Degradation of p53 mediated by MEX3A drives ovarian cancer growth by circumventing p53 tumor suppressive functions, suggesting targeting MEX3A as a potential strategy for treating of ovarian cancer expressing WT p53.
Abstract
Epithelial ovarian cancer is a highly heterogeneous and malignant female cancer with an overall low survival rate. Mutations in p53 are prevalent in the major ovarian cancer histotype, high-grade serous ovarian carcinoma (HGSOC), while p53 mutations are much less frequent in other ovarian cancer subtypes, particularly in ovarian clear cell carcinoma (OCCC). Advanced stage OCCC with wild-type (WT) p53 has a worse prognosis and increased drug resistance, metastasis, and recurrence than HGSOC. The mechanisms responsible for driving the aggressiveness of WT p53-expressing ovarian cancer remain poorly understood. Here, we found that upregulation of MEX3A, a dual-function protein containing a RING finger domain and an RNA-binding domain, was critical for tumorigenesis in WT p53-expressing ovarian cancer. MEX3A overexpression enhanced the growth and clonogenicity of OCCC cell lines. In contrast, depletion of MEX3A in OCCC cells, as well as ovarian teratocarcinoma cells, reduced cell survival and proliferative ability. MEX3A depletion also inhibited tumor growth and prolonged survival in orthotopic xenograft models. MEX3A depletion did not alter p53 mRNA level but did increase p53 protein stability. MEX3A-mediated p53 protein degradation was crucial to suppress ferroptosis and enhance tumorigenesis. Consistently, p53 knockdown reversed the effects of MEX3A depletion. Together, our observations identified MEX3A as an important oncogenic factor promoting tumorigenesis in ovarian cancer cells expressing WT p53.
Significance:
Degradation of p53 mediated by MEX3A drives ovarian cancer growth by circumventing p53 tumor suppressive functions, suggesting targeting MEX3A as a potential strategy for treating of ovarian cancer expressing WT p53.
Introduction
Epithelial ovarian cancer is one of the leading causes of cancer death in women worldwide (1). The overall 5-year survival rate is lower than 50% (2). Due to a lack of specific symptoms and early detection methods, most ovarian cancer patients are diagnosed after the cancer has spread. The 5-year survival rate at this late stage is below 30% (3). With decades of effort, factors contributing to hereditary ovarian cancer have been identified, leading to specific prevention and early detection strategies. However, the majority of ovarian cancer cases (> 70%) are not associated with genetic predisposition (4). The causes of noninherited ovarian cancer initiation and the underlying mechanisms crucial for the disease progression and treatment remain to be determined.
Understanding how cancer initiates and progresses is essential to improve early diagnosis, outwit drug resistance and increase patient survival. However, this is particularly challenging for ovarian cancer due to its heterogeneity and complex biology. Ovarian cancer can be classified into high-grade serous ovarian carcinoma (HGSOC), low-grade serous ovarian carcinoma (LGSOC), mucinous carcinoma (MC), endometrioid carcinoma (EC), and ovarian clear cell carcinoma (OCCC; refs. 5–7). The prevalence of these subtypes can vary across race and ethnicity (8).
Mutations in tumor suppressor p53 are the most common genetic alternations in sporadic human ovarian cancer and likely are responsible for poor clinical outcome (9, 10). The p53 mutation frequency in HGSOC is as high as 96% (11). In contrast, p53 mutations are present in less than 20% of advanced OCCC cases (12, 13). OCCC tends to occur at a younger age and is frequently diagnosed at early stage. However, when OCCC is diagnosed at an advanced stage, it shows more severe prognosis, drug resistance, metastasis, and recurrence compared with other ovarian cancer subtypes (14, 15). How this occurs despite the fact that more than 80% of OCCC cases retain wild-type (WT) p53 is not known.
Several studies have found that altered expression of RNA-binding proteins (RBP) promotes tumorigenesis (16). Presumably, this occurs via posttranscriptional regulation, although the mechanisms involved are not clear for most RBPs. One such class of RBPs are the evolutionarily conserved MEX3 proteins. MEX3 proteins are orthologs of Caenorhabditis elegans MEX3, which functions as a translational repressor during embryogenesis (17). Among the MEX3s, MEX3A has multiple molecular functions including posttranscriptional regulation of gene expression through the RNA-binding activity of its KH1 and KH2 domains (17). However, MEX3A also regulates proteins posttranslationally as an ubiquitin E3-ligase via its C-terminal RING finger domain (17, 18). In normal tissues, MEX3A is critical to maintain stem cell self-renewal ability (19). Recent evidence has suggested that MEX3A upregulation is associated with malignancy in many cancers, such as glioblastoma (18) and breast cancer (20). For ovarian cancer, MEX3A upregulation correlates with poor prognosis (21) and immune infiltration in HGSOC (22), based on analysis of multiple omics databases. Whether MEX3A possesses oncogenic functions and directly influences ovarian cancer malignancy, particularly in ovarian cancer harboring WT p53, has not been reported.
We found that in WT p53-expressing ovarian cancer cells, MEX3A depletion increased p53 protein level, induced ferroptotic phenotypes and inhibited tumor growth. MEX3A facilitated WT p53 protein ubiquitination and degradation. p53 knockdown rescued the MEX3A-depletion phenotypes, thus demonstrating that MEX3A-mediated p53 degradation was key to prevent ferroptosis and enhance tumorigenesis. Together, our results identified MEX3A as an important oncogenic factor, which prevents p53-mediated ferroptosis and thus promotes ovarian cancer tumorigenesis.
Materials and Methods
Human tissue samples
Tissue specimens were from 5 endometriosis (benign), 15 OCCC, 5 EC, 13 HGSOC, and 4 MC patients treated at Taipei Veterans General Hospital (TPEVGH), Taiwan. Sample collection was conducted in accordance with the Declaration of Helsinki ethical guidelines and approved by the Institutional Review Board of Human Subjects Research Ethics Committee of TPEVGH (2016–07–006B, 2018–01–007C#1, and 2019–07–009C). Written informed consent was obtained from all patients and specimens were encoded to protect patients. Tumors were histologically classified according to the World Health Organization criteria.
Cell lines and cell culture
Human primary ovarian surface epithelial cells (HOEC, T4198) and immortalized human endometriotic cell line (IHEC) (12Z, T0764) from Applied Biological Materials were grown on Type I collagen coated plates in Prigrow X series (TM4198) and Prigrow III (TM003) medium, respectively. Ovarian teratocarcinoma cell line PA-1 was from the ATCC. TOV21G was from the Bioresource Collection and Research Center, Taiwan. OCCC cell lines OVCA-429, JHOC9, and RMG2 were kindly provided by Dr. Ruby Huang (National Taiwan University). OVISE and RMG-1 were from the Japanese Collection of Research Bioresources. All cancer cell lines were authenticated using short tandem repeat profiling (Supplementary Table S1) and Mycoplasma screened by a PCR-based approach (BIOTOOLS, TTB-GBC8). PA-1 was maintained in EMEM with 10% heat inactivated FBS, TOV21G in 1:1 mixture of MCDB 105 and Medium 199 with 15% FBS, OVCA-429 in DMEM with 10% FBS, JHOC9, RMG2, and OVISE in RPMI1640 with 10% FBS, and RMG-1 in Ham's F1 supplemented with 10% FBS. All media contained 1× Pen-Strep-Ampho B solution (Biological Industries, 03–033–1B) and all cells were cultured at 37°C in a humidified incubator supplemented with 5% CO2. HOEC and IHEC cells were used within 5 passages after thawing. Cancer cell lines were used within 25 passages.
Plasmids and reagents
The sh-Ctrl (TRC2-Scramble_ASN0003), sh-MEX3A#1 (TRCN230492), sh-MEX3A#2 (TRCN230494), sh-p53#1 (TRCN342332), sh-p53#2 (TRCN342334), and pLAS5w.pPuro vector were from the National RNAi Core Facility (Academia Sinica). pcDNA3.1.MEX3A.C(K)DYK was from GenScript (OHu05722D). pLAS5w-MEX3A-flag.pPuro was made by inserting MEX3A cDNA from pcDNA3.1.MEX3A.C(K)DYK. pCMV.tag2B.p53, pcDNA3.1–6XHisA.p53, and pcDNA3.0-HA.Ub were kindly provided by Dr. Hsiu-Ming Shih (Academia Sinica). All constructs were verified by DNA sequencing. In vitro ubiquitinylation kit (BML-UW9920–0001) was from Enzo Life Sciences, Inc. Purified recombinant protein His-GAPDH was from SinoBiological Inc (10094-H07E), His-MEX3A was from CUSABIO (CSB-EP013740HU) and p53 was from R&D Systems (SP-454–020). Cycloheximide (CHX; C1988), Z-Leu-Leu-Leu-al MG132 (C2211), and DMSO (D2650) were from Sigma-Aldrich. Liproxstatin-1 (SE-S7699) and Ferrostatin-1(SE-S7243) were from Selleckchem. Erastin (HY-15763) was from MedChemExpress, Z-VAD-FMK (M3143), Necrostatin-1 (M2315), and 3-Methyladenine (M2296) were from AbMole.
