Abstract
Wound healing is a well-organized dynamic process involving coordinated consecutive phases: homeostasis, inflammation, proliferation and resolution. Fibroblasts play major roles in skin wound healing such as in wound contraction and release of growth factors which are of importance in angiogenesis and tissue remodeling. Abnormal fibroblast phenotypes have been identified in patients with chronic wounds. In this work, we analyzed scRNA-seq datasets of normal and wounded skin from mice at day 4 post-wound to investigate fibroblast heterogeneity during the proliferative phase of wound healing. Compositional analysis revealed a specific subset of fibroblast (cluster 3) that primarily increased in wounded skin (14%) compared to normal skin (3.9%). This subset was characterized by a gene signature marked by the plasma membrane proteins Sfrp2 + Sfrp4 + Sfrp1 + and the transcription factors Ebf1 + Prrx1 + Maged1 + . Differential gene expression and enrichment analysis identified epithelial to mesenchymal transition (EMT) and angiogenesis to be upregulated in the emerging subset of fibroblasts of the wounded skin. Using two other datasets for murine wounded skin confirmed the increase in cluster 3-like fibroblasts at days 2, 7 and 14 post-wounding with a peak at day 7. By performing a similarity check between the differential gene expression profile between wounded and normal skin for this emerging fibroblast subset with drug signature from the ConnectivityMap database, we identified drugs capable of mimicking the observed gene expression change in fibroblasts during wound healing. TTNPB, verteprofin and nicotinic acid were identified as candidate drugs capable of inducing fibroblast gene expression profile necessary for wound healing. On the other hand, methocarbamol, ifosfamide and penbutolol were recognized to antagonize the identified fibroblast differential expression profile during wound healing which might cause delay in wound healing. Taken together, analysis of murine transcriptomic skin wound healing datasets suggested a subset of fibroblasts capable of inducing EMT and further inferred drugs that might be tested as potential candidates to induce wound closure.
Subject terms: Computational biology and bioinformatics, Medical research
Introduction
The wound healing process consists of four overlapping phases: hemostasis, inflammation, proliferation and resolution1,2. Immediately after wounding, the hemostasis phase begins with formation of fibrin clots and vascular constriction. Next, migration of inflammatory cells into the wound starts. Recruitment of inflammatory cells such as neutrophils, macrophages and lymphocytes initiate the inflammatory phase of the wound healing process3. The proliferation phase overlaps with the inflammatory phase in which epithelial cells proliferate and migrate within the wound area4. During the proliferative phase, fibroblasts and endothelial cells are the most prominent cell types which support collagen formation, blood vessel growth and granulation tissue formation at the injury site4. After proliferation and extracellular matrix synthesis are completed, the healing process enters the final resolution phase in which tissue remodeling occurs4. During the resolution phase, the wound density returns to normal by pruning of many of the newly formed blood vessels5. During the wound healing process, activation of well-organized biological processes, such as epithelial to mesenchymal transition (EMT), mesenchymal to epithelial transition (MET) and angiogenesis, occurs6,7. During the EMT process, epithelial cells acquire mesenchymal characteristics, such as enhanced motility, change their interactions with ECM and lose their cell–cell junctions8–11. The EMT process, which is necessary for normal embryonic development, is accelerated during the wound healing process8,12. The EMT was also found to be accelerated by fibroblast growth factor 2 during wound healing13. Next, MET helps to reconstitute the apical junctional complex (AJCs) restoring the barrier function of the repaired skin. Additionally, angiogenesis increases during the healing process14. New capillary sprouts invade the wound clot followed by their organization into a network through the granulation tissue before their regression during the resolution phase of wound healing4.
Fibroblasts are diverse and dynamic in nature and play major role in wound healing15. As it relates to wound contraction, they are present in two distinct states: contractile and noncontractile16. Fibroblasts are activated and differentiated into myofibroblasts at the wound site to generate contractile forces to bring the wound edges together and facilitate wound closure16. The EMT process plays significant role in myofibroblast transition16,17. Fibroblasts also support the wound angiogenesis process by the production of appropriate extracellular matrix (ECM) bed18. Fibroblast malfunction has been reported to cause wound chronicity19. These awry fibroblasts show impaired pattern of cytokine release and display abnormal phenotypes with defective proliferation and early senescence20. Fibroblast-focused therapies for wound care are currently under development21,22. Growth factor-based therapies such as platelet derived growth factor B subunit B seek to stimulate fibroblasts activation and differentiation23,24. However, the focus on any single growth factor to heal wound has not been productive25. The complexity of the in vivo wound healing microenvironment requires intelligent approaches to modify fibroblast state as would be needed for wound closure.
Defining Fibroblast heterogeneity is an important and emerging area of research in wound healing. The application of single cell RNA sequencing (scRNA-seq) has established different states (e.g. contractile, regenerative, adaptive or fibrotic) of skin fibroblasts during development, homeostasis and injury in both human and murine models26–33. A recently published article provided a systematic view of skin cellular dynamics at day 4 during dermal wound healing in mice at the single cell level34. In the current study, we investigated the same dataset (GSE142471) to identify fibroblast states during wound healing and to change normal skin fibroblast states necessary for physiological wound healing. Next, we verified the presence of the identified states using other datasets (GSE153596, GSE178758 and GSE188432). Such understanding will inform therapies targeting the change of fibroblast state in non-healing wound. The primary outcomes of this study include: (1) identification of skin cell types states in normal skin and during wound healing, (2) identification of the fibroblast state emerging during the wound healing process, (3) identification of the differentially expressed genes, altered pathways and biological processes in fibroblasts during wound healing, and (4) identification of the drugs which could induce changes in fibroblast gene expression consistent with that required for physiological wound repair.
Results
Murine skin scRNA-seq data were retrieved from GEO database (accession no. 142471) from two groups: normal skin and day 4 wounded skin. Cells were clustered and cell types were identified for each of the identified 19 clusters. Clusters identified as fibroblasts were analyzed to identify the subset of fibroblasts having higher abundance during wound healing. Differential expression analysis and Gene Set Enrichment Analysis (GSEA) were performed on the identified subset of fibroblasts (cluster 3) which revealed upregulation of angiogenesis and EMT. The resulting DEGs were used to identify drugs which have potential to induce fibroblast state transition from the resting state found in cluster 3 in normal skin towards the wound healing state found in cluster 3 in wounded skin (Fig. 1a).
