Abstract
Acetylation is a key post-translational modification (PTM) involved in the regulation of both histone and non-histone proteins. It controls cellular processes such as DNA transcription, RNA modifications, proteostasis, aging, autophagy, regulation of cytoskeletal structures, and metabolism. Acetylation is essential to maintain neuronal plasticity and therefore essential for memory and learning. Homeostasis of acetylation is maintained through the activities of histone acetyltransferases (HAT) and histone deacetylase (HDAC) enzymes, with alterations to these tightly regulated processes reported in several neurodegenerative diseases including Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD), and amyotrophic lateral sclerosis (ALS). Both hyperacetylation and hypoacetylation can impair neuronal physiological homeostasis and increase the accumulation of pathophysiological proteins such as tau, α-synuclein, and Huntingtin protein implicated in AD, PD, and HD, respectively. Additionally, dysregulation of acetylation is linked to impaired axonal transport, a key pathological mechanism in ALS. This review article will discuss the physiological roles of protein acetylation and examine the current literature that describes altered protein acetylation in neurodegenerative disorders.
Keywords: PTMs, acetylation, HDACs, HATs, proteostasis, cytoskeleton, neurodegenerative disease
Introduction
Post-translational modifications (PTMs) of proteins define the molecular complexity of our cells. Through mechanisms such as covalent modifications of proteins, PTMs change the properties of a protein to determine its activity, localization, or interaction with other proteins, cells, or systems (Mann and Jensen, 2003). Over 400 PTMs have been reported in the literature, including methylation, acetylation, ubiquitination, phosphorylation, and glycosylation (Duan and Walther, 2015), which change the function of the proteins, leading to altered gene expression, cellular signaling, protein trafficking, and cellular structure (Khoury et al., 2011; Duan and Walther, 2015). Given the importance of PTMs, our understanding of the role of these modifications in complex neurodegenerative diseases is critical to understanding the causes and finding treatments or cures.
Age-related neurodegenerative diseases, including Alzheimer’s disease (AD), Huntington’s disease (HD), prion diseases, Parkinson’s disease (PD), frontotemporal lobar degeneration (FTLD), and amyotrophic lateral sclerosis (ALS) are some of the leading causes of mortality and morbidity worldwide (Taylor et al., 2002; Erkkinen et al., 2018; Ashby, 2019). These diseases are progressive and heterogeneous in nature and involve adverse changes to the central and/or peripheral nervous system, including the degeneration of neurons resulting in the loss of cognitive and/or motor functions (Lin and Beal, 2006; Hrelia et al., 2020). Due to the impact of neurodegenerative disease, developing targeted treatments based on pathological mechanisms has become a major focus of research. Aggregation of misprocessed proteins is a common feature of the major types of neurodegenerative disorders (Merlini et al., 2001; Tutar et al., 2013; Hrelia et al., 2020), which can be predisposed by genetic mutations, but the majority of cases are of unknown etiology, or sporadic in nature (Tutar et al., 2013). We may be able to discern the driving factors behind neurodegenerative disease by understanding how PTMs of disease-associated proteins alter underlying pathological processes, through mechanisms such as impaired gene regulation, proteostasis, or alterations to the cytoskeleton.
The role of altered PTMs in neurodegenerative disease is highlighted by findings that many disease-associated aggregated proteins have abnormal PTMs, which can result in protein aggregation, mislocalization, and/or misprocessing (Schaffert and Carter, 2020). Such dysregulation may ultimately be linked to downstream pathogenesis such as glutamate excitotoxicity, mitochondrial dysfunction, and activation of caspases (Hall, 2011), suggesting that altered PTMs may be drivers of neurodegeneration, and potential targets for novel therapeutic strategies. Studies to date have predominantly focused on the role of altered phosphorylation in neurodegenerative diseases (Hanger et al., 2009; Rudrabhatla, 2014), however, alterations in acetylation, particularly of cytoskeletal proteins, may also play a role in driving neurodegeneration (Sternberger et al., 1985; Cavallarin et al., 2010; Noble et al., 2013; Rudrabhatla, 2014; Xu et al., 2015; Schaffert and Carter, 2020). This review will provide a general overview of the role of protein acetylation and how it can be dysregulated in neurodegenerative disease. As this is a vast topic, we will focus on the disease-associated proteins and the cytoskeleton.
Acetylation
Acetylation is a key biological process for regulating the function and viability of all mammalian cells, by adding acetyl groups to the structure of proteins. Acetylation can occur on different chemical groups including hydroxyl, thiol, or amino groups (Portaleone, 2004). Although acetylation was first identified as a modification of histone proteins within the nucleus, it also occurs on several non-histone proteins, where it plays an essential role in regulating cellular responses and signaling in response to different types of stressors, both internal and external (Drazic et al., 2016).
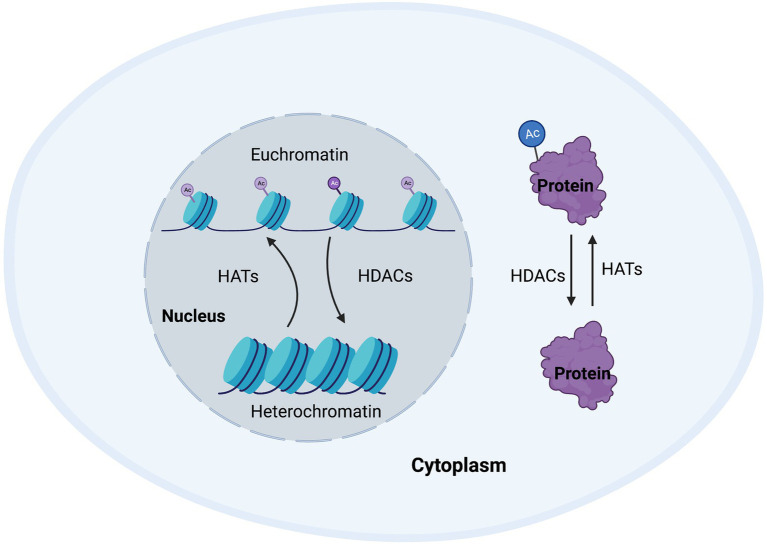
Around 80–90% of proteins are acetylated at the N-terminus of the polypeptide chain during translation (Drazic et al., 2016). This N-terminal acetylation is an abundant irreversible process and is carried out by enzymes referred to as N-terminal acetyltransferases (NATs), that transfer an acetyl group from acetyl-coenzyme A to the α-amino acid of proteins. Such modifications can alter the way the proteins form their tertiary structure, affect their half-life, or even their localization within the cell (Deng and Marmorstein, 2021). While NATs can regulate the acetylation of terminals mostly during translation, post-translational acetylation, which is the main subject of this review, occurs on the ε-amino group of lysine residues in a polypeptide chain and can occur as a reversible process. Post-translational acetylation is involved in a wide variety of processes, such as maintaining cellular homeostasis, protein folding, and protein localization, and is a tightly regulated process. The enzymes involve in maintaining this homeostasis are known as histone acetyltransferases (HATs) and histone deacetylases (HDACs; Figure 1; Peserico and Simone, 2010).
Figure 1.
Cellular homeostasis regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). Post-translational modifications regulate a wide range of proteins to enable appropriate transcription and maintain cellular function. HATs and HDACs are integral to this process by adding or removing acetyl groups to/from histone or non-histone proteins, thus changing chromatin accessibility, or protein structure, function, or half-life.
Histone acetyltransferases and histone deacetylases
HAT enzymes
Histone acetyltransferases are ubiquitously expressed and are broadly classified based on their localization within either the nucleus or cytoplasm. Nuclear HATs are chiefly responsible for the regulation of gene expression by acetylating the nuclear histones resulting in altered chromatin compaction and subsequent gene expression. Cytoplasmic HATs acetylate proteins in the cytoplasm; this includes cytoplasmic histone proteins, which are then transported into the nucleus for further modification. HATs regulate the acetylation process by transferring an acetyl group from acetyl-coenzyme A to the ε-amino group of lysine residues (Roth et al., 2001). Depending on their functional activities, structure, and sequential homology, HATs can be further grouped into three major families; (i) the GNAT family, (ii) the MYST family, and (iii) the EP300/CREBBP family (Kamei et al., 1996; Roth et al., 2001).
The GNAT family (or KAT2/GCN5-related N-acetyltransferases) acetylates both histones and non-histone proteins (Roth et al., 2001; Kim et al., 2010; Tapias and Wang, 2017) and is responsible for a wide variety of functions including transcription regulation, response to stress, and development (Tapias and Wang, 2017; Shirmast et al., 2021). Some of this superfamily is NATs, involved in translational modification, while the remaining enzymes are involved in post-translationally acetylating lysine residues of proteins (Deng and Marmorstein, 2021). The MYST family is named from its four core enzymes which are MOZ/histone acetyltransferase 6A, Ybf2/SaS3, SaS2, and Tip60/histone acetyltransferase 5 (Kim et al., 2010). The MYST family HATs interact with many of the core nuclear histones including H2A, H2AZ, H3, and H4 (Marmorstein, 2001; Roth et al., 2001). These enzymes are highly conserved in eukaryotes and regulate essential cellular functions of gene transcription, DNA replication and damage repair, and neurogenesis (Avvakumov and Côté, 2007; Tapias and Wang, 2017). The E1A binding protein p300/Cyclic adenosine monophosphate Response Element Binding protein (EP300/CREBBP) family includes the conserved HAT enzymes p300 and CREB binding protein (CBP), which are homologous and functionally similar (Roth et al., 2001; Yang, 2004). This family also has both nuclear histone and non-histone targets (Table 1; Roth et al., 2001; Yang, 2004). CBP and p300 are involved in gene expression by recruiting transcriptional machinery through acetylation and responding to cellular hypoxia, cellular differentiation, and early brain development (Arany et al., 1996; Tapias and Wang, 2017).
Table 1.
HAT enzymes and their roles in neurodegenerative disease (NDD).
| HAT family | Representative enzymes | Subcellular Localisation | Histone substrates | Examples of non-histone substrates | Identified eoles in brain | Identified involvement in *NDD |
|---|---|---|---|---|---|---|
| GNAT family (Roth et al., 2001; Yang, 2004; Kim et al., 2010; Wang Y. et al., 2014; Wang et al., 2020) | HAT1 | Nucleus/Cytoplasm | H4, H2A | HMG-N2/HMG17, AR, c-MYC, p53, and Ku70 | - | - |
| Gcn5 | Nucleus | H2B, H3 | Neural progenitor differentiation (Martínez-Cerdeño et al., 2012) | PD (Fan et al., 2020), *SCA7 (Chen et al., 2012) | ||
| PCAF | Nucleus | H3 | Regulates memories (Martínez-Cerdeño et al., 2012) | AD (Park et al., 2015), HD (Bodai et al., 2012) | ||
| HPA2 | Nucleus | H3, H4 | - | - | ||
| HPA3 | Nucleus | H4 | - | - | ||
| Elp3 | Nucleus/Cytoplasm | H3, H4 | Dendritic and axonal branching (Xu et al., 2000; Kim C. H. et al., 2012) | ALS (Simpson et al., 2009) | ||
| Elp1 | Nucleus | H3, H4 | - | |||
| MYST family (Roth et al., 2001; Yang, 2004; Kim et al., 2010; Wang Y. et al., 2014; Wang et al., 2020) | TIP60 | Nucleus/Cytoplasm | Embryonic neuronal development (Georgala et al., 2011; Kim C. H. et al., 2012) | AD (Müller et al., 2008), SCA1 (Gehrking et al., 2011) | ||
| MORF | Nucleus | H2A, H3, H4 | Adult neurogenesis (Thomas et al., 2000; Kueh et al., 2011) | AD (Li et al., 2021) | ||
| MOZ | Nucleus | H3, H4 | Regulation of neuronal progenitor cells & patterning genes (Katsumoto et al., 2006; Kueh et al., 2011; Perez-Campo et al., 2014; You et al., 2015) | AD (Li et al., 2021) | ||
| MOF | Nucleus/Mitochondria | H4 | AD (Li et al., 2021) | |||
| HBO1 | Nucleus | H4 | AD (Li et al., 2021) | |||
| Esa1 | Nucleus | H2AZ, H4 | - | - | ||
| Sas2 | Nucleus | H4 | - | - | ||
| Sas3 | Nucleus | H3 | - | - | ||
| p300/CREBBP (Roth et al., 2001; Yang, 2004; Kim et al., 2010; Wang Y. et al., 2014; Wang et al., 2020) | p300 | Nucleus/Cytoplasm | H2A, H2B, H3, H4 | HMG1, HMG-I(Y), p53, GATA-1, TCF, IF-2, ACTR (nuclear receptor coactivator), and SRC-1 | Neuronal tube development (Katsumoto et al., 2006; Kueh et al., 2011; Perez-Campo et al., 2014; You et al., 2015) and memory formation (Li et al., 2002; Korzus et al., 2004) | AD (Lu et al., 2014), PD (Kontopoulos et al., 2006), HD (Bodai et al., 2012) |
| CBP | Nucleus/Cytoplasm | H2A, H2B, H3, and H4 | AD (Caccamo et al., 2010), PD (Kontopoulos et al., 2006; Song et al., 2010), HD (Cong et al., 2005), ALS (Rouaux et al., 2003), and *SCA3 (Chai et al., 2002) |
AR, Androgen receptor; GATA-1, GATA-binding factor 1; HMG, high mobility group chromosomal protein; IF2, Initiation factor 2; SCA1, spinocerebellar ataxia type 1; SCA3, spinocerebellar ataxia type 3; SCA7, spinocerebellar ataxia type 7; SRC, Proto-oncogene tyrosine-protein kinase; and TCF, T-cell factor.
NDD: neurodegenerative disease.
Histone acetyltransferase enzymes have many critical roles in both the developing and adult brain. For example, in adult mice, PCAF, a member of the GNAT family has been shown to have a key regulatory function in the formation of both short-term and long-term memory following stress and anxiety (Martínez-Cerdeño et al., 2012). Moreover, knockdown of either or both Elp1 and Elp3 enzymes (GNAT family), was associated with defective dendritic and axonal branching in motor neurons along with poor migration and maturation of neuronal cells (Xu et al., 2000; Kim C. H. et al., 2012). The importance of these HATs can also be seen through their roles in the developing brain, as studies have shown conditional deletion of Gcn5 (GNAT family) resulted in a 26% loss of brain mass, due to dysregulation of neural progenitor differentiation (Martínez-Cerdeño et al., 2012). The MYST family enzymes are also vital for neuronal development and adult nervous system function. For instance, overexpression of TIP60 in mouse retinas at postnatal day 4 led to an increased level of PAX6, which plays an essential role in embryonic neuronal development (Georgala et al., 2011; Kim C. H. et al., 2012). Although there are few studies examining the role of MOZ, MOF, and HBO1 in the brain, the absence of these enzymes has been shown to result in impaired regulation of neuronal progenitor cells, embryonic lethality, and reduction of neuronal patterning genes (Katsumoto et al., 2006; Kueh et al., 2011; Perez-Campo et al., 2014; You et al., 2015). Another MYST family member, MORF, is highly expressed in both the embryonic and adult brain and it has been postulated that it plays a role in adult neurogenesis (Thomas et al., 2000; Kueh et al., 2011). Both, p300 and CBP have been shown to be expressed at a high level in the neural tube of mice and are essential for neuronal tube development (Katsumoto et al., 2006; Kueh et al., 2011; Perez-Campo et al., 2014; You et al., 2015). They are also thought to be critical to homeostasis and some aspects of memory formation in the adult brain, due to high expression in young and adult mice (Li et al., 2002; Korzus et al., 2004). Moreover, studies suggest that p300 may be involved in oligodendrocyte differentiation in the developing rat brain (Zhang et al., 2016). While it is important to note the varied functions of HATs in brain function, this process does not function in isolation and is tightly regulated by balancing acetylation and deacetylation.
HDAC enzymes
To allow regulation of the homeostasis of cellular processes controlled by lysine acetylation through HATs, the process of acetylation must be able to be reversed. This occurs through enzymes known as HDACs. HDACs were named after their initially identified role in deacetylating nuclear histone proteins to regulate transcription. However, HDACs are now recognized as playing important roles in deacetylating non-nuclear histones and non-histone proteins (Table 2) both in the nucleus and cytoplasm that have other cellular mechanisms including metabolism, protein degradation, and modulation, facilitating DNA damage repair, immune process, oxidative stress, angiogenesis, and apoptosis (Ruijter et al., 2003; Seto and Yoshida, 2014; Van Helleputte et al., 2014; Didonna and Opal, 2015; Li G. et al., 2020). Eighteen different HDACs have been identified in humans, which are known as HDAC1-11 and SIRT1-7. These are classified into four major classes named class I–IV depending on their distinguishable role in cellular processes and the subcellular regions in which they function (Glozak et al., 2005). The superfamily of HDAC proteins can be first divided into two classes depending on the type of cofactor involved in deacetylation; the zinc-dependent HDACs’ (also known as classical HDACs) and the sirtuin HDACs, which use nicotinamide adenine dinucleotide as a cofactor. The zinc-dependent HDACs are further subclassified into classes I, II, and IV whereas the sirtuin family makes up the class III HDAC enzymes depending on their functional and structural properties (Park and Kim, 2020).
Table 2.
