Abstract
Biomedical Entity Linking (BEL) is the task of mapping of spans of text within biomedical documents to normalized, unique identifiers within an ontology. This is an important task in natural language processing for both translational information extraction applications and providing context for downstream tasks like relationship extraction. In this paper, we will survey the progression of BEL from its inception in the late 80s to present day state of the art systems, provide a comprehensive list of datasets available for training BEL systems, reference shared tasks focused on BEL, discuss the technical components that comprise BEL systems, and discuss possible directions for the future of the field.
Keywords: Natural Language Processing, Entity Linking, Normalization
1. Introduction
Biomedical entity linking (BEL), also known as normalization or grounding, is a natural language processing (NLP) task dealing with the mapping of spans of text within biomedical documents to normalized, unique identifiers within an ontology. Associating these spans, known as mentions, with a discrete concept allows information extracted from the text to be easily filtered and aggregated. In this paper, we will survey the progression of BEL from its inception in the late 80s to present day state of the art systems, provide a comprehensive list of datasets available for training BEL systems, reference shared tasks focused on BEL, describe the technical components that comprise BEL systems, and discuss where the field needs to go from here.
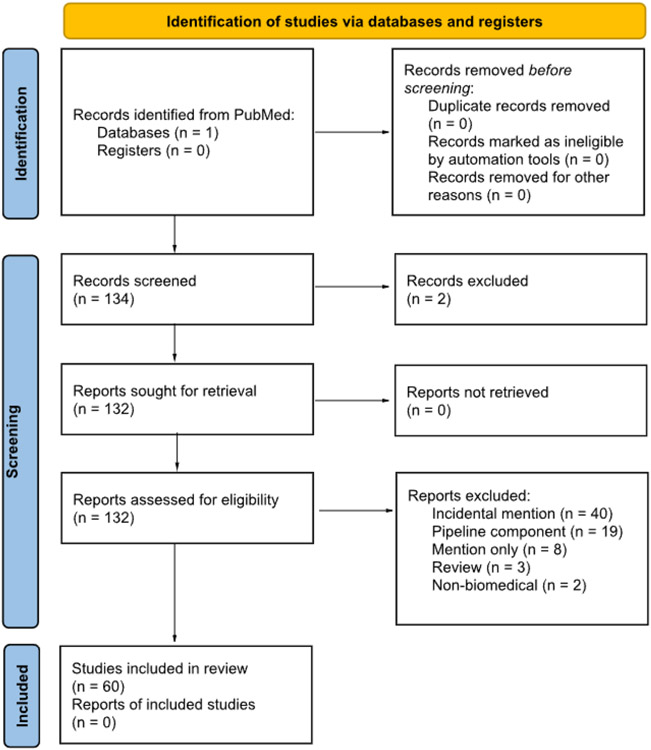
To conduct this systematic review, we queried PubMed for all full-text articles published between 1980 and August 2022 which contained any of the phrases “entity linking”, “entity normalization”, “concept linking”, or “concept normalization” in the abstract or title. This search yielded 134 results, of which, the earliest was published in 1985. We retrieved 132 of these for further review, excluding two results which corresponded to a book chapter and a corrigendum. After review, we excluded another 72 results for reasons such as the abstract or title incidentally containing one of the search phrases without referring to the NLP task (40), the primary focus of the paper being a pipeline which includes a non-novel BEL component (19), the paper mentioning entity linking in passing (11), the paper being a review which did not introduce any original contributions (3), and the paper focusing on non-biomedical applications of entity linking (2). A PRISMA diagram showing how we filtered the results of our search is shown in Figure 1. We have included some additional publications related to biomedical entity linking in the following sections which were not included in the PubMed results, but which we felt had substantively contributed to the field.
Figure 1:
PRISMA flow diagram of publications returned from a PubMed search for full-text articles published between 1980 and August 2022 which contained any of the phrases “entity linking”, “entity normalization”, “concept linking”, or “concept normalization” in the abstract or title.
2. History
2.1. Early Work
In the late 1980’s, medical literature was expanding rapidly, but physicians were unable to search it effectively due to unfamiliarity with the Medical Subject Headings (MeSH) vocabulary used to index citations in the MEDLINE database [1]. This impediment motivated the initial work on BEL. To improve search efficacy for non-expert users, two physicians at Massachusetts General Hospital proposed MicroMeSH in 1987, an “intelligent search assistant” for searching the MEDLINE database, which used a synonym, acronym, and abbreviation dictionary to map users’ search queries to a list of possible MeSH terms with wildcard matching [1]. The idea was later expanded to facilitate the MeSH indexing of articles directly with systems such as CLARIT (1991) [2], SAPHIRE (1995) [3], OSCAR4 (2011) [4], and MetaMap (2001) [5]. These subsequent systems used linguistic rules, patterns, and dictionaries to map concept mentions to MeSH terms. MetaMap became the backbone of the Medical Text Indexer (MTI) [6] in 2004. Today, the National Library of Medicine (NLM) at the National Institutes of Health (NIH) employs MTI as the automated first-line indexer for over 350 journals.
Application of BEL to clinical text was not far behind indexing publications. CHARTLINE (1992) [7] and MedLEE (1995) [8] used similar dictionary matching techniques to extract and link entities in clinical reports to the Unified Medical Language System (UMLS). REX (2006) [9], by physicians Friedlin and McDonald, linked mentions from clinical notes to ICD-9-CM codes to facilitate medical record coding and included the novel feature of negation recognition to mitigate false positives for negative mentions (i.e. patient denies smoking). Friedlin later adapted his REX system to identify adverse drug reactions (ADR) mentioned on drug labels and link them to the Medical Dictionary for Regulatory Activities (MedDRA) with a system called SPLICER [10]. Shortly after Friedlin’s publications, Savova et al. [11] also released an end-to-end clinical NLP system called cTAKES (2010), which included an entity linking component. QuickUMLS [12] (2016) addressed the computational performance limitations of its predecessors by using an approximate dictionary matching algorithm, CPMerge, to achieve higher F1 scores than both MetaMap and cTAKES while requiring only a fraction of their runtime. RysannMD [13] similarly created a fast and accurate system which used a probabilistic model based on the individual tokens in each mention to predict concept mappings.
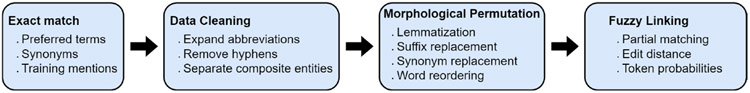
For developing the first generation of BEL systems, which relied exclusively on dictionary matching techniques and jointly performed NER and entity linking, researchers generally annotated their own training data from scratch. This changed in the mid-2010s with the release of prominent entity linking corpora, such as the ShARe/CLEF eHealth Challenge corpus[14] and the NCBI dataset [15] which provided a set of linked mentions out of the box. For the first time, researchers could model BEL as an independent task, limiting the scope of their work to matching a mention assumed to be an entity to its corresponding concept. This allowed for more complex perturbations of pre-extracted mentions, which would have been combinatorially intractable when considering a document in its entirety. D’Souza and Ng [16] broke ground with an influential sieve-based method that attempted to match mentions to concepts through ten progressively fuzzy layers of morphological permutations. Figure 2 illustrates potential layers in a rule-based BEL pipeline. Rather than permuting the limited set of mentions in an attempt to match concepts in the much larger ontology, Liu, et al. [17] created their own semantic lexicon based on knowledge from the UMLS and information mined from a large clinical corpus to maximize the probability of extracting mentions from a corpus which correspond to concepts in their MedLEx. Leal et al. [18] applied a rule-based similarity approach to the ShARe/CLEF dataset by searching for matches by minimizing Levenshtein distance to SNOMED-CT candidates and resolving ties by choosing the SNOMED-CT concept with the lowest Information Content (IC) [19]. While these systems were more sophisticated than their predecessors, they still shared many of the core limitations of the earliest work. Rule-based systems are generally fast, but they are unable to consider semantic meaning, so they struggle when linking mentions that require either context (i.e. does “depression” refer to a mood disorder or a sunken area?) or when vernacular for describing a concept is too lexically diverse (i.e. how many ways can you say “inadequate oral intake”?).