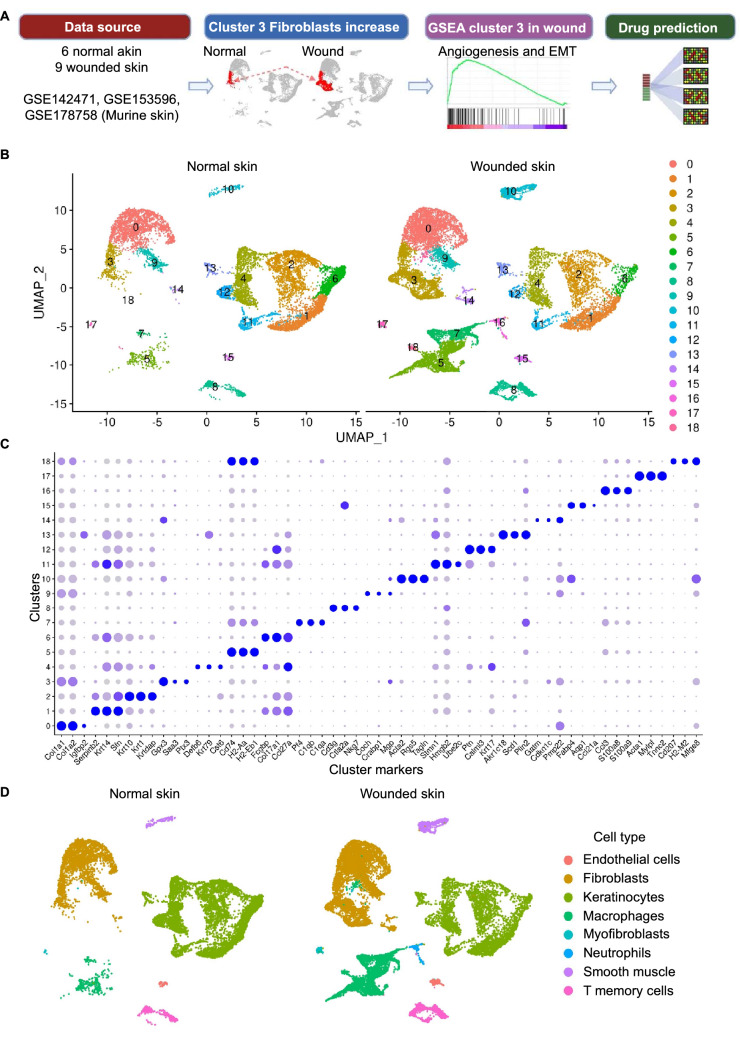
Figure 1.
Identification of 19 distinct clusters. (A) The analysis overview. (B) UMAP plot showing 19 distinct clusters. Each dot represents a single cell. Left panel represents normal skin cells and right panel represents wounded skin at day 4 post wounding (GSE14271). (C) Dotplot representing the top 3 markers in each cluster. (D) UMAP plots showing eight cell types for each condition (normal skin and wounded skin).
Identification of eight distinct cell types in murine skin
The initial data included 27,317 cells from 5 skin samples (2 normal skin samples and 3 skin samples at day 4 post wounding). After excluding low quality cells, 26,723 cells remained for the downstream analyses (Supplementary Fig. 1; Supplementary table 1). Clustering of single cells identified 19 distinct clusters (Fig. 1b,c). Cell type assignment identified eight cell types including keratinocytes (11,124 cells), fibroblasts (8,657 cells), macrophages (3,971 cells), T memory cells (1,214 cells), smooth muscle cells/ pericytes (868 cells), endothelial cells (319 cells), neutrophils (307 cells), myofibroblasts (263 cells) (Fig. 1c,d; Supplementary Fig. 2; Supplementary table 2).
Identification of wound healing-related genes and biological processes in the skin tissue
The transcriptional changes that occurred in skin at day 4 of wound healing were investigated by comparing genes from all cells in wounded skin and all cells from the normal skin samples (Supplementary table 3). Enrichment analysis using GSEA software identified affected pathways and biological processes in the wounded skin. The upregulated gene sets in skin during wound healing included granulocyte chemotaxis, neutrophil chemotaxis, IL6 Jak Stat3 signaling, IL18 signaling, myogenesis and EMT (Supplementary Fig. 3a and b; Supplementary table 4). Sixty genes were found to be upregulated with a log2FC > 0.25 involved in EMT. The top upregulated genes involved in epithelial mesenchymal transition included Cxcl1, Acta2, Thbs1, Timp1, Tpm1, Tpm2, Il6, Basp1 and Vim (Supplementary Fig. 3c). Genes involved in granulocytes chemotaxis in wounded skin included Cxcl2, Ccl4, Ccl3, Cxcl1, S100a9, Ccl2, S100a8, Cd74, Thbs1, Fcer1g, Ccl7, Ccl8, Rac2, Ccl11, Dpep1, Pde4b and Rarres2 with log2FC > 0.25 (Supplementary table 4). On the other hand, keratinization, cornification and keratinocyte differentiation were found to be downregulated in wounded skin (Supplementary table 4).
Identification of a subset of fibroblasts during wound healing with upregulated angiogenesis and EMT
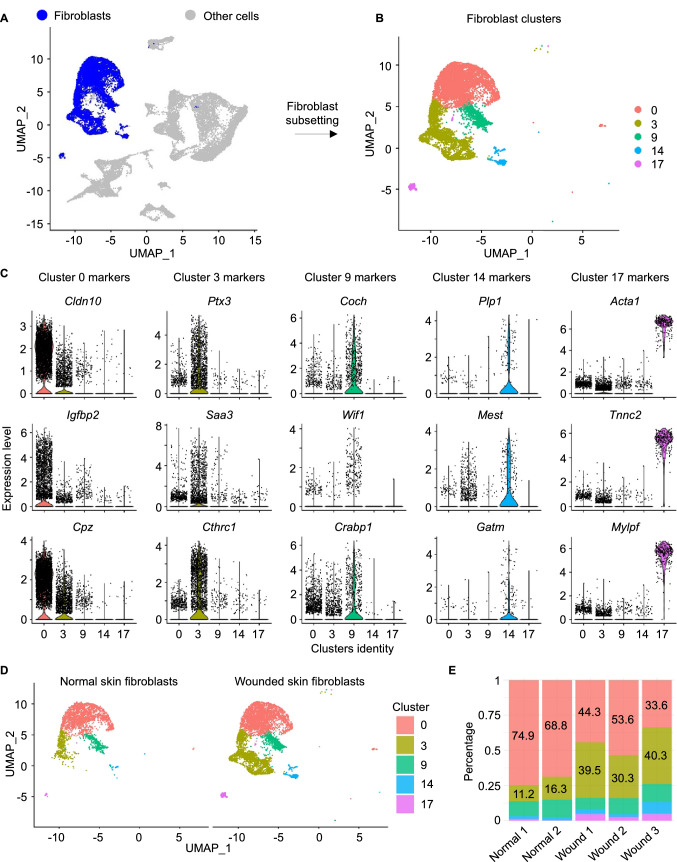
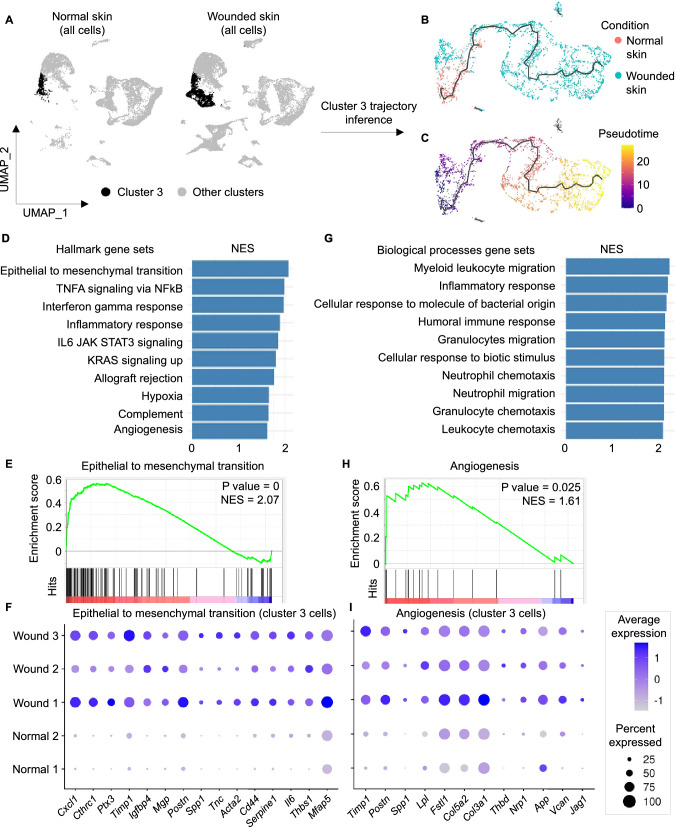
To identify the role of fibroblasts in the wound healing process, analysis of fibroblast clusters was performed. Cells identified as fibroblasts were found in five distinct states (clusters 0, 3, 9, 14 and 17) with specific upregulated markers for each (cluster 0: Igfbp2, Cpz and Cldn10; cluster 3: Saa3, Ptx3 and Cthrc1; cluster 9: Coch, Crabp1 and Wif1; cluster 14: Gatm, Plp1 and Mest; cluster 17: Acta1, Mylpf and Tnnc2) (Fig. 2a–c). Percentage of cells in clusters 0, 9 and 14 among all cell types was comparable between normal skin and wounded skin (cluster 0 cells: 18% in normal skin and 17.12% in wounded skin; cluster 9: 3.16% in normal skin and 3.98% in wounded skin; cluster 14: 0.53% in normal skin and 1.65% in wounded skin). On the other hand, cluster 3 had a higher percentage of cells in wounded skin relative to in normal skin, increasing from 3.93% to 14.09% (Supplementary table 1). To test if this increase in cluster 3 cells resulted from an increase in proliferation or differentiation, cell cycle analysis was performed using the function CellCycleScoring in Seurat to identify the ratio of dividing cells and non-dividing cells in cluster 3 from normal skin and wounded skin. The canonical marker genes for S, G2 and M phases were used to calculate cell cycle scores for each cell as reported previously35. Next, cells expressing markers for either S, G2 or M phases were assigned to proliferative cells, while cells not expressing mitotic marker genes were assigned to G1 phase. Analysis of cell cycle phase for cluster 3 cells identified that 54.3% and 55.15% of cells were assigned to G1 phase (non-proliferative cells) in normal skin and wounded skin respectively indicative of no proliferation differences. Also, myofibroblasts (cluster 17) increased from 0.1% to 1.56%. Fibroblasts in cluster 3 