Abstract
Background: Recently, increasing evidence has implicated methylenetetrahydrofolate reductase (MTHFR) gene mutation as a risk factor for ischemic stroke (IS) in the general population. However, studies have been inconclusive and lack evidence on specific populations. We aim to determine whether the rs1801133 (NC_000001.11 (MTHFR):g. 677C>T (p.Ala222Val) variant, we termed as MTHFR rs1801133 (677 C>T), is linked to an increased risk of IS in different age groups and ancestry groups.
Methods: The literature relevant to our study was found by searching the PubMed, Cochrane Library, Web of Science, EMBASE, and CNKI databases. A random effect model analysis was used to calculate the pooled odds ratio (OR) and 95% confidence interval (CI) to evaluate any possible association. We conducted a subgroup analysis based on the age and ancestry groups of the included populations.
Results: As of March 2022, 1,925 citations had been identified in electronic databases, of which 96 studies involving 34,814 subjects met our eligibility criteria. A strong link was found between IS and the MTHFR gene rs1801133 (677C>T) polymorphism in all genetic models [dominant genetic model (OR = 1.47; 95%CI = 1.33–1.61; p < 0.001), recessive genetic model (OR = 1.52; 95%CI = 1.36–1.71; p < 0.001), heterozygous model (OR = 1.36; 95%CI = 1.24–1.48; p < 0.001), homozygous model (OR = 1.82; 95%CI = 1.58–2.11; p < 0.001), and T allelic genetic model (OR = 1.37; 95%CI = 1.27–1.48; p < 0.001)]. Further subgroup analyses indicated that the MTHFR rs1801133 (677C>T) variant may increase the risk of IS in Asian, Hispanic, or Latin population, middle-aged, and elderly populations (p < 0.001).
Conclusion: Our results implied that mutation of the T allele of MTHFR rs1801133 (677C>T) could be a risk factor for IS. A significant association was found among Asian, Hispanic, or Latin population, middle-aged, and elderly people.
Keywords: polymorphism, ischemic stroke, meta-analysis, risk, MTHFR rs1801133 (677C>T)
1 Introduction
Ischemic stroke (IS) is an acute neurological deficit caused by vascular occlusion. It is one of the leading causes of death and disability worldwide (Francis et al., 2007; Malik and Dichgans, 2018; Phipps and Cronin, 2020) and is caused by a combination of environmental and genetic factors (Black et al., 2015; Prabhakaran et al., 2015; Malik et al., 2019). Several pathophysiological mechanisms are involved in the development of this condition. Hyperhomocysteinemia is reported to be independently associated with the risk of stroke (Linnebank et al., 2012). The 5,10-methylenetetrahydrofolate reductase (MTHFR) locus is mapped to chromosome 1 (1p36.3) which encodes for the dimeric proteins of 70–77 kDa subunits (Goyette et al., 1994). Folate metabolism is largely controlled by MTHFR, which catalyzes the conversion of 5,10-methylenetetrahydrofolate to 5-methylenetetrahydrofolate. 5-Methylenetetrahydrofolate provides a methyl group in the methylation reaction that transforms homocysteine into methionine (Figure 1), as well as the DNA methylation process (Brattström et al., 1998). Thus, the MTHFR enzyme activity is important for homeostasis of the serum homocysteine level.
FIGURE 1.
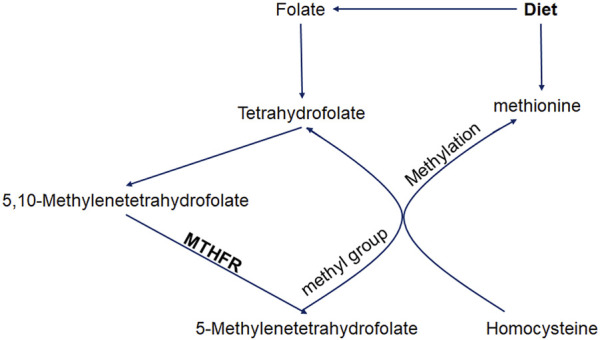
Roles of methylenetetrahydrofolate reductase in homocysteine metabolism.
The previous study had demonstrated that approximately 40% of the intragenic coding CpG islands were hyper-methylated, which had a higher C>T mutation rate. Also, the amino acid sequence of a protein and individual phenotypes could be changed by C>T substitutions at the CpG contexts in the protein-coding regions (Youk et al., 2020). The rs1801133 variant (NC_000001.11 (MTHFR):g. 677C>T (p.Ala222Val), also named MTHFR rs1801133 (677C>T), is a common mutant in MTHFR. The replacement of C with T at nucleotide 677 results in converting alanine to valine amino acid residue in the enzyme (Sharp and Little, 2004). Missense mutations cause a 50%–60% decrease in enzyme activity in patients who have the homozygous variant (TT) (Rozen, 199 7), which contributes to hyperhomocysteinemia (Castro et al., 2004). In addition, reduction of the MTHFR enzymatic activity would cause deficiency of folate, which is also an independent risk factor of IS (Qin et al., 2020). Moreover, when folic acid o is inadequate, the removal of homocysteine would be affected, leading to hyperhomocysteinemia and forming a vicious cycle (Liew and Gupta, 2015). Thus, it is important to determine the association between MTHFR rs1801133 (677C>T) polymorphism and the risk of IS for primary and secondary prevention of IS.
Many researchers have examined the relationship between MTHFR rs1801133 (677C>T) polymorphism and IS risk. However, there have been no definitive conclusions because different populations were examined and inconsistent results were obtained (Herak et al., 2017; Jiménez-González et al., 2021; Huang et al., 2022). Two meta-analyses were performed separately in 2016 and 2019 that reported a correlation between MTHFR rs1801133 (677C>T) polymorphism and IS (Song et al., 2016; Chang et al., 2019). However, only 22 studies were included in Song et al. (2016). Since then, many studies have been conducted in different populations. Moreover, the meta-analysis by Song et al. focused on the general population and did not consider whether MTHFR rs1801133 (677C>T) polymorphism might have varying effects on the risk of IS in different populations. Furthermore, Guilin Chang et al.‘s study, published in 2019, included only nine studies on the elderly population and did not consider young and middle-aged IS patients (Chang et al., 2019). Therefore, past meta-analyses were updated to investigate whether MTHFR rs1801133 (677C>T) polymorphism and stroke risk are related across age and ancestry groups in this study.
2 Materials and methods
The study follows the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines (Moher et al., 2009).
2.1 Literature search
A systematic search of the PubMed, EMBASE, Cochrane Library, Web of Science, and CNKI databases for relevant observational studies published until 15 March, 2022, was undertaken independently by two reviewers (Zhao and Li). We used the following search terms to identify eligible studies: (“methylenetetrahydrofolate reductase” OR “MTHFR” OR “C677T” OR “rs1801133”), (“ischemic stroke” OR “cerebral infarction” OR “stroke”), AND (“single nucleotide polymorphism” OR “SNP” OR “genetic polymorphism” OR “mutation” OR “variation”). We reviewed the full text of each study when abstracts and titles were insufficient to make a final determination regarding study inclusion. The reference lists of included studies and existing reviews were screened to identify additional eligible studies. Any disagreements in the study selection process were resolved by a third person (Dang).
2.2 Selection criteria
We included the studies according to the following inclusion criteria: 1) the full text could be searched in electronic databases; 2) the studies were case-control or cohort studies examining MTHFR rs1801133 (677C>T) and stroke susceptibility; 3) the study population was limited to patients diagnosed with stroke for the first time; 4) the MTHFR rs1801133 (677C>T) genotype frequency was provided; and 5) articles were published in English or Chinese. The main exclusion criteria included the following: 1) the studies were duplicate articles or non-original research (letters, commentaries, editorials, reviews, and meta-analyses); 2) the studies were case reports or involved animal experiments; 3) the genotype frequency of MTHFR rs1801133 (677C>T) was not provided; and 4) the p-value of the Hardy–Weinberg equilibrium (HWE) test was <0.05.
2.3 Data extraction and quality assessment
A pre-designed extraction form was used to extract the data. The extracted data included the name of the first author, type of stroke, publication date, ancestry groups, sample size (case and control), study design, mean or median age of the population, HWE, and the Newcastle–Ottawa scale score. Disagreements regarding data extraction were resolved by discussions among the two investigators (Zhao and Li), and a third reviewer (Dang) was consulted if necessary. The HWE p-value was also calculated using the genotypic frequencies of MTHFR polymorphisms, and the threshold of HWE deviation was set at 0.05. The quality of eligible publications was assessed using the Newcastle–Ottawa scale (Stang, 2010), and studies with a score of 7–9 were considered to be of good quality.
2.4 Statistical analysis
We used five genetic comparison models, the T allelic model (T vs. C), dominant model (TT + TC vs. CC), recessive model (TT vs. CC + TC), heterozygous model (TC vs. CC), and homozygous model (TT vs. CC), to estimate the relationship between MTHFR rs1801133 (677C>T) polymorphism and stroke susceptibility by calculating the odds ratio (OR) and 95% confidence interval (CI). The I2 statistic was used to evaluate heterogeneity among genetic comparison models, and the pooled OR was estimated via the Mantel-Haenszel random effect model. Heterogeneity between studies is indicated by an I2 > 50%. Moreover, the Bonferroni method was utilized to adjust for multiple comparisons to control the false positive error rate. As we performed multiple comparisons in this meta-analysis for 45 times, the p-value which was less than 0.05/50 (0.001) indicated statistical significance after Bonferroni correction. To determine the possible causes of heterogeneity, the ancestry groups (Asian, European, African, Hispanic, or Latin American (HLA), and other and not reported ancestries (ONR)) and the study population (young: <18 years; middle-aged: 18–60 years; elderly: >60 years) were analyzed in subgroups. In addition, we examined the impact of a single study on the pooled OR by performing sensitivity analyses on different genetic comparison models. Egger’s test, Begg’s test, and funnel plots were used to evaluate the potential publication bias in our study (Peters et al., 2006). Stata 17.0 was used to perform the statistical analysis of all genetic comparison models.
3 Results
3.1 Literature search and characteristics of the included studies
There were 1,925 articles initially identified after searching the databases. Among them, 692 articles were removed because of duplication, and 911 articles were removed after the titles and abstracts were screened. In total, 322 articles were further screened for eligibility by reviewing their full texts, and eventually, 96 articles that met the criteria were selected (Figure 2).
FIGURE 2.
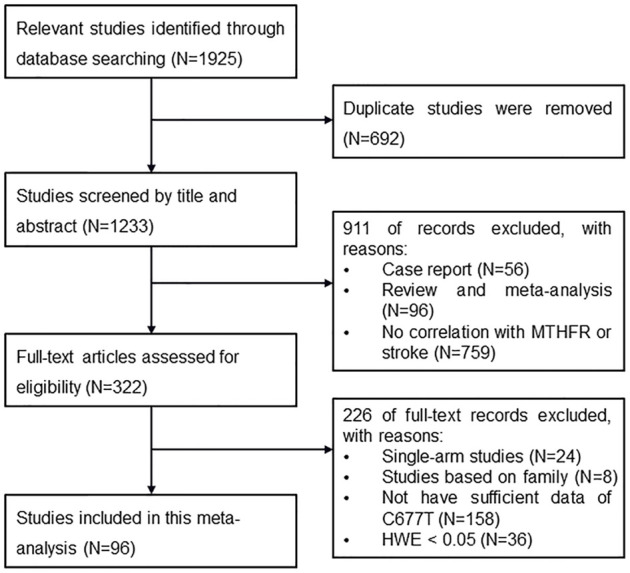
Flow chart of identification of eligible studies.
