ABSTRACT
Echinoderms represent a broad phylum with many tractable features to test evolutionary changes and constraints. Here, we present a single-cell RNA-sequencing analysis of early development in the sea star Patiria miniata, to complement the recent analysis of two sea urchin species. We identified 20 cell states across six developmental stages from 8 hpf to mid-gastrula stage, using the analysis of 25,703 cells. The clusters were assigned cell states based on known marker gene expression and by in situ RNA hybridization. We found that early (morula, 8-14 hpf) and late (blastula-to-mid-gastrula) cell states are transcriptionally distinct. Cells surrounding the blastopore undergo rapid cell state changes that include endomesoderm diversification. Of particular import to understanding germ cell specification is that we never see Nodal pathway members within Nanos/Vasa-positive cells in the region known to give rise to the primordial germ cells (PGCs). The results from this work contrast the results of PGC specification in the sea urchin, and the dataset presented here enables deeper comparative studies in tractable developmental models for testing a variety of developmental mechanisms.
Keywords: Inductive germline, Primordial germ cell (PGC), Vasa, Nanos, Sea star, Sea urchin, Cell fates, scRNA-seq
Highlighted Article: Single-cell RNA-sequencing analysis of early sea star development contrasts the results of primordial germ cell specification in the sea urchin, and enables deeper comparative studies in tractable developmental models.
INTRODUCTION
Revealing the genetic regulation of cell type identity leads to a deeper understanding of the mechanisms of embryogenesis and evolutionary diversity. Studies of developmental mechanisms between closely related vertebrate and invertebrate models have been fruitful in revealing evolutionary changes that include gene duplications followed by divergence, and changes in promoter elements that shift important kernels in gene regulatory networks (Briggs et al., 2018; Davidson, 2006). The sea star and sea urchin have recently been subject to comparative analyses of developmental heterochronies, gene regulatory networks and neurogenesis (Cary et al., 2020; Gildor et al., 2017; Hinman and Burke, 2018). Sea stars and sea urchins spawn millions of gametes, which, when fertilized, will develop external to the adults. The transparent embryos can be easily cultured, treated, dissociated and analyzed in great quantities. Single-cell atlases of development to larvae in echinoderm species have been reported for the sea urchin Strongylocentrotus purpuratus and Lytechinus variegatus (Foster et al., 2020; Massri et al., 2021; Paganos et al., 2021). Here, we set out to compile a single-cell dataset from a sister taxa of the sea urchins, the sea star Patiria miniata, as a resource for deeper analysis of development in this animal as well as a point of comparison for other echinoderms. Of particular interest to us is the difference in germ line specification methods used between the sea urchin and sea star. Known marker genes for this and other lineages are leveraged here to reveal gene activities that become targets for new functional analysis.
RESULTS AND DISCUSSION
Identification of cell states across early sea star development
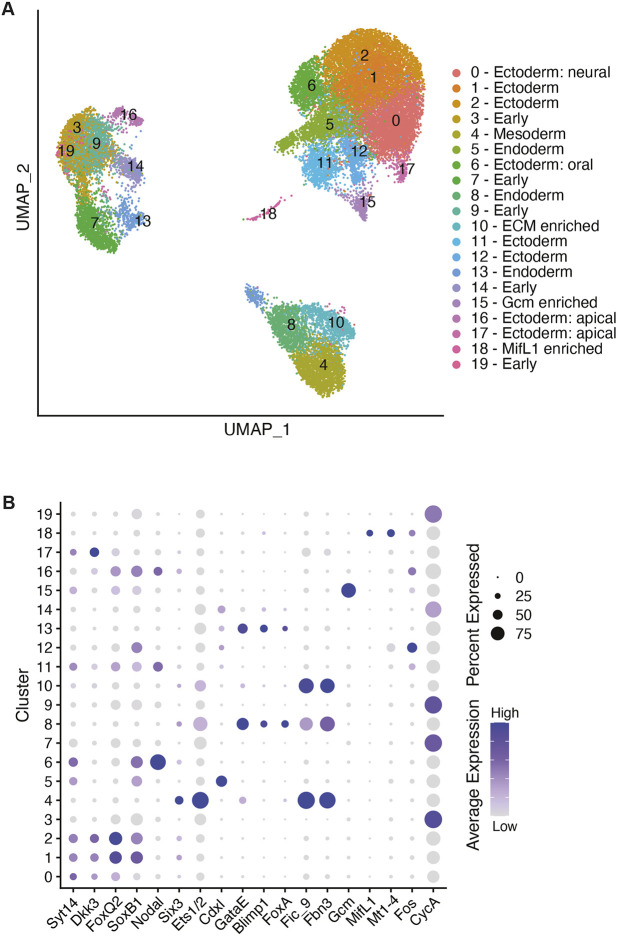
Sea star embryos were cultured to six key developmental stages; 8 h post-fertilization (hpf), 10 hpf, 14 hpf, blastula (B), early gastrula and mid-gastrula (MG) stage, at which point they were dissociated, processed for single-cell RNA-sequencing (scRNA-seq) via Drop-seq, and aligned to the P. miniata genome (Macosko et al., 2015). All six datasets were analyzed using Seurat and integrated using Harmony, an scRNA-seq data integration tool, to identify conserved cell types across all datasets (Korsunsky et al., 2019; Stuart et al., 2019). After quality control measures, cells from all datasets were included in the integration as follows: 8 hpf (868 cells), 10 hpf (1318 cells), 14 hpf (2448 cells), blastula (7272 cells), early gastrula (3349 cells) and mid-gastrula (10,448 cells). This analysis of 25,703 total cells yielded 20 clusters (cell states) across all time points identified by known marker gene expression and by in situ RNA hybridization (Fig. 1A,B; Table S6). In analyzing the six datasets together, it is evident that early (8-14 hpf) and late (B-MG) cell states are transcriptionally distinct. Cell clusters of early stages (clusters 3 and 7) decrease with development; these cells are presumably ‘lost’ to differentiation (Fig. S1). Clusters 3, 7 and 9, which make up the majority of the earlier stage datasets, showed enrichment of CyclinA and lack of distinct marker gene expression (Fig. 1B). This suggests that these early stage cell states harbor similar mRNA expression.
