Abstract
Chloroplasts have evolved from photosynthetic cyanobacteria-like progenitors through endosymbiosis. The chloroplasts of present-day land plants have their own transcription and translation systems that show several similarities with prokaryotic organisms. A remarkable feature of the chloroplast translation system is the use of non-AUG start codons in the protein synthesis of certain genes that are evolutionarily conserved from Algae to angiosperms. However, the biological significance of such use of non-AUG codons is not fully understood. The present study was undertaken to unravel the significance of non-AUG start codons in vivo using the chloroplast genetic engineering approach. For this purpose, stable transplastomic tobacco plants expressing a reporter gene i.e. uidA (GUS) under four different start codons (AUG/UUG/GUG/CUG) were generated and β-glucuronidase (GUS) expression was compared. To investigate further the role of promoter sequences proximal to the start codon, uidA was expressed under two different chloroplast gene promoters psbA and psbC that use AUG and a non-AUG (GUG) start codons, respectively, and also showed significant differences in the DNA sequence surrounding the start codon. Further, to delineate the role of RNA editing that creates AUG start codon by editing non-AUG codons, if any, which is another important feature of the chloroplast transcription and translation system, transcripts were sequenced. In addition, a proteomic approach was used to identify the translation initiation site(s) of GUS and the N-terminal amino acid encoded when expressed under different non-AUG start codons. The results showed that chloroplasts use non-AUG start codons in combination with the translation initiation site as an additional layer of gene regulation to over-express proteins that are required at high levels due to their high rates of turnover.
Subject terms: Biotechnology, Molecular biology, Plant sciences
Introduction
Chloroplasts are semi-autonomous organelles present in Algae to higher land plants and are generally considered to have originated from a photosynthetic cyanobacteria-like progenitor through endosymbiosis. Chloroplasts/plastids have their genome, although relatively small as compared to photosynthetic bacteria due to migration and subsequent integration of most of the genome into the nuclear genome1. The chloroplast genome of the present terrestrial plant codes for about 120–130 genes, mainly involved in photosynthesis, lipid metabolism, transcription, and translation mechanisms besides other cellular functions2. However, chloroplasts target the proteins encoded by nuclear-migrated genes for their functions through various mechanisms3. Due to the prokaryotic origin of chloroplasts, their transcription and translation closely resemble those of prokaryotic organisms, and studies have shown that chloroplast proteins can complement functionally in prokaryotes4,5. However, chloroplasts have evolved several other unique features involved in gene regulation at both transcription and translation stages6–10.
Similar to prokaryotes, translation initiation in chloroplasts is facilitated by the mRNA–rRNA interaction at the Shine–Dalgarno (SD) sequence and the key determinant in the process is the AUG start codon itself that interacts with N-formylmethionyl transfer RNA (tRNAfMet) to initiate the protein translation process11,12. Although AUG is the universal start codon for protein synthesis, several non-AUG codons have been shown to function as start codons13–21. For instance, ranslation of infA gene encoding initiation factor 1 has been shown to initiate from the UUG codon in green alga Chlorella vulgaris and tobacco22. Similarly, the translation of psbC gene in cyanobacterium Synechocystis has been shown to initiate from the GUG codon although an AUG codon was present just upstream of the GUG codon23. Such use of non-AUG codons is conserved across species and genera24, suggesting their biological significance. It has been estimated that more than ten Arabidopsis chloroplast genes are likely to be translated from non-AUG codons25. Recently, a systematic study conducted in E. coli involving the replacement of AUG with 63 non-AUG codons has shown that protein translation can initiate from a very large number of non-AUG codons, although the level of protein expression may vary significantly depending on the non-AUG codon used26. More recently, a proteomics-based approach has further revealed protein translation from non-canonical translation initiation sites (TIS) for several proteins in human cells21. In light of these findings, it is not clear whether transcription and translational mechanisms of the present-day terrestrial plant chloroplasts have evolved simply plasticity to accommodate any naturally occurring mutations or whether the chloroplasts used such evolved plasticity to select certain mutations to regulate gene expression further to meet the required levels of proteins for their functions.
The overall objective of this study is to unravel some of the regulatory features of transcription and translation mechanisms operating in the higher plant chloroplasts. For this purpose, systematic in vivo studies were conducted where the expression of the uidA (GUS) reporter gene was studied under the regulation of psbC promoter of tobacco that uses GUG as the start codon and compared its expression under four different start codons (AUG/UUG/GUG/CUG) that differed in their first base in the start codon. To ensure that the translation initiation context remained the same, the entire 5′ UTR and first six codons of psbC gene (codes for PSII 43KDa protein) were fused in frame with uidA gene. To assess further the role of sequence context surrounding the start codon in GUS expression, uidA with four different start codons (AUG/UUG/GUG/CUG) was also expressed under the regulation of a heterologous rice psbA (codes for PSII D1 protein) promoter that uses AUG as the start codon. In addition, to decipher the extent of changes that took place during the long course of evolution, all eight constructs tested for GUS expression in tobacco chloroplasts were also expressed in E. coli and compared the GUS protein expression/activity. The comparative GUS expression in tobacco chloroplasts suggests that plastid might have preferentially selected GUG over the universal AUG start codon to meet the required amount of PsbC protein, a component of the PSII complex that undergoes high turnover during photosynthesis. This view is further supported by the expression data of these constructs in E. coli where high levels of GUS were observed when AUG is the start codon, unlike the GUG start codon in chloroplasts. In addition, proteomics studies aimed at revealing protein translation initiation and N-terminal modifications showed that translation is initiated from multiple upstream sites in chloroplasts than from the predicted AUG/non-AUG sites. Another important observation was that the mRNA editing phenomenon commonly observed in chloroplasts that edit C base to U post transcription to create a start codon (ACG to AUG) did not alter any of the three non-AUG codons tested into an AUG codon. Taken together, all these pieces of experimental evidence underline significant evolutionary changes that chloroplasts of land plants have acquired to regulate the expression of selected genes to optimize the desired levels of protein using non-AUG start codons and other flanking sequences.
Results
psbC:uidA gene constructs designed to study protein expression under different start codons
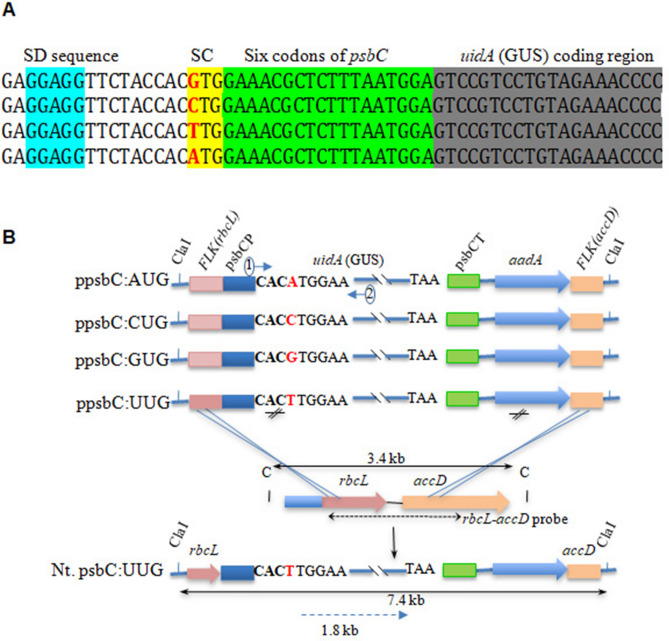
Four gene constructs that differed in the first nucleotide of the start codon (AUG/GUG/CUG/UUG) were created to study uidA expression under the tobacco psbC promoter (Fig. 1A). Based on previous studies by Kuroda and Maliga27 where it was shown that sequences downstream of the translation initiation codon play an important role in translation efficiency in chloroplasts, the sequence coding for the first six N-terminal amino acids of psbC was fused in frame with uidA to retain the translation initiation context of the native psbC (Fig. 1A). The tobacco partial rbcL:accD gene sequences were used to target site-specific integration of the chimeric psbC:uidA and selectable aadA genes through homologous recombination with the native plastid genome (Fig. 1B). The direction and the expected size of psbC:uidA transcripts and the location of the relevant restriction enzyme sites when stably integrated into the tobacco genome are shown in Fig. 1B.
