Abstract
Background and aims
Gastric cancer is the fifth most diagnosed malignant tumor worldwide with limited effective chemotherapy. Ferroptosis is a new type of programmed cell death, which is becoming as a novel therapeutic target for tumors. Arenobufagin (ArBu) is a bufadienolide isolated from toad skin and venom, which exhibits broad-spectrum anti-tumor activity. It is unclear whether ArBu causes ferroptosis, thereby exhibiting anti-tumor activity in gastric cancer. We aimed to determine whether ArBu causes ferroptosis in cultured human gastric cancer cells.
Experimental procedure
Different human gastric cancer cells were treated with ArBu (5–20 μM, 48 h). Indicators of apoptosis and ferroptosis were measured. CRISPR/Cas-9 system was employed to delete Nr1d1 gene.
Results
ArBu incubation reduced cell viability in a concentration-dependent manner. ArBu caused ferroptosis but not apoptosis at a lower concentration (10 μM), despite it caused both of them at a higher concentration (20 μM). Cotreatment with a selective ferroptosis inhibitor ferrostatin-1 protected against ArBu (10 μM)-induced reduction in cell viability. ArBu-mediated ferroptosis was associated with abnormal expression of genes involved in iron uptake, lipid peroxidation, and antioxidants. Particularly, Nr1d1 gene expression was most significantly increased after ArBu treatment. Furthermore, activating Rev-erbα encoded by Nr1d1 by a selective agonist GSK4112 (1 and 2 μM, 48 h) caused ferroptosis. In contrast, Rev-erbα knockout using the CRISPR/Cas-9 system diminished ArBu-induced ferroptosis in cultured human gastric cancer cells.
Conclusion
ArBu causes ferroptosis by increasing Rev-erbα expression in human gastric cancer cells. This has implications of ArBu as a promising therapy for gastric cancer.
Section
1. Natural Products;
Taxonomy (classification by EVISE)
Traditional medicine, pharmacology, gastric cancer, signal pathway.
Keywords: Traditional medicine, Lipid peroxidation, Iron metabolism, Antioxidants, CRISPR/cas9 system
Graphical abstract
Highlights
-
•
Gastric cancer cells are sensitive to ArBu-induced ferroptosis vs apoptosis.
-
•
ArBu alters ferroptosis-related gene and Nr1d1 expression.
-
•
Activating Rev-erbα encoded by Nr1d1causes ferroptosis.
-
•
Knockdown of Nr1d1 diminishes ArBu-induced ferroptosis.
-
•
The novelty is that ArBu causes ferroptosis by increasing Nr1d1 expression.
List of abbreviations
- Arenobufagin
ArBu
- coenzyme Q10
CoQ10
- ferroptosis suppressor protein 1
Fsp1
- glutathione peroxidase 4
Gpx4
- glutathione
GSH
- oxidized GSH
GSSG
- malondialdehyde
MDA
- retinoic acid-associated orphan receptor alpha
RORα
1. Introduction
Gastric cancer is the 5th most common cancer with high mortality and recurrence rate as well as poor survival worldwide. Surgical resection is the main curative approach of nonmetastatic gastric cancer. However, the 5-year survival rate of this disease declines quickly when the stage of disease increases. Although much research has been done on the pathogenic mechanism of gastric cancer, few novel chemotherapies have been developed for this disease. This may be due to lacking of understanding of signaling pathways for tumorigenesis and metastasis of gastric cancer.
Ferroptosis is a new type of programmed cell death, which is different from other forms of programmed cell death such as apoptosis.1 Ferroptosis can be induced by excessive iron and the accumulation of lipid peroxides. There are three hallmarks of ferroptosis: abnormal iron metabolism, decreased glutathione (GSH) biosynthesis and increased lipid peroxidation. Iron, as an essential reactive element, participates in numerous biological processes. It has two states including ferrous (Fe2+) or ferric (Fe3+). It has been shown that Fe2+ accumulation triggers and/or initiates ferroptosis. The most common cellular mechanism of protection against ferroptosis is mediated by glutathione peroxidase 4 (Gpx4), a GSH-dependent hydroperoxidase, which can convert lipid peroxides into non-toxic lipid alcohols.2 In this pathway, Slc7a11 is able to transport extracellular cystine into the cell,3 which is subsequently used for GSH synthesis. Gpx4 is able to decrease lipid hydroperoxides and protect against ferroptosis at the expense of reduced GSH.4 Thus, both Slc7a11 and Gpx4 suppress ferroptosis, and inhibition of Slc7a11or Gpx4 can trigger and initiate ferroptosis. Another separate pathway was recently discovered, which involves the oxidoreductase as a ferroptosis suppressor protein 1 (Fsp1).5,6 These studies demonstrate that Fsp1 decreases lipid peroxides by regenerating non-mitochondrial coenzyme Q10 (CoQ10).5,6 It has been shown that activation of ferroptosis causes mitochondrial dysfunction and increases toxic lipid peroxidation in cells, which inhibits tumor growth and progression. Thus, ferroptosis is a novel type of active cancer cell death, which can be induced by classic chemotherapeutic drugs.7 It has been shown that reduced ferroptosis is observed in gastric cancer, and this may result in tumor growth and decrease sensitivity to cisplatin (CDDP) and paclitaxel therapies.8, 9, 10 Therefore, upregulating ferroptosis could inhibit progression, and increase chemosensitivity for gastric cancer.
Toad venom, called Chansu, is obtained from the skin venom gland of the toad as a traditional Chinese medicine.11, 12, 13, 14, 15, 16 Chansu extracts have been reported to contain numerous constituents, including sterols, bufadienolides, indole alkaloids, and organic acids. Arenobufagin (ArBu, C24H32O6) is a bufadienolide compound that is considered as one of the major bioactive ingredients in the extracts.17 These extracts are clinically practiced in patients with non-small cell lung cancer, esophageal cancer, hepatocellular carcinoma, or pancreatic cancer.11, 12, 13, 14, 15, 16 In addition to the cardiotonic and analgesic effects, ArBu is also a broad-spectrum anti-tumor active compound against liver cancer, esophageal squamous cell carcinoma, and non-small cell lung cancer.18, 19, 20, 21, 22 This is associated with modulation of proliferation, apoptosis and autophagy in cancer cells. It is unclear whether ArBu regulates ferroptosis, thereby exhibiting anti-tumor activity in gastric cancer. We hypothesized that ArBu cause ferroptosis in human gastric cancer cells. To test this hypothesis, we treated different human gastric cell lines with ArBu, and determined the indicators of apoptosis and ferroptosis. In addition, we identified Rev-erbα as a target of ArBu in upregulating ferroptosis in cultured human gastric cancer cells.
