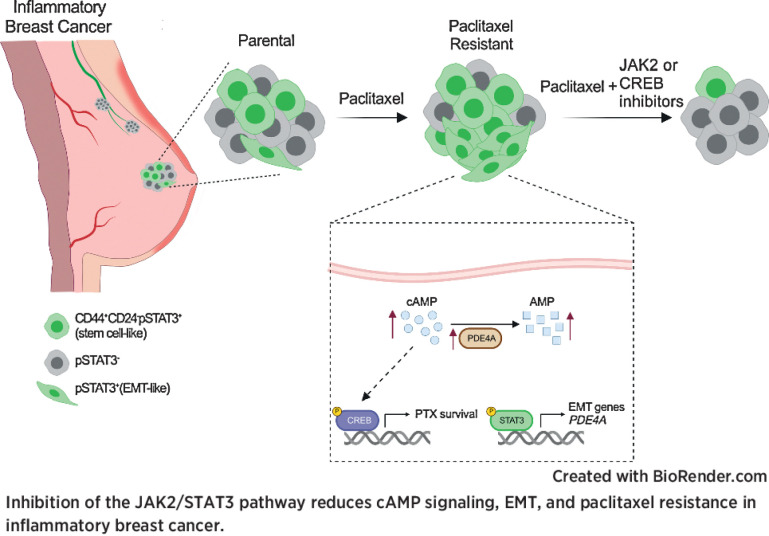
Chemotherapy resistance in inflammatory breast cancer is driven by the JAK2/STAT3 pathway, in part via cAMP/PKA signaling and a cell state switch, which can be overcome using paclitaxel combined with JAK2 inhibitors.
Abstract
Inflammatory breast cancer (IBC) is a difficult-to-treat disease with poor clinical outcomes due to high risk of metastasis and resistance to treatment. In breast cancer, CD44+CD24− cells possess stem cell-like features and contribute to disease progression, and we previously described a CD44+CD24−pSTAT3+ breast cancer cell subpopulation that is dependent on JAK2/STAT3 signaling. Here we report that CD44+CD24− cells are the most frequent cell type in IBC and are commonly pSTAT3+. Combination of JAK2/STAT3 inhibition with paclitaxel decreased IBC xenograft growth more than either agent alone. IBC cell lines resistant to paclitaxel and doxorubicin were developed and characterized to mimic therapeutic resistance in patients. Multi-omic profiling of parental and resistant cells revealed enrichment of genes associated with lineage identity and inflammation in chemotherapy-resistant derivatives. Integrated pSTAT3 chromatin immunoprecipitation sequencing and RNA sequencing (RNA-seq) analyses showed pSTAT3 regulates genes related to inflammation and epithelial-to-mesenchymal transition (EMT) in resistant cells, as well as PDE4A, a cAMP-specific phosphodiesterase. Metabolomic characterization identified elevated cAMP signaling and CREB as a candidate therapeutic target in IBC. Investigation of cellular dynamics and heterogeneity at the single cell level during chemotherapy and acquired resistance by CyTOF and single cell RNA-seq identified mechanisms of resistance including a shift from luminal to basal/mesenchymal cell states through selection for rare preexisting subpopulations or an acquired change. Finally, combination treatment with paclitaxel and JAK2/STAT3 inhibition prevented the emergence of the mesenchymal chemo-resistant subpopulation. These results provide mechanistic rational for combination of chemotherapy with inhibition of JAK2/STAT3 signaling as a more effective therapeutic strategy in IBC.
Significance:
Chemotherapy resistance in inflammatory breast cancer is driven by the JAK2/STAT3 pathway, in part via cAMP/PKA signaling and a cell state switch, which can be overcome using paclitaxel combined with JAK2 inhibitors.
Graphical Abstract
Introduction
Inflammatory breast cancer (IBC) is a rare and aggressive subtype of breast cancer with a unique clinical presentation, but despite improvements in multimodal therapy, survival rate remains low (1). Thus, a greater understanding of the molecular etiology of IBC is needed to develop more effective treatments.
Breast cancer is classified on the basis of the expression of estrogen (ER), progesterone (PR), and HER2 receptors into ER+, HER2+, and triple-negative (ER−PR−HER2−) subtypes (2). IBC encompasses these subtypes, although they are more commonly triple-negative (TN) or HER2+, and like non-IBC, subtype correlates with outcome and defines treatment strategy (3). However, IBC is considerably more aggressive than their non-IBC counterparts (4) and the search for IBC-specific molecular mechanisms is an intense area of investigation. Gene expression profiling of subtype-matched IBC and non-IBC revealed high E-cadherin and RhoC levels and low TGFβ signaling in IBC tumor cells (5–7). Targeted sequencing studies in IBC have not identified specifically-mutated cancer-driving genes (8–10), suggesting that the discovery of novel therapeutic targets in IBC will require alternative approaches.
Resistance to therapy and increased metastatic capacity are often attributed to the cancer stem cell phenotype and to high intratumor heterogeneity (11). In breast cancer, CD44+CD24− cells with stem cell–like features are associated with higher risk of metastasis and poor outcome (12–14). We previously described that CD44+CD24−pSTAT3+ cancer cells are more commonly present in TNBC compared with other breast cancer subtypes and that the IL6/JAK2/STAT3 signaling pathway is required for their survival (13, 15). STAT3 has known roles in therapeutic resistance, stem cell maintenance, metastasis, and inflammation (16). Furthermore, IL6, an inflammatory cytokine that activates the JAK/STAT signaling pathway, is upregulated in IBC compared with non-IBC tumors (17), implying a role for IL6/JAK/STAT3 signaling in IBC.
Here we show that on the basis of molecular profiling of patient samples and parental and chemotherapy-resistant IBC cell lines we developed, JAK2/STAT3 signaling is a key regulator of IBC growth and cellular identity, which can modulate response and resistance to chemotherapeutics. Furthermore, we showed that JAK2/STAT3 may mediate chemotherapy resistance by multiple mechanisms including switching cell state, as in epithelial-to-mesenchymal transition (EMT). We also identified cAMP signaling as a putative marker of both IBC and resistance. Finally, we found that combination of JAK2/STAT3 inhibition with chemotherapy was synergistic in resistant derivatives and suppressed EMT-associated resistance mechanisms. Our results will facilitate the stratification of patients with IBC to improve clinical outcomes.
Materials and Methods
Human breast cancer samples
All experiments with use of human tumor tissue were approved by the Dana-Farber Cancer Institute Institutional Review Board (protocol no. 14–400). Formalin-fixed paraffin-embedded breast tumor samples were collected from patients diagnosed with inflammatory breast cancer at Dana-Farber Harvard Cancer Center and Seoul National University Bundang Hospital using protocols approved by the Institutional Review Boards of the respective hospitals and following the Declaration of Helsinki ethical guidelines. Written informed consent was obtained from all patients and samples were de-identified prior to transfer to the laboratory. Tumor histology and expression of standard biomarkers (ER, PR, and HER2) were evaluated at the time of diagnosis according to ASCO/CAP guidelines (18).
Multicolor immunofluorescence
The triple immunofluorescence for CD44 (Neomarkers, clone 156–3C11, mouse monoclonal IgG2), CD24 (Neomarkers, clone SN3b, mouse monoclonal IgM), and pSTAT3 (Cell Signaling Technology, catalog no. 9145, Rabbit monoclonal) was performed on whole sections of primary tumors or xenografts as described previously (13, 15). Different immunofluorescence images from multiple areas of each sample were acquired with a Nikon Ti microscope attached to a Yokogawa spinning-disk confocal unit, 60× plan apo objective, and OrcaER camera controlled by Andor iQ software. The frequency of pSTAT3+ cells within each cell phenotype (i.e., CD44+CD24−, CD44+CD24+, CD44−CD24+, and CD44−CD24− cells) was calculated by counting an average of 100 cells in each sample. The frequency of each cell phenotype was calculated by counting an average of 300 cells in each sample. For immunofluorescence staining of xenografts with pHH3 or ClCasp3, following deparaffinization and antigen retrieval, slides were blocked with 10% goat serum in PBS for 10 minutes at room temperature. Cl-Casp3 (Cell Signaling Technology, catalog no. 9661, diluted 1:100) or pHH3 (Abcam, ab5176, diluted 1:200) were incubated in 5% goat serum in PBS-T for 1 to 2 hours at room temperature. Images were acquired with a Leica SP5X confocal microscope and pHH3 quantification was done using ImageJ to determine number of pHH3 positive cells out of total number of cells (DAPI) on three fields of view per sample.
Breast cancer cell lines
Inflammatory breast cancer cell lines were generously provided by Steve Ethier (SUM149 and SUM190 cell lines, University of Michigan) and Massimo Christofanelli (FCIBC02, Fox Chase Cancer Center). SUM149 cells were cultured in DMEM/F12 supplemented with 5% FBS, 10 mmol/L HEPES pH 7.4, 1 μg/mL hydrocortisone, 5 μg/mL insulin. SUM190 cells were grown in 50% DMEM/F12, supplemented with 10% FBS, and 50% Human Mammary Epithelial Cell Growth Media (Cell Applications, 815–500). FCIBC02 cells were grown in DMEM/F12 with 10% FBS. All cell lines were supplemented with 50 U/mL penicillin and 50 μg/mL streptomycin. The identities of the cell lines were confirmed by STR analysis, were regularly tested for Mycoplasma and were used within 20 passages. Paclitaxel- and doxorubicin-resistant derivatives were generated by treating 50% confluent cell lines with increasing concentrations of each drug starting at their IC90, during 24 hours for SUM149 and 48 hours for FCIBC02, followed by a holiday period until they were passaged and retreated.
Cellular viability, proliferation, and synergy assays
Cellular viability assays were performed in triplicate wells and repeated two to three times. Cells were plated in 96-well plates in 100 μL culture media (2,000 cells/well for SUM149, 4,000 cells/well for FCIBC02 cells, 5,000 cells/well for SUM190 cells). Treatments were started after an overnight incubation. Paclitaxel (Sigma T7191), Ruxolitinib (MedChemExpress, HY50858), Solcitinib (Selleckchem, S5917), Abrocitinib (MedChemExpress, HY-107429), AZD1480 (Selleckchem, S2162), Fedratinib (Selleckchem, S2736), C188–9 (Selleckchem, S8605), and SH-4–54 (Selleckchem, S7337) were dissolved in DMSO and doxorubicin (Sigma D1515) in water. Further dilutions were prepared in their respective solvents and then added to a 2× concentration of the final treatment into culture media. A 100 μL aliquot of these 2× dilutions were added to each well. Final solvent concentration was 0.1%. Cells were cultured at 37°C with 5% CO2. Seventy-two hours following treatment, cells were fixed by adding 100 μL of an ice-cold 3:1 mixture of methanol and glacial acetic acid, overnight at 4°C. Fixed cells were washed twice with 1× PBS and stained for 20 minutes with 1 μg/mL DAPI (Thermo Fisher Scientific, 62248). Cells were washed twice with 1× PBS and 100 μL PBS added to each well before cell counting. Numbers of DAPI stained cells were acquired with an automated Celigo microwell plate-based imaging cytometer. For shRNA dose curves and proliferation assays cells were plated in 96-well plates in 100 μL of culture media (1,000 cells/well). Treatments and doxycycline were started after an overnight incubation as above. Plates were fixed at day 3, 5, and 7 following treatment start and analyzed as above. For synergy studies, cells were seeded in 96-well plates as above. After an overnight incubation, 1,000× concentrations of a dilution series of paclitaxel, doxorubicin, ruxolitinib, or 3i (SelleckChem, S8846) were made and 0.1 μL of drugs were pin-transferred using the JANUS Automated Workstation. Five to seven concentrations for each drug were chosen, with three replicate wells per concentration. After a 72-hour incubation, plates were processed as above. The expected drug combination responses were calculated on the basis of ZIP reference model using SynergyFinder (19). Deviations between observed and expected responses with positive and negative values denote synergy and antagonism respectively. Cells were plated in technical triplicates and representative plots are shown for experiments that were repeated one to three times.
Lentiviral shRNA constructs and experiments
SMARTvector inducible shRNA lentiviral vectors were used to infect cells and the resulting cell lines were selected according to the protocol (Horizon). shRNAs used in this study: shCTRL: nontargeting Control 1 (VSC11653), shCREB#1: TATCTCACAACTCTTCACC (V3SH11252–224924211), shCREB#2: TTCTTCATTAGACGGACCT (V3SH11252–226763730), shPDE4A#1: TCGAGCACCGACTCATCGT (V3SH11252–225123696), shPDE4A#2: TTTCAACCCTGTGATTTGG (V3SH11252–229812765). Cells were the treated with 1 μg/mL doxycycline to induce shRNA expression.
qPCR
RNA was isolated with RNeasy Mini Kit (Qiagen) according to manufacturer's protocol. cDNA synthesis was performed with PrimeScript RT Master Mix (Takara) using 500 ng RNA per 10 μL reaction at 65°C for 15 minutes. For qPCR, 2 μL of this reaction was used with TB Green Premix Ex Taq II (Takara) master mix and 0.4 μmol/L primer in a 25 μL reaction. qPCR was performed with a CFX96 (Bio-Rad; denaturation at 95°C for 30 seconds, elongation at 95°C for 5 seconds, 60°C for 30 seconds with 40 cycles). Relative gene expression was calculated using 2−ΔΔCt method normalized to GAPDH expression. Every biological triplicate was run in two technical duplicates. Primers: PDE4A Fwd: GATGCCATGGACACCAGCGA, PDE4A Rev: ATTCTCTGCCTCGAAGCGCC, GAPDH Fwd: AGCCACATCGCTCAGACAC, GAPDH Rev: GCCCAATACGACCAAATCC.