Figure 1.
Schematic representation of expression cassettes used for chloroplast transformation. (A) Partial sequence of uidA (GUS) gene fused in frame with psbC gene 5′ untranslated region (5′UTR). SD and SC represent Shine–Dalgarno sequence (blue) and start codon (yellow), respectively. The first six codons (following start codon) are highlighted in green. (B) Partial restriction map showing the chimeric genes uidA and aadA. Also shown is the site of wild-type tobacco plastome where the chimeric uidA and aadA genes are expected to be integrated into the chloroplast genome (crossed lines) and the resulting transplastome with restriction sites and anticipated size of DNA fragments (solid line with double arrow) when restricted with ClaI restriction enzyme. Nt., psbCP, psbCT, FLK(rbcL), FLK(aacD) represent Nicotiana tabacum, psbC promoter, psbC terminator, partial sequences from rbcL and accD genes as flanking sequences (FLK) on either side of the transgene cassettes, respectively.
Site-specific integration of chimeric psbC:uidA gene into tobacco chloroplast genome
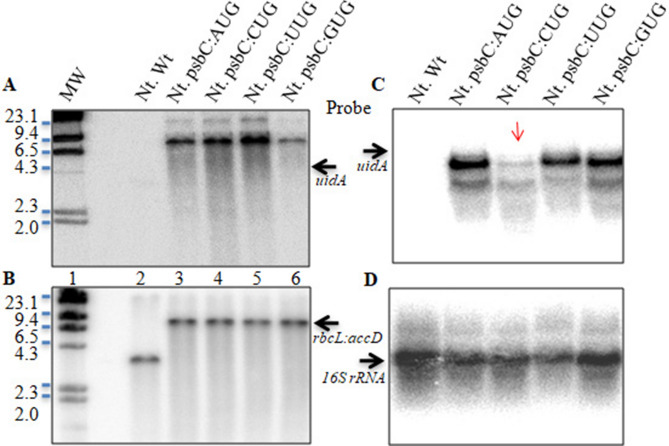
Southern hybridization showed the presence of ~ 7.4-kb band in the transplastomic plants (Fig. 2A) and not in wild-type plants when probed with the uidA gene sequence, which provided evidence for the stable and site-specific integration of psbC:uidA into the plastome (plastid/chloroplast genome). Furthermore, the presence of a 3.4 kb band in wild-type plants (Fig. 2B, lane 2) and its absence in transplastomic plants (Fig. 2B, lanes 3–6) when probed with rbcL:accD sequences further confirmed the stable integration of the transgene cassettes into the plastid genome. These results also showed the homoplasmic nature of transplastomes in the transformed plants that were selected for GUS expression and its activity studies.
Figure 2.
Stable integration and expression of GUS in tobacco chloroplasts under the tobacco psbC promoter with four different start codons. Southern hybridization of total genomic DNA probed with uidA (A) and partial gene sequences of rbcL:accD (B). Lane 1–6 correspond to size ladder (MW), wild type/control (Nt. Wt), psbC:AUG, psbC:CUG, psbC:TUG and psbC:GUG transplastomic Nicotiana tabacum (Nt.) plants, respectively. Note that in transcript/RNA Uracil (U) is used while in DNA/construct nucleotide Thymine (T) is used in place of Uracil. (C) Northern hybridization showing transcription of chimeric uidA under four different start codons in tobacco transplastomic plants. (D) The same blot reprobed with 16S rRNA to show equal loading of total RNA.
Transcription of psbC:uidA under psbC promoter with different start codons
Northern blot analysis showed the presence of uidA transcripts in all four transplastomic plants (Fig. 2C). In addition to the expected ~ 2.1 kb major band representing uidA transcript28, the presence of a minor band of lower size was also observed in all transplastomic lanes, which might represent the uidA transcripts that are not denatured completely at the time of gel loading. However, uidA transcript levels were significantly less where CUG was used as the start codon. To rule out any unequal loading of total RNA in the gel, the same blot was reprobed using the 16S rRNA sequence, and the results showed that RNA was loaded equally in all four lanes (Fig. 2D).
Role of sequence context flanking start codon in transcription and translation
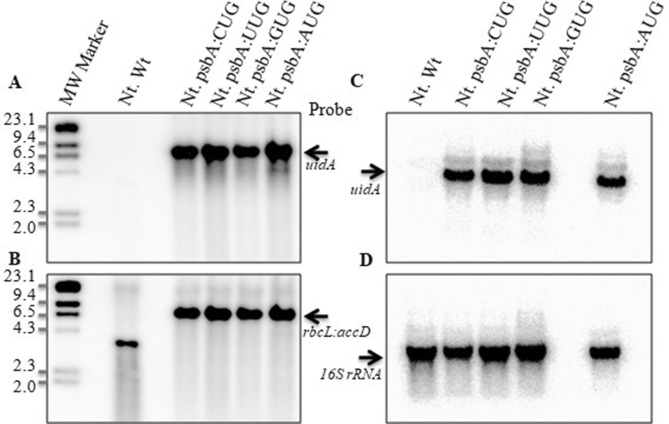
To assess the influence of the sequence around the start codon on transcription and protein synthesis, another set of four constructs was developed where uidA was placed under the heterologous rice chloroplast psbA promoter (Supplementary Fig. S1)29with different start codons (AUG, GUG, CUG and UUG) and stably transformed transplastomic tobacco plants were developed. The rice psbA promoter shared a high degree of homology with the tobacco psbA promoter (Supplementary Fig. S2A) and was shown to transcribe foreign genes efficiently in tobacco chloroplasts previously by our group30 and hence no fusions were made with psbA gene sequences coding N-terminal amino acids. The sequence homology between tobacco psbC and rice psbA promoters surrounding the start codon and 5′ UTR is shown in Supplementary Fig. S2B. While − 35 and − 10 sequences and ribosome binding site (SD) are well conserved between rice and tobacco for psbA promoters, the sequences between SD and the start codon differed significantly between psbC and psbA promoters (Supplementary Fig. S3). It is worth noting here that while AUG is the start codon for psbA in both rice29 and tobacco31, GUG is the start codon of psbC in tobacco23. Moreover, efficient expression of uidA gene under the heterologous rice psbA promoter in tobacco chloroplasts has been reported earlier30. Stable integration of transgene cassettes (psbA:uidA and aadA) was confirmed using Southern hybridization. An expected 7.4 kb band was observed in the transplastomic plants when probed with the uidA coding sequence (Fig. 3A). No corresponding band was observed in control plants, confirming the transplastomic nature of the plants regenerated under spectinomycin selection. Reprobing of the same blot with rbcL:accD flanking sequences further confirmed the site-specific integration of uidA and aadA genes into the tobacco plastome (Fig. 3B).
Figure 3.
Stable site-specific integration and expression of uidA under the rice psbA promoter with different start codons in tobacco chloroplasts. Southern hybridization of genomic DNA using uidA (A) and partial rbcL:accD (B) gene sequences to show the site-specific integration of transgenes into tobacco plastome. Northern hybridization showing the transcription of chimeric uidA in four different transplastomic plants (C) and the blot probed with 16S rRNA to show equal loading of total RNA (D).
Northern blot analysis confirmed efficient transcription of uidA under the rice psbA promoter, and an expected 1.8 kb band corresponding to uidA transcripts was observed in all four transplastomic plants when probed with uidA gene-specific sequences (Fig. 3C). Most importantly, the intensity of the band is comparable between all four transplastomic plants (Fig. 3C), unlike the transcription of uidA under the psbC promoter where transcript levels were significantly low when CUG was the start codon (Fig. 2C). Further, probing of the blot with 16S rRNA confirmed the equal loading of total RNA in all lanes (Fig. 3D).