2. Materials and methods
2.1. Cell culture and treatments
Human gastric adenocarcinoma cells (BGC-823 and MKN-4) were purchased from the ATCC company. These cells were cultured in RPMI 1640 Medium with fetal bovine serum (FBS, 10%) at 37 °C in a humidified atmosphere containing 5% CO2. Cells were passaged every 48 h.
The cells were seeded in a 6-well plate at a density of 1 × 106 cells/2 ml media, which were treated with ArBu (Cayman Chemical, Ann Harbor, MI; 5, 10, 20 μM), CDDP (2 μM),23 ferrostatin-1 (Fer-1, 1 μM),24 a selective Rev-erbα activator GSK4112 (1 μM and 2 μM, Sigma) or vehicle for 48 h.
2.2. Measuring cell viability
We used the Cell Counting Kit-8 Assay Kits (CCK-8; Dojindo, Kumamoto, Japan) to evaluate cell viability. In brief, cells were seeded in a 96-well plate with 1 × 104 cells per well. After treatments, CCK-8 solution (10 μl) was added to each well of the plate, which was then incubated for 1 h. The optical density of each well was determined by using a microplate reader at 450 nm. The relative cell viability was calculated and expressed as the percentage of live cells.
2.3. Evaluating caspase-3 activity
The caspase-3 colorimetric assay kit (BioVision, Milpitas, CA) was used to measure cellular caspase-3 activity according to the manufacturer's instructions. Briefly, cell pellets were lysed in the cell lysis buffer on ice. A total of 100–150 μg proteins were mixed with DEVD-p-nitroanilide substrate in reaction buffer, which contains 10 mM dithiothreitol. The p-nitroanilide light emission was detected using a colorimetric microplate reader with the absorbance at 405 nm.
2.4. Detecting apoptosis by flow cytometry
Annexin V-FITC Apoptosis Detection Kit (Sigma) was used to measure Annexin V+ cells according to the manufacturer's instructions. Briefly, cells were resuspended in Annexin V binding buffer after washing with cold PBS. The Annexin V binding buffer (100 μl) containing 100,000–150,000 cells was incubated with 6 μl of FITC conjugated Annexin V and 6 μl of PI at room temperature. After 10 min of incubation, the mixtures were added with Annexin V binding buffer (300 μl) for analysis using a FACSCalibur™ flow cytometer. The Annexin V+/PI− staining is regarded as apoptosis, and PI positive staining as necrosis. Thus, only annexin V+/PI− cells were counted and were expressed as a percentage of 10,000 events analyzed.
2.5. Iron assay
Intracellular ferrous iron level (Fe2+) was measured using the iron assay kit (#ab83366, Abcam). Briefly, we washed cells with cold PBS, and cell pellets were lysed in iron assay buffer (100 μl). The supernatants were collated and add, then iron reducer (5 μl) was added and mixed, which was incubated at 37 °C for 30 min. Finally, iron probe (100 μl) was added, mixed, and incubated for 1 h at 37 °C. The ferrous iron level was immediately detected in each well using a colorimetric microplate reader with the absorbance at 593 nm.
2.6. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA in cell pellets was extracted using a TRIzol (Life Technologies; Thermo Fisher Scientific, Inc.). A total of 400 ng RNA was used for the reverse transcription to synthesize cDNA using a PrimeScript RT Reagent kit (Perfect Real Time; Takara Bio, Inc., Otsu, Japan) under the condition of 37 °C for 30 min and 85 °C for 5 s qPCR was performed using a 7900 Thermal Cycler (ABI, Applied Biosystems; Thermo Fisher Scientific, Inc.) with GoTaq Green Master Mix (Promega Corporation, Madison, WI, USA). The PCR conditions were set up at an initial denaturation at 95 °C for 30 s, followed by 40 cycles of denaturation for 5 s at 95 °C, annealing for 30 s at 60 °C and extension for 15 s at 72 °C. The qPCR probes were shown in Table 1. Slc7a11, Gpx4, and Fsp1 can inhibit ferroptosis. Thus, we specifically measured their gene expression. The cycle threshold (Ct) values were obtained in each sample. Relative levels of mRNA were calculated using the 2−ΔΔCt method as described previously.25 We used 18S rRNA as a housekeeping gene for normalization.
Table 1.
Probes used in this study.