Chromatin immunoprecipitation sequencing
For pSTAT3 chromatin immunoprecipitation sequencing (ChIP-seq), cells were seeded in several 150 mm plates, 2.5 × 106 cells/plate for SUM149 and 2 × 106 cells/plate for FCIBC02, incubated for 24 and 48 hours respectively and then treatments added in 1 mL of non-FBS containing DMEM/F12. Plates were then double fixed in 1.5 mmol/L ethylene glycol bis(succinimidyl succinate) (EGS)/1% formaldehyde. Briefly, a 15 mmol/L EGS stock solution was freshly made in DMSO and a fixing solution prepared diluting 1:10 in room temperature PBS. Culture media was exhaustively removed from the plates and cell monolayers were fixed by adding 10 mL fixing solution to each plate and incubating for 30 minutes at room temperature in an orbital shaker. EGS was added to plates for 20 minutes, and then formaldehyde (32%, Electron Microscopy Sciences, catalog no. 15714) was added to the EGS at a final concentration of 1% for the last 10 minutes. Cross-linking was quenched by adding glycine to a final concentration of 0.125 M and further rocking the plates for 5 minutes. Cell monolayers were washed twice with ice-cold PBS and scrapped off the plate in 10 mL PBS, spun at 200 × g for 5 minutes, and flash frozen after removing the supernatant. Frozen pellets were resuspended in 1 mL of Cell Lysis Buffer (50 mmol/L HEPES pH 8, 140 mmol/L NaCl, 1 mmol/L EDTA pH 8, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100 and PPI) and incubated for 10 minutes at 4°C with gentle rotation. Nuclei were then centrifuged at 1,700 × g for 5 minutes at 4°C. Pelleted nuclei were washed twice with 1 mL Wash Buffer (10 mmol/L Tris-HCl pH 8, 200 mmol/L NaCl, 1 mmol/L EDTA pH 8, 0.5 mmol/L EGTA pH8 and PPI) and then resuspended in 1 mL of Shearing Buffer (10 mmol/L Tris-HCl pH 7.4, 1 mmol/L EDTA pH 8, 0.1% SDS, 1% Triton-X100, 0.1% sodium deoxycholate, 0.25% N-Lauroylsarcosine, 1 mmol/L DTT and PPI) and sonicated in a Covaris E220 focused-ultrasonicator (peak intensity: 140, 5% duty cycle, 200 cycles per burst, 10 minutes) using 1 mL AFA fiber tubes. An equivalent of 5 × 106 cells was sonicated in each tube and used for each ChIP. Lysates were centrifuged for 15 minutes at 10,000 × g in a refrigerated centrifuge. Cleared supernatants were transferred to new tubes and NaCl was added to a final concentration of 150 mmol/L. Samples were then incubated with Dynabeads Protein G (Life Technologies, 10003D, 40 μL/mL of lysate) for 1 hour at 4°C, with constant rotation (preclearing). After magnetic separation of the beads, the appropriate primary antibody was added to each tube for an overnight immunoprecipitation, at 4°C, with gentle rotation. Antibody-bound cross-linked chromatin was precipitated with prewashed Dynabeads Protein G for 2 hours at 4°C and with constant rotation. Beads were then washed sequentially with low salt wash buffer (20 mmol/L Tris-HCl pH 8, 150 mmol/L NaCl, 10 mmol/L EDTA, and 1% SDS), high salt wash buffer (50 mmol/L Tris-HCl pH 8, 10 mmol/L EDTA, and 1% SDS), LiCl wash buffer (50 mmol/L Tris-HCl pH 8, 10 mmol/L EDTA, and 1% SDS) and twice with 1× TE buffer. Each wash was performed for 5 minutes at 4°C with gentle rotation. DNA was eluted from the beads with 300 μL of 100 mmol/L sodium bicarbonate and 1% SDS for 30 minutes at room temperature with constant shaking. Cross-linking was reversed overnight at 65°C and RNA and protein were then sequentially digested by adding 0.2 mg/mL Rnase A for 30 minutes at 37°C, followed by 0.2 mg/mL Proteinase K for 1 hour at 55°C. DNA was extracted with 300 μL of phenol/chloroform/isoamyl-alcohol (Calbiochem, #516726) and centrifuged at 20,000 × g for 5 minutes at room temperature. DNA in the upper layer was then precipitated with 1 volume isopropanol, 1/3 volume of 2 M sodium perchlorate, and 5 μL glycogen and centrifuged at 20,000 × g for 10 minutes at room temperature. Pelleted DNA was washed twice with 70% ethanol and resuspended in 20 μL low TE. ChIP-seq libraries were prepared using the ThruPLEX DNA-seq Kit (Rubicon Genomics, R400427) according to the manufacturer's protocol.
RNA sequencing
For RNA sequencing (RNA-seq), cells were seeded in 100 mm plates, 1 × 106 cells/plate for SUM149 and 7.5 × 105 cells/plate for FCIBC02, incubated for 24 and 48 hours respectively and then treatments added in 0.5 mL of non-FBS containing DMEM/F12. Cells were incubated for further 72 hours and total RNA extracted using the Rneasy Mini Kit (Qiagen, #74106) with on-column DNA digestion. RNA-seq libraries were prepared using Illumina TruSeq Stranded mRNA sample preparation kits from 500 ng of purified total RNA according to the manufacturer's protocol. The finished dsDNA libraries were quantified by Qubit fluorimeter, Agilent TapeStation 2200, and RT-qPCR using the Kapa Biosystems Library Quantification Kit according to manufacturer's protocols. Uniquely indexed libraries were pooled in equimolar ratios and sequenced on an Illumina NextSeq500 with single-end 75 bp reads by the Dana-Farber Cancer Institute Molecular Biology Core Facility.
Single cell RNA-seq
For FCIBC02, parental cells were processed according to 10xGenomics v2 chemistry protocol. Briefly, cells were resuspended to a concentration of 1,000 cells/μL and 2,000 cells were targeted for recovery. For SUM149PR and FCIBC02PR cell lines, cells were prepared according to 10xGenomics v3 chemistry, and cells were resuspended at a concentration of 1,000 cells/μL and 5,000 cells were targeted for recovery and submitted to the Translational Immunogenomics Lab core for library preparation.
Xenografts assays
All animals were housed and maintained in Dana-Farber Cancer Institute LWC Assessment and Accreditation of Laboratory Animal Care-approved facility. All animal studies were conducted in accordance with the regulations formulated by the Dana-Farber Cancer Institute Animal Care and Use Committee (IACUC; protocol #11–023). Exponentially growing SUM149 or SUM190 cells (1 × 106) were resuspended in DMEM/F12 without FBS and 50% Matrigel (BD Biosciences) and injected orthotopically into the mammary fat pads of 6-week-old female NOD.Cg-PrkdcscidIl2rgtm1Sug/JicTac mice (Taconic). Treatments were started when tumors became palpable. Paclitaxel was delivered via intraperitoneal injection twice weekly at 10 mg/mL. Ruxolitinib was prepared for a dose of 60 mg/kg/day and delivered by subcutaneous osmotic pumps (ALZET, #1004). Briefly, ruloxitinib was first weighed in a 2 mL glass milliTube (Covaris, #520132) and solubilized in a volume of N,N-dymethylacetamide (DMAC, Sigma D5511) equivalent to 40% of the volume required to fill the pumps. The mixture was sonicated in a Covaris E220 focused-ultrasonicator (peak intensity: 200, 20% duty cycle, 200 cycles per burst, 60 minutes). Then a volume of propylene glycol (PG, Sigma P4347) equivalent to 60% of the pump-filling volume was added. Mixture was vortexed and used to fill the osmotic pumps. Two pumps per mouse were subcutaneously inserted, placing them dorsally on both sides away from the midline. Animals were euthanized and tumors were harvested when tumors in the control group reached ∼1.5 cm size. For histologic analyses, 5 μm sections of formalin-fixed paraffin-embedded (FFPE) tissue slides were stained with hematoxylin and eosin using standard protocols.
Immunoblotting
SUM149 cells were seeded in 100 mm plates at 5 × 105 cells/plate and FCIBC02 at 1 × 106 cells/plate in their respective complete media. After a 24-hour incubation, drugs were added at each cell lines respective IC50 value (paclitaxel (SUM149: 2.5 nmol/L, SUM149PR: 7.5 nmol/L, FCIBC02: 10 nmol/L, FCIBC02PR: 200 nmol/L) doxorubicin (SUM149: 10 nmol/L, SUM149DR: 200 nmol/L, FCIBC02: 100 nmol/L, FCIBC02DR: 1 μmol/L), ruxolitinib (SUM149s: 10 μmol/L, FCIBC02s: 15 μmol/L) or the combination were added in 1 mL media without FBS, to avoid pSTATs phosphorylation triggered by FBS. Cells were incubated for 72 hours and washed with ice-cold PBS. Depending on confluency, 200 to 400 μL RIPA buffer (50 mmol/L Tris-HCl pH 7.5, 150 mmol/L NaCl, 5 mmol/L EDTA pH8, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS and PPI) was added to the plates and cells were scraped and lysates were collected and incubated on ice for 30 minutes, and vortexed every 10 minutes. Samples were then sonicated in a cup horn sonicator (Qsonica Q500, 5 minutes net sonication time, 20 seconds On/10 seconds Off cycle with a 75% amplitude at 4°C). Sonicated lysates were centrifuged at 20,000 × g for 15 minutes at 4°C and supernatants transferred to new tubes. Protein concentration was determined by the Bradford assay. Samples (40 μg total protein per well) were resolved in 3% to 8% Tris-Acetate NuPAGE gels and transferred to PVDF membranes using a wet NuPAGE Transfer buffer system. Membranes were blocked with 5% nonfat dry milk in 0.1% Tween20 TBS pH7.4 (TBS-T) for 1 hour at room temperature followed by an overnight incubation with primary antibodies (in 5% BSA TBS-T). Membranes were washed and incubated for 1 hour at room temperature with appropriate secondary antibodies, then washed and developed with Clarity Western ECL substrate (Bio-Rad) and imaged using ChemiDoc MP imaging system (Bio-Rad).
Mass cytometry (CyTOF)
Antibodies used for CyTOF were purchased in carrier-free solutions and conjugated with the respective lanthanide metals by the CyTOF Antibody Resource and Core at Brigham and Women's Hospital. SUM149 and SUM149PR cells were continuously cultured in 10 cm dishes and split normally in each cell lines respective IC50 in either DMSO (0.1%), paclitaxel (SUM149: 2.5 nmol/L, SUM149PR: 7.5 nmol/L), ruxolitinib (10 μmol/L), or the combination for 14 days. Cells were refed every 3 days with fresh drug if they did not need to be split. At day 12, cells were plated in 10 cm at 2E6, and at day 14, cells were washed with PBS, then 5 mmol/L EDTA was added to plates for 15 minutes to detach cells without cleaving cell surface proteins. A total of 1 × 106 cells were then viably frozen in 90% FBS and 10% DMSO. Cells were then thawed and treated with 100 μmol/L intercalator-103Rh (Fluidigm) for 15 minutes at 37°C in their respective full medium without drug. A total of 1 × 106 cells per sample were barcoded using the Cell-ID 20-Plex Pd Barcoding Kit (Fluidigm) according to the manufacturer's instructions. Barcoded samples were pooled and stained simultaneously. Cells were fixed for 10 minutes with 1.6% paraformaldehyde (Electron Microscopy Sciences) and then incubated with Fc-receptor block (Human TruStain FcX, Biolegend) for 10 minutes, and then stained with surface antibody staining for 30 minutes at room temperature. Next, cells were permeabilized with methanol for 10 minutes on ice and stained with the antibody cocktail for intracellular epitopes for 30 minutes. Cells were kept at 4°C overnight in Fix and Perm Buffer (Fluidigm) with 62.5 nmol/L of Intercalator-IR (Fluidigm; ref. 20). Before analysis, cells were washed in CAS (Fluidigm), and then resuspended in CAS containing EQ Four Element Calibration Beads (Fluidigm; 1:10) and filtered through a 35 μm strainer. Samples were acquired using a CyTOF Helios instrument (Fluidigm), normalized as previously described (20), and analyzed with Cytobank. Cell Staining Media (PBS with 0.5% BSA, 0.02% NaN3) was used for all washes and antibody incubations. For analysis, the population was gated for intact cells, singlets, viable (defined as Rh-103 negative) and non-apoptotic (cleaved PARP negative). viSNE analysis was performed using the c-PARP negative population with the following markers for clustering: CDK4, HER2, CD24, CDK1, Vimentin, pS6, PTEN, CK5, p21, PR, CD10, CD44, CyclinD3, MUC1, E-Cadherin, CDK6, p63, TCF7, AR, GATA3, pAKT, pSTAT3, pSTAT5, EGFR, ER, HER3, Lamp2, EpCAM, Ki-67, SMA, pSMAD2–3, CD49f, and CK8–18.
Flow cytometry
For analysis of EpCAM levels in increasing doses of paclitaxel, SUM149PR cells were maintained in DMSO, 3 nmol/L, or 5 nmol/L of paclitaxel for 1 month before sorting. For EpCAM sorting and analysis, cells were collected and stained for EpCAM (Ber-EP4-FITC) at 1:50 dilution for 30 minutes on ice in the dark. For analysis, fluorescence intensities were acquired on an LSRFortessa cytometer (BD Biosciences) and analyzed using FlowJo. For sorting of EpCAM− and EpCAM+ populations, cells were sorted using BD FACSAria II SORP UV.