Of the 96 studies included, 95 were case-control studies, and one was a nested case-referent study. In total, 52 studies were conducted in the Asian population, 19 were in the European population, 1 was in the African population, 3 were in the Hispanic or Latin American population, and 21 were in other and not reported ancestry population. In total, 14 studies examined children, 27 examined middle-aged people, and 42 examined the elderly population. The other 13 studies examined the general population. The main features of the included studies are shown in Table 1.
TABLE 1.
Characteristics of studies included in the meta-analysis.
| Author | Year | Disease | Ancestry | Case | Control | Age | Design | HWE | NOS | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | CC | CT | TT | ||||||||
| Huang et al., (2022) | 2022 | IS | Asian | 94 | 72 | 32 | 101 | 62 | 5 | >18 | Case-control | 0.21 | 8 |
| Salomi et al., (2021) | 2021 | IS | Asian | 69 | 32 | 4 | 171 | 40 | 4 | 32.7±7.7 | Case-control | 0.36 | 8 |
| Cernera et al., (2021) | 2021 | IS | European | 64 | 140 | 76 | 138 | 206 | 85 | 8 | Case-control | 0.61 | 7 |
| Jiménez-González et al., (2021) | 2021 | IS | HLA | 35 | 105 | 38 | 60 | 83 | 40 | 34.1±5.4 | Case-control | 0.27 | 8 |
| Mazdeh et al., (2021) | 2021 | IS | ONR | 157 | 124 | 37 | 219 | 155 | 26 | 48±0.10 | Case-control | 0.84 | 7 |
| Hou et al., (2018) | 2018 | IS | Asian | 1063 | 793 | 138 | 1427 | 988 | 150 | 68.6±12.1 | Case-control | 0.22 | 9 |
| Mao and Han, (2018) | 2018 | IS | Asian | 73 | 189 | 27 | 82 | 100 | 16 | 68.42±13.26 | Case-control | 0.06 | 8 |
| Li et al., (2017) | 2017 | IS | Asian | 71 | 134 | 95 | 106 | 110 | 45 | 64.2±13.2 | Case-control | 0.08 | 8 |
| Kamberi et al., (2016) | 2017 | IS | European | 15 | 21 | 3 | 27 | 55 | 20 | 2.83–12.59 | Case-control | 0.40 | 8 |
| Ma et al., (2017) | 2017 | IS | Asian | 36 | 106 | 94 | 88 | 183 | 119 | Media 66.0 | Case-control | 0.27 | 7 |
| Lu et al., (2017) | 2017 | IS | Asian | 31 | 97 | 89 | 41 | 109 | 73 | NA | Case-control | 0.8 | 7 |
| Jiang et al., (2017) | 2017 | IS | Asian | 36 | 52 | 18 | 40 | 54 | 12 | 57.8±10.7 | Case-control | 0.33 | 8 |
| Kumar et al., (2016) | 2016 | IS | Asian | 161 | 84 | 5 | 183 | 65 | 2 | 52.83±12.5 | Case-control | 0.14 | 7 |
| Vijayan et al., (2016) | 2016 | IS | Asian | 164 | 35 | 1 | 185 | 8 | 0 | 57.74±13.84 | Case-control | 0.77 | 8 |
| Zhang et al., (2016) | 2016 | IS | Asian | 13 | 27 | 10 | 36 | 18 | 5 | NA | Case-Control | 0.23 | 8 |
| Herak et al., (2017) | 2017 | IS | European | 29 | 34 | 10 | 41 | 51 | 8 | 4.3(0.01–16.7) | Case-control | 0.15 | 7 |
| Ranellou et al., (2015) | 2015 | IS | European | 16 | 28 | 7 | 26 | 36 | 8 | 37.3±8.0 | Case-control | 0.40 | 8 |
| Wei et al., (2015) | 2015 | IS | Asian | 177 | 98 | 26 | 226 | 65 | 6 | 52.6±68.8 | Case-control | 0.60 | 8 |
| Luo et al., (2015) | 2015 | IS | Asian | 55 | 269 | 388 | 52 | 299 | 423 | 65.2±13.9 | Case-control | 0.93 | 8 |
| Lv et al., (2015) | 2015 | IS | Asian | 70 | 98 | 31 | 88 | 116 | 37 | 68.78 ± 10.63 | Case-control | 0.90 | 8 |
| Zhou et al., (2014) | 2014 | IS | Asian | 160 | 270 | 112 | 242 | 308 | 104 | Media 66 | Case-control | 0.72 | 8 |
| Atadzhanov et al., (2013) | 2014 | IS | African | 15 | 8 | 0 | 96 | 20 | 0 | 54 ± 16 | Case-control | 0.31 | 8 |
| Shi, (2014) | 2014 | IS | Asian | 18 | 36 | 31 | 33 | 35 | 14 | 77.89 ± 8.85 | Case-control | 0.38 | 8 |
| Supanc et al., (2014) | 2014 | IS | European | 41 | 78 | 36 | 75 | 59 | 16 | Middle-aged | Case-control | 0.40 | 8 |
| Fekih-Mrissa et al., (2013) | 2013 | IS | ONR | 35 | 43 | 6 | 60 | 35 | 5 | 56.06 ± 12.5 | Case-control | 0.97 | 8 |
| Zhang and Guo, (2012) | 2012 | IS | Asian | 9 | 19 | 12 | 10 | 20 | 10 | 39.25 ± 4.08 | Case-control | 1.00 | 8 |
| Djordjevic et al., (2012) | 2012 | IS | ONR | 24 | 50 | 6 | 33 | 54 | 13 | 6.7 ± 4.9 | Case-control | 0.21 | 8 |
| Djordjevic et al., (2012) | 2012 | IS | ONR | 39 | 23 | 11 | 59 | 47 | 14 | 40.3 ± 11.6 | Case-control | 0.33 | 88 |
| Somarajan et al., (2011) | 2011 | IS | Asian | 137 | 65 | 5 | 129 | 54 | 5 | 54 ± 15.9 | Case-control | 0.82 | 8 |
| Mohamed et al., (2011) | 2011 | IS | Asian | 69 | 60 | 21 | 85 | 48 | 9 | 61.0 ± 10.1 | Case-control | 0.53 | 8 |
| Hultdin et al., (2011) | 2011 | IS | ONR | 172 | 114 | 27 | 401 | 306 | 59 | 55.0 ± 8.0 | Nested | 0.95 | 8 |
| Salem-Berrabah et al., (2010) | 2011 | IS | ONR | 33 | 15 | 2 | 57 | 35 | 5 | Mean 57.62 | Case-control | 0.90 | 7 |
| Isordia-Salas et al., (2010) | 2010 | IS | HLA | 35 | 105 | 38 | 60 | 83 | 40 | 33.1 ± 5.8 | Case-control | 0.27 | 8 |
| Giusti et al., (2010) | 2010 | IS | European | 130 | 240 | 131 | 572 | 529 | 110 | Media 44.0 | Case-control | 0.437 | 7 |
| Tatarskyy et al., (2010) | 2010 | IS | ONR | 73 | 87 | 23 | 50 | 45 | 5 | 64.6 ± 9.1 | Case-control | 0.20 | 8 |
| Al-Allawi et al., (2009) | 2009 | IS | ONR | 26 | 30 | 14 | 27 | 20 | 3 | Media 60 | Case-control | 0.78 | 7 |
| Sun et al., (2009) | 2009 | IS | Asian | 45 | 36 | 38 | 49 | 37 | 10 | >60 | Case-control | 0.45 | 7 |
| Sabino et al., (2009) | 2009 | IS | HLA | 12 | 9 | 0 | 24 | 12 | 1 | 33.3 ± 16.3 | Case-control | 0.73 | 7 |
| Biswas et al., (2009) | 2009 | IS | Asian | 24 | 32 | 2 | 48 | 10 | 0 | <15 | Case-control | 0.47 | 7 |
| Biswas et al., (2009)) | 2009 | IS | Asian | 67 | 49 | 4 | 90 | 30 | 0 | Young | Case-control | 0.12 | 8 |
| (Morita et al., 2009) | 2009 | IS | European | 5 | 6 | 4 | 48 | 37 | 5 | Children | Case-control | 0.53 | 7 |
| Herak et al., (2009) | 2009 | IS | European | 11 | 17 | 5 | 46 | 56 | 10 | Children | Case-control | 0.22 | 7 |
| Goracy et al., (2009) | 2009 | IS | European | 65 | 69 | 18 | 83 | 46 | 6 | 66.7 ± 12.5 | Case-control | 0.91 | 8 |
| Zak et al., (2009) | 2009 | IS | European | 25 | 30 | 9 | 32 | 25 | 2 | Children | Case-control | 0.27 | 8 |
| Djordjevic et al., (2009) | 2009 | IS | ONR | 9 | 16 | 1 | 23 | 22 | 5 | Children | Case-control | 0.94 | 8 |
| Almawi et al., (2009) | 2009 | IS | ONR | 54 | 26 | 38 | 90 | 26 | 4 | NA | Case-control | 0.23 | 7 |
| Sawula et al., (2009) | 2009 | IS | European | 70 | 50 | 8 | 35 | 19 | 5 | 18–101 | Case-control | 0.31 | 8 |
| Yue et al., (2010) | 2009 | IS | Asian | 8 | 19 | 35 | 14 | 11 | 5 | 67 ± 9 | Case-control | 0.29 | 8 |
| Celiker et al., (2009) | 2009 | IS | ONR | 158 | 4 | 0 | 63 | 37 | 6 | Mean 69.8 | Case-control | 0.85 | 8 |
| Shi et al., (2008) | 2008 | IS | Asian | 23 | 45 | 29 | 20 | 45 | 34 | Young | Case-control | 0.07 | 8 |
| Moe et al., (2008) | 2008 | IS | Asian | 73 | 36 | 11 | 136 | 68 | 3 | 62.5 ± 1.1 | Case-control | 0.09 | 8 |
| Gao et al., (2008) | 2008 | IS | Asian | 4 | 20 | 18 | 6 | 11 | 13 | ≤45 | Case-control | 0.22 | 8 |
| Zhang et al., (2008) | 2008 | IS | Asian | 49 | 116 | 80 | 74 | 140 | 68 | 63.7 ± 10.4 | Case-control | 0.91 | 8 |
| Berge et al., (2007) | 2007 | IS | European | 29 | 125 | 182 | 25 | 141 | 163 | Old | Case-control | 0.47 | 7 |
| Nan et al., (2007) | 2007 | IS | Asian | 14 | 55 | 31 | 28 | 53 | 19 | <45 | case-control | 0.49 | 8 |
| Kim et al., (2007) | 2007 | IS | Asian | 81 | 113 | 43 | 66 | 119 | 38 | 61.18 ± 11.09 | Case-control | 0.21 | 8 |