| Probes | Gene | Protein | Cat# | Company |
|---|---|---|---|---|
| Probes | 18S | 18s RNA | Hs99999901_s1 | ThermoFisher |
| Tfrc | transferrin receptor | Hs00951083_m1 | ||
| Phkg2 | phosphorylase kinase, g2 | Hs04963859_m1 | ||
| Ireb2 | iron response element-binding protein 2 | Hs01021790_m1 | ||
| Hsbp1 | heat-shock 27-kDa protein 1 | Hs00833101_s1 | ||
| Hmox1 | heme oxygenase 1 | Hs01110250_m1 | ||
| Cisd1 | CDGSH iron-sulfur domain 1 | Hs05024086_m1 | ||
| Ncoa4 | nuclear receptor coactivator 4 | Hs00428331_g1 | ||
| Aco1 | aconitase 1 | Hs00158095_m1 | ||
| FTH1 | ferritin heavy chain 1 | Hs01694011_s1 | ||
| Steap3 | six-transmembrane epithelial antigen of prostate 3 | Hs00217292_m1 | ||
| Fancd2 | Fanconi anemia complementation group D2 | Hs00276992_m1 | ||
| Nfs1 | cysteine desulfurase | Hs00221619_m1 | ||
| Nrf2 | nuclear factor erythroid 2-related factor 2 | Hs00975961_g1 | ||
| Keap1 | kelch-like ECH- associated protein 1 | Hs00202227_m1 | ||
| Nqo1 | quinone oxidoreductase-1 | Hs01045993_g1 | ||
| Slc7a11 | solute carrier family 7 member 11 | Hs00921938_m1 | ||
| Gclc | glutamate-cysteine ligase catalytic subunit | Hs00155249_m1 | ||
| Cars | cysteinyl-tRNA synthetase | Hs00156305_m1 | ||
| Cbs | cystathionine-b-synthase | Hs01598251_m1 | ||
| Nox1 | NADPH oxidase 1 | Hs01071088_m1 | ||
| Abcc1 | multidrug-resistance protein 1 | Hs01561483_m1 | ||
| Slc1a5 | solute carrier family 1 member 5 | Hs01056542_m1 | ||
| Gls2 | glutaminase 2 | Hs00998733_m1 | ||
| Got1 | glutamic-oxaloacetic transaminase 1 | Hs00157798_m1 | ||
| G6pd | glucose-6-phosphate dehydrogenase | Hs00166169_m1 | ||
| Acsf2 | acyl-CoA synthetase family member 2 | Hs00910503_g1 | ||
| CS | citrate synthase | Hs02574374_s1 | ||
| Lpcat3 | Lysophosphatidylcholine acyltransferase 3 | Hs01553683_m1 | ||
| Acsl3 | acyl-CoA synthetase longchain family member 3 | Hs00244853_m1 | ||
| Acsl4 | acyl-CoA synthetase longchain family member 4 | Hs00244871_m1 | ||
| Acaca | Acetyl-CoA carboxylase alpha | Hs01046047_m1 | ||
| Hs07292504_s1 | ||||
| Akr1c | aldo-keto reductase family 1 member C1 | Hs00942483_m1 | ||
| Lox | lipoxygenase | Hs00831506_g1 | ||
| Pebp1 | Phosphatidylethanolaminebinding protein 1 | Hs01566408_m1 | ||
| Zeb1 | zinc finger E-box-binding homeobox 1 | Hs00926054_m1 | ||
| Sqs | squalene synthase/farnesyldiphosphate farnesyltransferase 1 | Hs00168352_m1 | ||
| Hmgcr | 3-hydroxy-3-methyl-glutarylcoenzyme | Hs00927433_m1 | ||
| A reductase | ||||
| Fads2 | fatty acid desaturase 2/acyl- | Hs00253876_m1 | ||
| CoA 6-desaturase | ||||
| Nr1d1 | Rev-erbα | Hs00253876_m1 |
2.7. GSH assay
Total GSH levels in cell lysates were evaluated as described previously.26 In brief, cells were collected and lysed using phosphate buffer (0.1 M, pH 7.5) which contained Triton X-100 (0.1%, v/v), EDTA (5 mM) and sulfosalicylic acid (0.6%,wt/vol). Cell lysates were centrifuged at 14,000 rpm for 5 min. Supernatant was added with glutathione reductase (1.67 U/ml) and 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB, 0.2 mg/ml) in phosphate buffer-EDTA, which was incubated for 30 s. In the reaction mixture, β-NADPH (0.2 mg/ml) was added, and the rate of DTNB reduction was detected using a microplate reader at a wavelength of 405 nm. Total GSH levels were calculated by comparing the DTNB reduction rate with known concentrations of GSH. For evaluating the levels of oxidized GSH (GSSG), cell lysates were incubated with 2-vinylpyridine (2%) to derivatize GSH. The levels of GSSG was assessed by detecting NADPH at 405 nm using a microplate reader.26
2.8. Evaluation of malondialdehyde (MDA) levels
After washing with PBS, cells were collected and lysed with ice-cold Tris-HCl (20 mM, pH 7.4) for 30 min. After centrifugation at 3000 rpm for 10 min at 4 °C, the supernatants were added with butylated hydroxytoluene (5 mM, Sigma-Aldrich, MO, USA). We measured MDA in the samples using a lipid peroxidation kit (Abcam, Waltham, MA). Briefly, 650 μl of 10.3 mM N-methyl-2-phenylindole in acetonitrile was added into the aforementioned supernatant mixture (200 μl), and this were further mixed with 150 μl of 15.4 mM methanesulfonic acid. This was then incubated at 45 °C for 40 min. The optical absorbance was detected at 586 nm using a microplate reader (Bio-Rad, San Diego, CA, USA). The protein levels in each sample were used for normalization.
2.9. Extraction and quantification of coenzyme Q10 (CoQ10)
Intracellular CoQ10 levels were determined as described previously.27 We prepared NP-40 lysis-buffer, which contained Nonidet P-40 (0.1%, v/v), EDTA (1 mM), NaCl (250 mM), glycerol (10%), Tris-HCl buffer (50 mM, pH 7.4), and protease inhibitor (1%). After washing with PBS, cells were collected and lysed in 100 μl ice-cold NP-40 lysis-buffer for 30 min. We then added ice-cold ethanol/n-hexane (2:5, v/v) mixture (700 μl) into cell lysate (50 μl) with incubation at 4 °C for 10 min. The CoQ10 was extracted and quantified according to the method of Okamoto et al.28 The protein levels in each sample were used to normalize total CoQ10 contents of cells.
2.10. Transfection
We used an RNA-guided CRISPR/Cas9-mediated genome editing approach to knockout Rev-erbα gene in BGC-823 cells. Rev-erbα targeting sgRNAs were purchased from Santa Cruz (Cat#: SC-401211), which was cloned into a lentiCRISPRv1 plasmid (Addgene, Cambridge, MA). This generated lentiCRISPR-Rev-erbα-sgRNA vector. We used a lentiCRISPRv1 plasmid which expressed an EGFP targeting sgRNA (Addgene) only to generate control lentiCRISPR-EGFP-sgRNA vector as previously described.29 Cells were seeded in a 6-well cell culture plate at 1 × 105 cells/well in medium and transfected with lentiCRISPR-Rev-erbα-sgRNA vector or control lentiCRISPR-EGFP-sgRNA vector for 48 h.29 Cells were then selected in medium containing puromycin (1 μg/ml) for 2 weeks. Rev-erbα expression was determined by qRT-PCR.
2.11. Statistical analyses
All experiments were carried out with at least three biological replicates. Data are expressed as the mean ± standard error of mean. For multiple comparisons, the statistical significance of the differences was evaluated by using one-way ANOVA followed by Tukey's post-test to specifically compare indicated groups. Data analysis was performed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). A value of p < 0.05 was considered statistically significant.