Antibodies
Western: STAT3 (Cell Signaling Technology, 9139, AB_331757, 1:1,000), pSTAT3 Tyr705 (Cell Signaling Technology, 9145, AB_2491009, 1:1,000); β-Actin (Merck, A2228, AB_476697, 1:10,000), E-cadherin (BD Biosciences, 610181, AB_397580, 1:2,000); pJAK2 Tyr1007/1008 (Cell Signaling Technology, 3776, AB_2617123 1:1000), JAK2 (Cell Signaling Technology, 3230, AB_2128522, 1:1,000), pJAK1 Tyr1034/1035 (Cell Signaling Technology, 74129, AB_2799851, 1:1,000), JAK1 (Cell Signaling Technology, 3332, AB_2128499, 1:1,000), pSTAT1 Tyr701 (Cell Signaling Technology, 9167, AB_561284, 1:1,000), STAT1 (Cell Signaling Technology, 14994, AB_2737027, 1:1,000), pSTAT3 Ser727 (Cell Signaling Technology, 9134, AB_331589, 1:1,000), pERK1/2 (Cell Signaling Technology, 9101, AB_331646, 1:1,000), ERK1/2 (Cell Signaling Technology, 9102, AB_330744, 1:1,000), CREB (Cell Signaling Technology, 9197, AB_331277, 1:1,000), Vimentin (Dako, GA630, AB_2827759, 1:1,000), HRP-conjugated antiMouse IgG (H+L; Thermo Fisher Scientific, 32430, AB_1185566), HRP-conjugated anti-Rabbit IgG (H+L; Thermo Fisher Scientific, 32460, AB_1185567). Immunofluoresence: CD44 (Neomarkers, clone 156–3C11, mouse monoclonal IgG2), CD24 (Neomarkers, clone SN3b, mouse monoclonal IgM), pSTAT3 Tyr705 (Cell Signaling Technology, 9145, AB_2491009,1:1,000), Cl-Casp3 (Cell Signaling Technology, 9661, AB_2341188, 1:100), pHH3 (Abcam, ab5176, AB_304763, 1:200). ChIP: pSTAT3 Tyr705, Cell Signaling Technology, 9131, AB_331586 (20uL/ChIP); CyTOF: Rabbit monoclonal anti-PR a/b (141Pr; Cell Signaling Technology, 8757, AB_2797144), mouse monoclonal anti-CD10 (142Nd; BD Biosciences, 555373, AB_395775), Rat monoclonal anti-CD44 (143Nd), (Biolegend, 103002, AB_312953), mouse monoclonal anti-cyclin D3 (144Nd), (Abcam, ab28283, AB_2070798), Mouse monoclonal anti-MUC1 (145Nd; Biolegend, 355602, AB_2561642), mouse monoclonal anti-LAMP2 (146Nd; Biolegend, 354302, AB_11204245), mouse monoclonal anti-CDK4 (147Sm; BD Biosciences, 559677, AB_397299), rabbit monoclonal anti-PTEN (148Nd; Cell Signaling Technology, 9559, AB_390810), rabbit monoclonal anti-E-Cadherin (149Sm; Cell Signaling Technology, 3195, AB_2291471), mouse monoclonal anti-EpCAM (150Nd; Biolegend, 324202, AB_756076), mouse monoclonal anti-Her2 (151Eu; BD Biosciences, 554299, AB_395352), rabbit polyclonal anti-CK5 (152Sm; Abcam, ab53121, AB_869889), mouse monoclonal anti-CD24 (153Eu; Biolegend, 311102, AB_314851), mouse monoclonal anti-CDK1 (154Sm; Biolegend, 626901, AB_2074779), rabbit monoclonal anti-CDK6 (155Gd; Cell Signaling Technology, 13331, AB_2721897), rabbit monoclonal anti-p63 (158Gd; Abcam, ab124762, AB_10971840), rabbit monoclonal anti-TCF7 (159Tb; Cell Signaling Technology, 2203, AB_2199302), rabbit monoclonal anti-AR (160Gd; Cell Signaling Technology, 5153, AB_10691711), goat polyclonal anti-HER3 (161Dy; R&D Systems, AF234, AB_355227), mouse monoclonal anti-Ki-67 (162Dy; BD Biosciences, 550609, AB_393778), mouse monoclonal anti-SMA (163Dy; Thermo Fisher Scientific, 14–9760–82, AB_2572996), mouse monoclonal anti-cPARP (164Dy; BD Biosciences, 552596, AB_394437), rabbit monoclonal anti-Vimentin (165Ho; Cell Signaling Technology, 5741, AB_10695459), rat monoclonal anti-GATA3 (166Er; eBioscience, 14–9966–80, AB_1210520), rabbit monoclonal anti-p21 (167Er; Cell Signaling Technology, 2947, AB_823586), rabbit monoclonal anti-phospho-AKT Ser473 (168Er; Cell Signaling Technology, 4060, AB_2315049), rabbit monoclonal anti-phospho-STAT3 Tyr705 (169Tm; Cell Signaling Technology, 9145, AB_2491009), rabbit monoclonal anti-EGFR (170Er; Cell Signaling Technology, 4267, AB_2246311), rabbit monoclonal anti-phospho-SMAD2 Ser465/467/SMAD3 Ser423/425 (171Yb; Cell Signaling Technology, 8828, AB_2631089), rabbit monoclonal anti-ERα (172Yb; Cell Signaling Technology, 13258, AB_2632959), rat monoclonal anti-CD49f (173Yb; Biolegend, 313602, AB_345296), rabbit monoclonal anti-phospho-STAT5 Tyr694 (174Yb; Cell Signaling Technology, 4322, AB_10548756), rabbit monoclonal anti-phospho-S6 Ser235/236 (175Lu; Cell Signaling Technology, 4858, AB_916156), mouse monoclonal anti-CK8/18 (176Yb; Cell Signaling Technology, 4546, AB_2134843).
ChIP-seq and RNA-seq analyses
Peak calling was performed using ChiLin pipeline 2.0.0 (21). Peak annotation was performed using annotatePeaks.pl of the HOMER package v4.9.1 with version hg19 of the human genome. Motif analysis was performed by ChiLin pipeline using the top 1,000 peak regions. RNA-seq datasets were aligned to the human reference GRCh37/hg19 genome using the STAR RNA-seq aligner (version STAR_2.5.1b; ref. 22). Two-pass mapping was performed using the following modified parameters: –outSAMstrandField intronMotif, –outFilterMultimapNmax 20, –alignSJoverhangMin 8, –alignSJDBoverhangMin 1, –outFilterMismatchNmax 999, –outFilterMismatchNoverLmax 0.1, –alignIntronMin 20, –alignIntronMax 1000000, – alignMatesGapMax 1000000, –outFilterType BySJout, –outFilterScoreMinOverLread 0.33, –outFilterMatchNminOverLread 0.33, –limitSjdbInsertNsj 1200000, –chimSegmentMin 15, –chimJunctionOverhangMin 15, –twopassMode Basic. The read counts for individual genes were generated using the htseq-count script of the HTSeq framework (version 0.6.1p1; ref. 23) using modified parameters (–stranded no) and the hg19 refGene annotation file available at the UCSC Genome Browser. Genes with 0 counts across all samples were filtered out. The gene-level counts from all studies were then normalized using TMM with edgeR (24). Log2 transformed TMM-normalized counts per million [log2(TMM-CPM + 1)] were used for analysis. Principle component analysis were conducted using prcomp function. Differentially expressing genes were identified by using DESeq2 (25) with cutoff of padj<0.05 and |FC|>2 (DR models) or |FC|>1.5 (PR models). Same threshold was applied to call DEGs between parental versus resistant derivatives or vehicle versus drug treatments for each derivative. De novo genes were defined as DEGs identified between parental versus resistant derivatives but not between the corresponding vehicle versus drug treatment comparison. The intersected counterparts were defined as adaptive genes. For gene set enrichment calculation, GSVA package (26) was used with predefined signatures provided in the Supplementary Table S1.
ChIP-seq and RNA-seq data integration: For a given factor peak, the distance to the nearest TSS was found and the target was defined as that gene. Differentially expressed targets of a factor were then defined as differentially expressed genes that were also associated to a factor peak. BETA analysis was performed as described previously (27). BETA basic modules were used to compute the statistical associations between DE genes and DB peaks using 100kb as the ranges to link gene TSS to each peak. P values were derived using one-tailed Kolmogorov–Smirnov test for upregulated and downregulated genes, respectively. For pathway map and process network analysis, differentially expressed gene lists were uploaded into Metacore. Genes were filtered for Padj < 0.01 and Log2FC > 1 for up genes, and < −1 for down genes. Only process networks that were FDR <0.1 were shown.
Single cell RNA-seq analyses
Data for SUM149, sequenced with 10× genomics v2 chemistry was obtained from ref. 28. We analyzed this cell line together with three additional cell lines sequenced across two batches with different chemistries (FCIBC02: batch 1, v2 chemistry, SUM149PR and FCIBC02PR batch 2, v3 chemistry). We also analyzed eight additional samples from four cell lines [CAL51, HCC1806, HCC1937, and Hs578 (replicate cell lines)] sequenced in both batch 1 and batch 2, which were used to correct batch effects and enable comparisons across samples (see below). The 12 single cell RNA-seq (scRNA-seq) samples were preprocessed using Cell Ranger (https://www.10xgenomics.com/) to obtain UMI counts for each gene in each cell. Cell filtering proceeded in two steps. First, for every cell in each sample, we calculated the logarithm of the proportion of UMI's from mitochondrial genes (mito_score). The median and median absolute deviation from the median (MAD) was calculated for the mito_score among cells with fewer than 50% of UMI's from mitochondrial genes, and cells with mito_scores greater than four MAD's above the median in each sample were excluded from downstream analysis. Second, from the remaining cells in each sample we excluded cells where the logarithm of the number of genes expressed was less than four MAD's below the median. Normalization and clustering for individual IBC cell lines was performed using the Seurat (29) standard log-normalization workflow based on genes detected in 10 or more cells. The top 10 principal components were used for SUM149, and the top 20 were used for FCIBC02, SUM149PR, and FCIBC02PR. The resolution parameter was set to 0.5 to run clustering with the Seurat function FindClusters. STAT3 signature activity was inferred using the Seurat function AddModuleScore. We used two approaches to compare expression between SUM149 and SUM149PR or between FCIBC02 and FCIBC02PR. Because both parental cell lines were sequenced using v2 chemistry, and both resistant cell lines were sequenced using v3 chemistry, and in different batches, we took steps to try to remove potential artefacts arising due to differences in chemistry, and (where possible) differences due to batch as described further below. scRNA-seq Cell Type Signature Analysis: Each pair of resistant and parental cell lines was normalized and analyzed separately. For each pair, we removed genes expressed in fewer than 10 cells across the resistant and parental cells. Resistant and parental cells were log-normalized in Seurat together with an equal number of cells from each replicate cell line from each of batches 1 and 2 (v2 and v3 chemistry, respectively). We used the function rescaleBatches from the R package “batchelor” (30), with the restrict parameter set, to estimate a scaling difference between batches for each gene based on the replicate samples only, and to correct the log-normalized data for the IBC samples accordingly. No correction was applied for genes with zero UMI counts in cells from the replicate cell lines in either batch. Single-cells were assigned to treatment types (parental, paclitaxel-treated, or resistant) based on the expression of gene signatures for each type. The positive and negative signature genes for each type were defined as the top n positive and negative differentially expressed genes for that type versus the other two types that were also present in the scRNA-seq data. Here, n was taken to be as large as possible given the number of differential genes in each direction that were present in the scRNA-seq data. The TNBC type signatures were defined in a similar way based on bulk RNA-seq data from 34 TNBC cell lines (Jovanović et al., in press). We assessed the evidence for each signature in each single cell based on a previously described method (28). Briefly, we considered there to be significant evidence for a signature in a cell when the average expression (gene centered or gene z-scored log-normalized data) for “up” genes minus the average expression for “down” genes exceeded the same statistic in 95% of randomly chosen signatures of the same size. Seurat Integration Analysis: We also applied an orthogonal approach to integrate single-cell data from resistant and parental cell lines pairs using Seurat's data integration functionality. As above we considered each resistant/parental pair separately, and only considered genes expressed in at least 10 cells in either cell line. For this analysis, we applied the Seurat integration standard workflow for log-normalized data, using a dataset dimensionality of 20. Integrated data for each pair of cell lines was clustered using FindClusters with the resolution parameter set as 0.5.
Mass spectrometry analysis of histone modifications
Cells were harvested while growing exponentially, pelleted, washed, and snap frozen. Global Chromatin profiling was performed as described in Creech and colleagues (31). Briefly, histones were isolated from cell nuclei using acid extraction and precipitated with trichloroacetic acid. Histones (10 μg each sample) were propionylated, desalted, and digested overnight with trypsin, following standard protocols. Histone peptides underwent a second round of propionylation, followed by desalting. Prior to MS analysis, a reference mixture of isotopically labeled synthetic peptides for histones H3 and H4 was added to each sample. Peptides were separated on a C18 column (EASY-nLC 1000, Thermo Fisher Scientific) and analyzed by MS using a PRM method (Q Exactive Plus Orbitrap, Thermo Fisher Scientific). Chromatographic peak areas of endogenous (light, L) and synthetic standard (heavy, H) peptides were extracted in Skyline and the ratios of light to heavy peak areas (L:H) were calculated. Ratios were log2 transformed, normalized to a typically unmodified region of H3 (41–49) or H4 (68–78) respectively, and row median normalized for each histone mark. Heat map was made using https://software.broadinstitute.org/morpheus.