Figure 2:
Possible steps in a rule-based BEL pipeline where the system attempts to match progressively more permuted versions of the initial mention.
2.2. Modern Era
While dictionary-based clinical NLP methods remain popular for production implementation because of their interpretability and configurability [20], learning-based methods have largely replaced them in informatics research because of their superior performance. This paradigm shift transitioned BEL from a matching problem to a mapping problem requiring successful systems to numerically represent mentions and concepts and train models to connect them. One of the best-known early attempts at applying machine learning to BEL was DNorm [21], which used TF-IDF representations of mentions and concepts to train a linear classifier to score pairs of mention and concept representations. DNorm demonstrated a nearly 10 point gain in F-measure performance over existing rule-based baselines, becoming the defacto baseline for subsequent systems. The author later incorporated DNorm into a joint NER and BEL model called TaggerOne [22], which considered the results of two scoring functions in semi-Markov models that determined both the mention boundaries of the entity and linked it to the appropriate concept.
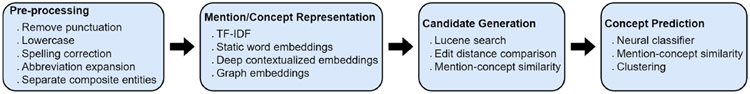
The first round of deep learning techniques applied to BEL represented tokens with static vector representations of words (such as TF-IDF and word embeddings [23]) and used architectures like CNN and BiLSTM to demonstrate improvement over classical machine learning (ML) baselines like DNorm [24, 25, 26]. The emergence of deep contextual embeddings, such as ELMo[27] and BERT[28], effected a sea change in natural language processing research, and BEL research has been no exception. While some researchers still investigate using static embeddings as their primary form of representation, all current state of the art systems use some form of deep contextualized embeddings, with BERT encoders pre-trained on clinical and/or biomedical text being the clear favorites [29, 30, 31]. As with classical ML BEL, both binary [32] and multi-class [33] classification models are popular, but the improved quality of representations and the ability to train the encoder has opened up other options as well, like similarity-based ranking [29] and clustering [31]. Figure 3 illustrates typical steps in a machine learning-based BEL pipeline and lists some of the configurable options for each step.
Figure 3:
Typical steps in a machine learning-based BEL pipeline.
3. Applications
The Apache Unstructured Information Management Architecture™ (UIMA) framework [34] is an interoperability platform developed to handle software systems that process large amounts of text. Its advantage is the plug-and-play aspect allowing different components to be pipelined together. The framework has been ported from general English to process large amounts of clinical and biomedical text. CLAMP [35], cTAKES [11], Leo [36], MedTagger [37], and NOBLE Coder [38] all utilize the UIMA framework. One key component to each of these frameworks with respect to this review is the addition of a BEL component into their information extraction pipelines. A typical pipeline includes components that 1) initially extract specific entity types (e.g., Diseases, Drugs) from the text, 2) determine the relationship between the entities (e.g., Treats, Reason), and 3) link the entities to their respective concept in an ontology (e.g., the UMLS). The BEL component normalizes synonymous terms (e.g., Heart Attack and Myocardial Infarction) allowing information across documents to be analyzed regardless of their lexical diversity. However, as with relation extraction, error propagation [39] becomes a challenge in real-world environments where any error that occurs when identifying the entities is propagated to downstream tasks including both the identification of the relations between the entities and the linking of those entities to their respective concepts.
These system have been used to develop information extraction pipelines to address use cases centered around the extraction of specific types of information from clinical notes [20]. For example, mapping clinical entities in notes to Fast Healthcare Interoperability Resources (FHIR) standards [40] to supplement discrete electronic health record data for purposes such as cohort identification and clinical monitoring. Another example includes automatically assigning International Codes for Diseases (ICD-9-CM/ICD-10-CM) to clinical records for automated billing [41, 42]. These codes are typically utilized for billing purposes but can also provide salient disease or symptom information about the patient [42].
4. Datasets
The set of biomedical corpora annotated for BEL continues to increase every year and this task continues to become a prominent research interest. Important dimensions for diversity of these datasets are the domain of the text corpus, target ontology for linking, and the types of entities being linked. Scientific literature, the original BEL domain, remains popular, with corpora often annotating broad ranges of biomedical concepts mapped to MeSH terms or UMLS concepts. Several BioCreative challenges have published corpora in this domain focused on niche entities like genes or chemicals, which sometimes map to smaller ontologies. Clinical domain datasets are often targeted to entities which provide clinical utility such as disorders, problems, tests, and treatments. These are generally mapped to either the UMLS or ICD codes. Other sources for datasets include online social media such as Tweets and discussion forum posts, as well as drug packaging labels, and Wikipedia. There is a particular research interest in using BEL to link adverse drug events (ADE) to either MedDRA or the UMLS. We identified at least seven datasets that have been curated for the sole purpose of linking drugs and ADEs. Table 1 shows for each dataset, the document type, entity types, the target ontology, the number of documents in the dataset, the number of mentions, and number of unique mentions (when provided).