HDAC enzymes and their roles in neurodegenerative disease (NDD).
| HDAC classes | Representative enzymes | Subcellular Localization | Histone substrates | Examples of non-histone substrates | Identified roles in brain | Identified involvement in NDD | Mislocalization in NDD |
|---|---|---|---|---|---|---|---|
| Class I (Ma and Schultz, 2008; Dovey et al., 2010; Yao and Yang, 2011; Lv et al., 2012; Wang et al., 2012; Chakrabarti et al., 2015; Park and Kim, 2020) | HDAC1 | Nucleus | H4 | MEF2, ATM, p53, MeCP2 | Neurogenesis in developing brains (Cunliffe, 2004; Yamaguchi et al., 2005; Ye et al., 2009; Harrison et al., 2011), dendritic growth (Tea et al., 2010) & microglial maturation (Alliot et al., 1999) | AD (Pao et al., 2020), HD (Jia et al., 2012b), FTLD (Pao et al., 2020) | Cytoplasm (Simões-Pires et al., 2013) |
| HDAC2 | Nucleus | H2A/2B, H3,H4 | NF-κB, MeCP. IRS-1 | AD (Pao et al., 2020), HD (Mielcarek et al., 2011), ALS (Janssen et al., 2010) | - | ||
| HDAC3 | Nucleus | H4 | NF-κB, HDAC4, HDAC5, HDAC7, HDAC9, pRb | Regulating neuronal homeostasis (Soriano and Hardingham, 2011; Norwood et al., 2014), axonal regeneration & memory formation (Fischle et al., 2002; McQuown et al., 2011) | AD (Pao et al., 2020), ALS (Janssen et al., 2010), HD (Jia et al., 2012b) | - | |
| HDAC8 | Nucleus/Cytoplasm | H2A/2B | HSP70 | Neuronal differentiation (Katayama et al., 2018) & skull development (Haberland et al., 2009a) | PD (Quan et al., 2021) | - | |
| Class IIa (Yao and Yang, 2011; Wang et al., 2012; Wang Z. et al., 2014; Caslini et al., 2019; Park and Kim, 2020; Wang et al., 2021) | HDAC4 | Nucleus/Cytoplasm | H4 | FOXO, p53, p21, HDAC3, Runx2, MEF2 | Memory formation and synaptic plasticity (Broide et al., 2007; Kim M. S. et al., 2012), Purkinje cell differentiation & postnatal cerebellum development (Majdzadeh et al., 2008) | AD (Chen Y.-A. et al., 2020), PD (Wu et al., 2017), HD (Mielcarek et al., 2011), ALS (Pegoraro et al., 2020), FTLD (Whitehouse et al., 2015) | Cytoplasm (Mielcarek et al., 2013)/Nucleus (Wu et al., 2017) |
| HDAC5 | Nucleus/Cytoplasm | H4 | HDAC3, MEF2, Runx2 | Differentiation of neuronal stem cells (Schneider et al., 2008) | AD (Agis-Balboa et al., 2013), PD (Mazzocchi, 2021), HD (Yeh et al., 2013), ALS (Janssen et al., 2010), FTLD (Whitehouse et al., 2015) | - | |
| HDAC7 | Nucleus/Cytoplasm | H3 | HDAC3, PML, Runx2, MEF2 | Dendric growth, neurogenesis, neuronal protection, & maturation (Lai et al., 2010; Sugo et al., 2010; Ma and D’Mello, 2011) | AD (Lu et al., 2019), PD (Mazzocchi, 2021) | - | |
| HDAC9 | Nucleus/Cytoplasm | H3, H4 | HDAC3, MEF2, CAM | - | |||
| Class IIb (Yao and Yang, 2011; Wang et al., 2012; Park and Kim, 2020) | HDAC6 | Cytoplasm | - | α-tubulin, Cortractin, HDAC11, HSP90, PP1 | Dendritic growth & branching (Kim et al., 2009) | AD (Simões-Pires et al., 2013), PD (Simões-Pires et al., 2013), HD (Simões-Pires et al., 2013), ALS-FTLD (Simões-Pires et al., 2013), CMT (d'Ydewalle et al., 2011) | Perinuclear (Simões-Pires et al., 2013) |
| HDAC10 | Cytoplasm | - | LcoR, and PP1 | Dendric growth, neurogenesis, neuronal protection, and maturation (Lai et al., 2010; Sugo et al., 2010; Ma and D’Mello, 2011) | - | ||
| Class III (Yao and Yang, 2011; Wang et al., 2012; Seto and Yoshida, 2014; Park and Kim, 2020) | SIRT1 | Nucleus/Cytoplasm | H3, H4 | NF-κB, p300, p53, Ku70, PCAF, TIP60 | Neuronal development, axonal growth, regeneration, & neuroprotection (Pandithage et al., 2008; Tiberi et al., 2012; Li X. H. et al., 2013; Kaluski et al., 2017; Sidorova-Darmos et al., 2018; Garcia-Venzor and Toiber, 2021) | AD (Bonda et al., 2011), PD (Li X. et al., 2020), HD (Jęśko et al., 2017), ALS (Outeiro et al., 2008) | Cytoplasm (Zschoernig and Mahlknecht, 2008) |
| SIRT2 | Cytoplasm | H3, H4 | α-tubulin | AD (Jęśko et al., 2017), PD (Outeiro et al., 2007), HD (Outeiro et al., 2007) | - | ||
| SIRT3 | Mitochondria | H4 | Ku70, GDH, SOD2 | AD (Jęśko et al., 2017), PD (Salvatori et al., 2017), HD (Salvatori et al., 2017), ALS (Salvatori et al., 2017) | - | ||
| SIRT4 | Mitochondria | - | GDH | AD (Jęśko et al., 2017), ALS (Körner et al., 2013) | - | ||
| SIRT5 | Mitochondria | - | Cytochrome c, CPS1 | AD (Akter et al., 2021) PD (Maszlag-Török et al., 2021), ALS (Harlan et al., 2019) | - | ||
| SIRT6 | Nucleus | H3 | TNF-α | ALS (Körner et al., 2013) | - | ||
| SIRT7 | Nucleus | H3 | p53 | - | - | ||
| Class IV (Wang et al., 2012; Núñez-Álvarez and Suelves, 2022) | HDAC11 | Nucleus/Cytoplasm | H3, H4 | HDAC6, Cdt1 | Maturation and development of neuronal cells and oligodendrocytes (Liu et al., 2008, 2009) | HD (Kumar et al., 2022), ALS (Janssen et al., 2010) | - |
ATM, Ataxia-telangiectasia-mutated; CAM, calmodulin; CPS1, Carbamoyl phosphate synthetase 1; Cdt1, chromatin licensing and DNA replication factor 1; FOXO, Forkhead box O; GDH, glutamate dehydrogenase; HSP, heat shock protein; IRS, insulin receptor substrate; LcoR, ligand-dependent receptor co-repressor; MeCP, methyl-CpG-binding domain protein; MEF, myocyte enhancer factor; NF-κB, nuclear transcription factor-kappa B; PP1, protein phosphatase, pRb, retinoblastoma protein; Runx, Runt-related transcription factor; SOD, superoxide dismutase; and TNF, tumor necrosis factor.
The class I HDACs includes HDAC1, 2, 3, and 8, and while most function within the nucleus, HDAC3 shuttles between the nucleus and cytoplasm for transcriptional regulation (Yao and Yang, 2011; Didonna and Opal, 2015; Park and Kim, 2020). Class II HDACs are further classified into class IIa and IIb based on their structural specifications. HDAC4, 5, 7, and 9 all contain a common N-terminal binding domain and belong to the class IIa HDACs, while HDACs 6 and 10 belong to the class IIb group. Class IIa HDACs shuttle between the nucleus and cytoplasm whereas class IIb enzymes are predominantly localized to the cytoplasm (Marsoni et al., 2008; Li G. et al., 2020). The class III HDACs (SIRT1-SIRT7) are structurally distinct from class I and class II HDACs. Among the SIRT proteins, SIRT1, 6, and 7 are abundant in the nucleus, although under certain conditions SIRT1 can be retained in the cytoplasm (Frye, 2000; Blander and Guarente, 2004; Michishita et al., 2005; Ramadori et al., 2008; Shoba et al., 2009). On the other hand, SIRT3, 4, and 5 are primarily present in mitochondria and SIRT2 is commonly found in the cytoplasm (Frye, 2000; Blander and Guarente, 2004; Michishita et al., 2005; Ramadori et al., 2008; Shoba et al., 2009). Lastly, class IV, HDACs contain only HDAC11. This enzyme carries homologous features to class I and II HDACS. HDAC11 is mainly found in the nucleus; however, it can be found co-localized with HDAC6 in the cytoplasm (Ruijter et al., 2003; Seto and Yoshida, 2014).
The function and localization of HDACs have been investigated in several different models. For example, class I HDAC enzymes are ubiquitously expressed at different levels in all tissues (Uhlén et al., 2015), whereas the other classes HDACs may have more tissue-specific expression (Kuwahara et al., 2003; Nakagawa et al., 2006; Zupkovitz et al., 2006; Montgomery et al., 2007; Knutson et al., 2008; Trivedi et al., 2008; Soriano and Hardingham, 2011). Multiple studies have shown the critical roles of these enzymes in developing the heart, skeletal muscle, liver, bone, vascular system, and immune system (Kuwahara et al., 2003; Chang et al., 2004; Méjat et al., 2005; Chang et al., 2006; Cohen, 2006; Nakagawa et al., 2006; Zupkovitz et al., 2006; Arnold et al., 2007; Montgomery et al., 2007; Knutson et al., 2008; Trivedi et al., 2008; Soriano and Hardingham, 2011; Seto and Yoshida, 2014; Parra, 2015; Li et al., 2016; Wang et al., 2017; Liu et al., 2020). All these HDACs are expressed in the brain and the class IV HDAC11 is expressed at the highest level in the brain compared to other classes (Frye, 2000; Blander and Guarente, 2004; Michishita et al., 2005; Broide et al., 2007; Ramadori et al., 2008; Haberland et al., 2009b; Shoba et al., 2009; Parra, 2015; Uhlén et al., 2015; Wey et al., 2016).
Like HATS, HDACs have been shown to be responsible for carrying out essential functions in both the developing and adult brains, such as maintaining synaptic plasticity, dendritic outgrowth, and axon regeneration (Morris and Monteggia, 2013). Class 1 HDACs may have a particularly important role in cortical development and function (Morris and Monteggia, 2013), due to their high expression in cortical tissue (Broide et al., 2007; Wey et al., 2016). Furthermore, studies have shown that both HDAC1 and HDAC2 have roles in neurogenesis in developing brains (Cunliffe, 2004; Yamaguchi et al., 2005; Ye et al., 2009; Harrison et al., 2011) as their deletion in glial fibrillary acidic protein (GFAP)-Cre transgenic mice led to an impairment in the generation of neurons from neural progenitors and increased neuronal death (Montgomery et al., 2009). Studies have also demonstrated that HDAC2 is involved in memory formation and regulating synaptic plasticity by restricting the maturation of adult neuronal synapses (Guan et al., 2009). Studies in the olfactory system of Drosophila suggest that both HDAC1 and HDAC2 are involved in the establishment of nervous system connections as they are required for appropriate dendritic growth (Tea et al., 2010) along with facilitating microglial maturation and function (Alliot et al., 1999). HDAC3, which is expressed at the highest level in the rat brain compared to other enzymes from class I (Broide et al., 2007) is responsible for regulating neuronal homeostasis (Soriano and Hardingham, 2011; Norwood et al., 2014). Studies in transgenic mice suggest that HDAC3 may also play a role in axon regeneration and negatively regulate long-term memory formation (Fischle et al., 2002; McQuown et al., 2011). Little is known about the regulatory roles of HDAC8 in brain function, but it is the least expressed from its class in the rat brain (Broide et al., 2007). However, it has been shown to negatively regulate neuronal differentiation (Katayama et al., 2018), and it regulates skull development in vertebrates, where the absence of this enzyme caused skull instability followed by perinatal lethality in a mouse model (Haberland et al., 2009b).
Although studies have revealed vital roles of class II HDACs in CNS development, most of the functions of these enzymes are still unknown. HDAC4 has been shown to impair memory formation and synaptic plasticity when knocked out of mice (Broide et al., 2007; Kim M. S. et al., 2012) and is essential for regulating postnatal cerebellum development and Purkinje cell differentiation in posterior lobes (Majdzadeh et al., 2008). However, Price and colleagues (2012) did not see abnormalities when HDAC4 was knocked out of forebrain neurons (Price et al., 2013) suggesting cell/region-specific roles for HDAC4 during early CNS development. HDAC5, the second most highly expressed class II enzyme in brain tissue, has been found to be involved in facilitating the differentiation of neuronal stem cells (Schneider et al., 2008). Several HDACs have roles in the cytoplasm of neurons and are involved in axonal integrity and dynamics. For example, HDAC5, much like HDAC3 and HDAC6, may be involved in axonal regeneration following injury by deacetylating microtubules, which is required for optimizing growth cone dynamics (Cho and Cavalli, 2012). Studies in cerebral and hippocampal cells have shown that HDAC6 like HDAC2 plays a crucial role in regulating the processes of dendritic growth and branching (Kim et al., 2009). Although HDAC6 was upregulated during axonal injury and neuronal oxidative stress (Witte et al., 2008), its role in development may be less important, as HDAC6 knockout mice have not shown any abnormalities other than increased microtubule acetylation (Zhang et al., 2008). The other HDACs from this class, HDAC7, 9, and 10, have also been shown to function in dendric growth, neurogenesis, neuronal protection, and maturation (Lai et al., 2010; Sugo et al., 2010; Ma and D'Mello, 2011). The class IV HDAC11 is yet to be studied widely, however, a few studies have demonstrated its function in the maturation and development of neuronal cells and oligodendrocytes (Liu et al., 2008, 2009).
Among the class III sirtuins, SIRT2, SIRT3, and SIRT5 are most abundantly expressed in the brain (Sidorova-Darmos et al., 2014) and enzymes from this class are also involved in regulating neuronal development, axonal growth, regeneration, and neuroprotection like other HDACs (Pandithage et al., 2008; Tiberi et al., 2012; Li X. H. et al., 2013; Kaluski et al., 2017; Sidorova-Darmos et al., 2018; Garcia-Venzor and Toiber, 2021). However, genetic modifications of most of these HDACs in animal models have been shown to be embryonically lethal, as such their core roles are remained to be elucidated.
Role of altered acetylation in neurodegenerative disease
A number of different PTMs have been implicated in the pathogenesis of neurodegenerative diseases (Didonna and Benetti, 2016). Most studies have focused on examining alterations to phosphorylation (Tenreiro et al., 2014); acetylation has been less well studied. However, dysregulation of acetylation has been reported to occur in several neurodegenerative diseases, such as AD, ALS, and HD (Saha and Pahan, 2006) and although not well characterized, some studies have postulated that dysregulation of HATs and HDACs may be paramount to the onset and/or progression of neurodegenerative disease (Saha and Pahan, 2006). For example, HATs and HDACs interact with a range of non-histone substrates that may be implicated in neurodegenerative diseases, such as p53, NF-KB, and STAT1 (Boutillier et al., 2003; Rouaux et al., 2003), while failing to maintain appropriate regulation of HATs and HDACs can result in activation of apoptotic pathways and widespread dysregulation in neuronal cells (Rouaux et al., 2003). Whether alterations to these enzymes are upstream drivers of neurodegeneration or secondary to other pathological processes is not well understood. Janssen et al. (2010) demonstrated that increased levels of HDAC2 and reduced HDAC11 mRNA were related to apoptotic neuronal death in human brain tissue from people diagnosed with ALS (Janssen et al., 2010). Additionally, accumulated mutant huntingtin (htt) protein in HD has also been shown to interact with the HAT domain of the CBP enzyme, decreasing HAT activity in post-mortem human brain tissue (Rouaux et al., 2004).
There is much work to do to fully understand alterations to the enzymes involved in acetylation as well as alteration to acetylation of proteins associated with neurodegenerative disease. Here we review the current literature on two key pathways implicated in neurodegenerative diseases; proteostasis (protein folding, aggregation, and degradation) as well as correct functioning of the cytoskeleton. We review both the role of acetylation in these physiological processes as well as reported alterations in disease since it is important to know how these physiological processes are regulated through acetylation to understand the dysregulations in neurodegenerative diseases.
Protein folding, aggregation, degradation, and metabolism
The prerequisite step for a functionally active protein is its folding into a three-dimensional structure. This protein folding process occurs in the endoplasmic reticulum (ER) and involves PTMs including acetylation (Stevens and Argon, 1999). Acetylation of proteins is also important for controlling their degradation. Each intracellular protein or protein complex has a specific lifespan after which it is degraded by the proteasome or by autophagy and acetylation has a crucial role in regulating both. For example, it has been shown that acetylated proteins are protected from ubiquitin-induced protein modification and degradation by the proteasome which increases the lifespan and functions of these proteins (Knorre et al., 2009). Conversely, studies have shown that histone degradation requires the addition of acetyl groups to undergo the proteasome activator PA200/Blm10-based degradation process (Qian et al., 2013). Furthermore, acetylation also controls aspects of the degradation machinery. Acetylation can aid in the regulation of autophagy from activation of core autophagic proteins, fusion of autophagosomes with lysosomes, and autophagic cargo assembly (Xu and Wan, 2022). ATG proteins, which play an essential role in phagophore formation, are maintained by HATs/HDACs to fine-tune the inhibition or activation of autophagy (Bánréti et al., 2013). For example, acetylation of ATG9A autophagic proteins in the lumen of the ER prevents activation of autophagosomes whereas deacetylation of ATG9A induces the formation of autophagosomes (Pehar and Puglielli, 2013). During stress conditions, like starvation, SIRT1 deacetylates these ATG proteins to facilitate autophagy (Bánréti et al., 2013). Additionally, acetylation of microtubules also occurs in response to stress and activates the MAPK/JNK autophagic signaling pathway (Bánréti et al., 2013). Alongside the established roles of acetylation as a post-translational modification, it is also linked to many aspects of metabolism. Studies have demonstrated that the acetyl-CoA metabolite availability is essential for acetylation to occur during histone or non-histone protein modifications (Peleg et al., 2016). This largely depends on the extent of mitochondrial production of acetyl-CoA available within the cell (Peleg et al., 2016). Additionally, other metabolite cofactors such as NAD+ are required for the deacetylase activities of sirtuins (Choudhary et al., 2014). While mitochondrial and metabolic activity is essential for acetylation to occur, studies have also shown that acetylation can regulate mitochondrial homeostasis, including storage and utilization of cellular energy. For example, a deficit of SIRT3 results in the production of reactive oxygen species along with altered oxidative metabolism (Guarente, 2011; Baeza et al., 2016). Interestingly, most of the enzymes involved in metabolic processes such as glycolysis, urea cycle, and gluconeogenesis have been found to be acetylated (Arif et al., 2010).
Acetylation and altered proteostasis in neurodegenerative disease
In neurodegenerative diseases, altered acetylation has been implicated in contributing to failure of protein clearance mechanisms through autophagy and the proteosome (Sambataro and Pennuto, 2017; Son et al., 2021). For instance, studies have reported that increased activity of p300/CBP enzyme altered autophagic flux resulting in excessive secretion of tau protein in transgenic AD mice (Chen X. et al., 2020). Furthermore, the HDAC inhibitor, 4b (preferentially inhibit HDAC1 and HDAC3 enzymes), was shown to improve cognitive function in a transgenic HD mouse model by clearing Huntingtin protein through proteasome and lysosome pathways (Jia et al., 2012a). Altered proteostasis may also result from protein aggregation, which is a pathophysiological hallmark for neurodegenerative diseases (Tutar et al., 2013). Aggregation is thought to result from several different conditions occurring in aggregate-prone proteins such as mutations, oxidative stress, or altered PTM causing proteins to misfold and generate insoluble aggregates (Tutar et al., 2013). These aggregates are thought to impair the structural and functional activities of neurons, which further facilitate the pathogenic process of disease conditions in AD, PD, FTLD, HD, and ALS (Tutar et al., 2013).
The majority of research investigating links between protein aggregation and PTMs has focused on abnormal phosphorylation, however, as most of these disease-related proteins can become acetylated, alterations to acetylation could contribute to protein misfolding and protein aggregate formation in neurodegenerative diseases (Schaffert and Carter, 2020). Here we focus on what we know about altered acetylation in some of the key proteins which become aggregated in neurodegenerative diseases.
Transactive response DNA binding protein 43 in ALS/FTD
Transactive response DNA binding protein 43, or TDP-43 is a transcription and RNA metabolism regulator protein primarily localized to the nucleus (Sephton et al., 2010). This protein is encoded by the human TARDBP gene and contains two RNA recognition motifs, a prion-like domain in the C-terminal region and a folded N-terminal domain. Like many proteins, TDP-43 also undergoes PTMs (François-Moutal et al., 2019). Protein pathology has been linked to hyperphosphorylation of the protein; however, more recent studies have implicated acetylation in the normal and pathological role of TDP-43. It has been shown that CBP-associated acetylation of lysine 145 and 192 regulates the binding of TDP-43 with target RNA and is a core site for regulating additional acetylation events (Cohen et al., 2015; Buratti, 2018). Another study demonstrated that the regulatory role of acetylation at lysine 136 site of TDP-43 where transfection with SIRT1 targeting lysine 136 sites reduced TDP-43 aggregation in studied shTDP-43-HEK293E cells (Garcia Morato et al., 2022). In ALS/FTD (frontotemporal dementia), studies have shown that hyperacetylation of TDP-43 caused a reduction in its splicing ability to targeted RNA and led to the aggregation of TDP-43 in the cytoplasm, inducing neuronal stress (Buratti, 2018). Acetylation of lysine 82 and lysine 192 of TDP-43 has also been associated with pathogenic mislocalization of TDP-43 to the cytoplasm (Kametani et al., 2016).
Fused in sarcoma in ALS/FTD
Fused in sarcoma (FUS), like TDP-43, is a predominantly nuclear RNA binding protein involved in RNA metabolism. This protein has been shown to contain N- and C-terminal nuclear localization signal (NLS) sites which undergo acetylation to regulate their function (Bock et al., 2021; Farina et al., 2021). Furthermore, acetylation at lysine 315/316 in the RNA recognition motif of FUS, regulates its binding with RNA whereas acetylation at lysine 510 of the C-terminal NLS may facilitate increased aggregation of the protein in the cytoplasm (Farina et al., 2021). Moreover, a liquid chromatography assay revealed that imbalance of N-terminal acetylation may also prompt FUS to aggregate in ALS/FTD diseases (Bock et al., 2021; Farina et al., 2021).