Clinical cohort analyses
Microarray data from four patient breast cancer cohorts were attained from GEO: the ISPY clinical trial for neoadjuvant chemotherapy (GSE32603; ref. 32; only patients with TNBC were selected for downstream analysis), Boersma cohort of laser capture micro-dissected stromal and epithelial cells of patients with IBC and nIBC (GSE5847; ref. 33), Woodward cohort of 20 IBC (GSE45584; ref. 7), Iwamoto cohort of fine needle aspirates of T4 IBC (GSE22597; ref. 34). Data were converted to log2 values and quantile normalized. Differential expression analysis on a probe level was performed using limma (35). Gene-set enrichment analysis (36) was conducted on log2-fold changes were used in using HTSAnalyzeR (37), and CAMP-related signaling pathways with FDR <0.1 were visualized in a heat map. Gene-signature scores for STAT (Supplementary Table S1) and PDE (PDE7B, PDE2A, PDE3A, PDE3B, PDE4A, PDE4B, PDE4C, PDE4D, PDE7A, PDE8A, PDE1B, PDE8B) family members were determined by averaging the expression of all probes that mapped to these genes. Differences in groups was tested by Wilcoxon rank sum test.
Exome sequencing
Samples were submitted to the DFCI Center for Cancer Genome Discovery (CCGD) for exome sequencing and analysis. Briefly, DNA yields were determined using PicoGreen. DNA was fragmented by Covaris sonication to 250bp and purified further with Agentcourt AMPure XP beads. Libraries were constructed using KAPA HTP Library Construction Kit. Libraries were pooled and sequenced on Illumina MiSeq to estimate the concentration based on the number of barcode reads per sample. Libraries were then pooled in equal mass (3 × 3-plex) and hybrid capture was performed using SureSelect Human All Exon v5 RNA baits and the Agilent SureSelect system. The captures were then pooled and sequenced over three lanes of the HiSeq2500 in Rapid Run mode. Pooled sample reads were deconvoluted and sorted using Picard tools. Reads were aligned to reference sequence b37 edition from the Human Genome Reference Consortium using “bwa aln” (http://bio-bwa.sourceforge.net/bwa.shtml) and the following parameters “-q 5 -l 32 -k 2 -o 1″ and duplicate reads were identified and removed using Picard tools (38). The alignments were further refined using the GATK for localized realignment around indel sites (https://www.broadinstitute.org/gatk/gatkdocs/org_broadinstitute_gatk_tools_walkers_indels_IndelRealigner.php). Recalibration of the quality scores was also performed using the GATK (http://gatkforums.broadinstitute.org/discussion/44/base-quality-score-recalibration-bqsr; refs. 39, 40). Quality control for sequencing was performed by generating metrics for the representation of each sample within the pool using unaligned reads after sorting on the barcode (http://broadinstitute.github.io/picard/picard-metric-definitions.html for a description of the metrics). All samples passed these quality control measures. Mutation analysis for single-nucleotide variants (SNV) was performed using MuTect v1.1.4 and annotated by variant effect predictor (VEP; refs. 41, 42). Insertions and deletions (InDel) were called using Indel Locator (http://www.broadinstitute.org/cancer/cga/indelocator). SomaticIndelDetector tool, which is part of GATK, was used for indel calling. MuTect was run in paired mode using CEPH as the project normal. Copy-number variants were identified using RobustCNV, an algorithm in development at CCGD. RobustCNV relies on localized changes in the mapping depth of sequenced reads to identify changes in copy number at the loci sampled during targeted capture. This strategy includes a normalization step in which systemic bias in mapping depth is reduced or removed using robust regression to fit the observed tumor mapping depth against a panel of normal (PON) sampled with the same capture bait set. Observed values are then normalized against predicted values and expressed as log2 ratios. A second normalization step is then done to remove GC bias using a loess fit. Finally, log2 ratios are centered on segments determined to be diploid based on allele fraction of heterozygous SNPs in the targeted panel. Normalized coverage data was next segmented using Circular Binary Segmentation (43) with the DNAcopy Bioconductor package. Finally, segments are assigned gain, loss, or normal-copy calls using a cutoff derived from the within-segment SD of postnormalized mapping depths and a tuning parameter which was set based on comparisons to array-CGH calls in separate validation experiments. Segment calls were then summarized to gene calls by assigning capture intervals and tallying up interval calls for each gene. Genes may contain multiple intervals with a combination of calls; therefore, a variety of gene calls are possible.
Metabolomic profiling and ELISA
SUM149 and FCIBC02 cell lines were plated in duplicate in three biological replicates and polar metabolites were extracted as described (44). Briefly, SUM149 and FCIBC02 cell lines were plated in duplicate and grown to approximately 80% confluency. Media was thoroughly removed by aspiration and an 80% methanol solution kept at −80°C was added to cell monolayers working on dry ice. Cells were then scraped off and transferred to conical tubes. The lysate/methanol mixture was centrifuged at full speed for 5 minutes at 4°C, and supernatant kept on dry ice. Solvent was evaporated under a stream of nitrogen. Samples from three independent experiments were submitted for polar metabolite profiling to the Beth Israel Deaconess Medical Center Mass Spectrometry Core Facility as described (44). Nonhierarchical clustering was performed using a Euclidean distance measure and Ward's clustering algorithm using MetaboAnalyst. Pathway and Joint Pathway analysis were performed using MetaboAnalyst (45) and only pathways that were FDR < 0.05 were shown. For ELISA SUM149 and FCIBC02 cells were plated in 6 cm dishes (2.5 × 105 for SUM149s, 4 × 105 for FCIBC02), and treatments were started as above for immunoblotting following an overnight incubation. Following 72-hour incubation, cells were lysed and the ELISA was performed according to the manufacture instructions (Abcam, ab65355).
Study approval
All animal studies described were carried out under protocol 11–023 approved by the Dana-Farber Cancer Institute Animal Care and Use Committee.
Data availability
The RNA-seq and ChIP-seq datasets have been deposited to Gene Expression Ominbus (GEO) with the accession number GSE163397. The original mass spectra and the protein sequence database used for searches have been deposited in the public proteomics repository MassIVE (http://massive.ucsd.edu) and are accessible at ftp://massive.ucsd.edu/MSV000090647/.
Results
High frequency of CD44+CD24−pSTAT3+ cells in IBC
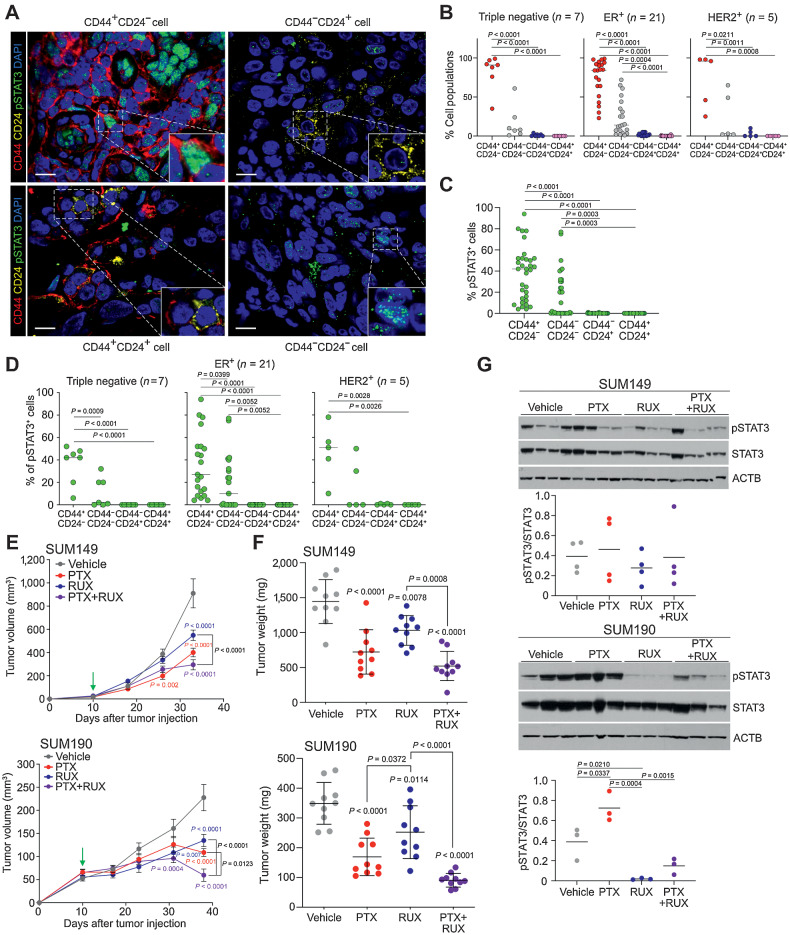
To investigate the frequency of CD44, CD24, and pSTAT3 in IBC, we performed multicolor immunofluorescence in IBC tumors from 33 patients (Supplementary Table S2) and quantified the frequency of pSTAT3+ cells in the four cell types (i.e., CD44+CD24−, CD44+CD24+, CD44−CD24+, CD44−CD24−; Fig. 1A). We found that CD44+CD24− was the most frequent cellular phenotype regardless of subtype, representing over 75% of cancer cells in most cases, whereas CD24+ cells were rarely detected (Fig. 1B). Within CD44+CD24− cells, a large fraction was pSTAT3+ and a subset of tumors had CD44−CD24−pSTAT3+ cells, which was not influenced by tumor subtype (Fig. 1C and D). We also compared pre- and post chemotherapy samples and observed a significant increase in the relative proportion of CD44+CD24− cells following treatment in the ER+ subtype, although patient numbers were low (Supplementary Fig. S1A). Although changes in the frequency of pSTAT3+ cells were not significant, pSTAT3 levels were more variable posttreatment, particularly in CD44+CD24− cells in ER+ tumors (Supplementary Figs. S1B–S1C). Therefore, in a subset of patients with IBC, treatment may select for stem-like CD44+CD24−pSTAT3+ cells increasing the risk of disease progression.
Figure 1.
Frequency of CD44+CD24−pSTAT3+ cells and dependency on JAK/STAT3 signaling in IBC. A, Representative immunofluorescence analysis of CD44, CD24, and pSTAT3 in IBC samples. Scale bars, 10 μm. Nuclei were stained with DAPI. B, Percent of the four cell types in each IBC tumor of the indicated subtype. C, Percent pSTAT3+ cells in the four cell types (n = 33 patients). D, Percent pSTAT3+ cells within the four cell types in each tumor classified by subtype. E, Graphs depicting SUM149 (top) and SUM190 (bottom) xenograft tumor volume in mice treated with vehicle, paclitaxel (PTX), ruxolitinib (RUX), or the combination, with green arrow depicting treatment start when tumors were first palpable (∼20 mm3 for SUM149 and ∼60 mm3 for SUM190). Error bars, SEM; n = 5 mice with 2 tumors/mouse. P values were calculated by two-way ANOVA. F, Graph of tumor weights at endpoint from experiment shown in E. Error bars, SD. G, Immunoblot analysis of pSTAT3 and STAT3 in xenografts. ACTB was used as a loading control. Dot plot depicts quantification of pSTAT3/STAT3 ratios. P values for B–D and F–G calculated by one-way ANOVA with Tukey multiple comparisons.
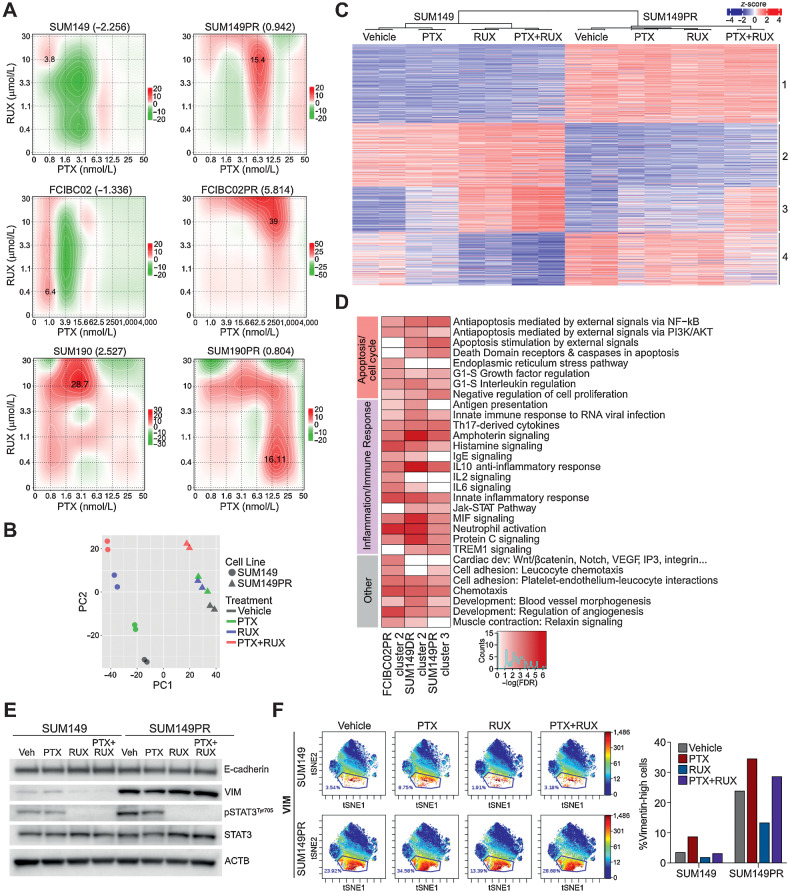
Dependency of IBCs on JAK2/STAT3 signaling
To explore whether the high fraction of pSTAT3+ cells reflects a dependency on JAK/STAT3 signaling, we analyzed the effects of ruxolitinib (RUX), a JAK1 and JAK2 inhibitor (46), on the growth of IBC cell lines in cell culture and xenograft assays. Because patients with IBC frequently receive neoadjuvant paclitaxel (PTX), we also tested paclitaxel alone and in combination with ruxolitinib. We found ruxolitinib alone decreased the viability and growth of both SUM149 and SUM190 cells in vitro, albeit at relatively high concentrations, and in immunodeficient mice (Fig. 1E and F; Supplementary Fig. S1D). Furthermore, in SUM190 xenografts, combination of ruxolitnib with paclitaxel significantly decreased tumor volume compared with either agent alone and SUM149 xenografts exhibited a similar trend (Fig. 1E and F; Supplementary Figs. S1E–S1F). Tumors treated with the combination had significantly higher phosphohistone H3+ (pHH3+) cells compared with vehicle and single agent treatments, suggesting mitotic arrest, whereas the frequency of cleaved caspase-3+ or CD44+ cells were not affected (Supplementary Figs. S1F–S1G).