Table 1:
Biomedical Entity Linking Datasets
| Domain | Doc Type | Citation | Date | Entity(ies) | Ontology | Doc Count | Mentions | Unique Concepts |
|---|---|---|---|---|---|---|---|---|
| Scientific Literature | Biomedical Abstract | GENIA [43] | 2003 | Biomedical (broad) | MeSH | 2,000 | 93,293 | – |
| NCBI Disease [15] | 2014 | Disorder | MeSH | 793 | 6,892 | 790 | ||
| MedMentions [44] | 2019 | Biomedical (broad) | UMLS | 4,392 | 352,496 | 34,724 | ||
| MM-ST21pv [44] | 2019 | Biomedical (broad) | UMLS | 4,392 | 203,282 | 25,419 | ||
| PubMedDS [45] | 2021 | Biomedical (broad) | MeSH | 13,197,430 | 57,943,354 | 44,881 | ||
| RegEl [46] | 2022 | DNA Regulatory Elements | Various | 419 | 8,369 | 2,947 | ||
| Biomedical Article | BC5CDR [47] | 2016 | Chemical, Disorder | MeSH | 1,500 | 10,227 | – | |
| CRAFT [48] | 2016 | Biomedical (broad) | Many– | 97 | – | – | ||
| BioNLP-2019 [49] | 2019 | Bacteria Biotope | NCBI | 392 | 7,232 | 1,072 | ||
| PhaarmaCoNER [50] (ESP) | 2019 | Chemical, Drug | UMLS | 1,000 | 7,624 | – | ||
| BC7NLMCHEM [51] | 2021 | Chemical | MeSH | 150 | 38,342 | 2,064 | ||
| Multi Source | Quaero [52] (FRA) | 2014 | Biomedical (broad) | UMLS | 2,538 | 26,407 | 5,796 | |
| Mantra [53] | 2014 | Biomedical (broad) | UMLS | 1,450 | 5,530 | 3,780 | ||
| Figure Caption | BC6BioID [54] | 2017 | Gene,Chemical | ChEBI,UniProt | 17,883 | 133,003 | 7,652 | |
| Clinical | Clinical Note | ShARe/CLEF [14] | 2013 | Disorder | UMLS | 431 | 19,557 | 1,871 |
| CUILESS2016 [55] | 2018 | Disorder | UMLS | 431 | 5,397 | 1,738 | ||
| n2c2 2019 [56] (Luo, 2019) | 2019 | Problem, Test, Treatment | UMLS | 100 | 10,919 | 3,792 | ||
| MADE [57] | 2019 | ADE, Drug, Indication | MedDRA | 1,089 | 43,000 | – | ||
| Cantemist [58] (ESP) | 2020 | Oncology | ICD-O | 1,301 | 16,030 | 850 | ||
| BRONCO [59] (DE) | 2021 | Oncology | ICD-10, OPS, ATC | 200 | 11,124 | 4,027 | ||
| Social Media/Online Literature | Drug Label | TAC2017 [60] | 2017 | ADE | MedDRA | 200 | 26,488 | – |
| Tweets | Twitter ADR [61] | 2015 | ADE, Indication | UMLS | 1,784 | 1,693 | – | |
| SMM4H-17 [62] | 2017 | ADE | MedDRA | 25,678 | – | – | ||
| TwADR-L [63] | 2016 | ADE | SIDER? | 1,436 | – | 273 | ||
| Drug Forum | DailyStrength ADR [61] | 2015 | ADE, Indication | UMLS | 6,279 | 4,929 | – | |
| CADEC [64] | 2015 | ADE,Disorder,Drug | AMT,MedDRA,SNOMED | 1,253 | 9,111 | 3,591 | ||
| PsyTAR [65] | 2019 | ADE,Disorder | UMLS | 891 | 7,414 | 1,671 | ||
| COMETA [66] | 2020 | Biomedical (broad) | UMLS | – | 20,000 | 3,645 | ||
| Wikipedia | WikiMed [45] | 2021 | Biomedical (broad) | UMLS | 393,618 | 1,067,083 | 57,739 |
International Classification of Diseases for Oncology (ICD-O); Operationen und Prozedurenschlüssel (OPS); Anatomical Therapeutic Chemical Classification System (ATC);
5. Performance Comparison
In Table 2, we compare the performance of various extant systems on six BEL datasets (BC5CDR Disease [47], BC5CDR Chemical [47], CADEC [64], NCBI Disease [15], n2c2 2019 [56], and ShARe/CLEF [14]) in terms of accuracy. These datasets were chosen because of their relative popularity and the number of authors choosing to evaluate their systems using accuracy. We chose accuracy as our common metric because it is reported for a plurality of systems. All results were reported by the respective authors, so it’s important to note that results may not be directly comparable due to differences in evaluation techniques. For example, Miftahutdinov and Tutubalina [67] evaluated their system using cross validation on the entire corpus rather than only the test partition. Some authors choose to remove conceptless annotations [68, 67]. Also, some systems [29] only require their systems to map mentions to a correct synonym for predictions to be considered correct, whereas other systems require the more stringent criteria of mapping to the correct concept ID [68, 69]. The latter specification generally results in lower accuracy because systems must solve the additional challenge of disambiguation.
Table 2:
Performance Comparison of Extant Systems
| BC5CDR (d) | BC5CDR (c) | CADEC | NCBI Disease | n2c2 2019 | ShARe/CLEF | |
|---|---|---|---|---|---|---|
| Chen, et al. [70] | - | - | - | - | 82.1 | - |
| D’Souza, et al. [16] | - | - | - | 84.7 | - | 90.8 |
| Ji, et al. [71] | - | - | - | 89.1 | - | 91.1 |
| Lee, et al. [72] | - | - | 65.0 | - | - | - |
| Li, et al. [73] | - | - | - | 86.1 | - | 90.3 |
| Liu, et al. (SAPBERT) [30] | 93.5 | 96.5 | - | 92.3 | - | - |
| Limsopatham and Collier [63] | - | - | 81.41 | - | - | - |
| Kalyan, et al. [74] | - | - | 82.6 | - | - | - |
| Miftahutdinov and Tutubalina (2018) [67] | - | - | 88.8 | - | - | - |
| Miftahutdinov, et al. (2021) [75] | 75.8 | 83.8 | - | - | - | - |
| Mondal, et al. [76] | - | - | - | 90.0 | - | - |
| Niu, et al. [26] | - | - | 84.7 | - | - | - |
| Pattisapu, et al. [77] | - | - | 76.7 | - | - | - |
| Phan, et al. [78] | 90.6 | 95.8 | - | 87.7 | - | - |
| Schumacher, et al. [68] | - | - | - | - | - | 62 |
| Silva, et al. [79] | - | - | - | - | 80.6 | - |
| Sung, et al. [29] | 93.2 | 96.6 | - | 91.1 | - | - |
| Tutubalina, et al. (2018) [25] | - | - | 70.1 | - | - | - |
| Wright, et al. [80] | 88 | - | - | 87.8 | - | - |
| Xu, et al. (2020) [81] | - | - | 87.5 | - | 83.6 | - |
| Xu and Miller (2022) [82] | - | - | - | - | 85.3 | 91.3 |
| Zhang, et al. (KRISSBERT) [69] | 90.7 | 96.9 | - | 89.9 | 80.2 | 90.4 |
| Zhao, et al. [83] | - | - | - | 88.2 | - | - |
Comparison of reported accuracies on six popular BEL datasets. The BC5CDR dataset contains partitions corresponding to disease (d) and chemical (c) normalization, which are often evaluated separately.
6. Shared Tasks
There have been a number of shared tasks focused on BEL, starting with the inaugural BioCreative challenge in 2004 [84]. Table 2 shows the different tasks that have been organized over the years. We classify these tasks into three categories based on the type of text that was annotated as outlined in the previous section. Within each category, the tasks are ordered based on their date. The table also includes the document source, entities and the associated ontology.
The majority of shared tasks focus on scientific literature with the early BioCreative tasks mapping a broad class of biomedical entities to concepts in the MeSH ontology[84]. Since that time, new shared tasks have been developed every four years or so, expanding from abstracts to full text, and incorporating new entity types. The clinical shared tasks began in 2013 [14] focusing on disorders with the most recent task [56] expanding to include both treatments and tests. The social media shared tasks both happened in 2017 and focused on adverse drug reactions(ADR).
7. Technical Discussion
All BEL systems are a pipeline of various components and techniques which can be mix and matched to fit a practitioner’s data and use case. Some potential applications of BEL are discussed in Section 3. In this section we will discuss the major categories of techniques, how they work, and where they’ve been applied.
7.1. Preprocessing
Many BEL publications make no mention of any pre-processing of the input corpus prior to training. Whether this step is implied or simply omitted is not entirely clear, but where mentioned, many systems follow standard pre-processing steps such as converting all text to lowercase and removing punctuation. Authors frequently correct spelling on the NCBI Disease dataset, for which D’Souza, et al. [16] curated a corpus-specific dictionary to this end, but we have not seen a generalizable tool in use for other datasets. Two additional common steps are expanding abbreviations to full form using the Abbreviation Plus Pseudo-Precision (Ab3P)[89] tool and separating composite mentions into distinct parts (i.e. “BRCA1/2” into “BRCA1” and “BRCA2”) using the SimConcept[90] tool. Finally, it is common practice to append the mentions from the training set to the synonym dictionary when evaluating performance on the test set [16, 29]. However, some have questioned whether this results in an unfair evaluation given the frequent overlap of mentions between training and test datasets [91].
