Abstract
Cancer has become one of the common causes of mortality around the globe due to mutations in the genome which allows rapid growth of cells uncontrollably without repairing DNA errors. Cancers could arise due alterations in DNA repair mechanisms (errors in mismatch repair genes), activation of oncogenes and inactivation of tumor suppressor genes. Each cancer type is different and each individual has a unique genetic change which leads them to cancer. Studying genetic and epigenetic alterations in the genome leads to understanding the underlying features. CAR T therapy over other immunotherapies such as monoclonal antibodies, immune checkpoint inhibitors, cancer vaccines and adoptive cell therapies has been widely used to treat cancer in recent days and gene editing has now become one of the promising treatments for many genetic diseases. This tool allows scientists to change the genome by adding, removing or altering genetic material of an organism. Due to advance in genetics and novel molecular techniques such as CRISPR, TALEN these genes can be edited in such a way that their original function could be replaced which in turn improved the treatment possibilities and can be used against malignancies and even cure cancer in future along with CAR T cell therapy due to the specific recognition and attacking of tumor.
Keywords: Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), Chimeric Antigen Receptor (CAR) T cell therapy, Cancer, Immunotherapy, Gene therapy, Gene editing
Introduction
Genetic disorders are not easy to cure like other diseases. Gene therapy has become the novel tool to treat and cure genetic diseases by improving the body’s ability to fight against diseases either by replacing the disease causing gene with a healthy gene, inactivating disease causing gene, or by introducing modified gene which could function normally. Gene therapy could be done using prokaryotic vectors, plasmid vectors, or other human gene editing tools. This review discusses the ability of gene therapy with immunotherapy specially with CAR T cells to treat hematological malignancies. With the promise of CRISPR as an effective editing tool and CAR T therapy as the latest effective immunotherapy technology, this combination can be used as a weapon to cure many malignancies [1].
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) discovered based on its initial findings by the discovery of E. coli type II acquired immune system, non-repeating, palindromic sequences by [2]. This is a cost efficient genome editing tool used by scientists around the globe which changed the field of molecular biology with its ability to treat genetic disorders. In cancer, previous studies highlighted the effect of CRISPR to target cancer cells and suppress tumor growth in the few years of its discovery [3]. Ability of bacteria to defend against bacteriophages brings the finding of CRISPR. This method was naturally used by bacteria as a defense mechanism in their adaptive immune system. Genes isolated from these bacteria was studied by the researchers, which brings the idea of using CRISPR to treat genetic diseases and carcinogenesis.
CRISPR/Cas9 system is composed of 2 components named guide RNA (gRNA) and CRISPR- Associated protein 9 (CAS 9) and the mechanism follows 3 main steps: recognition, cleavage, and repair. Delivery of Single guide-RNA (sgRNA) which designed specifically for target gene, sgRNA will recognize the target sequence in the genome, Cas9 is an endonuclease enzyme which cuts the DNA by creating double stranded breaks (DSB) upstream of protospacer adjacent motif (PAM) and upon damage repaired either by non-homologous end joining (NHEJ) or homology-directed repair (HDR) cellular mechanisms. 1st Cas9 extracted from Streptococcus pyogenes (SpCas-9) and belongs to type 11 of class 1 all CRISPR Cas systems have a unique Cas protein for each. Based on the number of Cas9 protein CRISPR/Cas9 systems are of 2 types, class I and II. By using conserved HNH and Ruvc nuclease domains, Cas9 could cleave strands specifically. By specifically modifying Cas9 protein activity this mechanism can be used to edit other genomes [4]. Cas9 specificity depends on sgRNA which has the guide sequence of 20 nucleotides. By minimizing the off target activity, 5`-NGG PAM should be taken into account for selection of guide sequence [5, 6]. Among all other gene editing technologies CRISPR is a novel tool for genome engineering in mammals due to minimum off target effects, high specificity, and accuracy. But still due to off target effects new mutations could arise. Conditions such as unaccepted concentrations of Cas9 protein and sgRNA levels, selection of target sites [7], unselective PAM sites could lead to undesirable cleavages, less Cas9 translation due to inadequate Cas9 codon optimization, epigenetic factors, method of delivery, and sgRNA position should be properly located at the 5`region of the gene of interest [8] should be properly optimized. Recent studies shows that optimizing the guide design and Cas 9 sequences such as SpCas9-HF1 with 4 residual mutations, evoCas9, and HiFiCas9 with amino acid changes, could reduce off target effects thus improve the potency [6]. CRISPR/Cas 9 systems can be delivered into cells by different means such as viral and non-viral methods like physical or chemical. Cas9 will be delivered with the help of cargos, mainly 3 forms which includes DNA, RNA, and protein (ribonucleoprotein) in which proteins allow faster gene editing compared to other two methods. Each method has its own pros and cons. Based on the cell type and other parameters cargo can be chosen. In case of viral delivery methods, Adeno-associated viral vectors (AAVVs), adenoviral vectors (AdVs) are used as delivery vectors more frequently due to the potential capability. Lentiviruses (LVs) on the other hand, have lavish cloning ability but these due to activation near oncogenes prevent the LV mediated gene editing I CRISPR/Cas9. Out of physical methods such as Electroporation, lipid nanoparticles, ligand fusion tags, and cell-penetrating peptides, electroporation is widely used due to the systematic delivery ability to many types of cells [9, 10]. As a result of advancement of nanotechnology, nanoparticle based delivery such as lipid, gold, polymer based, and exosome delivery is used in manly gene knockouts and further researches will prove a rapid development is the usage of nanoparticles [11].
Due to the fact that CRISPR is a good gene editing tool there are remarkable gene knocking out and knocking in (KO/KI) research which provides versatile applications by replacing, adding or removing a gene segment using sgRNA for a desired trait. For instance to bypass unexpected protein expression [12] to cancer immunotherapy [13], can be done by designing single guide RNA, transferring of gRNA and Cas9 followed by results analyzing. CRISPR is a high throughput gene editing tool for advanced genome editing from proteomics to genomics to study and treat several diseases which has been a dream over the years.
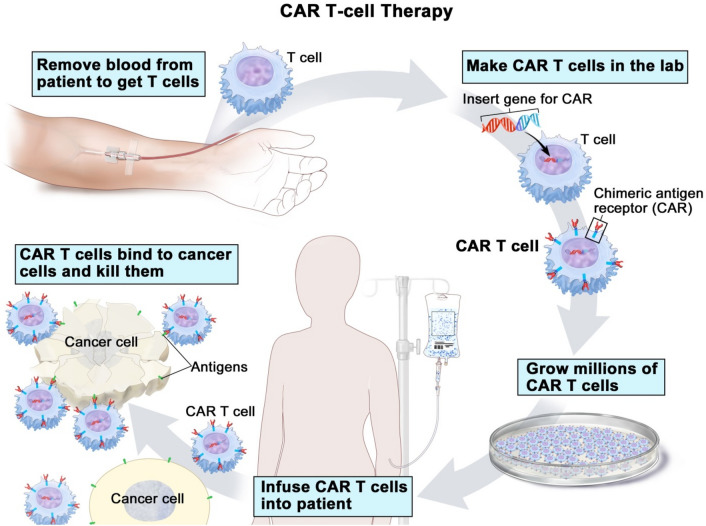
Chimeric Antigen Receptor T cells (CAR-T cells) are an immunotherapeutic tool that has been studied widely in cancer treatment research. These are T cells which are engineered in such a way that they structurally resemble TCR, could express chimeric antigen receptors that could recognize antigens, provide signaling and could target T cells to destroy tumor cells. CAR T cells therapy is an adoptive cell therapy where autologous T cells are genetically engineered which could exhibit CARs and programmed to kill antigen-expressing cells. To overcome the problem of destroying both healthy and tumor cells in radiation and chemotherapy can be answered by CAR T cell therapy thus the main severe side effect of cancer treatment can be eliminated. Conventional CRISPR technology could overcome the intrinsic defects and inadequate lymphocyte counts and inabilities of ordinary CAR T cells and exhibit optimal recognition and response even under the strong immunosuppressive tumor microenvironment. In this combination of techniques could ex vivo activate allogeneic T cells with endogenous knock-out genes to acknowledge the associated issues of immune rejection [9]. Using CRISPR to initiate CAR T cells by redirecting T cell antigen specificity provides precise targeting with less off target edits which relate to other gene editing methods like ZFNs and TALENs. CRISPR can even make CAR T cells to function more efficiently due to the simplicity, flexibility and the capability of editing multiplexable genome with its sgRNA [14, 15], by correcting damages on autologous T cells thus killing tumor cells and saving time and resources by saving patients [16] (Fig .1).
Fig. 1.
CAR-T cell therapy. T cells that were taken from people and edit in such a way that they could bind specifically with cancer cells. Blood taken via an apheresis machine in which only white blood cells will be taken and rest will be send back. T cells in these white bloods cells were inserted with a chimeric antigen receptor, and grown into millions of cells [17]
However, there are two options for CRISPR/Cas9 system introduction into the T cells. Option (a) is to deliver a CRISPR/Cas9 system and then transfer the CAR transgene into the T cells; or (b) first develop CAR‐T cells and then introduce the CRISPR/Cas9 system into the engineered‐T cells. Nevertheless, there are different approaches to deliver CRISPR/Cas9: (I) transfection with DNA plasmid encoding both Cas9 protein and sgRNA, (II) the viral delivery using lentivirus and retrovirus, and non-integrating viruses such as adenovirus and adenovirus‐associated virus (AAV) [18], (III) transfection with mRNA that encodes Cas9 or separate sgRNA, and (IV) CRISPR delivery via Cas9 protein with guide RNA (RNP complex) [19, 20].
Out of several immunotherapies such as monoclonal antibody therapy and checkpoint inhibitor therapy, CAR T cell therapy shows much effective therapeutic approach in hematologic tumors despite the gaps in understanding the overall. Due to the role of T cells in removing malignant cells and lack of MHC when recognized by tumor associated antigens (TAAs) in CAR T cells it stands out among other treatments. Blood stream cancers being one of the most difficult cancer types to treat, allogeneic stem cell transplantation with chemotherapy or radiotherapy was the conventional treatment process over the decades. Since most of hematologic malignancies caused by precursor B or T cells, T cell therapy using chimeric antigen receptors to target only those tumor cells such as CD19 CAR T cell therapy for CLL and ALL CD19, CD20, CD30 CAR T therapy for lymphoma, and CD19, BCMA for MM, CD30 in HL [21–24] shows an encouragement towards future oncogenic treatment processes.
CARs are composed of an antigen-binding domain, a hinge, a transmembrane domain, and an intracellular signaling domain. Extracellular single chain variable fragment (scFv) and chimeric signaling domain (antigen binding domain) which is responsible for T cell activation and destruction of tumor cells by binding to antigens on the surface of the tumor cells. Other domains responsible for anchoring of CARs, and T cell activation. For the CAR T cell therapy these domains undergo specific modification by gene modifications upon which tumor destruction occurs [25].
CRISPR as the stepping stone of next generation gene editing technology modification of several genes such as TCR genes, histocompatibility genes, signaling pathway components in CAR T cell therapy revealed notably hopeful results with healthier benefits. Apart from the fact that CRISPR/Cas 9 is used to produce CAR T cells, it can also be used to manipulate the genome in several other ways for better results. Studies focused on significant genes in tumor cells to study the behavior of cancer. CAR T cells produced normally by collecting T cells via apheresis, genetically engineering T cells to become CAR t cells, in vitro multiplication followed by dripping into the bloodstream so that these cells could attack cancer cells. In case of CRISPR, next generation CAR T cells with tumor targeting receptors are produced from healthy donors and not from each patient thus trouble of manufacturing for each patient can be eluded and immediate availability provides a great opportunity [26].
CRISPR/Cas9 demonstrates into human cells with plasmids which encodes Cas9 and sgRNA or with the help of lentivirus and retrovirus as a viral delivery method or by non-integrating viruses like adenovirus and adeno-associated virus (AAV). In fact AAV shows low immunogenicity thus commonly applied in somatic gene therapy [27]. CRISPR CAR T cell therapy mechanisms of action utilize different abilities of the tools. Basically the ability of knocking out genes is the targeted therapeutic application of CRISPR. In CAR T cells some genes are responsible and benefit the treatment while some genes could cause adverse effects. Upon studying on this, CRISPR used to knockout endogenous genes such as TCR, MHC for off-the-shelf CAR T cell production [28], inhibitory receptors like PD1 and TGF beta receptor [29], integration into specific genes like TRAC and TET2 followed by deleting off target genes to avoid self-killing [30, 31].