represent 11.2–16.3% of the fibroblast population in normal skin, while they represent 30.3–40.3% of the fibroblast population in wounded skin (Fig. 2d,e, Supplementary table 1). Trajectory inference of cluster 3 cells identified a trajectory capturing cluster 3 cells from both normal skin and wounded skin samples (Fig. 3a–c). Comparing genes between cluster 3 cells in wounded skin and normal skin resulted in identification of 1,650 DEGs with adjusted p value < 0.05. Among them, 506 genes were found to be upregulated and 282 genes were found to be downregulated using a cutoff log2FC ± 0.25. The resulting DEGs were classified based on their family into transcription factors, plasma membrane proteins, enzymes and other categories. The top upregulated plasma membrane proteins in cluster 3 in wounded skin included Sfrp2 and Sfrp4, while the top downregulated plasma membrane proteins included Il1r2 and Ghr (Supplementary Fig. 4). The transcription factors Nfkbia and Noct were upregulated, while Fosp, Ebf1, Klf4 and Aff3 were downregulated in wounded skin cluster 3. Regarding growth factors, Ptn and Mdk were upregulated, while Gas6 and Igf1 were downregulated in wounded skin cluster 3. All DEGs in cluster 3 of wounded skin samples are provided in supplementary table 5. Enrichment analysis using GSEA software resulted in identification of altered biological processes and pathways in cluster 3 in wounded skin compared to cluster 3 in normal skin (p value < 0.05 and FDR q-val < 0.1) (Fig. 3d; Supplementary table 6). The upregulated gene sets included epithelial mesenchymal transition (Fig. 3e,f), granulocyte chemotaxis (Fig. 3g) and angiogenesis (Fig. 3h,i). On the other hand, epidermal development, keratinocyte differentiation and oxidative phosphorylation were found to be downregulated in cluster 3 fibroblasts during wound healing (Supplementary table 6). To investigate the contribution of other fibroblast clusters (0,9,14 and 17), in this paradigm, we compared inter-cluster transcriptome profiles (Supplementary Fig. 5a and b). A total of 1,235 genes were found to be differentially expressed in fibroblast clusters when comparing each cluster from wounded skin with the corresponding cluster in normal skin with log2FC ± 0.25 and adjusted p value < 0.05. Among these, 255 genes were upregulated, and 105 genes were downregulated in more than one fibroblast cluster (Supplementary table 7). Other genes were differentially expressed in a cluster-specific basis (483 upregulated genes and 323 genes). A total of 69 genes had bidirectional behavior (upregulated and downregulated in distinct fibroblast clusters). Upregulated genes in wounded skin fibroblast clusters compared to normal skin that have role in EMT included 16 genes in cluster 0, 63 genes in cluster 3, 35 genes in cluster 9, 20 genes in cluster 14 and no genes in cluster 17 (Supplementary Fig. 6).
Figure 2.
Identification of distinct subsets of fibroblasts. (A) UMAP plot showing cells identified as fibroblasts or myofibroblasts in blue color and other cells in grey color. (B) Fibroblast clusters. (C) Top upregulated 3 genes in fibroblast clusters 0, 3, 9, 14 and 17. (D) UMAP plots showing cells identified as fibroblasts splitted by condition (normal skin: left panel and wounded skin: right panel). (E) Barplot representing percentage of each fibroblast cluster among the fibroblast population within each sample.
Figure 3.
Upregulation of EMT and angiogenesis in cluster 3 fibroblasts at day 4 post-wound in mice skin. (A) UMAP plot representing cluster 3 in a black color and other clusters in s grey color. (B, C) UMAP plots of cluster 3 cells colored by condition and pseudotime receptively. The black line represents the inferred trajectory. (D, G) Top 10 uppregulated hallmark and biological processes gene sets in cluster 3 in wounded skin compared to normal skin. Bars represent the normalized enrichment score (NES) of the gene set enrichment analysis. (E, H) GSEA enrichment plot for EMT and angiogenesis. Hits (black lines) represent the intersection between the DEGs and the enriched gene sets. (F, I) Dotplots representing relative expression level of the top upregulated genes having role in EMT and angiogenesis respectively with log2FC > 0.25 in cluster 3 during wound healing compared to cluster 3 in normal skin. Dot size represents percentage of cells within cluster 3 expressing the gene in each sample. Dot color represents the relative expression level of the gene.