RNA extraction and qRT-PCR
Total RNA was extracted using TRI Reagent (Merck, T9424), treated with DNase I (Thermo, EN0521) and cDNA reverse transcribed with Invitrogen SuperScript III Reverse Transcriptase (Thermo, 18–080–044). qRT-PCR was performed using Applied Biosystems QuantStudio 5 Real-Time PCR System with SYBR GREEN 2X master mix (KAPA Biosystems, KM4116). The mRNA relative quantities were determined using comparative cycle threshold methods with RNA18S5 as an internal control. Primers for qRT-PCR were 5′- GGCAGGCAAGGCTGCAAGA -3′ (forward) and 5′-GGGAGGCACGGATCATGGAG-3′ (reverse) for MEX3A (NM_001093725), 5′-AAGTCTGTGACTTGCACGTACTCC-3′ (forward) and 5′-GTCATGTGCTGTGACTGCTTGTAG-3′ (reverse) for p53 (NM_000546), 5′- TGCCTATAGTGGCGATGTCTCA-3′ (forward) and 5′-GTTCCATATCCATGGCTGACAA-3′ (reverse) for GLS2 (NM_013267), 5′-TCCTGCTTTGGCTCCATGAACG-3′ (forward), 5′-AGAGGAGTGTGCTTGCGGACAT-3′ (reverse) for SLC7A11 (NM_014331), and 5′-GTAACCCGTTGAACCCCATT-3′ (forward) and 5′-CCATCCAATCGGTAGTAGCG-3′ (reverse) for RNA18S5 (NR_003286).
RNA sequencing analysis
Total RNA was extracted and quantified using NanoDrop (Thermo, ND 1000). All samples with OD 260/280 ratio > 1.8 and OD 230/260 ratio > 2 were sent to BIOTOOLS Company in Taiwan for sequencing. RNA quality was measured by Qsep100 Bio-Fragment Analyzer. Samples with RQN > 6.8 were used for cDNA library construction. Paired-end sequencing (read length 150 bp) was performed on a NovaSeq 6000 platform and quality filtering and adaptor trimming of raw reads performed using Cutadapt (v1.12; ref. 23). Trimmed reads were aligned to Human genome (hg38) using STAR (v2.5.3a; ref. 24). After mapping to the reference genome, ∼20 million paired aligned reads were recovered from each RNA sequencing (RNA-seq) library. Expected read counts of each gene were estimated with RSEM (v1.2.31; ref. 25), followed by adjusting the CPM with the effective library size via the calcNormFactors function implemented in the R package edgeR (v.3.26.8; ref. 26). For each cell line, the expression profile of each sh-MEX3A clone was compared with the sh-Ctrl and gene expression fold-change calculated (Supplementary Table S2). To identify enriched functional pathways, we performed Gene Set Enrichment Analysis (GSEA; ref. 27) on the ranked gene list, determined by the Log Fold-Change.
Immunoblot assay
Whole cell lysate was prepared using RIPA lysis buffer (Millipore, 20–188) with 1 mmol/L PMSF, protease inhibitors (Sigma-Aldrich, S8830) and phosphatase inhibitor cocktail (BIOTOOLS, TAAR-BBI3) followed by sonication at 4°C using a UP50H (Hielscher). Protein concentration was determined by Bradford assay (Bio-Rad, 5000006). Immunoblotting (IB) was performed after SDS-PAGE, with overnight incubation with anti-MEX3A primary antibody (1:10,000 dilution, Genetex, customized, PJ90803), p53 (1:1,000, Santa Cruz, sc-126), phospho-p53 (Ser15; 1:1,000, Cell Signaling Technology, 9284), UbcH5B (1:1,000, Santa Cruz, sc-100617), GLS2 (1:1,000, ABClonal, A16029), SLC7A11 (1:2m000, ABClonal, A2413), His (1:1m000, Proteintech, 10001–0-AP), HA (1:1,000, BioLegend, 901503), or TUBULIN (1:10,000, Genetex, 628802), followed by 1:10,000 dilution of horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibody (Bethyl, A120–102P, A90–516D2). Streptavidin-HRP (SA00001–0, Proteintech) was detected using Western Chemiluminescent HRP Substrate (Millipore, WBKLS0500) and images captured by a UVP ChemStudio Plus BioImaging system.
Flow cytometry (FACS) for ROS, lipid peroxidation, cell death and cell-cycle analyses
For ROS detection, cells were stained for 30 min at 37 °C with 0.5–2 μmol/L CM-H2DCFDA (Thermo, C6827; ref. 28) in HBSS (Ca2+ and Mg2+-free) and DAPI. Oxidation of the probe was detected by monitoring the increase in fluorescence using excitation source and filter appropriate for FITC. For lipid peroxidation detection, cells were stained with 10 μmol/L BODIPY 581/591 C11 (Thermo, D3861; ref. 29) for 30 to 45 minutes at 37°C. Oxidation of the polyunsaturated butadienyl structure in BODIPY-C11 was measured by fluorescence emission at 600 nm with excitation at 570 nm. Cell death analysis was performed using eBioscience Fixable Viability Dye eFluor 455UV (65–0868-14) or BD FITC Annexin V Apoptosis Detection Kit I (556547). Cell-cycle analysis was performed using Invitrogen Click-iT EdU Flow Cytometry Assay Kits (C10634). FACS analysis was performed using a FACS Canto II system (BD Biosciences).
Coimmunoprecipitation
Whole cell lysate was obtained using RIPA buffer supplemented with PMSF, protease and phosphatase inhibitors followed by sonication. Normal rabbit (Abcam, ab172730) or mouse (Jackson ImmunoResearch, 015–000–003) IgG and A/G agarose (Thermo, 20421) were used to pre-clean the cell lysate before immunoprecipitation (IP) with 6-μg antibodies against MEX3A, p53 or the normal IgG overnight at 4°C. Thirty microliter prewashed protein A/G agarose was added followed by incubation at room temperature for 2 hours. After extensive washing with IP buffer (20 mmol/L HEPES, 20% glycerol, 2% NP-40, 300 mmol/L NaCl, 1% sodium deoxycholate, and 0.2% SDS) and PBS, interacting proteins were eluted, separated with SDS-PAGE and detected by IB. For IP-IB of ubiquitinated p53, PA-1, or TOV21G cells were transfected with tagged-MEX3A, -p53, and/or -ubiquitin for 48 hours and treated with MG132 (20 μmol/L) for 8 hours before harvest. IP was performed using anti-His or anti-p53 antibody to precipitate total p53 protein. IB was performed using anti-HA antibody to detect ubiquitin conjugated p53.
Cell growth, clonogenic and soft-agar colony forming assays
To determine growth curves, 105 cells were seeded in each well of a 6-well plate. Cell numbers were counted every other day until day 7. Two-way ANOVA was used to test for significant differences between control and MEX3A depleted or overexpressing cells, or between MEX3A-depleted cells and MEX3A/p53 double-knockdown cells. To evaluate cell survival ability, 2,000 (RMG-1 and OVISE), 5,000 (PA-1), or 104 (TOV21G) cells were seeded into 6-well plates and cultured in complete medium at 37°C for 2 weeks. Colonies (>50 cells) were stained using 1% crystal violet. Soft-agar colony forming assay was performed to evaluate anchorage independent growth ability. 2000 OVISE or 1000 OVCA-429 cells were mixed with a layer of 0.35% agar/complete growth medium over a layer of 0.5% agar/complete growth medium in a well of a 12-well plate. After culture at 37°C for 2 weeks, crystal violet-stained colonies were counted. Student t test was used to compare control and treatment groups.
IHC
Formalin-fixed, paraffin embedded primary and xenograft tumor sections were used for IHC.
Sections were dewaxed with xylene and rehydrated with descending ethanol series to water. Antigen retrieval was performed using Trilogy (920P-07) for 25 minutes under high pressure. Endogenous peroxidase was eliminated with 3% H2O2 for 20 minutes. Slides were blocked with 5% non-fat milk in PBST for 1hr at room temperature, followed by primary antibodies against p53 (Cell Signaling Technology, 2527) and SLC3A2 (Cell Signaling Technology, 47213) in blocking buffer overnight at 4°C. After washing, slides were incubated with HRP rabbit polymer for 60 minutes before visualization with liquid diaminobenzidine tetrahydrochloride plus substrate DAB chromogen from Dako REAL EnVision (Dako, K5007). All slides were counterstained with hematoxylin. The images were captured under 40X magnification by an Aperio GT450 scanner (Leica Biosystems).
RNAscope in situ hybridization
MEX3A mRNA expression in primary tissue sections were determined by RNAScope 2.5 HD Detection Reagent-Brown Kit with hs-Mex3a-O1 probe according to the manufacturer's instructions. The images were captured under 40× magnification by an Aperio GT450 scanner (Leica Biosystems).
Image quantification
For IHC and RNAscope analyses, the percentage of p53-positive tumor cells, the DAB optical density (OD) of SLC3A2 and quantification of MEX3A mRNA positive spots were analyzed using the QuPath digital analysis system. In brief, different cell types were classified using the cell detection annotation of the QuPath software. Tumor cells were determined on the basis of Nucleus/ Cell area ratio (0.68).