Expression of GUS under different start codons in transplastomic tobacco plants
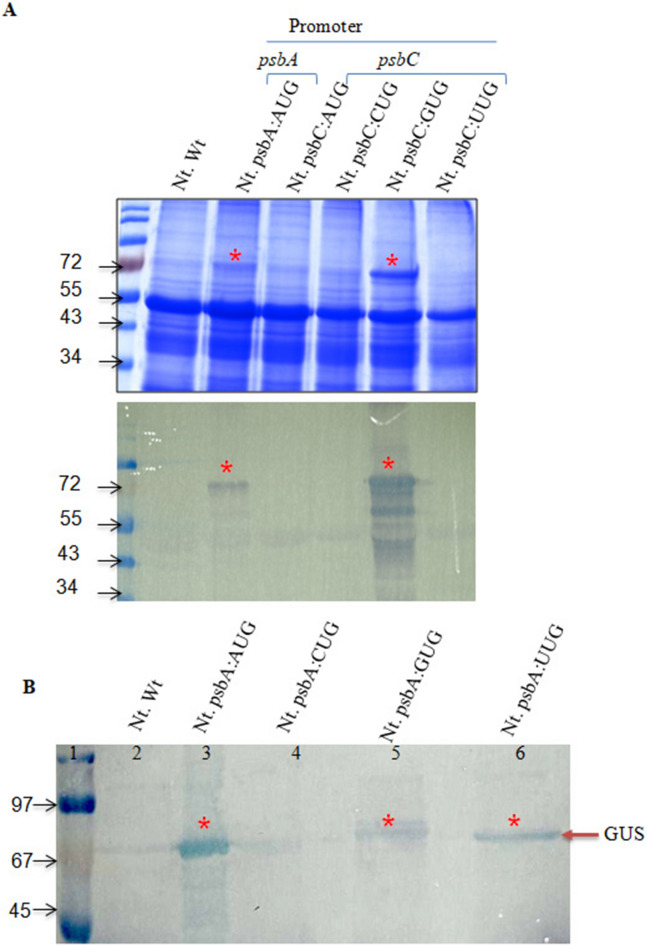
To assess the influence of the start codon on the translation efficiency, GUS expression under four-start codons (AUG, GUG, CUG, and UUG) driven by either psbC or psbA promoters was examined initially based on protein profile using SDS–PAGE (Fig. 4). A protein band corresponding to the expected size of GUS protein could be seen in the plants where uidA was placed under psbA:AUG, and psbC:GUG (Fig. 4A) when crude protein extract was loaded in the gel. These results were confirmed further using western blot analysis. Also, the presence of a GUS protein was observed in the transplastomic plants where AUG, GUG, and UUG start codons were utilized for uidA gene expression driven by psbA promoter (Fig. 4B). However, no GUS corresponding protein band was observed in the western blot in the transplastomic plants where CUG was the start codon. To confirm the presence of the GUS protein in the transplastomic plants transformed with psbA:AUG, psbA:GUG, psbA:UUG constructs, an immunoprecipitation experiment was carried out using anti-GUS antibodies. The expected size GUS band was observed in SDS-PAGE and western blot analysis with immunoprecipitated proteins from plants transformed with psbA:AUG, psbA:GUG, psbA:UUG constructs, while no corresponding band was found in the wild-type plant (Supplementary Fig. S4). Furthermore, the histochemical staining to detect β-glucuronidase (GUS) activity exhibited the presence of blue colur in transplastomic plants expressing these constructs (Supplementary Fig. S5).
Figure 4.
Comparative protein profile analyzed on (A) SDS-PAGE and corresponding western blot using anti-GUS antibodies from the transplastomic tobacco leaf. The details of promoter and start codon utilized for uidA expression are given just above the lane and wild type untransformed Nicotiana tabaccum (Nt. Wt) was used as a negative control. Note the high expression of GUS under psbC promoter having GUG as the start codon. (B) Western blot analysis of transplastomic tobacco plants expressing uidA with one of the four start codons (AUG, CUG, UUG, GUG) under psbA promoter. Asterisk symbol point out the expected size GUS protein band.
Comparison of GUS expression between psbC and psbA promoters in chloroplasts
To compare the GUS expression between the transplastomic lines developed using eight different constructs, a quantitative GUS assay based on the enzymatic activity was performed. The GUS expression under four start codons driven by the psbC promoter exhibited highest GUS activity when GUG was the translation initiation/start codon, followed by AUG and UUG (Fig. 5A). It is worth noting here that GUG is the start codon for psbC in wild-type tobacco plants. No detectable GUS activity was observed when CUG was the start codon, which could be attributed to the lack of sufficient mRNA to translate or possible rapid degradation. Unlike GUS expression under the psbC promoter, its expression under the psbA promoter showed significant changes depending upon the start codon used. Among the four transplastomic lines, the highest GUS activity was observed when AUG was the start codon, which is also the canonical start codon of psbA in both rice and tobacco (Fig. 5B). Although GUS activity was detected under GUG and UUG start codons, the level of expression was very low (669 and 3210 nmol of MU release/minute/microgram of protein, respectively) as compared to its activity under the AUG start codon. Again, GUS activity could not be detected when CUG was the start codon, irrespective of the promoters (psbA or psbC) used, which is similar to the results obtained with the psbC promoter.
Figure 5.
Comparison of GUS expression levels in transplastomic tobacco plants under different translation initiation codons (AUG, CUG, GUG, and UUG), driven by either tobacco psbC or rice psbA promoter and comparison of same constructs expressed in E. coli. Expression of GUS in tobacco chloroplasts under the regulation of native tobacco psbC promoter (A) and heterologous rice psbA promoter (B). Expression of GUS in E. coli under tobacco psbC promoter (D) and rice psbA promoter (E). Comparison of GUS expression under psbA and psbC promoters in N. tabaccum (C) and E. coli (F).
To delineate the role of non-AUG codons and the sequence context surrounding the translation initiation site in gene expression, GUS activity was compared between the psbC and psbA constructs having the same start codon but differed in their upstream and downstream sequences (Supplementary Figs. S2B and S6). The comparative GUS activity data showed that GUS expression was highest when uidA was expressed under the tobacco psbC promoter with GUG as the start codon, the native sequence under which psbC gene is expressed in wild-type tobacco plants (Fig. 5C). Interestingly, the level of GUS expression under the psbC promoter with the GUG start codon is more than double as compared to that of the GUS expression observed under the psbA promoter with AUG as the start codon (Fig. 5C).
Comparison of GUS expression under psbC and psbA promoters in E. coli
As transcription and translation mechanisms of chloroplasts share significant similarities with bacterial systems due to the prokaryotic origin, and yet chloroplasts have evolved several additional features, we were interested to assess the impact of evolutionary changes in the transcription and translation systems on gene expression due to usage of different start codons and the TIS sequence context in bacteria. For this purpose, we transformed all eight constructs (four each of psbC:uidA and psbA:uidA) into E. coli and compared GUS activity with chloroplast expression. The GUS expression could be seen in E. coli under both psbC and psbA promoters. The expression profile of GUS under the same start codon is comparable between psbC and psbA promoters in E. coli (Fig. 5D,E). These results are in contrast to the GUS expression profile observed in chloroplasts where the expression profile of GUS changed significantly between psbC and psbA promoters (Fig. 5A,B). Interestingly, there was no detectable GUS activity when CUG was the start codon in E. coli as well, similar to the results obtained in chloroplasts.
Comparison of GUS expression between chloroplasts and in E. coli
Significant differences between chloroplasts and E. coli in terms of GUS expression profile and levels of expression were observed depending on the start codon and the promoter combination used. While the highest GUS activity was observed in E. coli when AUG was the start codon, irrespective of the promoter used (Fig. 5D–F), its expression in chloroplasts was highest when uidA was expressed under the psbC promoter with GUG as the start codon (Fig. 5C). Again, significant differences were observed in the GUS activity when compared between those of chloroplasts and E. coli when UUG is used as the start codon (Fig. 5C,F). While the expression level of GUS with UUG as the start codon was similar in E. coli, its expression level in chloroplast was much higher under the psbC promoter as compared to the psbA promoter. The statistical analysis of the GUS enzyme activity based on three replicates showed that the GUS activity varied significantly depending on the start codon and the promoter combination used (P < 0.001).