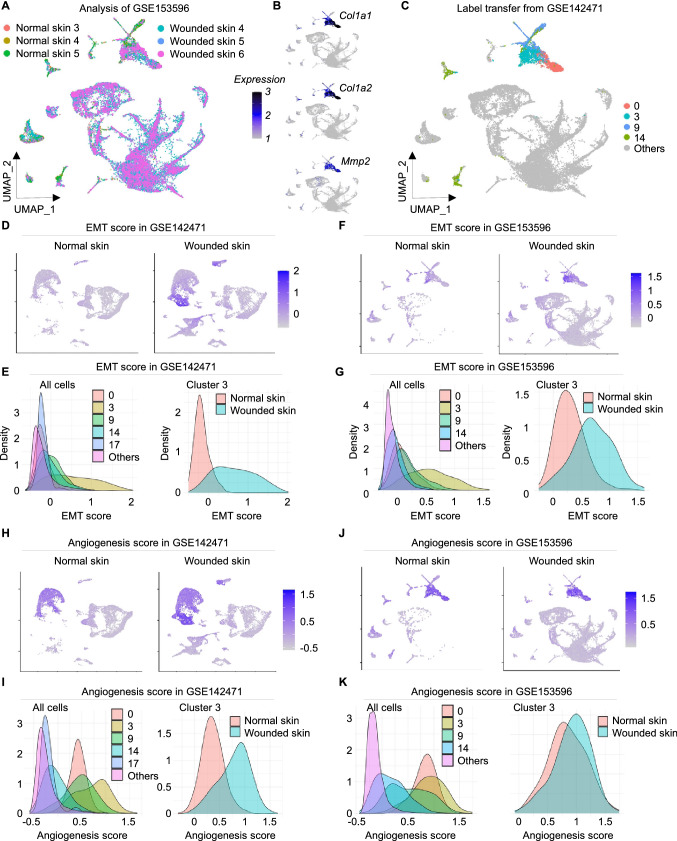
Validation of cluster 3-like cells in other datasets
Since our goal was to identify a fibroblast population that uniquely represented the ability to support EMT during wound healing, we focused on cluster 3 in our subsequent study. The dataset with accession number GSE153596 included 34,859 cells from 6 samples (3 normal skin and 3 wounded skin) at day 7 post-wound was thus retrieved and investigated for presence and quantification of cluster 3 cells. After excluding low quality cells 34,191 cells remained for downstream analysis among which 5,669 cells were labeled as fibroblasts similar to clusters 0, 3, 9 and 14 from the original dataset with high expression of Col1a1, Col1a2 and Mmp2 (Fig. 4a–c). Cells labeled as cluster 3 have higher EMT scores in both GSE153596 and GSE142471 compared to other fibroblast clusters and other cell types. Additionally, cluster 3 cells in wounded skin have higher EMT scores in wounded skin at day 4 and day 7 compared to normal skin (Fig. 4d–g). Angiogenesis scores were found to be higher in clusters 0, 3 and 9 in both GSE153596 and GSE142471 datasets (Fig. 4h–k). Next, to quantitate the abundance of cluster 3 fibroblasts at different days post-wounding, another dataset with accession number GSE178758 was investigated. This dataset included 19,968 cells from 4 samples at different time points post-wounding (days 0, 2,7 and 14). Cluster 3-like cells increased from 5.7% in normal skin to 27.7%, 40,8% and 26.6% in days 2, 7 and 14 respectively (Supplementary Fig. 7a, b and c). In conclusion, combining all three datasets it became clear that cluster 3 significantly increased post wounding in murine skin (p = 0.0003), while cluster 0 showed no significant alteration as compared to normal skin (p value = 0.99). Furthermore, the expression of EMT genes in these cluster 3-like cells remain low in the wound of aged mice (88 week old) as compared to young mice (7 week old) (adjusted p value < 0.05, supplementary Fig. 8).
Figure 4.
Upregulation of EMT and angiogenesis in cluster 3 fibroblasts at day 4 post-wound in mice skin. (A) UMAP representing cells from 3 normal skin and 3 wounded skin samples at day 7 post-wound retrieved from GSE153596. (B) Expression level of Col1a1, Col1a2 and Mmp2. (C) UMAP plot representing cells from GSE153596 labeled using mutual nearest neighbor and label transfer (see methods) from GSE142471. (D) EMT score in cells from GSE142471. (E) Left panel represents EMT score in fibroblast clusters and other cell types. Right panel represents EMT score in cluster 3-like cells from GSE142471 in normal and wounded skin. (F) EMT score in cells from GSE153596. (G) Left panel represents EMT score in fibroblast clusters and other cell types. Right panel represents EMT score in cluster 3-like cells from GSE153596 in normal and wounded skin. (H-I) Angiogenesis score in cells from GSE142471 in all cells and in cluster 3 fibroblasts. (J-K) Angiogenesis score in cells from GSE153596 in all cells and in cluster 3 fibroblasts.
Identification of drugs affecting the wound healing process
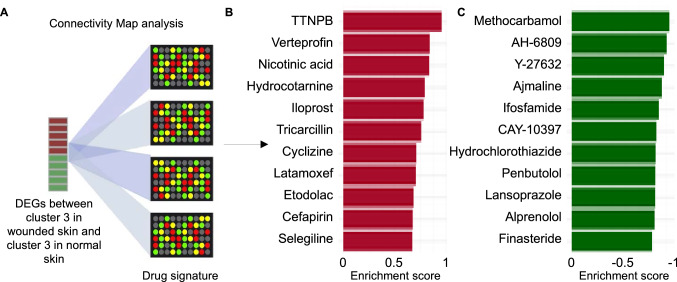
Prediction of drugs using Connectivity Map resulted in identification of 21 drugs with positive and 40 drugs with negative enrichment scores (P value < 0.05). Scores were based on the similarity between the effect of the drug on gene expression profile from the Connectivity Map database36 (drug signature) and the observed alteration in gene expression level in cluster 3 cells in wounded skin compared to cluster 3 in normal skin. The top 3 drugs with positive scores included TTNPB, verteporfin, and nicotinic acid (predicted to be capable of inducing fibroblast state change from the normal skin state towards the identified state in cluster 3 cells during wound healing). On the other hand, the top 3 drugs with negative scores included methocarbamol, AH-6809 and Y-27632 (predicted to be capable of inducing fibroblast state transition towards the resting state found in cluster 3 in normal skin) (Fig. 5a–c; Supplementary table 8).
Figure 5.
Identification of drugs mimicking or counteracting the differential expression pattern in cluster 3 during wound healing. (A) The similarity between the DEGs between cluster 3 cells in wounded skin and cluster 3 cells in normal skin with log2FC ± 0.25 were checked with the signature of each of the 6,100 drug/ compound instances in the Connectivity Map database. (B) The top identified drugs having signature that mimics the differential expression pattern in cluster 3 in wounded skin (positive enrichment). (C) The top identified drugs having signature that counteracts the differential expression pattern in cluster 3 in wounded skin (negative enrichment).
Methods
Data retrieval
scRNA-seq data was extracted from the gene expression omnibus (GEO) database ( GSE142471, GSE153596 and GSE178758). Those datasets were selected as they included murine small wound 2-6 mm at 2, 4, 7 and 14 post wound days. Besides, the mice in the included data have no diseases and received no treatments representing wound healing under normal physiological conditions. For the wound healing experiments in GSE142471, the authors of the original study used 5 mice skin samples where they performed scRNA-seq experiments using 7 week-old female K14-Cre; ROSAmTmG mice (C57BL/6 J background)34. They generated a full-thickness wound in the back skin of 3 mice using a 6-mm punch. Four days later, they excised the wound and the surrounding unwounded skin regions (1.5 cm in diameter) using a 10-mm punch. For the control group (2 mice), they shaved the mice, removed the back skin, and scrapped off the fat. Finally, they minced the skin from both groups into pieces of less than 1 mm in diameter where they were used for library generation using the Chromium Single Cell 3′ Reagents Kits v2. HiSeq 4000 platform was utilized to sequence samples and Cell Ranger 2.1.0. was used to map the resulting FASTQ reads to mm10 genome.
Single-cell RNA sequencing preprocessing
Seurat package (4.0.4) in R (v.4.1.1) was used for all the analyses except mentioned37–41. Firstly, the initial data in GSE142471, including a total of 27,317 cells from the 5 samples (2 normal skin samples and 3 skin samples during wound healing) was log normalized using 10,000 as a scaling factor for the total number of detected molecules per cell. Filtration criteria was determined to exclude low quality cells. Cells with less than 200 expressed genes, a total number of detected molecules less than 500 were excluded or more than 30,000 were excluded and cells with more than 10% of their genes originating from the mitochondrial genome were excluded. All downstream analyses were performed using the remaining 26,723 cells. After removing low quality cells, the top 2,000 genes exhibiting high cell-to-cell variation were identified for each sample. Next, highly variable genes were ranked based on their identification as variable genes in multiple samples and were used for integration of the 5 samples using canonical correlation analysis (CCA). To reduce the data dimensions, we performed a principal component analysis (PCA) and the top 25 principal components were chosen for dimensionality reduction and visualization using the UMAP algorithm.