Fig. 1.
Cell states identified across early sea star development. (A) UMAP visualization of 25,703 cells after integration of six datasets. Cells colored by cell state identity based on marker gene expression (Table S6). (B) Dot plot showing marker gene expression across clusters. Average gene expression level displayed by color intensity. Percentage of cells expressing the marker gene conveyed by circle size.
Blastula
We assessed the blastula and gastrula stage datasets individually in greater detail using expression of key regulatory genes characterized previously. We found the time and pattern of expression was consistent with previously reported in situ hybridizations. In blastulae, we detected ten cell states (blastula clusters B0-B9) which were identified by expression of such known marker genes (Fig. 2A,B; Tables S1 and S3). We identified ectodermal cells by expression of soxB1 (also known as sox2) (Yankura et al., 2013). foxq2 (foxe3l) and dkk3 are expressed in the animal pole domain, in which the apical organ of the nervous system will form (Cheatle Jarvela et al., 2016). Cluster B0 expresses high levels of soxB1, foxq2 and dkk3. Cluster B1 expresses foxq2 and dkk3, which we identify as the animal pole domain (Fig. S2). Although these apical ectodermal markers are widespread in the dataset, the vegetal markers are restricted to a subset of cell states. These vegetal markers include both mesodermal and endodermal progenitors. ets1/2 (ets1), a marker of the presumptive mesoderm, is expressed in the vegetal plate (Cary et al., 2020). gataE (Gata6), foxA (foxa1), brachyury (tbxt) and blimp1 (prdm1), all genes involved in endodermal development, mark different regions of the embryonic gut. However, in blastulae, they are all expressed in the vegetal region, surrounding the blastopore (Fresques et al., 2014; Hinman and Davidson, 2003). Cdxl (cdx1l), another endodermal marker, is also expressed as a vegetal ring in blastulae; it is later restricted to the blastopore region in mid-gastrulae (Annunziata et al., 2013). gataE, foxA and blimp1 are expressed in the same cluster as ets1/2 (cluster B2), whereas brachyury and cdxl are markers of cluster B3 (Fig. 2B; Fig. S2). At blastula stage, the transcription factor glial cells missing (gcm; gcml), which marks the pigment cells in the related purple sea urchin, shows no localized expression; gcm-positive cells are present throughout the ectoderm (Calestani et al., 2003; McCauley et al., 2010). In our dataset, cells with the highest gcm expression clustered into a unique population in cluster B8, with some gcm-positive cells present within the ectodermal clusters (Fig. 2B; Fig. S2).
Fig. 2.
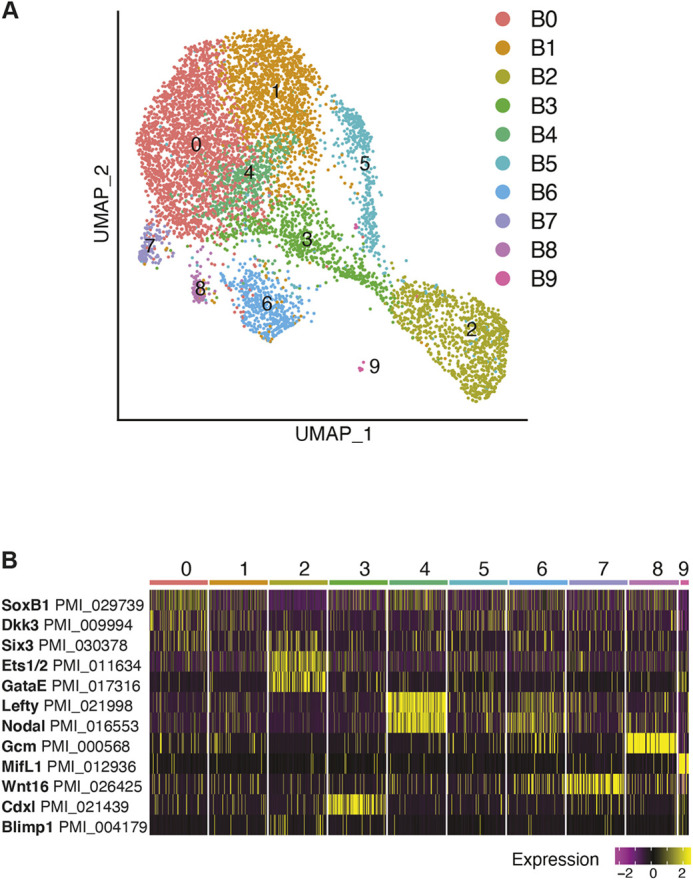
Gene expression at blastula stage. (A) UMAP plot of 7272 blastula stage cells. Clusters colored by cell state identity based on marker gene expression (Table S3). (B) Heatmap of blastula stage marker gene expression.