ScFv modifications using CRISPR
ScFv of CAR T cells which contains variable heavy and variable light chain antibodies linked by long flexible linker. This small portion is very critical in T cells efficacy and safety. Proper, applicable ScFv serves high avidity to CAR T cells [32]. By properly designing variable regions in ScFv, antitumor effect, proliferation, durable phenotypes and cytokine production of CAR T cells can be optimized.. Due to the effect of direct binding to tumor specific antigens ScFv can be designed using CRISPR to target specific antigens of interest [1, 33]. Reduction of CRS using optimized L17 CD19 CAR T cells generated via studies of ScFv activity shows a potential tool for tumor therapy. To treat MM optimized fully human ScFv CT053 has been studied along with clinical trials [34] proving the fact that ScFv could optimize the activity of CAR T cells. KHYG-1 NK/T cell line is important in case of cell culture and gene transduction. In the study by [35] use eight known CD19 antibodies out of which CD19 scFvs such as FMC63 CAR KHYG-1 and 4G7 CAR KHYG-1 produce with lentiviral packaging to lyse CD19 cell lines and conclude its importance in CAR T cell study with related to B cell lymphoma. Results demonstrate the interest of studying potential influence of transmembrane or spacer domain of CAR towards the function of CAR T cells due to the fact that these specific antibodies which gave positive results recognize similar conformational epitopes thus above mentioned factors could co relate with binding affinity to the epitope. According to [36] spacer domains could alter structural conformations of CARs used to measure and purify positive CARs, regulate synaptic cleft distance etc. which proves the above mentioned fact. By using CRISPR ScFv can be modified to optimize the activity of CAR T cells [14, 20, 34, 37].
How CRISPR is used in CAR T cell therapy
CAR T cell therapy targets different structures, genes, receptors for their mechanism of action as mentioned above. According to the research on inhibitory receptors and immune checkpoint such as PD1, TGF beta and CTLA-4 important hallmarks of those in cancer treatment were found out. Inhibitory receptors (iRs) are important in function of adaptive immune cells and in T cell exhaustion which is a common event in cancer [38, 39] by blocking checkpoint molecules re-establishing the potency of T cells is one important part in the cancer treatment process via immune-checkpoint therapy [40, 41]. CRISPR is a good safeguarding tool to derange immune checkpoint by protecting from checkpoint inhibition via knock out immune checkpoint molecules in CAR T cells. PD1 Programmed cell death protein 1, Transforming growth factor-beta (TGF β) and cytotoxic T lymphocyte protein 4 (CTLA4) are important checkpoints in case of tumor. First clinical trial of CRISPR/Cas 9 mediated PD1 knockout T cell therapy was done for lung cancer by interfering with the normal immune response of PD1 which takes advantage in cancer to proliferate [42]. Taken together there are several researches on CRISPR/Cas9-PD1 tumor-infiltrating lymphocytes (TIL) based adoptive T cell therapy (ACT) [43, 44] by blocking the checkpoint to restore the T cell exhaustion [45] PD-1/PD-1L1 related therapies for tumor immunity [46] demonstrate the effect of CRISPR/Cas9 in tumor therapy. By using Cas9 to disrupt CAR T cells, immune checkpoints such as PD-1 could overcome the major challenge of checkpoint inhibition in tumor cells [47] To reduce the alloreactivity, multiplex CRISPR CAR T cells being used as an anti-tumor agent by blocking PD1 inhibitory pathway with excision of TCR, B2M, and PD1 in T cells as a potential leukemic tumor treating model. using CRISPR, a major drawback of natural killer (NK) cell activation can be eliminated thus providing a potential target for cancer therapy [27]. Findings shows that the usage of CRISPR in knocking out the endogenous Transforming growth factor-beta receptor II (TGF-βR2) gene in anti-mesothelin CAR T cells could decrease the induction of Treg conversion thus prevent the exhaustion of CAR T cells to upgrade the utilization of CAR T cells in solid tumors [48]. Knock out of TCR α and β chains by CRISPR, was approved by FDA which is specific for NY-ESO-1 antigen and can be used as a therapy for relapsed tumors [6].
p53 protein which encodes by the TP53 gene, the cellular gatekeeper for cell growth and division plays a great role in preventing cells from tumor. In most of the tumors this gene get mutated, CRISPR shows a promising results in CRISPR/Cas 9 induced double strand breaks in p53 gene tumor suppressor interactome [49] and homologous recombination with functional cDNA to sustain p53 expression and tumor regression [50].
B cell maturation antigen (BCMA) and CD 19 are the most commonly targeted CAR T cell therapies due to their express levels in malignancies. In case of BCMA, its levels are higher in malignant myeloma cells thus has been a target for immunotherapy including CAR T therapy which is more reliable compared with others. According to American society of hematology (ASH) upon treating with BCMA targeted CAR T therapy there is a promising results on the patients with MM, however more in depth knowledge is needed about the recurrence rate and the long term usage of the therapy so far [51]. Due to the expression of CD19 in most B cell malignancies, CD19 gene is another targeted gene in CAR T therapy. By utilizing the aforementioned knock out and knock in ability of CRISPR able in uniform expression and of CAR and T cell potency by directing CD19 CAR to TRAC locus [52]. Multiplex gene editing via CRISPR-Cas 9 for CAR T, (TRAC/B2M) and (TRAC/B2M/PD-1) cells show superior antitumor activity and potency of CAR T cells [53]. According to [28] knocking out relevant genes in TRAC and CD 25 by sgRNA and Cas9 in human primary T cells could be done, even though the efficiency was low which has to be solved in future research to deliver better therapeutic implementation. Using human ScFv to develop anti CD19 CAR cells to answer the previous problem of human anti mouse immune response, while using murine products has been addressed, [54] with a hope of using it for anti-tumor activity of CD19 CAR T cells, along with further clinical trials and in vivo studies. CD19 being a biomarker in cancer and an attractive target in cancer therapy with CAR T cells being a good immunosuppressive, this combination is undoubtedly a good scope for B cell malignancies. Based on a research on bispecific CD19/CD22 CAR T cell therapy in B cell malignancies demonstrate the effect of the safety and feasibility of the above [55, 56]. According to the clinical trial on Cas9 CD19/CD22 targeting CAR T cell in case of relapsed and refractory B cells shows manageability and safety for treating r/r ALL patients [57]. Autologous anti CD19 CAR T cell KTE-X19 therapy used for patients with r/r mantle cell lymphoma which is a B cell non-Hodgkin’s lymphoma with aggressive clinical course but the therapy induced durable remission by initially eliminating circulating CD19 expressing malignant cells thus narrows the exhaustion of anti CD19 CAR T cells [58]. (Fig. 2).
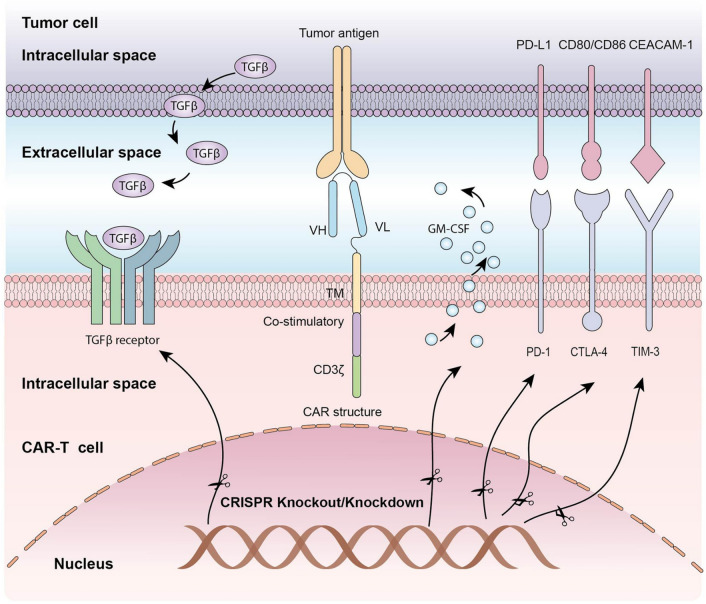
Fig. 2.
CRISPR gene editing mechanism. To boost the function of CAR-T cells, CRISPR can be used as a gene editing tool, via knocking out inhibitory molecules in T cells [13]
CRISPR to prevent GVHD of T cell therapy
However due to the alloantigens of the recipient T cell therapy endogenous genes/housekeeping genes such as T cell receptor (TCR), and major histocompatibility complex genes (MHC) could lead to graft-versus-host-disease (GVHD) due to HLA-mismatching, which could prevent by CRISPR gene editing, inactivation (knocking out) of endogenous T cell receptor (TCR) genes [14, 27]. Based on a research on DLBCL patients TCR and B2M double disrupted CAR T cells therapy by CRISPR, acknowledge the aforementioned problem of GVHD in CAR T cells therapy due to rapid reaction of Human Leukocyte Antigen (HLA) [51].
Anti-CD19 CAR T cells with KO TCRβ chain narrow down the alloreactivity and in contrast to the conventional CAR T cells show promising results in transduction rates and target killing ability, thus showing an encouraging era to cancer medicine [59]. CD7 CAR T cell named UCART7 is a fratricide resistant cell which could employ cytotoxicity without GVHD in malignant cells that express CD7 [20]. For reduction of off-the-shelf allogeneic CAR T cell therapy with sleeping beauty (SB) transposons and CRISPR Cas 9 with minicircle (mc) DNA plasmid for transfection increase efficacy. By inactivation of TCR by CRISPR Cas9-ribonucleoparticles (RNPs) for CD19 targeted CAR reduce allo-reactivity due to the reduction of endogenous TCR expression and SB-CD19-28z.CAR T/TCR KO T cells for a increase memory phenotype thus improve the chances of using CAR T cells for therapy with reduce GVHD [60].
TET2 gene a master regulator of hematopoiesis, which commonly gets dysfunction in CLL, and other hematopoietic cancers as inhibit potent CAR T cell properties of immunotherapy and finding Tet2 negative specific cells via above mechanisms could positively impact on tumor immune response [61, 62].
Over the years many researchers studied the effectiveness of gene editing tools like ZFNs and TALENs in producing allogeneic CAR T cells with knockout of multiple genes loci which could interfere with reactions, despite all the success CRISPR/Cas 9 system enhance the functional aspect compare to those with remarkable success in hematologic malignancies [13].
Production of CAR T cells using CRISPR
To treat several hematologic malignancies and solid tumors includes acute lymphoblastic leukemia (ALL), diffuse large B cell lymphoma (DLBCL), follicular lymphoma (FL), mantle cell lymphoma (MCL), and multiple myeloma (MM) CAR T cell therapy has been used in recent decades [24, 63] First attempt of CAR T cells targeted B cell antigen CD19 to treat CLL. CAR T cells against CART19 (Kymriah) received FDA approval in 2017 to use in pediatric relapsed or refractory ALL and CD19 (Yescarta) for adult relapsed or refractory B cell lymphoma [64]. All the FDA approved CAR T cell therapies derived from gene editing technologies like TALEN and CRISPR and there are around 6 CAR T cell based drugs which are approved by the US FDA. (Table 1)
Table 1.
List of FDA approved CAR T cell therapies
| Drug name | Targeted gene | Delivery Strategy | Targeted malignancy | Data source |
|---|---|---|---|---|
|
Kymriah (Tisagenlecleucel) |
CD19 | Lentiviral |
Refractory/relapsed B cell acute lymphoblastic leukemia B-cell nonHodgkin Lymphoma Relapsed/Refractory Diffuse Large B cell Lymphoma |
[21, 41, 65, 66] |
|
Yescarta (Axicabtagene ciloleucel) |
CD19 | Gamma-retroviral |
Relapsed/Refractory Large B cell Lymphoma including Diffuse Large B cell Lymphoma and follicular lymphoma Relapsed/Refractory Mantle Cell Lymphoma |
[67, 68] |
|
Tecartus (Brexucabtagene autoleucel) |
CD19 |
Relapsed/Refractory Mantle Cell Lymphoma (MCL) (adults) Relapsed/Refractory Acute Lymphocytic Leukemia |
[69–71] | |
|
Abecma (Idecabtagene vicleucel) |
B cell maturation antigen (BCMA) | Lentiviral vector | Refractory and relapsed multiple myeloma | [72, 73] |
|
Breyanzi (Lisocabtagene maraleucel) |
CD19 | Lentiviral vector | Refractory and relapsed large B cell lymphoma | [74, 75] |
|
Carvykti (ciltacabtagene autoleucel) |
B cell maturation antigen (BCMA) | Relapsed/Refractory Multiple Myeloma | [76, 77] |
Even though these drugs show potential impact on cancer several side effects are yet to be answered such as Yescarta shown cytokine release syndrome (CRS) [(FDA), U. S. Food and Drug Administration, 2017] Cilta-cel for hemophagocytic lymohohistiocytosis/macrophage activation syndrome and CRS (National Cancer Institute (NCI), 2022) upon use.