To confirm inhibition of JAK/STAT signaling by ruxolitinib in vivo, we performed immunoblot analysis of pSTAT3 in the xenografts. We found ruxolitinib significantly decreased pSTAT3 levels in SUM190 xenografts, and while we saw a similar trend in SUM149, these tumors tended to be heterogeneous for pSTAT3 (Fig. 1G). Paclitaxel induced a slight increase in pSTAT3 in some tumors, which was inhibited in the combination. Activation of pSTAT3 by paclitaxel may reflect an increased dependency of treatment-resistant samples on JAK/STAT signaling, which is in line with 9p24/JAK2 amplification being more common in residual TNBC after neoadjuvant chemotherapy (47, 48).
Cell line models of chemotherapy-resistant IBC
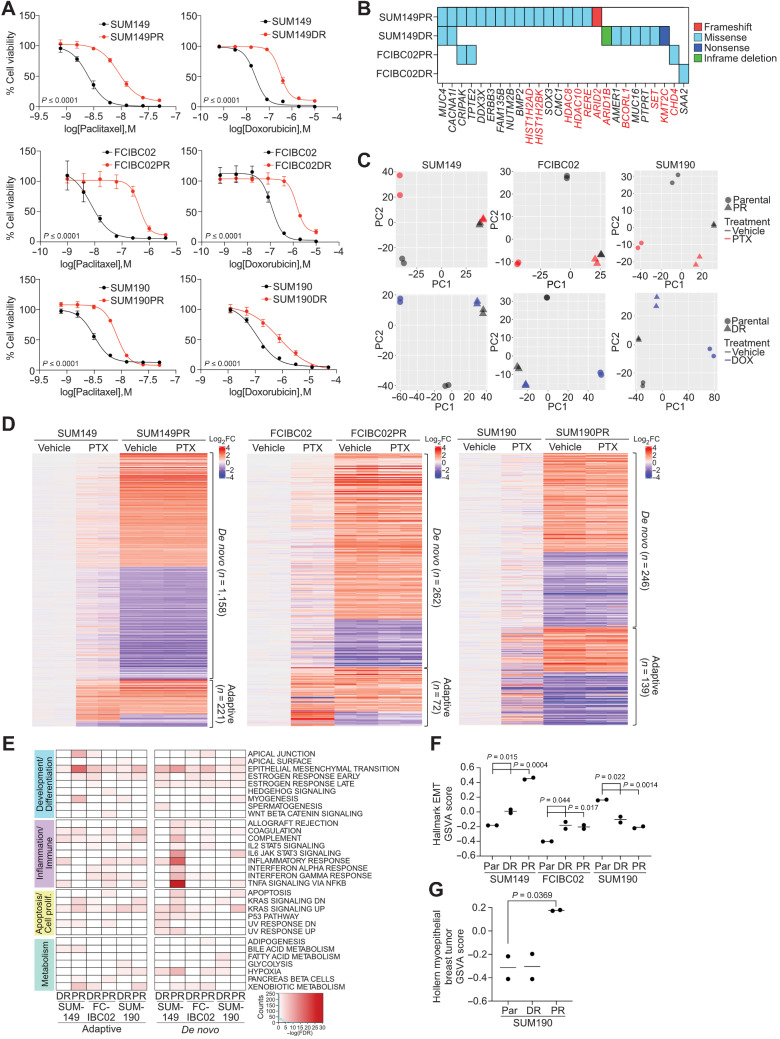
To explore mechanisms of chemotherapy resistance in IBC, we generated derivatives of IBC cell lines resistant to paclitaxel and doxorubicin, commonly used chemotherapeutic agents, by prolonged culture in the presence of drug. We used both triple-negative (SUM149 and FCIBC02) and HER2+ (SUM190) IBC cell lines to assess resistance mechanisms across subtypes. Compared with parental, paclitaxel (SUM149PR, FCIBC02PR, and SUM190PR) and doxorubicin (SUM149DR, FCIBC02DR, and SUM190DR) resistant cells displayed an increase in IC50 to the respective drug (Fig. 2A).
Figure 2.
Characterization of chemotherapy-resistant IBC cell lines. A, Cellular viability after paclitaxel or doxorubicin treatment of parental and resistant cells. Error bars, SD (n = 3). P values determined by comparison of curves using F test. B, Selected genes from Supplementary Fig. S2A, which were mutated in resistant derivatives and either overlapped with the COSMIC cancer database, were differentially expressed in pathway analysis, or were mutated in both paclitaxel-resistant cell lines. Highlighted genes in red correspond to chromatin modifiers. C, Principal component analysis of gene expression of parental and resistant cell lines treated with vehicle, paclitaxel (PTX), or doxorubicin (DOX). D, Heat map of significant differentially expressed genes between parental and resistant derivatives clustered by either their presence (adaptive) or absence (de novo) in parental paclitaxel treated cells compared with vehicle. E, Gene set enrichment analysis for Hallmark gene sets in the indicated resistant versus parental differential gene lists that were significantly enriched (FDR < 0.05). Color scale corresponds to −log(FDR q-value). F, Gene set variation analysis (GSVA) score showing relative enrichment of the Hallmark EMT gene set in the indicated cell lines. G, GSVA analysis as in F for enrichment of the Hollern Myoepithelial Breast Tumor gene signature in SUM190 cell lines. P values for F and G calculated by Student t test.
To determine if resistance was due to genetic alterations, we performed whole-exome sequencing on parental and resistant cell lines. We filtered for mutations that are only present in resistant derivatives and are most likely to impact cellular phenotypes (e.g., frameshift, missense, nonsense or in-frame deletions and insertions). Most mutations were unique to each cell line (Supplementary Fig. S2A; Supplementary Table S3) and some overlapped with the COSMIC database, implying clinical relevance (Fig. 2B). We also examined copy-number variations but found limited differences (Supplementary Fig. S2B). Functional analysis of mutant genes in resistant cells using Metacore (49) revealed genes were enriched in chromatin modification pathways in SUM149PR (Supplementary Fig. S2C; Supplementary Table S4), and mutations in these genes (Fig. 2B) highlight the importance of epigenetic changes in therapeutic resistance. Therefore, we analyzed histone modification patterns by mass spectrometry, but did not identify consistent resistance-associated differences (Supplementary Fig. S2D).
Next, we performed RNA-seq to identify treatment and resistance-associated gene expression changes that could reveal common targetable pathways. Differences were the largest between parental and resistant derivatives, except for SUM190DR where treatment of parental cells accounted for the most variation. In all other cell lines, treatment in parental cell lines was the source of the second highest variation, and in all cell lines, there were limited drug-induced differences in resistant derivatives (Fig. 2C; Supplementary Table S5). We focused on differentially expressed genes between parental and resistant derivatives, and classified them as either “adaptive,” which were differentially expressed following drug treatment in parental cells or “de novo,” which were not significantly affected by drug. Both gene sets were represented in all cell lines, but there were more de novo than adaptive genes. SUM149PR cells had the largest gene expression changes compared with parental cells, most of which were de novo genes (Fig. 2D; Supplementary Fig. S2E). To identify pathways associated with response and resistance to chemotherapy, we performed GSEA and identified EMT as a top enriched gene set across resistant derivatives in both adaptive and de novo gene lists with the most significant enrichment in SUM149PR cells. De novo genes were specifically associated with the “estrogen response late” gene set, whereas adaptive genes were more enriched for inflammation and immune-response related pathways, except for SUM149PR (Fig. 2E; Supplementary Table S4). Finally, the IL6/JAK/STAT3 gene set was enriched only in de novo genes in SUM149PR and SUM190PR cells (Fig. 2E).
Further analysis of the EMT pathway revealed a significant increase in the EMT signature in resistant TN-IBC and a decrease in HER2+ luminal SUM190s (Fig. 2F; Supplementary Table S1), which was validated by immunoblot for the mesenchymal marker vimentin (Supplementary Fig. S2F). We also found SUM190PR cells have an increase in the myoepithelial breast tumor signature (50), suggesting a shift from luminal to myoepithelial cell state (Fig. 2G; Supplementary Table S1). These data identified cell state switching as a potential mechanism of chemotherapy resistance in IBC.
Genomic targets of STAT3 in chemotherapy-sensitive and resistant IBC cells
Next, given the efficacy of ruxolitinib and paclitaxel combination in vivo (Fig. 1), we characterized STAT3 signaling in more detail. First, we found enrichment of the Hallmark JAK/STAT signature from GSEA in SUM149PR and SUM149DR RNA-seq data with an opposite trend in SUM190 (Supplementary Figs. S3A; Supplementary Table S1). Furthermore, we found paclitaxel-resistant and paclitaxel-treated SUM149 cells had higher pSTAT3Tyr705 levels compared with parental, whereas paclitaxel decreased pSTAT3Tyr705 in SUM149PR cells (Fig. 3A). FCIBC02 cells had high basal levels of pSTAT3Tyr705 that decreased after paclitaxel treatment, whereas doxorubicin-resistant lines had lower pSTAT3Tyr705 that did not change by doxorubicin treatment (Supplementary Fig. S3B). We also examined other components of the JAK/STAT pathway but found relatively few changes (Supplementary Figs. S3C–S3D). The variability in pSTAT3 depending on chemotherapeutic agent suggests a context-dependent role for JAK/STAT signaling in treatment response and resistance. To confirm the importance of JAK2/STAT3 in IBC cell survival, we tested specific inhibitors for JAK1, JAK2, and STAT3. JAK1 inhibitors had no effect on viability even at high concentrations, whereas JAK2 and STAT3 inhibitors decreased cell viability (Supplementary Figs. S3E–S3F). Resistant derivatives behaved similar to parental cells, except SUM149-resistant derivatives were more sensitive to the JAK2 inhibitor AZD1480 and more resistant to the STAT3 inhibitor SH-4–54.
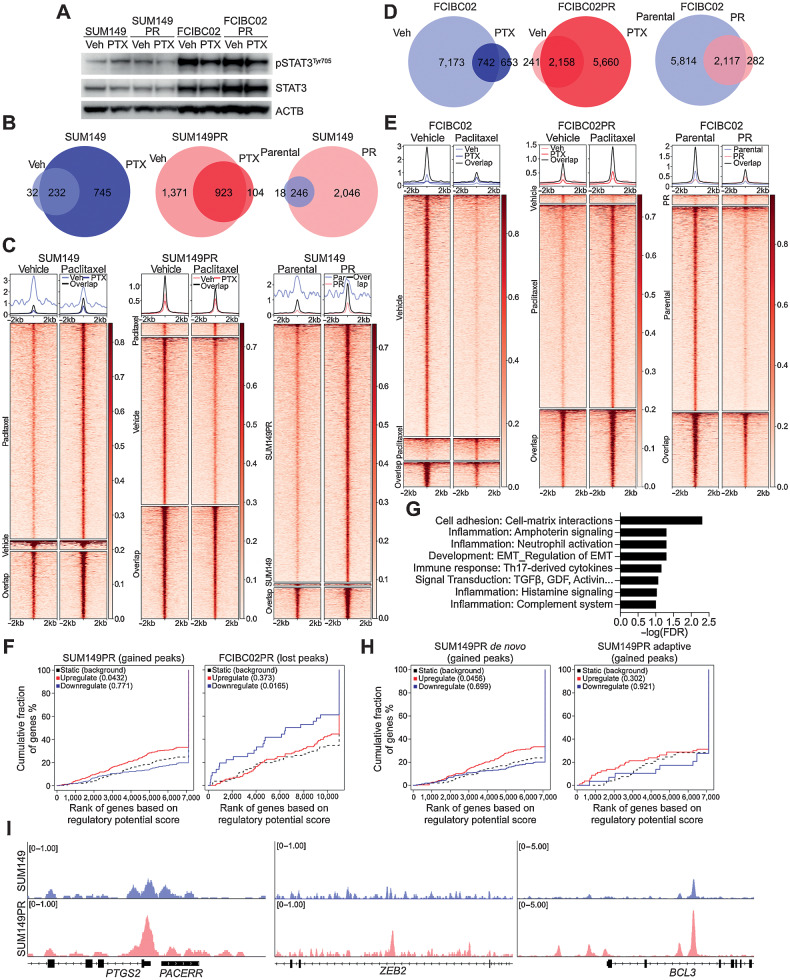
Figure 3.
pSTAT3 chromatin binding patterns in drug-sensitive and -resistant IBC cells. A, Western blot analysis of pSTAT3Tyr705, and STAT3 in the indicated cell lines following 24 hours of paclitaxel (PTX) treatment. ACTB was used as loading control B, Venn diagram depicting overlap of pSTAT3Tyr705 ChIP-seq peaks between vehicle (Veh) and paclitaxel (PTX) treatment of SUM149 and SUM149PR cell lines. C, Heat map depicting pSTAT3Tyr705 peaks, which are unique in vehicle (Veh)- and paclitaxel (PTX)-treated SUM149 and SUM149PR cells and the overlap between groups. The color key is the score of ChIP-seq signal over the selected genomic region; the signals across different genomic regions have been scaled to the same length. D, Venn diagram depicting overlap of pSTAT3Tyr705 peaks between Veh and PTX treatment of FCIBC02 and FCIBC02PR. E, Heat map of pSTAT3Tyr705 peaks in FCIBC02 and FCIBC02PR as shown in C. F, Integration of differential gene expression and pSTAT3Tyr705 targets by BETA analysis. The P value listed in the top left represents the significance of the up or down group relative to the unchanged (NON) group as determined by the Kolmogorov–Smirnov test. G, Process networks significantly enriched (FDR < 0.1) in genes that are upregulated in SUM149PR compared with SUM149 and are pSTAT3 targets only in SUM149PR cells. FDR calculated by MetaCore Enrichment Analysis test. H, BETA analysis as shown in F of integration of pSTAT3 targets and differentially expressed genes in either adaptive or de novo clusters as defined in Fig. 2D. I, Gene tracks depicting pSTAT3Tyr705 signal at selected genomic loci. X-axis shows position along the chromosome with gene structures drawn below. Y-axis shows genomic occupancy in units of rpm/bp.