Expression of the Wnt signaling ligands and receptors has previously been characterized in blastula and gastrula stages (McCauley et al., 2013). As signaling plays an important role in development and inductive germline specification, we sought to include the Wnt ligand expression in our analysis. Six Wnt ligands and three Wnt Frizzled receptors are encoded in the P. miniata transcriptome (McCauley et al., 2013). Our dataset showed co-expression of genes encoding ligands Wnt3, Wnt16 and the Wnt receptor Frizzled1 (Fzd2) in cluster B2 (Fig. S4). These three genes are known, using in situ hybridization, to be expressed in the vegetal pole at blastula stage (McCauley et al., 2013). wnt8 (wnt8a) is also expressed in the vegetal pole, not overlapping with frizzled1 expression. Our analysis revealed the wnt8 expression pattern showed overlap with wnt3 (cluster B3) and no overlap with wnt3 (cluster B7), mirroring what has been previously reported (Fig. S4) (McCauley et al., 2013). We also sought to identify expression of the highly conserved germ cell markers nanos genes (nanos2 and nanos3) and vasa (ddx4), the expression of which has been characterized by in situ hybridization (Fresques et al., 2014, 2016). vasa mRNA is expressed in the vegetal plate of blastulae and is particularly enriched in cluster B2, like the wnt3 and wnt16 ligands, and the receptor frizzled1 (Fig. S6H) (Fresques et al., 2014). Results from both scRNA-seq and in situ hybridization show low or no detection of nanos in blastulae based on the threshold of these techniques (Fig. S6A) (Fresques et al., 2016).
Mid-gastrula
Analysis of the mid-gastrula stage dataset reveals 11 cell states (mid-gastrula clusters, MG0-MG10) (Fig. 3A; Tables S1 and S4). At this stage, many characteristics of the animal ectoderm remain the same. Ectodermal cell types (clusters MG0,1,2,4) are still distinguished by expression of soxB1, foxQ2 and dkk3 (Fig. S3; Table S4). Expression of several vegetal markers detected at blastula stage now changes from overlapping to marking either vegetal ectoderm or different regions of the gut. In blastula stage, both ets1/2 and gataE were expressed in the same cluster (cluster B2). At gastrula stage, ets1/2 is particularly enriched in cluster MG3, and gataE marks a different cluster (cluster MG6) (Fig. 3B). ets1/2 at gastrula stage is present at the tip of the archenteron, where it marks presumptive mesoderm and ingressing mesenchyme cells (Cary et al., 2020; Hinman and Davidson, 2007). We see low level gataE expression in the archenteron, although gataE is enriched in the mid- to hindgut region in mid-gastrula (Hinman and Davidson, 2003). foxA, which is expressed in the mid/hindgut region, is enriched in the same cluster as gataE (Hinman et al., 2003). In mid-gastrulae, brachyury expression surrounds the blastopore but is not within the blastopore (Hinman et al., 2003). cdxl expression is restricted to the region around the blastopore (Annunziata et al., 2013). brachyury and cdxl are both enriched in cluster MG8, reflecting their expression pattern seen via in situ hybridization (Fig. S3).
Fig. 3.
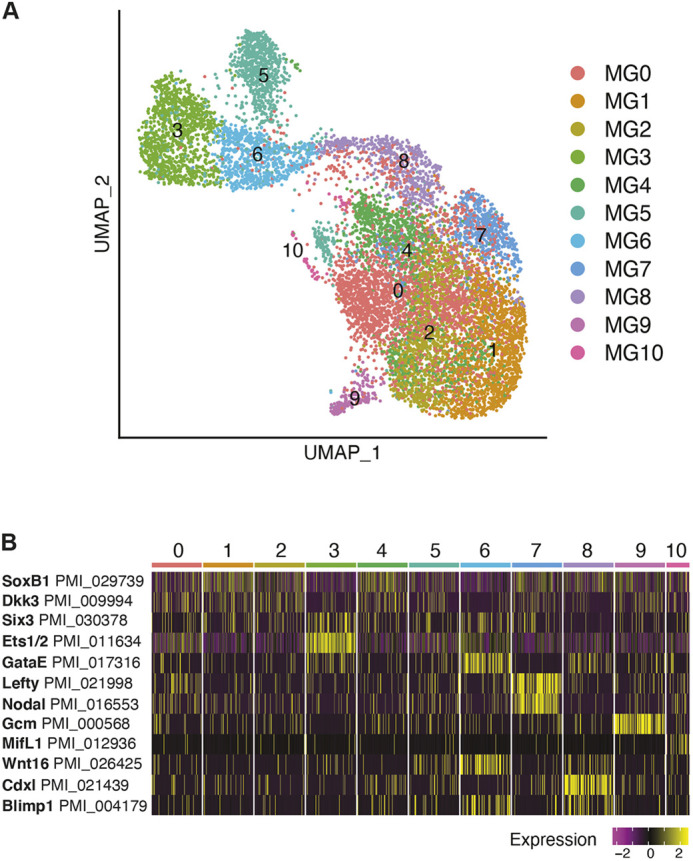
Gene expression at mid-gastrula stage. (A) UMAP plot of 10,448 mid-gastrula stage cells. Clusters colored by cell state identified by marker gene expression (Table S4). (B) Heatmap of mid-gastrula stage marker gene expression.
Lack of unique marker gene expression makes cluster 5 difficult to identify. Cluster MG5 showed enrichment of extracellular matrix protein expression, including fibrinogen (PMI_003448 Fic_9) and fibrillin (PMI_026729 Fbn3) (Table S4). Similar to blastula stage, one cluster is marked by high gcm expression, cluster MG9. A small number of cells cluster on their own (cluster MG10) and these cells are marked by macrophage inhibitory factor like 1 (mifl1) (Fig. 3B). MIF plays a role in immune function in vertebrates (Hibino et al., 2006; Nishihira, 2000). Cluster MG10 also expresses mt1-4, a metalloprotease, and fos (fosl), a transcription factor, both enriched in the sea urchin primary mesenchyme cells (PMCs) (Fig. S5A) (Rafiq et al., 2012; Sun and Ettensohn, 2014). mt1-4 is seen at the tip of the archenteron in gastrulae by in situ hybridization (Fig. S5B). Further studies are necessary to reveal what role these genes play in sea star development; this is especially poignant as sea star embryos lack PMCs and pigment cells.