CRISPR editing to reinforce CAR T cell therapy
The therapeutic efficacy of CAR T cell therapy is dependent upon several factors involving intrinsic and extrinsic T cell related mechanisms, limitations with CAR T cell manufacturing and poor CAR T cell expansion and persistence upon infusion. Combating cancer in a hostile, immunosuppressive tumor microenvironment and the associated toxicities of CAR T cell therapy are few of the major hurdles faced. Autologous CAR T therapy has been highly successful in treating hematopoietic malignancies. On the contrary, the therapeutic efficacy against solid tumors has shown to be less than modest due to the attributes of solid tumors. They are disorganized, cancerous cellular structures surrounded by stromal cells of the TME and vasculature that nourishes them and are connected to an immune filtrate consisting of both innate and adaptive immune cells. Unlike B cell malignancies, solid tumors either lack tumor specific antigens or are composed of tumor associated antigens and antigens expressed in healthy cells. This heterogeneous expression makes inefficient trafficking and homing of CAR T cells to tumor site and on target, off tumor toxicity all the more challenging [91]. Therefore, CRISPR as a gene editing tool can be used to circumvent these obstacles faced by CAR T cell therapy (Table 2).
Table 2.
List of CRISPR editing strategies used in CAR T cell therapy
| Purpose of strategy | Targeted proteins | KI/KO via CRISPR | Reference |
|---|---|---|---|
| Generation of allogeneic CART cells | TCR | KO | [78] |
| HLA | KO | [78] | |
| Protection from immunosuppression by blocking pre-apoptotic signaling | Fas | KO | [79] |
| Protection from immunosuppression by knocking out genes | PD-1 | KO | [80] |
| CTLA-4 | KO | [81] | |
| LAG-3 | KO | [82] | |
| TIM-3 | KO | [83] | |
| DGK | KO | [84] | |
| A2AR | KO | [85, 86] | |
| PTPN2 | KO | [87] | |
| TGFβR | KO | [48, 88, 89] | |
| Reduction of CAR T cell associated toxicities | GM-CSF | KO | [90] |
CRISPR editing of inhibitory molecules
Enhancing CAR T cell effector function and its persistence while mitigating T cell exhaustion caused due to chronic antigen stimulation and an immunosuppressive tumor microenvironment subsequently resulting in CAR T cell dysfunction, is a challenge. T cell dysfunction defined by the diminished proliferative capacity and antitumor activity, causes disease relapse within a few years in a majority of patients receiving CAR T cell therapy [92, 93]. Therefore, CAR T cells with reduced levels of exhaustion and differentiation generally result in a better therapeutic efficacy as exhausted T cells are very different from memory and effector T cells due to alterations in underlying transcriptional and epigenetic mechanisms involving TCF1, T-bet and TOX [94, 95]. The upregulation of inhibitory receptors such as PD1, CTLA4, TIM3, LAG3 and CD160 is a hallmark of exhausted T cells [40]. Therapeutic strategies blocking these immune checkpoint molecules are currently under extensive investigation as it would restore the potency and persistence of effector T cells [96].
PD1 is a key inhibitory receptor significantly expressed on exhausted CD8 T cells, mediating immune escape in tumor cells by limiting the capabilities of TCR and CD28, upon interactions with its ligands PD-L1 or PD-L2 on T cells [97]. In general, PD1 regulates the adaptive immune response. Furthermore, IFN-γ has been discovered as a key player in inducing PD-L1 expression [98].
Systemic administration of checkpoint blockade has reportedly generated immune related adverse events in patients and therefore to circumvent this issue, Rafiq et al. [99] engineered CART cells to secrete PD1 blocking scFv which demonstrated improved antitumor activity and persistence at the tumor site thereby evading from systemic immune toxicities. CRISPR genome editing platform was effectively used to knockdown the expression of PD-L1 in primary human T cells by electroporation of plasmids encoding sgRNA and Cas9 thereby impeding the PD1/PD-L1 axis. These engineered autologous T cells exhibited potent antitumor activity and augmented IFN-γ production which supplemented tumor cell lysis [100]. Furthermore, Cas9 RNP gene editing and lentiviral transduction were combined to generate PD1 disrupted anti-CD19 CAR T cells which dampened PD/PD-L1 axis and displayed enhanced antitumor activity in vitro and tumor cell lysis in PDL1 + tumor xenografts in myelogenous leukemia murine models, in comparison to normal anti-CD19 CAR T cells [80].
Also, with respect to solid tumors, Hu et al. [101] reported effective Cas9 RNP gene editing mediated knockdown of PD1 in Mesothelin CAR T cells and augmented cytotoxic properties and induced production of cytokines, IL-2 and IFN-γ against Triple Negative Breast Cancer (TNBC), in mammary gland tumor murine model. In fact, the antitumor potency of these Mesothelin CAR T cells was relatively stronger than α-PD1 antibody mediated immune blockade. A phase I clinical trial investigating the therapeutic efficacy and safety of CRISPR mediated multiplex genome editing of autologous CAR T cells was tested in three patients with refractory cancer. Modified CAR T cells were deficient in two genes encoding TCR, namely TCRα and TCRβ and PD1 to restrict T cell exhaustion and increase antitumor efficacy and safety, thereby facilitating the expression of cancer specific TCR transgene (NY-ESO-1). Genetic manipulation was exerted with the combination of Cas9 RNP electroporation and lentiviral transduction of the engineered TCR. Relatively higher off target mutations were observed for the sgRNA for TCRβ compared to other loci. Chromosomal translocations were identified, which also reduced over time following infusion. Heightened levels of engraftment and persistence of the engineered CAR T cells (until a period of nine months) were unprecedented outcomes of this study and it managed to validate the feasibility of CRISPR as a promising gene editing tool to be employed in CAR T cell therapy [102].
To minimize immunogenicity in allogeneic CAR T cell therapy, Liu et al. [53] knocked down genes encoding TCRα, and beta-2 microglobulin (B2M) which regulate expression of HLA-1, and thirdly PD1 in anti-CD19 CAR T cells. The team generated double (TCRα/B2M) and triple (TCRα/B2M/PD1) knockout CAR T cells respectively and the latter displayed a more potent antitumor function in vitro. Ren et al. [79] also reported that triple knockout CD19 CAR T cells (TCRα/B2M/PD1) resulted in complete eradication of tumor cells in a leukemia murine model and further validated the clinical efficacy of CRISPR mediated triple knockout CAR T cells and this was consistent with the findings reported by Choi et al. [103]. The study conducted by Nakazawa et al. [104] engineered PD1 deficient third generation EGFRvIII targeting CAR T using CRISPR which impeded growth of human glioblastoma cell lines in vitro. This corroborated the importance and therapeutic efficacy of PD1 as a target in CAR T cell therapy in the battle against solid tumors [105].
Another immunosuppressive component is the Fas receptor alternatively known as CD95 and APO-1, is a cell surface protein of the tumor necrosis factor (TNF) receptor family. Interactions between Fas and Fas ligand (FasL) trigger apoptosis in exhausted T cells via a mechanism known as activation induced cell death (AICD). Up-regulation of Fas receptors has been reported in CD8 T cells exposed to prolonged antigen stimulation [106]. CRISPR mediated triple knockout of TCR, HLA-I and Fas CAR T cells showed elevated levels of T cell expansion coupled with increased degranulation, cytokine production and potent antitumor activity in vitro, while Fas deficiency contributed to prolonged survival of CAR T cells in leukemia murine model, indicative of an attenuation in AICD. Ren et al. [79] also attempted quadruple genetic knockdown (TCR, HLA-1, PD1 and CTLA-4) with the one-shot system which however led to lowered efficiency in gene editing. The authors stated that targeting multiple genes eventually resulted in increased competition of the gRNAs for Cas9, in addition to the restricted packaging size of the lentiviral vector, attributed to the reduced efficiency in multiplex genome editing. Therefore, an improved delivery strategy needs to be investigated to exert CRISPR mediated simultaneous genetic ablation of four or more genes in CAR T cell therapy.
Less research has been carried out in CAR T therapy with respect to CTLA-4, a cell surface receptor dampening T cell activation and generation of effector T cells. Anti-CTLA4 checkpoint blockade immunotherapy is used in the clinical management of malignant melanomas with the aim of suppressing T cell inhibitory activity. Lin et al. [81] attempted a distinct approach in targeting tumors by appreciating and utilizing the inhibitory effects of CTLA-4. The study engineered chimeric CTLA-4/CD28/CD3z (CTLA-4 CAR T cells). The inhibitory effects of CTLA-4 are exerted upon the interactions between CTLA-4 and its ligands, CD80 and CD86 which are prevalent in many tumors and expressed in cells found in the TME. As CD80 and CD86 ligands are highly expressed in B cell malignancies, this study harnessed their established interaction with CTLA-4, to facilitate recognition of such tumor cells and induce targeted killing. In vitro and in vivo findings in a B cell lymphoma mouse model suggest that these engineered T cells showed strong antitumor effects and tumor growth suppression. The team also created an autologous transfer setting which involved the transfer of murine CTLA-4 chimeric T cells to a murine melanoma model. Results indicated higher levels of persistence of these cells and elevated antitumor efficacy against myeloid derived suppressor (MDSC) cells and presented toxicity against nonmalignant CD80/CD86 expressing cells as well, highlighting the possibility of CRS as well. Therefore, the safety of CTLA-4 chimeric T cells is yet to be investigated in preclinical and clinical settings.
Lymphocyte activation gene-3 (LAG-3), another hallmark of T cell exhaustion is generally expressed in activated CD4 and CD8 T cells, regulatory T cells, B cells and natural killer (NK) cells, and upregulated upon persistent antigen stimulation caused in cancer and chronic infections. Recently, LAG-3 is under clinical investigation as a candidate in immunotherapy against cancer [107]. CRISPR mediated LAG-3 genetic ablation of CD19 CAR T cells induced in vitro cytotoxic activity and showed significant elimination of tumor cells in a murine xenograft model and augmented engraftment in vivo [82].
Ciralo et al. [83] generated triple knockout CD8 T cells constituting genetic deficiencies in immune checkpoint inhibitors PD1, LAG-3 and TIM-3. In vitro findings were representative of efficient genome editing, no reduction in IFN-γ production and consequently enhanced cytotoxic activity exerted by the CD8 edited T cells. Improved persistence and higher levels of tumor infiltrating edited CD8 T cells in comparison to the impact of unedited CD8 T cells as per the in vivo analysis in the melanoma murine model, led to tumor regression in the mice.
In addition to the immune checkpoint molecules, other T cell inhibitory molecules such as diacylglycerol kinase (DGK) are emerging as potential candidates in cancer immunotherapy. However, there is limited research with respect to CAR T cell therapy. DGK is an enzyme that acts as a critical regulator of diacylglycerol (DAG) and phosphatidic acid (PA) levels by phosphorylating DAG to PA, which is part of the phosphatidylinositol (PI) cycle and are two key lipids acting as second messengers in immune cell signaling in T cells [108]. DGK acts as a switch; regulating crucial proteins involved in T cell development, survival, activation, anergy, secretion, and effector function [109]. Two DGK isoforms, the type-I α isoform, and the type-IV ζ isoform, are predominant in T cells [110]. Upon antigen presentation and consequent TCR stimulation, several downstream signaling pathways are triggered and signaling molecules are recruited. Among this is phospholipase, PLCγ1, which is recruited to the cell membrane. PLCγ1 metabolizes phospholipid PIP2 generating DAG and inositol triphosphate. DAG results in the downregulation of downstream signaling pathways including the RasGRP1/Ras/ERK and PKCθ/IKK/NF-κB pathways. On the other hand, PA is known to interact with mammalian targets of rapamycin (mTOR), SHP-1, RasGAP, Sos, PI5Kα, and p47(phox) [111].