To explore how STAT3 activation influences gene expression, we performed pSTAT3Tyr705 ChIP-seq in SUM149 and FCIBC02 parental and resistant cells in the presence and absence of paclitaxel or doxorubicin. Similar to the immunoblot, pSTAT3 chromatin binding showed remarkable differences between the two cell lines with increased binding in paclitaxel-treated SUM149 cells and a decrease in FCIBC02 cells (Fig. 3B–E; Supplementary Table S6). SUM149PR cells had more pSTAT3Tyr705 peaks than parental cells, which were lost following paclitaxel treatment (Fig. 3B and C). Conversely, FCIBC02PR and FCIBC02DR cells had fewer pSTAT3Tyr705 peaks than parental cells but gained peaks after drug treatment (Fig. 3D and E; Supplementary Figs. S4A–S4B). Given the higher basal level of STAT3 in FCIBC02 compared with SUM149 cells based on immunoblot and ChIP-seq, we hypothesized that these cells already have an activated STAT pathway even before paclitaxel treatment. Indeed, we found a large fraction of pSTAT3Tyr705 peaks that were gained after paclitaxel treatment or resistance in SUM149 cells were already present in FCIBC02 cells (Supplementary Fig. S4C). To examine whether pSTAT3 has different functional roles in these cell lines, we analyzed pSTAT3 chromatin targets using Metacore (49). We found significant enrichment in known STAT3-related signaling pathways, such as cell adhesion, development, regulation of EMT, Wnt, and Notch signaling, and cell-cycle networks (Supplementary Figs. S4D–S4E; Supplementary Table S4). Many of these pathways were similar to those found by RNA-seq (Fig. 2E), suggesting that pSTAT3 may facilitate chemotherapy resistance through its direct transcriptional targets.
To assess how pSTAT3 binding affects gene expression associated with chemotherapy resistance, we performed binding and expression target analysis (BETA; ref. 51) using pSTAT3Tyr705 ChIP-seq and genes differentially expressed between parental and resistant cells. We found that genes upregulated in SUM149PR cells were significantly enriched in pSTAT3Tyr705 peaks, indicating that increased pSTAT3Tyr705 binding in resistant cells correlates with higher expression of upregulated genes (Fig. 3F). Similarly, the decrease of pSTAT3Tyr705 peaks observed in FCIBC02-resistant derivatives correlated with decreased gene expression, supporting that pSTAT3 acts as a transcriptional activator in both cell lines (Fig. 3F; Supplementary Fig. S4F). These data highlight the context dependence of pSTAT3 signaling, as FCIBC02 cells with high basal levels of pSTAT3 may not be as dependent on pSTAT3 for the acquisition of resistance as SUM149 cells, but inhibition of pSTAT3 may sensitize cells to chemotherapy in both lines.
Next, to assess the functional impact of pSTAT3Tyr705 on resistance, we identified ChIP-seq targets found only in resistant lines and whose gene expression was significantly upregulated in resistant cells (Supplementary Table S5). Only SUM149PR had sufficient differentially expressed genes (>100) for pathway analysis, and we again saw enrichment for EMT, inflammation, development, and cell adhesion processes (Fig. 3G; Supplementary Table S4). BETA analysis of SUM149PR adaptive or de novo gene sets revealed a significant enrichment of pSTAT3 peaks in upregulated genes from the de novo cluster, which correlates with our RNA-seq data where we found enrichment of EMT and the IL6/JAK/STAT pathway (Fig. 3H). In line with this, EMT-related genes (e.g., ZEB2, PTGS2, and BCL3) upregulated in SUM149PR cells were enriched for pSTAT3Tyr705 peaks (Fig. 3I). Conversely, in FCIBC02DR cells loss of pSTAT3Tyr705 binding significantly correlated only with downregulated genes in the adaptive cluster (Supplementary Fig. S4G).
Finally, to identify transcriptional targets of pSTAT3 important for resistance across cell lines, we examined the overlap of resistant-specific pSTAT3Tyr705 targets with increased gene expression in resistant cells. The only gene that was shared was PDE4A (cAMP-specific 3′, 5′-cyclic phosphodiesterase 4A; Supplementary Fig. S5A). We found pSTAT3Tyr705 ChIP-seq peaks were increased in resistant derivatives near exon 1 of two PDE4A isoforms, PDE4A10, and PDE4A11 (Supplementary Fig. S5B). Overall, we identified a link between pSTAT3 and metabolic genes, cell state, response to treatment and acquisition of resistance in IBC cell lines.
Metabolic reprogramming and cAMP signaling in IBC and chemotherapy resistance
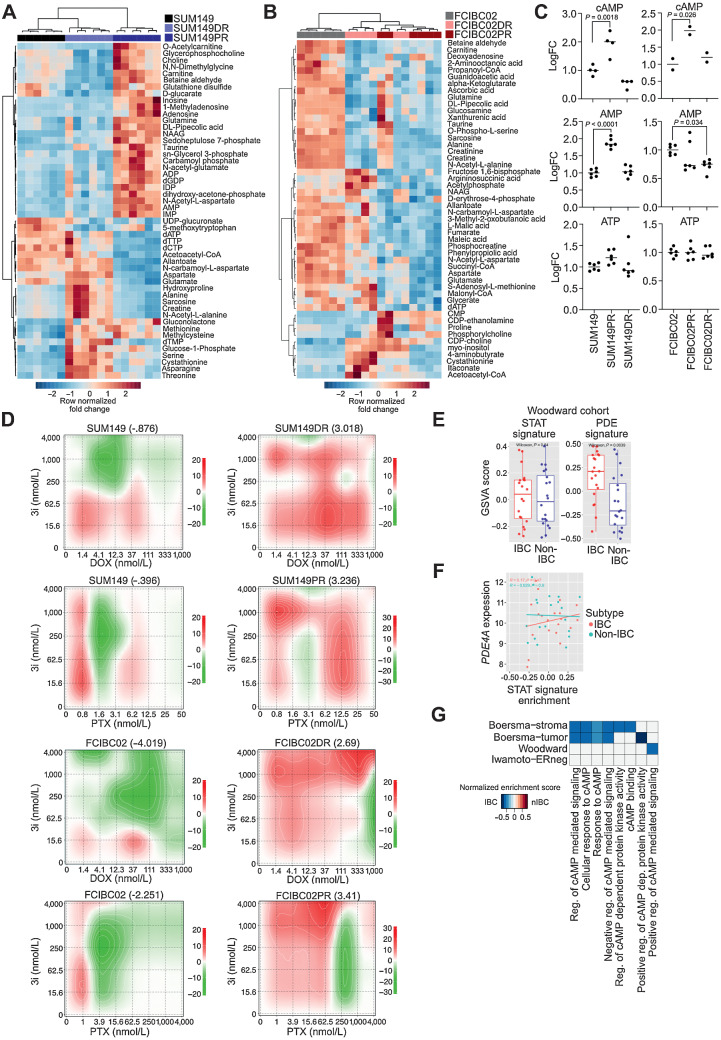
The efficacy of chemotherapeutic agents is influenced by cellular metabolism and abnormal levels of certain metabolites can confer treatment resistance (44). We identified PDE4A, which hydrolyzes the secondary messenger cAMP, as the only pSTAT3Tyr705 target common in all chemo-resistant derivatives (Supplementary Fig. S5A). To assess if upregulation of PDE4A reflects overall metabolic differences between parental and chemo-resistant cells, we performed metabolomic profiling by LC/MS-MS. We found that parental and resistant cell lines were metabolically distinct (Fig. 4A and B; Supplementary Table S7). In SUM149 cells, the top 50 differential metabolites separately clustered the three cell lines, whereas in FCIBC02, parental and resistant cells were clustered by metabolites, but FCIBC02DR and FCIBC02PR were intermingled (Fig. 4A and B).
Figure 4.
Metabolic reprogramming and upregulation of cAMP signaling in IBC resistance. A and B, Fold change metabolite abundance over SUM149 Parental (A) or FCIBC02 Parental (B) for top 50 differential metabolites in doxorubicin-resistant and paclitaxel-resistant cells as measured by LC/MS-MS. C, Relative abundance of cAMP, AMP, and ATP by LC/MS-MS (n = 6). P values calculated by one-way ANOVA. D, Synergy scores for cell lines treated with paclitaxel (PTX) or doxorubicin (DOX) in combination with the CREB inhibitor 3i (n = 3). Synergy was calculated using ZIP model where a score of 0 indicates an additive response and areas of red and green indicate synergistic and antagonistic dose regions, respectively. E, PDE gene-family and STAT (generated from KEGG) gene signature scores in a patient cohort (Woodward) featuring IBC and non-IBC patient samples. F, Correlation between STAT signature generated from KEGG and PDE4A expression in the Woodward cohort. G, Gene set enrichment scores of gene ontology cAMP-related pathways in IBC versus non-IBC (nIBC) patient samples (FDR < 0.1).
Next, we examined the levels of cAMP pathway components in our cell lines as measured by LC/MS-MS and found they were significantly increased in both paclitaxel-resistant derivatives (Fig. 4C). This increase in both AMP and cAMP as well as increased PDE4A expression suggests that the entire cAMP pathway may be elevated in paclitaxel-resistant derivatives. To determine how treatment changes cAMP levels, we performed a cAMP ELISA following treatment with paclitaxel or doxorubicin in parental and resistant cell lines. In SUM149 parental cells, paclitaxel treatment decreased cAMP levels, whereas in SUM149PR cells treatment had no effect. We also found cAMP was significantly decreased only in doxorubicin treated FCIBC02 parental cells, with the same trend in paclitaxel treated cells (Supplementary Fig. S5C).
Higher cAMP levels are associated with increased resistance to apoptosis (52), therefore, components of the cAMP pathway may be upregulated in resistant derivatives to maintain higher levels of cAMP signaling to increase survival. To test this hypothesis, we investigated whether inhibiting CREB, the main downstream mediator of cAMP signaling (52), would increase sensitivity to chemotherapeutic drugs. When we treated parental and resistant derivatives with the CREB inhibitor 3i (53), we found that both FCIBC02DR and FCIBC02PR were more resistant to CREB inhibition alone (Supplementary Fig. S5D). However, when we performed synergy studies with chemotherapeutic drugs and CREB inhibition, we found that in all resistant derivatives combination with 3i was more synergistic across a broad range of chemotherapeutic doses compared with parental cells (Fig. 4D).
To validate these findings, we generated doxycycline-inducible knockdowns for both PDE4A and CREB in FCIBC02 cells (Supplementary Figs. S5E and S5F). We first examined the role of these genes in cell proliferation and found CREB knockdown significantly decreased cell growth in all cell lines (Supplementary Fig. S5G). PDE4A knockdown also resulted in a significant decrease in proliferation, but this effect was much less pronounced than CREB, likely due to compensation from other PDE enzymes (Supplementary Fig. S5G). Downregulation of CREB also increased sensitivity to paclitaxel in both parental and resistant derivatives (Supplementary Fig. S5H). Although this effect was only significant for shCREB1#2, this hairpin had the greatest loss of CREB following doxy-induction (Supplementary Fig. S5F). Again, downregulation of PDE4A had less pronounced effects than loss of CREB and had little effect on paclitaxel sensitivity (Supplementary Fig. S5H). We also tested how combination treatment of the CREB inhibitor with ruxolitinib would impact sensitivity to paclitaxel. In the SUM149PR cell line, we found that increasing concentrations of 3i had little impact on synergy between ruxolitinib and paclitaxel at lower concentrations, but was antagonistic at the highest concentration, suggesting that cAMP may be acting downstream of STAT3, and the combination of these three drugs has limited added benefit (Supplementary Fig. S5I).