3. Results
3.1. ArBu incubation reduces viability and causes apoptosis in cultured human gastric cancer cells
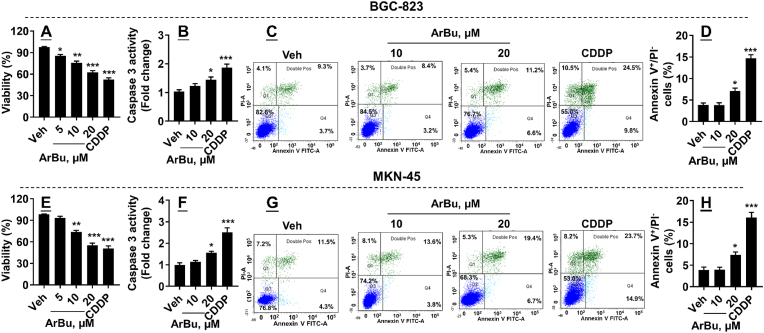
We first measured cell viability in undifferentiated BGC-823 cells incubated with ArBu (5–20 μM, 48 h). As shown in Fig. 1A, ArBu significantly reduced cell viability in a concentration-dependent manner. We then chose ArBu (10 and 20 μM) to treat cells, and measured caspase-3 activity and annexin V+/PI− cells, parameters of apoptosis, in these cells. As shown in Fig. 1B–D, ArBu increased caspase-3 activity and annexin V+/PI− cells at 20 μM but not at 10 μM in BGC-823 cells. Similarly, ArBu significantly reduced cell viability in a concentration-dependent manner in poorly differentiated MKN-45 cells (Fig. 1E). Additionally, ArBu increased caspase-3 activity and annexin V+/PI− cells in MKN-45 cells (Fig. 1F–H). As a positive control, CDDP incubation reduced viability and caused apoptosis in both BGC-823 and MKN-45 cells (Fig. 1). These results suggest that ArBu causes apoptosis in human gastric cancer at a higher concentration.
Fig. 1.
ArBu treatment reduced viability and caused apoptosis in BGC-823 and MKN-45 cells. BGC-823 and MKN-45 cells were treated with ArBu (5, 10, and 20 μM) for 48 h. (A, E) Cell viability was detected by using a Cell Counting Kit-8 Assay Kit. (B, F) Intracellular caspase-3 activity was measured using commercial kits. (C, D, G, H) Flow cytometry was performed to detect Annexin V+/PI− cells. N = 6. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs vehicle.
3.2. ArBu incubation causes ferroptosis at a lower concentration
As show in Fig. 1, ArBu reduced viability at 10 μM, and this dose of Arbu did not cause apoptosis, suggesting other types of cell death exist. Thus, we measured indicators of ferroptosis in these cells incubated with ArBu (10 and 20 μM). As shown in Fig. 2A, the content of cellular iron was increased in cells treated with ArBu at both 10 and 20 μM compared to vehicle group in BGC-823 cells. Incubation of ArBu significantly increased level of MDA, the end product of lipid peroxidation (Fig. 2B). GSSG/GSH ratio was increased in cells treated with ArBu (Fig. 2C).
Fig. 2.
ArBu treatment caused ferroptosis in BGC-823 and MKN-45 cells. BGC-823 and MKN-45 cells were treated with ArBu (10 and 20 μM) for 48 h (A, B, G, H) Intracellular iron and MDA levels were measured using commercial kits. (C, I) Cellular GSH and GSSG were measured and the ratio of GSSG to GSH was calculated. (D, E, J) qRT-PCR was performed to measure Slc7a11, Gpx4 and Fsp1 gene expression. (F, K) Intracellular CoQ10 was measured in cells treated with ArBu. N = 6. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs vehicle.
As shown in Fig. 2D and E, ArBu incubation reduced expression of Slc7a11 and Gpx4 in BGC-823 cells. In addition, the levels of Fsp1 and CoQ10 were decreased by ArBu treatment in BGC-823 cells (Fig. 2F). Similarly, ArBu incubation both 10 and 20 μM increased levels of iron, MDA, and GSSG/GSH ratio, but reduced levels of Slc7a11, Gpx4, Fsp1 and CoQ10 in MKN-45 cells (Fig. 2G–K). As expected,23 CDDP treatment also caused ferroptosis in both BGC-823 and MKN-45 cells. Altogether, these results demonstrate that ArBu treatment causes ferroptosis in human gastric cancer cells at a lower concentration.
3.3. Inhibition of ferroptosis protects against ArBu-mediated reduction in cell viability
To determine whether ferroptosis contributes to ArBu-mediated cell death, we chose 10 μM of ArBu to treat cells, as this concentration caused ferroptosis but not apoptosis (Fig. 1, Fig. 2). These cells were also co-treated with a selective ferroptosis inhibitor Fer-1 (1 μM). As expected, ArBu treatment at 10 μM caused ferroptosis in both BGC-823 and MKN-45 cells (Fig. 3A–D, E-I). Co-treatment with Fer-1 significantly attenuated ArBu-induced ferroptosis in these cells (Fig. 3A–D, E-I). Importantly, ArBu-mediated reduction of cell viability was significantly restored by Fer-1 co-treatment (Fig. 3E, J). Altogether, ferroptosis contributes to ArBu-mediated cell death in human gastric cancer cells.
Fig. 3.
Inhibiting ferroptosis protected against ArBu-mediated reduction in viability in BGC-823 and MKN-45 cells. BGC-823 and MKN-45 cells were treated with ArBu (10 μM) in the absence or presence of Fer-1 (1 μM) for 48 h (A, B, F, G) Intracellular iron and MDA levels were measured using commercial kits. (C, D, H, I) qRT-PCR was performed to measure Slc7a11 and Gpx4 gene expression. (E, J) Cell viability was detected by using a Cell Counting Kit-8 Assay Kit. N = 6. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs vehicle; †P < 0.05, ††P < 0.01, †††P < 0.001 vs veh/ArBu group.