Tau (MAPT) in AD and PD
Tauopathy is one of the major pathophysiological signatures of both AD and FTLD. Tau is a microtubule-associated protein (MAP) that has a crucial role in assembling and stabilizing microtubules (Weingarten et al., 1975; Goedert et al., 1989). Phosphorylation of tau and its links to neurodegenerative diseases have been extensively studied (Brotzakis et al., 2021), however acetylation of tau also occurs at the N-terminus and lysine residues and may regulate the binding of tau with microtubules (Derisbourg et al., 2015; Brotzakis et al., 2021). For example, in post-mortem AD tissue as well as in transgenic mouse model of tauopathy, the amino acid KXGS motif, which resides in the microtubule-binding region of tau protein was hypoacetylated which was shown to impair tau activity and result in accumulation in neurofibrillary tangles (Cook et al., 2014a). Additionally, hyperphosphorylation of this motif prevents tau acetylation which again causes dissociation of tau from microtubules, reducing the stability of microtubules and axonal transport (Cook et al., 2014b). While hypoacetylation of tau has been associated with increased toxicity, research also points to a relationship between elevated acetylated tau and tauopathies in AD (Min et al., 2010). A study by Sohn et al. (2016) demonstrated that hyperacetylation of tau was involved in the pathogenesis of AD, by impairing axonal initial segment and microtubule dynamics. The axon initial segment functions to maintain neuronal polarity between the axonal domain and somatic dendritic domain. Sohn demonstrated an increased acetylated level of tau at lysines 274 and 281 and destabilization of the axon initial segment in the superior temporal gyrus of human AD brain tissue (Sohn et al., 2016). In the human neuroblastoma-derived SHSY5Y PD model, both tau and α-synuclein, a key pathological PD protein, were found to be hyperacetylated, which was linked to HAT p300 modulation of the deacetylase enzymes SIRT2 and HDAC6 (Esteves et al., 2019). Beyond functional modifications related to altered tau acetylation itself in neurodegenerative disease, altered acetylated tau has been related to neurofibrillary tangle formation in AD through reduced clearance of tau protein. It has been shown that acetylation at lysine 311 of the human leukocyte antigen DRB1*04, which specifically binds with tau, is associated with the clearance of tau by T cells, and subsequent slowing down of neurodegeneration (Le Guen et al., 2021). Altered acetylation of tau has also been associated with neurodegenerative diseases like Pick’s disease and corticobasal degeneration in human tissue. It was reported that insoluble and aggregated neurofibrillary tangles in human tissue from these diseases had altered the acetylation of the K280 lysine residues of tau (Cohen et al., 2011). Additionally, there are other examples where acetylation of microtubules has altered binding of molecules such as of motor proteins dynein/dynactin and kinesin-1 to microtubules, or alter axonal transport such as of BDNF in HD (Dompierre et al., 2007). Acetylation of microtubules may further facilitate binding of microtubule-associated proteins such as tau, which can interact with tubulin and alter the pathophysiological events of accumulated tau proteins in neurodegenerative diseases (Selenica et al., 2014; Saunders et al., 2022).
Alpha-synuclein in PD and dementia with Lewy bodies
Alpha-synuclein aggregates are core pathology in PD and dementia with Lewy bodies. Under normal conditions, this protein is predominantly found in neuronal presynaptic terminals and is involved in regulating synaptic vesicle trafficking for the release of neurotransmitters (Vargas et al., 2014). Alpha-synuclein undergoes N-terminal acetylation which modulates its binding with tubulin, actin, and lipids (Iyer et al., 2016; Deng et al., 2020). Additionally, in studies of PD, altered acetylation of α-synuclein at lysine 6 and 10 resulted in aggregation leading to synucleinopathies and neuronal toxicity (De Oliveira et al., 2017; Vinueza-Gavilanes et al., 2020).
Huntingtin in Huntington’s disease
The function of htt protein which is aggregated in Huntington’s disease is not well understood, however, it has been implicated in axonal transport and vesicle trafficking (DiFiglia et al., 1995; Vitet et al., 2020). Mass spectroscopy of the protein synthesized in HEK 293 T cell lines demonstrated five acetylation sites including acetylation at lysine 9, 178, 236, 345, and 444 (Cong et al., 2011). These acetylation sites along with other PTMs have been implicated in the physiological and pathological functions of huntingtin protein, such as modifying its structure, oligomerization, modulating the binding with membrane and other proteins, and formation of fibrils (Chiki et al., 2017). Further research has shown that huntingtin binding protein, HYPK, is involved in regulating N-terminal acetylation, while mutation of HYPK in HD reduces the aggregation of huntingtin protein through altered N-terminal acetylation (Arnesen et al., 2010; Gottlieb et al., 2021). In addition, acetylation of lysine 444 promotes the removal of aggregated huntingtin protein through autophagy which is altered in HD pathophysiology (Jeong et al., 2009).
Beta-amyloid in Alzheimer’s disease
The beta-amyloid (Aβ) peptide, which is a cleavage product of the amyloid precursor protein, is the core component of amyloid plaques in AD. It contains two key acetylation sites; lysine 16 and 28. Acetylation at these sites has been implicated in reducing aggregation, oligomerization, and fibril formation of Aβ peptides (Pilkington et al., 2019; Guha and Subramaniyam, 2021). In contrast, another study suggested acetylated Aβ created amorphous aggregates resulting in increased reactive oxygen species and cytotoxicity in studied SH-SY5Y neuronal cells (Adhikari et al., 2020).
Superoxide dismutase 1 in ALS
The reactive oxygen species scavenging enzyme superoxide dismutase 1 (SOD1) is a key pathological protein linked to neurodegeneration in ALS (Ralph et al., 2005; Barber et al., 2006). Studies have suggested acetylation of lysine 70, which is regulated by SIRT1, caused the inactivation of the antioxidant function of SOD1 (Banks and Andersen, 2019). Furthermore, acetylation at lysine site 122 suppressed mitochondrial respiration which subsequently increased the oxidative stress scavenging activity of SOD1 (Banks and Andersen, 2019). Acetylation at lysine 123 of SOD1 has been shown to have regional and cell type specificity in the healthy adult mouse CNS which may indicate its essential function in these cells (Kaliszewski et al., 2016). One study suggested alteration of acetylation at lysine 123 facilitated protein aggregation and pathogenesis in a SOD1 knockout ALS model. Not only was acetylation of lysine 123 increased, but high levels of acetylated SOD1 misfolded protein were found to be aggregated in primary cilia of astrocytes and in the vesicles which were derived from these primary cilia (Kaliszewski, 2016). These vesicles containing misfolded acetylated SOD1 were taken up by neurons, leading to neurodegeneration (Kaliszewski, 2016).
Clinical acetylation sites in neurodegenerative disease
Despite much research into the role of acetylation in NDD, there have been few studies to investigate sites of clinical relevance to limit neurotoxic proteins (Min et al., 2015, 2018; Dave et al., 2021). Of note, hyperacetylation of tau at lysine 174 was an early occurrence in post-mortem human AD brains and in the hippocampus of PS19 AD mice (Min et al., 2015). Acetylation at this site slowed tau turn over, promoted accumulation of the protein, and resulted in reduced hippocampal volume in vivo. Moreover, a model of tau lys174 deacetylation led to improved cognitive performance in behavioral tasks. Small molecule inhibition of p300 also improved cognitive outcomes in PS19 AD mice (Min et al., 2015).
The cytoskeleton
Cytoskeletal alterations have been implicated in many neurodegenerative diseases (Cairns et al., 2004) and acetylation of many cytoskeletal proteins plays a key role in their regulation (Scott et al., 2012; Latario et al., 2020). An understanding of the normal role of acetylation in the regulation of the cytoskeleton is important to understand changes in neurodegenerative disease.
Regulation of actin filaments
Actin filaments are involved in numerous cellular processes including maintaining cell shape and mobility and are important in neurons for neurite outgrowth, maintaining the structure of the axon as well as cytoplasmic transport (Kirkpatrick and Brady, 1999). To regulate these cellular activities, all three types of actin isoforms (alpha, beta, and gamma) have been reported to be acetylated along with actin regulatory proteins such as the actin-related protein (Arp) 2/3 complex involved in the regulation of actin filaments, and cortactin, which recruits the Arp 2/3 complex to the cortical actin cytoskeleton (Choudhary et al., 2009).
Although the impact of acetylation on these proteins is yet to be fully understood, some functional information has been demonstrated. For instance, acetylation at lysine 61 of the gamma isoform relates to the stabilization of stress fiber (Kim et al., 2006; Choudhary et al., 2009) and six subunits of the Arp2/3 complex have been shown to be acetylated to regulate actin nucleation, the first step in polymerization to F-actin (Choudhary et al., 2009), a critical component of structures such as dendritic filopodia and synapses. The F-actin regulatory protein cortactin has also been reported to be acetylated through p300/CBP (Weaver, 2008; Zhang et al., 2009) and deacetylated via HDAC6 and SIRT2 (Li Y. et al., 2013; Kim et al., 2020). Overexpression of HDAC6 also hinders the association of cortactin with F-actin, leading to lower levels of polymerized and branched actin (Zhang et al., 2007). Additionally, altered acetylation of cortactin by HDAC6 is involved in the fusion of autophagosomes and lysosomes, alterations to which contribute to neurodegeneration (Li Y. et al., 2013; Kim et al., 2020).
Regulation of microtubules
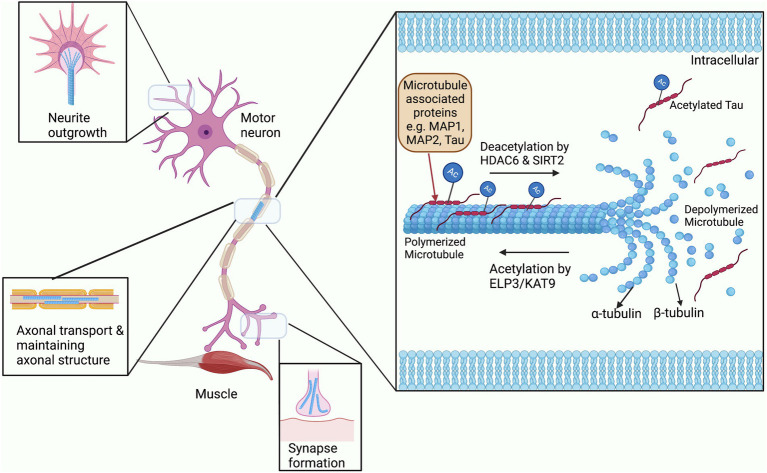
Acetylation of the cytoskeleton has been most widely studied in microtubules, and interestingly the first ever cytoplasmic acetylation protein that was studied was in association with microtubules (Sadoul et al., 2010). Microtubules are hollow tubular structures that act as tracks for cargo to move down the axon as required. These hollow tubes consist of two heterodimer subunits; α- and β-tubulin, and undergo continuous structural modification allowing growth and shrinkage according to the cellular demand (Figure 2; Guo and Van Den Bosch, 2018). In addition, microtubules are found in specialized structures such as mitotic spindles which are required for cell division. These functional activities of microtubules are widely regulated by PTM processes, particularly through acetylation of their α subunit (Janke, 2014). Acetylation of microtubules has mostly been studied at the lysine residue 40 of α-tubulin, although it also occurs at other lysine residues of both α- and β-tubulin (Choudhary et al., 2009). Acetylation of tubulin is related to the formation and stabilization of microtubule bundles and increasing the level of microtubule polymerization protein TPPP/p25 (Ogawa-Goto et al., 2007; Tőkési et al., 2010). These acetylated microtubule bundles are not only found in the cytoplasm of major cellular types like neurons, but also in the microtubular substructures such as flagella, mitotic spindles, and cilia (Reed et al., 2006; Sadoul et al., 2010). In addition, acetylation aids the binding and mobility of the axonal motor proteins kinesin and dynein to the microtubules during axonal transport (Reed et al., 2006; Sadoul et al., 2010).
Figure 2.
The role of acetylation and deacetylation on microtubule stability. Microtubules are constantly being modified through the addition and removal of acetylation by HATs and HDACs. Binding of Microtubule associated proteins or MAPs to microtubules is also regulated by acetylation and also affects their stability (Cohen et al., 2011). These modifications allow for the rapid expansion and removal of microtubule structures for neurite outgrowth, synapse formation, transport through the axon, and maintaining the overall structure of the axon.
As mentioned above, HDAC6 plays an essential role in regulating the acetylation of both the α and β-tubulins of microtubules. Both in vivo and in vitro studies have shown that hypoacetylation or hyperacetylation of α-tubulin occurs by overexpressing or inhibiting HDAC6, respectively (Zhang et al., 2003). HDAC6 is present in the perinuclear region and is co-localized with microtubule-associated motor complexes, particularly with dynactin p150glued suggesting a role in microtubule-associated transport (Hubbert et al., 2002). Osseni et al. (2020) demonstrated that HDAC6 function is critical for neuromuscular junction stability and organization, involving not only the structure of microtubules but also the maintenance of the acetylcholine receptor clusters (Osseni et al., 2020). The class III HDAC SIRT2 also predominantly localizes to the cytoplasm and is involved in deacetylating α-tubulin at lysine-40. SIRT2 and HDAC6, when overexpressed, have been shown to coimmunoprecipitate suggesting that they belong to the same multiprotein network. However, they can be inhibited independently to achieve the hyperacetylation of α-tubulin (North et al., 2003).
Regulation of intermediate filaments
The acetylation of intermediate filaments has been less well studied, however, both vimentin and cytokeratin 8 have been shown to be acetylated in their lysine residues destabilizing the polymer structure and affecting their ability to maintain the cellular shape and rigidity (Leech et al., 2008; Drake et al., 2009). Less is known about the acetylation of intermediate filaments expressed in the brain such as the neurofilament proteins, and astrocytes expressing GFAP (Gao et al., 2007). A study examining the effect of two global HDACs inhibitors, Trichostatin A, and Sodium Butyrate on primary cultured astrocytes demonstrated that HDACs are responsible for maintaining the ratio of the two isoforms of GFAP; GFAPδ, and GFAPα (Gao et al., 2007), through transcription and splicing. Increased levels of GFAPδ resulted in a collapse of the GFAP cytoskeletal network (Kanski et al., 2014).
Altered acetylation of cytoskeletal proteins in neurodegenerative diseases
There is growing evidence of the links between altered cytoskeletal proteins and neurodegenerative disease, including alterations to microtubule stability which can be detrimental to axonal transport, as well as impairments in actin dynamics potentially leading to altered plasticity and accumulation of intermediate filament proteins (Luo, 2002; Saha and Pahan, 2006; Kapitein and Hoogenraad, 2015; Esteves et al., 2019). While there has been a focus on examining alterations to phosphorylation, a growing body of evidence has also implicated altered acetylation of these proteins in disease (Zhang and Benson, 2001; Cartelli et al., 2010; Esteves et al., 2019; Kim et al., 2020). The acetylation of the microtubule-associated protein tau has been discussed in the previous section; below we expand on other cytoskeletal proteins that may be implicated as contributing to neurodegenerative disease.
Altered acetylation of microtubules
Several studies have demonstrated altered acetylation of microtubules in neurodegenerative diseases and this has been linked to facilitating the aggregation of toxic proteins in axons (Liu et al., 2012; Brunden et al., 2017). Kim C. H. et al. (2012) reported a decrease in acetylated α-tubulin in the frontal cortex of 13-month-old 5XFAD AD mice, which overproduced Aβ proteins. The decreased acetylated α-tubulin was linked to alterations in axonal transport. Multiple studies have shown that increased acetylation of microtubules can rescue axonal transport in AD mouse models (Kim C. et al., 2012; Brunden et al., 2017). Inhibiting deacetylation in an AD mouse model restored axonal transport of mitochondria, which was associated with improved cognition (Kim C. et al., 2012). Another study by Zhang et al. (2014) used the APPswe/PS1ΔE9 mouse model of AD and showed that HDAC6 was increased in this model leading to higher levels of microtubule instability. Treatment with HDAC6 inhibitors (Tubastatin A & ACY-1215) resulted in increased acetylation of α-tubulin and improvement of mitochondrial transport due to recruitment of more kinesin-1 and dynein to the microtubules, which facilitated fusion of lysosomes and autophagosomes enhancing the removal of Aβ plaques from neuronal cells. This study suggested the connection between impaired cognitive function in AD and altered acetylation of microtubules (Zhang et al., 2014). Further studies support this hypothesis, as decreased microtubule acetylation in AD has also been associated with increased microtubule severing by the severing protein katanin, destabilizing the microtubules and affecting the transport of axonal cargo (Mao et al., 2017).
In PD, a number of proteins implicated in disease, including parkin (a ubiquitin-protein ligase protein), TPPP/p25 (involved in microtubule bundle formation), PINK1 (autophagy triggering protein), LRRK2 (regulates immune response), and α-synuclein, are linked to microtubules and have been shown to be involved in altered microtubule acetylation and PD pathogenesis (Bonifati, 2014; Oláh et al., 2017). Esteves et al. (2019) demonstrated that there was decreased microtubule acetylation in a cybrid cell model involving PD patient mitochondrial DNA that was linked to impaired axonal transport and altered cellular distribution of mitochondria (Esteves et al., 2014). Conversely, mutant α-synuclein has been shown to hyperacetylate microtubules, which accumulated in dopaminergic neurons and result in altered binding to kinesin 1 and destabilization (Smith and Stillman, 1991; Alim et al., 2004). Several studies have linked altered acetylation of microtubules with altered interaction between LRRK2 with microtubules in growth cones. As LLRK2 is involved in stabilizing microtubule dynamics, genetic mutations to LLRK2 impact the homeostasis of microtubule acetylation and axonal transport system leading to motor impairment (Esteves and Cardoso, 2017). In addition, parkin and α-synuclein have also been linked to increased accumulation of acetylated microtubules in dopaminergic neurons of the midbrain (Feng, 2006).
Like PD and AD, neuronal damage in ALS has also been linked to altered cytoskeletal properties, axon transport, and axonal degeneration (Peters et al., 2015) with altered acetylation of microtubules of particular importance (Gal et al., 2013). Over-expression of mutant SOD1 in animal models has been proposed to alter the acetylation of microtubules by interacting with the HDAC6 enzyme (Gal et al., 2013). One study in three SOD1 transgenic mouse models (A4V, G93A, and G85R) showed that mutant SOD1 and HDAC6 formed a complex, which promoted intraneuronal aggregation of HDAC6, hindering the enzymes’ microtubule deacetylation activity and leading to hyperacetylated microtubules in neurons. The hyperacetylated microtubules were related to increased levels of axonal transport and promoted the spread of pathology in the models (Gal et al., 2013). On the contrary, other studies suggest that decreased microtubule acetylation is implicated in mutant SOD1-associated ALS. For example, an in vitro study demonstrated that the microtubule-dependent ER-Golgi transport system was impaired by mutant SOD1 due to decreased microtubule acetylation and reduced microtubule stability in cultured motor neuron cells (Soo et al., 2015). Additionally, acetylated tubulin inclusions were found in mutant SOD1 aggregates (Soo et al., 2015). Other aggregated proteins present in the cytoplasm of induced pluripotent stem cells (iPSCs) derived from ALS patients, including TDP-43 and FUS, have also been linked with altered HDAC6 activity leading to impaired microtubule stability, ER vesicle dynamics, and mitochondria-dependent axonal transport (Fiesel et al., 2010; Guo et al., 2017; Naumann et al., 2018). Fiesel et al. (2010) investigated the effects of altered TDP-43 activity on HDAC6 and microtubule stability. They silenced the TDP-43 gene in both HEK293E and SH-SY5Y cell lines to simulate the disease condition where the nuclear activity of TDP-43 is downregulated and showed that this resulted in reduced HDAC6 mRNA and protein synthesis which was confirmed by the presence of hyperacetylated tubulin (Fiesel et al., 2010). Another study examined iPSC-derived motor neurons from ALS patients with point mutations in the FUS gene, demonstrating impaired axonal transport and lowered ER vesicle transportation which was rescued by HDAC6 inhibitors (Guo et al., 2017).