Finally, we evaluated the expression of PDE gene family members in 3 patient cohorts of IBC and non-IBC (nIBC; refs. 7, 33, 34) to determine if PDE expression could be a biomarker of IBC or IBC-associated chemotherapeutic resistance. Although the STAT signature was unable to significantly differentiate IBC from non-IBC, the PDE gene signature was significantly higher in IBC in one of the three cohorts with an increased trend in one other cohort (Fig. 4E; Supplementary Fig. S5J; Supplementary Table S1). Furthermore, we found that PDE4A expression was significantly correlated with the STAT signature in one IBC cohort and had a positive trend in other cohorts, which was only seen in IBC samples (Fig. 4F; Supplementary Fig. S5K). These data suggest that STAT3 regulation of PDE4A may be unique to IBC and metabolomic profiling might identify novel IBC biomarkers. Finally, GSEA for genes differentially expressed between IBC and non-IBC patient samples also demonstrated an enrichment of cAMP-related signaling pathways in IBC (FDR < 0.1; Fig. 4G). Although these patient cohorts are relatively small and contain both treated and untreated samples, our results suggest that elevated cAMP–PKA–CREB signaling, in part due to activated JAK/STAT3, might distinguish IBC from non-IBC and contribute to chemotherapeutic resistance in IBC.
Cellular heterogeneity and therapeutic resistance in IBC
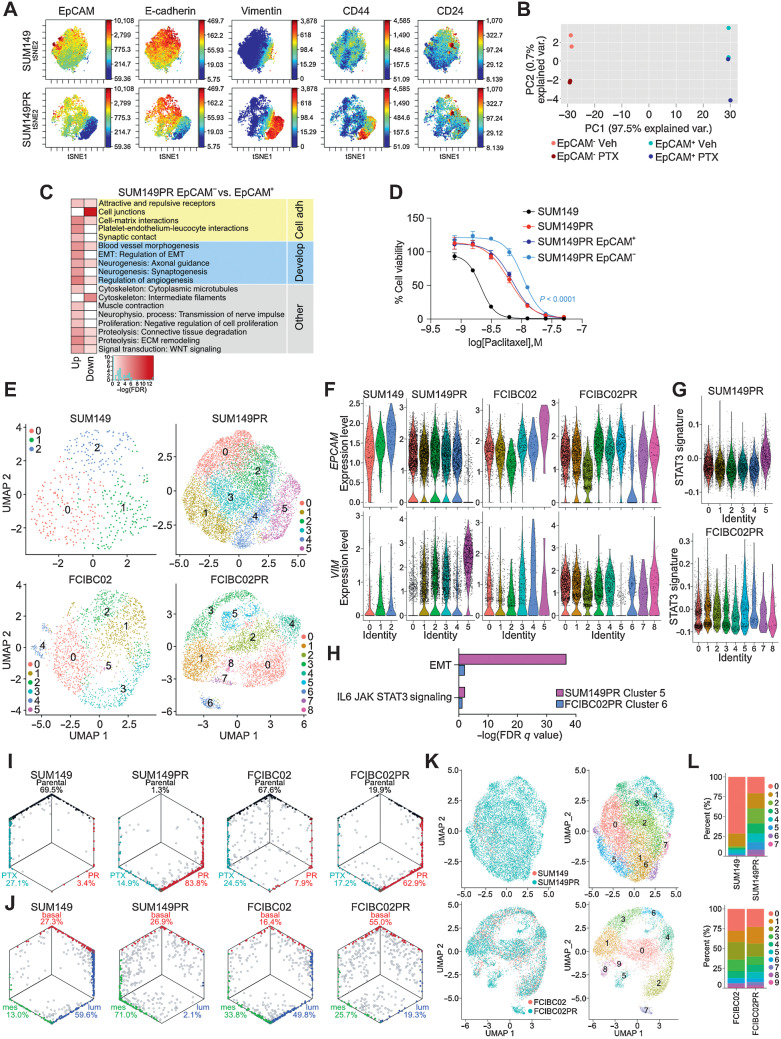
Next, we explored cellular heterogeneity in parental and resistant cell lines using CyTOF (54) and scRNA-seq. CyTOF profiles, using a panel of 34 markers, were similar between parental and chemo-resistant lines in the FCIBC02 series, whereas in the SUM149 set, SUM149PR was clearly distinct from parental (Supplementary Figs. S6A–S6B). FCIBC02DR cells showed a reduction of CD24 with concomitant increase in CD44, implying selection for CD44+CD24− cells. However, short-term treatment with chemotherapeutic agents had no discernable effects on the cellular protein levels of the markers tested in any cell line (Supplementary Figs. S6A–S6B). SUM149PR cells contained two subpopulations, one with high levels of luminal markers (e.g., EpCAM) and low levels of mesenchymal markers (e.g., vimentin) and a smaller subset with high mesenchymal and low luminal marker levels consistent with EMT (Fig. 5A; Supplementary Fig. S6B). Further analysis of SUM149PR cells by gating for EpCAM high or low expression showed that EpCAMlow cells had lower expression of the proliferation markers Ki67 and CDK1 and were mainly composed of stem cell–like CD44+CD24− cells (Supplementary Fig. S6C).
Figure 5.
Cellular heterogeneity and dynamics in the development of resistance. A, Selected viSNE maps of CyTOF analysis from Supplementary Fig. S6B of SUM149 and SUM149PR cells colored for expression of EpCAM, E-cadherin, vimentin, CD44, and CD24. Color scale indicates minimum and maximum values of expression. B, Principal component analysis plot depicting gene expression of SUM149PR EpCAM− and EpCAM+ cells treated with vehicle (Veh) or paclitaxel (PTX). C, Process networks significantly enriched (FDR < 0.005) in genes up- or downregulated between SUM149PR EpCAM− and EpCAM+ cells. Color scale corresponds to –log(FDR) of significance of enrichment, calculated by MetaCore Enrichment Analysis test. D, Cellular viability after paclitaxel treatment of indicated cell lines. Error bars, SD (n = 3). E, Uniform Manifold Approximation and Projection plots of cells from indicated cell lines by scRNA-seq, colored by cluster. Each point represents a single cell. F, Violin plots of EpCAM (top) and vimentin (bottom) expression levels in single cells clustered as shown in E. G, Violin plot of single cell expression of a STAT3 signature, generated by combination of differential gene expression between resistant and parental cells and pSTAT3 ChIP-seq resistant-only targets. Single cells clustered as in E. H, Subset of gene set enrichment analysis from Supplementary Fig. S7E of differentially expressed genes in EMT-like clusters (SUM149PR cluster 5, FCIBC02PR cluster 6) that were significantly enriched in the hallmark gene set for EMT and IL6/JAK/STAT3 signaling. I, Hexagonal plots showing classification of single cells as either Parental (black), Parental + paclitaxel treatment (PTX; teal), or paclitaxel resistant (PR; red) populations. J, Hexagonal plots depicting classification of single cells as either basal (red), mesenchymal (green), or luminal (blue), as defined by differential expression of bulk RNA-seq data from 34 TNBC cell lines. For I and J, gray cells are unclassified and mixed colors represent cells classified in both categories. Classifications were based on gene-centered expression data. K, Integrated scRNA-seq data colored by cell line (left) or by cluster (right). L, Bar plot depicting the percent of cells that belong to each cluster shown in K in parental and resistant cell lines.
To assess whether these distinct populations have different therapeutic sensitivities, we sorted EpCAM+ and EpCAM− cells from both SUM149 and SUM149PR cells and tested their response to paclitaxel. In the SUM149PR cell line, treatment with increasing doses of paclitaxel led to a progressive increase in EpCAM− cells (Supplementary Fig. S6D). However, the EpCAM− population appeared to be unstable and required the continuous presence of paclitaxel, since culturing EpCAM− cells sorted from SUM149 led to the reappearance and dominance of EpCAM+ cells. In contrast, EpCAM− cells sorted from SUM149PR and cultured in the presence of paclitaxel remained EpCAM− (Supplementary Fig. S6E). Gene expression profiles of EpCAM+ and EpCAM− fractions sorted from SUM149PR cells confirmed their more epithelial and more mesenchymal features, respectively, with differentially expressed genes enriched in cell adhesion and developmental pathways, such as EMT (Fig. 5B and C). EpCAM− cells sorted from the SUM149PR line were significantly more resistant to paclitaxel compared with SUM149PR and EpCAM+ SUM149PR cells (Fig. 5D).
To explore subclonal heterogeneity in more detail, we performed scRNA-seq on both parental and paclitaxel-resistant cell lines. In concordance with CyTOF, scRNA-seq identified a distinctly EPCAM−VIM+ cluster (cluster 5) in SUM149PR (Fig. 5E and F; Supplementary Table S8). In SUM149 parental, there was a small cluster of cells that were EPCAMlow and VIMhigh, suggesting a selection for these cells during acquired resistance to paclitaxel (Supplementary Fig. S7A). In FCIBC02PR, we detected a similar emergence of an EPCAMlow cluster (cluster 6), implying a loss of some epithelial-like features (Fig. 5E and F). We also detected clusters within both resistant derivatives (clusters 0 and 4 in SUM149PR and cluster 5 in FCIBC02PR), which have lower relative expression of VIM, but EPCAM remained unchanged, which may reflect cells that have retained epithelial features. Overall, in both resistant lines, we observed multiple luminal-mesenchymal cell states compared with parental cells, consistent with hybrid-EMT cell states associated with high intratumor heterogeneity and therapeutic resistance (55). Furthermore, we identified cells that coexpress epithelial and mesenchymal markers characteristic of hybrid-EMT cells in both resistant derivatives, and SUM149PR cells were particularly heterogeneous along the EMT spectrum (Supplementary Fig. S7B).
We next analyzed the expression of STAT3 to determine whether any cluster reflects increased STAT3 transcriptional activity. Although STAT3 expression was generally low by scRNA-seq, in FCIBC02PR cells, the EPCAMlow cluster 6 had higher expression of STAT3 (Supplementary Fig. S7C). We then generated a STAT3 signature based on genes that were both differentially expressed between resistant and parental cells and direct pSTAT3 targets only in resistant cells. This STAT3-driven-resistance signature was enriched in both EMT-like clusters in the resistant lines (Fig. 5G). Many clusters correlated with cell cycle phase in all cell lines, but each cell line also contained cell-cycle independent clusters (Supplementary Fig. S7D). GSEA analysis of differentially expressed genes in these clusters (FCIBC02PR cluster 5–7, and SUM149PR cluster 4–5) revealed that in FCIBC02PR, cells in cluster 6 and 7 had genes enriched in metabolic pathways and interferon response, respectively (Supplementary Fig. S7E; Supplementary Table S4). Furthermore, in the EMT-like clusters (SUM149PR cluster 5 and FCIBC02 cluster 6), we observed enrichment for EMT genes and IL6/JAK/STAT3 signaling (Fig. 5H). Overall, these single cell profiles demonstrate a high degree of cellular heterogeneity in both cell lines and a selection for subpopulations with more active STAT3 and mesenchymal features during acquisition of paclitaxel resistance.
Changes in IBC cellular dynamics during acquired resistance to paclitaxel
To explore how resistance and treatment alter population dynamics, we created signatures based on differentially expressed genes between parental, resistant, and treated cells based on bulk RNA-seq data. We then classified single cells according to these signatures to investigate whether they were transcriptionally like parental, resistant, or paclitaxel treated groups. In both SUM149 and FCIBC02 lines, we detected a small subpopulation of cells (3.4% and 7.9%, respectively), which was classified as having a resistant profile, whereas in both resistant lines most cells were classified as resistant (Fig. 5I). These data suggest small preexisting paclitaxel-resistant subpopulations in parental lines are selected for during treatment, but there may also be drug-induced changes underlying acquired resistance.
Because of the EMT-driven resistance mechanism observed in SUM149PR cells, and the presence of hybrid-EMT states, we also investigated differentiation state-related heterogeneity by classifying cells as basal, luminal, and mesenchymal based on signatures derived from RNA-seq of TNBC cell lines (Jovanovic et al. in press). Single cells were then classified on the basis of their transcriptional similarity to these subtypes within each cell line. Both SUM149 and FCIBC02 are basal, so the classification of single cells into the three subtypes reflects a relative scale. In SUM149 cells, we observed a shift from basal and luminal signatures in parental cells towards mesenchymal in the resistant derivative (Fig. 5J). FCIBC02 cells were more evenly distributed among the three subtypes with a higher fraction of unclassified cells. In FCIBC02PR cells, there was a similar decrease in luminal-like cells, but they instead shifted towards basal rather than mesenchymal (Fig. 5J). The general trends observed were robust enough to be maintained even when different statistical tests were used to assign cells to groups (Supplementary Fig. S7F).
To further examine how subpopulations in parental and resistant cells may be related to each other, we performed integrated clustering analysis. In SUM149/SUM149PR, the mesenchymal cell-enriched cluster (Cluster 4) was also detected in parental cells, but represented a relatively small proportion of cells, and most cells were classified as Cluster 0. In the resistant derivative, cells were more evenly distributed across clusters (Fig. 5K and L; Supplementary Fig. S7G). Cells in cluster 0 have higher expression of genes involved in mitosis and cell cycle, which may be more likely to be targeted by paclitaxel (Supplementary Table S8). In contrast, although FCIBC02 and FCIBC02PR cells were intermingled, Cluster 7 was only found in resistant cells. There was also an expansion of a rare population of parental cells in Cluster 5 (Fig. 5L), which had decreased VIM expression and increased ELF5 and MSX1 expression (Supplementary Table S8). Overall, our single cell analysis by CyTOF and scRNA-seq show that in the SUM149 cell line, one mechanism of resistance is through selection for rare preexisting EMT-like cells, whereas in FCIBC02 cells, resistance may be due to the emergence of new cell clusters with perturbed lineage programs. Finally, we observed a shift from luminal towards basal and mesenchymal phenotypes in both chemotherapy-resistant cell lines, which may in part be mediated through JAK/STAT signaling.