3.4. ArBu incubation alters ferroptosis-related gene expression in human gastric cancer cells
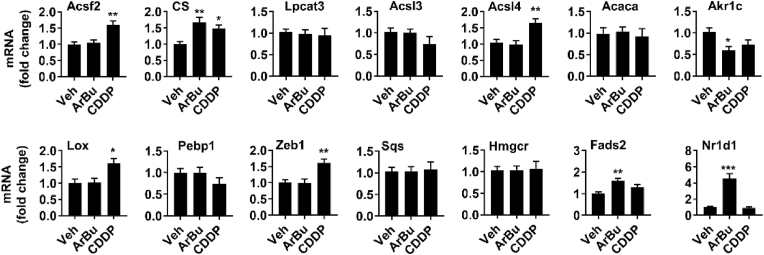
There are ∼40 identified genes which are associate with iron or energy metabolism, lipid synthesis, and oxidative stress, and they could regulate the sensitivity to ferroptosis.30 Thus, we characterized expression of these genes in BGC-823 cells incubated with ArBu (20 μM, 48 h). As shown in Fig. 4, Fig. 5, expression of genes involved in iron metabolism (Tfrc, Ireb2, Cisd1, Ncoa4, Fth1, and Nsf1), (anti)oxidant metabolism (Nrf2, Gclc, and Nox), energy metabolism (Slc1a5, Gls2, and Got1), and lipid metabolism (CS, Akr1c, Fads2, and Nr1d1) was altered in BGC-823 cells incubated with ArBu. CDDP treatment also altered expression related to iron, (anti)oxidant and lipid metabolism. Altogether, ArBu increases iron uptake, lipid peroxidation, and lowers antioxidants in cultured human gastric cancer cells.
Fig. 4.
ArBu treatment altered expression of genes involved in iron, energy and (anti)oxidant metabolism in BGC-823 cells. BGC-823 cells were treated with ArBu (20 μM) for 48 h qRT-PCR was performed to measure expression of genes involved in iron, energy (A) and (anti)oxidant (B) metabolism. N = 6. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs vehicle.
Fig. 5.
ArBu treatment altered expression of genes involved in lipid metabolism in BGC-823 cells. BGC-823 cells were treated with ArBu (20 μM) for 48 h qRT-PCR was performed to measure gene expression in lipid metabolism. N = 6. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs vehicle.
3.5. Activating Rev-erbα causes ferroptosis in gastric cancer cells
We have shown that Rev-erbα reduction correlates with clinicopathological features and prognosis in human gastric cancer.31,32 Rev-erbα has been shown to modulate ferroptosis in folic acid-induced ferroptosis in the kidney.33,34 Importantly, Rev-erbα was highly increased in cells treated with ArBu compared to other genes. Thus, we investigated whether Rev-erbα activation causes ferroptosis in BCG-823 cells. To this effect, we treated cells with a selective Rev-erbα agonist GSK4112 (1 and 2 μM) for 48 h. Rev-erbα represses expression of target genes including Bmal1, Cry1 and PGC1 through the RORE-containing regulatory regions.35 As shown in Fig. 6A, GSK4112 treatment significantly decreased expression of these target genes, suggesting the functional consequence of Rev-erbα activation. In addition, cell viability was significantly decreased after GSK4112 treatment (Fig. 6B). Furthermore, incubation with GSK4112 significantly increased the content of cellular iron and lipid peroxidation product MDA as well as reduced antioxidant levels (GSH, Gpx4, and CoQ10) in BCG-823 cells (Fig. 6C–J). These results suggest that activating Rev-erbα by GSK4112 causes ferroptosis in gastric cancer cells.
Fig. 6.
Activating Rev-erbα reduced viability and caused ferroptosis in BGC-823 cells. BGC-823 cells were treated with GSK4112 (1 and 2 μM) for 48 h. (A) qRT-PCR was performed to measure Bmal1, Cry1, and Pgc1α gene expression. (B) Cell viability was detected by using a Cell Counting Kit-8 Assay Kit. (C, D) Intracellular iron and MDA levels were measured using commercial kits. (E–G) Cellular GSH and GSSG were measured and the ratio of GSSG to GSH was calculated. (H, I) qRT-PCR was performed to measure Slc7a11, GPX, and Fsp1 gene expression. (J) Intracellular CoQ10 was measured in cells treated with ArBu. N = 6. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs vehicle.
3.6. Knockdown of Rev-erbα diminishes ArBu-induced ferroptosis in gastric cancer cells
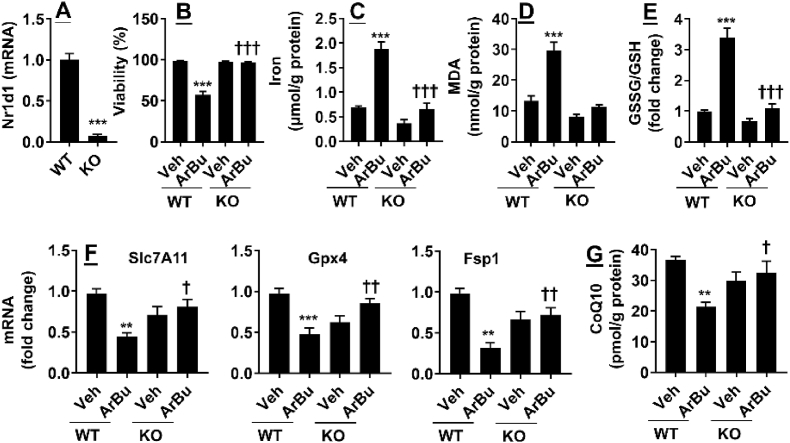
To determine whether ArBu causes ferroptosis by increasing Rev-erbα, we knocked out Rev-erbα using the CRISPR/cas9 system in BCG-823 cells. As shown in Fig. 7A, Nr1d1 gene expression was significantly reduced in cells transfected with lentiCRISPR-Rev-erbα-sgRNA. Cell viability was significantly decreased after ArBu treatment (20 μM, 48 h), and these effects were abolished in Rev-erbα knockout cells (Fig. 7B). As expected, ArBu treatment (20 μM, 48 h) significantly increased iron content and MDA, and reduced antioxidant (GSH, Gpx4, Fsp1, and CoQ10) (Fig. 7C–G). These effects were diminished in Rev-erbα knockout cells treated with ArBu (Fig. 7C–G). These results demonstrate that Nr1d1 gene deletion abolishes ArBu-mediated ferroptosis in cultured human gastric cancer cells.
Fig. 7.