For IHC, three xenograft tumors from each group were analyzed. For each tumor, three regions were arbitrarily selected and the numbers of total tumor cells (>10,000 cells) and p53-positive tumor cells were counted. To determine the expression level of SLC3A2, DAB OD of SLC3A2 positive cells was measured. The setting of intensity threshold was determined by the histogram generated from the OD value of all detected tumor cells, and the intensity score was set from 0 to 3 (0, no staining; 1, weak; 2, mild; 3, strong staining). Analysis procedures were done according to tutorials of the QuPath software. IHC mean intensity was defined as the multiplication of a percentage of positively stained tumor cell score (0, no staining; 1, < 30%; 2, 31%–60%; 3, > 61% of positively stained tumor cells) and an intensity score (0–3; ref. 30). The final score ranged from 0 to 9.
For each MEX3A-positive patient sample, the numbers of MEX3A-positive tumor cells were counted and the DAB OD of MEX3A mRNA spots measured. The intensity threshold of MEX3A mRNA spots was determined by the mean mRNA OD per MEX3A-positive tumor cell. The intensity of MEX3A mRNA spot above 0.05 was set as positive. The number of MEX3A mRNA spots per tumor cell was counted by the subcellular detection annotation of the QuPath software based on this threshold. Analysis procedures followed tutorials of the QuPath software and the Quantitative RNAscope Image Analysis Guide provided by Advanced Cell Diagnostics.
Intra bursa and subcutaneous xenograft models
Animal care and experiments were approved by the Institutional Animal Care and Utilization Committee of Academia Sinica (IACUC# 19–11–1354). NOD/SCID were anaesthetized and an incision made at the dorsomedial position directly above the ovarian fat pad. 1×105 GFP-Luc-labeled PA-1, 3×105 GFP-Luc-labeled TOV21G or 3×105 OVCA-429 cells were injected into the oviduct tubule leading to the bursa. The mice were closely monitored after the incision was closed. Progression of luciferase‐expressing tumors was monitored by intraperitoneal injection of 200 μL d‐luciferin in 30 mg/mL of PBS and bioluminescent imaging (IVIS Spectrum Lumina II; Perkin Elmer). Bioluminescence was quantified using Living Images software version 4.5.5. (Perkin Elmer) in photons (ph)/s/cm2/sr]. Subcutaneous tumor xenograft models were performed with injection of 3×106 cells into the lower flanks of each mouse. Tumor size was measured every other day using a Peira TM900 tumor volume-measuring device. The experiment was terminated 3 weeks after injection or when tumors reached 2-cm diameter.
Statistics
All data are presented as means ± SD (n = 3 unless otherwise noted). T test or two-way ANOVA was used to compare control and treatment groups. The Kaplan–Meier estimation method was used for overall survival analysis, and a log-rank test was used to compare differences. Statistical analyses were performed using Prism 9.
Data availability
The RNA-seq data generated in this study is publicly available in Gene Expression Omnibus at GSE200509.
Results
MEX3A upregulation is observed in EC and OCCC subtypes, as well as HGSOC
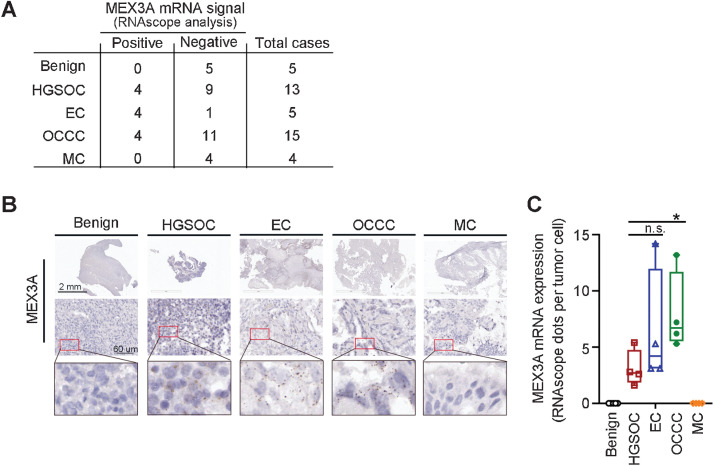
MEX3A is upregulated in HGSOC and its expression associated with poor prognosis (21, 22). To determine whether MEX3A upregulation also occurs in other ovarian cancer subtypes, we examined MEX3A mRNA level in a small cohort consisting of 5 endometrioma (as benign controls), 15 OCCC, 5 EC, 13 HGSOC and 4 MC patient samples collected in Taiwan (Fig. 1A). RNAscope analysis showed that 12 ovarian cancer samples were MEX3A-positive while none of the benign samples had detectable MEX3A mRNA spots (Fig. 1A and B). Among the MEX3A-positive samples, 8 were endometriosis-associated ovarian cancer (4 OCCC and 4 EC; ref. 31) while 4 were HGSOC. Thus, our results indicated that upregulation of MEX3A can also occur in endometriosis-associated ovarian cancer.
Figure 1.
Upregulation of MEX3A is observed in EC and OCCC subtypes, as well as HGSOC. A, Summary table of MEX3A mRNA expression detected using RNAscope analysis in an ovarian cancer cohort. B, Representative images of MEX3A RNAscope using benign and ovarian cancer tumor sections from patients. Red boxes indicate enlarged regions. Scale bars indicate 2 mm or 60 μm as shown in the images. C, Quantification and comparison of MEX3A mRNA (dots) detected by RNAscope in MEX3A-positive HGSOC, EC, and OCCC samples. Significant differences are based on unpaired t test. *, P < 0.05; n.s., not significant.
MEX3A promotes tumorigenesis in WT p53 ovarian cancer, in part, by preventing cell death
Among the MEX3A-positive patients, MEX3A expression was higher in OCCC compared with HGSOC or EC (Fig. 1B and C). As a high portion of OCCC cases retain WT p53 (12, 13) and it is unclear how these ovarian cancer cells circumvent WT p53 during tumorigenesis, we sought to understand the tumorigenesis function of MEX3A in ovarian cancer cells expressing WT p53.
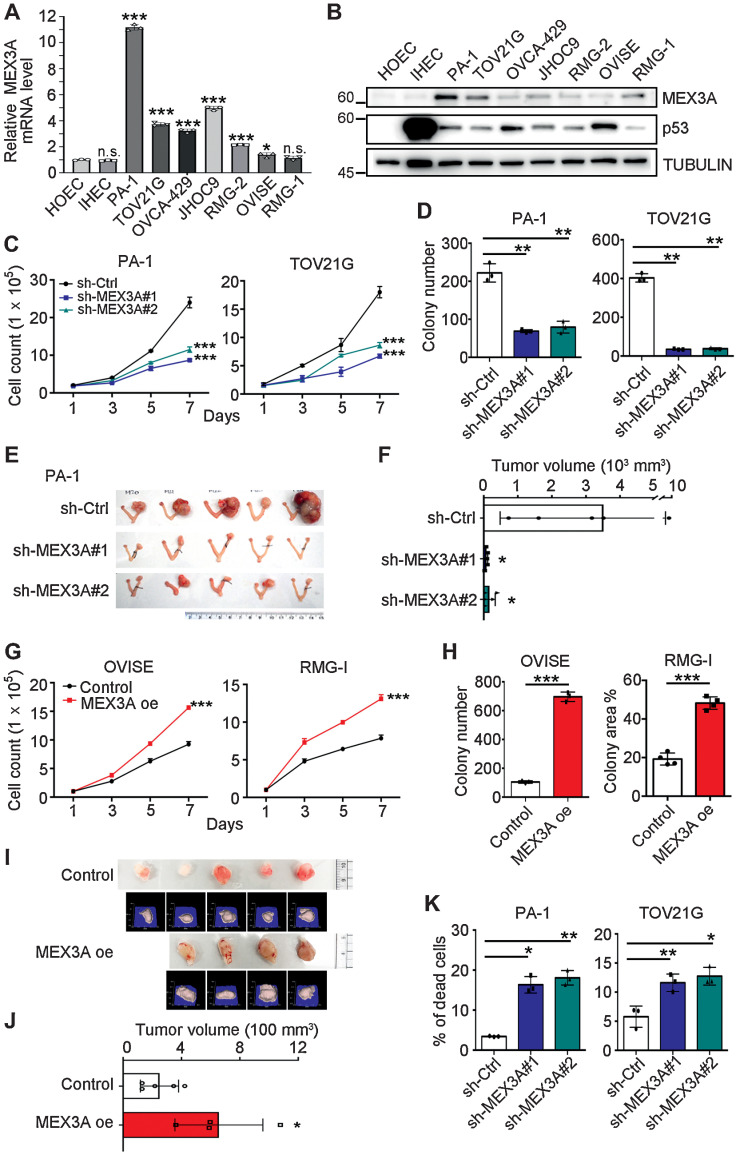
Examination of MEX3A expression in a panel of WT p53 ovarian cancer cell lines found that PA-1, an ovarian teratocarcinoma cell line expressing WT p53 protein and often used as a model for cancer stemness, had the highest MEX3A mRNA and protein expression (Fig. 2A and B). Five out of six OCCC cell lines expressed significantly higher MEX3A mRNA than the normal controls (Fig. 2A; note that primary HOECs and an IHEC served as normal controls). TOV21G had the highest MEX3A protein level and other OCCC cell lines (OVCA-429, JHOC9 and RMG-2) had moderately higher MEX3A protein expression than the normal controls (Fig. 2B). In OVISE and RMG-1 cells, MEX3A mRNA expression was low compared with other OCCC cells but was slightly higher than the HOEC and IHEC while MEX3A protein level was elevated in RMG-1 (Fig. 2A and B). We noted that the identities of OVISE and RMG-1 are controversial in the literature (32–35). Nevertheless, these two ovarian cancer cell lines both express WT p53 based on IHC or IB (Fig. 2B; refs. 32, 36). Because of their high MEX3A expression, PA-1 and TOV21G were used for loss-of-function assays where MEX3A was depleted using lentivirus-shRNA (Supplementary Fig. S1A and S1B). OVISE and RMG-1 were used for gain-of-function assays where MEX3A was ectopically expressed using lentiviral-overexpression (Supplementary Fig. S1C and S1D).