| Komitopoulou et al., (2006) | 2006 | IS | European | 36 | 46 | 8 | 46 | 39 | 18 | 5.55 ± 0.48 | Case-control | 0.06 | 8 |
| Sazci et al., (2006) | 2006 | IS | ONR | 42 | 41 | 9 | 115 | 119 | 25 | 53.45 ± 9.21 | Case-control | 0.47 | 8 |
| Li et al., (2006) | 2006 | IS | Asian | 235 | 184 | 35 | 190 | 128 | 16 | 65.20 ± 12.75 | Case-control | 0.34 | 8 |
| Dikmen et al., (2006) | 2006 | IS | ONR | 75 | 57 | 14 | 32 | 21 | 2 | 63.4 ± 0.87 | Case-control | 0.52 | 8 |
| Gao et al., (2006) | 2006 | IS | Asian | 30 | 49 | 21 | 32 | 44 | 24 | 61.08 ± 10.77 | Case-control | 0.25 | 8 |
| Yanqun et al., (2006) | 2006 | IS | Asian | 49 | 73 | 40 | 49 | 41 | 10 | 55 ± 6 | Case-control | 0.74 | 7 |
| Hermans et al., (2006) | 2006 | IS | ONR | 4 | 13 | 6 | 62 | 58 | 22 | 71.7 ± 9.3 | Case-control | 0.18 | 9 |
| Pezzini et al., (2005) | 2005 | IS | European | 46 | 83 | 34 | 60 | 75 | 23 | 35.0 ± 7.5 | Case-control | 0.96 | 9 |
| En et al., (2005) | 2005 | IS | Asian | 88 | 43 | 5 | 51 | 17 | 2 | 65 ± 10 | Case-control | 0.69 | 8 |
| Yan et al., (2006) | 2005 | IS | Asian | 34 | 24 | 3 | 37 | 17 | 3 | Old | Case-control | 0.58 | 8 |
| Jinhuan et al., (2005) | 2005 | IS | Asian | 55 | 28 | 4 | 53 | 24 | 3 | Mean 65 | Case-control | 0.89 | 8 |
| Alluri et al., (2005) | 2005 | IS | ONR | 47 | 21 | 1 | 48 | 1 | 0 | 7–78 | Case-control | 0.94 | 8 |
| Kawamoto et al., (2005) | 2005 | IS | Asian | 33 | 43 | 21 | 91 | 110 | 40 | 78 ± 8.3 | Case-control | 0.49 | 8 |
| Yi et al., (2005) | 2005 | IS | Asian | 27 | 42 | 9 | 22 | 25 | 3 | 68.3 ± 7.6 | Case-control | 0.23 | 8 |
| Lingling et al., (2004) | 2004 | IS | Asian | 26 | 18 | 3 | 12 | 15 | 5 | 67.16 ± 10.11 | Case-control | 0.93 | 8 |
| Choi et al., (2003) | 2003 | IS | Asian | 62 | 97 | 36 | 73 | 100 | 25 | 61.4 ± 10.9 | Case-control | 0.30 | 7 |
| Jin et al., (2004) | 2003 | IS | Asian | 21 | 59 | 14 | 40 | 49 | 11 | 69.68 ± 8.9 | Case-control | 0.48 | 8 |
| Jin et al., (2004) | 2003 | IS | Asian | 15 | 23 | 21 | 15 | 11 | 7 | NA | Case-control | 0.09 | 7 |
| Yan et al., (2003) | 2003 | IS | Asian | 55 | 28 | 4 | 53 | 24 | 3 | 66.13 ± 12.54 | Case-control | 0.89 | 8 |
| Li et al., (2002) | 2002 | IS | Asian | 58 | 65 | 20 | 97 | 49 | 8 | 40–90 | Case-control | 0.58 | 8 |
| Chuanqing et al., (2002) | 2002 | IS | Asian | 24 | 35 | 10 | 25 | 35 | 7 | 62.3 ± 12.9 | Case-control | 0.30 | 8 |
| Wenping et al., (2003) | 2002 | IS | Asian | 8 | 34 | 19 | 25 | 46 | 15 | 61–79 | Case-control | 0.43 | 8 |
| Huang et al., (2002) | 2002 | IS | Asian | 11 | 25 | 13 | 16 | 24 | 10 | 55.5 ± 13.0 | Case-control | 0.85 | 8 |
| Guangsen and Chongwen, (2002) | 2002 | IS | Asian | 40 | 47 | 15 | 37 | 47 | 16 | 65 | Case-control | 0.87 | 8 |
| Akar et al., (2001) | 2001 | IS | ONR | 24 | 18 | 4 | 39 | 23 | 6 | Children | Case-control | 0.34 | 7 |
| Wu et al., (2001) | 2001 | IS | Asian | 23 | 40 | 14 | 92 | 113 | 24 | 61.4 ± 6.8 | Case-control | 0.21 | 8 |
| Yaqin et al., (2001) | 2000 | IS | Asian | 15 | 23 | 21 | 15 | 11 | 7 | NA | Case-control | 0.09 | 7 |
| Zheng et al., (2000) | 2000 | IS | Asian | 43 | 62 | 10 | 62 | 45 | 15 | Mean 59 | Case-control | 0.14 | 8 |
| Cumming et al., (1999) | 1999 | IS | ONR | 41 | 6 | 1 | 42 | 6 | 0 | 7–36 | Case-control | 0.64 | 7 |
| Harmon et al., (1999) | 1999 | IS | European | 74 | 73 | 27 | 86 | 78 | 19 | >60 | Case-control | 0.83 | 7 |
| Gaustadnes et al., (1999) | 1999 | IS | European | 97 | 88 | 22 | 545 | 449 | 90 | 25–68 | Case-control | 0.85 | 8 |
| Akar et al., (1999) | 1999 | IS | ONR | 14 | 10 | 4 | 63 | 37 | 6 | Children | Case-control | 0.85 | 8 |
| Press et al., (1999) | 1999 | IS | ONR | 72 | 85 | 10 | 50 | 57 | 8 | 65 ± 8 | Case-control | 0.12 | 8 |
| Lalouschek et al., (1999) | 1999 | IS | ONR | 35 | 37 | 9 | 32 | 40 | 9 | 64.76 ± 13.2 | Case-control | 0.50 | 8 |
| Xinliang et al., (1999) | 1999 | IS | Asian | 10 | 45 | 25 | 58 | 32 | 20 | 60.2 ± 6.1 | Case-control | 0.48 | 8 |
| De Stefano et al., (1998) | 1998 | IS | European | 28 | 27 | 17 | 65 | 98 | 35 | Mean 33.9 | Case-control | 0.85 | 8 |
| Kostulas et al., (1998) | 1998 | IS | ONR | 50 | 40 | 10 | 50 | 40 | 10 | NA | Case-control | 0.63 | 7 |
| Salooja et al., (1998) | 1998 | IS | European | 81 | 76 | 16 | 114 | 107 | 21 | 68 (22–94) | Case-control | 0.56 | 8 |
| Nakata et al., (1998) | 1998 | IS | Asian | 19 | 23 | 6 | 35 | 51 | 19 | 30–80 | Case-control | 0.96 | 8 |
| Markus et al., (1997) | 1997 | IS | European | 162 | 146 | 37 | 76 | 63 | 22 | 65.7 (10.5) | Case-control | 0.13 | 7 |
3.2 Meta-analysis results
There were 34,814 participants (15,569 cases and 19,245 controls) in the 96 studies included in the meta-analysis.
3.2.1 Dominant model
The TT + CT genotype showed significant heterogeneity compared with the CC genotype in the dominant genetic model (I2 = 69.9%; p < 0.001) (Table 2). The MTHFR rs1801133 (677C>T) mutation significantly increased the IS risk under the dominant genetic model (OR = 1.47; 95%CI = 1.33–1.61; p < 0.001). In the ancestry subgroup analysis, the rs1801133 (677C>T) polymorphism of MTHFR was evidently linked to an increased risk of IS in Asian (OR = 1.59; 95%CI = 1.41–1.80; p < 0.001) and Hispanic or Latin population (OR = 1.93; 95%CI = 1.39–2.67; p < 0.001). MTHFR rs1801133 (677C>T) gene polymorphism was associated with IS susceptibility in all age groups except young populations (middle-aged: OR = 1.59, 95%CI = 1.33–1.90, and p < 0.001; elderly: OR = 1.34, 95%CI = 1.17–1.53, and p < 0.001).
TABLE 2.
Pooled odds ratios (ORs) and 95% confidence intervals (CIs) of the association between C677T polymorphism and stroke in the dominant model.
| Model | OR (95% CI) | PZ | I2 | PH |
|---|---|---|---|---|
| Dominant | ||||
| All IS | 1.47 (1.33–1.61) | <0.001 a | 69.9% | <0.001 |
| Ancestry | ||||
| Asian | 1.59 (1.41–1.80) | <0.001 a | 67.2% | <0.001 |
| European | 1.35 (1.10–1.65) | 0.004 | 69.9% | <0.001 |
| African | 2.56 (0.96–6.85) | 0.061 | — | — |
| Hispanic or Latin American | 1.93 (1.39–2.67) | <0.001 a | 0.0% | 0.824 |
| Other and not reported ancestries | 1.20 (0.91–1.57) | 0.190 | 76.4% | <0.001 |
| Age | ||||
| Young | 1.46 (1.12–1.91) | 0.005 | 52.2% | 0.012 |
| Middle-aged | 1.59 (1.33–1.90) | <0.001 a | 69.8% | <0.001 |
| Elderly | 1.34 (1.17–1.53) | <0.001 a | 71.3% | <0.001 |
| NA | 1.77 (1.30–2.41) | <0.001 a | 69.3% | <0.001 |
Bonferroni correction for multiple testing was applied (p-value threshold, 0.001);
PZ, p-value for the Z-test; PH, p-value for heterogeneity;
Association was still significant after Bonferroni correction for multiple testing.
3.2.2 Recessive model
The TT genotype showed significant heterogeneity compared with the CC + CT genotype in the recessive genetic model (I2 = 55.7%; p < 0.001) (Table 3). MTHFR rs1801133 (677C>T) polymorphism was associated with an increased risk of stroke under the recessive model (OR = 1.52; 95%CI = 1.36–1.71; p < 0.001). The ancestry subgroup analysis showed a significant difference in the Asian populations, with combined ORs of 1.58 (95% CI = 1.38–1.81; p < 0.001). The middle-aged and elderly people had an increased risk of stroke according to the subgroup analysis (middle-aged: OR = 1.55, 95%CI = 1.22–1.96, and p < 0.001; elderly: OR = 1.47, 95%CI = 1.28–1.68, and p < 0.001)
TABLE 3.