7.2. Mention/Concept Representation
Rule-based systems represent mentions using tokens[5, 16], in other words, actual human-readable words and phrases. These representations can do fairly well given that many mentions are morphologically similar to known synonyms of their corresponding concept, but this technique has a real upper bound when mentions differ significantly from documented synonyms, and as Blair, et al. [92] note, synonym coverage for biomedical entities is far from complete. Representing mentions numerically opens up a world of possibilities for choosing sophisticated learning algorithms. The simplest such representation is Term Frequency-Inverse Document Frequency (TF-IDF) vectors, used in the first machine learning-based BEL system, DNorm[21]. This technique scores tokens with a ratio its frequency in a mention by its overall frequency in the set of concept synonyms. While this technique is intuitive, it fails to capture semantic meaning and shares many shortcomings with token representation. Word embeddings, which project tokens into a latent semantic vector space, do address the problem capturing semantic meaning. The first iteration of such techniques, led by Word2Vec[23], created static vector representations of tokens which effectively aggregated the contextual usage of a given token within a corpus and embedded it in the semantic space. For the first time, word embeddings allowed us to mathematically compare the similarity of two given tokens without requiring any additional knowledge. The improved quality of these representations correlated with a higher quality output from the systems which incorporated them. The primary downside to these static representations is that they cannot capture the nuance of words that have different meanings in different contexts. Deep contextualized embeddings such as ELMo[27] and BERT[28] capture not only aggregate semantic meaning, but also take into account a token’s context within a specific sentence. These techniques provide unquestionably state of the art embedding quality embeddings, which are the foundation of all the current top performing BEL systems. However, quality comes at a computational cost and generating deep contextualized embeddings at any practical scale requires access to a GPU. The final major category of representations is graph-based techniques, such as concept vectors. Node2Vec [93], as employed by Ferré, et al. [94] in their CONTES system, models concepts in an ontology as nodes in a graph and relationships between concepts as edges, it then generates a vector space which embeds concepts such that connected nodes in the graph correspond to closeness within the vector space. CONTES used these concept vectors only to represent concepts, and learned a mapping between the semantic space representing mentions and the ontology space generated by Node2Vec. They also note that this technique may not scale well to large ontologies.
7.3. Linking Algorithms
The crux of any BEL system is the algorithm which links the representation of a mention to a concept in the target ontology. The most basic implementation of this mapping is a dictionary lookup, which checks if the mention is an exact match of some known concept synonym. To increase recall, systems [16] may create morphological permutations of the mention and check if the permutations match any known synonyms, but the expression of natural language is diverse and any system which generates enough blind permutations to achieve respectable recall will inevitably generate a huge number of false positives. But there is still a place for morphological feature extraction in sophisticated BEL systems, some have used Lucene search to select a small set of candidate concepts prior to using deep learning techniques to make a final prediction [95].
Learning algorithms train systems find mappings between mentions and concepts in a vector space, which allows them to achieve both higher recall and precision. BEL systems incorporating classical machine learning started with linear classifiers to learn positive and negative correlations between tokens in mentions and concept synonyms [21]. As the quality of word representations improved and access to GPUs became widespread in the 2010s, deep learning techniques such as CNN [63], RNN [63], GRU [25], and BiLSTM [96] came into vogue. Other systems have trained lesser known learning algorithms such as RankSVM [33] and TreeLSTM [97], but neither of these have achieved widespread adoption.
As expected, using a BERT for BEL performs quite well. Typically, researchers use BERT classifiers [30], but sequence-to-sequence translation models have been explored as well [98]. Other models have leveraged the high quality of BERT embeddings to rely on simple similarity measures to perform their mapping [29], training only the encoder and omitting a secondary neural architecture entirely. PageRank, an algorithm originally designed for scoring the relevance of search engine results, has been used to link entities when using graph-based representations [99].
One technique uncommon in BEL that deserves more attention is clustering, which Angell, et al. [31] employed following candidate generation by creating an affinity graph with mention-mention and mention-concept connections for all mentions and candidates in a given document. They iteratively pruned connections in the graph to create clusters until each cluster contained exactly one concept linked one or more mentions. This approach is especially helpful for disambiguating mentions of generic phrases which corresponded to entities described more specifically elsewhere in the document and yielded the current state of the art performance for few-shot entity linking.
7.4. Training Techniques
In addition to the building blocks described in the previous sections, we noted several training techniques commonly employed by successful BEL systems. The most common of these is a two step process in which a system first uses a high-recall technique to select a small pool of candidate concepts from the target ontology, followed by a higher precision technique to select a single concept for prediction out of the pool of candidates. The algorithms used for candidate generation vary widely, but recurring solutions include search engine-style algorithms like bag-of-words retrieval function BM25 [33] or lucene [95], similarity of mention representations [76, 29], and edit distance [99]. A related strategy for narrowing the range of possible candidates is to predict the semantic type of the mention and only consider candidates of the predicted semantic type. The MedType [45] system was created to perform this type of semantic type prediction in entity linking pipelines. Another way that semantic types have been used to augment BEL pipelines is to train the prediction step to rank all candidates with the correct semantic type over those with the wrong semantic type [95, 81], as opposed to loss functions which only consider the top-ranked candidate. External knowledge bases such as Wikipedia [100, 30] have also shown promise as valuable sources of information for inclusion in BEL systems.
The state of the art SAPBERT model [30] attributed its success to a self-alignment pre-training strategy in which only difficult positive and negative examples for a given gold concept in each mini-batch are used for training. The subsequent multi-similarity loss function simultaneously pushes negative examples away from the gold concept, while pulling the positive examples closer. Finally, it is also common to perform entity linking jointly with other NLP tasks, in particular, named entity recognition [83, 101, 22].
7.5. Multilingual-based Approaches
Entity linking in non-English corpora presents additional challenges and several non-English corpora[52, 58, 50] exist to train systems to tackle these challenges. The most straightforward approach is to link directly from the source documents to an ontology in the same language. This can work well if the ontology has good coverage, but in the UMLS, there are many times more English synonyms available than those in non-English target language, even in the best cases (Spanish and French with more than six times and twenty-four times respectively[102]). Non-uniform distribution of non-English synonyms does allow that there are cases in which this strategy could still work for specific languages and problems, such as identifying disorders in Italian clinical notes[103], but for other languages and use cases, the scarcity of target language synonyms can be a insurmountable obstacle for this strategy. A naive approach for overcoming these challenges is to simply translate the non-English mentions into English using standard translation software and perform BEL on the translations. This works reasonably well, but is limited by the quality of the translation, which may struggle to properly translate medical jargon[103]. Roller, et al., 2018[104] combined these two approaches sequentially, first looking for a match for a given mention in the target language UMLS, then English language UMLS, and finally searching English UMLS for the translation of the mention. Deep learning-based approaches[26] favoring encoder models learning a direct mapping from non- English mentions to English synonyms[105] have performed well. The current best performing model for multilingual BEL adapts the SAPBERT[30] system to map mentions in any language to language-agnostic CUIs in the UMLS. This system augments the cross-lingual links between CUIs by leveraging the titles of Wikipedia articles available in multiple languages where the article title can be mapped to the UMLS for at least one language. The authors found that performance for a given language generally correlated with its similarity to English, likely because more general translation knowledge could be incorporated into the model[102].
8. Discussion
A fundamental limitation of BEL is that treating the task as a classification problem with a learning-based approach requires the output space to be at minimum equal to the number of concepts to be predicted. While this works well when the output space tends to be small [63], these approaches struggle as the size of the taxonomy increases [95], particularly with concepts that have only a few example mentions in the training data. While current state of the art accuracies greater than 90% on many of the most common BEL datsets would seem to indicate that the problem is largely solved, Tutubalina, et al. [91] found that approximately 80% of entity mentions in the test datasets they analyzed were either duplicated within the test set or replicated exactly in the training dataset. Because many systems [16, 95] add training mentions to their synonym dictionaries used for inference against test data, this unrealistically inflates the actual abilities of a system to link mentions in a corpus with higher variability. They supported this hypothesis by creating a “refined” version of five popular BEL datasets, removing all duplication of mentions in the test sets, and comparing a state of the art BEL system’s performance on the original and refined test sets. Their results showed a substantial performance impact from the de-duplication, indicating that developing effective solutions to BEL as a zero or few-shot learning problem is an area ripe for future improvements. Developing effective techniques for distant [77] and self-supervision [69] will be crucial to scaling BEL systems to perform well when linking mentions to concepts which are dissimilar to annotated data.