Non-AUG start codon tested was not subject to RNA editing in chloroplasts
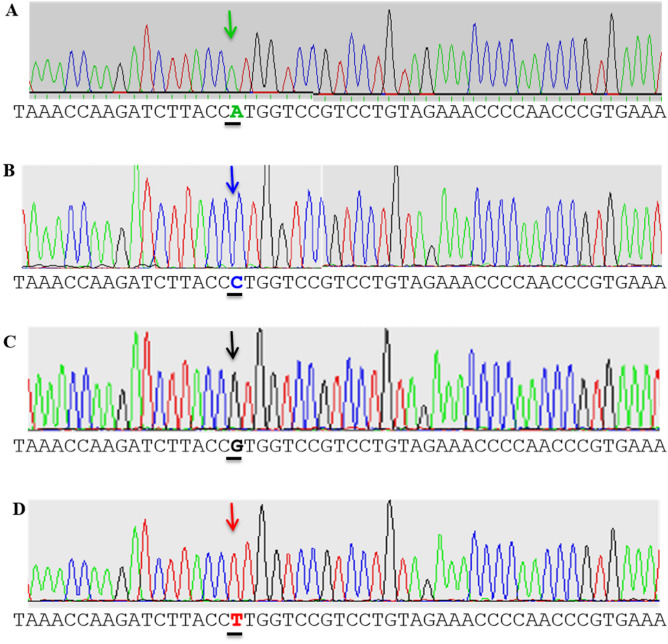
As RNA editing is a common phenomenon in chloroplasts, especially since start codons are created by editing transcripts post-transcriptionally, we were interested to see whether the uidA transcripts with modified start codons are subject to RNA editing to revert to the native start codon before translation in the chloroplasts. To verify this, we analyzed the uidA transcripts expressed under the psbA promoter with different start codons and by sequencing respective cDNAs. Sequence analysis showed that there were no changes in the cDNA sequence in any of the four transplastomic plants when compared to the corresponding DNA sequence present in the vector used to transform the chloroplasts (Fig. 6). These results suggest that mRNA with mutations in the first nucleotide of the start codon tested in this study has not been subjected to RNA editing phenomenon present in the chloroplasts.
Figure 6.
Partial sequence of uidA transcripts isolated from transplastomic plants that had different translation initiation start (TIS) codons. Transplastomic tobacco plants expressing psbA:uidA gene under AUG (A), CUG (B), GUG (C) and UUG (D) start codons, respectively. Sequencing data showed that the transcripts had the same base as per the gene construct in each of the transplastomic plants, ruling out any RNA editing of uidA transcripts in these plants.
Translation initiates from multiple sites in chloroplasts and it varies depending on the sequence context of TIS
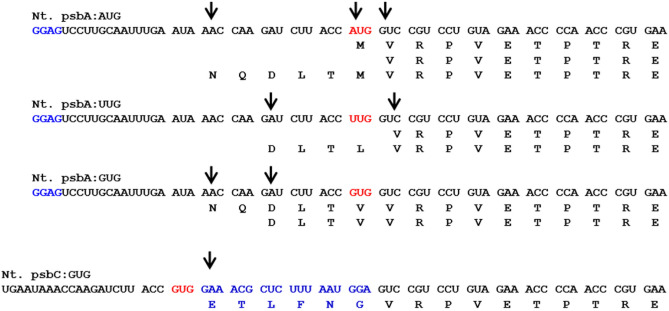
Although the AUG start codon plays an important role in the initiation of protein translation, we were interested to see how tRNAs and ribosomes recognize and initiate protein synthesis when AUG is replaced with a non-AUG codon. The proteomics-based N-terminal sequence data revealed several interesting aspects of protein translation initiation and post-translational modifications in the chloroplasts. As can be seen from the summarized proteomics data (Fig. 7, Supplementary Table S1 and Supplementary Fig. S7), translation is initiated from multiple and, in some cases, novel sites that are upstream to the expected start codon. As can be seen from Fig. 7, translation is initiated from two or more sites, and the sites of initiation varied depending on the non-AUG start codon. In contrast, translation was initiated from a single site when uidA was expressed under the psbC promoter with GUG as the start codon, which is also the native start codon for psbC. Further, the N-terminal sequence of GUS revealed a number of post-translational modifications that included N-terminal methionine excision and N-terminal methionine formylation (Supplementary Table S1).
Figure 7.
Translation initiation of GUS protein expressed under different start codons in transplastomic tobacco chloroplasts. Each arrow indicates the translation initiation site as determined by mass spectrometry-based peptide sequencing (MS/MS). Note that GUS protein is translated from multiple sites that are upstream to the expected start codon (shown in red) and even when the universal start codon (AUG) was present in the transcript downstream to the SD sequence. Note that whereas protein translation initiated from three and five upstream codons when GUS was expressed with the GUG start codon under the psbA promoter, it initiated from the single site when expressed under the psbC promoter, indicating the role of the sequence context besides the start codon.
Discussion
Expression of uidA under tobacco psbC and rice psbA promoters with different start codons
A number of genes encoded by the chloroplast genome in terrestrial plants were shown to be translated from non-AUG start codons32. Conversion of AUG codon to AUU or AUC in chloroplast petD gene in the green alga Chlamydomonas was shown to reduce the translation initiation rate by 10–20% as compared to AUG33. On the other hand, the conversion of UUG, a non-AUG codon, to AUG was shown to enhance the translation efficiency of chloroplast infA gene in another green alga Chlorella vulgaris by 300-fold22. Further, the change of the AUG start codon to non-AUG codons was shown to influence the protein translation efficiency in Chlamydomonas34. For example, conversion of the AUG start codon of petA gene (encoding cytochrome f) to AUU, ACG, ACC, ACU, and UUC was shown to influence cytochrome-f levels in Chlamydomonas chloroplast significantly34. In addition, sequence downstream to the AUG start codon was also shown to influence the translation initiation and elongation of protein synthesis in tobacco chloroplasts27. The present study was undertaken to understand the role of the start codon and its flanking region in the gene regulation at transcription and translation stages in chloroplasts of higher plants. For this purpose, we chose the promoter of tobacco psbC gene having GUG as the native start codon35 and another promoter of rice psbA gene with AUG as the native start codon30 to express uidA (GUS) reporter gene under four different start codons (GUG, AUG, UUG and CUG) and assessed the role of start codons and surrounding sequence context in gene expression at transcription and translation in vivo in the stably transformed tobacco chloroplasts. The constructs having rice psbA promoter and terminators were used to compare gene expression under a different sequence context surrounding the start codon and, also to avoid any possible homologous recombinations between native and introduced promoter/terminator, if any other than the desired locus (rbcL-accD), that may, leading to large DNA deletions and rearrangements in the plastid genome28.
An important finding in the present study is the very low level of transcription of uidA under the psbC promoter with CUG as the start codon (Fig. 2C) as compared to other start codons (AUG, GUG or UUG) tested. On the other hand, there are no significant differences in the transcript levels of uidA when expressed under the psbA promoter irrespective of the start codon used (Fig. 3C). It is possible that the transcription factors that bind the promoter region and recruit RNA polymerase to initiate transcription are no longer able to recognize the promoter element, hence leading to the lack of efficient transcription of uidA. These results further emphasize the previous finding that the overall sequence context around the start codon is important than the start codon itself for efficient transcription of genes in chloroplasts27, similar to prokaryotes36–38 and eukaryotes39.
Another important finding in this study is the higher expression of GUS with GUG as the start codon than with the AUG start codon when expressed under the psbC promoter, while the expression is just reverse when expressed under the psbA promoter (Fig. 5A,B). Importantly, the expression level of GUS under the psbC promoter with AUG and UUG start codons is almost double compared to the highest expression level observed under the psbA promoter with AUG as the start codon (Fig. 5C), suggesting that the sequence context of the psbC promoter surrounding the translation initiation is the most favorable for protein translation in chloroplasts. However, the same sequence context is not favorable in E. coli, where the sequence context with AUG is the most favorable under both psbA and psbC promoters (Fig. 5D,E). The comparative expression data between chloroplasts and E. coli suggest the significance of not only the sequence context and start codons in protein translation efficiency but also the evolutionary changes that took place in the protein translation machinery of chloroplasts from where they evolved. It is possible that chloroplasts might have selected and enriched different mutations that have a cumulative effect on gene expression over a period of time to express certain genes like psbC, a chlorophyll-binding protein of PSII complex that undergoes light-mediated turnover during photosynthesis40, as another layer of gene regulation as observed in other organisms41 including bacteria26,42–46.