Figure 2.
MEX3A promotes cell survival and tumorigenesis in ovarian cancer cells expressing WT p53. A, qRT-PCR analysis of MEX3A level in HOEC, IHEC, PA-1, and OCCC cells. RNA18S5 was used as an internal control. Three independent experiments were performed and data are means ± SD from one representative experiment (n = 3). *, P < 0.05; ***, P < 0.001. n.s., not significant. Significant differences are based on unpaired t test. B, IB of MEX3A and p53 protein expression with TUBULIN as a loading control. Blots shown are from one representative experiment of three replicates. C, Cell growth assays using control (sh-Ctrl) or MEX3A-depleted (sh-MEX3A #1 or #2) PA-1 and TOV21G cells. Data are shown as mean ± SD with P value based on two-way ANOVA test (n = 3). ***, P < 0.001. The experiments were repeated 3 times. D, Clonogenic assays using sh-Ctrl or sh-MEX3A PA-1 and TOV21G cells. Data are shown as mean ± SD with P value based on unpaired t test (n = 3). **, P < 0.01. The experiments were repeated 3 times. E and F, Orthotopic xenograft models in NOD/SCID mice using sh-Ctrl or sh-MEX3A PA-1 cells. Five mice were used for each group. Data are means ± SD, significant difference is based on unpaired T-test of the tumor size 8 weeks after the intra bursa injection. G, Cell growth assays using control or MEX3A overexpressing (oe) OVISE and RMG-1 cells. Data are shown as mean ± SD with p value based on two-way ANOVA-test (n = 3). ***, P < 0.001. The experiments were repeated 3 times. H, Clonogenic assays using control or MEX3A oe OVISE and RMG-1 cells. Data are shown as mean ± SD with p value based on unpaired t test (n = 4). ***, P < 0.001. The experiments were repeated 3 times. I and J, Subcutaneous xenograft models in NOD/SCID mice using control (n = 5) or MEX3A oe (n = 4) RMG-1 cells. Photographs and 3D reconstruction from stereographic tumor images are shown. Data are means ± SD, significant difference is based on unpaired t test of the tumor size 3 weeks after the injection. K, Annexin V/ PI staining assays using sh-Ctrl or sh-MEX3A PA-1 and TOV21G cells. Data are shown as mean ± SD with P value based on unpaired t test (n = 3). *, P < 0.05; **, P < 0.01. The experiments were repeated 3 times.
Upon MEX3A depletion (sh-MEX3A), cell proliferation (Fig. 2C), and colony forming ability (Fig. 2D; Supplementary Fig. S1E) of PA-S1 and TOV21G cells were significantly reduced compared with the control (sh-Ctrl). Orthotopic xenograft assay was then performed by injecting EGFP-Luciferase (EGFP-LUC) expressing PA-1 cells, without or with MEX3A depletion, into the ovarian bursa of NOD/SCID mice. Eight weeks after injection, mice were euthanized and tumor size evaluated. Tumors derived from sh-Ctrl cells were significantly larger than the sh-MEX3A cell-derived tumors (Fig. 2E and F). Similar tumorigenic function of MEX3A was also observed in OVCA-429 cells (Supplementary Fig. S1F–S1J). MEX3A knockdown also inhibited cell proliferation (Supplementary Fig. S1G), anchorage independent growth ability (Supplementary Fig. S1H) and tumorigenesis (Supplementary Fig. S1I and S1J) in OVCA-429 cells.
In contrast, MEX3A overexpression in OVISE and RMG-1 cells increased cell proliferation and colony formation (Fig. 2G and H; Supplementary Fig. S1K). MEX3A overexpression also promoted anchorage-independent growth ability in OVISE cells (Supplementary Fig. S1L) and facilitated tumor growth in an RMG-1 subcutaneous xenograft model (Fig. 2I and J). These results indicated that MEX3A expression promotes tumorigenesis.
Interestingly, despite observation that cell proliferation, colony-forming ability, and tumorigenesis were impeded by MEX3A depletion, cell-cycle progression was not considerably affected in PA-1 cells (Supplementary Fig. S1M). Instead, the percentage of cell death was significantly increased upon MEX3A knockdown (Fig. 2K). In TOV21G, MEX3A depletion increased cell death (Fig. 2K), and also inhibited the cell cycle to retain more cells at the G1 stage (Supplementary Fig. S1N). Elevated cell death was also observed in MEX3A depleted OVCA-429 cells (Supplementary Fig. S1O). Together, these observations indicated that MEX3A is required for cell survival and proliferation of ovarian cancer harboring WT p53 during tumorigenesis.
MEX3A inhibits ferroptosis in WT p53 ovarian cancer cells
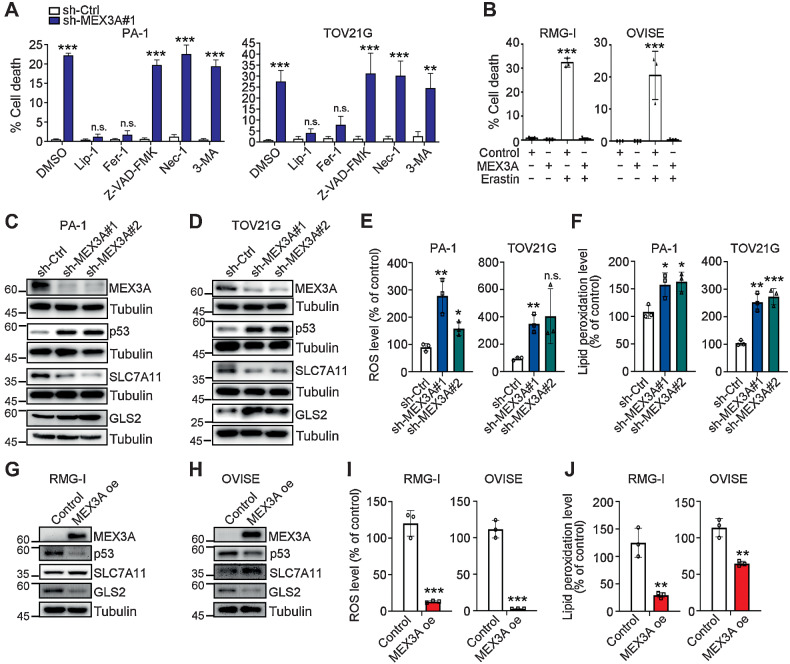
To identify MEX3A downstream targets involved in MEX3A-dependent survival and tumorigenesis of WT p53 ovarian cancer cells, RNA-seq analysis was performed. GSEA of the ranked gene list reflecting expression fold-change showed that the p53 pathway was enriched among genes upregulated in MEX3A-depleted PA-1 and TOV21G cells (Supplementary Fig. S2A; Supplementary Table S2). We therefore hypothesized that MEX3A upregulation may prevent cell death by inhibiting p53 function. WT p53 is known to regulate many cell death pathways including necrosis (37), apoptosis (38), autophagy (39), and ferroptosis (40). We therefore treated MEX3A-depleted PA-1 and TOV21G cells with the inhibitor of ferroptosis [Liproxstatin-1 (Lip-1) and Ferrostatin-1 (Fer-1)], apoptosis (Z-VAD-FMK), necrosis [Necrostatin-1 (Nec-1)], or autophagy [3-Methyladenine (3-MA)]. Cell death induced by MEX3A-depletion was rescued by ferroptosis inhibitors, but not others (Fig. 3A), indicating that MEX3A may prevent ferroptosis. Consistent with this hypothesis, we found that MEX3A-overexpressing RMG-1 and OVISE cells were resistant to the ferroptosis inducer Erastin (Fig. 3B).
Figure 3.