The sea star embryo is known to have left/right morphological asymmetry by gastrula stage. A Nodal signaling gradient on the right side of the embryo is responsible for restricting the posterior enterocoel (PE), which houses the presumptive germ line, to the left side, as well as restricting nanos and vasa expression to this region (Fresques and Wessel, 2018). In our dataset, nodal (nodall) expression, along with its target lefty (lefty1), is enriched in one cluster (cluster MG7) which we identify as the right-side ectoderm (Fig. 3B) (Duboc et al., 2005). Although Nodal has been identified to inhibit germ line gene expression, the signal (or signals) that activate germ line gene expression remains unknown. In contrast to Nodal signaling, Wnt pathway component expression is present in the same region as nanos and vasa RNA expression in the early embryo and in the forming gut during the gastrula stage (Fresques and Wessel, 2018; Fresques et al., 2014, 2016; McCauley et al., 2013). In situ hybridization shows enrichment of nanos and vasa transcripts at the top of the archenteron and in a vegetal ring in the hindgut region at mid-gastrula stage (Fresques and Wessel, 2018; Fresques et al., 2014, 2016). Among the six Wnt ligands present in the sea star transcriptome, RNA expression patterns of wnt3, wnt8 and wntA/4 show overlap with the region of nanos and vasa RNA expression in the gastrula stage embryo (Fresques and Wessel, 2018; McCauley et al., 2013). Among the three Wnt receptors, frizzled1/2/7 (fzd2) RNA expression is concentrated in the forming gut of the early gastrula (McCauley et al., 2013). The three signaling ligands, wnt3, wnt8 and wntA/4, show appropriate expression patterns to be candidates for initiating germ line specification by interacting with the Frizzled1/2/7 receptor, due to their tight colocalization with the nanos and vasa RNA expression pattern in the early embryo. At mid-gastrula, the Wnt signaling ligands have been described as having a nested gene expression pattern. wnt16 shows most vegetal expression followed by wnt3 and then wnt8 along the ectoderm. In our dataset, wnt16 enrichment is seen in cluster MG6 with wnt3 expression. wnt3 is also co-expressed with Wnt8 in cluster MG8. frizzled1 is present in cluster MG3, which we identify as the tip of the archenteron (Fig. S4) (McCauley et al., 2013).
Derivation of the germ line cells
Origins of the germ line have a particularly important regulatory node in the mid-gastrula stage, where nanos and vasa are both expressed and will be restricted to the region of PE formation. The PE is posited to be the source of the germ line, as it is the source of restricted and specific germline gene enrichment in the sea star larva (Fresques et al., 2014; Fresques and Wessel, 2018). Moreover, removal of the PE via micropipetting results in larvae with significantly fewer germline cells (Inoue et al., 1992). Because of this, we sought to characterize the expression pattern of nanos and vasa-positive cells in mid-gastrulae. Nanos expression is detected in 267 total cells, co-expression of nanos and vasa is seen in 212 cells. Nanos expression is much less abundant compared with vasa; vasa expression is seen in all clusters at mid-gastrula stage, but expression in the hindgut cluster (cluster MG8) was of greatest interest to us (Fig. S6H). We took a closer look at the vasa-positive cells in the hindgut cluster and compared gene expression in the hindgut cells that express vasa with those that do not. In extracting a list of differentially expressed genes in vasa-positive cells of the hindgut cluster, we found the transcription factor foxY3 (Tables S2, S5). Delta/Notch signaling in the purple sea urchin was found to regulate nanos expression through the FoxY transcription factor in somatic cells adjacent to the PGCs (Oulhen et al., 2019; Materna et al., 2013). P. miniata has three FoxY transcription factors and we sought to characterize their expression patterns via in situ hybridization. Expression of all three FoxY genes was present at mid-gastrula, in the same region as nanos and vasa expression (Fig. 4; Fig. S7A). By late gastrula stage, expression of foxY1 and foxY2 was lost (Fig. 4). foxY3 was expressed in the same domain as nanos and vasa at mid- and late gastrula stage (Fig. 4). foxY3 expression was enriched in clusters MG3, MG6 and MG8, which correspond to the archenteron and gut region (Fig. S6H,I). Furthermore, we detected foxY3 and vasa co-expression at mid-gastrula using scRNA-seq (Fig. S7B). wnt3 was also expressed in the vasa, foxY3-positive cells, along with wnt16 and wnt1; wnt8 and wnt3 were expressed in the cells neighboring the vasa, foxY3-positive cells (Fig. S7C; Table S5). This suggests differential expression of Wnt ligands in cells of the mid/hindgut region. Moreover, at this stage, vasa-positive cells expressed hindgut identity genes such as gataE and foxA, and the homeobox gene hox11/13b (hbox7) (Table S3; Fig. S7D). These findings are of particular importance for understanding how the germ line is formed, and would be difficult to ascertain using only in situ hybridization.
Fig. 4.