Knockdown of Rev-erbα abolished ArBu-induced viability reduction and ferroptosis in BGC-823 cells. BGC-823 cells were treated with ArBu (20 μM) for 48 h after Rev-erbα knockout using the CRISPR/Cas-9 system. (A) qRT-PCR was performed to measure Nr1d1 gene expression. (B) Cell viability was detected by using a Cell Counting Kit-8 Assay Kit. (C, D) Intracellular iron and MDA levels were measured using commercial kits. (E) Cellular GSH and GSSG were measured and the ratio of GSSG to GSH was calculated. (F) qRT-PCR was performed to measure Slc7a11, Gpx4, and Fsp1 gene expression. (G) Intracellular CoQ10 was measured in cells treated with ArBu. N = 6. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs WT/vehicle; †P < 0.05, ††P < 0.01, †††P < 0.001 vs WT/ArBu group.
4. Discussion
Few novel chemotherapies for gastric cancer have been developed due to the difficulties in target identification and validation. Ferroptosis is significantly inhibited in gastric cancer, which results in tumor growth. Therefore, upregulating ferroptosis is a promising approach of chemotherapy for gastric cancer. Here we reported that ArBu treatment increased ferroptosis in cultured human gastric cancer cells. This was associated with increased Rev-erbα expression. Furthermore, activating Rev-erbα caused ferroptosis, whereas knockout of Rev-erbα diminished ArBu-induced ferroptosis in cultured human gastric cancer cells. Therefore, ArBu causes ferroptosis by increasing Rev-erbα expression in human gastric cancer cells.
ArBu possesses significant antineoplastic activity in vitro and in vivo.18, 19, 20,22,36,37 This is associated the regulation of proliferation, cell cycle, apoptosis, autophagy, and migration. There are no reports on the modulation of ArBu on ferroptosis. The present study for the first time revealed that ArBu treatment caused ferroptosis in cultured human gastric cancer cells. Compared to apoptosis, ArBu caused ferroptosis at a lower concentration in these cells. This suggests that these cells are more sensitive to ArBu-induced ferroptosis. Autophagy is a cellular process and promotes cell survival following stress or nutrient limitation by recycling and/or degrading cellular components. Although autophagy also accompanies cell death following many toxic insults, the requirement of autophagy for apoptosis or ferroptosis is highly contextual.38,39 Further study is warranted to determine the role of autophagy in ArBu-induced apoptosis and ferroptosis through specific genetic or chemical inhibition of the process in human gastric cancer cells.
Tfrc gene encodes the transferrin receptor, which is responsible for iron uptake, while Cisd1 is iron-containing outer mitochondrial membrane protein. Tfrc is a specific ferroptosis marker and Cisd1 knockdown augments iron-induced lipid peroxidation and erastin-induced ferroptosis.40,41 Genetic inhibition of CS inhibits erastin-induced ferroptosis, whereas Akr1c protects against stress-induced ferroptosis.42,43 ArBu increased expression of Tfrc and CS genes, and reduced Cisd1 and Akr1c gene expression. In addition, ArBu also altered other gene expression involved in iron metabolism (Ireb2, Ncoa4, Fth1, and Nsf1), (anti)oxidant metabolism (Nrf2, Gclc, and Nox), energy metabolism (Slc1a5, Gls2, and Got1), and lipid metabolism (Fads2, and Nr1d1). This agrees with the findings that ArBu augmented MDA but reduced GSH levels in adenocarcinomic human alveolar basal epithelial cells (A549).44 Further study is required to determine the beneficial effects of ArBu against gastric cancer using animal models. Ferroptosis is aberrant expressed in gastric tumors.45 Recently, some reports suggested that ferroptosis could be an effective therapeutic strategy to ameliorate chemotherapy resistance.45 We noticed that CDDP affected different gene expression (e.g., Ireb2, Hmox1, Nqo1, and Zeb2) compared to ArBu. This suggests that CDDP and ArBu cause ferroptosis through different mechanisms. Therefore, ArBu could be a promising therapy for preventing the progression, alleviating chemotherapy resistance, enhancing CDDP chemotherapy sensitivity in gastric cancer. A previous report has shown that ArBu can enter the nucleus, and intercalates with DNA leading to G2 cell cycle arrest.46 Further study is warranted to determine whether ArBu enters the nucleus and modulates transcription factors, therefore altering ferroptosis-related gene transcription in gastric cancer cells.
Rev-erbα encoded by Nr1d1 gene is a nuclear receptor, which is an essential molecular clock that drives the daily rhythms of metabolism. Rev-erb family members play pivotal roles in modulating pathological processes including sleep disorders, diabetes, atherosclerosis, Alzheimer's disease, and tumors by regulating the biological clock, inflammatory/immune responses, and lipid metabolism. Rev-erbα mediates transcription repression via two pathways. Firstly, Rev-erbα binds to the ROR response element (RORE) of targeting genes, including Bmal1 gene, by competing with the retinoic acid-associated orphan receptor alpha (RORα), leading to inhibition of these gene transcription.47 Additionally, Rev-erbα could repress gene transcription through directly binding to target response elements and/or by recruiting the corepressor, including histone deacetylases, on targeting gene promoters.48 The expression of genes involved in fatty acid metabolism (CD36, Fabp3, and Fabp4) were decreased in cells containing Rev-erb mutants.47 Rev-erbα activation may increase mitochondrial respiration, leading to increased reactive oxygen species generation and subsequent lipid peroxidation. Further study is warranted to determine how Rev-erbα altered expression of genes involved in iron and energy metabolism as well as (anti)oxidant stress. Recently, Rev-erb-α/β causes ferroptosis by inhibiting the expression of Slc7a11 and Hmox1 genes by direct binding to a RORE cis-element.33,34 Consistently, Slc7a11 gene expression was deceased by GSK4112 treatment, whereas Rev-erbα deficiency increased Slc7a11 gene expression in human gastric cancer cells. Therefore, Rev-erbα activation reduced intracellular GSH synthesis by repressing Slc7a11 and subsequent cystine import. As the target genes of Rev-erbα and circadian clock, Bmal1 protein degradation promotes ferroptosis along with Cry1 reduction.49 Pgc1α is an essential transcription co-activator which plays important roles in mitochondrial energy metabolism. Increased Pgc1α is associated with puerarin's protection against ferroptosis after subarachnoid hemorrhage in rats.50 Therefore, Rev-erbα may promote ferroptosis by modulating clock machinery or Pgc1α in human gastric cancer cells. Since Hmox1 was unchanged by ArBu treatment, we did not measure it gene expression in cells treated with a selective Rev-erbα agonist or deficient of Rev-erbα.