MEX3A depletion leads to ferroptosis phenotypes in WT p53 ovarian cancer cells. A, Fixable Viability Dye staining assays using sh-Ctrl or sh-MEX3A PA-1 and TOV21G cells treated with ferroptosis inhibitors (400 nmol/L Lip-1 and 5 μmol/L Fer-1), apoptosis inhibitor (10 μmol/L Z-VAD-FMK), necrosis inhibitor (10 μmol/L Nec-1), or autophagy inhibitor (1 mmol/L 3-MA). Data are shown as mean ± SD with P value based on unpaired t test (n = 4). *, P < 0.05; **, P < 0.01. n.s., not significant. The experiments were repeated 3 times. B, Fixable Viability Dye staining assays using control or MEX3A oe RMG-1 and OVISE cells treated with ferroptosis inducer (25 μmol/L and 0.1 μmol/L Erastin, respectively). Data formatting is as described for A. (n = 3). ***, P < 0.001. C and D, IB of p53, SLC7A11, and GLS2 protein expression using sh-Ctrl or sh-MEX3A PA-1 (C) and TOV21G (D) cells. TUBULIN was used as a loading control. Blots shown are from one representative experiment of three replicates. E, Reactive oxygen species (ROS) level in the sh-Ctrl or sh-MEX3A PA-1 and TOV21G cells were detected by CM-H2DCFDA. Relative CM-H2DCFDA mean fluorescence intensity is presented as percent of control. Three independent experiments were performed and data are means ± SD from one representative experiment (n = 3). Significant differences are based on unpaired t test (n = 3). *, P < 0.05; **, P < 0.01. n.s., not significant. F, Lipid peroxidation level in the sh-Ctrl or sh-MEX3A PA-1 and TOV21G cells detected by BODIPY-C11 staining. Relative BODIPY-C11 mean fluorescence intensity is presented as percent of control. Three independent experiments were performed and data are means ± SD from one representative experiment (n = 3). Significant differences are based on unpaired t test (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001. G and H, IB of p53, SLC7A11, and GLS2 protein expression using control or MEX3A oe RMG-1 (G) and OVISE (H) cells. TUBULIN was used as a loading control. Blots shown are from one representative experiment of three replicates. I, ROS level in the control or MEX3A oe RMG-1 and OVISE cells detected by CM-H2DCFDA. Data formatting is as described for E. J, Lipid peroxidation level in the control or MEX3A oe RMG-1 and OVISE cells detected by BODIPY-C11 staining. Data formatting is as described for F.
p53 sensitizes cells to ferroptosis by transcriptionally suppressing SLC7A11, a gene encoding the cystine/glutamate antiporter (40), or by upregulating expression of glutaminase2 (GLS2) (41). Differential expression of SLC7A11 and GLS2 was also found in our RNA-seq analysis and confirmed by qRT-PCR in MEX3A-depleted cells (Supplementary Fig. S2B and S2C). Similarly, at the protein level, MEX3A-depleted PA-1 and TOV21G cells had upregulation of p53 and GLS2 but downregulation of SLC7A11 (Fig. 3C and D). Note that different GLS2 isoforms were detected in PA-1 and TOV21G cells (42). Consistent with this altered expression of ferroptosis factors, elevated cellular ROS and lipid peroxidation levels were also observed in these MEX3A-depleted cells (Fig. 3E and F; ref. 43). In contrast, MEX3A overexpressing RMG-1 and OVISE cells showed reduced p53 and GLS2, increased SLC7A11 protein level (Fig. 3G and H), and decreased ROS and lipid peroxidation (Fig. 3I and J). Together, these results indicated that MEX3A inhibited ferroptosis in WT p53 ovarian cancer cells.
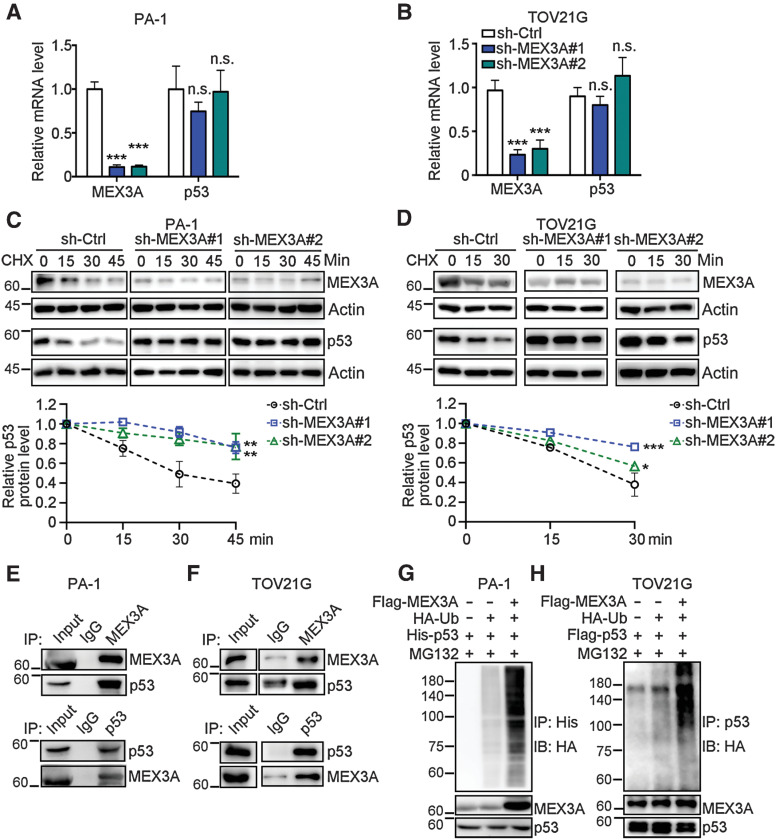
MEX3A facilitates p53 protein degradation
MEX3A has been reported to facilitate mRNA decay or protein degradation of tumor suppressors involved in cell cycle, cell survival and apoptosis via its RNA-binding capacity or E3 ligase activity, respectively (44). We found that depletion of MEX3A did not affect p53 mRNA level (Fig. 4A and B) but did increase p53 protein level in PA-1 and TOV21G cells (Fig. 3C and D). This indicated that MEX3A may regulate p53 posttranslationally. Consistent with this idea, IB analysis using whole cell lysates from CHX-treated PA-1 and TOV21G cells without or with MEX3A depletion further indicated that MEX3A expression facilitated p53 protein degradation (Fig. 4C and D). Additional experiments demonstrated MEX3A-p53 protein–protein interaction (Fig. 4E and F) and MEX3A-mediated poly-ubiquitination of p53 (Fig. 4G and H). In vitro ubiquitination assay further showed that MEX3A-mediated ubiquitination of p53 used the E2 ubiquitin-conjugating enzyme UBCH5B (Supplementary Fig. S3A). Together these experiments indicated that the ubiquitin E3 ligase function of MEX3A, rather than its RNA-binding function, was responsible for regulation of p53.
Figure 4.
MEX3A-promotes ubiquitination to destabilize p53. A and B, qRT-PCR analysis of MEX3A and p53 expression in sh-Ctrl or sh-MEX3A PA-1 (A) and TOV21G (B) cells. RNA18S5 was used as an internal control. Three independent experiments were performed and data are means ± SD from one representative experiment. Significant differences are based on unpaired t test (n = 3). ***, P < 0.001. n.s., not significant. C and D, Time course assay using CHX (20 μmol/L) treated sh-Ctrl or sh-MEX3A PA-1 (C) and TOV21G (D) cells. The levels of endogenous MEX3A and p53 were determined using IB analysis. ACTIN was used as a loading control. For each cell, quantification of relative endogenous p53 protein levels from three independent experiments was shown. Data are means ± SD. Significant differences are based on two-way ANOVA test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. E and F, Coimmunoprecipitation and reciprocal coimmunoprecipitation assays of MEX3A and p53 using PA-1 (E) and TOV21G (F) cells. Cell lysates were IP with indicated antibodies and analyzed by IB assay. Normal IgG was used as an IP control. G, IP-IB of ubiquitinated p53 using PA-1 cells transfected with Flag-tagged MEX3A, His-tagged p53 and/or HA-tagged ubiquitin and treated with MG132 (20 μmol/L). Total. His-p53 proteins were immunoprecipitated and ubiquitinated His-p53 species detected. H, IP-IB of ubiquitinated p53 using TOV21G cells transfected with Flag-tagged MEX3A, Flag-tagged p53 and/or HA-tagged ubiquitin and treated with MG132 (20 μmol/L). Total p53 proteins were immunoprecipitated and ubiquitinated p53 species detected.
It is worth noting that the increased level of p53 seen in MEX3A-depleted cells was in the active state as phosphorylation of p53 serine 15 could be detected (Supplementary Fig. S3B and S3C). Moreover, MEX3A-mediated p53 protein degradation occurred in WT p53 expressing ovarian cancer cells, but not in p53 mutated cells. MEX3A did not interact with the p53 missense mutant in KURAMOCHI, a cell line that highly recapitulates human HGSOC morphology (Supplementary Fig. S3D; ref. 45), and depletion of MEX3A did not have significant effect on mutated p53 protein level (Supplementary Fig. S3E). Overall, these results indicated that MEX3A downregulated WT p53 by facilitating p53 protein ubiquitination and degradation.