The development of non-English BEL corpora [52, 58, 50] and recent multilingual systems [105, 102] are a great start for expanding BEL to be a truly multilingual task, but BEL performance on non-English texts trails far behind the state of the art performance on English texts, especially for languages which are absent or severely underrepresented in the UMLS. More work is needed specifically to develop more non-English BEL corpora and to find new strategies for overcoming the difficulties of mapping underrepresented languages to the UMLS.
Newman-Griffis, et al. [106], demonstrate that existing BEL datasets do not sufficiently capture the ambiguity resulting from unique strings mapping to multiple possible CUIs in the UMLS. Polysemy, where a word can have multiple senses, can harm the generalizability of models when the training data exposes models to only one sense a word, erroneously causing it to appear unambiguous. This phenomenon can be especially prevalent in datasets which annotate only narrow slices of clinical entities, such as diseases. In such a case, “cold” may be annotated several times in reference to a viral infection, but never as a relative perception of temperature, though both senses of the word may appear in actual notes. Another source of ambiguity common to telegraphic clinical language is metonymy, in which one concept is used as shorthand for a related concept. Without properly understanding context, BEL systems can easily conflate devices for procedures (i.e. “stent”), substances for lab measurements (i.e. “potassium”), and diagnoses for symptoms. A final source of ambiguity can result from the level of specificity in the annotation, such as whether an instance was noted to be a sequela, whether multiple were specified (i.e. “injuries” vs. “injury”), or hierarchical specificity (i.e. “hemiplegia” vs. “left hemiplegia”). They recommend developing ambiguity-focused datasets to train systems to capture a more nuanced contextual understanding of ambiguous mentions.
A subsequent paper by Newman-Griffis [107] introduces the research paradigm of “translational NLP”, in which basic and applied NLP research inform one another. Under this paradigm, we can see a potential way to mitigate at least one class of ambiguity. When researchers query structured clinical data, which may include the discretized results of a BEL algorithm, they rarely search for a single concept in isolation, rather they curate a set of concepts [108], often hierarchically related, which correspond to a more general clinical phenotype. In such cases, hierarchical classification errors could in many cases be close enough to the gold concept to still be included in the correct phenotype. Evaluating performance with respect to ontological similarity rather than solely considering a binary measure of whether a prediction exactly matches the gold concept could be a productive line of inquiry for future BEL research.
9. Conclusions
In this paper, we reviewed previous work on BEL providing an overview of the progression of historical approaches (Section 2) and providing a reference for the BEL datasets (Section 4) and shared tasks (Section 6) that have been developed. We then discussed salient challenges and opportunities for future work, highlighting four areas specifically:
Reported results are inflated by overlap between training and test mentions and duplication within test datasets. Evaluating systems performance on datasets without the benefit of overlap and duplication makes it clear that there is much work to be done with BEL as a zero or few-shot learning problem.
BEL performance on non-English mentions is significantly lower than on English, especially for those languages absent or severely underrepresented in the UMLS.
Current BEL datasets do not sufficiently capture the ambiguity resulting from unique strings mapping to multiple distinct concepts.
Alternative performance metrics like ontological similarity should be explored in order to develop systems which best meet real world use cases.
Table 3:
Entity Linking Shared Tasks
| Domain | Year | Task | Document Source | Entity Type(s) | Ontology |
|---|---|---|---|---|---|
| Scientific Literature | 2004 | BC I (1b)[84] | MEDLINE | Fly, mouse, and yeast genes | Organizer provided |
| 2006 | BC II (1b)[85] | MEDLINE | Human genes | EntrezGene | |
| 2010 | BC III GN[86] | PMC full text | Genes | EntrezGene | |
| 2016 | BC V CDR (3a)[47] | PubMed | Chemicals, diseases, chemical-disease interactions | MeSH | |
| 2017 | BC VI Bio-ID (1)[54] | Figure captions | Genes, chemicals, cell type, subcellular location, tissue, organism | ||
| 2019 | BioNLP 2019 (1)[49] | PubMed | Microorganism, habitat, phenotype | NCBI, OntoBiotope | |
| 2021 | BC VII NLMCHEM (1b)[51] | PubMed | Chemicals | MeSH | |
| clinical | 2013 | ShARe/CLEF 2013 (1b,2)[14] | Clinical records | Disorders | SNOMED CT |
| 2014 | SE-2014 (7b)[87] | Disorders | SNOMED CT | ||
| 2015 | SE-2015 Task 14 (1,2a)[88] | Disorders | SNOMED CT | ||
| 2019 | 2019 n2c2 (3)[56] | Problems, treatments, tests | SNOMED CT, RxNorm | ||
| 2019 | PharmaCoNER[50] | Clinical records | Drugs, chemicals | SNOMED CT | |
| 2020 | IberLEF CANTEMIST-NORM[58] | (ESP) | Tumor morphology | ICD-O | |
| social Media | 2017 | SMM4H 2017 (3)[62] | ADRs | MedDRA | |
| 2017 | TAC 2017[60] | Drug labels | ADRs | MedDRA |
BioCreative (BC); SemEval (SE); Task/Track number in parentheses
Acknowledgements
The authors thank Amy Olex for her extensive review of the paper.