Altered acetylation of microtubules and resultant alterations in axonal transport is also implicated in HD pathogenesis (Mac Donald et al., 2003; Gauthier et al., 2004; Lee et al., 2004). Trushina et al. (2003) reported increased microtubule deacetylation after binding of mutant huntingtin protein with microtubules in primary neuronal culture models and transgenic mice (Trushina et al., 2003). Altered acetylation of microtubules in HD has been linked with impaired transport of brain-derived neurotrophic factor (BDNF), along with decreased recruitment of motor complexes such as dynein/dynactin and kinesin-1 to microtubules in studied primary cell culture and transgenic HD mouse models (Dompierre et al., 2007). Such deficits could be rescued by pharmacological intervention to increase microtubule acetylation and prevent neuronal damage (Dompierre et al., 2007). Furthermore, in vitro studies have also linked hyperacetylated microtubules with reduced vulnerability of striatal cells in HD, by improving autophagic flux and preventing mutant huntingtin diffusing into the neurons (Guedes-Dias et al., 2015).
Altered acetylation in microtubule-associated transport has been linked to a number of other neurodegenerative diseases such as Charcot–Marie–Tooth disease (CMT) and Rett syndrome (RTT) (Xu et al., 2014; Picci et al., 2020). A recent study of an inducible CMT type-2A (CMT2A) mutant MFN2R94Q mouse model with progressive motor and sensory neuronal degeneration demonstrated decreased acetylated α-tubulin in the distal sciatic nerves (Picci et al., 2020). Additionally, CMT type-2D (CMT2D) has been shown to have hypoacetylated α-tubulin-related axonal transport deficits in human stem cell models with the mutant glycyl-tRNA synthetase protein (gene mutation linked to CMT pathophysiology) found to bind to HDAC6 and increase its activity, causing a subsequent reduction in acetylated α-tubulin in peripheral nerves (Smith et al., 2022). The dynamin 2 gene (another gene mutation linked to CMT pathophysiology) is also a microtubule-associated protein, and mutations in the encoded protein have been shown to alter microtubule acetylation in patient-derived cells of CMT. Mutations of dynamin 2 resulted in the altered formation of the Golgi apparatus and microtubule-dependent transport, causing neurodegeneration and neuropathies (Tanabe and Takei, 2009). In RTT, lower levels of acetylated microtubules were present in cultured neuronal cells derived from the MeCP2 knock-out transgenic RTT mouse model, again altering BDNF vesicle transport, which was linked to altered dendritic growth and synaptic activity (Xu et al., 2014). Lebrun et al. (2021) recently linked loss of motor function and seizures in the Mecp2308/y transgenic RTT mouse model, where altered MeCP2 levels either caused overexpression of HDAC6 or impaired expression of microtubule-associated proteins, which was rescued with HDAC6 inhibitors (Lebrun et al., 2021). Another study used a MeCP2 deficient cell culture model which also resulted in overexpression of HDAC6, decreasing microtubule acetylation and reducing the structural stability of cilia (Frasca et al., 2020), and impairing the cilium-related Sonic Hedgehog signaling cascade pathway, which is required for forebrain development, The phenotype was improved by using HDAC6 inhibitor, tubacin (Frasca et al., 2020).
Role of altered acetylation in glial cells in neurodegenerative diseases
While there is a wide base of literature surrounding the role of acetylation in neurons during NDD, altered acetylation has been also studied in glial cells, and has been associated with neurodegeneration. Microglia have been implicated in the progression of several neurodegenerative diseases through inflammatory responses, synapse loss, failure to clear protein aggregates, or by activating neurotoxic astrocytes (Hansen et al., 2018; Madore et al., 2020). Studies have demonstrated the regulatory role of acetylation in the activation of microglia and their inflammatory responses in neurodegenerative diseases. For example, the pan HDAC inhibitor trichostatin A (TSA) potentiated lipopolysaccharide-induced inflammatory responses of microglia in murine N9 and rat primary cultured cells (Suuronen et al., 2003). On the contrary, in a transgenic Cx3cr1CreERT2 Hdac1fl/flHdac2fl/fl mouse model, deletion of HDAC1 and HDAC2 from microglial cells enhanced microglial phagocytic activity, which aided in clearing amyloid plaques and improved cognitive function (Datta et al., 2018). Another study demonstrated that acetylated and phosphorylated STAT3 (transcription factor) further activated the studied primary microglial cells (Eufemi et al., 2015).
Additionally, astrocytes have been shown to be regulated through post-translational acetylation processes. In a glial cell culture model, HDAC inhibitors such as TSA upregulated release of neurotrophic factors including GDNF and BDNF from astrocytes followed by protecting dopaminergic neurons (Chen et al., 2006; Wu et al., 2008). On the other hand, SIRT2 inhibitor AGK2 reduced the astrocyte activation level as well as pro-inflammatory factors in a primary cell culture AD model (Scuderi et al., 2014). These data indicate the complex regulatory function of acetylation in glial cells in both healthy and disease states (Neal and Richardson, 2018).
The future of PTMs as therapeutics for neurodegenerative disease
In this review, we have discussed the vital role of acetylation in the nervous system, and how dysregulation can contribute to the pathogenesis or degeneration seen in neurodegenerative disease. Although we are still in the early stages of research into therapeutics targeting HATs, HDAC inhibitors (HDACi) have been developed in both the preclinical and clinical settings for several years. Treatments targeting HDACs have been widely implicated in cancer therapies, possibly due to an imbalance in acetylation levels closely related to the occurrence of cancers (Mohamed et al., 2007; Chen et al., 2013; Hashimoto et al., 2013; Xu et al., 2019; Wu et al., 2020). Indeed, multiple pan-HDACi have been approved by the FDA for the treatment of cancer, including vorinostat/suberoylanilide hydroxamic acid (SAHA; Marks and Breslow, 2007), belinostat (PXD-101; Foss et al., 2015), and panobinostat (LBH589; Moore, 2016). SAHA was the first FDA-approved HDACi in 2006 and has been successful in treating T-cell lymphomas by reducing the expression of mutant regulatory proteins such as oncogene mutant p53 (Foggetti et al., 2019). Additionally, both pan-HDACi belinostat and panobinostat have been used to treat cutaneous T-cell lymphoma and myeloma (Eckschlager et al., 2017).
Given the success of HDACi in cancer treatment, researchers have sought to repurpose these FDA-approved drugs for neurodegenerative disease, with some FDA-approved HDACi being effective for treating models of HD (Hockly et al., 2003), ALS, PD, and AD (Athira et al., 2021). However, due to the broad range of pan-HDACi targets within cells, more targeted HDACi are being tested pre-clinically such as the HDAC6 specific molecules like ACY-738 or tubastatin A. ACY-1215/Ricolinostat was one of the first HDAC6 specific inhibitors to enter clinical trials for myeloma, lymphoma and metastatic breast cancer (Vogl et al., 2017; Silva et al., 2020; Amengual et al., 2021). The same compound is also in phase II clinical trials for diabetic neuropathy (NCT03176472) and CMT (Benoy et al., 2017). Recently discovered HDACi such as EVP-0334, RDN-929, and CKD-504 are under different phases of clinical trials for the treatment of FTLD, AD, PD, and HD (Rodrigues et al., 2020), and similar compounds such as ACY-738 and Tubastatin A have also shown promising results ameliorating the disease progression in preclinical studies of AD, ALS-FUS, and MS (Multiple Sclerosis) (Kim C. et al., 2012; Guo et al., 2017; LoPresti, 2019). Despite the promise shown by HDACi in treating neurodegenerative diseases, a few caveats need to be considered, including the use of therapies that can cause global hypo or hyper-acetylation in the nervous system.
Although many studies show hypoacetylation to be critical to neurodegeneration processes (Saha and Pahan, 2006), other studies have demonstrated that hyperacetylation is implicated in the pathogenesis of neurodegenerative diseases (Bonet-Ponce et al., 2016; Sohn et al., 2016). In the healthy nervous system, acetylation is highly redundant and exquisitely regulated, however, the use of HAT modifying enzymes or HDACi may lead to off-target effects. The use of HDACi like sodium butyrate has led to hyperacetylation of histone H4 which subsequently facilitated the expression of an oxidative stress-sensitive protein known as PKCδ and caused neurotoxicity in human dopaminergic neuronal cells (Jin et al., 2014). Additionally, oxidative stress molecules that are produced during the progression of these neurodegenerative diseases have been shown to hyperacetylate microtubules and disrupt autophagic trafficking in studied ARPE-19 cells which were exposed to Rotenone drug to induce oxidative stress. This stress-induced microtubule hyperacetylation was successfully reduced by using free radical scavenger drugs like N-acetylcysteine (Bonet-Ponce et al., 2016). Further studies are required to elucidate whether hypoacetylation or hyperacetylation has therapeutic benefits for axons in neurodegenerative diseases.
There are several hurdles to overcome for the use of HDACi in treating neurodegenerative diseases, that focus on two core themes: target preference (developing HDACi for selective isoforms or families), and selective delivery (the ability to target specific tissue or cell type for therapy). Small molecule inhibitors, such as ACY molecules, ACY-738, and ACY-1215 are prime examples of target preference and specificity, as both compounds have been reported to be HDAC6-specific inhibitors (Santo et al., 2012). However, their specificity depends greatly on the tissue uptake and overall dose. For example, high levels of ACY-738 have been reported to alter H3 acetylation levels in mesangial cell lines and the spinal cord of WT mice (Regna et al., 2015; Rossaert et al., 2019). Target specificity also remains a caveat in the use of PTM modifying drugs in the clinic for neurodegenerative diseases. For instance, reports have suggested that HDAC6i ACY-738 increased the life span by 41 days in transgenic FUS+/+ HDAC6 knock-out mice, suggesting potential off-target activity of ACY-738 drug (Rossaert et al., 2019). Additionally, some other off-target effects of these inhibitors can be cellular apoptosis or T-cell-mediated immune responses (Majid et al., 2015).
Further difficulty for potential drug candidates of NDD is the ability for novel small molecule inhibitors to cross the blood brain barrier (BBB). HDACi such as MS-275, SAHA, valproic acid and Tubastatin A showed low BBB permeability in studies (Choi et al., 2019). As a result, achieving the required therapeutic efficacy requires higher dosing, which can lead to potential off-target effects (Choi et al., 2019). Literature suggests that benzylic amide derivatives showed higher BBB permeability and inhibitory activity in the baboon model against HDAC1 and 2. The BBB permeability of these drug derivatives were optimized through image guided synthetic process (Seo et al., 2014). Another recent study designed benzoheterocycle derivatives that were structurally different from the available HDACi and identified benzothiazole derivative 9b which showed higher BBB permeability than SAHA. These approaches for developing HDACi should be further studied to achieve the desired therapeutic treatment for neurodegenerative diseases (Choi et al., 2019). Further refinement of selectivity and delivery methods remain to be developed. To overcome the limitations of tissue specificity and off-target effects of HDACi, advanced genomic targeting methodologies such as CRISPR-Cas has been used to control targeted HAT/HDAC activity. Studies have successfully fused dCas9 to the p300 acetyltransferase to catalyze acetylation of H3 followed by transcriptional activation of the targeted genes (Hilton et al., 2015; Shrimp et al., 2018), however, methods for cell-type specific treatment remain to be developed.
Conclusion
In conclusion, acetylation plays a critical role in maintaining the homeostasis of cellular proteins and cytoskeleton. This tightly regulated process has been shown to be dysregulated in neurodegenerative disease; however, the underlying pathophysiological mechanisms are yet to be fully understood. Nevertheless, through emerging therapeutics, altered acetylation can be a promising target to limit or prevent the pathological processes that lead to protein aggregation or defects of axonal transport in neurodegenerative diseases.
Author contributions
FK, RA, AP, and AK: conceptualization. FK: writing - original draft. FK, RA, AP, AC, and AK: writing - reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by FightMND, MND Research Australia, the National Health and Medical Research Council of Australia, and the JO and JR Wicking Trust (equity trustees). AK NHMRC fellowship number is APP1136913.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the authors, reviewers, and editors who helped to prepare and publish this manuscript.
References
- Adhikari R., Yang M., Saikia N., Dutta C., Alharbi W. F., Shan Z., et al. (2020). Acetylation of Aβ42 at lysine 16 disrupts amyloid formation. ACS Chem. Neurosci. 11, 1178–1191. doi: 10.1021/acschemneuro.0c00069, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agis-Balboa R. C., Pavelka Z., Kerimoglu C., Fischer A. (2013). Loss of HDAC5 impairs memory function: implications for Alzheimer's disease. J. Alzheimers Dis. 33, 35–44. doi: 10.3233/JAD-2012-121009, PMID: [DOI] [PubMed] [Google Scholar]
- Akter R., Afrose A., Rahman M. R., Chowdhury R., Nirzhor S. S. R., Khan R. I., et al. (2021). A comprehensive analysis into the therapeutic application of natural products as SIRT6 modulators in Alzheimer’s disease, aging, cancer, inflammation, and diabetes. Int. J. Mol. Sci. 22:4180. doi: 10.3390/ijms22084180, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alim M. A., Ma Q.-L., Takeda K., Aizawa T., Matsubara M., Nakamura M., et al. (2004). Demonstration of a role for α-synuclein as a functional microtubule-associated protein. J. Alzheimers Dis. 6, 435–442. doi: 10.3233/JAD-2004-6412, PMID: [DOI] [PubMed] [Google Scholar]
- Alliot F., Godin I., Pessac B. (1999). Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Dev. Brain Res. 117, 145–152. doi: 10.1016/S0165-3806(99)00113-3, PMID: [DOI] [PubMed] [Google Scholar]
- Amengual J. E., Lue J. K., Ma H., Lichtenstein R., Shah B., Cremers S., et al. (2021). First-in-class selective HDAC6 inhibitor (ACY-1215) has a highly favorable safety profile in patients with relapsed and refractory lymphoma. Oncologist 26, 184–e366. doi: 10.1002/onco.13673, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z., Huang L. E., Eckner R., Bhattacharya S., Jiang C., Goldberg M. A., et al. (1996). An essential role for p 300/CBP in the cellular response to hypoxia. Proc. Natl. Acad. Sci. 93, 12969–12973. doi: 10.1073/pnas.93.23.12969, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif M., Selvi B. R., Kundu T. K. (2010). Lysine acetylation: the tale of a modification from transcription regulation to metabolism. Chembiochem 11, 1501–1504. doi: 10.1002/cbic.201000292, PMID: [DOI] [PubMed] [Google Scholar]
- Arnesen T., Starheim K. K., Van Damme P., Evjenth R., Dinh H., Betts M. J., et al. (2010). The chaperone-like protein HYPK acts together with Nat a in cotranslational N-terminal acetylation and prevention of huntingtin aggregation. Mol. Cell. Biol. 30, 1898–1909. doi: 10.1128/MCB.01199-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold M. A., Kim Y., Czubryt M. P., Phan D., McAnally J., Qi X., et al. (2007). MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev. Cell 12, 377–389. doi: 10.1016/j.devcel.2007.02.004, PMID: [DOI] [PubMed] [Google Scholar]
- Ashby M. (2019). Dementia: Unwelcome Change Has Arrived and We Are Not Ready! J. Bioeth. Inq. 16, 143–146. doi: 10.1007/s11673-019-09921-5, PMID: [DOI] [PubMed] [Google Scholar]
- Athira K., Sadanandan P., Chakravarty S. (2021). Repurposing Vorinostat for the treatment of disorders affecting brain. NeuroMolecular Med. 23, 449–465. doi: 10.1007/s12017-021-08660-4, [DOI] [PubMed] [Google Scholar]
- Avvakumov N., Côté J. (2007). Functions of myst family histone acetyltransferases and their link to disease. Chrom. Dis. 41, 301–322. doi: 10.1007/1-4020-5466-1_13 [DOI] [PubMed] [Google Scholar]
- Baeza J., Smallegan M. J., Denu J. M. (2016). Mechanisms and dynamics of protein acetylation in mitochondria. Trends Biochem. Sci. 41, 231–244. doi: 10.1016/j.tibs.2015.12.006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks C., Andersen J. (2019). Mechanisms of SOD1 regulation by post-translational modifications. Redox Biol. 26:101270. doi: 10.1016/j.redox.2019.101270, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bánréti Á., Sass M., Graba Y. (2013). The emerging role of acetylation in the regulation of autophagy. Autophagy 9, 819–829. doi: 10.4161/auto.23908, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber S. C., Mead R. J., Shaw P. J. (2006). Oxidative stress in ALS: a mechanism of neurodegeneration and a therapeutic target. BBA Mol. Basis Dis. 1762, 1051–1067. doi: 10.1016/j.bbadis.2006.03.008, PMID: [DOI] [PubMed] [Google Scholar]
- Benoy V., Vanden Berghe P., Jarpe M., Van Damme P., Robberecht W., Van Den Bosch L. (2017). Development of improved HDAC6 inhibitors as pharmacological therapy for axonal Charcot–Marie–tooth disease. Neurotherapeutics 14, 417–428. doi: 10.1007/s13311-016-0501-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G., Guarente L. (2004). The sir 2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417–435. doi: 10.1146/annurev.biochem.73.011303.073651, PMID: [DOI] [PubMed] [Google Scholar]
- Bock A. S., Murthy A. C., Tang W. S., Jovic N., Shewmaker F., Mittal J., et al. (2021). N-terminal acetylation modestly enhances phase separation and reduces aggregation of the low-complexity domain of RNA-binding protein fused in sarcoma. Protein Sci. 30, 1337–1349. doi: 10.1002/pro.4029, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodai L., Pallos J., Thompson L. M., Marsh J. L. (2012). Pcaf modulates polyglutamine pathology in a drosophila model of Huntington’s disease. Neurodegener. Dis. 9, 104–106. doi: 10.1159/000330505, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonda D. J., Lee H.-g., Camins A., Pallàs M., Casadesus G., Smith M. A., et al. (2011). The sirtuin pathway in ageing and Alzheimer disease: mechanistic and therapeutic considerations. Lancet Neurol. 10, 275–279. doi: 10.1016/S1474-4422(11)70013-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonet-Ponce L., Saez-Atienzar S., Da Casa C., Sancho-Pelluz J., Barcia J. M., Martinez-Gil N., et al. (2016). Rotenone induces the formation of 4-hydroxynonenal aggresomes. Role of ROS-mediated tubulin hyperacetylation and autophagic flux disruption. Mol. Neurobiol. 53, 6194–6208. doi: 10.1007/s12035-015-9509-3, PMID: [DOI] [PubMed] [Google Scholar]
- Bonifati V. (2014). Genetics of Parkinson's disease–state of the art, (2013). Parkinsonism Relat. Disord. 20, S23–S28. doi: 10.1016/S1353-8020(13)70009-9, PMID: [DOI] [PubMed] [Google Scholar]
- Boutillier A. L., Trinh E., Loeffler J. P. (2003). Selective E2F-dependent gene transcription is controlled by histone deacetylase activity during neuronal apoptosis. J. Neurochem. 84, 814–828. doi: 10.1046/j.1471-4159.2003.01581.x, PMID: [DOI] [PubMed] [Google Scholar]
- Broide R. S., Redwine J. M., Aftahi N., Young W., Bloom F. E., Winrow C. (2007). Distribution of histone deacetylases 1–11 in the rat brain. J. Mol. Neurosci. 31, 47–58. doi: 10.1007/BF02686117, PMID: [DOI] [PubMed] [Google Scholar]
- Brotzakis Z. F., Lindstedt P. R., Taylor R. J., Rinauro D. J., Gallagher N. C., Bernardes G. J., et al. (2021). A structural ensemble of a tau-microtubule complex reveals regulatory tau phosphorylation and acetylation mechanisms. ACS Centr. Sci. 7, 1986–1995. doi: 10.1021/acscentsci.1c00585, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunden K. R., Lee V. M., Smith A. B., III, Trojanowski J. Q., Ballatore C. (2017). Altered microtubule dynamics in neurodegenerative disease: therapeutic potential of microtubule-stabilizing drugs. Neurobiol. Dis. 105, 328–335. doi: 10.1016/j.nbd.2016.12.021, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E. (2018). TDP-43 post-translational modifications in health and disease. Expert Opin. Ther. Targets 22, 279–293. doi: 10.1080/14728222.2018.1439923, PMID: [DOI] [PubMed] [Google Scholar]
- Caccamo A., Maldonado M. A., Bokov A. F., Majumder S., Oddo S. (2010). CBP gene transfer increases BDNF levels and ameliorates learning and memory deficits in a mouse model of Alzheimer's disease. Proc. Natl. Acad. Sci. 107, 22687–22692. doi: 10.1073/pnas.1012851108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns N. J., Lee V. M. Y., Trojanowski J. Q. (2004). The cytoskeleton in neurodegenerative diseases. J. Pathol. 204, 438–449. doi: 10.1002/path.1650, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartelli D., Ronchi C., Maggioni M. G., Rodighiero S., Giavini E., Cappelletti G. (2010). Microtubule dysfunction precedes transport impairment and mitochondria damage in MPP+-induced neurodegeneration. J. Neurochem. 115, 247–258. doi: 10.1111/j.1471-4159.2010.06924.x, PMID: [DOI] [PubMed] [Google Scholar]
- Caslini C., Hong S., Ban Y. J., Chen X. S., Ince T. A. (2019). HDAC7 regulates histone 3 lysine 27 acetylation and transcriptional activity at super-enhancer-associated genes in breast cancer stem cells. Oncogene 38, 6599–6614. doi: 10.1038/s41388-019-0897-0, PMID: [DOI] [PubMed] [Google Scholar]
- Cavallarin N., Vicario M., Negro A. (2010). The role of phosphorylation in synucleinopathies: focus on Parkinson's disease. CNS Neurol. Disord. Drug Targets 9, 471–481. doi: 10.2174/187152710791556140, PMID: [DOI] [PubMed] [Google Scholar]
- Chai Y., Shao J., Miller V. M., Williams A., Paulson H. L. (2002). Live-cell imaging reveals divergent intracellular dynamics of polyglutamine disease proteins and supports a sequestration model of pathogenesis. Proc. Natl. Acad. Sci. 99, 9310–9315. doi: 10.1073/pnas.152101299, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A., Oehme I., Witt O., Oliveira G., Sippl W., Romier C., et al. (2015). HDAC8: a multifaceted target for therapeutic interventions. Trends Pharmacol. Sci. 36, 481–492. doi: 10.1016/j.tips.2015.04.013, PMID: [DOI] [PubMed] [Google Scholar]
- Chang S., McKinsey T. A., Zhang C. L., Richardson J. A., Hill J. A., Olson E. N. (2004). Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 24, 8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Young B. D., Li S., Qi X., Richardson J. A., Olson E. N. (2006). Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cells 126, 321–334. doi: 10.1016/j.cell.2006.05.040, PMID: [DOI] [PubMed] [Google Scholar]
- Chen Y. C., Gatchel J. R., Lewis R. W., Mao C.-A., Grant P. A., Zoghbi H. Y., et al. (2012). Gcn 5 loss-of-function accelerates cerebellar and retinal degeneration in a SCA7 mouse model. Hum. Mol. Genet. 21, 394–405. doi: 10.1093/hmg/ddr474, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. W., Kao S. Y., Wang H. J., Yang M. H. (2013). Histone modification patterns correlate with patient outcome in oral squamous cell carcinoma. Cancer 119, 4259–4267. doi: 10.1002/cncr.28356, PMID: [DOI] [PubMed] [Google Scholar]
- Chen X., Li Y., Wang C., Tang Y., Mok S.-A., Tsai R. M., et al. (2020). Promoting tau secretion and propagation by hyperactive p 300/CBP via autophagy-lysosomal pathway in tauopathy. Mol. Neurodegener. 15, 1–19. doi: 10.1186/s13024-019-0354-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-A., Lu C.-H., Ke C.-C., Chang C.-W., Yang B.-H., Gelovani J.G., et al. (eds.) (2020). “Monitoring HDAC4 expression in Alzheimer’s disease using [18 F] TFAHA-PET.” in International Conference of FASMI (FEDERATION OF ASIAN SOCIETIES FOR MOLECULAR IMAGING). Springer.