DGK attenuates DAG levels, restricting the function of the TCR signaling cascade and ultimately serving as a braking mechanism in the immune synapse [108, 112]. DKGα has been recognized as an inhibitor in the tumor milieu and therefore recent investigations into its potential as a target for cancer immunotherapy are underway [113]. Nevertheless, Jung et al. [84] knocked out both DGKα and DGK ζ isoforms using CRISPR in CAR T cells which led to heightened TCR signaling and sustained and robust effector function in vitro, while in vivo findings reported that DGK deficiency resulted in remarkable tumor eradication of U87MGvIII glioblastoma tumors and resistance to immunosuppressive molecules including prostaglandin E2 and TGF-β.
Although transforming growth factor-β (TGF-β) is a tumor suppressor in pre-malignant cells, inhibiting cancer cell proliferation and inducing apoptosis, and over the course of tumor progression, TGF-β transforms to a multipotent cytokine responsible for creating an immunosuppressive tumor microenvironment via the and reprogramming cellular metabolism and the modulation of the proliferation and differentiation of immune cells and the matrix composition [114–116]. TGF-β is secreted by stromal cells and exists as three ligands: TGF-β1, TGF-β2 and TGF-β3 which exerts actions upon interactions with its receptors, TGF-βRI and TGF-βRII [116].
TGF-β secreted by CD4(+) T cells has been reported to mediate tumor immune escape and immunosuppression, while promoting the differentiation of CD4(+) T cells to FOXP3 dependent regulatory T cells leading to the execution of immune tolerance [117–119]. Moreover, TGF-β is known to hinder CD8 T cell cytotoxic function via a SMAD dependent inhibition of target genes encoding perforins, granzymes and interferon and restrict trafficking of immune cells to the tumor through the downregulation of CD8+ T cell expression of CXCR3 [120].
As a result, there is ongoing research on targeting TGF-β and TGF-β receptors to enhance antitumor efficacy of CAR T cell therapy against solid tumors [48, 88, 89]. Interestingly, Hu et al. [101] demonstrated that engineered TGF-β CAR T cells engaged in shielding tumor targeting cells from TGF-β mediated immunosuppression concomitantly enhancing their antitumor function, and attenuated the differentiation into T reg cells. The team also discovered that TGF‐β CAR‐Treg cells do not cause CAR mediated suppression of effector T cells. Although the team harnessed the potential of the immunosuppressive cytokine as a T cell stimulant, the fact whether TGF-β CAR T cells will execute the same effect in a clinical setting is a concern requiring elucidation with additional studies. Tang et al. [48] designed CRISPR knocked down TGF-βRII CAR T cells, decreased the differentiation into T reg cells and inhibited CAR T cell exhaustion. In addition, these CAR T cells exhibited remarkable antitumor efficacy, sustained proliferation and complete tumor cell clearance in cell line derived and patient derived xenograft models expressing mesothelin. Moreover, Alishah et al. [89] efficiently knocked down TGF-βRII in CAR T cells via CRISPR and their findings were also consistent with Tang et al. [48].
Adenosine is another potent immunosuppressive agent found in the tumor milieu known to interact with four types of receptors: A1R, A2AR, A2BR, and A3R, out of which A2AR and A2BR contribute to immunosuppressive effects through adenosine [121]. More specifically, A2AR acts as a critical regulator of CD8+ T cells in the TME [122]. Adenosine is predominantly expressed in solid tumors and generated from extracellular ATP in a CD73 and CD39 dependent manner [123]. In vitro and in vivo findings elucidated that targeting A2AR in CAR T cells exhibited enhanced antitumor efficacy, while the double blockade of A2AR and PD-1 in CAR T cells augmented cytokine generation in vivo, and all in all, A2AR genetic and pharmacologic inhibition did provide protection from adenosine mediated immunosuppression, which is a challenge in solid tumors [124].
CRISPR gene editing technology has also been utilized to knockdown A2AR in CAR T cells. Li et al. [85] knocked down both A2AR and A2BR in CAR T cells via CRISPR and demonstrated strong antitumor efficacy in vitro and a further heightened efficacy in vivo in two patient derived (pancreatic cancer) xenograft models resulting in a low tumor load. It was also discovered that A2AR knockout significantly contributed to immunosuppressive effects in CAR T cells, compared to A2BR genetic abrogation, rendering A2AR as the major target for tumor immunotherapy. Moreover, Guiffrida et al. [86] reported that the knockdown of A2AR in CAR T cells via CRISPR surpasses the efficiency of shRNA and pharmacologic inhibition. It led to an enhanced effector T cell activity in vitro and improved antitumor efficacy in vivo.
Protein Tyrosine Phosphatase N2 (PTPN2) hinders cytokine signaling which is a critical regulator of T cell function, homeostasis and differentiation by attenuating signal transducer and activator of transcription 5–1 (STAT-1), STAT-3 and STAT-5 [125]. Consistent with these findings, Wiede et al. [126] reported that PTPN2 genetic abrogation in CD8+T cells targeting oncoprotein HER2 upgraded antitumor immunity in vitro and in vivo. PTPN2 deficiency in CAR T cells promoted the homing of adoptive CAR T cells to solid tumors and stimulated cytokine production [126]. Moreover, PTPN2 suppression improved expansion and survival of effector T cells [127]. CRISPR mediated deficiency of PTPN22 significantly enhanced in vitro and in vivo antitumor activity [87].
PAK4 is serine/threonin protein kinase enzyme which has been reported to be a critical regulator of mesenchymal related transcriptional activation in tumor ECs and thus reduces T cell adhesion due to consequent improved vessel permeability. CRISPR mediated PAK4 knockdown results in inhibited cell proliferation, migration and invasion and reduced monolayer permeability in human glioblastoma (GBM) derived tumor endothelial cells (EC) and leads to a normalization of the tumor vasculature and enhanced T cell infiltration. PAK4 deficiency improved CAR T immunotherapy in mouse GBM models [128].
Disialoganglioside GD2 antigen, demonstrating restricted expression on normal tissues and increased expression levels in human cancers such as glioma, neuroblastoma and melanoma, has become an attractive and valuable target in CAR T cell therapy against solid tumors. Prapa et al. [129] reported that intracerebral administration of anti-GD2 CAR T cells shows promise in the combat against glioblastoma, in addition to showing potential as a therapeutic target for CAR T cell therapy in lung cancer [130].
CRISPR editing of transcription factors
IKZF3 is a lymphocyte maturation driving transcription factor predominantly expressed in certain hematological malignancies and solid tumors, and its overexpression reportedly promotes metastasis in vivo [131]. CRISPR mediated IKZF3 deficiency in HER2 CAR T cells resulted in improved antitumor activity in vitro and in vivo, along with increased T cell activation and proliferation and augmented cytokine production [132].
The NR4A family of nuclear receptor transcription factors are associated with diminished T cell activity, in addition to being linked to the expression of inhibitory receptors such as PD-1 and TIM-3. Studies have found that the triple knockout of NR4A1, NR4A2 and NR4A3 transcription factors promoted tumor regression and survival of tumor murine models [133]. Furthermore, double knockout of TOX and TOX2 transcription factors in CAR T cells which are also known to play a vital role in CD8+T cell exhaustion in association with N4RA, resulted in increased cytokine expression and reduced expression of surface inhibitory receptors, suggesting that the genetic disruption of the aforementioned transcription factors could be promising strategies in improving therapeutic efficacy of CAR T cells [134]. As another strategy to counter T cell exhaustion, Lynn et al. [135] showed that canonical AP-1 factor (c-Jun) overexpression in CAR T cells led to an augmentation in antitumor activity, improved expansion and reduced terminal differentiation in vivo.
CRS and ICANS
There are two types of toxicities associated with CART therapy that dampen their therapeutic efficacy. The most frequent toxicity is cytokine release syndrome (CRS) which is a systemic inflammatory response occurring due to the interplay between target tumor cells and the antigen expressing CART cells upon its infusion and migration to the tumor site [136]. Upon antigen recognition of CART cells at the tumor site, CART cells proliferate and induce an in situ inflammatory cytokine cascade in the tumor microenvironment resulting in direct and indirect mechanisms of tumor killing. CART cells secrete perforin, granzymes, granulocyte macrophage-colony stimulating factor (GM-CSF) and inflammatory cytokines such as IFN-γ and TNF-α to induce pyroptosis of tumor cells, releasing large amounts of DAMPs which activate bystander effector cells such as macrophages [137–139]. In addition to the activation by cytokines and catecholamine, interactions between the macrophage expressed CD40 and CART cell expressed ligand CD40L, lead to macrophage activation [140]. Consequently, this results in increased levels of inflammatory mediators including IL-6, IL-1 and nitric oxide, marking the onset of CRS [141, 142]. This event triggers the endothelium and further results in vascular leakage in several organs and tissues, ultimately causing hypotension and organ damage (Gust et al., 2017). Moreover, the amplified production of cytokines and chemokines and its diffusion along with the concomitant migration of CART cells, peripheral monocytes and T cells to the central nervous system and cerebrospinal fluid, crossing the blood–brain barrier, marks the onset of immune effector cell associated neurotoxicity syndrome (ICANS). ICANS, a neuropsychiatric disorder which usually peaks several days after the onset of CRS, is the second common toxicity associated with CART therapy [143, 144].
CRISPR gene editing platform can be utilized to tackle these obstacles in CART therapy. Sterner et al. [145] generated GM-CSF deficient anti CD19 CART cells using CRISPR knockout technology which significantly reduced GM-CSF level. When its antitumor efficacy was tested in a xenograft model, it exhibited a relatively prominent antitumor activity than wild type CART cells and further demonstrated that the GM-CSF knockout did not impair normal T cell functions vital for antitumor activity.
Yi et al. [90] conducted a study where CRISPR edited GM-CSF KO CART cells along with co-expression of anti-IL6 scFv and IL1RA (IL6 and IL1 blockers, respectively) and additional TCR KO for tracing edited CART. One patient with non-Hodgkin lymphoma and two patients with multiple myelomas T cells were edited using CRISPR technology ex vivo and infused back into the patients. CART secreted cytokine blockers successfully antagonized the actions of IL6 and IL1 by releasing large amounts of anti-IL6 scFv and IL1RA, and all patients showed complete response, only one patient developed grade 2 CRS and none developed any neurotoxicity symptoms. The study reported that GM-CSF KO did not impair CART proliferation and clinical efficacy of the treatment, and further the deficiency of TCR had no influence on its anti-tumor efficacy; results suggested long term persistence upon infusion and were indicative of re-expansion upon antigen exposure. It was evident from this study that CRISPR editing platform did not hinder CART functions in any way and did not pose any concerns threatening the safety of patients, while it further emphasized how CRISPR could successfully tackle cytokine toxicity and neurotoxicity via the autonomous secretion of cytokine antagonists in patients with refractory hematologic malignancies. In addition, Cox et al. [146] also proved that antitumor efficacy of engineered T cells can be enhanced by the neutralization of GM-CSF.
Nevertheless, the study done by Yi et al. [90] has to be expanded and include a larger number of patients and further elucidation of the specific molecular mechanism of each cytokine playing a role in CRS is required. Moreover, the genetic landscape and T cell memory phenotype of GM-CSF KO CART cells need to be explored.
Another approach to control and reverse toxicity effects in the presence of autonomous uncontrolled T cell growth, is the use of inducible caspase 9, a suicide gene which effectively eliminates engineered CART cells when needed. Hoyos et al. [147] incorporated an IL15 gene and an inducible caspase 9 (iC9) based suicide gene to CAR.19 cells and demonstrated that within 24 h of pharmacologic activation of the suicide gene, > 95% of CART cells were effectively eliminated in vitro, and this was successful in vivo as well. Budde et al. [148] introduced inducible caspase 9 suicide gene to CD20CAR19 T cells which also demonstrated 90% elimination within 24 h of activating the suicide gene and reached a success of 98% within the next 24 h. Diaconu et al. [149] confirmed that while higher doses of CID (a molecule targeting iC9 engineered T cells) successfully eliminate majority of CART cells, lower doses selectively eradicate only iC9 engineered T cells with enhanced transcriptional activity in a humanized mouse model while mediating the control of tumor growth and protection from tumor rechallenge.
Safety concerns associated with CRISPR gene editing
Prior to application of CRISPR edited cells in a clinical setting, there are safety concerns that need to be addressed first. Cas9 activity, sgRNA design, delivery method and target site selection are several factors which determine the therapeutic efficacy and safety of the CRISPR gene editing platform. Among these, a main concern is the off target toxicity effect caused by unintended genetic cleavage and mutations at genomic sites related to the targeted sites, resulting in chromosomal rearrangements such as deletions and translocations, which potentially could be oncogenic depending on the genetic sequence affected [150]. Furthermore, CRISPR Cas 9 genome editing induced a p53 mediated DNA damage response subsequently causing cell cycle arrest in human retinal pigment epithelial cells [151]. CRISPR gRNAs have the potential to trigger unintended innate immune responses in T cells resulting in cytotoxicity [152]. Although such events may be rare as CRISPR has largely enhanced its specificity via recent advances in optimizing multiple elements of CRISPR technology including modifications of the gRNA, improved Cas9 variants, base editing and prime editing, the potential adverse side effects of off target mutagenesis is still unknown [153–155].