Pooled odds ratios (ORs) and 95% confidence intervals (CIs) of the association between C677T polymorphism and stroke in the recessive model.
| Model | Or (95% CI) | PZ | I2 | PH |
|---|---|---|---|---|
| Recessive | ||||
| All IS | 1.52 (1.36–1.71) | <0.001 a | 55.7% | <0.001 |
| Ancestry | ||||
| Asian | 1.58 (1.38–1.81) | <0.001 a | 47.1% | <0.001 |
| European | 1.48 (1.11–1.97) | 0.007 | 73.1% | <0.001 |
| African | — | — | — | — |
| Hispanic or Latin American | 0.96 (0.68–1.37) | 0.839 | 0.0% | 0.949 |
| Other and not reported ancestries | 1.47 (1.05–2.05) | 0.026 | 49.6% | 0.005 |
| Age | ||||
| Young | 1.25 (0.81–1.92) | 0.313 | 53.7% | 0.009 |
| Middle-aged | 1.55 (1.22–1.96) | <0.001 a | 57.5% | <0.001 |
| Elderly | 1.47 (1.28–1.68) | <0.001 a | 46.8% | 0.001 |
| NA | 1.99 (1.29–3.07) | <0.001 a | 64.7% | 0.001 |
Bonferroni correction for multiple testing was applied (p-value threshold 0.001);
PZ, p-value for the Z-test; PH, p-value for heterogeneity;
Association was still significant after Bonferroni correction for multiple testing.
3.2.3 Heterozygous model
The TC genotype showed significant heterogeneity compared with the CC genotype in the heterozygous genetic model (I2 = 60.4%; p < 0.001) (Table 4). There was an obvious association between MTHFR rs1801133 (677C>T) polymorphism and increased risk of stroke under the heterozygous model (OR = 1.36; 95%CI = 1.24–1.48; p < 0.001). In the subgroup analysis, MTHFR rs1801133 (677C>T) gene polymorphism was associated with stroke susceptibility in Asian (OR = 1.46; 95%CI = 1.30–1.63; p < 0.001), Hispanic or Latin populations (OR = 2.09; 95%CI = 1.50–2.95; p < 0.001), middle-aged (OR = 1.50; 95%CI = 1.27–1.78; p < 0.001), and elderly groups (OR = 1.23; 95%CI = 1.08–1.40; p = 0.001).
TABLE 4.
Pooled odds ratios (ORs) and 95% confidence intervals (CIs) of the association between C677T polymorphism and stroke in the heterozygous model.
| Model | OR (95% CI) | PZ | I2 | PH |
|---|---|---|---|---|
| Heterozygote | ||||
| All IS | 1.36 (1.24–1.48) | <0.001 a | 60.4% | <0.001 |
| Ancestry | ||||
| Asian | 1.46 (1.30–1.63) | <0.001 a | 58.2% | <0.001 |
| European | 1.27 (1.08–1.51) | 0.005 | 50.4% | 0.006 |
| African | 2.56 (0.96–6.85) | 0.061 | — | — |
| Hispanic or Latin American | 2.09 (1.50–2.95) | <0.001 a | 0.0% | 0.825 |
| Other and not reported ancestries | 1.10 (0.86–1.40) | 0.453 | 67.6% | <0.001 |
| Age | ||||
| Young | 1.44 (1.13–1.84) | 0.003 | 38.7% | 0.069 |
| Middle-aged | 1.50 (1.27–1.78) | <0.001 a | 62.5% | <0.001 |
| Elder | 1.23 (1.08–1.40) | 0.001 a | 62.8% | 0.001 |
| NA | 1.48 (1.14–1.92) | 0.003 | 50.2% | 0.020 |
Bonferroni correction for multiple testing was applied (p-value threshold 0.001);
PZ, p-value for the Z-test; PH, p-value for heterogeneity;
Association was still significant after Bonferroni correction for multiple testing.
3.2.4 Homozygous model
The TT genotype showed significant heterogeneity compared with the CC genotype in the homozygous genetic model (I2 = 64.2%; p < 0.001) (Table 5). There was also a significant association between MTHFR rs1801133 (677C>T) polymorphism and an increased risk of stroke under this model (OR = 1.82; 95%CI = 1.58–2.11; p < 0.001). However, the stratification analysis results were similar to those of the recessive model. Significant correlation was detected between MTHFR rs1801133 (677C>T) polymorphisms and the increased risk of stroke in the Asian population (OR = 1.98; 95%CI = 1.66–2.37; p < 0.001). Furthermore, the middle-aged and elderly people had an increased risk of stroke in the subgroup analysis (middle-aged: OR = 1.92, 95%CI = 1.45–2.53, and p < 0.001; elderly: OR = 1.74, 95%CI = 1.44–2.11, and p < 0.001).
TABLE 5.
Pooled odds ratios (ORs) and 95% confidence intervals (CIs) of the association between C677T polymorphism and stroke in the homozygous model.
| Model | Or (95% CI) | PZ | I2 | PH |
|---|---|---|---|---|
| Homozygote | ||||
| All IS | 1.82 (1.58–2.11) | <0.001 a | 64.2% | <0.001 |
| Ancestry | ||||
| Asian | 1.98 (1.66–2.37) | <0.001 a | 59.4% | <0.001 |
| European | 1.65 (1.14–2.39) | 0.008 | 79.3% | <0.001 |
| African | — | — | — | — |
| Hispanic or Latin American | 1.60 (1.05–2.46) | 0.030 | 0.0% | 0.863 |
| Other and not reported ancestries | 1.59 (1.09–2.32) | 0.017 | 56.2% | 0.001 |
| Age | ||||
| Young | 1.43 (0.88–2.33) | 0.146 | 55.6% | 0.006 |
| Middle-aged | 1.92 (1.45–2.53) | <0.001 a | 62.7% | <0.001 |
| Elder | 1.74 (1.44–2.11) | <0.001 a | 62.3% | <0.001 |
| NA | 2.45 (1.47–4.01) | 0.001 a | 69.1% | <0.001 |
Bonferroni correction for multiple testing was applied (p-value threshold 0.001);
PZ, p-value for the Z-test; PH, p-value for heterogeneity;
Association was still significant after Bonferroni correction for multiple testing.
3.2.5 Allelic model
The T allele showed significant heterogeneity compared with the C allele in the allelic genetic model (I2 = 75.7%; p < 0.001) (Table 6). There was an obvious association between MTHFR rs1801133 (677C>T) polymorphism and an increased risk of stroke under the allelic model (OR = 1.37; 95%CI = 1.27–1.48; p < 0.001). In the subgroup analysis, MTHFR rs1801133 (677C>T) gene polymorphism was associated with stroke susceptibility in Asian populations (OR = 1.46; 95%CI = 1.33–1.60; p < 0.001). The middle-aged and elderly people with T allele mutation had a higher risk of stroke (middle-aged: OR = 1.42, 95%CI = 1.24–1.64, and p < 0.001; elderly: OR = 1.30, 95%CI = 1.18–1.43, and p < 0.001).
TABLE 6.
Pooled odds ratios (ORs) and 95% confidence intervals (CIs) of the association between C677T polymorphism and stroke in the allelic model.
| Model | Or (95% CI) | PZ | I2 | PH |
|---|---|---|---|---|
| Allele | ||||
| All IS | 1.37 (1.27–1.48) | <0.001 a | 75.7% | <0.001 |
| Ancestry | ||||
| Asian | 1.46 (1.33–1.60) | <0.001 a | 72.6% | <0.001 |
| European | 1.29 (1.09–1.53) | 0.003 | 79.5% | <0.001 |
| African | 2.51 (1.06–5.93) | 0.036 | — | — |
| Hispanic or Latin American | 1.28 (1.05–1.57) | 0.016 | 0.0% | 0.981 |
| Other and not reported ancestries | 1.26 (0.96–1.52) | 0.116 | 81.4% | <0.001 |
| Age | ||||
| Young | 1.31 (1.05–1.64) | 0.016 | 65.5% | <0.001 |
| Middle-aged | 1.42 (1.24–1.64) | <0.001 a | 73.7% | <0.001 |
| Elder | 1.30 (1.18–1.43) | <0.001 a | 75.0% | <0.001 |
| NA | 1.70 (1.28–2.26) | <0.001 a | 80.6% | <0.001 |
Bonferroni correction for multiple testing was applied (p-value threshold 0.001);
PZ, p-value for the Z-test; PH p-value for heterogeneity;
Association was still significant after Bonferroni correction for multiple testing.
3.3 Sensitivity analysis
A sensitivity analysis was conducted to compare the pooled ORs after individually excluding each included study. There was no significant change in the results (Supplementary Figure S1).
3.4 Publication bias
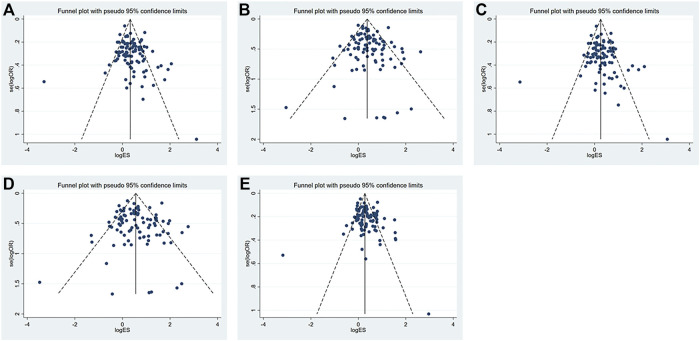
The funnel plot is shown in Figure 3. All research studies included in this study distributed above the funnel plots, which indicated that variability of the effect size was low and the results were reliable. The Egger’s funnel plots for these five models were basically symmetrical though Egger’s test, indicating there was publication bias in the dominant (p = 0.04) and heterozygous models (p = 0.03) (Table 7). Nonetheless, we did not find any publication bias in all genetic models using Begg’s tests. The correlation between the lnOR and its variance and the level of heterogeneity across studies might contribute to the discrepancy between Egger’s and Begg’s tests. Actually, Begg’s test is more robust and has the appropriate type I error rates despite the sample size, the number of included studies, and the level of heterogeneity. Furthermore, when the summary estimates are ORs or RRs and there is obvious heterogeneity between studies (Schwarzer et al., 2002; Peters et al., 2006), type I error rates for Egger’s test are higher than those for Begg’s test.
FIGURE 3.
Funnel plots of the (A) dominant model; (B) recessive model; (C) heterozygous model; (D) homozygous model; and (E) allelic model.
TABLE 7.
Publication bias of included studies.
| IS | Egger’s test | Begg’s test | |||
|---|---|---|---|---|---|
| Co-efficiency | Standard error | p-value | Z | p-value | |
| Dominant model | 0.83 | 0.39 | 0.04 a | 1.56 | 0.12 |
| Recessive model | 0.37 | 0.29 | 0.20 | 1.28 | 0.20 |
| Heterozygote model | 0.74 | 0.34 | 0.03 a | 1.83 | 0.07 |
| Homozygote model | 0.28 | 0.36 | 0.43 | 1.18 | 0.24 |
| Allele model | 0.76 | 0.44 | 0.09 | 1.71 | 0.09 |
p < 0.05.