Such a selection process would have also been facilitated by the plasticity built in the translation mechanism of chloroplasts with the parallel evolution of their ribosome and/or factors interacting with TIS and near-cognate start codons with mismatches. This view is supported by the observation that a number of genes with important functions in the bacteria42,47,48 and certain viral proteins required for the rapid multiplication of viruses are translated from non-AUG start codons49,50.
CUG has been shown to function as the start codon in several eukaryotic organisms20,21,51,52 and the second most efficient start codon after the universal AUG codon20. In contrast, in prokaryotes, nearly ~ 80% of annotated genes initiate at AUG codons, ~ 12% at GUG, and ~ 8% at UUG, with variable incidences of AUU, CUG, and AUC across species47. For instance, in 69 model bacterial genomes, CUG has been reported as a start codon in 20 annotated open reading frames/ ORFs/genes (0.024% ORFs suggesting negligible translation initiation frequency and less efficient start codon)26. However, in the present study the lack of GUS expression in both chloroplasts and E. coli with CUG as the start codon, irrespective of the promoter used, suggests organism-specific preferences.
Transcripts having non-AUG start codons are not subject to RNA editing in chloroplasts
RNA editing, a phenomenon present in chloroplasts that creates the start codon or changes the amino acid sequence in the proteins by base editing at selected positions in the transcript, has not been found in non-AUG start codons with an altered nucleotide at the first base. RNA editing is a significant feature of gene regulation in terrestrial plant chloroplasts, by which the mRNA sequence is selectively altered post-transcriptionally, which is not observed in the algal chloroplasts or the bacteria53. RNA editing is used either to create a start codon or to alter the amino acid in the protein sequence. For instance, an AUG start codon is shown to be created before translation in the mRNA of rpl2, psbL and ndhD genes53–56 in chloroplast through the conversion of C to U through RNA editing. In our study, the sequencing results of cDNAs corresponding to uidA transcripts representing all four transplastomic lines (Fig. 6) did not reveal any changes related to RNA editing, suggesting that RNA editing did not recognize the first base edited start codon, possibly due to the high selectivity of RNA editing, which is limited to certain codons only in selected genes57.
Change in start codon leads to shift in protein translation initiation site (TIS)
Identification of TIS when the start codon is altered is another objective of the present study. Studies by Sasaki and Nakashima58 showed that the translation of an insect viral RNA is initiated with glutamine, without the participation of the initiator methionine tRNA. The tRNA that reads the AUG codon in eubacteria also reads GUG and UUG codons and translates the codons as formylmethionine59. Determination of N-terminal ends of GUS expressed under four different start codons showed protein translation from different codons upstream to the typical start codon, resulting in a heterogeneous population (isoforms) of proteins differing in their N-terminal ends, which in turn might be allowing organisms to select a particular isoform with different turnover rates, localization and biological function. Protein synthesis from codons other than AUG and amino acids other than methionine has been reported earlier20,26,60,61. Also, the CUG codon was shown to code for serine in vivo in Candida albicans instead of leucine as predicted by the genetic code62. In addition, our data showed that N-terminal methionine got modified and/or processed similar to that in prokaryotic and eukaryotic organisms21,63,64, suggesting a high degree of conservation in post-translational modifications across organisms and organelles.
Conclusion
The comparative GUS expression under four-start codons that differed in their first base (AUG, UUG, GUG and CUG) under psbC and psbA promoters in tobacco chloroplasts and its expression under the same combination of the translation initiation site and start codons in E. coli revealed that the chloroplasts of the present-day land plants have acquired an additional layer of regulatory feature as compared to prokaryotic progenitors during the long course of evolution. In addition, proteomics-based N-terminal sequencing of GUS expressed in chloroplasts showed that protein translation initiates from multiple novel sites depending on the start codon used and TIS flanking it, again revealing that chloroplast has evolved enough plasticity that allows them not only to use non-AUG start codon to express particular gene but also continue to select and enrich a combination of mutations as a post-transcriptional regulatory mechanism to attain the required levels of proteins that undergo light-mediated rapid turnover during photosynthesis.
Methods
Vector construction, chloroplast transformation, and molecular and GUS assays
The plastid transformation vector pVSR326 (Fig. 1, GenBank Acc. No. AF527485, 28) was used to construct four vectors to express uidA under the psbC promoter (ppsbC:AUG, ppsbC:CUG, ppsbC:GUG and ppsbC:UUG) and another four vectors to express uidA under the psbA promoter (ppsbA:AUG, ppsbA:CUG, ppsbA:GUG and ppsbA:UUG) (Supplementary Fig S1). Note that in transcript/RNA Uracil (U) is used while in DNA/construct nucleotide Thymine (T) is used in place of Uracil. For consistency in terminology in figures and text, we have used the transcript nomenclature throughout the manuscript i.e. U (not T). These plastid transformation vectors are designed to contain flanking sequences (FLK) on either side of the transgene cassettes, derived from the host plant chloroplast genome, to facilitate two homologous recombinations for site-specific integration of transgenes. The vectors used in this study contained partial sequences rbcL and accD genes as flanking sequences for homologous recombination that integrate uidA (encoding ß-Glucuronidase) and aadA gene (encoding aminoglycoside adenine transferase) cassettes in the non-coding region present between rbcL and accD genes65. A three-step PCR-based procedure was followed to generate four psbC:uidA chimeric genes that differed in the first base of the start codon. In the first step, the psbC promoter of tobacco was amplified using the oligonucleotides psbC-SalI and psbCGUS-ATG/psbCGUS-CTG/psbCGUS-GTG/psbCGUS-TTG (Supplementary Table S2) and Nicotiana tabcum (tobacco) total genomic DNA as the template in 25 µl reaction volume for 30 cycles. In the second step, 5 µl of PCR mix from the first step was added to the second PCR mix that contained pVSR32630 plasmid DNA as the template, without any additional primers and amplified for 10 cycles. In the third step, 5 µl of PCR mix from the second step was used as the template in the third PCR mix that contained psbC-SalI and GUS3-SacI primers. The PCR-amplified psbC:uidA was digested with SalI and SacI enzymes and cloned into pVSR326 (GenBank Acc. No.AF527485.1) at the same sites to create ppsbC:AUG, ppsbC:CUG, ppsbC:GUG and ppsbC:UUG vectors.
The ppsbA:CUG, ppsbA:GUG and ppsbA:UUG vectors were generated using QuikchangeH Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer’s instructions. Primer pairs used in the mutagenesis are listed in Supplementary Table S2. Chloroplast transformation vector pVSR326, where uidA is placed under the psbA promoter of rice with the AUG start codon, was used to compare GUS expression under the psbA promoter with CUG/GUG and UUG start codons. All introduced mutations were verified by sequencing DNA.
All vectors contained rrn:aadA:rbcL (aadA gene driven by rrn promoter and rbcL terminator) selectable gene cassette imparting resistance to spectinomycin antibiotic. The tobacco plastid DNA sequences spanning rbcL:accD genes (nucleotides 58,056–60,627; EMBL, Z00044) were used for site-specific integration of transgenes into the plastome. Standard procedures were followed for cloning, Southern, Northern, and protein analysis66. A method based on the particle delivery system (Gene Gun method) was followed for tobacco (Nicotiana tabacum cv. Petit Havana) chloroplast transformation65. All eight constructs were also transformed into β-glucuronidase-deficient E.coli strain GMS407 using the method of Sambrook et al.66.
GUS activity and histochemical assays
GUS activity assay and histochemical staining of β-glucuronidase (GUS) expression was performed as described elsewhere67. Briefly, for the GUS assay, about 100 mg transplastomic tobacco leaf tissue was homogenized in 300 µl of GUS extraction buffer containing 50 mM NaH2PO4, pH 7.0, 10 mM EDTA, 10 mM beta-mercaptoethanol followed by centrifugation at 10,000 rpm for 10 min. In the case of E. coli GMS407, cells were sonicated using the same GUS extraction buffer followed by separating the extracts using a centrifuge. The protein concentration in the extracts was estimated using the Bradford method68. A 4 µg of total protein was added to GUS assay buffer containing 1 mM 4-methyl umbelliferyl beta-d-glucuronide (MUG; Sigma M-9130), the total volume of extraction buffer was adjusted to 200 µl and incubated at 37 °C. After 10 min of reaction, 50 µl aliquot was taken and added to 950 µl stop buffer (0.2 M Na2CO3) to terminate the reaction. The amount of 4-methyl umbelliferone released was estimated by measuring the fluorescence emission at 360 nm excitation and 460 nm emission using the spectrofluorimeter. The fluorimeter was calibrated with freshly prepared 1 µM, 10 µM, and 100 µM methyl umbelliferone (MU) standards. GUS activity was calculated as Nano moles of MU release/minute/microgram protein. The level of nanomoles of MU released directly correlates with the level of GUS expression. The statistical analysis for the GUS activity assay was done using SAS statistical software version 9.3 (https://sscnars.icar.gov.in/) to see the level of significance. Standard errors and standard deviation were calculated from the replicates.