- Chen P. S., Peng G., Li G., Yang S., Wu X., Wang C., et al. (2006). Valproate protects dopaminergic neurons in midbrain neuron/glia cultures by stimulating the release of neurotrophic factors from astrocytes. Mol. Psychiatry 11, 1116–1125. doi: 10.1038/sj.mp.4001893, PMID: [DOI] [PubMed] [Google Scholar]
- Chiki A., DeGuire S. M., Ruggeri F. S., Sanfelice D., Ansaloni A., Wang Z. M., et al. (2017). Mutant exon 1 huntingtin aggregation is regulated by T3 phosphorylation-induced structural changes and crosstalk between T3 phosphorylation and acetylation at K6. Angew. Chem. Int. Ed. 56, 5202–5207. doi: 10.1002/anie.201611750, PMID: [DOI] [PubMed] [Google Scholar]
- Cho Y., Cavalli V. (2012). HDAC5 is a novel injury-regulated tubulin deacetylase controlling axon regeneration. EMBO J. 31, 3063–3078. doi: 10.1038/emboj.2012.160, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M. A., Park S. Y., Chae H. Y., Song Y., Sharma C., Seo Y. H. (2019). Design, synthesis and biological evaluation of a series of CNS penetrant HDAC inhibitors structurally derived from amyloid-β probes. Sci. Rep. 9:13187. doi: 10.1038/s41598-019-49784-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., et al. (2009). Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840. doi: 10.1126/science.1175371, PMID: [DOI] [PubMed] [Google Scholar]
- Choudhary C., Weinert B. T., Nishida Y., Verdin E., Mann M. (2014). The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 15, 536–550. doi: 10.1038/nrm3841, PMID: [DOI] [PubMed] [Google Scholar]
- Cohen T. J., Guo J. L., Hurtado D. E., Kwong L. K., Mills I. P., Trojanowski J. Q., et al. (2011). The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat. Commun. 2:252. doi: 10.1038/ncomms1255, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T. J., Hwang A. W., Restrepo C. R., Yuan C.-X., Trojanowski J. Q., Lee V. M. (2015). An acetylation switch controls TDP-43 function and aggregation propensity. Nat. Commun. 6:5845. doi: 10.1038/ncomms6845, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M. M., Jr. (2006). The new bone biology: pathologic, molecular, and clinical correlates. Am. J. Med. Genet. A 140, 2646–2706. doi: 10.1002/ajmg.a.31368, PMID: [DOI] [PubMed] [Google Scholar]
- Cong X., Held J. M., DeGiacomo F., Bonner A., Chen J. M., Schilling B., et al. (2011). Mass spectrometric identification of novel lysine acetylation sites in huntingtin. Mol. Cell. Proteomics 10:M111.009829. doi: 10.1074/mcp.M111.009829, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong S.-Y., Pepers B. A., Evert B. O., Rubinsztein D. C., Roos R. A., van Ommen G.-J. B., et al. (2005). Mutant huntingtin represses CBP, but not p 300, by binding and protein degradation. Mol. Cell. Neurosci. 30, 12–23. doi: 10.1016/j.mcn.2005.05.003, PMID: [DOI] [PubMed] [Google Scholar]
- Cook C., Carlomagno Y., Gendron T. F., Dunmore J., Scheffel K., Stetler C., et al. (2014a). Acetylation of the KXGS motifs in tau is a critical determinant in modulation of tau aggregation and clearance. Hum. Mol. Genet. 23, 104–116. doi: 10.1093/hmg/ddt402, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook C., Stankowski J. N., Carlomagno Y., Stetler C., Petrucelli L. (2014b). Acetylation: a new key to unlock tau’s role in neurodegeneration. Alzheimers Res. Ther. 6:29. doi: 10.1186/alzrt259, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe V. T. (2004). Histone deacetylase 1 is required to repress notch target gene expression during zebrafish neurogenesis and to maintain the production of motoneurones in response to hedgehog signalling. Development 131, 2983–2995. doi: 10.1242/dev.01166 [DOI] [PubMed] [Google Scholar]
- Datta M., Staszewski O., Raschi E., Frosch M., Hagemeyer N., Tay T. L., et al. (2018). Histone deacetylases 1 and 2 regulate microglia function during development, homeostasis, and neurodegeneration in a context-dependent manner. Immunity 48, 514–529.e6. doi: 10.1016/j.immuni.2018.02.016, PMID: [DOI] [PubMed] [Google Scholar]
- Dave N., Vural A. S., Piras I. S., Winslow W., Surendra L., Winstone J. K., et al. (2021). Identification of retinoblastoma binding protein 7 (Rbbp 7) as a mediator against tau acetylation and subsequent neuronal loss in Alzheimer’s disease and related tauopathies. Acta Neuropathol. 142, 279–294. doi: 10.1007/s00401-021-02323-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Oliveira R. M., Vicente Miranda H., Francelle L., Pinho R., Szegö É. M., Martinho R., et al. (2017). The mechanism of sirtuin 2–mediated exacerbation of alpha-synuclein toxicity in models of Parkinson disease. PLoS Biol. 15:e2000374. doi: 10.1371/journal.pbio.2000374, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S., Marmorstein R. (2021). Protein N-terminal acetylation: structural basis, mechanism, versatility, and regulation. Trends Biochem. Sci. 46, 15–27. doi: 10.1016/j.tibs.2020.08.005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S., Pan B., Gottlieb L., Petersson E. J., Marmorstein R. (2020). Molecular basis for N-terminal alpha-synuclein acetylation by human Nat B. elife 9:e57491. doi: 10.7554/eLife.57491, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derisbourg M., Leghay C., Chiappetta G., Fernandez-Gomez F.-J., Laurent C., Demeyer D., et al. (2015). Role of the tau N-terminal region in microtubule stabilization revealed by newendogenous truncated forms. Sci. Rep. 5:9659. doi: 10.1038/srep09659, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didonna A., Benetti F. (2016). Post-translational modifications in neurodegeneration. Aims Biophys. 3, 27–49. doi: 10.3934/biophy.2016.1.27 [DOI] [Google Scholar]
- Didonna A., Opal P. (2015). The promise and perils of HDAC inhibitors in neurodegeneration. Ann. Clin. Trans. Neurol. 2, 79–101. doi: 10.1002/acn3.147, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M., Sapp E., Chase K., Schwarz C., Meloni A., Young C., et al. (1995). Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron 14, 1075–1081. doi: 10.1016/0896-6273(95)90346-1, PMID: [DOI] [PubMed] [Google Scholar]
- Dompierre J. P., Godin J. D., Charrin B. C., Cordelieres F. P., King S. J., Humbert S., et al. (2007). Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington's disease by increasing tubulin acetylation. J. Neurosci. 27, 3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey O. M., Foster C. T., Cowley S. M. (2010). Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc. Natl. Acad. Sci. 107, 8242–8247. doi: 10.1073/pnas.1000478107, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake P. J., Griffiths G. J., Shaw L., Benson R. P., Corfe B. M. (2009). Application of high-content analysis to the study of post-translational modifications of the cytoskeleton. J. Proteome Res. 8, 28–34. doi: 10.1021/pr8006396, PMID: [DOI] [PubMed] [Google Scholar]
- Drazic A., Myklebust L. M., Ree R., Arnesen T. (2016). The world of protein acetylation. Biochim. Biophys. Acta Proteins Proteom. 1864, 1372–1401. doi: 10.1016/j.bbapap.2016.06.007, PMID: [DOI] [PubMed] [Google Scholar]
- Duan G., Walther D. (2015). The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput. Biol. 11:e1004049. doi: 10.1371/journal.pcbi.1004049, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Ydewalle C., Krishnan J., Chiheb D. M., Van Damme P., Irobi J., Kozikowski A. P., et al. (2011). HDAC6 inhibitors reverse axonal loss in a mouse model of mutant HSPB1–induced Charcot-Marie-tooth disease. Nat. Med. 17, 968–974. doi: 10.1038/nm.2396, PMID: [DOI] [PubMed] [Google Scholar]
- Eckschlager T., Plch J., Stiborova M., Hrabeta J. (2017). Histone deacetylase inhibitors as anticancer drugs. Int. J. Mol. Sci. 18:1414. doi: 10.3390/ijms18071414, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkkinen M. G., Kim M.-O., Geschwind M. D. (2018). Clinical neurology and epidemiology of the major neurodegenerative diseases. Cold Spring Harb. Perspect. Biol. 10:a033118. doi: 10.1101/cshperspect.a033118, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves A. R., Cardoso S. M. (2017). LRRK2 at the crossroad between autophagy and microtubule trafficking: insights into Parkinson’s disease. Neuroscientist 23, 16–26. doi: 10.1177/1073858415616558, PMID: [DOI] [PubMed] [Google Scholar]
- Esteves A., Gozes I., Cardoso S. (2014). The rescue of microtubule-dependent traffic recovers mitochondrial function in Parkinson's disease. BBA Mol. Basis Dis. 1842, 7–21. doi: 10.1016/j.bbadis.2013.10.003, PMID: [DOI] [PubMed] [Google Scholar]
- Esteves A., Palma A., Gomes R., Santos D., Silva D., Cardoso S. (2019). Acetylation as a major determinant to microtubule-dependent autophagy: relevance to Alzheimer's and Parkinson disease pathology. BBA Mol. Basis Dis. 1865, 2008–2023. doi: 10.1016/j.bbadis.2018.11.014, PMID: [DOI] [PubMed] [Google Scholar]
- Eufemi M., Cocchiola R., Romaniello D., Correani V., Di Francesco L., Fabrizi C., et al. (2015). Acetylation and phosphorylation of STAT3 are involved in the responsiveness of microglia to beta amyloid. Neurochem. Int. 81, 48–56. doi: 10.1016/j.neuint.2015.01.007, PMID: [DOI] [PubMed] [Google Scholar]
- Fan F., Li S., Wen Z., Ye Q., Chen X., Ye Q. (2020). Regulation of PGC-1α mediated by acetylation and phosphorylation in MPP+ induced cell model of Parkinson’s disease. Aging (Albany NY) 12, 9461–9474. doi: 10.18632/aging.103219, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina S., Esposito F., Battistoni M., Biamonti G., Francia S. (2021). Post-translational modifications modulate proteinopathies of TDP-43, FUS and hn RNP-A/B in amyotrophic lateral sclerosis. Front. Mol. Biosci. 8:585. doi: 10.3389/fmolb.2021.693325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J. (2006). Microtubule: a common target for parkin and Parkinson’s disease toxins. Neuroscientist 12, 469–476. doi: 10.1177/1073858406293853, PMID: [DOI] [PubMed] [Google Scholar]
- Fiesel F. C., Voigt A., Weber S. S., Van den Haute C., Waldenmaier A., Görner K., et al. (2010). Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 29, 209–221. doi: 10.1038/emboj.2009.324, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W., Dequiedt F., Hendzel M. J., Guenther M. G., Lazar M. A., Voelter W., et al. (2002). Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell 9, 45–57. doi: 10.1016/S1097-2765(01)00429-4, PMID: [DOI] [PubMed] [Google Scholar]
- Foggetti G., Ottaggio L., Russo D., Mazzitelli C., Monti P., Degan P., et al. (2019). Autophagy induced by SAHA affects mutant P 53 degradation and cancer cell survival. Biosci. Rep. 39:BSR20181345. doi: 10.1042/BSR20181345, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss F., Advani R., Duvic M., Hymes K. B., Intragumtornchai T., Lekhakula A., et al. (2015). A phase II trial of Belinostat (PXD 101) in patients with relapsed or refractory peripheral or cutaneous T-cell lymphoma. Br. J. Haematol. 168, 811–819. doi: 10.1111/bjh.13222, PMID: [DOI] [PubMed] [Google Scholar]
- François-Moutal L., Perez-Miller S., Scott D. D., Miranda V. G., Mollasalehi N., Khanna M. (2019). Structural insights into TDP-43 and effects of post-translational modifications. Front. Mol. Neurosci. 12:301. doi: 10.3389/fnmol.2019.00301, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasca A., Spiombi E., Palmieri M., Albizzati E., Valente M. M., Bergo A., et al. (2020). MECP2 mutations affect ciliogenesis: a novel perspective for Rett syndrome and related disorders. EMBO Mol. Med. 12:e10270. doi: 10.15252/emmm.201910270, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye R. A. (2000). Phylogenetic classification of prokaryotic and eukaryotic sir 2-like proteins. Biochem. Biophys. Res. Commun. 273, 793–798. doi: 10.1006/bbrc.2000.3000, PMID: [DOI] [PubMed] [Google Scholar]
- Gal J., Chen J., Barnett K. R., Yang L., Brumley E., Zhu H. (2013). HDAC6 regulates mutant SOD1 aggregation through two SMIR motifs and tubulin acetylation. J. Biol. Chem. 288, 15035–15045. doi: 10.1074/jbc.M112.431957, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.-s., Hubbert C. C., Lu J., Lee Y.-S., Lee J.-Y., Yao T.-P. (2007). Histone deacetylase 6 regulates growth factor-induced actin remodeling and endocytosis. Mol. Cell. Biol. 27, 8637–8647. doi: 10.1128/MCB.00393-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Morato J., Hans F., von Zweydorf F., Feederle R., Elsässer S. J., Skodras A. A., et al. (2022). Sirtuin-1 sensitive lysine-136 acetylation drives phase separation and pathological aggregation of TDP-43. Nat. Commun. 13:1223. doi: 10.1038/s41467-022-28822-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Venzor A., Toiber D. (2021). SIRT6 through the brain evolution, development, and aging. Front. Aging Neurosci. 13:747989. doi: 10.3389/fnagi.2021.747989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier L. R., Charrin B. C., Borrell-Pagès M., Dompierre J. P., Rangone H., Cordelières F. P., et al. (2004). Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cells 118, 127–138. doi: 10.1016/j.cell.2004.06.018, PMID: [DOI] [PubMed] [Google Scholar]
- Gehrking K. M., Andresen J. M., Duvick L., Lough J., Zoghbi H. Y., Orr H. T. (2011). Partial loss of tip 60 slows mid-stage neurodegeneration in a spinocerebellar ataxia type 1 (SCA1) mouse model. Hum. Mol. Genet. 20, 2204–2212. doi: 10.1093/hmg/ddr108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgala P. A., Carr C. B., Price D. J. (2011). The role of Pax 6 in forebrain development. Dev. Neurobiol. 71, 690–709. doi: 10.1002/dneu.20895, PMID: [DOI] [PubMed] [Google Scholar]
- Glozak M. A., Sengupta N., Zhang X., Seto E. (2005). Acetylation and deacetylation of non-histone proteins. Gene 363, 15–23. doi: 10.1016/j.gene.2005.09.010, PMID: [DOI] [PubMed] [Google Scholar]
- Goedert M., Spillantini M., Jakes R., Rutherford D., Crowther R. (1989). Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer's disease. Neuron 3, 519–526. doi: 10.1016/0896-6273(89)90210-9, PMID: [DOI] [PubMed] [Google Scholar]
- Gottlieb L., Guo L., Shorter J., Marmorstein R. (2021). N-alpha-acetylation of huntingtin protein increases its propensity to aggregate. J. Biol. Chem. 297:101363. doi: 10.1016/j.jbc.2021.101363, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J.-S., Haggarty S. J., Giacometti E., Dannenberg J.-H., Joseph N., Gao J., et al. (2009). HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459, 55–60. doi: 10.1038/nature07925, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. (2011). The logic linking protein acetylation and metabolism. Cell Metab. 14, 151–153. doi: 10.1016/j.cmet.2011.07.007, PMID: [DOI] [PubMed] [Google Scholar]
- Guedes-Dias P., de Proença J., Soares T. R., Leitão-Rocha A., Pinho B. R., Duchen M. R., et al. (2015). HDAC6 inhibition induces mitochondrial fusion, autophagic flux and reduces diffuse mutant huntingtin in striatal neurons. BBA Mol. Basis Dis. 1852, 2484–2493. doi: 10.1016/j.bbadis.2015.08.012, PMID: [DOI] [PubMed] [Google Scholar]
- Guha S., Subramaniyam R. (2021). Amyloid beta peptides, tau protein acetylation and therapeutic strategies for treating Alzheimer’s disease: a review. Life Res. 4:9. doi: 10.53388/life2021-0121-305, PMID: 35777803 [DOI] [Google Scholar]
- Guo W., Naujock M., Fumagalli L., Vandoorne T., Baatsen P., Boon R., et al. (2017). HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat. Commun. 8, 209–221. doi: 10.1038/emboj.2009.324, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Van Den Bosch L. (2018). Therapeutic potential of HDAC6 in amyotrophic lateral sclerosis. Cell Stress 2, 14–16. doi: 10.15698/cst2018.01.120, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M., Mokalled M. H., Montgomery R. L., Olson E. N. (2009a). Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 23, 1625–1630. doi: 10.1101/gad.1809209, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M., Montgomery R. L., Olson E. N. (2009b). The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 10, 32–42. doi: 10.1038/nrg2485, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall G. F. (2011). “What is the link between protein aggregation and interneuronal lesion propagation in neurodegenerative disease,” in Neurodegenerative Diseases–Processes, Prevention, Protection and Monitoring Rijeka. ed. Chang R. C.-C. (Croatia: InTech; ), 1–17. [Google Scholar]
- Hanger D. P., Anderton B. H., Noble W. (2009). Tau phosphorylation: the therapeutic challenge for neurodegenerative disease. Trends Mol. Med. 15, 112–119. doi: 10.1016/j.molmed.2009.01.003, PMID: [DOI] [PubMed] [Google Scholar]
- Hansen D. V., Hanson J. E., Sheng M. (2018). Microglia in Alzheimer’s disease. J. Cell Biol. 217, 459–472. doi: 10.1083/jcb.201709069, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan B. A., Pehar M., Killoy K. M., Vargas M. R. (2019). Enhanced SIRT6 activity abrogates the neurotoxic phenotype of astrocytes expressing ALS-linked mutant SOD1. FASEB J. 33, 7084–7091. doi: 10.1096/fj.201802752R, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M. R., Georgiou A. S., Spaink H. P., Cunliffe V. T. (2011). The epigenetic regulator histone deacetylase 1 promotes transcription of a core neurogenic programme in zebrafish embryos. BMC Genomics 12, 1–18. doi: 10.1186/1471-2164-12-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T., Yamakawa M., Kimura S., Usuba O., Toyono M. (2013). Expression of acetylated and dimethylated histone H3 in colorectal cancer. Dig. Surg. 30, 249–258. doi: 10.1159/000351444, PMID: [DOI] [PubMed] [Google Scholar]
- Hilton I. B., Dippolito A. M., Vockley C. M., Thakore P. I., Crawford G. E., Reddy T. E., et al. (2015). Epigenome editing by a CRISPR-Cas 9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33, 510–517. doi: 10.1038/nbt.3199, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockly E., Richon V. M., Woodman B., Smith D. L., Zhou X., Rosa E., et al. (2003). Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc. Natl. Acad. Sci. 100, 2041–2046. doi: 10.1073/pnas.0437870100, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrelia P., Sita G., Ziche M., Ristori E., Marino A., Cordaro M., et al. (2020). Common protective strategies in neurodegenerative disease: focusing on risk factors to target the cellular redox system. Oxidative Med. Cell. Longev. 2020, 1–18. doi: 10.1155/2020/8363245, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., et al. (2002). HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458. doi: 10.1038/417455a, PMID: [DOI] [PubMed] [Google Scholar]