Conclusion
Cancer has become one of the most difficult diseases to treat worldwide with a high fatality rate. Finding cures and treatment options are very important these days. In this review we discussed the upcoming treatment possibility for cancer, which is CAR T cell therapy with the help of CRISPR/Cas 9 system. CRISPR is a gene editing tool which uses recombinant T cells to treat cancer by interfering with immune cells. Different aspects of these cells can be targeted in cancer therapy such as knocking out endogenous genes, inhibitory receptors, and integration of specific genes. Among other gene editing tools, CRISPR shows immense possibilities to edit T cells either by adding, removing or altering genes to change its genome for good. Due to the fact that these cells target and attack only tumor cells major interference of chemo and radiotherapy which is healthy cell attacking can be avoided. With the usage of healthy donor cells can cause GVHD, which is one such obstacle of CAR T cells which could be overcome with the use of CRISPR. There are several FDA approved CAR T cell therapies which use gene editing tools such as CRISPR for the production process and these findings prove the fact that CRISPR can be used as a promising treatment option to treat blood cancer.
Major safety concerns associated with CRISPR include off target toxicity, chromosomal breakage, limited efficiency of gRNA and concerns of delivery strategies. Furthermore, concerns regarding limited therapeutic efficacy of CAR T cell therapy include issues arising with the immunosuppressive tumor microenvironment, intricacy and expense associated with manufacturing autologous CAR T cell products and associated toxicity conditions (cytokine release syndrome and associated neurotoxicity). There are ongoing clinical trials of CRISPR modified CAR T cells as well as for solid tumors. Strategies such as whole exome and whole genome sequencing can be used to look out for any unwanted gene editing effects caused by CRISPR modified CAR T cell products.
Abbreviations
- AAVVs
Adeno-associated viral vectors
- AdVs
Adenoviral vectors
- AICD
Activation induced cell death
- ALL
Acute lymphoblastic leukemia
- CAR T cell
Chimeric Antigen Receptor T cell
- CRISPR
Clustered Regularly Interspaced Palindromic Repeats
- CRS
Cytokine Release Syndrome
- DAG
Diacylglycerol
- DGK
Diacylglycerol kinase
- DLBCL
Diffuse large B cell lymphoma
- DSB
Double Stranded Break
- FL
Follicular Lymphoma
- gRNA
Guide RNA
- GBM
Glioblastoma
- GVHD
Graft-versus-host-disease
- HDR
Homology Directed Repair
- HLA
Human Leukocyte Antigen
- ICANS
Immune effector cell associated neurotoxicity syndrome
- KI
Knocking In
- KO
Knocking Out
- LV
Lentivirus
- MCL
Mantle Cell Lymphoma
- MM
Multiple Myeloma
- MHC
Major Histocompatibility Complex
- NHEJ
Non Homologous End Joining Repair
- NK
Natural Killer
- PA
Phosphatidic Acid
- PAM
Protospacer Adjacent Motif
- PI
Phosphatidylinositol
- PTPN2
Protein Tyrosine Phosphatase N2
- scFv
Single chain variable fragment
- sgRNA
Single Guide RNA
- TAA
Tumor Associated Antigen
- TCR
T Cell Receptor
- TGF-β
Transforming growth factor-β
- TME
Tumor Microenvironment
- TNBC
Triple Negative Breast Cancer
- TNF
Tumor Necrosis Factor
Funding
The authors have not disclosed any funding.
Declarations
Competing Interests
The authors declare that no competing interest exists.
Footnotes
This article has been retracted. Please see the retraction notice for more detail:https://doi.org/10.1007/s12032-025-02692-7
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
3/24/2025
This article has been retracted. Please see the Retraction Notice for more detail: 10.1007/s12032-025-02692-7
References
- 1.Zhao L, Cao YJ. Engineered T cell therapy for cancer in the clinic. Front Immunol. 2019. 10.3389/fimmu.2019.02250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishino Y, Shinagawa H, Makina K, Amemura M, Nakata A. Nucleotide Sequence of the iap Gene, Responsible for Alkaline Phosphatase Isozyme Conversion in Escherichia coli, and Identification of the Gene Product. J Bacteriol. 1987;169(12):5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landhuis E. The definition of gene therapy has changed. Nature. 2021. 10.1038/D41586-021-02736-8. [DOI] [PubMed] [Google Scholar]
- 4.Mengstie MA, Wondimu BZ. Mechanism and applications of CRISPR/Cas-9-mediated genome editing. Biologics. 2021. 10.2147/BTT.S326422. [Google Scholar]
- 5.Johansen KH. How CRISPR/Cas9 gene editing is revolutionizing T cell research. DNA Cell Biol. 2022. 10.1089/dna.2021.0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uddin F, Rudin CM, Sen T. CRISPR Gene Therapy: Applications, Limitations, and Implications for the Future. Front Oncol. 2020. 10.3389/fonc.2020.01387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013. 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song G, Jia M, Chen K, Kong X, Khattak B, Xie C, Li A, Mao L. CRISPR/Cas9: a powerful tool for crop genome editing. Crop J. 2016. 10.1016/j.cj.2015.12.002. [Google Scholar]
- 9.Roberts R. CRISPR CAR-T cells: edited T Cells Are Revolutionizing Cancer Treatment. 2021. https://www.synthego.com/blog/car-t-crispr-cancer
- 10.Yip BH. Recent advances in CRISPR/Cas9 delivery strategies. Biomolecules. 2020;10(6):839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan L, Ouyang K, Xu X, Xu L, Wen C, Zhou X, Qin Z, Xu Z, Sun W, Liang Y. Nanoparticle delivery of CRISPR/Cas9 for genome editing. Front Genetics. 2021. 10.3389/fgene.2021.673286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishibashi A, Saga K, Hisatomi Y, Li Y, Kaneda Y, Nimura K. A simple method using CRISPR-Cas9 to knock-out genes in murine cancerous cell lines. Sci Rep. 2020;10(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ou X, Ma Q, Yin W, Ma X, He Z. CRISPR/Cas9 gene-editing in cancer immunotherapy: promoting the present revolution in cancer therapy and exploring more. Front Cell Dev Biol. 2021. 10.3389/fcell.2021.674467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, Mei H, Hu Y. Applications and explorations of CRISPR/Cas9 in CAR T-cell therapy. Brief Funct Genom. 2020. 10.1093/BFGP/ELZ042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quazi S. Elucidation Of CRISPR-Cas9 application in novel cellular immunotherapy. Mol Biol Rep. 2022;49:7069–77. [DOI] [PubMed] [Google Scholar]
- 16.Dimitri A, Herbst F, Fraietta JA. Engineering the next-generation of CAR T-cells with CRISPR-Cas9 gene editing. Mol Cancer. 2022. 10.1186/s12943-022-01559-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Institute NC, Health at the NI of. Definition of CAR T-cell therapy—NCI Dictionary of Cancer Terms—NCI. Published 2022. https://www.cancer.gov/publications/dictionaries/cancer-terms/def/car-t-cell-therapy.Accessed Dec 9 2022.
- 18.Quazi S. Telomerase gene therapy: a remission toward cancer. Med Oncol. 2022. 10.1007/s12032-022-01702-2. [DOI] [PubMed] [Google Scholar]
- 19.Freen-van Heeren JJ. Using CRISPR to enhance T cell effector function for therapeutic applications. Cytokin: X. 2021. 10.1016/j.cytox.2020.100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Razeghian E, Nasution MKM, Rahman HS, Gardanova ZR, Abdelbasset WK, Aravindhan S, Bokov DO, Suksatan W, Nakhaei P, Shariatzadeh S, Marofi F, Yazdanifar M, Shamlou S, Motavalli R, Khiavi FM. A deep insight into CRISPR/Cas9 application in CAR-T cell-based tumor immunotherapies. Stem Cell Res Ther. 2021;12(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han D, Xu Z, Zhuang Y, Ye Z, Qian Q. Current progress in CAR-T cell therapy for hematological malignancies. J Cancer. 2021. 10.7150/jca.48976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haslauer T, Greil R, Zaborsky N, Geisberger R. CAR T-cell therapy in hematological malignancies. Int J Mol Sci. 2021. 10.3390/ijms22168996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Z, Yu C, Francisco NM, Zhang Y, Wu M. The application of CAR-T cell therapy in hematological malignancies: advantages and challenges. Acta Pharmaceutica Sinica B. 2018. 10.1016/j.apsb.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quazi S. An overview Of CAR T cell mediated B Cell maturation antigen therapy. Clin Lymphoma Myeloma Leuk. 2022;22:e392-404. [DOI] [PubMed] [Google Scholar]
- 25.Xin T, Cheng L, Zhou C, Zhao Y, Hu Z, Wu X. In-vivo induced CAR-T cell for the potential breakthrough to overcome the barriers of current CAR-T cell therapy. Front Oncol. 2022. 10.3389/fonc.2022.809754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson K. Developing Novel Cell Therapies by Rewiring T-Cell Genomes | Technology Networks. 2022. https://www.technologynetworks.com/biopharma/articles/developing-novel-cell-therapies-by-rewiring-t-cell-genomes-363596
- 27.Ren J, Zhao Y. Advancing chimeric antigen receptor T cell therapy with CRISPR/Cas9. Protein Cell. 2017. 10.1007/s13238-017-0410-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamali E, Rahbarizadeh F, Hojati Z, Frödin M. CRISPR/Cas9-mediated knockout of clinically relevant alloantigenes in human primary T cells. BMC Biotechnol. 2021. 10.1186/s12896-020-00665-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sow HS, Ren J, Camps M, Ossendorp F, Ten Dijke P. Combined inhibition of TGF-β signaling and the PD-L1 immune checkpoint is differentially effective in tumor models. Cells. 2019. 10.3390/cells8040320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain N, Zhao Z, Iyer A, Lopez M, Feucht J, Koche R, Yang J, Zhan Y, Sadelain M. Emergence of a hyper-proliferative phenotype in TET2 edited human CAR T cells. Cancer Res. 2021. 10.1158/1538-7445.AM2021-LB153.34167950 [Google Scholar]
- 31.Shah NN, Fry TJ. Mechanisms of resistance to CAR T cell therapy. Nat Rev Clin Oncol. 2019. 10.1038/s41571-019-0184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujiwara K, Masutani M, Tachibana M, Okada N. Impact of scFv structure in chimeric antigen receptor on receptor expression efficiency and antigen recognition properties. Biochem Biophys Res Commun. 2020;527(2):350. [DOI] [PubMed] [Google Scholar]
- 33.Ochi T, Maruta M, Tanimoto K, Kondo F, Yamamoto T, Kurata M, Fujiwara H, Masumoto J, Takenaka K, Yasukawa M. A single-chain antibody generation system yielding CAR-T cells with superior antitumor function. Commun Biol. 2021. 10.1038/s42003-021-01791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang M, Zhang W, Yu K, Wang P, Jiang H, Chen L, Meng H, Weng Y, Tao R, Huang X, Xing C, Wang H, Wan J, Wang S, Dai L, Hendrix AY, Xiao J, Wang W, Ma H, Jiang S. A novel BCMA CAR-T-cell therapy with optimized human scFv for treatment of relapsed/refractory multiple myeloma: results from phase I clinical trials. Haematologica. 2022;107(8):1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang CH, Kim Y, Lee HK, Lee SM, Jeong HG, Choi SU, Park CH. Identification of potent CD19 scFv for CAR T cells through scFv screening with NK/T-cell line. Int J Mol Sci. 2020. 10.3390/ijms21239163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jayaraman J, Mellody MP, Hou AJ, Desai RP, Fung AW, Pham AHT, Chen YY, Zhao W. CAR-T design: elements and their synergistic function. EBioMedicine. 2020;58:102931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong M, Clubb JD, Chen YY (n.d.). Engineering CAR-T cells for next-generation cancer therapy [DOI] [PubMed]
