]
- Roussel E., Tran-Nguyen V.-K., Bouhedjar K., Dems M.A., Belaidi A., Matougui B., Peres B., Azioune A., Renaudet O., Falson P., Boumendjel A. Optimization of the chromone scaffold through QSAR and docking studies: Identification of potent inhibitors of ABCG2. Eur. J. Med. Chem. 2019;184:111772. doi: 10.1016/j.ejmech.2019.111772. [DOI] [PubMed] [Google Scholar]
- Sang Z., Wang K., Zhang P., Shi J., Liu W., Tan Z. Design, synthesis, in-silico and biological evaluation of novel chalcone derivatives as multi-function agents for the treatment of Alzheimer's disease. Eur. J. Med. Chem. 2019;180:238–252. doi: 10.1016/j.ejmech.2019.07.021. [DOI] [PubMed] [Google Scholar]
- Shin J.W., Chun K.S., Kim D.H., Kim S.J., Kim S.H., Cho N.C., Na H.K., Surh Y.J. Curcumin induces stabilization of Nrf2 protein through Keap1 cysteine modification. Biochem. Pharmacol. 2020;173 doi: 10.1016/j.bcp.2020.113820. [DOI] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Traboulsi H., Cloutier A., Boyapelly K., Bonin M.-A., Marsault É., Cantin A.M., Richter M.V. The Flavonoid Isoliquiritigenin Reduces Lung Inflammation and Mouse Morbidity during Influenza Virus Infection. Antimicrob. Agents Chemother. 2015;59(10):6317–6327. doi: 10.1128/AAC.01098-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdameri G., Gauthier C., Terreux R., Kachadourian R., Day B.J., Winnischofer S.M.B., Rocha M.E.M., Frachet V., Ronot X., Di Pietro A., Boumendjel A. Investigation of chalcones as selective inhibitors of the breast cancer resistance protein: critical role of methoxylation in both inhibition potency and cytotoxicity. J. Med. Chem. 2012;55(7):3193–3200. doi: 10.1021/jm2016528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanaparthi S., Bantu R., Jain N., Janardhan S., Nagarapu L. Synthesis and anti-proliferative activity of a novel 1,2,3-triazole tethered chalcone acetamide derivatives. Bioorg. Med. Chem. Lett. 2020;30(16) doi: 10.1016/j.bmcl.2020.127304. [DOI] [PubMed] [Google Scholar]
- Wang Y.u., Zhang W., Dong J., Gao J. Design, synthesis and bioactivity evaluation of coumarin-chalcone hybrids as potential anticancer agents. Bioorg. Chem. 2020;95:103530. doi: 10.1016/j.bioorg.2019.103530. [DOI] [PubMed] [Google Scholar]
- Yan J., Chen J., Zhang S., Hu J., Huang L., Li X. Synthesis, evaluation, and mechanism study of novel indole-chalcone derivatives exerting effective antitumor activity through microtubule destabilization in vitro and in vivo. J. Med. Chem. 2016;59(11):5264–5283. doi: 10.1021/acs.jmedchem.6b00021. [DOI] [PubMed] [Google Scholar]
- Zhang B., Duan D., Ge C., Yao J., Liu Y., Li X., Fang J. Synthesis of xanthohumol analogues and discovery of potent thioredoxin reductase inhibitor as potential anticancer agent. J. Med. Chem. 2015;58(4):1795–1805. doi: 10.1021/jm5016507. [DOI] [PubMed] [Google Scholar]
- Zhou B., Xing C. Diverse Molecular Targets for Chalcones with Varied Bioactivities. Med. Chem. 2015;5(8):388–404. doi: 10.4172/2161-0444.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Yang S., Li S.M. Naturally occurring prenylated chalcones from plants: structural diversity, distribution, activities and biosynthesis. Nat. Prod. Rep. 2021;38(12):2236–2260. doi: 10.1039/d0np00083c. [DOI] [PubMed] [Google Scholar]
- Zhu Y.-J., Zheng B.o., Wang H.-Y., Chen L. New knowledge of the mechanisms of sorafenib resistance in liver cancer. Acta Pharmacol. Sin. 2017;38(5):614–622. doi: 10.1038/aps.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang C., Zhang W., Sheng C., Zhang W., Xing C., Miao Z. Chalcone: A Privileged Structure in Medicinal Chemistry. Chem. Rev. 2017;117(12):7762–7810. doi: 10.1021/acs.chemrev.7b00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.