Funding
EF was supported by CTSA award No. UL1TR002649 from the National Center for Advancing Translational Sciences. BTM was partially supported by the National Science Foundation under Grant No. 1939951.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Lowe HJ, Barnett GO. MicroMeSH: a microcomputer system for searching and exploring the National Library of Medicine’s Medical Subject Headings (MeSH) Vocabulary. In: Proceedings of the Annual Symposium on Computer Application in Medical Care. American Medical Informatics Association; 1987. p. 717. [Google Scholar]
- [2].Evans DA, Ginther-Webster K, Hart M, Lefferts RG, Monarch IA. Automatic indexing using selective NLP and first-order thesauri. In: Intelligent Text and Image Handling-Volume 2; 1991. p. 624–643. [Google Scholar]
- [3].Hersh W, Leone T. The SAPHIRE server: a new algorithm and implementation. In: Proceedings of the Annual Symposium on Computer Application in Medical Care. American Medical Informatics Association; 1995. p. 858. [PMC free article] [PubMed] [Google Scholar]
- [4].Jessop DM, Adams SE, Willighagen EL, Hawizy L, Murray-Rust P. OSCAR4: a flexible architecture for chemical text-mining. Journal of cheminformatics. 2011;3(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. In: Proceedings of the AMIA Symposium. American Medical Informatics Association; 2001. p. 17. [PMC free article] [PubMed] [Google Scholar]
- [6].Aronson AR, Mork JG, Gay CW, Humphrey SM, Rogers WJ, et al. The NLM indexing initiative’s medical text indexer. Medinfo. 2004;89. [PubMed] [Google Scholar]
- [7].Miller RA, Gieszczykiewicz FM, Vries JK, Cooper GF. CHARTLINE: providing bibliographic references relevant to patient charts using the UMLS Metathesaurus Knowledge Sources. In: Proceedings of the Annual Symposium on Computer Application in Medical Care. American Medical Informatics Association; 1992. p. 86. [PMC free article] [PubMed] [Google Scholar]
- [8].Friedman C, Hripcsak G, DuMouchel W, Johnson SB, Clayton PD. Natural language processing in an operational clinical information system. Natural Language Engineering. 1995;1(1):83–108. [Google Scholar]
- [9].Friedlin FJ, McDonald C. A Natural Language Processing System to Extract and Code Concepts Relating to Congestive Heart Failure from Chest Radiology Reports. AMIA Annual Symposium proceedings AMIA Symposium. 2006:269–73. [PMC free article] [PubMed] [Google Scholar]
- [10].Friedlin J, Duke J. Applying natural language processing to extract codify adverse drug reaction in medication labels; 2010. [Google Scholar]
- [11].Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 2010;17(5):507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Soldaini L, Goharian N. Quickumls: a fast, unsupervised approach for medical concept extraction. In: MedIR workshop, sigir; 2016. p. 1–4. [Google Scholar]
- [13].Cuzzola J, Jovanović J, Bagheri E. RysannMD: a biomedical semantic annotator balancing speed and accuracy. Journal of Biomedical Informatics. 2017;71:91–109. [DOI] [PubMed] [Google Scholar]
- [14].Pradhan S, Elhadad N, South B, Martinez D, Christensen L, Vogel A, et al. Task 1: ShARe/CLEF eHealth evaluation lab 2013; 2013. p. 1–6. CLEF 2013 Conference - Working notes ; Conference date: 23-09-2013 Through 26-September-2013. [Google Scholar]
- [15].Doğan RI, Leaman R, Lu Z. NCBI disease corpus: A resource for disease name recognition and concept normalization. Journal of biomedical informatics. 2014;47:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].D’Souza J, Ng V. Sieve-based entity linking for the biomedical domain. In: Proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing (Volume 2: Short Papers); 2015. p. 297–302. [Google Scholar]
- [17].Liu H, Wu ST, Li D, Jonnalagadda S, Sohn S, Wagholikar K, et al. Towards a semantic lexicon for clinical natural language processing. In: AMIA Annual Symposium Proceedings. vol. 2012. American Medical Informatics Association; 2012. p. 568. [PMC free article] [PubMed] [Google Scholar]
- [18].Leal A, Martins B, Couto FM. ULisboa: Recognition and normalization of medical concepts. In: Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015); 2015. p. 406–411. [Google Scholar]
- [19].McInnes BT, Pedersen T, Pakhomov SV. UMLS-Interface and UMLS-Similarity: open source software for measuring paths and semantic similarity. In: AMIA annual symposium proceedings. vol. 2009. American Medical Informatics Association; 2009. p. 431. [PMC free article] [PubMed] [Google Scholar]
- [20].Wen A, Fu S, Moon S, El Wazir M, Rosenbaum A, Kaggal VC, et al. Desiderata for delivering NLP to accelerate healthcare AI advancement and a Mayo Clinic NLP-as-a-service implementation. NPJ digital medicine. 2019;2(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Leaman R, Islamaj Doğan R, Lu Z. DNorm: disease name normalization with pairwise learning to rank. Bioinformatics. 2013;29(22):2909–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Leaman R, Lu Z. TaggerOne: joint named entity recognition and normalization with semi-Markov Models. Bioinformatics. 2016;32(18):2839–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mikolov T, Chen K, Corrado G, Dean J. Efficient estimation of word representations in vector space. arXiv preprint arXiv:13013781. 2013. [Google Scholar]
- [24].Limsopatham N, Collier N. Normalising Medical Concepts in Social Media Texts by Learning Semantic Representation. In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers); 2016. p. 1014–1023. [Google Scholar]
- [25].Tutubalina E, Miftahutdinov Z, Nikolenko S, Malykh V. Medical concept normalization in social media posts with recurrent neural networks. Journal of biomedical informatics. 2018;84:93–102. [DOI] [PubMed] [Google Scholar]
- [26].Niu J, Yang Y, Zhang S, Sun Z, Zhang W. Multi-task character-level attentional networks for medical concept normalization. Neural Processing Letters. 2019;49(3):1239–1256. [Google Scholar]
- [27].Peters ME, Neumann M, Iyyer M, Gardner M, Clark C, Lee K, et al. Deep contextualized word representations. In: Proc. of NAACL; 2018. [Google Scholar]
- [28].Devlin J, Chang MW, Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:181004805. 2018. [Google Scholar]
- [29].Sung M, Jeon H, Lee J, Kang J. Biomedical Entity Representations with Synonym Marginalization. In: Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics; 2020. p. 3641–3650. [Google Scholar]
- [30].Liu F, Shareghi E, Meng Z, Basaldella M, Collier N. Self-alignment Pre-training for Biomedical Entity Representations. arXiv e-prints. 2020:arXiv-2010. [Google Scholar]
- [31].Angell R, Monath N, Mohan S, Yadav N, McCallum A. Clustering-based Inference for Biomedical Entity Linking. In: Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; 2021. p. 2598–2608. [Google Scholar]
- [32].Dong H, Suárez-Paniagua V, Zhang H, Wang M, Whitfield E, Wu H. Rare Disease Identification from Clinical Notes with Ontologies and Weak Supervision. In: 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC). IEEE; 2021. p. 2294–2298. [DOI] [PubMed] [Google Scholar]
- [33].Wang Q, Ji Z, Wang J, Wu S, Lin W, Li W, et al. A study of entity-linking methods for normalizing Chinese diagnosis and procedure terms to ICD codes. Journal of Biomedical Informatics. 2020;105:103418. [DOI] [PubMed] [Google Scholar]
- [34].Ferrucci D, Lally A, Verspoor K, Nyberg E. Unstructured Information Management Architecture (UIMA) Version 1.0. 2008. [Google Scholar]
- [35].Soysal E, Wang J, Jiang M, Wu Y, Pakhomov S, Liu H, et al. CLAMP–a toolkit for efficiently building customized clinical natural language processing pipelines. Journal of the American Medical Informatics Association. 2018;25(3):331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cornia R, Patterson OV, Ginter T, DuVall SL. Rapid NLP Development with Leo. In: AMIA; 2014. . [Google Scholar]
- [37].Liu H, Bielinski SJ, Sohn S, Murphy S, Wagholikar KB, Jonnalagadda SR, et al. An information extraction framework for cohort identification using electronic health records. AMIA Summits on Translational Science Proceedings. 2013;2013:149. [PMC free article] [PubMed] [Google Scholar]