- 38.Himmel ME, Saibil SD, Saltman AP. Immune checkpoint inhibitors in cancer immunotherapy. Can Med Assoc J. 2020;192(24):E651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wherry EJ. T cell exhaustion. Nat Immunol. 2011. 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 40.Fuertes Marraco S, Neubert N, Verdeil G, Speiser D. Inhibitory receptors beyond T cell exhaustion. Front Immunol. 2015. 10.3389/fimmu.2015.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rezalotfi A, Fritz L, Förster R, Bošnjak B. Challenges of CRISPR-Based Gene Editing in Primary T Cells. Int J Mol Sci. 2022;23(3):1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cyranoski D. CRISPR gene-editing tested in a person for the first time. Nature. 2016;539(7630):479. [DOI] [PubMed] [Google Scholar]
- 43.Chamberlain CA, Bennett EP, Kverneland AH, Svane IM, Donia M, Met O. Highly efficient PD-1-targeted CRISPR-Cas9 for tumor-infiltrating lymphocyte-based adoptive T cell therapy. Mol Ther Oncolytics. 2022. 10.1016/j.omto.2022.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmgren G. News: PD-1 Targeted Cancer Immunotherapy Meets CRISPR—CRISPR Medicine. 2022. https://crisprmedicinenews.com/news/pd-1-targeted-cancer-immunotherapy-meets-crispr/
- 45.Al Saber M, Biswas P, Dey D, Kaium MA, Islam MA, Tripty MIA, Hasanur Rahman MD, Rahaman TI, Biswas MY, Paul P, Rahman MA, Hasan MN, Kim B. A comprehensive review of recent advancements in cancer immunotherapy and generation of CAR T cell by CRISPR-Cas9. Processes. 2022. 10.3390/pr10010016. [Google Scholar]
- 46.Xu Y, Chen C, Guo Y, Hu S, Sun Z. Effect of CRISPR/Cas9-edited PD-1/PD-L1 on tumor immunity and immunotherapy. Front Immunol. 2022. 10.3389/fimmu.2022.848327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghaffari S, Khalili N, Rezaei N. CRISPR/Cas9 revitalizes adoptive T-cell therapy for cancer immunotherapy. J Exp Clin Cancer Res. 2021;40(1):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang N, Cheng C, Zhang X, Qiao M, Li N, Mu W, Wei X, Wang H. TGF-β inhibition via CRISPR promotes the long-term efficacy of car T cells against solid tumors. JCI Insight. 2020. 10.1172/jci.insight.133977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang L, Ingelshed K, Shen Y, Boddul SV, Iyer VS, Kasza Z, Sedimbi S, Lane DP, Wermeling F. CRISPR/Cas9-induced dna damage enriches for mutations in a p53-linked interactome: implications for CRISPR-based therapies. Cancer Res. 2021. 10.1158/0008-5472.can-21-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azangou-Khyavy M, Ghasemi M, Khanali J, Boroomand-Saboor M, Jamalkhah M, Soleimani M, Kiani J. CRISPR/Cas: from tumor gene editing to t cell-based immunotherapy of cancer. Front Immunol. 2020. 10.3389/fimmu.2020.02062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo R, Lu W, Zhang Y, Cao X, Jin X, Zhao M. Targeting BCMA to treat multiple myeloma: updates from the 2021 ASH annual meeting. Front Immunol. 2022. 10.3389/fimmu.2022.839097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eyquem J, Mansilla-Soto J, Giavridis T, Van Der Stegen SJC, Hamieh M, Cunanan KM, Odak A, Gönen M, Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543(7643):113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu X, Zhang Y, Cheng C, Cheng A, Zhang X, Li N, Xia C, Wei X, Liu X, Wang H. CRISPR-Cas9-mediated multiplex gene editing in car-T cells. Cell Res. 2016;27(1):154–7. 10.1038/cr.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wutti-in Y, Sujjitjoon J, Sawasdee N, Panya A, Kongkla K, Yuti P, Yongpitakwattana P, Thepmalee C, Junking M, Chieochansin T, Pongvarin N, Yamabhai M, Yenchitsomanus PT. Development of a novel anti-CD19 CAR containing a fully human scFv and three costimulatory domains. Front Oncol. 2022. 10.3389/fonc.2021.802876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dai H, Wu Z, Jia H, Tong C, Guo Y, Ti D, Han X, Han Y, Zhang W, Wang C, Zhang Y, Chen M, Yang Q, Wang Y, Han W. Bispecific CAR-T cells targeting both CD19 and CD22 for therapy of adults with relapsed or refractory B cell acute lymphoblastic leukemia. J Hematol Oncol. 2020;13(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spiegel JY, Patel S, Muffly L, Hossain NM, Oak J, Baird JH, Frank MJ, Shiraz P, Sahaf B, Craig J, Iglesias M, Younes S, Natkunam Y, Ozawa MG, Yang E, Chinnasamy H, Ehlinger Z, Reynolds W, Lynn R, et al. CAR T cells with dual targeting of CD19 and CD22 in adult patients with recurrent or refractory B cell malignancies: a phase 1 trial. Nat Med. 2021. 10.1038/s41591-021-01436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu Y, Zhou Y, Zhang M, Ge W, Li Y, Wang L, Wei G, Han L, Wang H, Yu S, Chen Y, Wang Y, He X, Zhang X, Gao M, Yang J, Li X, Ren J, Huang H. CRISPR/Cas9-Engineered Universal CD19/CD22 Dual-Targeted CAR-T Cell Therapy for Relapsed/Refractory B-cell Acute Lymphoblastic Leukemia. Clin Cancer Res. 2021;27(10):2764. [DOI] [PubMed] [Google Scholar]
- 58.Wang M, Munoz J, Goy A, Locke FL, Jacobson CA, Hill BT, Timmerman JM, Holmes H, Jaglowski S, Flinn IW, McSweeney PA, Miklos DB, Pagel JM, Kersten MJ, Milpied N, Fung H, Topp MS, Houot R, Beitinjaneh A, Reagan PM. KTE-X19 CAR T-cell therapy in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2020. 10.1056/NEJMoa1914347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stenger D, Stief TA, Kaeuferle T, Willier S, Rataj F, Schober K, Vick B, Lotfi R, Wagner B, Grünewald TGP, Kobold S, Busch DH, Jeremias I, Blaeschke F, Feuchtinger T. Endogenous TCR promotes in vivo persistence of CD19-CAR-T cells compared to a CRISPR/Cas9-mediated TCR knockout CAR. Blood. 2020;136(12):1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tipanee J, Samara-Kuko E, Gevaert T, Chuah MK, VandenDriessche T. Universal allogeneic CAR T cells engineered with Sleeping Beauty transposons and CRISPR-CAS9 for cancer immunotherapy. Mol Ther. 2022. 10.1016/j.ymthe.2022.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fraietta JA, Nobles CL, Sammons MA, Lundh S, Carty SA, Reich TJ, Cogdill AP, Morrissette JD, DeNizio JE, Reddy S, Hwang Y, Gohil M, Kulikovskaya I, Nazimddin F, Gupta M, Chen F, Everett JK, Alexander KA, Lin-Shiao E, et al. Disruption of TET2 Promotes the Therapeutic Efficacy of CD19-targeted T-cells. Nature. 2018;558(7709):307–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang S. Tet2 at the interface between cancer and immunity. Communications Biology. 2022. 10.1038/s42003-020-01391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salas-Mckee J, Kong W, Gladney WL, Jadlowsky JK, Plesa G, Davis MM, Fraietta JA. CRISPR/Cas9-based genome editing in the era of CAR T cell immunotherapy. Hum Vaccines Immunother. 2019. 10.1080/21645515.2019.1571893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martinez, M., & Moon, E. K. (n.d.). CAR T cells for solid tumors: New strategies for finding, infiltrating, and surviving in the tumor microenvironment. Frontiers in Immunology, 10(Feb), 128. 10.3389/FIMMU.2019.00128/BIBTEX [DOI] [PMC free article] [PubMed]
- 65.Schuster S, Bishop M, Tam C, Waller E, Borchmann P, McGuirk J, Jäger U, Jaglowski S, Andreadis C, Westin J, Fleury I, Bachanova V, Mielke S, Magenau M, Holte H, Holte L, Maziarz R. Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. N Engl J Med. 2019;380(1):45–56. 10.1056/nejmoa1804980. [DOI] [PubMed] [Google Scholar]
- 66.Pasquini M, Hu Z, Curran K, Laetsch T, Locke F, Rouce R, Pulsipher M, Phillips C, Keating A, Frigault A, Salzberg D, Jaglowski S, Sasine J, Rosenthal J, Ghosh M, Landsburg D, Margossian S, Martin P, Kamdar M, Grupp S. Real-world evidence of Tisagenlecleucel for Pediatric Acute lymphoblastic leukemia and non-Hodgkin lymphoma. Blood Adv. 2020;4(21):5414–24. 10.1182/bloodadvances.2020003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Locke F, Ghobadi A, Jacobson C, Miklos D, Lekakis L, Oluwole O, Lin Y, Braunschweig I, Hill B, Timmerman J, Deol A, Reagan P, Stiff P, Flinn I, Farooq U, Goy A, McSweeney P, Munoz J, Siddiqi T, Neelapu S. Long-term safety and activity of Axicabtagene Ciloleucel in refractory large B-cell lymphoma (Zuma-1): a single-arm, multicentre, phase 1–2 trial. Lancet Oncol. 2019;20(1):31–42. 10.1016/s1470-2045(18)30864-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neelapu S, Dickinson M, Munoz J, Ulrickson M, Thieblemont C, Oluwole O, Herrera A, Ujjani C, Lin Y, Riedell P, Kekre N, Vos S, Lui C, Milletti F, Dong J, Xu H, Chavez J. Axicabtagene CILOLEUCEL as first-line therapy in high-risk large B-cell lymphoma: the phase 2 zuma-12 trial. Nat Med. 2022;28(4):735–42. 10.1038/s41591-022-01731-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson M, Torosyan A, Halford Z. Brexucabtagene Autoleucel: a novel chimeric antigen receptor T-cell therapy for the treatment of mantle cell lymphoma. Ann Pharmacother. 2021;56(5):609–19. 10.3389/fimmu.2022.830292. [DOI] [PubMed] [Google Scholar]
- 70.Ball, G., Lemieux, C., Cameron, D., & Seftel, M. (n.d.). Cost-effectiveness of Brexucabtagene Autoleucel versus best supportive care for the treatment of relapsed/refractory mantle cell lymphoma following treatment with a Bruton’s tyrosine kinase inhibitor in Canada. Current Oncology, 29(3), 2021–2045. 10.3390/curroncol29030164. [DOI] [PMC free article] [PubMed]
- 71.Frey N. Approval of Brexucabtagene Autoleucel for adults with relapsed and refractory acute lymphocytic leukemia. Blood. 2022;140(1):11–5. 10.1182/blood.2021014892. [DOI] [PubMed] [Google Scholar]
- 72.Mann H, Comenzo R. Evaluating the therapeutic potential of Idecabtagene Vicleucel in the treatment of multiple myeloma: evidence to date. Onco Targets Ther. 2022;15(1):799–813. 10.2147/ott.s305429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Munshi N, Anderson L, Shah N, Madduri D, Berdeja J, Lonial S, Raje N, Lin Y, Siegel D, Oriol A, Moreau P, Yakoub-Agha I, Delforge M, Cavo M, Einsele H, Goldschmidt H, Weisel K, Rambaldi A, Reece D, San-Miguel J. Idecabtagene Vicleucel in relapsed and refractory multiple myeloma. N Engl J Med. 2021;384(8):705–16. 10.1056/nejmoa2024850. [DOI] [PubMed] [Google Scholar]
- 74.Iragavarapu C, Hildebrandt G. Lisocabtagene Maraleucel for the treatment of B-cell lymphoma. Expert Opin Biol Ther. 2021;21(9):1151–6. 10.1080/14712598.2021.1933939. [DOI] [PubMed] [Google Scholar]
- 75.Kamdar M, Solomon S, Arnason J, Johnston P, Glass B, Bachanova V, Ibrahimi S, Mielke S, Mutsaers P, Hernandez-Ilizaliturri F, Izutsu K, Morschhauser F, Lunning M, Maloney D, Crotta A, Montheard S, Previtali A, Stepan L, Ogasawara K, Abramson J. Lisocabtagene Maraleucel versus standard of care with salvage chemotherapy followed by autologous stem cell transplantation as second-line treatment in patients with relapsed or refractory large B-cell lymphoma (transform): results from an interim analysis. Lancet. 2022;399(10343):2294–308. 10.1016/s0140-6736(22)00662-6. [DOI] [PubMed] [Google Scholar]
- 76.Berdeja J, Madduri D, Usmani S, Jakubowiak A, Agha M, Cohen A, Stewart A, Hari P, Htut M, Lesokhin A, Deol A, Munshi N, O’Donnell E, Avigan D, Singh I, Zudiare E, Yeh T, Allred A, Olyslager Y, Jagannath S. Ciltacabtagene autoleucel, a B-cell maturation antigen-directed chimeric antigen receptor T-cell therapy in patients with relapsed or refractory multiple myeloma (cartitude-1): a phase 1b/2 open-label study. Lancet. 2021;398(10297):314–24. 10.1016/s0140-6736(21)00933-8. [DOI] [PubMed] [Google Scholar]