4 Discussion
This meta-analysis demonstrates associations between MTHFR rs1801133 (677C>T) genetic polymorphism and susceptibility to IS under all genetic models. Our results were consistent with a previous meta-analysis performed in 2016, wherein this polymorphism was found to be potentially involved in the development of IS (Song et al., 2016). This suggests the MTHFR C677T mutation is a genetic risk factor of IS, and primary and secondary prevention should be initiated in a timely manner in population with this mutation.
Several factors might explain the association of the MTHFR rs1801133 (677C>T) mutation and increased IS risk. Most importantly, the MTHFR rs1801133 (677C>T) mutation leads to decreased MTHFR activity and elevated homocysteine levels (Castro et al., 2004). Hyperhomocysteinemia is linked to the overproduction of free radicals (Xi et al., 2016), induction of oxidative stress (Esse et al., 2019; Tchantchou et al., 2021), endothelial injury (Salvio et al., 2021), coagulation, and lipid metabolism disturbance (Herrmann, 2001), which all contribute to the incidence of IS. Meanwhile, previous studies demonstrated that people with the MTHFR rs1801133 (677C>T) mutation show a poor response to homocysteine-lowering treatment (Qin et al., 2020). Furthermore, a meta-analysis published in the current year showed that MTHFR rs1801133 (677C>T) polymorphism is related to susceptibility to H-type hypertension (Liao et al., 2022), which is a traditional risk factor for IS.
We also conducted a subgroup analysis on the basis of age and ancestry of the study population to further understand the significance of the MTHFR rs1801133 (677C>T) mutation in various populations. The result of subgroup analysis showed increased IS risk in populations with the MTHFR rs1801133 (677C>T) mutation in middle-aged and elderly groups. It was consistent with the fact that older people were more susceptible to atherosclerosis, which was an important cause of IS. Previous studies had demonstrated that the MTHFR rs1801133 (677C>T) mutation increased the risk of IS in patients with large-artery atherosclerosis (Cui, 2016) and adults (Xin et al., 2009).
Another important finding of this meta-analysis was the stable association between IS risk and MTHFR rs1801133 (677C>T) polymorphism in Asian populations in all genetic models and Hispanic or Latin American population in dominant and heterozygous models. Similar trends were also found among other populations, although no statistically significant difference was found in some genetic models. This may be related to the following reasons: 1) the frequency of the MTHFR 677T gene variant differs among ethnic groups due to different genetic backgrounds. Previous studies indicated that the frequency of the MTHFR rs1801133 T allele was 24–40% in Europeans, 40% in Koreans, and 26–37% in Japanese (Shao et al., 2017); 2) MTHFR rs1801133 (677C>T) was associated with increased coronary heart disease only when the folate level was low (Klerk et al., 2002). Thus, various dietary habits and differences in folate intake may also contribute to this difference; 3) the difference in the power of included studies may be another cause of this result. Nonetheless, the results for these ancestry groups need to be interpreted with caution, and more high-quality studies are still required to explore the correlation between MTHFR rs1801133 (677C>T) polymorphism and IS risk in these ancestry groups.
This study has several strengths. First, we included the most recent and relevant studies in this meta-analysis. In addition, we further analyzed the association between MTHFR rs1801133 (677C>T) polymorphism and IS risk in different populations. Finally, this meta-analysis included high-quality observational studies using real-world data with a large number of patients.
However, this meta-analysis also has some limitations. First, the study was based on the secondary study-level data. Age groups were defined according to the mean or median age of study subjects, and some studies did not provide clear information on age. Thus, the age stratification of subgroups might not be accurate. Second, the findings of this meta-analysis were mainly based on case-control studies and lacked prospective research; therefore, they should be interpreted with caution.
5 Conclusion
Our findings showed that the MTHFR rs1801133 (677C>T) variant may contribute to an increased risk of IS. This association was statistically significant in the Asian and Hispanic or Latin American cohorts and showed a similar trend in the populations of other ancestries. For middle-aged and elderly people, MTHFR rs1801133 (677C>T) might be a promising biomarker for early detection and prediction of the prognosis of IS. However, high-quality, prospective studies are needed in the future.
Acknowledgments
The authors would like to thank all of our teammates for contributing to this work. They also thank Lisa Kreiner, PhD, from Liwen Bianji, (Edanz) (www.liwenbianji.cn/), for editing the English text of a draft of this manuscript.
Author contributions
All authors contributed to the data analysis and drafting or revision of the manuscript, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Funding
This work was supported by the National Natural Science Foundation of China under the Grant No. 81971116 and the Key Research and Development Program of Shaanxi under the Grant No. 2019ZDLSF01–04.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1021423/full#supplementary-material
References
- Akar N., Akar E., Deda G., Sipahi T., OrsAl A. (1999). Factor V1691 G-A, prothrombin 20210 G-A, and methylenetetrahydrofolate reductase 677 C-T variants in Turkish children with cerebral infarct. J. Child. Neurol. 14 (11), 749–751. 10.1177/088307389901401113 [DOI] [PubMed] [Google Scholar]
- Akar N., Akar E., Ozel D., Deda G., Sipahi T. (2001). Common mutations at the homocysteine metabolism pathway and pediatric stroke. Thromb. Res. 102 (2), 115–120. 10.1016/s0049-3848(01)00226-2 [DOI] [PubMed] [Google Scholar]
- Al-Allawi N. S., Avo A. S., Jubrael J. S. (2009). Methylenetetrahydrofolate reductase C677T polymorphism in Iraqi patients with ischemic stroke. Neurol. India 57 (5), 631–634. [DOI] [PubMed] [Google Scholar]
- Alluri R. V., Mohan V., Komandur S., Chawda K., Chaudhuri J. R., Hasan Q. (2005). MTHFR C677T gene mutation as a risk factor for arterial stroke: A hospital based study. Eur. J. Neurol. 12 (1), 40–44. 10.1111/j.1468-1331.2004.00938.x [DOI] [PubMed] [Google Scholar]
- Almawi W. Y., Khan A., Al-Othman S. S., Bakhiet M. (2009). Case-control Study of methylenetetrahydrofolate reductase mutations and hyperhomocysteinemia and risk of stroke. J. Stroke Cerebrovasc. Dis. 18 (5), 407–408. 10.1016/j.jstrokecerebrovasdis.2008.12.003 [DOI] [PubMed] [Google Scholar]
- Atadzhanov M., Mwaba M. H., Mukomena P. N., Lakhi S., Rayaprolu S., Ross O. A., et al. (2013). Association of the APOE, MTHFR and ACE genes polymorphisms and stroke in Zambian patients. Neurol. Int. 5 (4), e20–e72. 10.4081/ni.2013.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berge E., Haug K. B. F., Sandset E. C., Haugbro K. K., Turkovic M., Sandset P. M. (2007). The factor V Leiden, prothrombin gene 20210GA, methylenetetrahydrofolate reductase 677CT and platelet glycoprotein IIIa 1565TC mutations in patients with acute ischemic stroke and atrial fibrillation. Stroke 38 (3), 1069–1071. 10.1161/01.STR.0000258076.04860.8e [DOI] [PubMed] [Google Scholar]
- Biswas A., Ranjan R., Meena A., Akhter M. S., Yadav B. K., Munisamy M., et al. (2009). Homocystine levels, polymorphisms and the risk of ischemic stroke in young Asian Indians. J. Stroke Cerebrovasc. Dis. 18 (2), 103–110. 10.1016/j.jstrokecerebrovasdis.2008.09.014 [DOI] [PubMed] [Google Scholar]
- Black M., Wang W., Wang W. (2015). Ischemic stroke: From next generation sequencing and GWAS to community genomics? Omics 19 (8), 451–460. 10.1089/omi.2015.0083 [DOI] [PubMed] [Google Scholar]
- Brattström L., Wilcken D. E., Ohrvik J., Brudin L. (1998). Common methylenetetrahydrofolate reductase gene mutation leads to hyperhomocysteinemia but not to vascular disease: The result of a meta-analysis. Circulation 98 (23), 2520–2526. 10.1161/01.cir.98.23.2520 [DOI] [PubMed] [Google Scholar]
- Castro R., Rivera I., Ravasco P., Camilo M. E., Jakobs C., Blom H. J., et al. (2004). 5, 10-methylenetetrahydrofolate reductase (MTHFR) 677C-->T and 1298A-->C mutations are associated with DNA hypomethylation. J. Med. Genet. 41 (6), 454–458. 10.1136/jmg.2003.017244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celiker G., Can U., Verdi H., Yazici A. C., Ozbek N., Atac F. B. (2009). Prevalence of thrombophilic mutations and ACE I/D polymorphism in Turkish ischemic stroke patients. Clin. Appl. Thromb. Hemost. 15 (4), 415–420. 10.1177/1076029608315163 [DOI] [PubMed] [Google Scholar]
- Cernera G., Comegna M., Gelzo M., Savoia M., Bruzzese D., Mormile M., et al. (2021). Molecular analysis of prothrombotic gene variants in patients with acute ischemic stroke and with transient ischemic attack. Med. Kaunas. Lith. 57, 723. 10.3390/medicina57070723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang G., Kuai Z., Wang J., Wu J., Xu K., Yuan Y., et al. (2019). The association of MTHFR C677T variant with increased risk of ischemic stroke in the elderly population: A meta-analysis of observational studies. BMC Geriatr. 19 (1), 331. 10.1186/s12877-019-1304-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B. O., Kim N. K., Kim S. H., Kang M. S., Lee S., Ahn J. Y., et al. (2003). Homozygous C677T mutation in the MTHFR gene as an independent risk factor for multiple small-artery occlusions. Thromb. Res. 111 (1-2), 39–44. 10.1016/j.thromres.2003.08.022 [DOI] [PubMed] [Google Scholar]