p53 downregulation is required for MEX3A to inhibit ferroptosis and promote cell survival and tumorigenesis in ovarian cancer expressing WT p53
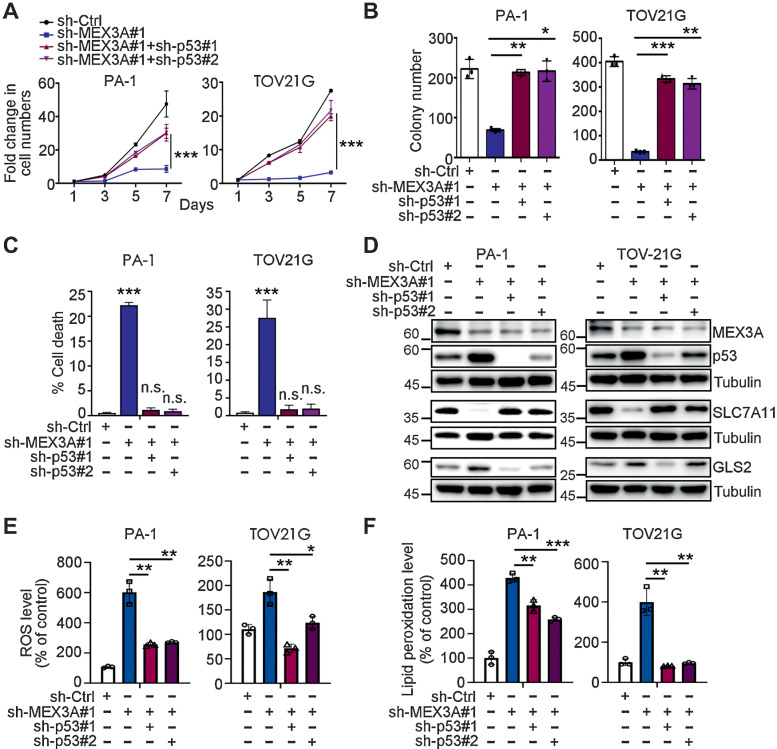
The data described above suggested that p53 degradation may be an essential step for MEX3A to inhibit ferroptosis and facilitate tumorigenesis. Consistent with this hypothesis, p53 knockdown rescued the proliferation and clonogenic ability (Fig. 5A and B), as well as the elevated cell death (Fig. 5C) of MEX3A-depleted cells. In contrast, knockdown of p53 in WT p53 ovarian cancer cells with high MEX3A expression did not substantially affect cell proliferation or cell death (Supplementary Fig. S4A and S4B); albeit that there was a small decrease (∼0.5%) in cell death for p53-depleted PA-1 cells. Consistent with these results, the elevated GLS2 and reduced SLC7A11 resulting from MEX3A depletion (Fig. 3C and D) could also be reversed by p53 knockdown (Fig. 5D). Consistent with these molecular changes, the elevated ROS and lipid peroxidation level caused by MEX3A-depletion was significantly reduced in MEX3A/p53-double knockdown cells (Fig. 5E and F).
Figure 5.
p53 knockdown reverses the increased ferroptosis phenotype of MEX3A-depleted WT p53 ovarian cancer cells. A, Cell growth assays using sh-Ctrl, sh-MEX3A or MEX3A/p53-double knockdown PA-1 and TOV21G cells. Data are shown as mean ± SD with P value based on two-way ANOVA test (n = 3). ***, P < 0.001. The experiments were repeated 3 times. B, Clonogenic assays using sh-Ctrl, sh-MEX3A, or MEX3A/p53-double knockdown PA-1 and TOV21G cells. Data are shown as mean ± SD with P value based on unpaired t test (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001. The experiments were repeated 3 times. C, Fixable Viability Dye staining assays using sh-Ctrl, sh-MEX3A, or MEX3A/p53-double knockdown PA-1 and TOV21G cells. Data are shown as mean ± SD with P value based on unpaired t test (n = 3). ***, P < 0.001. n.s., not significant. The experiments were repeated 3 times. D, IB of MEX3A, p53, SLC7A11, and GLS2 protein expression using sh-Ctrl, sh-MEX3A, or MEX3A/p53-double knockdown PA-1 and TOV21G cells. TUBULIN was used as a loading control. Blots shown are from one representative experiment of three replicates. E, ROS level in sh-Ctrl, sh-MEX3A or MEX3A/p53-double knockdown PA-1 and TOV21G cells detected by CM-H2DCFDA. Relative CM-H2DCFDA mean fluorescence intensity is presented as percent of control. Three independent experiments were performed and data are means ± SD from one representative experiment. Significant differences are based on unpaired t test (n = 3). *, P < 0.05; **, P < 0.01. F, Lipid peroxidation level in sh-Ctrl, sh-MEX3A, or MEX3A/p53-double knockdown PA-1 and TOV21G detected by BODIPY-C11 staining. Relative BODIPY-C11 mean fluorescence intensity is presented as percent of control. Three independent experiments were performed and data are means ± SD from one representative experiment. Significant differences are based on unpaired t test (n = 3). **, P < 0.01; ***, P < 0.001.
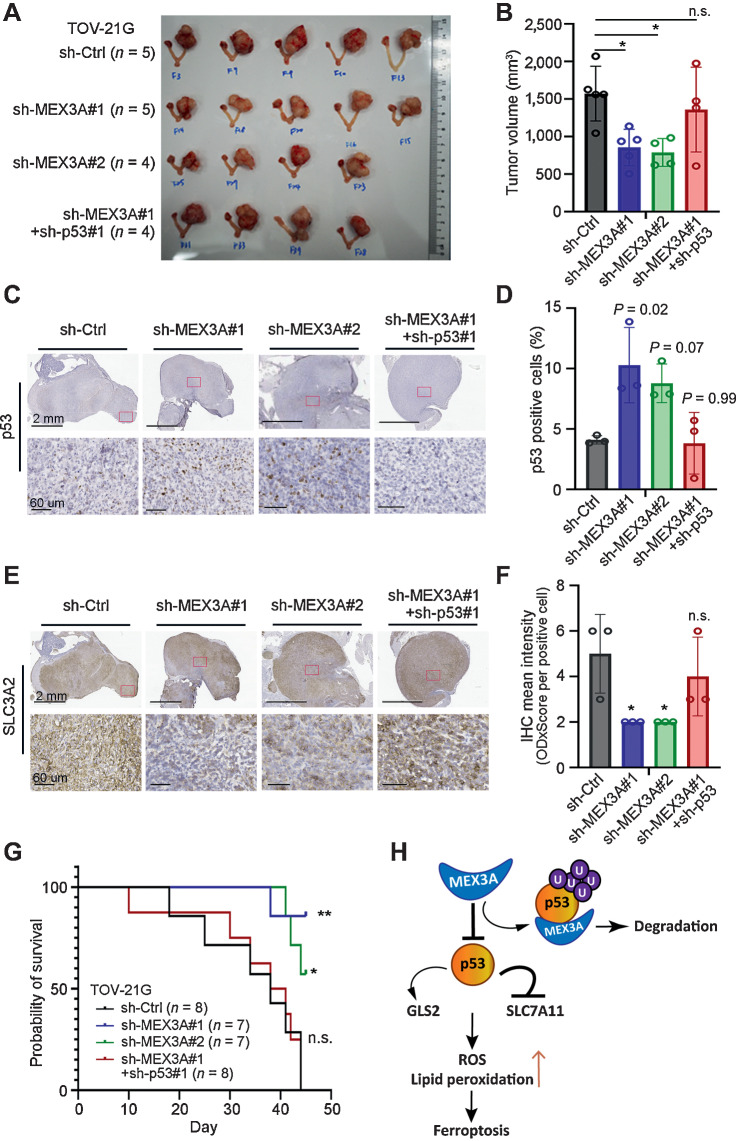
To evaluate whether MEX3A-mediated p53 degradation is critical for tumorigenesis of ovarian cancer harboring WT p53, EGFP-LUC expressing TOV21G cells with MEX3A depletion alone (sh-MEX3A) or MEX3A/p53-double knockdown (sh-MEX3A + sh-p53) were used for intra bursa xenograft assays, along with sh-Ctrl cells for comparison. Tumors derived from MEX3A-depleted TOV21G cells, but not MEX3A/p53 double knockdown cells, were significantly smaller than the control (Fig. 6A and B). This was consistent with cell-based assays (Figs. 2, 3, 5; Supplementary Fig. S1) and xenograft models using PA-1 and OVCA-429 cells (Fig. 2E and F; Supplementary Fig. S1I and S1J). IHC analysis found an increased proportion of p53-positive cells in tumors derived from MEX3A-depleted TOV21G cells compared with sh-Ctrl or MEX3A/p53-double knockdown tumors (Fig. 6C and D). We further used SLC3A2, whose expression is correlated with SLC7A11 (46) and acts as a chaperone for SLC7A11 membrane anchoring (47), as a marker to examine whether ferroptosis was increased in the MEX3A-depleted tumor cells. Consistent with our cell-based assays, the MEX3A-depleted TOV21G tumors showed lower SLC3A2 expression indicating these tumor cells had elevated ferroptosis compared with the sh-Ctrl. This phenotype was also reversed in the MEX3A/p53-double knockdown tumors (Fig. 6E and F). Furthermore, mice bearing tumors derived from MEX3A-depleted TOV21G cells had longer survival compared with those bearing tumors expressing MEX3A (sh-Ctrl; Fig. 6G). This in vivo phenotype resulting from MEX3A depletion was also reversed by p53 knockdown. Survival of the MEX3A/p53-double knockdown group was significantly shorter than the sh-MEX3A groups (Fig. 6G). Together, these results demonstrated that MEX3A acts as a potent oncogenic factor by downregulating p53 to promote tumorigenesis in ovarian cancer expressing WT p53.