- 77.Martin T, Usmani S, Berdeja J, Agha M, Cohen A, Hari P, Avigan D, Deol A, Htut M, Lesokhin A, Munshi N, O’Donnell E, Stewart A, Schecter J, Goldberg J, Jackson C, Yeh T, Banerjee A, Allred A, Jagannath S. Ciltacabtagene Autoleucel, an anti–B-cell maturation antigen chimeric antigen receptor T-cell therapy, for relapsed/refractory multiple myeloma: Cartitude-1 2-year follow-up. J Clin Oncol. 2022. 10.1200/jco.22.00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kagoya Y, Guo T, Yeung B, Saso K, Anczurowski M, Wang C, Murata K, Sugata K, Saijo H, Matsunaga Y, Ohashi Y, Butler M, Hirano N. Genetic ablation of HLA class I, class II, and the T-cell receptor enables allogeneic T cells to be used for adoptive T-cell therapy. Cancer Immunol Res. 2020;8(7):26–936. 10.1158/2326-6066.cir-18-0508. [DOI] [PubMed] [Google Scholar]
- 79.Ren J, Liu X, Fang C, Jiang S, June C, Zhao Y. Multiplex genome editing to generate universal car T cells resistant to PD1 inhibition. Clin Cancer Res. 2017;23(9):2255–66. 10.1158/1078-0432.ccr-16-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rupp LJ, Schumann K, Roybal KT, Gate RE, Ye CJ, Lim WA, Marson A. CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci Rep. 2017. 10.1038/s41598-017-00462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin S, Cheng L, Ye W, Li S, Zheng D, Qin L, Wu Q, Long Y, Lin S, Wang S, Huang G, Li P, Yao Y, Sun X. Chimeric CTLA4-CD28-CD3Z T cells potentiate antitumor activity against CD80/CD86–positive B cell malignancies. Front Immunol. 2021. 10.3389/fimmu.2021.642528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Y, Zhang X, Cheng C, Wei M, Liu X, Li N, Wei X, Liu X, Xia C, Wang H. CRISPR-Cas9 mediated lag-3 disruption in car-T cells. Front Med. 2017;11(4):554–62. 10.1007/s11684-017-0543-6. [DOI] [PubMed] [Google Scholar]
- 83.Ciralo E, Althoff S, Ru J, Rosney S, Butze M, Puhl M, Frentsch M, Bullinger L, Na I. Simultaneous genetic ablation of PD-1, LAG-3, and tim-3 in CD8 T cells delays tumor growth and improves survival outcome. Int J Mol Sci. 2022;23(6):3207. 10.3390/ijms23063207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jung I, Kim Y, Yu H, Lee M, Kim S, Lee J. CRISPR/Cas9-mediated knockout of DGK improves antitumor activities of human T cells. Can Res. 2018;78(16):4692–703. 10.1158/0008-5472.can-18-0030. [DOI] [PubMed] [Google Scholar]
- 85.Li N, Tang N, Cheng C, Hu T, Wei X, Han W, Wang H. Improving the anti-solid tumor efficacy of CAR-T cells by inhibiting adenosine signaling pathway. OncoImmunology. 2020. 10.1080/2162402x.2020.1824643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Giuffrida L, Sek K, Henderson M, Lai J, Chen A, Meyran D, Todd K, Petley E, Mardiana S, Molck C, Stewart G, Solomon B, Parish I, Neeson P, Harrison S, Kats L, House I, Darcy P, Baevis P. CRISPR/Cas9 mediated deletion of the adenosine A2A receptor enhances car T cell efficacy. Nat Commun. 2021. 10.1038/s41467-021-23331-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Du X, Wiede F, Darcy P, Tiganis T. Abstract PR04: CRISPR-mediated PTPN2 deletion in car T cells enhances anti-tumor efficacy. Cancer Immunol Res. 2022. 10.1158/2326-6074.tumimm21-pr04. [Google Scholar]
- 88.Stüber T, Monjezi R, Wallstabe L, Kühnemundt J, Nietzer S, Dandekar G, Wöckel A, Hudecek M. Inhibition of TGF-β-receptor signaling augments the antitumor function of ROR1-specific car T-cells against triple-negative breast cancer. J ImmunoTher Cancer. 2020. 10.1136/jitc-2020-000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alishah K, Birtel M, Masoumi E, Jafarzadeh L, Mirzaee H, Hadjati J, Voss R, Diken M, Asad S. CRISPR/Cas9-mediated TGFβRII disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells in vitro. J Transl Med. 2021;19(1):1. 10.1186/s12967-021-03146-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yi Y, Chai X, Zheng L, Zhang Y, Shen J, Hu B, Tao G. CRISPR-edited cart with GM-CSF knockout and auto secretion of IL6 and IL1 blockers in patients with hematologic malignancy. Cell Discov. 2021. 10.1038/s41421-021-00255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Andrea A, Chiron A, Mallah S, Bessoles S, Sarrabayrouse G, Hacein-Bey-Abina S (n.d.). Advances in car-T cell genetic engineering strategies to overcome hurdles in solid tumors treatment. Front Immunol. 10.3389/fimmu.2022.830292. [DOI] [PMC free article] [PubMed]
- 92.Park J, Rivière I, Gonen M, Wang X, Sénéchal B, Curran K, Sauter C, Wang Y, Santomasso B, Mead E, Roshal M, Maslak P, Davila M, Brentjens R, Sadelain M. Long-term follow-up of CD19 car therapy in Acute lymphoblastic leukemia. N Engl J Med. 2018;378(5):449–59. 10.1056/nejmoa1709919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maude S, Laetsch T, Buechner J, Rives S, Boyer M, Bittencourt H, Bader P, Verneris M, Stefanski H, Myers G, Qayed M, Moerloose B, Hiramatsu H, Schlis K, Davis K, Martin P, Nemecek E, Yanik G, Peters C, Grupp S. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N Engl J Med. 2018;378(5):439–48. 10.1056/nejmoa1709866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alizadeh D, Wong R, Yang X, Wang D, Pecoraro J, Kuo C, Aguilar B, Qi Y, Ann D, Starr R, Urak R, Wang X, Forman S, Brown C. IL15 enhances car-T cell antitumor activity by reducing mtorc1 activity and preserving their stem cell memory phenotype. Cancer Immunol Res. 2019;7(5):759–72. 10.1158/2326-6066.cir-18-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Beltra J, Manne S, Abdel-Hakeem M, Kurachi M, Giles J, Chen Z, Casella V, Ngiow S, Khan O, Huang Y, Yan P, Xu W, Amaravadi R, Xu X, Karakousis G, Mitchell T, Schuchter L, Huang A, Wherry E. Developmental relationships of four exhausted CD8+ T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity. 2020. 10.1016/j.immuni.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li J, Li W, Huang K, Zhang Y, Kupfer G, Zhao Q. Chimeric antigen receptor T cell (CAR-T) immunotherapy for solid tumors: lessons learned and strategies for moving forward. J Hematol Oncol. 2018. 10.1186/s13045-018-0568-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zheng B, Ren T, Huang Y, Sun K, Wang S, Bao X, Liu K, Guo W. PD-1 axis expression in musculoskeletal tumors and antitumor effect of nivolumab in osteosarcoma model of humanized mouse. J Hematol Oncol. 2018. 10.1186/s13045-018-0560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qin W, Hu L, Zhang X, Jiang S, Li J, Zhang Z, Wang X. The diverse function of pd-1/PD-L pathway beyond cancer. Front Immunol. 2019. 10.3389/fimmu.2019.02298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rafiq S, Yeku OO, Jackson HJ, Purdon TJ, van Leeuwen DG, Drakes DJ, Song M, Miele MM, Li Z, Wang P, Yan S, Xiang J, Ma X, Seshan VE, Hendrickson RC, Liu C, Brentjens RJ. Targeted delivery of a pd-1-blocking scfv by car-T cells enhances anti-tumor efficacy in vivo. Nat Biotechnol. 2018;36(9):847–56. 10.1038/nbt.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Su S, Hu B, Shao J, Shen B, Du J, Du Y, Zhou J, Yu L, Huang X, Liu B. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci Rep. 2016. 10.1038/srep20070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hu W, Zi Z, Jin Y, Li G, Shao K, Cai Q, Ma X, Wei F. CRISPR/Cas9-mediated PD-1 disruption enhances human mesothelin-targeted car T cell effector functions. Cancer Immunol Immunother. 2018;68(3):365–77. 10.1007/s00262-018-2281-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stadtmauer E, Fraietta J, Davis M, Cohen A, Weber K, Lancaster E, Mangan P, Kulikovskaya I, Gupta M, Chen F, Tian L, Gonzalez V, Xu J, Melenhorst J, Plesa G, Shea J, Matlawski T, Cervini A, June C. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020. 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Choi B, Yu X, Castano A, Darr H, Henderson D, Bouffard A, Larson R, Scarfo I, Bailey S, Gerhard G, Frigault M, Leick M, Schmidt A, Sagert J, Curry W, Carter B, Maus M. CRISPR-Cas9 disruption of PD-1 enhances activity of universal EGFRVIII car T cells in a preclinical model of human glioblastoma. J ImmunoTherapy Cancer. 2019. 10.1186/s40425-019-0806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakazawa T, Natsume A, Nishimura F, Morimoto T, Matsuda R, Nakamura M, Yamada S, Nakagawa I, Motoyama Y, Park Y-S, Tsujimura T, Wakabayashi T, Nakase H. Effect of CRISPR/cas9-mediated PD-1-disrupted primary human third-generation CAR-T cells targeting EGFRVIII on in vitro human glioblastoma cell growth. Cells. 2020;9(4):998. 10.3390/cells9040998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Qin L, Zhao R, Chen D, Wei X, Wu Q, Long Y, Jiang Z, Li Y, Wu H, Zhang X, Cui S, Wei W, Yao H, Liu Z, Cao S, Yao Y, Zhang Z, Li P. Chimeric antigen receptor T cells targeting PD-L1 suppress tumor growth. Biomark Res. 2020. 10.1186/s40364-020-00198-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yajima T, Hoshino K, Muranushi R, Mogi A, Onozato R, Yamaki E, Yoshikai Y, Kuwano H. FAS/FASL signaling is critical for the survival of exhausted antigen-specific CD8+ T cells during tumor immune response. Mol Immunol. 2019;107:97–105. 10.1016/j.molimm.2019.01.01. [DOI] [PubMed] [Google Scholar]
- 107.Ruffo E, Wu R, Bruno T, Workman C, Vignali D. Lymphocyte-activation gene 3 (LAG3): the next immune checkpoint receptor. Semin Immunol. 2019;42:101305. 10.1016/j.smim.2019.101305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Almena M, Andrada E, Liebana R, Merida I. Diacylglycerol metabolism attenuates T-cell receptor signaling and alters thymocyte differentiation. Trends Biochem Sci. 2011;36(11):593–603. 10.1016/j.tibs.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen S, Hu Z, Zhong X. Diacylglycerol kinases in T cell tolerance and effector function. Front Cell Dev Biol. 2016. 10.3389/fcell.2016.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Krishna S, Zhong X. Role of diacylglycerol kinases in T cell development and function. Crit Rev Immunol. 2013;33(2):97–118. 10.1615/critrevimmunol.2013006696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhong X, Shin J, Gorentla B, O’Brien T, Srivatsan S, Xu L, Chen Y, Xie D, Pan H. Receptor signaling in immune cell development and function. Immunol Res. 2010;49(13):109–23. 10.1007/s12026-010-8175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Watternberg B, Raben D. Diacylglycerol kinases put the brakes on immune function. Sci STKE. 2007;2007:398. 10.1126/stke.3982007pe43. [DOI] [PubMed] [Google Scholar]