- Chuanqing T., Minaqing H., Jianzeng W., Sizhong X., Chunying X., Chuyan P., et al. (2002). The relation of methylenetetradrofolate reductase gene Mutation and Cystathionine β-synthase Gene Mutation Exist in cerebral infarction. Thrombosis hemostasis 04, 149–151. [Google Scholar]
- Cui T. (2016). MTHFR C677T mutation increased the risk of ischemic stroke, especially in large-artery atherosclerosis in adults: An updated meta-analysis from 38 researches. Int. J. Neurosci. 126 (1), 10–19. 10.3109/00207454.2014.990559 [DOI] [PubMed] [Google Scholar]
- Cumming A. M., Olujohungbe A., Keeney S., SingH H., Hay C. R., Serjeant G. R. (1999). The methylenetetrahydrofolate reductase gene C677T polymorphism in patients with homozygous sickle cell disease and stroke. Br. J. Haematol. 107 (3), 569–571. 10.1046/j.1365-2141.1999.01728.x [DOI] [PubMed] [Google Scholar]
- De Stefano V., Chiusolo P., Paciaroni K., CasorellI I., Rossi E., MolinariM., et al. (1998). Prothrombin G20210A mutant genotype is a risk factor for cerebrovascular ischemic disease in young patients. Blood 91 (10), 3562–3565. 10.1182/blood.v91.10.3562.3562_3562_3565 [DOI] [PubMed] [Google Scholar]
- Dikmen M., Ozbabalik D., Gunes H. V., DegIrmencI I., Bal C., Ozdemir G., et al. (2006). Acute stroke in relation to homocysteine and methylenetetrahydrofolate reductase gene polymorphisms. Acta Neurol. Scand. 113 (5), 307–314. 10.1111/j.1600-0404.2005.00556.x [DOI] [PubMed] [Google Scholar]
- Djordjevic V., Stankovic M., Brankovic-Sreckovic V., Rakicevic L., Damnjanovic T., Antonijevic N., et al. (2012). Prothrombotic genetic risk factors in stroke: A possible different role in pediatric and adult patients. Clin. Appl. Thromb. Hemost. 18 (6), 658–661. 10.1177/1076029611432136 [DOI] [PubMed] [Google Scholar]
- Djordjevic V., Stankovic M., Brankovic-Sreckovic V., Rakicevic L., Radojkovic D. (2009). Genetic risk factors for arterial ischemic stroke in children: A possible MTHFR and eNOS gene-gene interplay? J. Child. Neurol. 24 (7), 823–827. 10.1177/0883073808330164 [DOI] [PubMed] [Google Scholar]
- En X., Bingmei D., Shengqiang C., Haifeng X., Xuefen L. (2005). The relationship between Gene Polymorphisms of MTRR A66G, MS D919G, MTHFR C677T and Cerebral Infarction. Chin. J. Neuromed 09, 902–904+907. [Google Scholar]
- Esse R., Barroso M., Tavares de Almeida I., Castro R. (2019). The contribution of homocysteine metabolism disruption to endothelial dysfunction: State-of-the-Art. Int. J. Mol. Sci. 20, 867. 10.3390/ijms20040867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekih-Mrissa N., et al. (2013). Role of methylenetetrahydrofolate reductase A1298C polymorphism in cerebral venous thrombosis. Blood Coagulation Fibrinolysis 24 (2), 118–119. [DOI] [PubMed] [Google Scholar]
- Francis J., Raghunathan S., Khanna P. (2007). The role of genetics in stroke. Postgrad. Med. J. 83 (983), 590–595. 10.1136/pgmj.2007.060319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Sun Y., Li Y., Su L., Guo L. (2008). Hyperhomocysteine genetic polymorphism of methylenetetrahydrofolate reductase in young adults with ischemic stroke. J. Zhengzhou Uiversity Med. Sci. 03, 570–573. 10.13705/j.issn.1671-6825.2008.03.052 [DOI] [Google Scholar]
- Gao X., Yang H., ZhiPing T. (2006). Association studies of genetic polymorphism, environmental factors and their interaction in ischemic stroke. Neurosci. Lett. 398 (3), 172–177. 10.1016/j.neulet.2005.12.078 [DOI] [PubMed] [Google Scholar]
- Gaustadnes M., RudigerN., Moller J., Rasmussen K., Bjerregaard Larsen T., Ingerslev J. (1999). Thrombophilic predisposition in stroke and venous thromboembolism in Danish patients. Blood Coagul. Fibrinolysis 10 (5), 251–259. 10.1097/00001721-199907000-00006 [DOI] [PubMed] [Google Scholar]
- Giusti B., Saracini C., Bolli P., Magi A., Martinelli I., Peyvandi F., et al. (2010). Early-onset ischaemic stroke: Analysis of 58 polymorphisms in 17 genes involved in methionine metabolism. Thromb. Haemost. 104 (2), 231–242. 10.1160/TH09-11-0748 [DOI] [PubMed] [Google Scholar]
- Goracy I., Cyrylowski L., KaczMarczykM., FabiAn A., Koziarska D., Goracy J., et al. (2009). C677T polymorphism of the methylenetetrahydrofolate reductase gene and the risk of ischemic stroke in Polish subjects. J. Appl. Genet. 50 (1), 63–67. 10.1007/BF03195654 [DOI] [PubMed] [Google Scholar]
- Goyette P., Sumner J. S., Milos R., Duncan A. M., Rosenblatt D. S., Matthews R. G., et al. (1994). Human methylenetetrahydrofolate reductase: Isolation of cDNA, mapping and mutation identification. Nat. Genet. 7 (2), 195–200. 10.1038/ng0694-195 [DOI] [PubMed] [Google Scholar]
- Guangsen Z., Chongwen D. (2002). Correlation analysis between plasma homocysteine level and polymorphism of homocysteine metabolism re- lated enzymes in ischemic cerebrovascular or cardiovascular diseases. Chin. J. Hematol., 13–16. [PubMed] [Google Scholar]
- Harmon D. L., Doyle R. M., Meleady R., DoyleM., Shields D. C., BaRRy R., et al. (1999). Genetic analysis of the thermolabile variant of 5, 10-methylenetetrahydrofolate reductase as a risk factor for ischemic stroke. Arterioscler. Thromb. Vasc. Biol. 19 (2), 208–211. 10.1161/01.atv.19.2.208 [DOI] [PubMed] [Google Scholar]
- Herak D. C., Antolic M. R., Krleza J. L., Pavic M., Dodig S., Duranovic V., et al. (2009). Inherited prothrombotic risk factors in children with stroke, transient ischemic attack, or migraine. Pediatrics 123, e653–e660. 10.1542/peds.2007-3737 [DOI] [PubMed] [Google Scholar]
- Herak D. C., Lenicek Krleza J., Radic Antolic M., Horvat I., Djuranovic V., Zrinski Topic R., et al. (2017). Association of polymorphisms in coagulation factor genes and enzymes of homocysteine metabolism with arterial ischemic stroke in children. Clin. Appl. Thromb. Hemost. 23 (8), 1042–1051. 10.1177/1076029616672584 [DOI] [PubMed] [Google Scholar]
- Hermans M. P., Gala J. L., Buysschaert M. (2006). The MTHFR CT polymorphism confers a high risk for stroke in both homozygous and heterozygous T allele carriers with Type 2 diabetes. Diabet. Med. 23 (5), 529–536. 10.1111/j.1464-5491.2006.01841.x [DOI] [PubMed] [Google Scholar]
- Herrmann W. (2001). The importance of hyperhomocysteinemia as a risk factor for diseases: An overview. Clin. Chem. Lab. Med. 39 (8), 666–674. 10.1515/CCLM.2001.110 [DOI] [PubMed] [Google Scholar]
- Hou J., Zeng X., Xie Y., Wu H., Zhao P. (2018). Genetic polymorphisms of methylenetetrahydrofolate reductase C677T and risk of ischemic stroke in a southern Chinese Hakka population. Med. (United States) 97, e13645. 10.1097/MD.0000000000013645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. W., Li L. L., Li J., Chen X. R., Yu M. (2022). Association of the methylenetetrahydrofolate reductase (MTHFR) gene variant C677T with serum homocysteine levels and the severity of ischaemic stroke: A case-control study in the southwest of China. J. Int. Med. Res. 50 (2), 3000605221081632. 10.1177/03000605221081632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Zhao Yl Y., Li S. (2002). Hyperhomocysteine, methylenetetrahydrofolate reductase gene, and other risk factors in ischemic stroke. Zhonghua Yi Xue Za Zhi 82 (2), 119–122. [PubMed] [Google Scholar]
- Hultdin J., Van Guelpen B., Winkvist A., Hallmans G., Weinehall L., Stegmayr B., et al. (2011). Prospective study of first stroke in relation to plasma homocysteine and MTHFR 677C>T and 1298A>C genotypes and haplotypes - evidence for an association with hemorrhagic stroke. Clin. Chem. Lab. Med. 49 (9), 1555–1562. 10.1515/CCLM.2011.234 [DOI] [PubMed] [Google Scholar]
- Isordia-Salas I., Barinagarrementería-Aldatz F., Leaños-Miranda A., Borrayo-Sánchez G., Vela-Ojeda J., García-Chávez J., et al. (2010). The C677T polymorphism of the methylenetetrahydrofolate reductase gene is associated with idiopathic ischemic stroke in the young Mexican-Mestizo population. Cerebrovasc. Dis. 29 (5), 454–459. 10.1159/000289349 [DOI] [PubMed] [Google Scholar]
- Jiang S., Li J., Zhang Y., Venners S. A., Tang G., Wang Y., et al. (2017). Methylenetetrahydrofolate reductase C677T polymorphism, hypertension and risk of stroke: A prospective, nested case-control study. Int. J. Neurosci. 127 (3), 253–260. 10.1080/00207454.2016.1183126 [DOI] [PubMed] [Google Scholar]
- Jiménez-González M. C., Santiago-German D., Castillo-Henkel E. F., Alvarado-Moreno J. A., Hernandez-Juarez J., Leanos-Miranda A., et al. (2021). Identification of genetic risk factors associated with ischaemic stroke in young Mexican patients. Neurologia 36 (5), 337–345. 10.1016/j.nrleng.2018.01.011 [DOI] [PubMed] [Google Scholar]
- Jin Z., Lei L., Hong S., Qingqing J., Qing W., Weiping J., et al. (2004). The relationship between MTHFR gene polymorphism and cerebral hemorrhage. J. Clin. Neurol. 4, 267–269. [Google Scholar]
- Jin Z., Lei L., Hong S., Qingqing J., Qing W., Weiping J., et al. (2004). Relation between MTHFR and brain infarction. Med. J. CASC, 267–269. 10.13705/j.issn.1671-6825.2008.03.052 [DOI] [Google Scholar]
- Jinhuan C., Guang Y., Yan S. (2005). The study of MTHFR gene C677T polymorphism in the case of cerebral thrombosis. JP Mt., 968–970. [Google Scholar]
- Kamberi B., Kamberi F., Spiroski M. (2016). Vascular genetic variants and ischemic stroke susceptibility in albanians from the Republic of Macedonia. Open Access Maced. J. Med. Sci. 4 (4), 556–564. 10.3889/oamjms.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto R., Kohara K., Oka Y., Tomita H., Tabara Y., Miki T. (2005). An association of 5, 10-methylenetetrahydrofolate reductase (MTHFR) gene polymorphism and ischemic stroke. J. Stroke Cerebrovasc. Dis. 14 (2), 67–74. 10.1016/j.jstrokecerebrovasdis.2004.12.003 [DOI] [PubMed] [Google Scholar]