For histochemical GUS assay, 10 days old seedlings and leaf discs from 5-week-old transplastomic lines were incubated in 1 mM X-Gluc substrate in 50 mM NaH2PO4, pH 7.0 at 37 °C for 12–16 h. Photographs were taken under the binocular microscope after rinsing the seedlings/leaf disc 3 times in 70% ethanol for 5 min each. GUS expression was categorized based on the intensity of the blue color (low, medium, and high) in the leaf discs/seedlings observed under the microscope.
cDNA synthesis and sequencing
The total RNA from psbA:AUG, psbA:CUG psbA:GUG, and psbA:UUG construct expressing transplastomic plants leaves were isolated using TRIzol reagent (Invitrogen, Life Technologies, USA) as per manufacturer’s instructions. The quality and quantity of total RNA were assessed by NanoDrop 1000 spectrophotometer. The RNA samples with A260/A280 ratio of ~ 2.0 and A260/A230 ratio of 2.0–2.2 were considered pure. The total RNA was stored at − 70 °C. The DNase-treated 1 µg total RNA was used for the synthesis of first strand cDNA using AffinityScript qRT-PCR cDNA synthesis kit (Stratagene, Agilent Technologies, USA) according to the manufacturer’s instructions. PCR amplification was performed in BIORAD thermal cycler using 25 µl of total volume reaction containing 100 ng template cDNA, 1X Taq Polymerase buffer, 1.5 mM magnesium chloride, 1 mM dNTPs, 0.4 µM of each forward and reverse primer pair and 2U of Taq DNA polymerase. At least two cDNAs were sequenced per line from Macrogen Corporation, South Korea.
Protein extraction and SDS–PAGE analysis
Total protein from 0.5 g tobacco transplastomic plant leaf was extracted with 500 µl 2X PBS (pH 7.4) containing 0.1% Tween 20 and proteinase inhibitor cocktail (Roche) by grinding using mortar and pestle. The supernatant was obtained after centrifugation at 10,000 rpm for 10 min. The concentration of the protein in the supernatant was estimated by the Bradford method68. The protein samples were subjected to 12% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) separation69. The gels were either stained with Coomassie Blue to visualize the proteins or used for Western blotting. For psbA:AUG 20 ug total protein while for remaining samples 50 ug total protein was taken for SDS-PAGE experiment. Similarly, total protein from overnight grown untransformed E. coli GMS407 (negative control), a mutant strain of beta-D-glucuronidase, and transformed E. coli GMS407 cells expressing GUS (using different translation initiation codons such as AUG, CUG, GUG and UUG) either under tobacco psbC promoter or rice psbA promoter were extracted by breaking cells via sonication and subjected to SDS-PAGE analysis.
Immunoprecipitation
Protein A Sepharose 4 Fast Flow beads (GE Healthcare) were washed thrice at 1200 rpm with 1X PBS. 10 µl rabbit anti-GUS antibody (Sigma Aldrich, catalog number G5420) was added in 200 µl washed protein A beads in 1X PBS and allowed to bound at 4 °C for 4 h under shaking. The antibody-protein A bead complex was washed thrice with 1X PBS. The total protein from the tobacco transplastomic plant leaf was added to the antibody-protein A complex and incubated in a cold room overnight under shaking. Three washes were performed with 1X PBS at 1200 rpm for 5 min. The antigen (GUS protein)-antibody-protein A bead complex was resuspended in 15 µl 4X SDS PAGE sample buffer by boiling. After immunoprecipitation, the samples were separated on 10% SDS–PAGE mini gels as described in the instruction manual for the Mini-Protean III Electrophoresis Cell (BioRad).
Western blot analysis
After separating proteins using SDS-PAGE analysis, proteins were transferred/blotted onto a polyvinylidene difluoride (PVDF) membrane (Millipore) to facilitate antibody probing. The PVDF membrane was blocked in 4% BSA (bovine serum albumin) in TBS-Tween 20 (TBST) for 30 min, followed by washing thrice in TBST for 10 min. The membrane was incubated with rabbit-raised anti-GUS antibody (1:10,000 dilution) in TBST for 1 h. The membrane was then washed thrice for 10 min each in TBST and incubated with an anti-rabbit alkaline phosphatase-conjugated secondary antibody (1:10,000 dilution) (Sigma) for 30 min. The membrane was washed thrice for 10 min each and developed with Western Blue® Stabilized Substrate for alkaline phosphatase (Promega). Please note that the lack of protein detection in western blot analysis despite high GUS enzyme activity in psbC:AUG and psbC:UUG expressing plants could be because the anti-GUS antibody somehow did not recognize GUS translated from these two constructs as the presence of different forms of GUS protein is possible due to initiation from multiple sites. Perhaps the majority of proteins in these two cases are initiated at some other sites downstream/upstream to UUG or AUG.
In-gel digestion of proteins
After electrophoresis, the gel was stained with Coomassie Brilliant Blue. Protein bands were excised, chopped into small pieces, and destained with 200 µl 50% methanol/10% acetic acid. The gel pieces were then rinsed with autoclaved MilliQ water and equilibrated with 100 mM ammonium bicarbonate (NH4HCO3) for 15 min at room temperature with gentle agitation. The gel pieces were washed with 200 µl 1:1 (v/v) NH4HCO3 and acetonitrile, dehydrated with 100% acetonitrile for 15 min, and then vacuum-dried. The sample was subjected to reduction with 10 mM dithiothreitol (DTT) in 25 mM NH4HCO3 for 45 min at 56 °C followed by alkylation with 50 mM iodoacetamide (IAA) in 25 mM NH4HCO3 for 30 min in the dark at room temperature. The gel pieces were rinsed briefly with 1:1 NH4HCO3 and acetonitrile solution, dehydrated with 100% acetonitrile for 15 min, dried, and digested with 50 µl enzyme solution containing 15 ng/μl Asparagine-N (Cat no. V1621, Promega) or Glu-C (Cat no. V165A, Promega) or trypsin in 25 mM NH4HCO3 for 18 to 20 h at 37 °C. Following digestion, peptides were extracted twice with 100 μl of 1% trifluoroacetic acid in 60% acetonitrile, and the extracts were concentrated under a vacuum. The peptide pellet was stored at − 80 °C until further use.
Nano-LC-based reverse-phase separation
Peptide pellets were redissolved in 20 μl of 0.1% TFA, and 15 μl of the sample was bound onto a 100 μmi.d. × 20 mm Easy-nLC precolumn (Proxeon Biosystems) at a flow rate of 5 μl/min. Reverse-phase separation of the peptide mixture was performed in a nano-LC system (Proxeon Biosystems) using a C-18 analytical column of 75 μmi.d. X 100 mm length at a flow rate of 300 nL/min with a solvent system comprising 0.1% TFA in 5% ACN , v/v (solvent A) and 0.1% TFA in 90% ACN (solvent B). The gradient was 0% B for 5 min, followed by a linear gradient of 0% to 45% B for 65 min, 45% to 100%B for 1 min, and 100%B for 10 min. The column eluates were directed to a Proteineer fc fraction collector (Bruker Daltonics), and fractions were spotted every 10 s onto a pre-spotted anchor chip 384/96 (PAC) target plates.