- Iyer A., Roeters S. J., Schilderink N., Hommersom B., Heeren R. M., Woutersen S., et al. (2016). The impact of N-terminal acetylation of α-synuclein on phospholipid membrane binding and fibril structure. J. Biol. Chem. 291, 21110–21122. doi: 10.1074/jbc.M116.726612, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C. (2014). The tubulin code: molecular components, readout mechanisms, and functions. J. Cell Biol. 206, 461–472. doi: 10.1083/jcb.201406055, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen C., Schmalbach S., Boeselt S., Sarlette A., Dengler R., Petri S. (2010). Differential histone deacetylase mRNA expression patterns in amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 69, 573–581. doi: 10.1097/NEN.0b013e3181ddd404, PMID: [DOI] [PubMed] [Google Scholar]
- Jeong H., Then F., Melia T. J., Jr., Mazzulli J. R., Cui L., Savas J. N., et al. (2009). Acetylation targets mutant huntingtin to autophagosomes for degradation. Cells 137, 60–72. doi: 10.1016/j.cell.2009.03.018, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jęśko H., Wencel P., Strosznajder R. P., Strosznajder J. B. (2017). Sirtuins and their roles in brain aging and neurodegenerative disorders. Neurochem. Res. 42, 876–890. doi: 10.1007/s11064-016-2110-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Kast R. J., Steffan J. S., Thomas E. A. (2012a). Selective histone deacetylase (HDAC) inhibition imparts beneficial effects in Huntington's disease mice: implications for the ubiquitin–proteasomal and autophagy systems. Hum. Mol. Genet. 21, 5280–5293. doi: 10.1093/hmg/dds379, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Pallos J., Jacques V., Lau A., Tang B., Cooper A., et al. (2012b). Histone deacetylase (HDAC) inhibitors targeting HDAC3 and HDAC1 ameliorate polyglutamine-elicited phenotypes in model systems of Huntington's disease. Neurobiol. Dis. 46, 351–361. doi: 10.1016/j.nbd.2012.01.016, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Kanthasamy A., Harischandra D. S., Kondru N., Ghosh A., Panicker N., et al. (2014). Histone hyperacetylation up-regulates protein kinase Cδ in dopaminergic neurons to induce cell death: relevance to epigenetic mechanisms of neurodegeneration in Parkinson disease. J. Biol. Chem. 289, 34743–34767. doi: 10.1074/jbc.M114.576702, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliszewski M. (2016). The role of SOD1 acetylation in neurodegeneration.
- Kaliszewski M., Kennedy A. K., Blaes S. L., Shaffer R. S., Knott A. B., Song W., et al. (2016). SOD1 lysine 123 acetylation in the adult central nervous system. Front. Cell. Neurosci. 10:287. doi: 10.3389/fncel.2016.00287, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaluski S., Portillo M., Besnard A., Stein D., Einav M., Zhong L., et al. (2017). Neuroprotective functions for the histone deacetylase SIRT6. Cell Rep. 18, 3052–3062. doi: 10.1016/j.celrep.2017.03.008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y., Xu L., Heinzel T., Torchia J., Kurokawa R., Gloss B., et al. (1996). A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cells 85, 403–414. doi: 10.1016/s0092-8674(00)81118-6, PMID: [DOI] [PubMed] [Google Scholar]
- Kametani F., Obi T., Shishido T., Akatsu H., Murayama S., Saito Y., et al. (2016). Mass spectrometric analysis of accumulated TDP-43 in amyotrophic lateral sclerosis brains. Sci. Rep. 6, 1–15. doi: 10.1038/srep23281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanski R., Sneeboer M. A., van Bodegraven E. J., Sluijs J. A., Kropff W., Vermunt M. W., et al. (2014). Histone acetylation in astrocytes suppresses GFAP and stimulates a reorganization of the intermediate filament network. J. Cell Sci. 127, 4368–4380. doi: 10.1242/jcs.145912, PMID: [DOI] [PubMed] [Google Scholar]
- Kapitein L. C., Hoogenraad C. C. (2015). Building the neuronal microtubule cytoskeleton. Neuron 87, 492–506. doi: 10.1016/j.neuron.2015.05.046, PMID: [DOI] [PubMed] [Google Scholar]
- Katayama S., Morii A., Makanga J. O., Suzuki T., Miyata N., Inazu T. (2018). HDAC8 regulates neural differentiation through embryoid body formation in P 19 cells. Biochem. Biophys. Res. Commun. 498, 45–51. doi: 10.1016/j.bbrc.2018.02.195, PMID: [DOI] [PubMed] [Google Scholar]
- Katsumoto T., Aikawa Y., Iwama A., Ueda S., Ichikawa H., Ochiya T., et al. (2006). MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev. 20, 1321–1330. doi: 10.1101/gad.1393106, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G. A., Baliban R. C., Floudas C. A. (2011). Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci. Rep. 1:90. doi: 10.1038/srep00090, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. S., Akhtar M. W., Adachi M., Mahgoub M., Bassel-Duby R., Kavalali E. T., et al. (2012). An essential role for histone deacetylase 4 in synaptic plasticity and memory formation. J. Neurosci. 32, 10879–10886. doi: 10.1523/JNEUROSCI.2089-12.2012, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Choi H., Jung E.S., Lee W., Oh S., Jeon N. L., et al. (2012). HDAC6 inhibitor blocks amyloid beta-induced impairment of mitochondrial transport in hippocampal neurons. PLoS ONE 7: e42983. doi: 10.1371/journal.pone.0042983, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-y., Hwang H.-G., Lee J.-Y., Kim M., Kim J.-Y. (2020). Cortactin deacetylation by HDAC6 and SIRT2 regulates neuronal migration and dendrite morphogenesis during cerebral cortex development. Mol. Brain 13:105. doi: 10.1186/s13041-020-00644-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. H., Kim J.-W., Jang S.-M., An J.-H., Song K.-H., Choi K.-H. (2012). Transcriptional activity of paired homeobox Pax 6 is enhanced by histone acetyltransferase tip 60 during mouse retina development. Biochem. Biophys. Res. Commun. 424, 427–432. doi: 10.1016/j.bbrc.2012.06.126, PMID: [DOI] [PubMed] [Google Scholar]
- Kim A. H., Puram S. V., Bilimoria P. M., Ikeuchi Y., Keough S., Wong M., et al. (2009). A centrosomal Cdc 20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cells 136, 322–336. doi: 10.1016/j.cell.2008.11.050, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-H., Saraf A., Florens L., Washburn M., Workman J. L. (2010). Gcn 5 regulates the dissociation of SWI/SNF from chromatin by acetylation of SWI 2/SNF 2. Genes Dev. 24, 2766–2771. doi: 10.1101/gad.1979710, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., et al. (2006). Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618. doi: 10.1016/j.molcel.2006.06.026, PMID: [DOI] [PubMed] [Google Scholar]
- Kirkpatrick L., Brady S. (1999). “Molecular components of the neuronal cytoskeleton,” in Basic Neurochemistry: Molecular, Cellular and Medical Aspects. 6th Edn. eds. Siegel G. J., Agranoff B. W., Albers R. W., Fisher S. K., Uhler M. D.. (Philadelphia: Lippincott-Raven; ). [Google Scholar]
- Knorre D., Kudryashova N., Godovikova T. (2009). Chemical and functional aspects of posttranslational modification of proteins. Acta Theriol. 1, 29–51. doi: 10.32607/20758251-2009-1-3-29-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson S. K., Chyla B. J., Amann J. M., Bhaskara S., Huppert S. S., Hiebert S. W. (2008). Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 27, 1017–1028. doi: 10.1038/emboj.2008.51, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontopoulos E., Parvin J. D., Feany M. B. (2006). α-Synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum. Mol. Genet. 15, 3012–3023. doi: 10.1093/hmg/ddl243, PMID: [DOI] [PubMed] [Google Scholar]
- Körner S., Böselt S., Thau N., Rath K. J., Dengler R., Petri S. (2013). Differential sirtuin expression patterns in amyotrophic lateral sclerosis (ALS) postmortem tissue: neuroprotective or neurotoxic properties of sirtuins in ALS? Neurodegener. Dis. 11, 141–152. doi: 10.1159/000338048, PMID: [DOI] [PubMed] [Google Scholar]
- Korzus E., Rosenfeld M. G., Mayford M. (2004). CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron 42, 961–972. doi: 10.1016/j.neuron.2004.06.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueh A. J., Dixon M. P., Voss A. K., Thomas T. (2011). HBO1 is required for H3K14 acetylation and normal transcriptional activity during embryonic development. Mol. Cell. Biol. 31, 845–860. doi: 10.1128/MCB.00159-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Kaur S., Kapil L., Singh C., Singh A. (2022). HDAC11: a novel inflammatory biomarker in Huntington's disease. EXCLI J. 21, 647–650. doi: 10.17179/excli2022-4741, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara K., Saito Y., Takano M., Arai Y., Yasuno S., Nakagawa Y., et al. (2003). NRSF regulates the fetal cardiac gene program and maintains normal cardiac structure and function. EMBO J. 22, 6310–6321. doi: 10.1093/emboj/cdg601, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai I.-L., Lin T.-P., Yao Y.-L., Lin C.-Y., Hsieh M.-J., Yang W.-M. (2010). Histone deacetylase 10 relieves repression on the melanogenic program by maintaining the deacetylation status of repressors. J. Biol. Chem. 285, 7187–7196. doi: 10.1074/jbc.M109.061861, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latario C. J., Pickrell L. E., Higgs H. N. (2020). Lysine acetylation of cytoskeletal proteins: emergence of an actin code. J. Cell Biol. 219:e202006151. doi: 10.1083/jcb.202006151, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guen Y., Luo G., Ambati A., Damotte V., Jansen I., Yu E., et al. (2021). Protective effects of HLA-DRB1* 04 subtypes in Parkinson's and Alzheimer's diseases implicate acetylated tau PHF6 sequences. med Rxiv [Preprint]. doi: 10.1101/2021.12.26.21268354v1 [DOI]
- Lebrun N., Delépine C., Selloum M., Meziane H., Nectoux J., Herault Y., et al. (2021). HDAC inhibitor ameliorates behavioral deficits in Mecp 2308/y mouse model of Rett syndrome. Brain Res. 1772:147670. doi: 10.1016/j.brainres.2021.147670, PMID: [DOI] [PubMed] [Google Scholar]
- Lee W.-C. M., Yoshihara M., Littleton J. T. (2004). Cytoplasmic aggregates trap polyglutamine-containing proteins and block axonal transport in a drosophila model of Huntington's disease. PNAS Nexus 101, 3224–3229. doi: 10.1073/pnas.0400243101, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech S. H., Evans C. A., Shaw L., Wong C. H., Connolly J., Griffiths J. R., et al. (2008). Proteomic analyses of intermediate filaments reveals cytokeratin 8 is highly acetylated–implications for colorectal epithelial homeostasis. Proteomics 8, 279–288. doi: 10.1002/pmic.200700404, PMID: [DOI] [PubMed] [Google Scholar]
- Li X. H., Chen C., Tu Y., Sun H.-t., Zhao M.-l., Cheng S.-x., et al. (2013). Sirt 1 promotes axonogenesis by deacetylation of Akt and inactivation of GSK3. Mol. Neurobiol. 48, 490–499. doi: 10.1007/s12035-013-8437-3, PMID: [DOI] [PubMed] [Google Scholar]
- Li X., Feng Y., Wang X.-X., Truong D., Wu Y.-C. (2020). The critical role of SIRT1 in Parkinson’s disease: mechanism and therapeutic considerations. Aging Dis. 11, 1608–1622. doi: 10.14336/AD.2020.0216, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Huang H., Zhu M., Bai H., Huang X. (2021). Roles of the MYST family in the pathogenesis of Alzheimer’s disease via histone or non-histone acetylation. Aging Dis. 12, 132–142. doi: 10.14336/AD.2020.0329, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Shin D., Kwon S. H. (2013). Histone deacetylase 6 plays a role as a distinct regulator of diverse cellular processes. FEBS J. 280, 775–793. doi: 10.1111/febs.12079, PMID: [DOI] [PubMed] [Google Scholar]
- Li G., Tian Y., Zhu W.-G. (2020). The roles of histone deacetylases and their inhibitors in cancer therapy. Front. Cell Dev. Biol. 8:1004. doi: 10.3389/fcell.2020.576946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Xiao H., Isobe K.-i. (2002). Histone acetyltransferase activities of cAMP-regulated enhancer-binding protein and p 300 in tissues of fetal, young, and old mice. J. Gerontol. Ser. A Biol. Med. Sci. 57, B93–B98. doi: 10.1093/gerona/57.3.B93, PMID: [DOI] [PubMed] [Google Scholar]
- Li M., Zhuang Y., Shan B. (2016). Analysis of expression and functions of histone deacetylase 6 (hdac 6). Histone deacetylases. Methods Mol. Biol. 1436, 85–94. doi: 10.1007/978-1-4939-3667-0_6, PMID: [DOI] [PubMed] [Google Scholar]
- Lin M. T., Beal M. F. (2006). Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443, 787–795. doi: 10.1038/nature05292, PMID: [DOI] [PubMed] [Google Scholar]
- Liu H., Hu Q., D'ercole A. J., Ye P. (2009). Histone deacetylase 11 regulates oligodendrocyte-specific gene expression and cell development in OL-1 oligodendroglia cells. Glia 57, 1–12. doi: 10.1002/glia.20729, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Hu Q., Kaufman A., D'Ercole A. J., Ye P. (2008). Developmental expression of histone deacetylase 11 in the murine brain. J. Neurosci. Res. 86, 537–543. doi: 10.1002/jnr.21521, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.-A., Rizzo V., Puthanveettil S. (2012). Pathologies of axonal transport in neurodegenerative diseases. Transl. Neurosci. 3, 355–372. doi: 10.2478/s13380-012-0044-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wang Y., Zhang R., Jin T., Qu L., Jin Q., et al. (2020). HDAC10 is positively associated with PD-L1 expression and poor prognosis in patients with NSCLC. Front. Oncol. 10:485. doi: 10.3389/fonc.2020.00485, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoPresti P. (2019). The selective HDAC6 inhibitor ACY-738 impacts memory and disease regulation in an animal model of multiple sclerosis. Front. Neurol. 10:519. doi: 10.3389/fneur.2019.00519, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Deng Y., Yu D., Cao H., Wang L., Liu L., et al. (2014). Histone acetyltransferase p 300 mediates histone acetylation of PS1 and BACE1 in a cellular model of Alzheimer's disease. PLoS One 9:e103067. doi: 10.1371/journal.pone.0103067, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Tan L., Wang X. (2019). Circular HDAC9/micro RNA-138/Sirtuin-1 pathway mediates synaptic and amyloid precursor protein processing deficits in Alzheimer’s disease. Neurosci. Bull. 35, 877–888. doi: 10.1007/s12264-019-00361-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L. (2002). Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu. Rev. Cell Dev. Biol. 18, 601–635. doi: 10.1146/annurev.cellbio.18.031802.150501, PMID: [DOI] [PubMed] [Google Scholar]
- Lv W.-W., Wei H.-M., Wang D.-L., Ni J.-Q., Sun F.-L. (2012). Depletion of histone deacetylase 3 antagonizes PI3K-mediated overgrowth of drosophila organs through the acetylation of histone H4 at lysine 16. J. Cell Sci. 125, 5369–5378. doi: 10.1242/jcs.106336, PMID: [DOI] [PubMed] [Google Scholar]
- Ma C., D'Mello S. R. (2011). Neuroprotection by histone deacetylase-7 (HDAC7) occurs by inhibition of c-Jun expression through a deacetylase-independent mechanism. J. Biol. Chem. 286, 4819–4828. doi: 10.1074/jbc.M110.146860, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P., Schultz R. M. (2008). Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev. Biol. 319, 110–120. doi: 10.1016/j.ydbio.2008.04.011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mac Donald M. E., Gines S., Gusella J. F., Wheeler V. C. (2003). Huntington’s disease. NeuroMolecular Med. 4, 7–20. doi: 10.1385/NMM:4:1-2:7, PMID: [DOI] [PubMed] [Google Scholar]
- Madore C., Yin Z., Leibowitz J., Butovsky O. (2020). Microglia, lifestyle stress, and neurodegeneration. Immunity 52, 222–240. doi: 10.1016/j.immuni.2019.12.003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdzadeh N., Wang L., Morrison B. E., Bassel-Duby R., Olson E. N., D'Mello S. R. (2008). HDAC4 inhibits cell-cycle progression and protects neurons from cell death. Dev. Neurobiol. 68, 1076–1092. doi: 10.1002/dneu.20637, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid T., Griffin D., Criss Z., II, Jarpe M., Pautler R. G. (2015). Pharmocologic treatment with histone deacetylase 6 inhibitor (ACY-738) recovers Alzheimer's disease phenotype in amyloid precursor protein/presenilin 1 (APP/PS1) mice. Alzheimer's Dement. 1, 170–181. doi: 10.1016/j.trci.2015.08.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M., Jensen O. N. (2003). Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255–261. doi: 10.1038/nbt0303-255, PMID: [DOI] [PubMed] [Google Scholar]
- Mao C.-X., Wen X., Jin S., Zhang Y. Q. (2017). Increased acetylation of microtubules rescues human tau-induced microtubule defects and neuromuscular junction abnormalities in drosophila. Dis. Model. Mech. 10, 1245–1252. doi: 10.1242/dmm.028316, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks P. A., Breslow R. (2007). Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 25, 84–90. doi: 10.1038/nbt1272, PMID: [DOI] [PubMed] [Google Scholar]
- Marmorstein R. (2001). Structure of histone acetyltransferases. J. Mol. Biol. 311, 433–444. doi: 10.1006/jmbi.2001.4859, PMID: [DOI] [PubMed] [Google Scholar]
- Marsoni S., Damia G., Camboni G. (2008). A work in progress: the clinical development of histone deacetylase inhibitor. Epigenetics 3, 164–171. doi: 10.4161/epi.3.3.6253 [DOI] [PubMed] [Google Scholar]
- Martínez-Cerdeño V., Lemen J. M., Chan V., Wey A., Lin W., Dent S. R., et al. (2012). N-Myc and GCN5 regulate significantly overlapping transcriptional programs in neural stem cells. PLoS One 7:e39456. doi: 10.1371/journal.pone.0039456, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maszlag-Török R., Boros F. A., Vécsei L., Klivényi P. (2021). Gene variants and expression changes of SIRT1 and SIRT6 in peripheral blood are associated with Parkinson’s disease. Sci. Rep. 11:10677. doi: 10.1038/s41598-021-90059-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzocchi M. (2021). Defining the potential of class-IIa histone deacetylases as a therapeutic target for Parkinson’s disease. PhD Thesis. University College Cork.