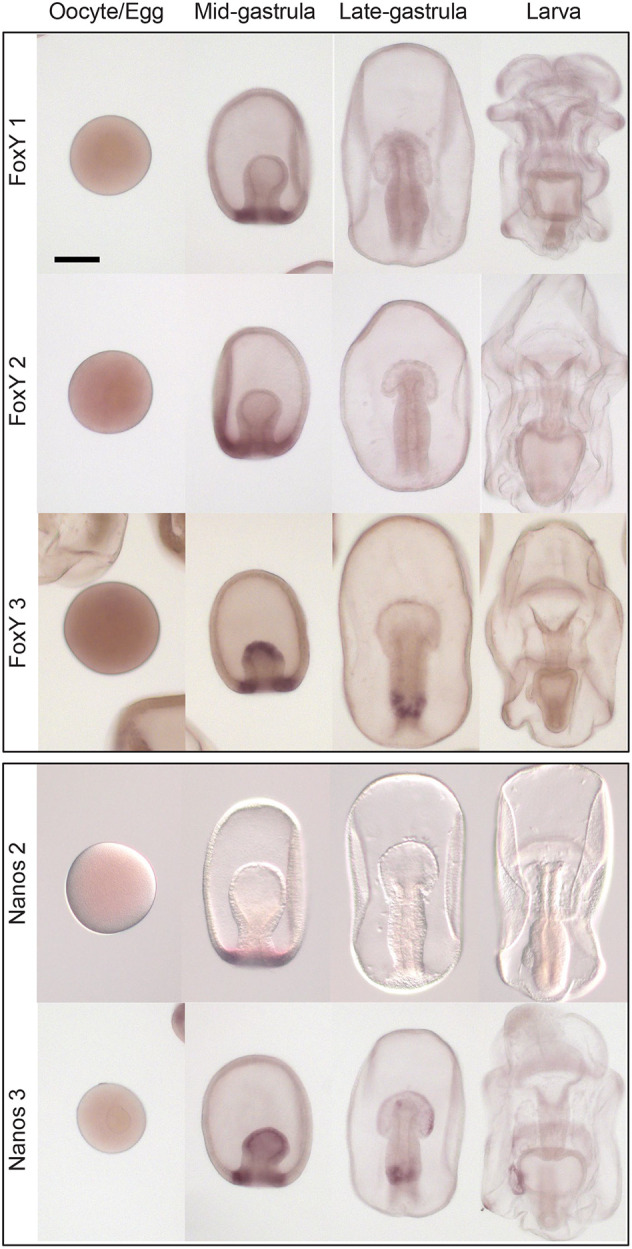
FoxY and Nanos gene expression. Whole mount in situ hybridizations for Nanos and the FoxY transcription factors during early sea star embryogenesis. nanos3 (PMI_027092; the focus of this study) expression is seen in mid-gastrula at the tip of the archenteron and the hindgut region. nanos3 expression persists as a ring in the gut in late gastrula and becomes restricted to the PE; some expression is detected in the anterior pouches. nanos2 (PMI_029792) is expressed transiently as a ring around the blastopore in mid-gastrula. foxY1 (PMI_028394) and foxY2 (PMI_008472) are also transiently expressed at mid-gastrula stage. foxY3 (PMI_000733) is expressed in the same domain as nanos3 at mid and late gastrula stages. At least 50 embryos from each developmental stage from three parental matings were tested. Scale bar: 100 μm.
At the sea star mid-gastrula stage, the vasa transcript is expressed more broadly than in the future germ line. This contrasts its expression in the sea urchin, where it is restricted to the small micromeres in gastrulae, and in the mouse, where it is expressed only after the PGCs have colonized the embryonic gonad (Fujiwara et al., 1994; Juliano et al., 2006). This is a distinct strategy of cell-specific germ cell factor accumulation in which expression is spread broadly and then cleared from cells to yield highly localized expression. In the sea star, Vasa protein is also under strict control through selective post-translational degradation by Gustavus (Gus), an E3 ligase (Perillo et al., 2022). In the future, investigating how Nanos protein localization is regulated will inform us whether these transcripts are also under distinct regulation at the protein level. Here, we report co-expression of vasa-positive cells with the foxY3 transcription factor. We also note Wnt ligand expression in vasa-positive cells and in cells of the same cluster identity that do not show vasa expression. We hypothesize that both Wnt and Delta/Notch signaling may regulate nanos and vasa expression at gastrula stage. These pathways could be active concurrently with Nodal restriction, which originates from a gradient on the right side of the embryo.
Echinoderms also offer an opportunity to investigate two modes of germ cell development in closely related species and uncover evolutionary differences. Nanos and Vasa eventually localize to putative PGC precursors in both species (the small micromeres in sea urchins and in the PE in the sea star), yet their embryonic expression patterns are vastly different. Early in sea urchin embryogenesis, nanos and vasa expression is restricted to the small micromeres (Juliano et al., 2006). In the sea urchin, nanos transcription is regulated by direct action of β-catenin in the small micromeres or the Delta/Notch transcription factor, FoxY, in the adjacent somatic Veg2 mesoderm cells (Oulhen et al., 2019). Whether canonical Wnt signaling plays a role in activating expression of nanos and vasa in sea star, and whether the foxY3 transcription factor co-expressed with the germ line genes in sea star regulates their expression, could point to a regulatory node conserved between both species. Notably, in sea urchin, removal of the micromeres, the parent cells of the small micromeres, results in adults with developed gametes (Yajima and Wessel, 2011). Whether nanos expression, downstream of FoxY, in the Veg2 cells plays a role in reconstituting the germ line remains an interesting question and could point to an auxiliary, inductive mechanism present in the sea urchin. Testing effects of cell signaling perturbation in micromere-deleted embryos will help to decipher this process.
Further cell type comparisons between species by scRNA-seq mechanisms
In the future, comparison of scRNA-seq datasets of different echinoderm species will likely reveal species-specific cell type differences. For example, at early blastula stage in the sea urchin, unique skeleton and pigment cell states are identified (Foster et al., 2020). Of note, the skeletogenic and pigment cell lineages are mesodermal cell types of the sea urchin, absent in the sea star embryo. However, here we report expression of gcm, a marker for pigment, and co-expression of fos and mt1-4 markers expressed in primary mesenchyme cells present in sea urchin and not sea star embryos. Multi-species comparison could further reveal both similarities and differences between the transcriptomic profile of similar cell states between sea urchin and sea star.