Protein identification from 1D gel phase digested samples
Peptide samples were injected into a nano-LC system, fractionated, and spotted onto PAC (Pre-Anchored Chip) targets70. Sample plates were subjected to MADLI TOF/TOF-based acquisition in the automated mode through WARP-LC 1.2 (Workflow Administration by Result driven Processing, Bruker Daltonics) software tool. The MALDI TOF/TOF instrument parameters mentioned in Kumar et al.70 were employed for automatic acquisition: carrier plates with samples were subjected to pre-teaching to ensure the optimum and complete acquisition of 448 sample spots. Mass list calculation was done through the WARP-LC interface. A manually updated background list containing trypsin autolysis peaks, matrix peaks, and keratin peaks was used during mass list generation. Spectral peaks (m/z) corresponding to background peaks were excluded for MS/MS measurement. Only those peaks (precursors) that have an S/N > 20 were included in the measurement. Post-acquisition processes including mass annotation, baseline subtraction, and smoothening were performed using Flex Analysis software, version 3.0, through WARP-LC. Protein identification was achieved using Biotools, version 3.2, through an in-house-licensed Mascot server (version 2.3, March 2010).
Supplementary Information
Acknowledgements
We thank Abhishek Kumar Jha for his help with GUS assay experiments.
Author contributions
Conceptualization: S.L., V.S.R., K.K., A.B., S.K. Writing original draft: V.S.R. and S.L. Review and editing: K.K., S.K., A.B. Overall investigation: V.S.R. and S.L. Conducting experiments: K.K., S.K., A.B., A.D., R.P., P.P., N.V.C., B.S.R. All authors read and approved the manuscript.
Funding
We gratefully acknowledge the financial support of ICGEB and Department of Biotechnology (DBT), Government of India for this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Leelavathi Sadhu, Email: leelavathisadhu@gmail.com.
Krishan Kumar, Email: krishan.kumar6@icar.gov.in.
Vanga Siva Reddy, Email: vsreddy@gmail.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-27347-9.
References
- 1.Martin W. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- 2.Daniell H, Lin C-S, Yu M, Chang W-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jarvis P, Robinson C. Mechanisms of protein import and routing in chloroplasts. Curr. Biol. 2004;14:1064–1077. doi: 10.1016/j.cub.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 4.Gold JC, Spremulli LL. Euglena gracilis chloroplast initiation factor 2. Identification and initial characterization. J. Biol. Chem. 1985;260:14897–14900. doi: 10.1016/S0021-9258(18)95677-2. [DOI] [PubMed] [Google Scholar]
- 5.Kraus BL, Spremulli LL. Chloroplast initiation factor 3 from Euglena gracilis: Identification and initial characterization. J. Biol. Chem. 1986;261:4781–4784. doi: 10.1016/S0021-9258(19)89172-X. [DOI] [PubMed] [Google Scholar]
- 6.Rochaix JD. Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Plant Mol. Biol. 1996;32:327–341. doi: 10.1007/BF00039389. [DOI] [PubMed] [Google Scholar]
- 7.Sugita M, Sugiura M. Regulation of gene expression in chloroplasts of higher plants. Plant Mol. Biol. 1996;32:315–326. doi: 10.1007/BF00039388. [DOI] [PubMed] [Google Scholar]
- 8.Danon A. Translational regulation in the chloroplast. Plant Physiol. 1997;115:1293–1298. doi: 10.1104/pp.115.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stern DS, Higgs DC, Yang J. Transcription and translation in chloroplasts. Trends Plant Sci. 1997;2:308–315. doi: 10.1016/S1360-1385(97)89953-0. [DOI] [Google Scholar]
- 10.Tiller N, Bock R. The translational apparatus of plastids and its role in plant development. Mol. Plant. 2014;7:1105–1120. doi: 10.1093/mp/ssu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuda D, Mauro VP. Determinants of initiation codon selection during translation in mammalian cells. PLoS ONE. 2010;5:e15057. doi: 10.1371/journal.pone.0015057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belinky F, Rogozin IB, Koonin EV. Selection on start codons in prokaryotes and potential compensatory nucleotide substitutions. Sci. Rep. 2017;7:12422. doi: 10.1038/s41598-017-12619-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zitomer RS, Walthall DA, Rymond BC, Hollenberg CP. Saccharomyces cerevisiae ribosomes recognize non-AUG initiation codons. Mol. Cell. Biol. 1984;4:1191–1197. doi: 10.1128/mcb.4.7.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peabody DS. Translation initiation at an ACG triplet in mammalian cells. J. Biol. Chem. 1987;262:11847–11851. doi: 10.1016/S0021-9258(18)60891-9. [DOI] [PubMed] [Google Scholar]
- 15.Clements JM, Laz TM, Sherman F. Efficiency of translation initiation by non-AUG codons in Saccharomyces cerevisiae. Mol. Cell. Biol. 1988;8:4533–4536. doi: 10.1128/mcb.8.10.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hann SR, King MW, Bentley DL, Anderson CW, Eisenman RN. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt’s lymphomas. Cell. 1988;52:185–195. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- 17.Varshney U, RajBhandary UL. Initiation of protein synthesis from a termination codon. Proc. Natl. Acad. Sci. U. S. A. 1990;87:1586–1590. doi: 10.1073/pnas.87.4.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang K-J, Wang C-C. Translation initiation from a naturally occurring non-AUG codon in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:13778–13785. doi: 10.1074/jbc.M311269200. [DOI] [PubMed] [Google Scholar]
- 19.Tang H-L. Translation of a yeast mitochondrial tRNA synthetase initiated at redundant non-AUG codons. J. Biol. Chem. 2004;279:49656–49663. doi: 10.1074/jbc.M408081200. [DOI] [PubMed] [Google Scholar]
- 20.Kearse MG, Wilusz JE. Non-AUG translation: A new start for protein synthesis in eukaryotes. Genes Dev. 2017;31:1717–1731. doi: 10.1101/gad.305250.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Na CH. Discovery of noncanonical translation initiation sites through mass spectrometric analysis of protein N termini. Genome Res. 2018;28:25–36. doi: 10.1101/gr.226050.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirose T, Ideue T, Wakasugi T, Sugiura M. The chloroplast infA gene with a functional UUG initiation codon. FEBS Lett. 1999;445:169–172. doi: 10.1016/S0014-5793(99)00123-4. [DOI] [PubMed] [Google Scholar]
- 23.Kuroda H. Translation of psbC mRNAs starts from the downstream GUG, not the upstream AUG, and requires the extended Shine-Dalgarno sequence in tobacco chloroplasts. Plant Cell Physiol. 2007;48:1374–1378. doi: 10.1093/pcp/pcm097. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K. Evolutionarily conserved non-AUG translation initiation in NAT1/p97/DAP5 (EIF4G2) Genomics. 2005;85:360–371. doi: 10.1016/j.ygeno.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 25.Depeiges A, Degroote F, Espagnol MC, Picard G. Translation initiation by non-AUG codons in Arabidopsis thaliana transgenic plants. Plant Cell Rep. 2006;25:55–61. doi: 10.1007/s00299-005-0034-0. [DOI] [PubMed] [Google Scholar]
- 26.Hecht A. Measurements of translation initiation from all 64 codons in E. coli. Nucleic Acids Res. 2017;45:3615–3626. doi: 10.1093/nar/gkx070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuroda H, Maliga P. Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol. 2001;125:430–436. doi: 10.1104/pp.125.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staub JM, Maliga P. Expression of a chimeric uidA gene indicates that polycistronic mRNAs are efficiently translated in tobacco plastids. Plant J. 1995;7(5):845–848. doi: 10.1046/j.1365-313X.1995.07050845.x. [DOI] [PubMed] [Google Scholar]
- 29.Hayashi K. A role of the -35 element in the initiation of transcription at psbA promoter in tobacco plastids. Plant Cell Physiol. 2003;44:334–341. doi: 10.1093/pcp/pcg041. [DOI] [PubMed] [Google Scholar]
- 30.Reddy VS. Analysis of chloroplast transformed tobacco plants with cry1Ia5 under rice psbA transcriptional elements reveal high level expression of Bt toxin without imposing yield penalty and stable inheritance of transplastome. Mol. Breed. 2002;9:259–269. doi: 10.1023/A:1020357729437. [DOI] [Google Scholar]