- 113.Noessner E. DGK-α: A checkpoint in cancer-mediated immuno-inhibition and target for immunotherapy. Front Cell Dev Biol. 2017. 10.3389/fcell.2017.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Batlle E, Massague J. Transforming growth factor-β signaling in immunity and cancer. Immunity. 2019;50(4):924–40. 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hua W, Dijke P, Kostidis S, Giera M, Hornsveld M. TGFΒ-induced metabolic reprogramming during epithelial-to-mesenchymal transition in cancer. Cell Mol Life Sci. 2019;77(11):2103–23. 10.1007/s00018-019-03398-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stuelten C, Zhang Y. Transforming growth factor-β: An agent of change in the tumor microenvironment. Front Cell Dev Biol. 2021. 10.3389/fcell.2021.764727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Donkor M, Sarkar A, Li M. TGF-β1 produced by activated CD4+T cells antagonizes T cell surveillance of tumor development. OncoImmunology. 2012;1(2):162–71. 10.4161/onci.1.2.18481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Freudenberg K, Lindner N, Dohnke S, Garbe A, Schallenberg S, Kretschmer K. Critical role of TGF-β and IL-2 receptor signaling in Foxp3 induction by an inhibitor of DNA methylation. Front Immunol. 2018. 10.3389/fimmu.2018.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Folmont A, Bourhis J-H, Chouaib S, Terry S. Multifaceted role of the transforming growth factor β on effector T cells and the implication for car-T cell therapy. Immuno. 2021;1(3):160–73. 10.3390/immuno1030010. [Google Scholar]
- 120.Gunderson A, Yamazaki T, McCarty K, Fox N, Phillips M, Alice A, Blair T, Whiteford M, O’Brien D, Ahmad R, Kiely M, Hayman A, Crocenzi T, Gough M, Crittenden M, Young K. TGFΒ suppresses CD8+ T cell expression of CXCR3 and tumor trafficking. Nat Commun. 2020. 10.1038/s41467-020-15404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sitkovsky M, Hatfield S, Abbott R, Belikoff B, Lukashev D, Ohta A. Hostile, hypoxia–A2-adenosinergic tumor biology as the next barrier to overcome for tumor immunologists. Cancer Immunol Res. 2014;2(7):598–605. 10.1158/2326-6066.cir-14-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cekic C, Linden J. Adenosine A2A receptors intrinsically regulate CD8+ T cells in the tumor microenvironment. Can Res. 2014;74(24):7239–49. 10.1158/0008-5472.can-13-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Allard B, Turcotte M, Stagg J. CD73-generated adenosine: Orchestrating the tumor-stroma interplay to promote cancer growth. J Biomed Biotechnol. 2012;2012(485156):1–8. 10.1155/2012/485156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Beavis P, Henderson M, Guiffrida L, Mills J, Sek K, Cross R, Davenport A, John L, Mardiana S, Slaney C, Johnstone R, Trapani J, Stagg J, Loi S, Kats L, Gyorki D, Kershaw M, Darcy P. Targeting the adenosine 2A receptor enhances chimeric antigen receptor T cell efficacy. J Clin Investig. 2017;127(3):929–41. 10.1172/jci89455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wiede F, Sacirbegovic F, Leong Y, Yu D, Tiganis T. PTPN2-deficiency exacerbates T follicular helper cell and B cell responses and promotes the development of autoimmunity. J Autoimmun. 2017;76:85–100. 10.1016/j.jaut.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 126.Wiede F, Lu K, Du X, Liang S, Hochheiser K, Dodd G, Kearney C, Beavis P, Gebhardt T, Darcy P, Tiganis T, Zhang S. ptpn2 phosphatase deletion in T cells promotes anti-tumour immunity and T-cell efficacy in solid tumours. EMBO J. 2019. 10.15252/embj.2019103637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Flosbach M, Oberle S, Scherer S, Zecha J, von Hoesslin M, Wiede F, Chennupati V, Cullen J. PTPN2 deficiency enhances programmed T cell expansion and survival capacity of activated T cells. Cell Rep. 2020;32(4):107957. 10.1016/j.celrep.2020.107957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ma W, Wang Y, Zhang R, Yang F, Zhang D, Huang M, Zhang L, Dorsey J, Binder Z, O’Rourke D, Fraietta J, Gong Y, Fan Y. Targeting PAK4 to reprogram the vascular microenvironment and improve CAR-T immunotherapy for glioblastoma. Nat Cancer. 2020;2(1):83–97. 10.1038/s43018-020-00147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Prapa M, Chiavelli C, Golinelli G, Grisendi G, Bestagno M, Tinco R, Dall’Ora M, Neri G, Candini O, Petrachi T, Bertoni L, Carnevale G, Depenni R, Feletti A, Iaccarino C, Pavesi G, Dominici M. GD2 car T cells against human glioblastoma. npj Precis Oncol. 2021. 10.1038/s41698-021-00233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Reppel L, Tsahouridis O, Akulian J, Davis I, Lee H, Fuca G, Weiss J, Dotti G, Pecot C, Savoldo B. Targeting disialoganglioside GD2 with chimeric antigen receptor-redirected T cells in lung cancer. J Immunother Cancer. 2022;10(1):3897. 10.1136/jitc-2021-003897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hung J, Kao Y, Huang C, Hsu W. Overexpression of Aiolos promotes epithelial-mesenchymal transition and cancer stem cell-like properties in lung cancer cells. Sci Rep. 2019. 10.1038/s41598-019-39545-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zou Y, Liu B, Li L, Yin Q, Tang J, Huang X, Zhu X, Chi T. IKZF3 deficiency potentiates chimeric antigen receptor T cells targeting solid tumors. Cancer Lett. 2022;524:121–30. 10.1016/j.canlet.2021.10.016. [DOI] [PubMed] [Google Scholar]
- 133.Chen J, López-Moyado I, Seo H, Lio C, Hempleman L, Sekiya T, Yoshimura A, Scott-Browne J, Rao A. NR4A transcription factors limit car T cell function in solid tumours. Nature. 2019;567(7749):530–4. 10.1038/s41586-019-0985-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Seo H, Chen J, González E, Samaniego-Castruita D, Das A, Wang Y, Moyado I, Georges R, Zhang W, Onodera A, Wu C, Lu F, Hogan P, Bhandoola A, Rao A. Tox and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8 + T cell exhaustion. Proc Natl Acad Sci USA. 2019;116(25):12410–5. 10.1073/pnas.1905675116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lynn R, Weber E, Sotillo E, Gennert D, Xu P, Good Z, Anbunathan H, Lattin J, Jones R, Tieu V, Nagaraja S, Granja J, Bourcy C, Majzner R, Satpathy A, Quake S, Monje M, Chang H, Mackall C. C-jun overexpression in car T cells induces exhaustion resistance. Nature. 2019;576(7786):293–300. 10.1038/s41586-019-1805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Titov A, Petukhov A, Staliarova A, Motorin D, Bulatov E, Shuvalov O, Soond S, Melino G, Zaritskey A, Barlev N. The biological basis and clinical symptoms of CAR-T therapy-associated toxicites. Cell Death Dis. 2018. 10.1038/s41419-018-0918-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gong T, Liu L, Jiang W, Zhou R. Damp-sensing receptors in sterile inflammation and inflammatory diseases. Nat Rev Immunol. 2019;20(2):95–112. 10.1038/s41577-019-0215-7. [DOI] [PubMed] [Google Scholar]
- 138.Liu Y, Fang Y, Chen X, Wang Z, Liang X, Zhang T, Liu M, Zhou N, Lv J, Tang K, Xie J, Gao Y, Cheng F, Zhou Y, Zhen Zhang Y, Zhang X, Gao Q, Zhang Y, Huang B. Gasdermin E–mediated target cell pyroptosis by car T cells triggers cytokine release syndrome. Sci Immunol. 2020. 10.1126/sciimmunol.aax7969. [DOI] [PubMed] [Google Scholar]
- 139.Zhou Z, He H, Wang K, Shi X, Wang Y, Su Y, Wang Y, Li D, Liu W, Zhang Y, Shen L, Ding J, Shao F. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science. 2020. 10.1126/science.aaz7548. [DOI] [PubMed] [Google Scholar]
- 140.Staedtke V, Bai R, Kim K, Darvas M, Davila M, Riggins G, Rothman P, Papadopoulos N, Vogelstein B, Zhou S. Disruption of a self-amplifying catecholamine loop reduces cytokine release syndrome. Nature. 2018;564(7735):273–7. 10.1038/s41586-018-0774-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Giavridis T, van der Stegen S, Eyquem J, Hamieh M, Piersigilli A, Sadelain M. Car T cell–induced cytokine release syndrome is mediated by macrophages and abated by IL-1 blockade. Nat Med. 2018;24(6):731–8. 10.1038/s41591-018-0041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Norelli M, Camisa B, Barbiera G, Falcone L, Purevdorj A, Genua M, Sanvito F, Ponzoni M, Doglioni C, Cristofori P, Traversari C, Bordignon C, Ciceri F, Ostuni R, Bonini C, Casucci M, Bondanza A. Monocyte-derived IL-1 and IL-6 are differentially required for cytokine-release syndrome and neurotoxicity due to car T cells. Nat Med. 2018;24(6):739–48. 10.1038/s41591-018-0036-4. [DOI] [PubMed] [Google Scholar]
- 143.Morris E, Neelapu S, Giavridis T, Sadelain M. Cytokine release syndrome and associated neurotoxicity in cancer immunotherapy. Nat Rev Immunol. 2021;22(2):85–96. 10.1038/s41577-021-00547-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xiao X, Huang S, Chen S, Wang Y, Sun Q, Xu X, Li Y. Mechanisms of cytokine release syndrome and neurotoxicity of car T-cell therapy and associated prevention and management strategies. J Exp Clin Cancer Res. 2021. 10.1186/s13046-021-02148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sterner R, Sakemura R, Cox M, Yang N, Khadka R, Forsman C, Hansen M, Kenderian S. GM-CSF inhibition reduces cytokine release syndrome and neuroinflammation but enhances car-T cell function in Xenografts. Blood. 2019;133(7):697–709. 10.1182/blood-2018-10-881722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cox M, Manriquez Roman C, Tapper E, Siegler E, Chappell D, Durrant C, Ahmed O, Sinha S, Mwangi R, Scott N, Hefazi M, Schick K, Horvei P, Ruff M, Can I, Adada M, Bezerra E, Kankeu Fonkoua L, Parikh S, Kenderian S. GM-CSF disruption in CART cells modulates T cell activation and enhances cart cell anti-tumor activity. Leukemia. 2022;36(6):1635–45. 10.1038/s41375-022-01572-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hoyos V, Savoldo B, Quintarelli C, Mahendravada A, Zhang M, Vera J, Heslop H, Rooney C, Brenner M, Dotti G. Engineering CD19-specific T lymphocytes with interleukin-15 and a suicide gene to enhance their anti-lymphoma/leukemia effects and safety. Leukemia. 2010;24(6):1160–70. 10.1038/leu.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Budde L, Berger C, Lin Y, Wang J, Lin X, Frayo S, Brouns S, Spencer D, Till B, Jensen M, Riddell S, Press O. Combining a CD20 chimeric antigen receptor and an inducible caspase 9 suicide switch to improve the efficacy and safety of T cell adoptive immunotherapy for lymphoma. PLoS ONE. 2013. 10.1371/journal.pone.0082742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Diaconu I, Ballard B, Zhang M, Chen Y, West J, Dotti G, Savoldo B. Inducible caspase-9 selectively modulates the toxicities of CD19-specific chimeric antigen receptor-modified T cells. Mol Ther. 2017;25(3):580–92. 10.1016/j.ymthe.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fu Y, Foden J, Khayter C, Maeder M, Reyon D, Joung J, Sander J. High-frequency off-target mutagenesis induced by CRISPR-cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–6. 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR–cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;54(7):927–30. 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 152.Kim S, Koo T, Jee H, Cho H, Lee G, Lim D, Shin H, Kim J. CRISPR RNAS trigger innate immune responses in human cells. Genome Res. 2018;28(3):367–73. 10.1101/gr.231936.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Naeem M, Majeed S, Hoque M, Ahamad I. Latest developed strategies to minimize the off-target effects in CRISPR-cas-mediated genome editing. Cells. 2020;9(7):1608. 10.3390/cells9071608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rees HA, Komor AC, Yeh W-H, Caetano-Lopes J, Warman M, Edge AS, Liu DR. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat Commun. 2017. 10.1038/ncomms15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Anzalone A, Randolph P, Davis J, Sousa A, Koblan L, Levy J, Chen P, Wilson C, Newby G, Rauguram A, Liu D. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576(7785):149–57. 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]