- Kim O. J., Hong S. P., Ahn J. Y., Hong S. H., Hwang T. S., Kim S. O., et al. (2007). Influence of combined methionine synthase (MTR 2756A > G) and methylenetetrahydrofolate reductase (MTHFR 677C > T) polymorphisms to plasma homocysteine levels in Korean patients with ischemic stroke. Yonsei Med. J. 48 (2), 201–209. 10.3349/ymj.2007.48.2.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klerk M., Verhoef P., Clarke R., Blom H. J., Kok F. J., Schouten E. G., et al. (2002). MTHFR 677C-->T polymorphism and risk of coronary heart disease: A meta-analysis. JAMA 288 (16), 2023–2031. 10.1001/jama.288.16.2023 [DOI] [PubMed] [Google Scholar]
- Komitopoulou A., Kapsimali Z., Pergantou H., Adamtziki E., AroniS S. (2006). Mutations and polymorphisms in genes affecting hemostasis proteins and homocysteine metabolism in children with arterial ischemic stroke. Cerebrovasc. Dis. 22 (1), 13–20. 10.1159/000092332 [DOI] [PubMed] [Google Scholar]
- Kostulas K., CrisbyM., Huang W. X., LannfeLt L., HagenfeLdt L., Eggertsen G., et al. (1998). A methylenetetrahydrofolate reductase gene polymorphism in ischaemic stroke and in carotid artery stenosis. Eur. J. Clin. Invest. 28 (4), 285–289. 10.1046/j.1365-2362.1998.00281.x [DOI] [PubMed] [Google Scholar]
- Kumar A., Misra S., Hazarika A., Kumar P., Sagar R., Pathak A., et al. (2016). Association between methylenetetrahydrofolate reductase (MTHFR) C677T gene polymorphism and risk of ischemic stroke in north Indian population: A hospital based case–control study. Egypt. J. Med. Hum. Genet. 17 (4), 359–365. 10.1016/j.ejmhg.2016.01.001 [DOI] [Google Scholar]
- Lalouschek W., Aull S., Serles W., Schnider P., Mannhalter C., PabInger-FaschIng I., et al. (1999). C677T MTHFR mutation and factor V leiden mutation in patients with TIA/minor stroke: A case-control study. Thromb. Res. 93 (2), 61–69. 10.1016/s0049-3848(98)00154-6 [DOI] [PubMed] [Google Scholar]
- Li A., Shi Y., Xu L., Zhang Y., Zhao H., Li Q., et al. (2017). A possible synergistic effect of MTHFR C677T polymorphism on homocysteine level variations increased risk for ischemic stroke. Med. (United States) 96, e9300. 10.1097/MD.0000000000009300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. M., Zhang C., Lu X. l., Feng H. y., Su Q. x., Zeng Y., et al. (2006). Relationship between methylenetrahydrofolate reductase gene and ischemic stroke. Chin. Crit. Care Med. 18 (5), 264–267. [PubMed] [Google Scholar]
- Li C., Zhang C., Qiu S., Lu X., Zeng Y., Wu H., et al. (2002). Polymorphisms of ACE-1 and MTHFR genes and genetic susceptibility of ischemic stroke. Zhonghua Yi Xue Za Zhi 82 (15), 1046–1049. [PubMed] [Google Scholar]
- Liao S., Guo S., Ma R., He J., Yan Y., Zhang X., et al. (2022). Association between methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and H-type hypertension: A systematic review and meta-analysis. Ann. Hum. Genet. 86, 278–289. 10.1111/ahg.12468 [DOI] [PubMed] [Google Scholar]
- Liew S. C., Gupta E. D. (2015). Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism: Epidemiology, metabolism and the associated diseases. Eur. J. Med. Genet. 58 (1), 1–10. 10.1016/j.ejmg.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Lingling H., et al. (2004). Correlation between C677T mutation of MT HF R gene and cerebral infarction. Jiangsu Med. J., 861–862. 10.19460/j.cnki.0253-3685.2004.11.031 [DOI] [Google Scholar]
- Linnebank M., Moskau S., Semmler A., Hoefgen B., Bopp G., Kallweit U., et al. (2012). A possible genetic link between MTHFR genotype and smoking behavior. PLoS One 7, e53322. 10.1371/journal.pone.0053322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Xiaoyan C., Boai Z. (2017). Analysis of relationship of plasma homocysteine level and polymorphism of methylenetetrahydrofolate reductase with ischemic stroke. Chin. J. Stroke 12 (05), 404–409. [Google Scholar]
- Luo M., Li J., Sun X., Lai R., Wang Y., Xu X., et al. (2015). Interactions among candidate genes selected by meta-analyses resulting in higher risk of ischemic stroke in a Chinese population. PLoS ONE 10, e0145399. 10.1371/journal.pone.0145399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q. Q., Lu J., Sun H., Zhang J. S. (2015). Association of methylenetetrahydrofolate reductase (MTHFR) gene polymorphism with ischemic stroke in the Eastern Chinese Han population. Genet. Mol. Res. 14 (2), 4161–4168. 10.4238/2015.April.27.31 [DOI] [PubMed] [Google Scholar]
- Ma L., Jiang Y., Kong X., Yan M., Zhao T., Zhao H., et al. (2017). Synergistic effect of the MTHFR C677T and EPHX2 G860A polymorphism on the increased risk of ischemic stroke in Chinese type 2 diabetic patients. J. Diabetes Res. 2017, 6216205. 10.1155/2017/6216205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik R., Chauhan G., Traylor M., Sargurupremraj M., Okada Y., Mishra A., et al. (2019). Publisher Correction: Multiancestry genome-wide association study of 520, 000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 51 (7), 1192–1193. 10.1038/s41588-019-0449-0 [DOI] [PubMed] [Google Scholar]
- Malik R., Dichgans M. (2018). Challenges and opportunities in stroke genetics. Cardiovasc. Res. 114 (9), 1226–1240. 10.1093/cvr/cvy068 [DOI] [PubMed] [Google Scholar]
- Mao X., Han L. (2018). The relationship of methylenetetrahydrofolate reductase gene C677T polymorphism and ischemic stroke in Chinese han population. Ann. Clin. Lab. Sci. 48 (2), 242–247. [PubMed] [Google Scholar]
- Markus H. S., AliN., Swaminathan R., SankArAlingAm A., Molloy J., Powell J. (1997). A common polymorphism in the methylenetetrahydrofolate reductase gene, homocysteine, and ischemic cerebrovascular disease. Stroke 28 (9), 1739–1743. 10.1161/01.str.28.9.1739 [DOI] [PubMed] [Google Scholar]
- Mazdeh M., Khazaie M., Omrani M. D., Noroozi R., Komaki A., Karimi M., et al. (2021). Association between methylene tetrahydrofolate reductase polymorphisms and risk of ischemic stroke. Int. J. Neurosci. 131 (1), 44–48. 10.1080/00207454.2020.1733554 [DOI] [PubMed] [Google Scholar]
- Moe K. T., Woon F. P., De Silva D. A., Wong P., Koh T. H., Kingwell B., et al. (2008). Association of acute ischemic stroke with the MTHFR C677T polymorphism but not with NOS3 gene polymorphisms in a Singapore population. Eur. J. Neurol. 15 (12), 1309–1314. 10.1111/j.1468-1331.2008.02308.x [DOI] [PubMed] [Google Scholar]
- Mohamed E. H. M., Tan K. S., Ali J. M., Mohamed Z. (2011). TT genotype of the methylenetetrahydrofolate reductase C677T polymorphism is an important determinant for homocysteine levels in multi-ethnic Malaysian ischaemic stroke patients. Ann. Acad. Med. Singap. 40 (4), 186–191. [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D. G. (2009). Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. Bmj 339, b2535. 10.1136/bmj.b2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita D. C., Donaldson A., Butterfield R. J., Benedict S. L., Bale J. F., Jr (2009). Methylenetetrahydrofolate reductase gene polymorphism and childhood stroke. Pediatr. Neurol. 41 (4), 247–249. 10.1016/j.pediatrneurol.2009.04.017 [DOI] [PubMed] [Google Scholar]
- Nakata Y., KaTsuya T., Takami S., SatoN., Fu Y., IshiKawa K., et al. (1998). Methylenetetrahydrofolate reductase gene polymorphism: Relation to blood pressure and cerebrovascular disease. Am. J. Hypertens. 11 (8), 1019–1023. 10.1016/s0895-7061(98)00046-6 [DOI] [PubMed] [Google Scholar]
- Nan G., Wang L., Bu S. (2007). The association of MTHFR A1298C and CBS G919A gene mutations and ischemic stroke in young adults. Chin. J. Lab. Diagn, 1697–1699. [Google Scholar]
- Peters J. L., Sutton A. J., Jones D. R., Abrams K. R., Rushton L. (2006). Comparison of two methods to detect publication bias in meta-analysis. Jama 295 (6), 676–680. 10.1001/jama.295.6.676 [DOI] [PubMed] [Google Scholar]
- Pezzini A., Grassi M., Del Zotto E., Archetti S., Spezi R., Vergani V., et al. (2005). Cumulative effect of predisposing genotypes and their interaction with modifiable factors on the risk of ischemic stroke in young adults. Stroke 36 (3), 533–539. 10.1161/01.STR.0000155741.31499.c2 [DOI] [PubMed] [Google Scholar]
- Phipps M. S., Cronin C. A. (2020). Management of acute ischemic stroke. Bmj 368, l6983. 10.1136/bmj.l6983 [DOI] [PubMed] [Google Scholar]
- Prabhakaran S., Ruff I., Bernstein R. A. (2015). Acute stroke intervention: A systematic review. JAMA 313 (14), 1451–1462. 10.1001/jama.2015.3058 [DOI] [PubMed] [Google Scholar]
- Press R. D., BeamerN., EvAns A., DeLoughery T. G., Coull B. M. (1999). Role of a common mutation in the homocysteine regulatory enzyme methylenetetrahydrofolate reductase in ischemic stroke. Diagn. Mol. Pathol. 8 (1), 54–58. 10.1097/00019606-199903000-00009 [DOI] [PubMed] [Google Scholar]
- Qin X., Spence J. D., Li J., Zhang Y., Li Y., Sun N., et al. (2020). Interaction of serum vitamin B(12) and folate with MTHFR genotypes on risk of ischemic stroke. Neurology 94, e1126–e1136. 10.1212/WNL.0000000000008932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranellou K., Paraskeva A., Kyriazopoulos P., Batistatou A., Evangelou A., El-Aly M., et al. (2015). Polymorphisms in prothrombotic genes in young stroke patients in Greece: A case-controlled study. Blood Coagul. Fibrinolysis 26 (4), 430–435. 10.1097/MBC.0000000000000274 [DOI] [PubMed] [Google Scholar]
- Rozen R. (1997). Genetic predisposition to hyperhomocysteinemia: Deficiency of methylenetetrahydrofolate reductase (MTHFR). Thromb. Haemost. 78 (1), 523–526. 10.1055/s-0038-1657581 [DOI] [PubMed] [Google Scholar]
- Sabino A., Fernandes A. P., Lima L. M., Ribeiro D. D., Sousa M. O., de Castro Santos M. E. R., et al. (2009). Polymorphism in the methylenetetrahydrofolate reductase (C677T) gene and homocysteine levels: A comparison in Brazilian patients with coronary arterial disease, ischemic stroke and peripheral arterial obstructive disease. J. Thromb. Thrombolysis 27 (1), 82–87. 10.1007/s11239-007-0172-z [DOI] [PubMed] [Google Scholar]