Figure 6.
p53 downregulation is required for MEX3A to promote tumorigenesis and drug resistance in WT p53 ovarian cancer. A and B, Orthotopic xenograft tumors derived from TOV21G cells without or with MEX3A depletion or MEX3A/p53-double knockdown (B). Tumor volume was plotted (C). Data are presented as mean ± SD. Significant difference is based on unpaired t test. *, P < 0.05. n.s., not significant. C–F, Representative images of p53 (C and D) and SLC3A2 (E and F) IHC staining using serial tumor sections from the orthotopic xenograft mice in A. Red boxes indicate the enlarged regions. Scale bars, 2 mm or 60 μm as shown in the images. For each group, numbers of p53-positive cells (D) and SLC3A2 expression level (F) from three tumors were evaluated. Data are presented as mean ± SD. Significant difference compared with the sh-Ctrl group is based on unpaired t test. *, P < 0.05. n.s., not significant. G, Kaplan–Meier overall survival analysis of NOD/SCID mice intrabursally injected with sh-Ctrl (black line), sh-MEX3A (blue and green lines), or MEX3A/p53-double knockdown (sh-MEX3A#1+sh-p53#1, red line) TOV21G cells. Significant differences are based on log-rank test. *, P < 0.05; **, P < 0.01. n.s., not significant. H, Model of MEX3A-mediated ferroptosis inhibition in ovarian cancer harboring WT p53. p53 induces ferroptosis by repressing SLC7A11 and upregulating GLS2 expression. In WT p53 ovarian cancer cells, MEX3A destabilize p53 protein by ubiquitination. Downregulation of WT p53 protein is essential for MEX3A to inhibit ferroptosis-induced cell death.
Discussion
Our study demonstrated that the dual-function RBP MEX3A is critical for cell survival and tumorigenesis in ovarian cancer expressing WT p53. Instead of regulating p53 RNA, MEX3A destabilizes p53 protein by ubiquitination. Downregulation of WT p53 is essential for MEX3A to inhibit ferroptosis-induced cell death and thus promote tumorigenesis (Fig. 6H). In ovarian cancer with mutated p53, such as HGSOC, MEX3A upregulation also promotes tumorigenesis and is associated with poor prognosis (21, 22). However, MEX3A neither interacts with nor affects mutant p53 protein level (Supplementary Fig. S3). How MEX3A selectively interacts with WT p53 and while still having tumorigenic effect in different ovarian cancer subtypes requires further study.
Ferroptosis, an iron-dependent programmed cell death distinct from apoptosis, is characterized by lipid peroxide accumulation, shrunken mitochondria and increased membrane density (43, 48). Although mechanisms underlying ferroptosis in many cancers are under intensive investigation, it has remained unclear how WT p53 ovarian cancer cells keep ferroptosis at bay during tumorigenesis. p53 promotes ferroptosis by repressing SLC7A11 to inhibit cysteine uptake (40), transactivating GLS2 to regulate glutaminolysis (41, 49), and regulating spermidine/spermine N1-acetyltransferase 1 (SAT1) to increase lipid peroxide accumulation (50). In terms of ovarian cancer, a significant association of ferroptosis with chemotherapeutic response and resistance has been suggested (51, 52). Thus, induction of ferroptosis in WT p53 ovarian cancer cells may be a good way to mitigate chemoresistance.
It is worthwhile to mention that some commonly altered genes in ovarian cancers also play roles in ferroptosis. For example, ARID1A, a chromatin remodeling factor often mutated in OCCC (53), has been shown to facilitate NRF2-mediated transcriptional activation of SLC7A11 to maintain glutathione (GSH) homeostasis (54). Thus, ARID1A-deficient cells can be sensitive to cysteine level and vulnerable to oxidative stress. This could explain, in part, why OCCC cells greatly rely on cysteine for survival (55). Another example is proto-oncogene KRAS. KRAS has been shown to either upregulate SLC7A11 transcription or stabilize its protein in cancer to suppress ferroptosis to enhance tumor development (56). These KRAS mutated tumors are dependent on SLC7A11-mediated cystine uptake, and therefore could be particularly sensitive to SLC7A11 inhibitors, such as Erastin.
Among the OCCC cell lines used in our study, TOV21G and OVISE are ARID1A deficient, while RMG-1 expresses WT ARID1A (32, 33). TOV21G also carries a KRAS mutation (32, 33). As for the teratocarcinoma cell line PA-1, it does not carry mutations in either ARID1A or KRAS, but has a NRAS mutation (33). How NRAS contributes to ferroptosis regulation is currently unclear. Nevertheless, our results suggested that MEX3A is a key oncoprotein for WT p53 ovarian cancer cells to avoid death from ferroptosis regardless of the status of ARID1A and KRAS. MEX3A expression keeps p53 protein at low level, which maintains the expression of SLC7A11 and consequently inhibits ferroptosis (Figs. 3 and 5). Interestingly, we noticed that genes involved in KRAS signaling were differentially expressed upon MEX3A depletion in both PA1 and TOV21G cells (Supplementary Fig. S2). Whether MEX3A inhibits ferroptosis via KRAS or other commonly altered genes in ovarian cancers, in addition to p53, remains to be determined.
In ovarian cancer, most p53 detected by IHC analysis is mutated while WT p53 protein is suppressed to an undetectable level in most cases. In many cancers, MDM2 is upregulated to facilitate p53 ubiquitination and degradation (57). In ovarian cancer, however, MDM2 is rarely amplified (58). What keeps WT p53 protein level low in malignant ovarian cancers was unclear. Our study provides an additional regulatory mechanism by which MEX3A serves as an E3 ligase to facilitate WT p53 protein degradation in ovarian cancer. Interestingly, our RNA-seq results found MDM2 upregulation in MEX3A-depleted PA-1 and TOV21G cells (Supplementary Fig. S5A). The upregulated genes were also significantly enriched for genes encoding MDM2 interacting proteins (Supplementary Fig. S5B). Whether MDM2 and MEX3A may functionally compensate for one another to suppress WT p53 protein level in ovarian cancer or other types of cancer expressing WT p53 is worth further investigation.
In summary, our study identified MEX3A as a tumor-promoting factor, which plays an essential role to inhibit p53-dependent ferroptosis in ovarian cancer. These results provide insights into the mechanisms of ovarian tumorigenesis and could facilitate development of effective strategies for cancer therapy selectively targeting ovarian cancer expressing WT p53.
Supplementary Material
Table S1. Short tandem repeat of ovarian teratocarcinoma and clear cell carcinoma cell line. Figure S1. MEX3A expression promoted tumorigenic ability and cell survival in WT p53 OC cells. Figure S2. The p53 pathway was enriched in upregulated genes in MEX3A-depleted OC cells expressing WT p53. Figure S3. Stabilized WT p53 protein in MEX3A-depleted cells was activated. MEX3A did not interact with or affect the level of mutant p53 protein. Figure S4. p53 depletion alone did not significantly affect cell proliferation or cell death in MEX3A expressing WT p53 OC cells. Figure S5. MEX3A and MDM2 in OC.
RNAseq: Gene expression fold-change between sh-MEX3A and sh-Ctrl
Acknowledgments
The authors would like to thank Dr. Michael Hsiao (Genomics Research Center, Academia Sinica, Taiwan) for providing NOD/SCID mice, the Bioinformatics Core at Institute of Molecular Biology, Academia Sinica for providing the RNA-seq analysis services and the DNA Sequencing Core Facility of the Institute of Biomedical Sciences, Academia Sinica for DNA sequencing analysis.
This work was supported by Academia Sinica [AS-GCS-111-L01 and AS-KPQ-109-BioMed] and the Taiwan National Science and Technology Council (Ministry of Science and Technology) [MOST 109–0210–01–18–02 and MOST111–2314-B-001–012] to Wendy W. Hwang-Verslues; [MOST 109–2314-B-010–042] andTPEVGH [V107C-30, V108C-065, V109C-097 and V110C-027] to Yi-Jen Chen.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
Y.J. Chen reports grants from Ministry of Science and Technology, Taiwan; and grants from TPEVGH during the conduct of the study. W.W. Hwang-Verslues reports grants from National Science and Technology Council (Ministry of Science and Technology), Taiwan, and grants from Academia Sinica during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
C.-K. Wang: Formal analysis, validation, investigation. T.-J. Chen: Formal analysis, validation, investigation, visualization. G.Y.T Tan: Formal analysis, validation, investigation. F.-P. Chang: Formal analysis, validation, investigation. S. Sridharan: Formal analysis, validation, investigation. C.-H.A. Yu: Formal analysis, validation, investigation, visualization. Y.-H. Chang: Resources. Y.-J. Chen: Resources. L.-T. Cheng: Formal analysis. W.W. Hwang-Verslues: Conceptualization, formal analysis, supervision, funding acquisition, validation, visualization, writing–original draft, project administration, writing–review and editing.