In conclusion, ArBu treatment causes ferroptosis in human gastric cancer cells, which is associated with dysregulation of iron and lipid metabolism as well as reduced antioxidants. Co-treatment with a selective ferroptosis inhibitor ferrostatin-1 protected against ArBu-induced reduction in cell viability. Activating Rev-erbα causes ferroptosis, whereas Rev-erbα diminishes ArBu-induced ferroptosis in human gastric cancer cells. Thus, ArBu causes ferroptosis by increasing Rev-erbα in gastric cancer. This has implications of ArBu as a promising therapy for gastric cancer.
Footnotes
Peer review under responsibility of The Center for Food and Biomolecules, National Taiwan University.
Author contributions
CK, LA, WJ, LD, WX, LC, and WZ performed experiments and data analysis. WZ designed the study and drafted the manuscript.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
This study was supported by the Anhui Provincial Natural Science Foundation (2008085MH294), and the Cultivating Program of National Natural Science Foundation for Young Scholars of the First Affiliated Hospital of Anhui Medical University (Grant No. 2019KJ19).
Footnotes
Peer review under responsibility of The Center for Food and Biomolecules, National Taiwan University.
References
- 1.Dixon S.J., Lemberg K.M., Lamprecht M.R., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang W.S., Stockwell B.R. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 2016;26(3):165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conrad M., Sato H. The oxidative stress-inducible cystine/glutamate antiporter, system x (c) (-) : cystine supplier and beyond. Amino Acids. 2012;42(1):231–246. doi: 10.1007/s00726-011-0867-5. [DOI] [PubMed] [Google Scholar]
- 4.Koppula P., Zhuang L., Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 2021;12(8):599–620. doi: 10.1007/s13238-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bersuker K., Hendricks J.M., Li Z., et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575(7784):688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doll S., Freitas F.P., Shah R., et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575(7784):693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 7.Wu Y., Yu C., Luo M., et al. Ferroptosis in cancer treatment: another way to rome. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.571127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H., Deng T., Liu R., et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol Cancer. 2020;19(1):43. doi: 10.1186/s12943-020-01168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hao S., Yu J., He W., et al. Cysteine dioxygenase 1 mediates erastin-induced ferroptosis in human gastric cancer cells. Neoplasia. 2017;19(12):1022–1032. doi: 10.1016/j.neo.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao S., Liu X., Yuan L., Chen X., Wang F. Expression of ferroptosis-related genes shapes tumor microenvironment and pharmacological profile in gastric cancer. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.694003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma J., Lu M. Clinical research on 109 cases of non-small cell Lung cancer treated by cinobutacini injection plus gemcitabine and cisplatin. J Tradit Chin Med. 2011;52:2115–2118. [Google Scholar]
- 12.Cao J., Wang Y., Ge X., Zhou J. Clinical observation on toad venom injection in combination with chemotherapy treatment of advanced malignant tumor. Liaoning J Tradit Chin Med. 2005;32:36–37. [Google Scholar]
- 13.Zhao J., Xing L., Li W., Zhang S., Li G., Qian H. Clinical study on efficacy of bufalin injection combined with chemotherapy in the treatment of advanced non small cell lung cancer. Lishizhen Med and Mate Med Res. 2006;17:1604–1605. [Google Scholar]
- 14.Shang A., Deng B., Li C. Toad venom injection treatment of advanced esophageal cancer with radiotherapy 31 cases. China Naturopa. 2005;13:51–52. [Google Scholar]
- 15.Yang Y., Xiong Y. Clinical observation of toad venom Jieninggao treatment of 120 advanced cancer cases of severe pain. Chin West Med J. 2006;4:58–60. [Google Scholar]
- 16.Meng Z., Yang P., Shen Y., et al. Pilot study of huachansu in patients with hepatocellular carcinoma, nonsmall-cell lung cancer, or pancreatic cancer. Cancer. 2009;115(22):5309–5318. doi: 10.1002/cncr.24602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J.H., Cao Y.T., Pan H.Y., Wang L.H. Identification of antitumor constituents in toad venom by spectrum-effect relationship analysis and investigation on its pharmacologic mechanism. Molecules. 2020;25(18) doi: 10.3390/molecules25184269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang D.M., Liu J.S., Deng L.J., et al. Arenobufagin, a natural bufadienolide from toad venom, induces apoptosis and autophagy in human hepatocellular carcinoma cells through inhibition of PI3K/Akt/mTOR pathway. Carcinogenesis. 2013;34(6):1331–1342. doi: 10.1093/carcin/bgt060. [DOI] [PubMed] [Google Scholar]
- 19.Lv J., Lin S., Peng P., et al. Arenobufagin activates p53 to trigger esophageal squamous cell carcinoma cell apoptosis in vitro and in vivo. OncoTargets Ther. 2017;10:1261–1267. doi: 10.2147/OTT.S104767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma L., Zhu Y., Fang S., Long H., Liu X., Liu Z. Arenobufagin induces apoptotic cell death in human non-small-cell lung cancer cells via the noxa-related pathway. Molecules. 2017;22(9) doi: 10.3390/molecules22091525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei X., Yang J., Mao Y., et al. Arenobufagin inhibits the phosphatidylinositol 3-kinase/Protein kinase B/mammalian target of rapamycin pathway and induces apoptosis and autophagy in pancreatic cancer cells. Pancreas. 2020;49(2):261–272. doi: 10.1097/MPA.0000000000001471. [DOI] [PubMed] [Google Scholar]
- 22.Zhao L.J., Zhao H.Y., Wei X.L., et al. The lipid homeostasis regulation study of arenobufagin in zebrafish HepG2 xenograft model and HepG2 cells using integrated lipidomics-proteomics approach. J Ethnopharmacol. 2020;260 doi: 10.1016/j.jep.2020.112943. [DOI] [PubMed] [Google Scholar]