The sea star (P. miniata) and sea urchin (Paracentrotus lividus) have been compared to assess heterochronies in marker gene expression, revealing a later onset of maternal to embryonic transition in the sea star compared with this sea urchin (Gildor et al., 2017). It is possible that this temporal difference plays a role in the diversity of cell states present or differences in lineage mapping of early developmental stages when comparing two species. An alternative hypothesis is that cell fates in the sea star rely more on cell signaling for fate decisions than in the sea urchin, and that this difference is revealed by scRNA-seq analysis.
Evolution of germ line specification
Evidence suggests that the inductive mode (epigenesis) in metazoans, such as sponges, jellyfish and hydra, is the ancestral mechanism of germ cell specification (Extavour and Akam, 2003). Discovery of an inductive mode for germ line specification in the sea star, an echinoderm species, presents an opportunity for comparative analysis with the mouse, a mammalian species for which we have the most comprehensive description of the inductive mechanism. Mouse PGCs acquire their fate in response to signaling and arise from mesoderm differentiating cells. Single-cell analysis revealed that early germ cells express homeobox genes, including Hoxb1 and Hoxa1, and the mesodermal marker brachyury (T), at the same level as their somatic cell neighbors. The early germ cells then initiate transcriptional repression of homeobox genes, which remain highly expressed in the neighboring somatic cells as the embryo develops (Saitou et al., 2003). Here, we see a similarity in the sea star, with co-expression of hox11/13b and brachyury with nanos-positive cells (Fig. S7D). In the mouse, expression of germline determinants with mesodermal markers is resolved by repression of the mesodermal markers in the PGC precursor cells; Stella (Dppa3)-positive cells repress homeobox genes (Saitou et al., 2003). Cell lineage analysis of scRNA-seq data in mice also reveals a lack of germ cell determinants in somatic cells and vice versa (Chan et al., 2019). Notably, the axolotl homologue of brachyury, a well conserved mesodermal marker, Axbra, is seen in all cells of the mesodermal region known to include PGC precursors, suggesting that the PGC precursors express brachyury and are specified from early mesoderm (Johnson et al., 2003). If and how expression of mesodermal markers such as brachyury affects germ cell development in mice, axolotls and sea stars may reveal whether there is a conserved relationship between mesodermal and germ cell factor expression in inductive germ line specification mechanisms. Thus, in the sea star, apart from restricting nanos and vasa transcript expression, repression of mesodermal markers in nanos- and vasa-positive cells remains an important step in germ line specification, which has yet to be identified.
MATERIALS AND METHODS
Embryo culture and dissociation for single cell analysis
Adult P. miniata animals were collected by either Peter Halmay (San Diego Fishermen's Working Group) or Josh Ross (South Coast Bio-Marine) off the Californian coast. Embryos were cultured essentially as described previously (Fresques et al., 2016). Embryos were cultured in filtered (0.2 μm) sea water collected at the Marine Biological Laboratories in Woods Hole, MA, USA, until the appropriate stage for dissociation. All embryos used in the study resulted from the mating of one male and one female to ensure complete comparative capability. Multiple fertilizations were initiated in this study and timed such that the appropriate stages of embryonic development were reached at a common endpoint. The embryos were then collected and washed twice with calcium-free sea water, and then suspended in hyalin-extraction media (HEM) for 10-15 min, depending on the stage of dissociation. When cells were beginning to dissociate, the embryos were collected and washed in 0.5 M NaCl, gently sheared with a pipette, run through a 40 μm Nitex mesh, counted on a hemocytometer and diluted to reach the appropriate concentration for the scRNA-seq protocol. Equal numbers of embryos were used in each time point and at no time were cells or embryos pelleted in a centrifuge (Oulhen et al., 2019).
In situ hybridization
DIG-labeled RNA probes were made using a Roche DIG probe synthesis kit as described previously (Fresques et al., 2016). Probe-hybridized embryos were developed with NBT+BCIP for purple staining. Embryos were incubated with a probe for 1 week and were developed essentially as described previously (Fresques et al., 2016).
scRNA-seq
Single cell encapsulation was performed using the Chromium Single Cell Chip B kit on the 10x Genomics Chromium Controller. Single cell cDNA and libraries were prepared using the Chromium Single Cell 3′ Reagent kit v3 Chemistry. Libraries were sequenced by Genewiz on the Illumina Hiseq (2×150 bp paired-end runs). Single-cell unique molecular identifier (Fujii et al., 2009) counting (counting of unique barcodes given to individual transcript molecules) was performed using Cell Ranger Single Cell Software Suite 3.0.2 from 10x Genomics. The custom transcriptome reference was generated from P. miniata assembly V2.0 (echinobase.org) using CellRanger mkref. The P. miniata V2.0 assembly was used for analysis owing to a higher rate of genome mapping compared with V3.0. In addition, the germ line genes analyzed in this study are annotated in V2.0. The annotation for Pm-nanos was manually edited to account for a longer 3′ untranslated region. Duplicate blastula and gastrula stage libraries were aggregated using the cellranger aggr function. Cellranger gene expression matrices were further analyzed using the R package Seurat v 3.2.2 (Stuart et al., 2019). Cells of 8-14 hpf stages with at least 1000 and at most 6000 genes (features), and cells with at least 1000 and at most 2500 genes in blastula to mid-gastrula stages were included in downstream analysis. Individual datasets were normalized by scaling gene expression in each cell by total gene expression and then log transformed. The top 2000 highly variable genes across the datasets were then used to integrate the datasets. Individual time point datasets were integrated using the Seurat toolkit Harmony to remove batch effects and identify conserved cell populations across the datasets. The clustering parameters used were: dimensions, 20; resolution, 1.0. Cluster markers were found using FindConservedMarkers and FindMarkers functions. The UMI count and gene number per cell are presented in Figs S8 and S9. See Supplementary Data 1 for the code used (PmAnalysis.txt).