- [38].Tseytlin E, Mitchell K, Legowski E, Corrigan J, Chavan G, Jacobson RS. NOBLE-Flexible concept recognition for large-scale biomedical natural language processing. BMC bioinformatics. 2016;17(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Luo Y, Uzuner Ö, Szolovits P. Bridging semantics and syntax with graph algorithms—state-of-the-art of extracting biomedical relations. Briefings in bioinformatics. 2017;18(1):160–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Peterson K A Corpus-Driven Standardization Framework for Encoding Clinical Problems with SNOMED CT Expressions and HL7 FHIR. University of Minnesota; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chen P, Barrera A, Rhodes C. Semantic analysis of free text and its application on automatically assigning ICD-9-CM codes to patient records. In: 9th IEEE International Conference on Cognitive Informatics (ICCI’10). IEEE; 2010. p. 68–74. [Google Scholar]
- [42].Goldstein I, Arzumtsyan A, Uzuner Ö. Three approaches to automatic assignment of ICD-9-CM codes to radiology reports. In: AMIA Annual Symposium Proceedings. vol. 2007. American Medical Informatics Association; 2007. p. 279. [PMC free article] [PubMed] [Google Scholar]
- [43].Kim JD, Ohta T, Tateisi Y, Tsujii J. GENIA corpus—a semantically annotated corpus for bio-textmining. Bioinformatics. 2003;19(suppl_1):i180–i182. [DOI] [PubMed] [Google Scholar]
- [44].Mohan S, Li D. MedMentions: A Large Biomedical Corpus Annotated with {UMLS} Concepts. In: Automated Knowledge Base Construction (AKBC); 2019. . [Google Scholar]
- [45].Vashishth S, Joshi R, Dutt R, Newman-Griffis D, Rose C. MedType: Improving Medical Entity Linking with Semantic Type Prediction. arXiv e-prints. 2020:arXiv-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Garda S, Lenihan-Geels F, Proft S, Hochmuth S, Schülke M, Seelow D, et al. RegEl corpus: identifying DNA regulatory elements in the scientific literature. Database. 2022;2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Li J, Sun Y, Johnson RJ, Sciaky D, Wei CH, Leaman R, et al. BioCreative V CDR task corpus: a resource for chemical disease relation extraction. Database: The Journal of Biological Databases and Curation. 2016;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cohen KB, Verspoor K, Fort K, Funk C, Bada M, Palmer M, et al. The Colorado Richly Annotated Full Text (CRAFT) Corpus: Multi-Model Annotation In The Biomedical Domain; 2016. [Google Scholar]
- [49].Bossy R, Deléger L, Chaix E, Ba M, Nédellec C. Bacteria biotope at BioNLP open shared tasks 2019. In: Proceedings of the 5th workshop on BioNLP open shared tasks; 2019. p. 121–131. [Google Scholar]
- [50].Gonzalez-Agirre A, Marimon M, Intxaurrondo A, Rabal O, Villegas M, Krallinger M. PharmaCoNER: Pharmacological Substances, Compounds and proteins Named Entity Recognition track. In: Proceedings of The 5th Workshop on BioNLP Open Shared Tasks. Hong Kong, China: Association for Computational Linguistics; 2019. p. 1–10. [Google Scholar]
- [51].Islamaj R, Leaman R, Kim S, Kwon D, Wei CH, Comeau DC, et al. NLM-Chem, a new resource for chemical entity recognition in PubMed full text literature. Scientific Data. 2021;8(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Névéol A, Grouin C, Leixa J, Rosset S, Zweigenbaum P. The QUAERO French medical corpus: A ressource for medical entity recognition and normalization. In: In proc biotextm, reykjavik. Citeseer; 2014. . [Google Scholar]
- [53].Kors JA, Clematide S, Akhondi SA, Van Mulligen EM, Rebholz-Schuhmann D. A multilingual gold-standard corpus for biomedical concept recognition: the Mantra GSC. Journal of the American Medical Informatics Association. 2015;22(5):948–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Arighi C, Hirschman L, Lemberger T, Bayer S, Liechti R, Comeau D, et al. Bio-ID track overview. In: Proc. BioCreative Workshop. vol. 482; 2017. p. 376. [Google Scholar]
- [55].Osborne JD, Neu MB, Danila MI, Solorio T, Bethard SJ. CUILESS2016: a clinical corpus applying compositional normalization of text mentions. Journal of biomedical semantics. 2018;9(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Luo YF, Sun W, Rumshisky A. MCN: A comprehensive corpus for medical concept normalization. Journal of Biomedical Informatics. 2019;92:103132. [DOI] [PubMed] [Google Scholar]
- [57].Jagannatha A, Liu F, Liu W, Yu H. Overview of the first natural language processing challenge for extracting medication, indication, and adverse drug events from electronic health record notes (MADE 1.0). Drug safety. 2019;42(1):99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Miranda-Escalada A, Farré E, Krallinger M. Named entity recognition, concept normalization and clinical coding: Overview of the cantemist track for cancer text mining in spanish, corpus, guidelines, methods and results. In: Proceedings of the Iberian Languages Evaluation Forum (IberLEF 2020), CEUR Workshop Proceedings; 2020. . [Google Scholar]
- [59].Kittner M, Lamping M, Rieke DT, Götze J, Bajwa B, Jelas I, et al. Annotation and initial evaluation of a large annotated German oncological corpus. JAMIA open. 2021;4(2):ooab025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Roberts K, Demner-Fushman D, Tonning JM. Overview of the TAC 2017 Adverse Reaction Extraction from Drug Labels Track. Theory and Applications of Categories. 2017. [Google Scholar]
- [61].Nikfarjam A, Sarker A, O’connor K, Ginn R, Gonzalez G. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. Journal of the American Medical Informatics Association. 2015;22(3):671–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sarker A, Belousov M, Friedrichs J, Hakala K, Kiritchenko S, Mehryary F, et al. Data and systems for medication-related text classification and concept normalization from Twitter: insights from the Social Media Mining for Health (SMM4H)-2017 shared task. Journal of the American Medical Informatics Association. 2018;25(10):1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Limsopatham N, Collier N. Normalising Medical Concepts in Social Media Texts by Learning Semantic Representation. In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). Berlin, Germany: Association for Computational Linguistics; 2016. p. 1014–1023. [Google Scholar]
- [64].Karimi S, Metke-Jimenez A, Kemp M, Wang C. Cadec: A corpus of adverse drug event annotations. Journal of Biomedical Informatics. 2015;55:73–81. [DOI] [PubMed] [Google Scholar]
- [65].Zolnoori M, Fung KW, Patrick TB, Fontelo P, Kharrazi H, Faiola A, et al. The PsyTAR dataset: From patients generated narratives to a corpus of adverse drug events and effectiveness of psychiatric medications. Data in Brief. 2019;24:103838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Basaldella M, Liu F, Shareghi E, Collier N. COMETA: A Corpus for Medical Entity Linking in the Social Media. In: Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP). Online: Association for Computational Linguistics; 2020. p. 3122–3137. [Google Scholar]
- [67].Miftahutdinov Z, Tutubalina E. Deep Neural Models for Medical Concept Normalization in User-Generated Texts. In: Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics: Student Research Workshop; 2019. p. 393–399. [Google Scholar]
- [68].Schumacher E, Dredze M. Learning unsupervised contextual representations for medical synonym discovery. JAMIA Open. 2019. November;2(4):538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhang S, Cheng H, Vashishth S, Wong C, Xiao J, Liu X, et al. Knowledge-rich self-supervised entity linking. arXiv preprint arXiv:211207887. 2021. [Google Scholar]
- [70].Chen L, Fu W, Gu Y, Sun Z, Li H, Li E, et al. Clinical concept normalization with a hybrid natural language processing system combining multilevel matching and machine learning ranking. Journal of the American Medical Informatics Association. 2020;27(10):1576–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ji Z, Wei Q, Xu H. BERT-based ranking for biomedical entity normalization. AMIA Summits on Translational Science Proceedings. 2020;2020:269. [PMC free article] [PubMed] [Google Scholar]
- [72].Lee K, Hasan SA, Farri O, Choudhary A, Agrawal A. Medical concept normalization for online user-generated texts. In: 2017 IEEE International Conference on Healthcare Informatics (ICHI). IEEE; 2017. p. 462–469. [Google Scholar]