Clustering of single cells and cell types assignment
Clustering of single cells was performed using a graph-based clustering approach. Next, the Louvain algorithm was applied to group cells together with different resolutions and a resolution of 0.25, which represents distinct states among each cell type in this dataset, was selected for downstream analysis. Cluster markers were identified by comparing genes from each of the resulting 19 clusters with the rest of the cells combined using the Wilcoxon Rank Sum test. For a gene to be considered for comparison, it has to be expressed in at least 20% of the cells of either the cluster or in 20% of the rest of the cells. Cell type assignment was performed using the PanglaoDB database using the top 10 upregulated genes in each cluster42. Cell type annotation was performed automatically using the PanglaoDB database, while manual annotation was performed for clusters which was not automatically assigned.
Differential expression analysis
To identify the effect of the wound healing process on gene expression profile of skin tissue, gene expression values were compared between all cells from the wounded skin and all cells from the normal skin. Wilcoxon Rank Sum Test was performed to identify the differentially expressed genes (DEGs) and for a gene to be considered for the comparison, it has to be expressed in at least 20% of cells in either of the 2 groups. Additionally, genes were compared between cluster 3 cells in wounded skin and normal skin, which were identified to have a higher percentage of cells in wounded skin samples compared to normal skin. Identified DEGs from all analyses were annotated using ingenuity pathways analysis (IPA) software43–45. In addition, 4 other comparisons were performed to compare between other fibroblasts subsets from wounded skin and normal skin in clusters 0, 9, 14 and 17. The resulting DEGs for all fibroblasts comparisons were classified into genes differentially expressed in one fibroblast subset, genes having similar change in multiple subsets and genes with bidirectional change (upregulated and downregulated in distinct fibroblast clusters).
Gene set enrichment analysis
The resulting DEGs from comparing all cells in wounded skin with all cells in normal skin and between cluster 3 cells in wounded skin with normal skin with adjusted p value < 0.05 were considered for enrichment analysis. Enrichment was performed using the GSEA software using the default parameters46,47. Enrichment was performed against hallmark gene sets and gene ontology biological processes gene sets. The genes identified to be involved in EMT from this analysis were checked for their differential expression in other fibroblast clusters 0, 9, 14 and 17 (Supplementary Fig. 6).
Trajectory inference for cluster 3 cells
Fibroblasts identified in cluster 3 from normal skin and wounded skin were used to infer a trajectory linking between them. Monocle3 package (v 1.0.0) in R was used to infer a trajectory based on the continuous change in gene expression profiles in cluster 3 cells48,49. PCA analysis was performed and the first 10 dimensions were used to reduce the data dimensions.
Cell cycle analysis
To test if the increase in cluster 3 cells resulted from an increase in proliferation or differentiation, cell cycle analysis was performed using the function CellCycleScoring in Seurat to identify the ratio of dividing cells and non-dividing cells in cluster 3 cells from normal skin and wounded skin. The canonical marker genes for S, G2 and M phases were used to calculate cell cycle scores for each cell35. Next, cells expressing markers for either S, G2 or M phases were assigned to proliferative cells, while cells not expressing mitotic marker genes were assigned to G1 phase.
Analysis of wound healing at day 7 post-wound
To verify the presence of cluster 3-like cells in wounded skin from other datasets, the dataset with GEO accession number GSE153596 was retrieved including 3 samples of normal skin and 3 samples of 2 mm wounded skin at day 7 post-wound in 21-day old mice27. Cells with more than 200 detected genes, between 500 and 30,000 total number of counts and less than 10% mitochondrial DNA expression were kept for downstream analysis. All samples were integrated using CCA and log normalization was performed with a scaling factor of 10,000. Next, PCA was performed to reduce data dimensions and UMAP was used for visualization. The functions FindTransferAnchors and Transfer Data in Seurat were used to map the cluster labels from the original dataset (GSE142471) into the new samples based on mutual nearest neighbors. The function AddModuleScore in Seurat was used to calculate scores for EMT and angiogenesis using the identified upregulated genes in cluster 3 in wounded skin.
Identification of cluster 3-like cells over time during wound healing
To further identify the dynamics of cluster 3 cells, we retrieved a spatial transcriptomic dataset (GSE178758) of wound healing at different time points (normal skin, day 2, 7 and 14 post wound)29. The authors performed splinted excisional 6 mm diameter wound in mice skin. Cells with more than 200 detected genes, between 500 and 30,000 total number of counts and less than 10% mitochondrial DNA expression were kept for downstream analysis. All samples were integrated using CCA and log normalization was performed with a scaling factor of 10,000. Then, PCA was performed to reduce data dimensions and UMAP was used for visualization. SingleR and celldex packages in R were used to identify fibroblasts based on Spearman correlation across marker genes between each cell and the reference samples50. Cell type assignment was computed for each cell against the reference labels including sorted 18 main cell types51. Cluster labels from GSE142471 were used to identify cells similar to cluster 3 cells using the functions FindTransferAnchors and TransferData in Seurat.
Analysis of wound healing in young and aged mice
To identify the effect of aging in cluster 3-like cells proportion and the accompanied EMT activation, 2 combined count matrices were retrieved from GSE188432 representing young and aged skin samples52. The data included normal skin and wounded skin from 7 young and 3 aged mice skin samples. The authors generated a full-thickness wound using 6-mm punch in 7-week-old and 88-week-old C57BL/6 J mice. Cells with more than 200 detected genes, between 500 and 30,000 total number of counts and less than 20% mitochondrial DNA expression were kept for downstream analysis (46,713 cells). All samples were integrated using CCA and log normalization was performed with a scaling factor of 10,000. Then, PCA was performed to reduce data dimensions and UMAP was used for visualization. Cluster labels from GSE142471 were used to identify cells similar to cluster 3 cells using the functions FindTransferAnchors and TransferData in Seurat. The function AddModuleScore in Seurat was used to calculate scores for EMT in young and aged skin samples. Additionally, FindMarkers function in Seurat was used to compare the EMT-related genes in cluster 3-like cells between aged and young mice.
Predictions of drugs for fibroblast shate change
To identify which drugs or compounds have an effect on gene expression profile mimicking that observed change in cluster 3 fibroblast in wounded skin compared to normal skin, a similarity check was performed using the Connectivity map software36. Connectivity map is a large public database of drugs and gene signatures which represent the effect of drugs on gene expression profile. The basic concept of Connectivity map is to compare between drug-specific gene expression profiles as references with a specific gene signature by submitting a list of upregulated and downregulated genes relevant to a particular biological condition. The connectivity map database includes 6,100 microarray experiments of 1,309 drugs on different cell lines such as MCF7, HL60 and PC3 and varying concentrations and time points against vehicle controls. The fold change of treatment to control was calculated and sorted into decreasing order and converted to a ranked list. DEGs with adjusted p value < 0.05 and log2FC ± 0.25 resulting from comparing genes from cluster 3 in wounded skin and normal skin were annotated with HT_HG-U133A IDs as a requirement for Connectivity Map enrichment. Next, the annotated genes were submitted as a signature query in Connectivity Map software to perform the enrichment. The query was compared to the ranked vector based on the fold changes of the probesets. The algorithm gives a connectivity score either positive or negative for each instance using a nonparametric rank-ordered Kolmogorov–Smirnov (KS) test. Next, the resulting connectivity scores were normalized using random permutation, so values close to + 1 reflects closeness or similarity between the submitted query and the effect of the drug on the gene expression profile using the ranked list. A score close to -1 emphasizes an inverse similarity between the submitted query and the reference profile. The algorithm detected the similarity between drug signatures from the database and the differential expression pattern in cluster 3 fibroblasts to unravel unexpected connections between them.