Supplementary Material
Acknowledgements
Part of this research was conducted using resources and services at the Computational Biology Core and Center for Computation and Visualization, Brown University. A special thanks to August Guang, Joselynn Wallace and Ashok Ragavendran of the Computational Biology Core.
Footnotes
Author contributions
Conceptualization: S.F., N.O., T.F., G.W.; Methodology: S.F., N.O., H.Z.; Validation: S.F., N.O., G.W.; Formal analysis: S.F., N.O., H.Z.; Investigation: S.F., N.O., T.F.; Resources: G.W.; Data curation: S.F., N.O.; Writing - review & editing: S.F., N.O., T.F., G.W.; Visualization: N.O., T.F., H.Z.; Supervision: G.W.; Project administration: G.W.; Funding acquisition: G.W.
Funding
We thank the National Institutes of Health (1R35GM140897, G.W. and 1P20GM119943, N.O.) and the National Science Foundation (IOS-1923445, G.W.). Deposited in PMC for release after 12 months.
Data availability
Datasets have been deposited in GEO under accession number GSE196654.
References
- Annunziata, R., Martinez, P. and Arnone, M. I. (2013). Intact cluster and chordate-like expression of ParaHox genes in a sea star. BMC Biol. 11, 68. 10.1186/1741-7007-11-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, J. A., Weinreb, C., Wagner, D. E., Megason, S., Peshkin, L., Kirschner, M. W. and Klein, A. M. (2018). The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360, eaar5780. 10.1126/science.aar5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calestani, C., Rast, J. P. and Davidson, E. H. (2003). Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening. Development 130, 4587-4596. 10.1242/dev.00647 [DOI] [PubMed] [Google Scholar]
- Cary, G. A., Mccauley, B. S., Zueva, O., Pattinato, J., Longabaugh, W. and Hinman, V. F. (2020). Systematic comparison of sea urchin and sea star developmental gene regulatory networks explains how novelty is incorporated in early development. Nat. Commun. 11, 6235. 10.1038/s41467-020-20023-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, M. M., Smith, Z. D., Grosswendt, S., Kretzmer, H., Norman, T. M., Adamson, B., Jost, M., Quinn, J. J., Yang, D., Jones, M. G.et al. (2019). Molecular recording of mammalian embryogenesis. Nature 570, 77-82. 10.1038/s41586-019-1184-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatle Jarvela, A. M., Yankura, K. A. and Hinman, V. F. (2016). A gene regulatory network for apical organ neurogenesis and its spatial control in sea star embryos. Development 143, 4214-4223. 10.1242/dev.134999 [DOI] [PubMed] [Google Scholar]
- Davidson, E. H. (2006). The Regulatory Genome. San Diego: Academic Press. [Google Scholar]
- Duboc, V., Rottinger, E., Lapraz, F., Besnardeau, L. and Lepage, T. (2005). Left-right asymmetry in the sea urchin embryo is regulated by nodal signaling on the right side. Dev. Cell 9, 147-158. 10.1016/j.devcel.2005.05.008 [DOI] [PubMed] [Google Scholar]
- Extavour, C. G. and Akam, M. (2003). Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130, 5869-5884. 10.1242/dev.00804 [DOI] [PubMed] [Google Scholar]
- Foster, S., Oulhen, N. and Wessel, G. (2020). A single cell RNA sequencing resource for early sea urchin development. Development 147, dev191528. 10.1242/dev.191528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresques, T. M. and Wessel, G. M. (2018). Nodal induces sequential restriction of germ cell factors during primordial germ cell specification. Development 145, dev155663. 10.1242/dev.155663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresques, T., Zazueta-Novoa, V., Reich, A. and Wessel, G. M. (2014). Selective accumulation of germ-line associated gene products in early development of the sea star and distinct differences from germ-line development in the sea urchin. Dev. Dyn. 243, 568-587. 10.1002/dvdy.24038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresques, T., Swartz, S. Z., Juliano, C., Morino, Y., Kikuchi, M., Akasaka, K., Wada, H., Yajima, M. and Wessel, G. M. (2016). The diversity of nanos expression in echinoderm embryos supports different mechanisms in germ cell specification. Evol. Dev. 18, 267-278. 10.1111/ede.12197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, T., Sakamoto, N., Ochiai, H., Fujita, K., Okamitsu, Y., Sumiyoshi, N., Minokawa, T. and Yamamoto, T. (2009). Role of the nanos homolog during sea urchin development. Dev. Dyn. 238, 2511-2521. 10.1002/dvdy.22074 [DOI] [PubMed] [Google Scholar]
- Fujiwara, Y., Komiya, T., Kawabata, H., Sato, M., Fujimoto, H., Furusawa, M. and Noce, T. (1994). Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Proc. Natl. Acad. Sci. USA 91, 12258-12262. 10.1073/pnas.91.25.12258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildor, T., Hinman, V. and Ben-Tabou-De-Leon, S. (2017). Regulatory heterochronies and loose temporal scaling between sea star and sea urchin regulatory circuits. Int. J. Dev. Biol. 61, 347-356. 10.1387/ijdb.160331sb [DOI] [PubMed] [Google Scholar]
- Hibino, T., Loza-Coll, M., Messier, C., Majeske, A. J., Cohen, A. H., Terwilliger, D. P., Buckley, K. M., Brockton, V., Nair, S. V., Berney, K.et al. (2006). The immune gene repertoire encoded in the purple sea urchin genome. Dev. Biol. 300, 349-365. 10.1016/j.ydbio.2006.08.065 [DOI] [PubMed] [Google Scholar]
- Hinman, V. F. and Burke, R. D. (2018). Embryonic neurogenesis in echinoderms. Wiley Interdiscip. Rev. Dev. Biol. 7, e316. 10.1002/wdev.316 [DOI] [PubMed] [Google Scholar]
- Hinman, V. F. and Davidson, E. H. (2003). Expression of a gene encoding a Gata transcription factor during embryogenesis of the starfish Asterina miniata. Gene Expr. Patterns 3, 419-422. 10.1016/S1567-133X(03)00082-6 [DOI] [PubMed] [Google Scholar]