- Salem-Berrabah O. B., Mrissa R., Machghoul S., Hamida A. B., N'siri B., Mazigh C., et al. (2010). Hyperhomocysteinemia, C677T MTHFR polymorphism and ischemic stroke in Tunisian patients. Tunis. Med. 88 (9), 655–659. [PubMed] [Google Scholar]
- Salomi B. S. B., Solomon R., Turaka V. P., Aaron S., Christudass C. S. (2021). Cryptogenic stroke in the young: Role of candidate gene polymorphisms in Indian patients with ischemic etiology. Neurol. India 69 (6), 1655–1662. 10.4103/0028-3886.333441 [DOI] [PubMed] [Google Scholar]
- Salooja N., Catto A., Carter A., Tudenham E. G., Grant P. J. (1998). Methylene tetrahydrofolate reductase C677T genotype and stroke. Clin. Lab. Haematol. 20 (6), 357–361. 10.1046/j.1365-2257.1998.00158.x [DOI] [PubMed] [Google Scholar]
- Salvio G., Ciarloni A., Cutini M., Balercia G. (2021). Hyperhomocysteinemia: Focus on endothelial damage as a cause of erectile dysfunction. Int. J. Mol. Sci. 22, 418. 10.3390/ijms22010418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawula W., Banecka-Majkutewicz Z., Kadzinski L., Jakobkiewicz-Banecka J., Wegrzyn G., Nyka W., et al. (2009). Homocysteine level and metabolism in ischemic stroke in the population of Northern Poland. Clin. Biochem. 42 (6), 442–447. 10.1016/j.clinbiochem.2008.12.019 [DOI] [PubMed] [Google Scholar]
- Sazci A., Ergul E., Tuncer N., Akpinar G., Kara I. (2006). Methylenetetrahydrofolate reductase gene polymorphisms are associated with ischemic and hemorrhagic stroke: Dual effect of MTHFR polymorphisms C677T and A1298C. Brain Res. Bull. 71 (1-3), 45–50. 10.1016/j.brainresbull.2006.07.014 [DOI] [PubMed] [Google Scholar]
- Schwarzer G., Antes G., Schumacher M. (2002). Inflation of type I error rate in two statistical tests for the detection of publication bias in meta-analyses with binary outcomes. Stat. Med. 21 (17), 2465–2477. 10.1002/sim.1224 [DOI] [PubMed] [Google Scholar]
- Shao W., Yuan Y., Li Y. (2017). Association between MTHFR C677T polymorphism and methotrexate treatment outcome in rheumatoid arthritis patients: A systematic review and meta-analysis. Genet. Test. Mol. Biomarkers 21 (5), 275–285. 10.1089/gtmb.2016.0326 [DOI] [PubMed] [Google Scholar]
- Sharp L., Little J. (2004). Polymorphisms in genes involved in folate metabolism and colorectal neoplasia: A HuGE review. Am. J. Epidemiol. 159 (5), 423–443. 10.1093/aje/kwh066 [DOI] [PubMed] [Google Scholar]
- Shi C., Kang X., Wang Y., Zhou Y. (2008). The coagulation factor V Leiden, MTHFRC677T variant and eNOS 4ab polymorphism in young Chinese population with ischemic stroke. Clin. Chim. Acta. 396 (1-2), 7–9. 10.1016/j.cca.2008.06.009 [DOI] [PubMed] [Google Scholar]
- Shi M. (2014). The correlation of homocysteine concentration and MTHFR gene polymorphism with ischemic stroke. China: Zhenzhou University. [Google Scholar]
- Somarajan B. I., Kalita J., Mittal B., Misra U. K. (2011). Evaluation of MTHFR C677T polymorphism in ischemic and hemorrhagic stroke patients. A case-control study in a Northern Indian population. J. Neurol. Sci. 304 (1-2), 67–70. 10.1016/j.jns.2011.02.010 [DOI] [PubMed] [Google Scholar]
- Song Y., Li B., Wang C., Wang P., Gao X., Liu G. (2016). Association between 5, 10-methylenetetrahydrofolate reductase C677T gene polymorphism and risk of ischemic stroke: A meta-analysis. J. Stroke Cerebrovasc. Dis. 25 (3), 679–687. 10.1016/j.jstrokecerebrovasdis.2015.11.041 [DOI] [PubMed] [Google Scholar]
- Stang A. (2010). Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25 (9), 603–605. 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- Sun J. Z., Xu Y., Lu H., Zhu Y. (2009). Polymorphism of the methylenetetrahydrofolate reductase gene association with homocysteine and ischemic stroke in type 2 diabetes. Neurol. India 57 (5), 589–593. 10.4103/0028-3886.57808 [DOI] [PubMed] [Google Scholar]
- Supanc V., Sonicki Z., Vukasovic I., Solter V. V., Zavoreo I., Kes V. B. (2014). The role of classic risk factors and prothrombotic factor gene mutations in ischemic stroke risk development in young and middle-aged individuals. J. Stroke Cerebrovasc. Dis. 23, e171–e176. 10.1016/j.jstrokecerebrovasdis.2013.09.025 [DOI] [PubMed] [Google Scholar]
- Tatarskyy P. F., Kucherenko A. M., Kravchenko S. A., Shulzenko D. V., Kuznetsova S. M., Livshits L. A. (2010). Ischemic stroke in Ukrainian population: Possible involvement of the F2 G20210A, F5 G1691A and MTHFR C677T gene variants. Biopolym. Cell 26, 299–305. 10.7124/bc.000163 [DOI] [Google Scholar]
- Tchantchou F., Goodfellow M., Li F., Ramsue L., Miller C., Puche A., et al. (2021). Hyperhomocysteinemia-induced oxidative stress exacerbates cortical traumatic brain injury outcomes in rats. Cell. Mol. Neurobiol. 41 (3), 487–503. 10.1007/s10571-020-00866-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan M., Chinniah R., Ravi P. M., Sivanadham R., Mosses Joseph A. K., Vellaiappan N. A., et al. (2016). MTHFR (C677T) CT genotype and CT-apoE3/3 genotypic combination predisposes the risk of ischemic stroke. Gene 591 (2), 465–470. 10.1016/j.gene.2016.06.062 [DOI] [PubMed] [Google Scholar]
- Wei L. K., Au A., Menon S., Gan S. H., Griffiths L. R. (2015). Clinical relevance of MTHFR, eNOS, ACE, and ApoE gene polymorphisms and serum vitamin profile among Malay patients with ischemic stroke. J. Stroke Cerebrovasc. Dis. 24 (9), 2017–2025. 10.1016/j.jstrokecerebrovasdis.2015.04.011 [DOI] [PubMed] [Google Scholar]
- Wenping S., Qi W., Mingquan S. (2003). Genetic polymorphisms of 5,10-methylenetetrahydrofolate reductase (MTHFR) in patients with cerebral infarction. Chin. J. Geriatr. Cardiovasc Cerebrovasc. D., 36–38. [Google Scholar]
- Wu Y., Tomon M., Sumino K. (2001). Methylenetetrahydrofolate reductase gene polymorphism and ischemic stroke: Sex difference in Japanese. Kobe J. Med. Sci. 47 (6), 255–262. [PubMed] [Google Scholar]
- Xi H., Zhang Y., Xu Y., Yang W. Y., Jiang X., Sha X., et al. (2016). Caspase-1 inflammasome activation mediates homocysteine-induced pyrop-apoptosis in endothelial cells. Circ. Res. 118 (10), 1525–1539. 10.1161/CIRCRESAHA.116.308501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X. Y., Song Y. Y., Ma J. F., Fan C. N., Ding J. Q., Yang G. Y., et al. (2009). Gene polymorphisms and risk of adult early-onset ischemic stroke: A meta-analysis. Thromb. Res. 124 (5), 619–624. 10.1016/j.thromres.2009.07.007 [DOI] [PubMed] [Google Scholar]
- Xinliang Z., et al. (1999). The relationship of polymorphisms of MTHFR gene and plasma homocysteine levels with stroke. Chin. J. Cardiol. 1, 60–62. [Google Scholar]
- Yan S., Chengguo Z., Xueqiang H. (2006). Relationship of plasma homocysteine polymorphism in its enzyme genes and cerebral infarction in the elderly. J. Clin. Neurol., 22–24. [Google Scholar]
- Yan S., Chengguo Z., Chijin S., Jinhuan C. (2003). Relationship between plasma HCY, polymorphism in MTHFR and cerebral thombosis. Henan J. Pract. Nerv. Dis. 3, 5–7. [Google Scholar]
- Yanqun X., Lingli J., Qing L., Yinhua L., Qinghui Z. (2006). The relationship between the gene polymorphism of methylenetetrahydrofolate reductase and plasma homocysteine level with cerebrovascular disease. Lab. Med. 3, 201–204. [Google Scholar]
- Yaqin Y., Bo L., Jieping S., Le Y., Liping Y. (2001). Study on the relationship between methylenetetrahydrofolate reductase gene polymorphism and acute cerebrovascular disease. J. N. BETHUNEUNIV.Med. Sci. 6, 623–625. 10.13481/j.1671-587x.2001.06.032 [DOI] [Google Scholar]
- Yi F., Jianrong L., Peihua N., Yayun Y., Shengdi C. (2005). The relationship of plasma homocysteine levels and polymorphism in homocysteine metabolism related enzymes with brain stroke. Chin. J. G. eriat, 6, 413–417. [Google Scholar]
- Youk J., An Y., Park S., Lee J. K., Ju Y. S. (2020). The genome-wide landscape of C: G > T: A polymorphism at the CpG contexts in the human population. BMC Genomics 21 (1), 270. 10.1186/s12864-020-6674-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue H., Wang Y., Zhang H., Liu J. (2010). Relationship between the plasma homocysteine levels and the polymorphisms of its metabolic enzymes and the brain infarction. Shanxi Med. J. 39 (02), 108–111. [Google Scholar]
- Zak I., Sarecka-Hujar B., Kopyta I., Emich-Widera E., Marszal E., Wendorff J., et al. (2009). The T allele of the 677C>T polymorphism of methylenetetrahydrofolate reductase gene is associated with an increased risk of ischemic stroke in Polish children. J. Child. Neurol. 24 (10), 1262–1267. 10.1177/0883073809333527 [DOI] [PubMed] [Google Scholar]
- Zhang P., Guo X. (2012). Qibi, correlational study among the polymorphism of MTHFR gene, the plasma tHcy level and acute ischemic stroke in young adult. Chin. J. Stroke 7 (04), 271–277. [Google Scholar]
- Zhang Y., Xie R. P., Shen Y., Fan D. S. (2008). Interaction between methylenetetrahydrofolate reductase C677T gene polymorphism and sleep duration on risk of stroke pathogenesis. Beijing Da Xue Xue Bao Yi Xue Ban. 40 (3), 262–269. [PubMed] [Google Scholar]
- Zhang Y., Zhao Y., Gu B. (2016). Role of Hcy-related C677T gene mutation in hemorrhagic cerebral infarction. Chin. J. Geriatric Heart Brain Vessel Dis. 18 (06), 577–580. [Google Scholar]
- Zheng Y. Z., Tong J., Do X. P., Pu X. Q., Zhou B. T. (2000). Prevalence of methylenetetrahydrofolate reductase C677T and its association with arterial and venous thrombosis in the Chinese population. Br. J. Haematol. 109 (4), 870–874. 10.1046/j.1365-2141.2000.02112.x [DOI] [PubMed] [Google Scholar]
- Zhou B.-S., Bu G. Y., Li M., Chang B. G., Zhou Y. P. (2014). Tagging SNPs in the MTHFR gene and risk of ischemic stroke in a Chinese population. Int. J. Mol. Sci. 15 (5), 8931–8940. 10.3390/ijms15058931 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.