- 23.Guo J., Xu B., Han Q., et al. Ferroptosis: a novel anti-tumor action for cisplatin. Cancer Res Treat. 2018;50(2):445–460. doi: 10.4143/crt.2016.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bi J., Yang S., Li L., et al. Metadherin enhances vulnerability of cancer cells to ferroptosis. Cell Death Dis. 2019;10(10):682. doi: 10.1038/s41419-019-1897-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao X., Huang X., Zhou Z., Lin X. An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath. 2013;3(3):71–85. [PMC free article] [PubMed] [Google Scholar]
- 26.Rahman I., Kode A., Biswas S.K. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat Protoc. 2006;1(6):3159–3165. doi: 10.1038/nprot.2006.378. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi T., Mine Y., Okamoto T. Extracellular coenzyme Q10 (CoQ10) is reduced to ubiquinol-10 by intact Hep G2 cells independent of intracellular CoQ10 reduction. Arch Biochem Biophys. 2019;672 doi: 10.1016/j.abb.2019.108067. [DOI] [PubMed] [Google Scholar]
- 28.Okamoto T., Fukunaga Y., Ida Y., Kishi T. Determination of reduced and total ubiquinones in biological materials by liquid chromatography with electrochemical detection. J Chromatogr. 1988;430(1):11–19. doi: 10.1016/s0378-4347(00)83129-1. [DOI] [PubMed] [Google Scholar]
- 29.Shalem O., Sanjana N.E., Hartenian E., et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassannia B., Vandenabeele P., Vanden Berghe T. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35(6):830–849. doi: 10.1016/j.ccell.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Wang X., Wang N., Wei X., Yu H., Wang Z. REV-ERBα reduction correlates with clinicopathological features and prognosis in human gastric cancer. Oncol Lett. 2018;16(2):1499–1506. doi: 10.3892/ol.2018.8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Jia R., Chen K., Wang J., Jiang K., Wang Z. RORalpha and REV-ERBalpha are associated with clinicopathological parameters and are independent biomarkers of prognosis in gastric cancer. Technol Cancer Res Treat. 2021;20 doi: 10.1177/15330338211039670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo L., Zhang T., Wang F., et al. Targeted inhibition of Rev-erb-alpha/beta limits ferroptosis to ameliorate folic acid-induced acute kidney injury. Br J Pharmacol. 2021;178(2):328–345. doi: 10.1111/bph.15283. [DOI] [PubMed] [Google Scholar]
- 34.Hu M., An S. Ruscogenin prevents folic acid-induced acute kidney damage by inhibiting rev-erbalpha/beta-mediated ferroptosis. Comput Intell Neurosci. 2022;2022 doi: 10.1155/2022/8066126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant D., Yin L., Collins J.L., et al. GSK4112, a small molecule chemical probe for the cell biology of the nuclear heme receptor Rev-erbalpha. ACS Chem Biol. 2010;5(10):925–932. doi: 10.1021/cb100141y. [DOI] [PubMed] [Google Scholar]
- 36.Delebinski C.I., Georgi S., Kleinsimon S., et al. Analysis of proliferation and apoptotic induction by 20 steroid glycosides in 143B osteosarcoma cells in vitro. Cell Prolif. 2015;48(5):600–610. doi: 10.1111/cpr.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao J., Zhang Q., Zou G., Gao G., Yue Q. Arenobufagin, isolated from toad venom, inhibited epithelial-to-mesenchymal transition and suppressed migration and invasion of lung cancer cells via targeting IKKbeta/NFkappaB signal cascade. J Ethnopharmacol. 2020;250 doi: 10.1016/j.jep.2019.112492. [DOI] [PubMed] [Google Scholar]
- 38.Gao M., Monian P., Pan Q., Zhang W., Xiang J., Jiang X. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26(9):1021–1032. doi: 10.1038/cr.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denton D., Kumar S. Autophagy-dependent cell death. Cell Death Differ. 2019;26(4):605–616. doi: 10.1038/s41418-018-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng H., Schorpp K., Jin J., et al. Transferrin receptor is a specific ferroptosis marker. Cell Rep. 2020;30(10):3411–3423. doi: 10.1016/j.celrep.2020.02.049. e3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan H., Li X., Zhang X., Kang R., Tang D. CISD1 inhibits ferroptosis by protection against mitochondrial lipid peroxidation. Biochem Biophys Res Commun. 2016;478(2):838–844. doi: 10.1016/j.bbrc.2016.08.034. [DOI] [PubMed] [Google Scholar]
- 42.Gagliardi M., Cotella D., Santoro C., et al. Aldo-keto reductases protect metastatic melanoma from ER stress-independent ferroptosis. Cell Death Dis. 2019;10(12):902. doi: 10.1038/s41419-019-2143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stockwell B.R., Friedmann Angeli J.P., Bayir H., et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kan J., Huang H., Jiang Z., et al. Arenobufagin promoted oxidative stress-associated mitochondrial pathway apoptosis in A549 non-small-cell lung cancer cell line. Evid Based Complement Alternat Med. 2020;2020 doi: 10.1155/2020/8909171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Si C., Zhou X., Deng J., et al. Role of ferroptosis in gastrointestinal tumors: from mechanisms to therapies. Cell Biol Int. 2022;46(7):997–1008. doi: 10.1002/cbin.11804. [DOI] [PubMed] [Google Scholar]
- 46.Deng L.J., Peng Q.L., Wang L.H., et al. Arenobufagin intercalates with DNA leading to G2 cell cycle arrest via ATM/ATR pathway. Oncotarget. 2015;6(33):34258–34275. doi: 10.18632/oncotarget.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho H., Zhao X., Hatori M., et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485(7396):123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jetten A.M., Kang H.S., Takeda Y. Retinoic acid-related orphan receptors alpha and gamma: key regulators of lipid/glucose metabolism, inflammation, and insulin sensitivity. Front Endocrinol. 2013;4:1. doi: 10.3389/fendo.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang M., Chen P., Liu J., et al. Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci Adv. 2019;5(7) doi: 10.1126/sciadv.aaw2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang Y., Wu H., Hu Y., et al. Puerarin attenuates oxidative stress and ferroptosis via AMPK/PGC1alpha/Nrf2 pathway after subarachnoid hemorrhage in rats. Antioxidants. 2022;11(7) doi: 10.3390/antiox11071259. [DOI] [PMC free article] [PubMed] [Google Scholar]