- Hinman, V. F. and Davidson, E. H. (2007). Evolutionary plasticity of developmental gene regulatory network architecture. Proc. Natl. Acad. Sci. USA 104, 19404-19409. 10.1073/pnas.0709994104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman, V. F., Nguyen, A. T., Cameron, R. A. and Davidson, E. H. (2003). Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc. Natl. Acad. Sci. USA 100, 13356-13361. 10.1073/pnas.2235868100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, C., Kiyomoto, M. and Shirai, H. (1992). Germ cell differentiation in starfish: the posterior enterocoel as the origin of germ cells in Asterina pectinifera. Dev. Growth Differ. 34, 413-418. 10.1111/j.1440-169X.1992.00413.x [DOI] [PubMed] [Google Scholar]
- Johnson, A. D., Crother, B., White, M. E., Patient, R., Bachvarova, R. F., Drum, M. and Masi, T. (2003). Regulative germ cell specification in axolotl embryos: a primitive trait conserved in the mammalian lineage. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 1371-1379. 10.1098/rstb.2003.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano, C. E., Voronina, E., Stack, C., Aldrich, M., Cameron, A. R. and Wessel, G. M. (2006). Germ line determinants are not localized early in sea urchin development, but do accumulate in the small micromere lineage. Dev. Biol. 300, 406-415. 10.1016/j.ydbio.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Korsunsky, I., Millard, N., Fan, J., Slowikowski, K., Zhang, F., Wei, K., Baglaenko, Y., Brenner, M., Loh, P. R. and Raychaudhuri, S. (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289-1296. 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko, E. Z., Basu, A., Satija, R., Nemesh, J., Shekhar, K., Goldman, M., Tirosh, I., Bialas, A. R., Kamitaki, N., Martersteck, E. M.et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202-1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massri, A. J., Greenstreet, L., Afanassiev, A., Berrio, A., Wray, G. A., Schiebinger, G. and Mcclay, D. R. (2021). Developmental single-cell transcriptomics in the Lytechinus variegatus sea urchin embryo. Development 148, dev198614. 10.1242/dev.198614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna, S. C., Swartz, S. Z. and Smith, J. (2013). Notch and Nodal control forkhead factor expression in the specification of multipotent progenitors in sea urchin. Development 140, 1796-1806. 10.1242/dev.091157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccauley, B. S., Weideman, E. P. and Hinman, V. F. (2010). A conserved gene regulatory network subcircuit drives different developmental fates in the vegetal pole of highly divergent echinoderm embryos. Dev. Biol. 340, 200-208. 10.1016/j.ydbio.2009.11.020 [DOI] [PubMed] [Google Scholar]
- Mccauley, B. S., Akyar, E., Filliger, L. and Hinman, V. F. (2013). Expression of wnt and frizzled genes during early sea star development. Gene Expr. Patterns 13, 437-444. 10.1016/j.gep.2013.07.007 [DOI] [PubMed] [Google Scholar]
- Nishihira, J. (2000). Macrophage migration inhibitory factor (MIF): its essential role in the immune system and cell growth. J. Interferon Cytokine Res. 20, 751-762. 10.1089/10799900050151012 [DOI] [PubMed] [Google Scholar]
- Oulhen, N., Swartz, S. Z., Wang, L., Wikramanayake, A. and Wessel, G. M. (2019). Distinct transcriptional regulation of Nanos2 in the germ line and soma by the Wnt and delta/notch pathways. Dev. Biol. 452, 34-42. 10.1016/j.ydbio.2019.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganos, P., Voronov, D., Musser, J. M., Arendt, D. and Arnone, M. I. (2021). Single-cell RNA sequencing of the Strongylocentrotus purpuratus larva reveals the blueprint of major cell types and nervous system of a non-chordate deuterostome. Elife 10, e70416. 10.7554/eLife.70416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo, M., Swartz, S. Z. and Wessel, G. M. (2022). A conserved node in the regulation of Vasa between an induced and an inherited program of primordial germ cell specification. Dev. Biol. 482, 28-33. 10.1016/j.ydbio.2021.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiq, K., Cheers, M. S. and Ettensohn, C. A. (2012). The genomic regulatory control of skeletal morphogenesis in the sea urchin. Development 139, 579-590. 10.1242/dev.073049 [DOI] [PubMed] [Google Scholar]
- Saitou, M., Payer, B., Lange, U. C., Erhardt, S., Barton, S. C. and Surani, M. A. (2003). Specification of germ cell fate in mice. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 1363-1370. 10.1098/rstb.2003.1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Butler, A., Hoffman, P., Hafemeister, C., Papalexi, E., Mauck, W. M., III, Hao, Y., Stoeckius, M., Smibert, P. and Satija, R. (2019). Comprehensive integration of single-cell data. Cell 177, 1888-1902. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z. and Ettensohn, C. A. (2014). Signal-dependent regulation of the sea urchin skeletogenic gene regulatory network. Gene Expr. Patterns 16, 93-103. 10.1016/j.gep.2014.10.002 [DOI] [PubMed] [Google Scholar]
- Yajima, M. and Wessel, G. M. (2011). Small micromeres contribute to the germline in the sea urchin. Development 138, 237-243. 10.1242/dev.054940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankura, K. A., Koechlein, C. S., Cryan, A. F., Cheatle, A. and Hinman, V. F. (2013). Gene regulatory network for neurogenesis in a sea star embryo connects broad neural specification and localized patterning. Proc. Natl. Acad. Sci. USA 110, 8591-8596. 10.1073/pnas.1220903110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.