References
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2. Doherty JA, Peres LC, Wang C, Way GP, Greene CS, Schildkraut JM. Challenges and opportunities in studying the epidemiology of ovarian cancer subtypes. Curr Epidemiol Rep 2017;4:211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reid BM, Permuth JB, Sellers TA. Epidemiology of ovarian cancer: a review. Cancer Biol Med 2017;14:9–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci USA 2011;108:18032–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Seidman JD, Horkayne-Szakaly I, Haiba M, Boice CR, Kurman RJ, Ronnett BM. The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int J Gynecol Pathol 2004;23:41–4. [DOI] [PubMed] [Google Scholar]
- 6. Koonings PP, Campbell K, Mishell DR Jr, Grimes DA. Relative frequency of primary ovarian neoplasms: a 10-year review. Obstet Gynecol 1989;74:921–6. [PubMed] [Google Scholar]
- 7. Kobel M, Bak J, Bertelsen BI, Carpen O, Grove A, Hansen ES, et al. Ovarian carcinoma histotype determination is highly reproducible, and is improved through the use of immunohistochemistry. Histopathology 2014;64:1004–13. [DOI] [PubMed] [Google Scholar]
- 8. Iida Y, Okamoto A, Hollis RL, Gourley C, Herrington CS. Clear cell carcinoma of the ovary: a clinical and molecular perspective. Int J Gynecol Cancer 2021;31:605–16. [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y, Cao L, Nguyen D, Lu H. TP53 mutations in epithelial ovarian cancer. Transl Cancer Res 2016;5:650–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ahmed AA, Etemadmoghadam D, Temple J, Lynch AG, Riad M, Sharma R, et al. Driver mutations in TP53 are ubiquitous in high-grade serous carcinoma of the ovary. J Pathol 2010;221:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cunningham JM, Winham SJ, Wang C, Weiglt B, Fu Z, Armasu SM, et al. DNA methylation profiles of ovarian clear cell carcinoma. Cancer Epidemiol Biomarkers Prev 2022;31:132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bolton KL, Chen D, Corona de la Fuente RI, Fu Z, Murali R, Köbel M, et al. Molecular subclasses of clear cell ovarian carcinoma and their impact on disease behavior and outcomes. Clin Cancer Res 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kommoss S, Gilks CB, du Bois A, Kommoss F. Ovarian carcinoma diagnosis: the clinical impact of 15 years of change. Br J Cancer 2016;115:993–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mabuchi S, Sugiyama T, Kimura T. Clear cell carcinoma of the ovary: molecular insights and future therapeutic perspectives. J Gynecol Oncol 2016;27:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pereira B, Billaud M, Almeida R. RNA-binding proteins in cancer: Old players and new actors. Trends Cancer 2017;3:506–28. [DOI] [PubMed] [Google Scholar]
- 17. Buchet-Poyau K, Courchet J, Le Hir H, Seraphin B, Scoazec JY, Duret L, et al. Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res 2007;35:1289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bufalieri F, Caimano M, Lospinoso Severini L, Basili I, Paglia F, Sampirisi L, et al. The RNA-binding ubiquitin ligate MEX3A affects glioblastoma tumorigenesis by inducing ubiquitylation and degradation of RIG-I. Cancers 2020;12:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pereira B, Amaral AL, Dias A, Mendes N, Muncan V, Silva AR, et al. MEX3A regulates Lgr5(+) stem cell maintenance in the developing intestinal epithelium. EMBO Rep 2020;21:e48938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oliemuller E, Newman R, Tsang SM, Foo S, Muirhead G, Noor F, et al. SOX11 promotes epithelial/mesenchymal hybrid state and alters tropism of invasive breast cancer cells. Elife 2020;9:e58374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li F, Zhao C, Diao Y, Wang Z, Peng J, Yang N, et al. MEX3A promotes the malignant progression of ovarian cancer by regulating intron retention in TIMELESS. Cell Death Dis 2022;13:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang P, Su T, Zhang S. Comprehensive analysis of prognostic value of MEX3A and its relationship with immune infiltrates in ovarian cancer. J Immunol Res 2021;2021:5574176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 2011;17:10–2. [Google Scholar]
- 24. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene Set Enrichment Analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oparka M, Walczak J, Malinska D, van Oppen L, Szczepanowska J, Koopman WJH, et al. Quantifying ROS levels using CM-H2DCFDA and HyPer. Methods 2016;109:3–11. [DOI] [PubMed] [Google Scholar]
- 29. Drummen GP, van Liebergen LC, Op den Kamp JA, Post JA. C11-BODIPY(581/591), an oxidation-sensitive fluorescent lipid peroxidation probe: (micro)spectroscopic characterization and validation of methodology. Free Radic Biol Med 2002;33:473–90. [DOI] [PubMed] [Google Scholar]
- 30. Klein M, Vignaud JM, Hennequin V, Toussaint B, Bresler L, Plenat F, et al. Increased expression of the vascular endothelial growth factor is a pejorative prognosis marker in papillary thyroid carcinoma. J Clin Endocrinol Metab 2001;86:656–8. [DOI] [PubMed] [Google Scholar]
- 31. Karnezis AN, Cho KR, Gilks CB, Pearce CL, Huntsman DG. The disparate origins of ovarian cancers: pathogenesis and prevention strategies. Nat Rev Cancer 2017;17:65–74. [DOI] [PubMed] [Google Scholar]
- 32. Anglesio MS, Wiegand KC, Melnyk N, Chow C, Salamanca C, Prentice LM, et al. Type-specific cell line models for type-specific ovarian cancer research. PLoS One 2013;8:e72162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell 2017;170:564–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan TZ, Ye J, Yee CV, Lim D, Ngoi NYL, Tan DSP, et al. Analysis of gene expression signatures identifies prognostic and functionally distinct ovarian clear cell carcinoma subtypes. EBioMedicine 2019;50:203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bitler BG, Wu S, Park PH, Hai Y, Aird KM, Wang Y, et al. ARID1A-mutated ovarian cancers depend on HDAC6 activity. Nat Cell Biol 2017;19:962–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Makii C, Oda K, Ikeda Y, Sone K, Hasegawa K, Uehara Y, et al. MDM2 is a potential therapeutic target and prognostic factor for ovarian clear cell carcinomas with wild-type TP53. Oncotarget 2016;7:75328–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell 2012;149:1536–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fridman JS, Lowe SW. Control of apoptosis by p53. Oncogene 2003;22:9030–40. [DOI] [PubMed] [Google Scholar]
- 39. Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 2008;10:676–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, et al. Ferroptosis as a p53-mediated activity during tumor suppression. Nature 2015;520:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A 2010;107:7461–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Katt WP, Lukey MJ, Cerione RA. A tale of two glutaminases: homologous enzymes with distinct roles in tumorigenesis. Future Med Chem 2017;9:223–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang WS, Stockwell BR. Ferroptosis: death by lipid peroxidation. Trends Cell Biol 2016;26:165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lederer M, Muller S, Glass M, Bley N, Ihling C, Sinz A, et al. Oncogenic potential of the dual-function protein MEX3A. Biology 2021;10:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Elias KM, Emori MM, Papp E, MacDuffie E, Konecny GE, Velculescu VE, et al. Beyond genomics: critical evaluation of cell line utility for ovarian cancer research. Gynecol Oncol 2015;139:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang Y, Dai Z, Barbacioru C, Sadee W. Cystine-glutamate transporter SLC7A11 in cancer chemosensitivity and chemoresistance. Cancer Res 2005;65:7446–54. [DOI] [PubMed] [Google Scholar]
- 47. Nakamura E, Sato M, Yang H, Miyagawa F, Harasaki M, Tomita K, et al. 4F2 (CD98) heavy chain is associated covalently with an amino acid transporter and controls intracellular trafficking and membrane topology of 4F2 heterodimer. J Biol Chem 1999;274:3009–16. [DOI] [PubMed] [Google Scholar]
- 48. Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012;149:1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gao M, Monian P, Quadri N, Ramasamy R, Jiang X. Glutaminolysis and transferrin regulate ferroptosis. Mol Cell 2015;59:298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ou Y, Wang SJ, Li D, Chu B, Gu W. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc Natl Acad Sci USA 2016;113:E6806–E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li L, Qiu C, Hou M, Wang X, Huang C, Zou J, et al. Ferroptosis in ovarian cancer: a novel therapeutic strategy. Front Oncol 2021;11:665945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hong T, Lei G, Chen X, Li H, Zhang X, Wu N, et al. PARP inhibition promotes ferroptosis via repressing SLC7A11 and synergizes with ferroptosis inducers in BRCA-proficient ovarian cancer. Redox Biol 2021;42:101928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 2010;330:228–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ogiwara H, Takahashi K, Sasaki M, Kuroda T, Yoshida H, Watanabe R, et al. Targeting the vulnerability of glutathione metabolism in ARID1A-deficient cancers. Cancer Cell 2019;35:177–90. [DOI] [PubMed] [Google Scholar]
- 55. Novera W, Lee ZW, Nin DS, Dai MZ, Binte Idres S, Wu H, et al. Cysteine deprivation targets ovarian clear cell carcinoma via oxidative stress and iron-sulfur cluster biogenesis deficit. Antioxid Redox Signal 2020;33:1191–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Badgley MA, Kremer DM, Maurer HC, DelGiorno KE, Lee HJ, Purohit V, et al. Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science 2020;368:85–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Manfredi JJ. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev 2010;24:1580–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Foulkes WD, Stamp GW, Afzal S, Lalani N, McFarlane CP, Trowsdale J, et al. MDM2 overexpression is rare in ovarian carcinoma irrespective of TP53 mutation status. Br J Cancer 1995;72:883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Short tandem repeat of ovarian teratocarcinoma and clear cell carcinoma cell line. Figure S1. MEX3A expression promoted tumorigenic ability and cell survival in WT p53 OC cells. Figure S2. The p53 pathway was enriched in upregulated genes in MEX3A-depleted OC cells expressing WT p53. Figure S3. Stabilized WT p53 protein in MEX3A-depleted cells was activated. MEX3A did not interact with or affect the level of mutant p53 protein. Figure S4. p53 depletion alone did not significantly affect cell proliferation or cell death in MEX3A expressing WT p53 OC cells. Figure S5. MEX3A and MDM2 in OC.
RNAseq: Gene expression fold-change between sh-MEX3A and sh-Ctrl
Data Availability Statement
The RNA-seq data generated in this study is publicly available in Gene Expression Omnibus at GSE200509.