Discussion
Understanding the dynamic cell biology of wound healing process in vivo in light of scRNA-seq data will provide critical insight into the molecular basis of wound closure. Fibroblasts have numerous roles in skin wound healing including deposition of new ECM of murine skin53. We analyzed scRNA-seq data of mice skin in normal state and during wound healing with the objective to identify and characterize the subset of fibroblasts playing a major role in the wound healing process. To be able to identify drugs that could induce fibroblasts to enable wound healing, genes from the emerging subset of fibroblasts in wound versus those in normal skin were compared followed by a similarity check between the resulting differential gene expression pattern and the drug effects on gene expression profile.
Compositional analysis identified a distinct percentage of cell types in normal and wounded skin. The observed early upregulation of neutrophil chemotaxis is consistent with the reported sequel of events wherein early recruitment of neutrophils is followed by monocytes which differentiate into wound macrophage54,55. Fibroblasts were identified in different states based on distinct gene expression profiles. This is consistent with previous reports that dermal fibroblasts include processes relevant to wound healing54,56,57. Among the identified fibroblast subsets, the increase in cluster 3 percentage from 3.93% in normal skin to 14.09% among all cells in wounded skin hints towards their potential role in the wound healing process. The top upregulated plasma membrane proteins found in wounded skin cluster 3 fibroblasts were Sfrp2, Sfrp4 and Sfrp1. Sfrp2 has been found to be essential for myocardial tissue survival and repair by enhancing engraftment of mesenchymal stem cells and formation of granulation tissue58,59. On the other hand, downregulation of Gpc4 (Glypican 4) induces cell migration and proliferation in breast cancer cell lines60. Upregulation of EMT process in cluster 3 fibroblasts during wound healing is consistent with previous reports13,61. During the EMT process, epithelial cells acquire mesenchymal features such as enhanced motility, change their interaction with ECM and undergo cytoskeletal rearrangement8–11. Cxcl1, Cthrc1, Ptx3 genes support EMT and were upregulated in cluster 3 wound fibroblasts. Cxcl1 promote EMT in cutaneous wound healing by mediating formation of new vessels62,63. Cthrc1 improves wound healing via regulating NOTCH and TGF-β pathways and recruiting M2 macrophages64. The deficiency of Ptx3 has pathological consequences such as epithelial hyperplasia, excessive accumulation of collagen and defective mature tissue formation65. Data analysis presented in this work identified multiple genes involved in angiogenesis to be upregulated in cluster 3 fibroblasts in wounded skin including Fstl1 and Nrp1. Fstl1 (Follistatin Like 1) is a known angiogenic factor that promotes ischemic tissue revascularization and endothelial cell function66. Nrp1 (Neuropilin 1) is critically important to drive angiogenesis67. Anti-Nrp1 treatment exhibited a significant reduction in the wound vascular density (67% decrease) during dermal wound healing67. Investigation of cluster 3 wound fibroblasts suggest their involvement in EMT and angiogenesis during the wound healing process. EMT upregulation was found to be cluster 3-specific, while angiogenesis score was found to be higher in clusters 0, 3 and 9 indicating that multiple subsets of fibroblasts are involved in the angiogenesis process. Cluster 3 cells were found to have a similar proportion of proliferative and non-proliferative cells in both normal skin and wounded skin indicating that the observed increase in cluster 3 cells resulted from cell differentiation. This indicates that some other mechanisms rather than proliferation might be involved in the rise of cluster 3 cells during wound healing. Trajectory inference also explained such differentiation of cluster 3 fibroblasts in normal skin (resting state) into cluster 3 fibroblasts in the wound healing state. These observations lead to the hypothesis that cluster 3 cells present in uninjured skin differentiate to the observed state during wound healing activating EMT.
Wound healing outcomes may be viewed as a product of the acquisition of appropriate cell state and fate by participating cell types. Such state and fate of any single cell depends on their gene expression profiles. In this work the differential gene profile of cluster 3 fibroblast can be considered as the signature profile requirement for physiological wound healing. Identification of drugs with the ability to induce pro-healing signature gene expression profile will help populate a drug candidate list for preclinical and clinical validation studies. The result of the similarity check compared to the signature of cluster 3 wound fibroblasts revealed a group of drugs with positive enrichment scores (pro-healing) and another group with negative enrichment scores (anti-healing). Anti-healing drugs will interfere with the healing process and should be useful to induce wound chronicity for experimental processes. Among the pro-healing drugs, TTNPB, an analog of retinoic acid, activates retinoic acid receptors potently and selectively68. TTNPB in combination with other small molecules has been reported to enable chemical reprogramming of mice embryonic fibroblasts to pluripotent stem cells69. Verteporfin induces skin regeneration without scar formation through Yap (Yes-associated protein) inhibition70. Additionally, verteporfin was found to inhibit fibrogenic genes and was suggested to be a novel antifibrotic agent71. The third drug identified to have a positive enrichment score was nicotinic acid which represents the active form of vitamin-B3. Nicotinic acid was reported to have anti-inflammatory and antioxidant effects72–74. Furthermore, nicotinic acid topical administration was found to improve tissue regeneration in excisional skin wounds through the increment of fibroblast proliferation, collagen synthesis, and vascularization75,76. Findings of this study provide a list of candidate drugs that should be preclinically and clinically tested for their effects on improving wound closure. In conclusion, findings of this work at transcriptome level underscore the diversity and dynamicity of wound-site fibroblasts and their potential responsiveness to drugs that may induce their change of state favoring wound closure. However, the results are solely based on analysis of transcriptomic datasets and further mechanistic studies are needed to support its inferences.
Supplementary Information
Acknowledgements
This study is based on a Master of Science thesis by F.Z.G. and supervised by H.M.E.A. at the American University in Cairo. This work was supported in part by grants from the American University in Cairo to F.Z.G. and H.M.E.A. This work was also supported by the US National Institutes of Health grant DK128845 and in part by Department of Defense (DoD) grants W81XWH-21-1-0033 to C.K.S., and DoD grant W81XWH-22-1-0146 to K.S. Research programs led by C.K.S. were supported by the Lilly Endowment INCITE (Indiana Collaborative Initiative for Talent Enrichment) program and Templeton foundation grant (61742).
Author contributions
F.Z.G. and A.S.A. contributed equally to this work. K.S. and C.K.S., with contributions from all other authors, wrote the manuscript. C.K.S. and H.M.E.A. guided and supervised the project. F.Z.G., A.S.A., Y.A.M., A.A.E., A.A., K.S., C.K.S. and H.M.E.A. reviewed and approved the final manuscript.
Data availability
The datasets are publicly available in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) repository (GSE142471, GSE153596, GSE178758 and GSE188432).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fatma Z. Gharbia and Ahmed S. Abouhashem.
Contributor Information
Chandan K. Sen, Email: cksen@iu.edu
Hassan M. E. Azzazy, Email: hazzazy@aucegypt.edu
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-27152-4.