- 31.Ruhlman T, Verma D, Samson N, Daniell H. The role of heterologous chloroplast sequence elements in transgene integration and expression. Plant Physiol. 2010;152(4):2088–2104. doi: 10.1104/pp.109.152017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simpson GG. Noncanonical translation initiation of the Arabidopsis flowering time and alternative polyadenylation regulator FCA. Plant Cell. 2010;22:3764–3777. doi: 10.1105/tpc.110.077990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X, Kindle K, Stern D. Initiation codon mutations in the Chlamydomonas chloroplast petD gene result in temperature-sensitive photosynthetic growth. EMBO J. 1993;12:3627–3635. doi: 10.1002/j.1460-2075.1993.tb06036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X, Kindle KL, Stern DB. The initiation codon determines the efficiency but not the site of translation initiation in Chlamydomonas chloroplasts. Plant Cell. 1995;7:1295–1305. doi: 10.1105/tpc.7.8.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiroshi K. Translation of psbC mRNAs starts from the downstream GUG, not the upstream AUG, and requires the extended shine-dalgarno sequence in tobacco chloroplasts. Plant Cell Physiol. 2007;48:1374–1378. doi: 10.1093/pcp/pcm097. [DOI] [PubMed] [Google Scholar]
- 36.Muralikrishna P, Wickstrom E. Inducible high expression of the Escherichia coli infC gene subcloned behind a bacteriophage T7 promoter. Gene. 1989;80(2):369–374. doi: 10.1016/0378-1119(89)90301-6. [DOI] [PubMed] [Google Scholar]
- 37.Stenstrom CM, Holmgren E, Isaksson LA. Cooperative effects by the initiation codon and its flanking regions on translation initiation. Gene. 2001;273:259–265. doi: 10.1016/S0378-1119(01)00584-4. [DOI] [PubMed] [Google Scholar]
- 38.Stenstrom CM, Isaksson LA. Influences on translation initiation and early elongation by the messenger RNA region flanking the initiation codon at the 3′ side. Gene. 2002;288:1–8. doi: 10.1016/S0378-1119(02)00501-2. [DOI] [PubMed] [Google Scholar]
- 39.Ivanov IP, Loughran G, Sachs MS, Atkins JF. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1) Proc. Natl. Acad. Sci. U. S. A. 2010;107:18056–18060. doi: 10.1073/pnas.1009269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christopher DA, Mullet JE. Separate photosensory pathways co-regulate blue light/ultraviolet-A- activated psbD-psbC transcription and light-induced D2 and CP43 degradation in barley (Hordeum vulgare) chloroplasts. Plant Physiol. 1994;104:1119–1129. doi: 10.1104/pp.104.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov, I. P. Translation initiation from conserved non-AUG codons provides additional layers of regulation and coding capacity. MBio8, (2017). [DOI] [PMC free article] [PubMed]
- 42.Gvozdjak, A. & Samanta, M. P. Genes preferring non-AUG start codons in bacteria. arXiv:2008.10758 [q-bio.GN] (2020).
- 43.Butler JS, Springer M, Grunberg-Manago M. AUU-to-AUG mutation in the initiator codon of the translation initiation factor IF3 abolishes translational autocontrol of its own gene (infC) in vivo. Proc. Natl. Acad. Sci. U. S. A. 1987;84(12):4022–4025. doi: 10.1073/pnas.84.12.4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brombach M, Pon CL. The unusual translational initiation codon AUU limits the expression of the infC (initiation factor IF3) gene of Escherichia coli. Mol. Gen. Genet. 1987;208:94–100. doi: 10.1007/BF00330428. [DOI] [PubMed] [Google Scholar]
- 45.Parsons GD, Donly BC, Mackie GA. Mutations in the leader sequence and initiation codon of the gene for ribosomal protein S20 (rpsT) affect both translational efficiency and autoregulation. J. Bacteriol. 1988;170:2485–2492. doi: 10.1128/jb.170.6.2485-2492.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haggerty TJ, Lovett ST. IF3-mediated suppression of a GUA initiation codon mutation in the recJ gene of Escherichia coli. J. Bacteriol. 1997;179:6705–6713. doi: 10.1128/jb.179.21.6705-6713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villegas A, Kropinski AM. An analysis of initiation codon utilization in the Domain Bacteria—Concerns about the quality of bacterial genome annotation. Microbiology. 2008;154:2559–2561. doi: 10.1099/mic.0.2008/021360-0. [DOI] [PubMed] [Google Scholar]
- 48.Cao X, Slavoff SA. Non-AUG start codons: Expanding and regulating the small and alternative ORFeome. Exp. Cell Res. 2020;391(1):111973. doi: 10.1016/j.yexcr.2020.111973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Curran J, Kolakofsky D. Ribosomal initiation from an ACG codon in the Sendai virus P/C mRNA. EMBO J. 1988;7:245–251. doi: 10.1002/j.1460-2075.1988.tb02806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dorn P, DaSilva L, Martarano L, Derse D. Equine infectious anemia virus tat: Insights into the structure, function, and evolution of lentivirus trans-activator proteins. J. Virol. 1990;64:1616–1624. doi: 10.1128/jvi.64.4.1616-1624.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwab SR, Shugart JA, Horng T, Malarkannan S, Shastri N. Unanticipated antigens: Translation initiation at CUG with leucine. PLoS Biol. 2004;2:e366. doi: 10.1371/journal.pbio.0020366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Medveczky P, Németh A, Gráf L, Szilágyi L. Methionine-independent translation initiation from naturally occurring non-AUG codons. Curr. Chem. Biol. 2007;1:129–139. [Google Scholar]
- 53.Bock R, Hagemann R, Kossel H, Kudla J. Tissue- and stage-specific modulation of RNA editing of the psbF and psbL transcript from spinach plastids—A new regulatory mechanism? Mol. Gen. Genet. 1993;240:238–244. doi: 10.1007/BF00277062. [DOI] [PubMed] [Google Scholar]
- 54.Maier RM. RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol. 1996;32:343–365. doi: 10.1007/BF00039390. [DOI] [PubMed] [Google Scholar]
- 55.Chateigner-Boutin AL, Small I. Plant RNA editing. RNA Biol. 2010;7:213–219. doi: 10.4161/rna.7.2.11343. [DOI] [PubMed] [Google Scholar]
- 56.Tsudzuki T, Wakasugi T, Sugiura M. Comparative analysis of RNA editing sites in higher plant chloroplasts. J. Mol. Evol. 2001;53:327–332. doi: 10.1007/s002390010222. [DOI] [PubMed] [Google Scholar]
- 57.Zoschke R, Bock R. Chloroplast translation: Structural and functional organization, operational control, and regulation. Plant Cell. 2018;30:745–770. doi: 10.1105/tpc.18.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sasaki J, Nakashima N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl. Acad. Sci. U. S. A. 2000;97(4):1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.RajBhandary UL. More surprises in translation: Initiation without the initiator tRNA. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1325–1327. doi: 10.1073/pnas.040579197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chattapadhyay R, Pelka H, Schulman LH. Initiation of in vivo protein synthesis with non-methionine amino acids. Biochemistry. 1990;29:4263–4268. doi: 10.1021/bi00470a001. [DOI] [PubMed] [Google Scholar]
- 61.Drabkin HJ, RajBhandary UL. Initiation of protein synthesis in mammalian cells with codons other than AUG and amino acids other than methionine. Mol. Cell. Biol. 1998;18:5140–5147. doi: 10.1128/MCB.18.9.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santos MA, Tuite MF. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 1995;23:1481–1486. doi: 10.1093/nar/23.9.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ivanov P, Firth AE, Michel AM, Atkins JF, Baranov PV. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. 2011;39:4220–4234. doi: 10.1093/nar/gkr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bonissone S, Gupta N, Romine M, Bradshaw RA, Pevzner PA. N-terminal protein processing: A comparative proteogenomic analysis. Mol. Cell. Proteomics. 2013;12:14–28. doi: 10.1074/mcp.M112.019075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. U. S. A. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 67.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 69.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 70.Kumar S, Kumar K, Pandey P, Rajamani V, Padmalatha KV, Dhandapani G, Kanakachari M, Leelavathi S, Kumar PA, Reddy VS. Glycoproteome of elongating cotton fiber cells. Mol. Cell Proteomics. 2013;12(12):3677–3689. doi: 10.1074/mcp.M113.030726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.