- [73].Li H, Chen Q, Tang B, Wang X, Xu H, Wang B, et al. CNN-based ranking for biomedical entity normalization. BMC bioinformatics. 2017;18(11):79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kalyan KS, Sangeetha S. Bertmcn: Mapping colloquial phrases to standard medical concepts using bert and highway network. Artificial Intelligence in Medicine. 2021;112:102008. [DOI] [PubMed] [Google Scholar]
- [75].Miftahutdinov Z, Kadurin A, Kudrin R, Tutubalina E. Medical concept normalization in clinical trials with drug and disease representation learning. Bioinformatics. 2021;37(21):3856–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mondal I, Purkayastha S, Sarkar S, Goyal P, Pillai J, Bhattacharyya A, et al. Medical Entity Linking using Triplet Network. In: Proceedings of the 2nd Clinical Natural Language Processing Workshop; 2019. p. 95–100. [Google Scholar]
- [77].Pattisapu N, Anand V, Patil S, Palshikar G, Varma V. Distant supervision for medical concept normalization. Journal of biomedical informatics. 2020;109:103522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Phan MC, Sun A, Tay Y. Robust representation learning of biomedical names. In: Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics; 2019. p. 3275–3285. [Google Scholar]
- [79].Pape-Haugaard L, et al. Clinical concept normalization on medical records using word embeddings and heuristics. Digital Personalized Health and Medicine: Proceedings of MIE 2020. 2020;270:93. [DOI] [PubMed] [Google Scholar]
- [80].Wright D, Katsis Y, Mehta R, Hsu CN. NormCo: Deep Disease Normalization for Biomedical Knowledge Base Construction. In: Automated Knowledge Base Construction (AKBC); 2018. . [Google Scholar]
- [81].Xu D, Gopale M, Zhang J, Brown K, Begoli E, Bethard S. Unified Medical Language System resources improve sieve-based generation and Bidirectional Encoder Representations from Transformers (BERT)–based ranking for concept normalization. Journal of the American Medical Informatics Association. 2020;27(10):1510–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Xu D, Miller T. A simple neural vector space model for medical concept normalization using concept embeddings. Journal of Biomedical Informatics. 2022;130:104080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Zhao S, Liu T, Zhao S, Wang F. A neural multi-task learning framework to jointly model medical named entity recognition and normalization. In: Proceedings of the AAAI Conference on Artificial Intelligence. vol. 33; 2019. p. 817–824. [Google Scholar]
- [84].Hirschman L, Colosimo M, Morgan A, Yeh A. Overview of BioCreAtIvE task 1B: normalized gene lists. BMC bioinformatics. 2005;6(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Morgan AA, Lu Z, Wang X, Cohen AM, Fluck J, Ruch P, et al. Overview of BioCreative II gene normalization. Genome biology. 2008;9(2):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Lu Z, Kao HY, Wei CH, Huang M, Liu J, Kuo CJ, et al. The gene normalization task in BioCreative III. BMC bioinformatics. 2011;12(8):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Pradhan S, Chapman W, Man S, Savova G. Semeval-2014 task 7: Analysis of clinical text. In: Proc. of the 8th International Workshop on Semantic Evaluation (SemEval 2014. Citeseer; 2014. . [Google Scholar]
- [88].Elhadad N, Pradhan S, Gorman S, Manandhar S, Chapman W, Savova G. SemEval-2015 task 14: Analysis of clinical text. In: proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015); 2015. p. 303–310. [Google Scholar]
- [89].Sohn S, Comeau DC, Kim W, Wilbur WJ. Abbreviation definition identification based on automatic precision estimates. BMC bioinformatics. 2008;9(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wei CH, Leaman R, Lu Z. SimConcept: a hybrid approach for simplifying composite named entities in biomedical text. IEEE journal of biomedical and health informatics. 2015;19(4):1385–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Tutubalina E, Kadurin A, Miftahutdinov Z. Fair evaluation in concept normalization: a large-scale comparative analysis for bert-based models. In: Proceedings of the 28th International Conference on Computational Linguistics; 2020. p. 6710–6716. [Google Scholar]
- [92].Blair DR, Wang K, Nestorov S, Evans JA, Rzhetsky A. Quantifying the impact and extent of undocumented biomedical synonymy. PLoS computational biology. 2014;10(9):e1003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Grover A, Leskovec J. Node2vec: Scalable Feature Learning for Networks. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. KDD ’16. New York, NY, USA: Association for Computing Machinery; 2016. p. 855–864. Available from: 10.1145/2939672.2939754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Ferré A, Ba M, Bossy R. Improving the CONTES method for normalizing biomedical text entities with concepts from an ontology with (almost) no training data. Genomics & informatics. 2019;17(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Xu D, Zhang Z, Bethard S. A Generate-and-Rank Framework with Semantic Type Regularization for Biomedical Concept Normalization. In: Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics; 2020. p. 8452–8464. [Google Scholar]
- [96].Wiatrak M, Iso-Sipila J. Simple Hierarchical Multi-Task Neural End-To-End Entity Linking for Biomedical Text. In: Proceedings of the 11th International Workshop on Health Text Mining and Information Analysis; 2020. p. 12–17. [Google Scholar]
- [97].Liu H, Xu Y. A deep learning way for disease name representation and normalization. In: National CCF conference on natural language processing and Chinese computing. Springer; 2017. p. 151–157. [Google Scholar]
- [98].Boguslav MR, Hailu ND, Bada M, Baumgartner WA, Hunter LE. Concept recognition as a machine translation problem. BMC bioinformatics. 2021;22(1):1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Ruas P, Lamurias A, Couto FM. Linking chemical and disease entities to ontologies by integrating PageRank with extracted relations from literature. Journal of Cheminformatics. 2020;12(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Wang Y, Fan X, Chen L, Chang EI, Ananiadou S, Tsujii J, et al. Mapping anatomical related entities to human body parts based on wikipedia in discharge summaries. BMC bioinformatics. 2019;20(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Xionga Y, Huanga Y, Chena Q, Wanga X, Nic Y, Tanga B. A Joint Model for Medical Named Entity Recognition and Normalization. Proceedings http://ceur-ws org ISSN. 2020;1613:0073. [Google Scholar]
- [102].Liu F, Vulić I, Korhonen A, Collier N. Learning Domain-Specialised Representations for Cross-Lingual Biomedical Entity Linking. arXiv preprint arXiv:210514398. 2021. [Google Scholar]
- [103].Chiaramello E, Pinciroli F, Bonalumi A, Caroli A, Tognola G. Use of “off-the-shelf” information extraction algorithms in clinical informatics: A feasibility study of MetaMap annotation of Italian medical notes. Journal of biomedical informatics. 2016;63:22–32. [DOI] [PubMed] [Google Scholar]
- [104].Roller R, Kittner M, Weissenborn D, Leser U. Cross-lingual Candidate Search for Biomedical Concept Normalization. MultilingualBIO: Multilingual Biomedical Text Processing. 2018:16. [Google Scholar]
- [105].Wajsbürt P, Sarfati A, Tannier X. Medical concept normalization in French using multilingual terminologies and contextual embeddings. Journal of Biomedical Informatics. 2021;114:103684. [DOI] [PubMed] [Google Scholar]
- [106].Newman-Griffis D, Divita G, Desmet B, Zirikly A, Rosé CP, Fosler-Lussier E. Ambiguity in medical concept normalization: An analysis of types and coverage in electronic health record datasets. Journal of the American Medical Informatics Association. 2021;28(3):516–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Newman-Griffis D, Lehman JF, Rosé C, Hochheiser H. Translational NLP: A New Paradigm and General Principles for Natural Language Processing Research. In: Proceedings of the conference. Association for Computational Linguistics. North American Chapter. Meeting. vol. 2021. NIH Public Access; 2021. p. 4125. [PMC free article] [PubMed] [Google Scholar]
- [108].Bennett TD, Moffitt RA, Hajagos JG, Amor B, Anand A, Bissell MM, et al. Clinical characterization and prediction of clinical severity of SARS-CoV-2 infection among US adults using data from the US National COVID Cohort Collaborative. JAMA network open. 2021;4(7):e2116901–e2116901. [DOI] [PMC free article] [PubMed] [Google Scholar]