- McQuown S. C., Barrett R. M., Matheos D. P., Post R. J., Rogge G. A., Alenghat T., et al. (2011). HDAC3 is a critical negative regulator of long-term memory formation. J. Neurosci. 31, 764–774. doi: 10.1523/JNEUROSCI.5052-10.2011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méjat A., Ramond F., Bassel-Duby R., Khochbin S., Olson E. N., Schaeffer L. (2005). Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat. Neurosci. 8, 313–321. doi: 10.1038/nn1408, PMID: [DOI] [PubMed] [Google Scholar]
- Merlini G., Bellotti V., Andreola A., Palladini G., Obici L., Casarini S., et al. (2001). Protein aggregation. Clin. Chem. Lab. Med. 39, 1065–1075. doi: 10.1515/CCLM.2001.172 [DOI] [PubMed] [Google Scholar]
- Michishita E., Park J. Y., Burneskis J. M., Barrett J. C., Horikawa I. (2005). Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 16, 4623–4635. doi: 10.1091/mbc.e05-01-0033, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielcarek M., Benn C. L., Franklin S. A., Smith D. L., Woodman B., Marks P. A., et al. (2011). SAHA decreases HDAC 2 and 4 levels in vivo and improves molecular phenotypes in the R6/2 mouse model of Huntington's disease. PLoS One 6:e27746. doi: 10.1371/journal.pone.0027746, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielcarek M., Landles C., Weiss A., Bradaia A., Seredenina T., Inuabasi L., et al. (2013). HDAC4 reduction: a novel therapeutic strategy to target cytoplasmic huntingtin and ameliorate neurodegeneration. PLoS Biol. 11:e1001717. doi: 10.1371/journal.pbio.1001717, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S.-W., Chen X., Tracy T. E., Li Y., Zhou Y., Wang C., et al. (2015). Critical role of acetylation in tau-mediated neurodegeneration and cognitive deficits. Nat. Med. 21, 1154–1162. doi: 10.1038/nm.3951, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S.-W., Cho S.-H., Zhou Y., Schroeder S., Haroutunian V., Seeley W. W., et al. (2010). Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron 67, 953–966. doi: 10.1016/j.neuron.2010.08.044, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S.-W., Sohn P. D., Li Y., Devidze N., Johnson J. R., Krogan N. J., et al. (2018). SIRT1 deacetylates tau and reduces pathogenic tau spread in a mouse model of tauopathy. J. Neurosci. 38, 3680–3688. doi: 10.1523/JNEUROSCI.2369-17.2018, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed M. A., Greif P. A., Diamond J., Sharaf O., Maxwell P., Montironi R., et al. (2007). Epigenetic events, remodelling enzymes and their relationship to chromatin organization in prostatic intraepithelial neoplasia and prostatic adenocarcinoma. BJU Int. 99, 908–915. doi: 10.1111/j.1464-410X.2006.06704.x, PMID: [DOI] [PubMed] [Google Scholar]
- Montgomery R. L., Davis C. A., Potthoff M. J., Haberland M., Fielitz J., Qi X., et al. (2007). Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 21, 1790–1802. doi: 10.1101/gad.1563807, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery R. L., Hsieh J., Barbosa A. C., Richardson J. A., Olson E. N. (2009). Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc. Natl. Acad. Sci. 106, 7876–7881. doi: 10.1073/pnas.0902750106, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore D. (2016). Panobinostat (Farydak): a novel option for the treatment of relapsed or relapsed and refractory multiple myeloma. Pharm. Therap. 41, 296–300. [PMC free article] [PubMed] [Google Scholar]
- Morris M. J., Monteggia L. M. (2013). Unique functional roles for class I and class II histone deacetylases in central nervous system development and function. Int. J. Dev. Neurosci. 31, 370–381. doi: 10.1016/j.ijdevneu.2013.02.005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller T., Meyer H. E., Egensperger R., Marcus K. (2008). The amyloid precursor protein intracellular domain (AICD) as modulator of gene expression, apoptosis, and cytoskeletal dynamics—relevance for Alzheimer's disease. Prog. Neurobiol. 85, 393–406. doi: 10.1016/j.pneurobio.2008.05.002, PMID: [DOI] [PubMed] [Google Scholar]
- Nakagawa Y., Kuwahara K., Harada M., Takahashi N., Yasuno S., Adachi Y., et al. (2006). Class II HDACs mediate CaMK-dependent signaling to NRSF in ventricular myocytes. J. Mol. Cell. Cardiol. 41, 1010–1022. doi: 10.1016/j.yjmcc.2006.08.010, PMID: [DOI] [PubMed] [Google Scholar]
- Naumann M., Pal A., Goswami A., Lojewski X., Japtok J., Vehlow A., et al. (2018). Impaired DNA damage response signaling by FUS-NLS mutations leads to neurodegeneration and FUS aggregate formation. Nat. Commun. 9:335. doi: 10.1038/s41467-017-02299-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal M., Richardson J. R. (2018). Epigenetic regulation of astrocyte function in neuroinflammation and neurodegeneration. BBA Mol. Basis Dis. 1864, 432–443. doi: 10.1016/j.bbadis.2017.11.004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble W., Hanger D. P., Miller C. C., Lovestone S. (2013). The importance of tau phosphorylation for neurodegenerative diseases. Front. Neurol. 4:83. doi: 10.3389/fneur.2013.00083, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- North B. J., Marshall B. L., Borra M. T., Denu J. M., Verdin E. (2003). The human sir 2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11, 437–444. doi: 10.1016/S1097-2765(03)00038-8, PMID: [DOI] [PubMed] [Google Scholar]
- Norwood J., Franklin J. M., Sharma D., D'Mello S. R. (2014). Histone deacetylase 3 is necessary for proper brain development. J. Biol. Chem. 289, 34569–34582. doi: 10.1074/jbc.M114.576397, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez-Álvarez Y., Suelves M. (2022). HDAC11: a multifaceted histone deacetylase with proficient fatty deacylase activity and its roles in physiological processes. FEBS J. 289, 2771–2792. doi: 10.1111/febs.15895, PMID: [DOI] [PubMed] [Google Scholar]
- Ogawa-Goto K., Tanaka K., Ueno T., Tanaka K., Kurata T., Sata T., et al. (2007). P 180 is involved in the interaction between the endoplasmic reticulum and microtubules through a novel microtubule-binding and bundling domain. Mol. Biol. Cell 18, 3741–3751. doi: 10.1091/mbc.e06-12-1125, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oláh J., Bertrand P., Ovádi J. (2017). Role of the microtubule-associated TPPP/p 25 in Parkinson’s and related diseases and its therapeutic potential. Expert Rev. Proteomics 14, 301–309. doi: 10.1080/14789450.2017.1304216, PMID: [DOI] [PubMed] [Google Scholar]
- Osseni A., Ravel-Chapuis A., Thomas J.-L., Gache V., Schaeffer L., Jasmin B. J. (2020). HDAC6 regulates microtubule stability and clustering of AChRs at neuromuscular junctions. J. Cell Biol. 219:e201901099. doi: 10.1083/jcb.201901099, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outeiro T. F., Kontopoulos E., Altmann S. M., Kufareva I., Strathearn K. E., Amore A. M., et al. (2007). Sirtuin 2 inhibitors rescue α-synuclein-mediated toxicity in models of Parkinson's disease. Science 317, 516–519. doi: 10.1126/science.1143780, PMID: [DOI] [PubMed] [Google Scholar]
- Outeiro T. F., Marques O., Kazantsev A. (2008). Therapeutic role of sirtuins in neurodegenerative disease. Biochim Biophys Acta Proteins Proteom. 1782, 363–369. doi: 10.1016/j.bbadis.2008.02.010, PMID: [DOI] [PubMed] [Google Scholar]
- Pandithage R., Lilischkis R., Harting K., Wolf A., Jedamzik B., Lüscher-Firzlaff J., et al. (2008). The regulation of SIRT2 function by cyclin-dependent kinases affects cell motility. J. Cell Biol. 180, 915–929. doi: 10.1083/jcb.200707126, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao P.-C., Patnaik D., Watson L. A., Gao F., Pan L., Wang J., et al. (2020). HDAC1 modulates OGG1-initiated oxidative DNA damage repair in the aging brain and Alzheimer’s disease. Nat. Commun. 11:2484. doi: 10.1038/s41467-020-16361-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-Y., Kim J.-S. (2020). A short guide to histone deacetylases including recent progress on class II enzymes. Exp. Mol. Med. 52, 204–212. doi: 10.1038/s12276-020-0382-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-Y., Kim M.-J., Kim Y. J., Lee Y.-H., Bae D., Kim S., et al. (2015). Selective PCAF inhibitor ameliorates cognitive and behavioral deficits by suppressing NF-κB-mediated neuroinflammation induced by Aβ in a model of Alzheimer's disease. Int. J. Mol. Med. 35, 1109–1118. doi: 10.3892/ijmm.2015.2099, PMID: [DOI] [PubMed] [Google Scholar]
- Parra M. (2015). Class II a HDAC s–new insights into their functions in physiology and pathology. FEBS J. 282, 1736–1744. doi: 10.1111/febs.13061, PMID: [DOI] [PubMed] [Google Scholar]
- Pegoraro V., Marozzo R., Angelini C. (2020). Micro RNAs and HDAC4 protein expression in the skeletal muscle of ALS patients. Clin. Neuropathol. 39, 105–114. doi: 10.5414/NP301233, PMID: [DOI] [PubMed] [Google Scholar]
- Pehar M., Puglielli L. (2013). Lysine acetylation in the lumen of the ER: a novel and essential function under the control of the UPR. Biochim. Biophys. Acta, Mol. Cell Res. 1833, 686–697. doi: 10.1016/j.bbamcr.2012.12.004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S., Feller C., Ladurner A. G., Imhof A. (2016). The metabolic impact on histone acetylation and transcription in ageing. Trends Biochem. Sci. 41, 700–711. doi: 10.1016/j.tibs.2016.05.008, PMID: [DOI] [PubMed] [Google Scholar]
- Perez-Campo F. M., Costa G., Lie-a-Ling M., Stifani S., Kouskoff V., Lacaud G. (2014). MOZ-mediated repression of p16INK4a is critical for the self-renewal of neural and hematopoietic stem cells. Stem Cells 32, 1591–1601. doi: 10.1002/stem.1606, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peserico A., Simone C. (2010). Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J. Biomed. Biotechnol. 2011:371832. doi: 10.1155/2011/371832, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters O. M., Ghasemi M., Brown R. H. (2015). Emerging mechanisms of molecular pathology in ALS. J. Clin. Invest. 125, 1767–1779. doi: 10.1172/JCI71601, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picci C., Wong V. S., Costa C. J., McKinnon M. C., Goldberg D. C., Swift M., et al. (2020). HDAC6 inhibition promotes α-tubulin acetylation and ameliorates CMT2A peripheral neuropathy in mice. Exp. Neurol. 328:113281. doi: 10.1016/j.expneurol.2020.113281, PMID: [DOI] [PubMed] [Google Scholar]
- Pilkington A. W., Schupp J., Nyman M., Valentine S. J., Smith D. M., Legleiter J. (2019). Acetylation of Aβ40 alters aggregation in the presence and absence of lipid membranes. ACS Chem. Neurosci. 11, 146–161. doi: 10.1021/acschemneuro.9b00483, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portaleone P. (2004). Encyclopedia of Endocrine Diseases. Milan, Italy: Academic Press, 9–12. [Google Scholar]
- Price V., Wang L., D'Mello S. R. (2013). Conditional deletion of histone deacetylase-4 in the central nervous system has no major effect on brain architecture or neuronal viability. J. Neurosci. Res. 91, 407–415. doi: 10.1002/jnr.23170, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian M.-X., Pang Y., Liu C. H., Haratake K., Du B.-Y., Ji D.-Y., et al. (2013). Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cells 153, 1012–1024. doi: 10.1016/j.cell.2013.04.032, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan P., Wang K., Yan S., Wen S., Wei C., Zhang X., et al. (2021). Integrated network analysis identifying potential novel drug candidates and targets for Parkinson's disease. Sci. Rep. 11:13154. doi: 10.1038/s41598-021-92701-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph G. S., Radcliffe P. A., Day D. M., Carthy J. M., Leroux M. A., Lee D. C., et al. (2005). Silencing mutant SOD1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nat. Med. 11, 429–433. doi: 10.1038/nm1205, PMID: [DOI] [PubMed] [Google Scholar]
- Ramadori G., Lee C. E., Bookout A. L., Lee S., Williams K. W., Anderson J., et al. (2008). Brain SIRT1: anatomical distribution and regulation by energy availability. J. Neurosci. 28, 9989–9996. doi: 10.1523/JNEUROSCI.3257-08.2008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed N. A., Cai D., Blasius T. L., Jih G. T., Meyhofer E., Gaertig J., et al. (2006). Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 16, 2166–2172. doi: 10.1016/j.cub.2006.09.014, PMID: [DOI] [PubMed] [Google Scholar]
- Regna N. L., Vieson M. D., Gojmerac A. M., Luo X. M., Caudell D. L., Reilly C. M. (2015). HDAC expression and activity is upregulated in diseased lupus-prone mice. Int. Immunopharmacol. 29, 494–503. doi: 10.1016/j.intimp.2015.10.006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues D. A., Pinheiro P. S., Sagrillo F. S., Bolognesi M. L., Fraga C. A. (2020). Histone deacetylases as targets for the treatment of neurodegenerative disorders: challenges and future opportunities. Med. Res. Rev. 40, 2177–2211. doi: 10.1002/med.21701, PMID: [DOI] [PubMed] [Google Scholar]
- Rossaert E., Pollari E., Jaspers T., Van Helleputte L., Jarpe M., Van Damme P., et al. (2019). Restoration of histone acetylation ameliorates disease and metabolic abnormalities in a FUS mouse model. Acta Neuropathol. Commun. 7:107. doi: 10.1186/s40478-019-0750-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. Y., Denu J. M., Allis C. D. (2001). Histone acetyltransferases. Annu. Rev. Biochem. 70, 81–120. doi: 10.1146/annurev.biochem.70.1.81, PMID: [DOI] [PubMed] [Google Scholar]
- Rouaux C., Jokic N., Mbebi C., Boutillier S., Loeffler J. P., Boutillier A. L. (2003). Critical loss of CBP/p 300 histone acetylase activity by caspase-6 during neurodegeneration. EMBO J. 22, 6537–6549. doi: 10.1093/emboj/cdg615, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouaux C., Loeffler J.-P., Boutillier A.-L. (2004). Targeting CREB-binding protein (CBP) loss of function as a therapeutic strategy in neurological disorders. Biochem. Pharmacol. 68, 1157–1164. doi: 10.1016/j.bcp.2004.05.035, PMID: [DOI] [PubMed] [Google Scholar]
- Rudrabhatla P. (2014). Regulation of neuronal cytoskeletal protein phosphorylation in neurodegenerative diseases. J. Alzheimers Dis. 41, 671–684. doi: 10.3233/JAD-130794, PMID: [DOI] [PubMed] [Google Scholar]
- Ruijter A. J., Ahv G., Caron H. N., Kemp S., ABV K. (2003). Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370, 737–749. doi: 10.1042/bj20021321, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoul K., Wang J., Diagouraga B., Khochbin S. (2010). The tale of protein lysine acetylation in the cytoplasm. J. Biomed. Biotechnol. 2011:970382. doi: 10.1155/2011/970382, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R., Pahan K. (2006). HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 13, 539–550. doi: 10.1038/sj.cdd.4401769, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatori I., Valle C., Ferri A., Carrì M. T. (2017). SIRT3 and mitochondrial metabolism in neurodegenerative diseases. Neurochem. Int. 109, 184–192. doi: 10.1016/j.neuint.2017.04.012, PMID: [DOI] [PubMed] [Google Scholar]
- Sambataro F., Pennuto M. (2017). Post-translational modifications and protein quality control in motor neuron and polyglutamine diseases. Front. Mol. Neurosci. 10:82. doi: 10.3389/fnmol.2017.00082, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo L., Hideshima T., Kung A. L., Tseng J.-C., Tamang D., Yang M., et al. (2012). Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood 119, 2579–2589. doi: 10.1182/blood-2011-10-387365, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders H. A., Johnson-Schlitz D. M., Jenkins B. V., Volkert P. J., Yang S. Z., Wildonger J. (2022). Acetylated α-tubulin K394 regulates microtubule stability to shape the growth of axon terminals. Curr. Biol. 32, 614–630.e5. doi: 10.1016/j.cub.2021.12.012, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffert L.-N., Carter W. G. (2020). Do post-translational modifications influence protein aggregation in neurodegenerative diseases: a systematic review. Brain Sci. 10:232. doi: 10.3390/brainsci10040232, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J. W., Gao Z., Li S., Farooqi M., Tang T.-S., Bezprozvanny I., et al. (2008). Small-molecule activation of neuronal cell fate. Nat. Chem. Biol. 4, 408–410. doi: 10.1038/nchembio.95, PMID: [DOI] [PubMed] [Google Scholar]
- Scott I., Zencheck W. D., Xiao H., Weiss L. M. (2012). Lysine post-translational modifications and the cytoskeleton. Essays Biochem. 52, 135–145. doi: 10.1042/bse0520135, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuderi C., Stecca C., Bronzuoli M. R., Rotili D., Valente S., Mai A., et al. (2014). Sirtuin modulators control reactive gliosis in an in vitro model of Alzheimer’s disease. Front. Pharmacol. 5:89. doi: 10.3389/fphar.2014.00089, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenica M.-L., Benner L., Housley S. B., Manchec B., Lee D. C., Nash K. R., et al. (2014). Histone deacetylase 6 inhibition improves memory and reduces total tau levels in a mouse model of tau deposition. Alzheimers Res. Ther. 6:12. doi: 10.1186/alzrt241, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y. J., Kang Y., Muench L., Reid A., Caesar S., Jean L., et al. (2014). Image-guided synthesis reveals potent blood-brain barrier permeable histone deacetylase inhibitors. ACS Chem. Neurosci. 5, 588–596. doi: 10.1021/cn500021p, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sephton C. F., Good S. K., Atkin S., Dewey C. M., Mayer P., Herz J., et al. (2010). TDP-43 is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 285, 6826–6834. doi: 10.1074/jbc.M109.061846, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E., Yoshida M. (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 6:a018713. doi: 10.1101/cshperspect.a018713, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirmast P., Ghafoori S., Irwin R., Abendroth J., Mayclin S., Lorimer D., et al. (2021). Structural characterization of a GNAT family acetyltransferase from Elizabethkingia anophelis bound to acetyl-CoA reveals a new dimeric interface. Sci. Rep. 11, 1–9. doi: 10.1038/s41598-020-79649-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoba B., Lwin Z. M., Ling L. S., Bay B. H., Yip G. W., Kumar S. D. (2009). Function of sirtuins in biological tissues. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 292, 536–543. doi: 10.1002/ar.20875, PMID: [DOI] [PubMed] [Google Scholar]