References
- 1.Gosain A, DiPietro LA. Aging and wound healing. World J. Surg. 2004;28:321–326. doi: 10.1007/s00268-003-7397-6. [DOI] [PubMed] [Google Scholar]
- 2.Singh K, Singh K. Carcinogenesis and diabetic wound healing: evidences of parallelism. Curr. Diabetes Rev. 2015;11:32–45. doi: 10.2174/1573399811666150109122205. [DOI] [PubMed] [Google Scholar]
- 3.Broughton G, 2nd, Janis JE, Attinger CE. The basic science of wound healing. Plast. Reconstr. Surg. 2006;117:12S–34S. doi: 10.1097/01.prs.0000225430.42531.c2. [DOI] [PubMed] [Google Scholar]
- 4.Campos AC, Groth AK, Branco AB. Assessment and nutritional aspects of wound healing. Curr. Opin. Clin. Nutr. Metab. Care. 2008;11:281–288. doi: 10.1097/MCO.0b013e3282fbd35a. [DOI] [PubMed] [Google Scholar]
- 5.Singh K, et al. Epigenetic modification of MicroRNA-200b contributes to diabetic vasculopathy. Mol. Ther. 2017;25:2689–2704. doi: 10.1016/j.ymthe.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sen CK, Roy S. The hyperglycemia stranglehold stifles cutaneous epithelialmesenchymal plasticity and functional wound closure. J. Invest. Dermatol. 2021;141:1382–1385. doi: 10.1016/j.jid.2020.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh K, et al. Cutaneous epithelial to mesenchymal transition activator ZEB1 regulates wound angiogenesis and closure in a glycemic status-dependent manner. Diabetes. 2019;68:2175–2190. doi: 10.2337/db19-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 9.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim J, Thiery JP. Epithelial-mesenchymal transitions: insights from development. Development. 2012;139:3471–3486. doi: 10.1242/dev.071209. [DOI] [PubMed] [Google Scholar]
- 11.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Haensel D, Dai X. Epithelial-to-mesenchymal transition in cutaneous wound healing: Where we are and where we are heading. Dev. Dyn. 2018;247:473–480. doi: 10.1002/dvdy.24561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koike Y, Yozaki M, Utani A, Murota H. Fibroblast growth factor 2 accelerates the epithelial-mesenchymal transition in keratinocytes during wound healing process. Sci. Rep. 2020;10:18545. doi: 10.1038/s41598-020-75584-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tonnesen MG, Feng X, Clark RA. Angiogenesis in wound healing. J. Investig. Dermatol. Symp. Proc. 2000;5:40–46. doi: 10.1046/j.1087-0024.2000.00014.x. [DOI] [PubMed] [Google Scholar]
- 15.Muhl L, et al. Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat. Commun. 2020;11:3953. doi: 10.1038/s41467-020-17740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat. Rev. Mol. Cell Biol. 2002;3:349–363. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Liu Y. Dissection of key events in tubular epithelial to myofibroblast transition and its implications in renal interstitial fibrosis. Am. J. Pathol. 2001;159:1465–1475. doi: 10.1016/S0002-9440(10)62533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayrand D, et al. Angiogenic properties of myofibroblasts isolated from normal human skin wounds. Angiogenesis. 2012;15:199–212. doi: 10.1007/s10456-012-9253-5. [DOI] [PubMed] [Google Scholar]
- 19.Wall IB, et al. Fibroblast dysfunction is a key factor in the non-healing of chronic venous leg ulcers. J. Invest. Dermatol. 2008;128:2526–2540. doi: 10.1038/jid.2008.114. [DOI] [PubMed] [Google Scholar]
- 20.Cook H, Davies KJ, Harding KG, Thomas DW. Defective extracellular matrix reorganization by chronic wound fibroblasts is associated with alterations in TIMP-1, TIMP-2, and MMP-2 activity. J. Invest. Dermatol. 2000;115:225–233. doi: 10.1046/j.1523-1747.2000.00044.x. [DOI] [PubMed] [Google Scholar]
- 21.Domaszewska-Szostek A, Krzyzanowska M, Siemionow M. Cell-based therapies for chronic wounds tested in clinical studies: Review. Ann. Plast. Surg. 2019;83:e96–e109. doi: 10.1097/SAP.0000000000001947. [DOI] [PubMed] [Google Scholar]
- 22.desJardins-Park, H. E., Foster, D. S., & Longaker, M. T. Fibroblasts and wound healing: an update. Regen. Med.13, 491–495, 10.2217/rme-2018-0073 (2018). [DOI] [PubMed]
- 23.Barrientos S, Brem H, Stojadinovic O, Tomic-Canic M. Clinical application of growth factors and cytokines in wound healing. Wound Repair Regen. 2014;22:569–578. doi: 10.1111/wrr.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.So K, et al. Avotermin for scar improvement following scar revision surgery: a randomized, double-blind, within-patient, placebo-controlled, phase II clinical trial. Plast. Reconstr. Surg. 2011;128:163–172. doi: 10.1097/PRS.0b013e318217429b. [DOI] [PubMed] [Google Scholar]
- 25.Frykberg RG, Banks J. Challenges in the treatment of chronic wounds. Adv. Wound Care (New Rochelle) 2015;4:560–582. doi: 10.1089/wound.2015.0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerrero-Juarez CF, Dedhia PH, Jin S, et al. Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat. Commun. 2019;10(1):650. doi: 10.1038/s41467-018-08247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phan QM, Fine GM, Salz L, et al. Left expression in fibroblasts maintains developmental potential in adult skin to regenerate wounds. Elife. 2020;9:e60066. doi: 10.7554/eLife.60066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phan QM, Sinha S, Biernaskie J, Driskell RR. Single-cell transcriptomic analysis of small and large wounds reveals the distinct spatial organization of regenerative fibroblasts. Exp. Dermatol. 2021;30(1):92–101. doi: 10.1111/exd.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foster DS, Januszyk M, Yost KE, et al. Integrated spatial multiomics reveals fibroblast fate during tissue repair. Proc. Natl. Acad. Sci. USA. 2021;118(41):e2110025118. doi: 10.1073/pnas.2110025118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gay D, Ghinatti G, Guerrero-Juarez CF, et al. Phagocytosis of Wnt inhibitor SFRP4 by late wound macrophages drives chronic Wnt activity for fibrotic skin healing. Sci. Adv. 2020;6(12):eaay3704. doi: 10.1126/sciadv.aay3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen CJ, Kajita H, Takaya K, et al. Single-Cell RNA-seq analysis reveals cellular functional heterogeneity in dermis between fibrotic and regenerative wound healing fates. Front. Immunol. 2022;13:875407. doi: 10.3389/fimmu.2022.875407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rustagi Y, Abouhashem AS, Verma P, et al. Endothelial phospholipase Cγ2 improves outcomes of diabetic ischemic limb rescue following VEGF therapy. Diabetes. 2022;71(5):1149–1165. doi: 10.2337/db21-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh K, Rustagi Y, Abouhashem AS, et al. Genome-wide DNA hypermethylation opposes healing in patients with chronic wounds by impairing epithelial-mesenchymal transition. J. Clin. Invest. 2022;132(17):157279. doi: 10.1172/JCI157279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haensel D, et al. Defining epidermal basal cell states during skin homeostasis and wound healing using single-cell transcriptomics. Cell Rep. 2020;30:3932–3947. doi: 10.1016/j.celrep.2020.02.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nestorowa, S., Hamey, F.K., & Pijuan Sala, B., et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 128(8): e20–e31. 10.1182/blood-2016-05-716480 (2016) [DOI] [PMC free article] [PubMed]