- Shrimp J. H., Grose C., Widmeyer S. R., Thorpe A. L., Jadhav A., Meier J. L. (2018). Chemical control of a CRISPR-Cas 9 acetyltransferase. ACS Chem. Biol. 13, 455–460. doi: 10.1021/acschembio.7b00883, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova-Darmos E., Sommer R., Eubanks J. H. (2018). The role of SIRT3 in the brain under physiological and pathological conditions. Front. Cell. Neurosci. 12:196. doi: 10.3389/fncel.2018.00196, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova-Darmos E., Wither R. G., Shulyakova N., Fisher C., Ratnam M., Aarts M., et al. (2014). Differential expression of sirtuin family members in the developing, adult, and aged rat brain. Front. Aging Neurosci. 6:333. doi: 10.3389/fnagi.2014.00333, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Kalinsky K., Chiuzan C., Zeleke T., Qingfei P., Yu J. (2020). Abstract CT107: phase IB trial of ACY-1215 (ricolinostat) combined with nab-paclitaxel in metastatic breast cancer (MBC). Cancer Res. 80:CT107-CT. doi: 10.1158/1538-7445.AM2020-CT107 [DOI] [Google Scholar]
- Simões-Pires C., Zwick V., Nurisso A., Schenker E., Carrupt P.-A., Cuendet M. (2013). HDAC6 as a target for neurodegenerative diseases: what makes it different from the other HDACs? Mol. Neurodegener. 8, 1–16. doi: 10.1186/1750-1326-8-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson C. L., Lemmens R., Miskiewicz K., Broom W. J., Hansen V. K., van Vught P. W., et al. (2009). Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum. Mol. Genet. 18, 472–481. doi: 10.1093/hmg/ddn375, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. S., Kim J. H., Chun C., Gharai A., Moon H. W., Kim E. Y., et al. (2022). HDAC6 inhibition corrects electrophysiological and axonal transport deficits in a human stem cell-based model of Charcot-Marie-tooth disease (type 2D). Adv. Biol. 6:2101308. doi: 10.1002/adbi.202101308, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S., Stillman B. (1991). Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 10, 971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn P. D., Tracy T. E., Son H.-I., Zhou Y., Leite R. E., Miller B. L., et al. (2016). Acetylated tau destabilizes the cytoskeleton in the axon initial segment and is mislocalized to the somatodendritic compartment. Mol. Neurodegener. 11:47. doi: 10.1186/s13024-016-0109-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S. M., Park S. J., Fernandez-Estevez M., Rubinsztein D. C. (2021). Autophagy regulation by acetylation—implications for neurodegenerative diseases. Exp. Mol. Med. 53, 30–41. doi: 10.1038/s12276-021-00556-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Kanthasamy A., Anantharam V., Sun F., Kanthasamy A. G. (2010). Environmental neurotoxic pesticide increases histone acetylation to promote apoptosis in dopaminergic neuronal cells: relevance to epigenetic mechanisms of neurodegeneration. Mol. Pharmacol. 77, 621–632. doi: 10.1124/mol.109.062174, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo K. Y., Halloran M., Sundaramoorthy V., Parakh S., Toth R. P., Southam K. A., et al. (2015). Rab 1-dependent ER–Golgi transport dysfunction is a common pathogenic mechanism in SOD1, TDP-43 and FUS-associated ALS. Acta Neuropathol. 130, 679–697. doi: 10.1007/s00401-015-1468-2, PMID: [DOI] [PubMed] [Google Scholar]
- Soriano F. X., Hardingham G. E. (2011). In cortical neurons HDAC3 activity suppresses RD4-dependent SMRT export. PLoS One 6:e21056. doi: 10.1371/journal.pone.0021056, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberger N. H., Sternberger L. A., Ulrich J. (1985). Aberrant neurofilament phosphorylation in Alzheimer disease. Proc. Natl. Acad. Sci. 82, 4274–4276. doi: 10.1073/pnas.82.12.4274, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens F. J., Argon Y. (1999). Protein folding in the ER. Semin. Cell Dev. Biol. 10, 443–454. doi: 10.1006/scdb.1999.0315 [DOI] [PubMed] [Google Scholar]
- Sugo N., Oshiro H., Takemura M., Kobayashi T., Kohno Y., Uesaka N., et al. (2010). Nucleocytoplasmic translocation of HDAC9 regulates gene expression and dendritic growth in developing cortical neurons. Eur. J. Neurosci. 31, 1521–1532. doi: 10.1111/j.1460-9568.2010.07218.x, PMID: [DOI] [PubMed] [Google Scholar]
- Suuronen T., Huuskonen J., Pihlaja R., Kyrylenko S., Salminen A. (2003). Regulation of microglial inflammatory response by histone deacetylase inhibitors. J. Neurochem. 87, 407–416. doi: 10.1046/j.1471-4159.2003.02004.x, PMID: [DOI] [PubMed] [Google Scholar]
- Tanabe K., Takei K. (2009). Dynamic instability of microtubules requires dynamin 2 and is impaired in a Charcot-Marie-tooth mutant. J. Cell Biol. 185, 939–948. doi: 10.1083/jcb.200803153, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapias A., Wang Z.-Q. (2017). Lysine acetylation and deacetylation in brain development and neuropathies. Genomics Proteomics Bioinformatics 15, 19–36. doi: 10.1016/j.gpb.2016.09.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. P., Hardy J., Fischbeck K. H. (2002). Toxic proteins in neurodegenerative disease. Science 296, 1991–1995. doi: 10.1126/science.1067122, PMID: [DOI] [PubMed] [Google Scholar]
- Tea J. S., Chihara T., Luo L. (2010). Histone deacetylase Rpd 3 regulates olfactory projection neuron dendrite targeting via the transcription factor Prospero. J. Neurosci. 30, 9939–9946. doi: 10.1523/JNEUROSCI.1643-10.2010, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenreiro S., Eckermann K., Outeiro T. F. (2014). Protein phosphorylation in neurodegeneration: friend or foe? Front. Mol. Neurosci. 7:42. doi: 10.3389/fnmol.2014.00042, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T., Voss A. K., Chowdhury K., Gruss P. (2000). Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development 127, 2537–2548. doi: 10.1242/dev.127.12.2537, PMID: [DOI] [PubMed] [Google Scholar]
- Tiberi L., Van Den Ameele J., Dimidschstein J., Piccirilli J., Gall D., Herpoel A., et al. (2012). BCL6 controls neurogenesis through Sirt 1-dependent epigenetic repression of selective notch targets. Nat. Neurosci. 15, 1627–1635. doi: 10.1038/nn.3264, PMID: [DOI] [PubMed] [Google Scholar]
- Tőkési N., Lehotzky A., Horváth I., Szabó B., Oláh J., Lau P., et al. (2010). TPPP/p 25 promotes tubulin acetylation by inhibiting histone deacetylase 6. J. Biol. Chem. 285, 17896–17906. doi: 10.1074/jbc.M109.096578, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi C. M., Lu M. M., Wang Q., Epstein J. A. (2008). Transgenic overexpression of Hdac 3 in the heart produces increased postnatal cardiac myocyte proliferation but does not induce hypertrophy. J. Biol. Chem. 283, 26484–26489. doi: 10.1074/jbc.M803686200, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trushina E., Heldebrant M. P., Perez-Terzic C. M., Bortolon R., Kovtun I. V., Badger J. D., et al. (2003). Microtubule destabilization and nuclear entry are sequential steps leading to toxicity in Huntington's disease. Proc. Natl. Acad. Sci. 100, 12171–12176. doi: 10.1073/pnas.2034961100, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutar Y., Özgür A., Tutar L. (2013). Role Of Protein Aggregation In Neurodegenerative Diseases. Washington, DC: National Academies Press. [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B. M., Lindskog C., Oksvold P., Mardinoglu A., et al. (2015). Tissue-based map of the human proteome. Science 347:1260419. doi: 10.1126/science.1260419, PMID: [DOI] [PubMed] [Google Scholar]
- Van Helleputte L., Benoy V., Van Den Bosch L. (2014). The role of histone deacetylase 6 (HDAC6) in neurodegeneration. Res. Rep. Biol. 5, 1–13. doi: 10.2147/RRB.S35470 [DOI] [Google Scholar]
- Vargas K. J., Makani S., Davis T., Westphal C. H., Castillo P. E., Chandra S. S. (2014). Synucleins regulate the kinetics of synaptic vesicle endocytosis. J. Neurosci. 34, 9364–9376. doi: 10.1523/JNEUROSCI.4787-13.2014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinueza-Gavilanes R., Íñigo-Marco I., Larrea L., Lasa M., Carte B., Santamaría E., et al. (2020). N-terminal acetylation mutants affect alpha-synuclein stability, protein levels and neuronal toxicity. Neurobiol. Dis. 137:104781. doi: 10.1016/j.nbd.2020.104781, PMID: [DOI] [PubMed] [Google Scholar]
- Vitet H., Brandt V., Saudou F. (2020). Traffic signaling: new functions of huntingtin and axonal transport in neurological disease. Curr. Opin. Neurobiol. 63, 122–130. doi: 10.1016/j.conb.2020.04.001, PMID: [DOI] [PubMed] [Google Scholar]
- Vogl D. T., Raje N., Jagannath S., Richardson P., Hari P., Orlowski R., et al. (2017). Ricolinostat, the first selective histone deacetylase 6 inhibitor, in combination with Bortezomib and dexamethasone for relapsed or refractory multiple myeloma Ricolinostat, Bortezomib, and dexamethasone for myeloma. Clin. Cancer Res. 23, 3307–3315. doi: 10.1158/1078-0432.CCR-16-2526, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cheng F., Chen J., Li Y., Villagra A., Smith M. R., et al. (2017). A novel role for histone deacetylase 10 (HDAC10) in the regulation of PD-L1 expression and immune tolerance mediated by antigen presenting cells (APCs). Blood 130:3561. doi: 10.1182/blood.V130.Suppl_1.3561.3561, PMID: 15711539 [DOI] [Google Scholar]
- Wang Y., Li W., Schulz V. P., Zhao H., Qu X., Qi Q., et al. (2021). Impairment of human terminal erythroid differentiation by histone deacetylase 5 deficiency. Blood 138, 1615–1627. doi: 10.1182/blood.2020007401, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Miao X., Liu Y., Li F., Liu Q., Sun J., et al. (2014). Dysregulation of histone acetyltransferases and deacetylases in cardiovascular diseases. Oxidative Med. Cell. Longev. 2014, 1–11. doi: 10.1155/2014/641979, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Qin G., Zhao T. C. (2014). HDAC4: mechanism of regulation and biological functions. Epigenomics 6, 139–150. doi: 10.2217/epi.13.73, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Sun H., Wang G., Ren H. (2020). Imbalance of lysine acetylation contributes to the pathogenesis of Parkinson’s disease. Int. J. Mol. Sci. 21:7182. doi: 10.3390/ijms21197182, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wei X., Pang Q., Yi F. (2012). Histone deacetylases and their inhibitors: molecular mechanisms and therapeutic implications in diabetes mellitus. Acta Pharm. Sin. B 2, 387–395. doi: 10.1016/j.apsb.2012.06.005, PMID: 34071497 [DOI] [Google Scholar]
- Weaver A. M. (2008). Cortactin in tumor invasiveness. Cancer Lett. 265, 157–166. doi: 10.1016/j.canlet.2008.02.066, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten M. D., Lockwood A. H., Hwo S.-Y., Kirschner M. W. (1975). A protein factor essential for microtubule assembly. Proc. Natl. Acad. Sci. 72, 1858–1862. doi: 10.1073/pnas.72.5.1858, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wey H.-Y., Gilbert T. M., Zürcher N. R., She A., Bhanot A., Taillon B. D., et al. (2016). Insights into neuroepigenetics through human histone deacetylase PET imaging. Sci. Transl. Med. 8:351ra106. doi: 10.1126/scitranslmed.aaf7551, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse A., Doherty K., Yeh H. H., Robinson A. C., Rollinson S., Pickering-Brown S., et al. (2015). Histone deacetylases (HDACs) in frontotemporal lobar degeneration. Neuropathol. Appl. Neurobiol. 41, 245–257. doi: 10.1111/nan.12153, PMID: [DOI] [PubMed] [Google Scholar]
- Witte H., Neukirchen D., Bradke F. (2008). Microtubule stabilization specifies initial neuronal polarization. J. Cell Biol. 180, 619–632. doi: 10.1083/jcb.200707042, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Chen P. S., Dallas S., Wilson B., Block M. L., Wang C.-C., et al. (2008). Histone deacetylase inhibitors up-regulate astrocyte GDNF and BDNF gene transcription and protect dopaminergic neurons. Int. J. Neuropsychopharmacol. 11, 1123–1134. doi: 10.1017/S1461145708009024, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Qiu Y., Jiao Y., Qiu Z., Liu D. (2020). Small molecules targeting HATs, HDACs, and BRDs in cancer therapy. Front. Oncol. 10:560487. doi: 10.3389/fonc.2020.560487, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Yang X., Zhang L., Zhang Y., Feng L. (2017). Nuclear accumulation of histone deacetylase 4 (HDAC4) exerts neurotoxicity in models of Parkinson’s disease. Mol. Neurobiol. 54, 6970–6983. doi: 10.1007/s12035-016-0199-2, PMID: [DOI] [PubMed] [Google Scholar]
- Xu Y., Deng Y., Qing H. (2015). The phosphorylation of α-synuclein: development and implication for the mechanism and therapy of the Parkinson's disease. J. Neurochem. 135, 4–18. doi: 10.1111/jnc.13234, PMID: [DOI] [PubMed] [Google Scholar]
- Xu W., Edmondson D. G., Evrard Y. A., Wakamiya M., Behringer R. R., Roth S. Y. (2000). Loss of Gcn 5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26, 229–232. doi: 10.1038/79973, PMID: [DOI] [PubMed] [Google Scholar]
- Xu X., Kozikowski A. P., Pozzo-Miller L. (2014). A selective histone deacetylase-6 inhibitor improves BDNF trafficking in hippocampal neurons from Mecp 2 knockout mice: implications for Rett syndrome. Front. Cell. Neurosci. 8:68. doi: 10.3389/fncel.2014.00068, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wan W. (2022). Acetylation in the regulation of autophagy. Autophagy doi: 10.1080/15548627.2022.2062112 [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- Xu X., Yu H., Xu Y. (2019). Ras-ERK1/2 signaling promotes the development of osteosarcoma by regulating H2BK12ac through CBP. Cancer Manag. Res. 11, 9153–9163. doi: 10.2147/CMAR.S219535, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Tonou-Fujimori N., Komori A., Maeda R., Nojima Y., Li H., et al. (2005). Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and notch signaling pathways. Development. 132, 3027–3043. doi: 10.1242/dev.01881, PMID: [DOI] [PubMed] [Google Scholar]
- Yang X. J. (2004). Lysine acetylation and the bromodomain: a new partnership for signaling. BioEssays 26, 1076–1087. doi: 10.1002/bies.20104, PMID: [DOI] [PubMed] [Google Scholar]
- Yao Y.-L., Yang W.-M. (2011). Beyond histone and deacetylase: an overview of cytoplasmic histone deacetylases and their nonhistone substrates. J. Biomed. Biotechnol. 2011, 1–15. doi: 10.1155/2011/146493, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F., Chen Y., Hoang T., Montgomery R. L., Zhao X.-h., Bu H., et al. (2009). HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the β-catenin–TCF interaction. Nat. Neurosci. 12, 829–838. doi: 10.1038/nn.2333, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh H. H., Young D., Gelovani J. G., Robinson A., Davidson Y., Herholz K., et al. (2013). Histone deacetylase class II and acetylated core histone immunohistochemistry in human brains with Huntington’s disease. Brain Res. 1504, 16–24. doi: 10.1016/j.brainres.2013.02.012, PMID: [DOI] [PubMed] [Google Scholar]
- You L., Yan K., Zhou J., Zhao H., Bertos N. R., Park M., et al. (2015). The lysine acetyltransferase activator Brpf 1 governs dentate gyrus development through neural stem cells and progenitors. PLoS Genet. 11:e1005034. doi: 10.1371/journal.pgen.1005034, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Benson D. L. (2001). Stages of synapse development defined by dependence on F-actin. J. Neurosci. 21, 5169–5181. doi: 10.1523/JNEUROSCI.21-14-05169.2001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., He X., Liu L., Jiang M., Zhao C., Wang H., et al. (2016). Hdac 3 interaction with p 300 histone acetyltransferase regulates the oligodendrocyte and astrocyte lineage fate switch. Dev. Cell 36, 316–330. doi: 10.1016/j.devcel.2016.01.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Kwon S., Yamaguchi T., Cubizolles F., Rousseaux S., Kneissel M., et al. (2008). Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol. Cell. Biol. 28, 1688–1701. doi: 10.1128/MCB.01154-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li N., Caron C., Matthias G., Hess D., Khochbin S., et al. (2003). HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO J. 22, 1168–1179. doi: 10.1093/emboj/cdg115, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Liu C., Wu J., Tao J.-J., Sui X.-l., Yao Z.-G., et al. (2014). Tubastatin A/ACY-1215 improves cognition in Alzheimer's disease transgenic mice. J. Alzheimers Dis. 41, 1193–1205. doi: 10.3233/JAD-140066, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang X., Yuan Z., Zhang Y., Yong S., Salas-Burgos A., Koomen J., et al. (2007). HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol. Cell 27, 197–213. doi: 10.1016/j.molcel.2007.05.033, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang M., Dong H., Yong S., Li X., Olashaw N., et al. (2009). Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene 28, 445–460. doi: 10.1038/onc.2008.388, PMID: [DOI] [PubMed] [Google Scholar]
- Zschoernig B., Mahlknecht U. (2008). SIRTUIN 1: regulating the regulator. Biochem. Biophys. Res. Commun. 376, 251–255. doi: 10.1016/j.bbrc.2008.08.137, PMID: [DOI] [PubMed] [Google Scholar]
- Zupkovitz G., Tischler J., Posch M., Sadzak I., Ramsauer K., Egger G., et al. (2006). Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol. Cell. Biol. 26, 7913–7928. doi: 10.1128/MCB.01220-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]