- 36.Lamb J, et al. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 37.Hao Y, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abouhashem AS, Singh K, Azzazy HME, Sen CK. Is low alveolar Type II cell SOD3 in the lungs of elderly linked to the observed severity of COVID-19? Antioxid Redox Signal. 2020;33:59–65. doi: 10.1089/ars.2020.8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gordillo GM, Guda PR, Singh K, Biswas A, Abouhashem AS, Rustagi Y, Sen A, Kumar M, Das A, Ghatak S, Khanna S, Sen CK, Roy S. Tissue nanotransfection causes tumor regression by its effect on nanovesicle cargo that alters microenvironmental macrophage state. Mol. Ther. 2022 doi: 10.1016/j.ymthe.2022.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franzen, O., Gan, L. M., & Bjorkegren, J. L. M. PanglaoDB: A web server for exploration of mouse and human single-cell RNA sequencing data. Database Oxford10.1093/database/baz046 (2019). [DOI] [PMC free article] [PubMed]
- 43.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghosh N, et al. Urolithin A augments angiogenic pathways in skeletal muscle by bolstering NAD(+) and SIRT1. Sci. Rep. 2020;10:20184. doi: 10.1038/s41598-020-76564-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wisler JR, et al. Proteomic pathway analysis of monocyte-derived exosomes during surgical sepsis identifies immunoregulatory functions. Surg. Infect. (Larchmt) 2020;21:101–111. doi: 10.1089/sur.2019.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 48.Cao J, et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–667. doi: 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aran D, Looney AP, Liu L, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20(2):163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benayoun BA, Pollina EA, Singh PP, et al. Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res. 2019;29(4):697–709. doi: 10.1101/gr.240093.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vu R, Jin S, Sun P, et al. Wound healing in aged skin exhibits systems-level alterations in cellular composition and cell-cell communication. Cell Rep. 2022;40(5):111155. doi: 10.1016/j.celrep.2022.111155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bainbridge P. Wound healing and the role of fibroblasts. J. Wound Care. 2013;22(407–408):410–412. doi: 10.12968/jowc.2013.22.8.407. [DOI] [PubMed] [Google Scholar]
- 54.Sinha M, et al. Direct conversion of injury-site myeloid cells to fibroblast-like cells of granulation tissue. Nat. Commun. 2018;9:936. doi: 10.1038/s41467-018-03208-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sindrilaru A, Scharffetter-Kochanek K. Disclosure of the culprits: Macrophages-versatile regulators of wound healing. Adv. Wound Care (New Rochelle) 2013;2:357–368. doi: 10.1089/wound.2012.0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng CC, et al. Single-cell RNA-seq reveals fibroblast heterogeneity and increased mesenchymal fibroblasts in human fibrotic skin diseases. Nat. Commun. 2021;12:3709. doi: 10.1038/s41467-021-24110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guerrero-Juarez CF, et al. Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat. Commun. 2019;10:650. doi: 10.1038/s41467-018-08247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alfaro MP, et al. The Wnt modulator sFRP2 enhances mesenchymal stem cell engraftment, granulation tissue formation and myocardial repair. Proc. Natl. Acad. Sci. USA. 2008;105:18366–18371. doi: 10.1073/pnas.0803437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mirotsou M, et al. Secreted frizzled related protein 2 (Sfrp2) is the key Akt-mesenchymal stem cell-released paracrine factor mediating myocardial survival and repair. Proc. Natl. Acad. Sci. USA. 2007;104:1643–1648. doi: 10.1073/pnas.0610024104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munir J, Van Ngu T, Na Ayudthaya PD, Ryu S. Downregulation of glypican-4 facilitates breast cancer progression by inducing cell migration and proliferation. Biochem. Biophys. Res. Commun. 2020;526:91–97. doi: 10.1016/j.bbrc.2020.03.064. [DOI] [PubMed] [Google Scholar]
- 61.Yan C, et al. Epithelial to mesenchymal transition in human skin wound healing is induced by tumor necrosis factor-alpha through bone morphogenic protein-2. Am. J. Pathol. 2010;176:2247–2258. doi: 10.2353/ajpath.2010.090048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Y, et al. CXCL1-LCN2 paracrine axis promotes progression of prostate cancer via the Src activation and epithelial-mesenchymal transition. Cell Commun. Signal. 2019;17:118. doi: 10.1186/s12964-019-0434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ding J, Tredget EE. The role of chemokines in fibrotic wound healing. Adv. Wound Care (New Rochelle) 2015;4:673–686. doi: 10.1089/wound.2014.0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qin S, et al. CTHRC1 promotes wound repair by increasing M2 macrophages via regulating the TGF-beta and notch pathways. Biomed. Pharmacother. 2019;113:108594. doi: 10.1016/j.biopha.2019.01.055. [DOI] [PubMed] [Google Scholar]
- 65.Rua R, McGavern DB. Pentraxin 3 innately preps damaged tissue for wound healing. J. Exp. Med. 2015;212:829. doi: 10.1084/jem.2126insight2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ouchi N, et al. Follistatin-like 1, a secreted muscle protein, promotes endothelial cell function and revascularization in ischemic tissue through a nitric-oxide synthase-dependent mechanism. J. Biol. Chem. 2008;283:32802–32811. doi: 10.1074/jbc.M803440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matthies AM, Low QE, Lingen MW, DiPietro LA. Neuropilin-1 participates in wound angiogenesis. Am. J. Pathol. 2002;160:289–296. doi: 10.1016/S0002-9440(10)64372-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beard RL, et al. Synthesis and structure-activity relationships of stilbene retinoid analogs substituted with heteroaromatic carboxylic acids. J. Med. Chem. 1995;38:2820–2829. doi: 10.1021/jm00015a004. [DOI] [PubMed] [Google Scholar]
- 69.Kim Y, Jeong J, Choi D. Small-molecule-mediated reprogramming: a silver lining for regenerative medicine. Exp. Mol. Med. 2020;52:213–226. doi: 10.1038/s12276-020-0383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang D, Rinkevich Y. Converting fibroblastic fates leads to wound healing without scar. Signal Transduct. Target Ther. 2021;6:332. doi: 10.1038/s41392-021-00738-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi-Wen X, et al. Verteporfin inhibits the persistent fibrotic phenotype of lesional scleroderma dermal fibroblasts. J. Cell. Commun. Signal. 2021;15:71–80. doi: 10.1007/s12079-020-00596-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Digby JE, et al. Anti-inflammatory effects of nicotinic acid in human monocytes are mediated by GPR109A dependent mechanisms. Arterioscler. Thromb. Vasc. Biol. 2012;32:669–676. doi: 10.1161/ATVBAHA.111.241836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lappas M, Permezel M. The anti-inflammatory and antioxidative effects of nicotinamide, a vitamin B(3) derivative, are elicited by FoxO3 in human gestational tissues: Implications for preterm birth. J. Nutr. Biochem. 2011;22:1195–1201. doi: 10.1016/j.jnutbio.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 74.Kamat JP, Devasagayam TP. Nicotinamide (vitamin B3) as an effective antioxidant against oxidative damage in rat brain mitochondria. Redox Rep. 1999;4:179–184. doi: 10.1179/135100099101534882. [DOI] [PubMed] [Google Scholar]
- 75.Ashkani Esfahani S, et al. Topical nicotinamide improves tissue regeneration in excisional full-thickness skin wounds: A stereological and pathological study. Trauma Mon. 2015;20:18193. doi: 10.5812/traumamon.18193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rink C, et al. Transcriptome of the subcutaneous adipose tissue in response to oral supplementation of type 2 Leprdb obese diabetic mice with niacin-bound chromium. Physiol. Genom. 2006;27:370–379. doi: 10.1152/physiolgenomics.00071.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets are publicly available in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) repository (GSE142471, GSE153596, GSE178758 and GSE188432).