Molecular mechanism of ruxolitinib-chemotherapy synergy in IBC
To investigate whether ruxolitinib treatment can increase sensitivity to chemotherapies in resistant derivatives and prevent a shift in lineage programs, we performed synergy studies. In general, combination of ruxolitinib with paclitaxel was synergistic around each cell line's IC50. In both SUM149 and FCIBC02 parental cells, combination had mostly additive or antagonistic effects, whereas in paclitaxel-resistant cells, combination was synergistic around each cell line's respective IC50 dose of paclitaxel (6.25 nmol/L PTX, maximum delta score = 15.4 in SUM149PR and 250 nmol/L PTX, maximum delta score = 39 in FCIBC02PR; Fig 6A). In the SUM190 HER2+ cell line, we observed synergy in both parental and resistant lines, although SUM190PR cells had a lower synergy score (Fig. 6A). Combination of RUX with doxorubicin was mostly additive across a broad range of doses (Supplementary Fig. S8A). We also tested synergy using a more specific JAK2 inhibitor, Fedratinib, and found similar results (Supplementary Fig. S8B).
Figure 6.
Mechanism of synergy of JAK inhibition with chemotherapeutic agents in drug-sensitive and -resistant IBC cells. A, Synergy scores for combination of paclitaxel (PTX) and ruxolitinib (RUX) in the indicated cell lines. Calculated using ZIP model where a score of 0 indicates an additive response and areas of red and green indicate synergistic and antagonistic dose regions, respectively. B, PCA plot of gene expression of SUM149 and SUM149PR cells treated with vehicle, PTX, RUX, or the combination. C, Heat map depicting gene expression changes in SUM149 and SUM149PR cells after PTX, RUX, or PTX+RUX treatment. D, Process network enrichment analysis (FDR < 0.01) of up- and downregulated genes in the indicated clusters as defined by C and Supplementary Figs. S8D–S8E. Color scale corresponds to −log(FDR) of significance of enrichment, calculated by MetaCore Enrichment Analysis test. E, Western blot analysis of the indicated proteins in cell lines treated with PTX, RUX, or the combination for 72 hours. F, Selected viSNE maps from Supplementary Fig. S9 of CyTOF analysis of SUM149 and SUM149PR cells treated with indicated drugs for 14 days. Plots are colored for expression of vimentin (VIM) and gated for VIMhigh cells. Bar-plot is quantification of VIMhigh cells from gates shown in viSNE plots.
To investigate the molecular basis of the observed synergies, we performed RNA-seq on cells treated with ruxolitinib or each chemotherapeutic agent alone, and in combination. We found that in TN-IBC cell lines the most significant expression changes were induced by combination treatments with some variability in the degree of response between parental and resistant cells and the overlap with single agent-induced genes (Fig. 6B; Supplementary Fig. S8C; Supplementary Table S9). For example, paclitaxel-ruxolitinib (PTX+RUX) combination induced changes were more pronounced in parental SUM149 than in SUM149PR cells (Fig. 6B and C), but the opposite was observed in FCIBC02 (Supplementary Figs. S8C–S8D). In both SUM149PR and FCIBC02PR derivatives however, single agents had limited effects, and combination was necessary to produce significant gene expression changes (Fig. 6B and C; Supplementary Figs. S8C–S8D). In the doxorubicin–ruxolitinib (DOX+RUX) combination in SUM149, both parental and resistant cells had similar levels of combination induced changes (Supplementary Figs. S8C and S8E). In contrast, ruxolitinib alone had the most significant gene expression changes in SUM190 cells and combination with paclitaxel did not induce many further changes. (Supplementary Figs. S8C and S8F). In SUM149PR and SUM149DR cells, combination treatment induced genes that were upregulated compared with both vehicle and single agents, and in parental cells, were also upregulated by RUX alone, suggesting that in resistant derivatives combination treatment may be necessary for these RUX-specific effects (Fig. 6C, cluster 3; Supplementary Fig. S8E, cluster 2). In both FCIBC02 and FCIBC02PR cells, ruxolitinib and paclitaxel combination induced unique gene expression changes (Supplementary Fig. S8D, cluster 2). Analysis of process networks by Metacore demonstrated that genes within these clusters were most significantly enriched in apoptosis and cell-cycle networks as well as inflammation and immune response-related functions (Fig. 6D; Supplementary Table S4). Many of the top activated networks include IRAK1/2, JAK2, STAT5, IL6, and NF-κB implying autoregulatory feedback loops and crosstalk with NF-κB signaling.
To explore the clinical relevance of our findings, we investigated the expression of JAK/STAT signaling components following chemotherapy treatment. We assessed the levels of our IBC-STAT signature, defined by genes downregulated following RUX treatment in both SUM149 and FCIBC02 (Supplementary Table S1), in an ISPY clinical trial with neoadjuvant chemotherapy (32) in patients with TNBC due to lack of similar data availability in patients with IBC. Although numbers were low, we detected a decrease in the IBC–STAT signature 48 hours after treatment (T2, P = 0.015) in patients who had a pathologic complete response, but not in nonresponders (Supplementary Fig. S8G).
Finally, we tested whether treatment with ruxolitinib prevents the emergence of mesenchymal VIM+EpCAM− cells. We found that ruxolitinib alone and in combination with paclitaxel was able to decrease VIM protein expression in parental cells following short-term (72 hours) treatment (Fig. 6E). However, in SUM149PR cells, VIM remained unchanged potentially indicating stable mesenchymal cell states. To test this further, we performed CyTOF on SUM149 parental or resistant cells treated with single agents or the combination for 14 days. Concordant with our previous data, we saw that prolonged exposure to paclitaxel increased the proportion of VIM+EpCAM− cells in both cell lines (Fig. 6F; Supplementary Fig. S9). Following long-term culture in ruxolitinib alone, both parental and resistant cells had a decreased VIM+EpCAM− population compared with vehicle. Importantly, in the parental cells, combination treatment prevented the expansion of this resistant mesenchymal subpopulation. In resistant derivatives, there is still a slight expansion of this population, but it does not reach the same level as paclitaxel alone (Fig. 6F). Therefore, a short period of ruxolitinib alone before combination treatment with paclitaxel may help delay or prevent the emergence of a more resistant mesenchymal cell population.
Discussion
IBC remains a poorly defined breast cancer subtype with unique features and resistance to treatment. Thus, there is an urgent need to improve our understanding of drivers of IBC to design more effective therapies. Here, we found a central role for JAK2/STAT3 signaling in regulating IBC phenotypes and response to common chemotherapeutic agents. We demonstrated that inhibition of the JAK2/STAT3 pathway in combination with chemotherapy is a new candidate treatment option for IBC.
We previously characterized stem cell-like CD44+CD24− cells in non-IBC and found that these cells are associated with poor outcome, commonly pSTAT3+ and dependent on JAK/STAT signaling (12, 15, 56). Here, we found that most IBCs are composed of CD44+CD24− cells regardless of tumor subtype and a large fraction of these cells are pSTAT3+. Serum levels of IL6 are higher in patients with IBC compared with those with non-IBC (57) potentially contributing to the high frequency of pSTAT3+ cells in IBC. We also found that inhibition of JAK/STAT3 in combination with chemotherapy significantly decreased IBC tumor volume, suggesting that this combination may be more effective in patients with IBC, a hypothesis currently being tested in clinical trials (NCI.gov: NCT02623972). Inhibition of STAT3 to overcome therapeutic resistance is also applicable to other cancer types, and there are currently clinical trials investigating different types of STAT3 inhibition strategies (e.g., direct STAT3 inhibitors and antisense oligonucletides) in multiple malignancies (58).
IBC is relatively resistant to therapies, which may be due to the high frequency of stem cell–like cells with activated JAK/STAT3 signaling (59, 60). In prostate cancer, JAK2/STAT3 activation led to lineage plasticity, whereby switching from a luminal androgen receptor positive phenotype to mesenchymal/neuroendocrine state was associated with resistance to anti-androgens and metastatic progression (61, 62). We also observed substantial differentiation state-related heterogeneity within our IBC cell lines and treatment with paclitaxel and the development of resistance increased the relative frequencies of EpCAMlowVIMhighCD44highCD24lowpSTAT3+ cells in TN-IBC lines. Similarly, paclitaxel-resistant derivates of the SUM190 HER2+ IBC line shifted from luminal to a more myoepithelial phenotype. These results suggest that the increased frequency of stem cell-like cells in IBC with activated JAK2/STAT3 signaling leads to cellular plasticity that promotes treatment resistance, but resistance mechanisms are heterogeneous and depend on tumor subtype.
One of the long-standing questions in IBC is the identification of molecular markers that are specific to IBC compared with non-IBC. Here we identified the PDE family of cAMP phosphodiesterases as candidate biomarkers for IBC, which may also be more resistant to chemotherapy. We found elevated cAMP signaling in paclitaxel-resistant cells and in IBC patient samples compared with non-IBC tumors. One of the major downstream targets of cAMP is PKA, which activates CREB and RAF to regulate gene expression and cell survival (63). cAMP/PKA signaling is required for the survival of many normal stem cells and has been implicated in cancer stem cells and therapeutic resistance (64). PKA also activates PDEs as part of a negative feedback loop to maintain cAMP homeostasis (63, 65), which could explain the elevated PDE expression and high cAMP levels in our resistant cell lines. We found that inhibition and knockdown of CREB sensitized cells to paclitaxel identifying PDEs and the cAMP/PKA signaling pathway as candidate biomarkers and novel therapeutic targets in IBC.
pSTAT3Tyr705 functions as a transcriptional activator inducing tumor-promoting gene expression programs (66). Our analysis of pSTAT3Tyr705 chromatin binding in TN-IBC cell lines revealed divergent changes in pSTAT3 peaks following paclitaxel treatment and acquisition of resistance. In SUM149 cells with lower basal pSTAT3 compared with FCIBC02, there were fewer pSTAT3 peaks at baseline compared with paclitaxel-resistant cells, and paclitaxel treatment increased the number of pSTAT3 peaks in parental but not in resistant cells. In contrast, parental FCIBC02 cells had higher levels of chromatin-bound pSTAT3 than resistant cells, but paclitaxel treatment increased the number of pSTAT3 in resistant derivates, which could contribute to the more significant synergy in combination treatment in FCIBC02. The heterogeneity of these responses highlights the need for predictive biomarkers to select patients with IBC for JAK2/STAT3-targeting combination therapy.
To examine how IBC cells respond to paclitaxel and ruxolitinib combination therapy, we performed synergy studies with RNA-seq in vitro. Potential effects of treatment on the microenvironment, which is out of the scope of this study, is warranted given that synergy was more pronounced in vivo than in vitro. The importance of JAK2/STAT3 signaling in the stroma, for example in angiogenesis, likely explains these differences. Considering the unique IBC microenvironment is not reproduced in any preclinical models, the impact of inhibiting JAK2/STAT3 signaling in IBC would have to be dissected in clinical trials. Interestingly, when cells were treated with the combination of chemotherapy and ruxolitinib, cells paradoxically had increased inflammation and immune response pathways, suggesting possible compensatory mechanisms. Thus, our data suggest that patients with IBC receiving the combination of JAK2/STAT3 inhibitors with chemotherapy may benefit from the addition of immunotherapy such as PD-L1 immune checkpoint inhibitors. Recent data support this, where inhibition of STAT3 made mammary tumor cells more immunogenic in syngeneic models (67).
In summary, our analysis of clinical IBC samples in combination with comprehensive profiling of chemotherapy-sensitive and -resistant IBC cell line models emphasizes a key role for JAK2/STAT3 signaling and identified novel biomarkers and candidate therapeutic targets in IBC to explore in future preclinical and clinical studies.
Supplementary Material
Supplementary Figures
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Table S5
Supplementary Table S6
Supplementary Table S7
Supplementary Table S8
Supplementary Table S9
Acknowledgments
The authors thank members of the Polyak laboratory for their critical reading of the manuscript and useful advice. The authors thank Incyte for providing ruxolitinib for some of the initial experiments. Graphical Abstract created with BioRender.com This research was funded by the NCI R35 CA197623 (to K. Polyak), R01CA202634 (to P. Sicinski), P50 CA168504 (to P. Sicinski), P01CA250959 (to K. Polyak, P. Sicinski), NIH U54-HG008097 (to J.D. Jaffe and M. Papanastasiou), T32CA236754 (to L.E. Stevens), DOD W81XWH-14–1-0240 (to K. Polyak), the Claudia Adams Barr Program for Innovative Basic Cancer Research Award (to A. Fassl), the Ludwig Center at Harvard (to A. Toker, K. Polyak, F. Michor), the Susan G. Komen Foundation PDF14302777 (to S. Shu), DFCI Helen Gurley Brown Presidential Initiative Award (to A. Trinh and L.E. Stevens), and the V Foundation (to K. Polyak).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
L.E. Stevens reports grants from NIH (T32CA236754) and Helen Gurley Brown Presidential Initiative Award during the conduct of the study. A. Fassl reports grants from Claudia Adams Barr Award/Dana-Farber Cancer Institute during the conduct of the study and grants from Claudia Adams Barr Award/Dana-Farber Cancer Institute outside the submitted work. A. Trinh reports grants from Helen Gurley Brown Foundation during the conduct of the study and reports employment with GenieUs Genomics. M. Seehawer reports grants from EMBO Postdoctoral Fellowships during the conduct of the study and grants from EMBO Postdoctoral Fellowships outside the submitted work. M. Aleckovic reports other support from Shasqi Inc. outside the submitted work. D.A. Frank reports grants from Roche Genentech and Kymera Therapeutics and personal fees from Vigeo and Revitope outside the submitted work. A. Meissner reports a patent related to hypermethylated CGI targets in cancer and is a cofounder and scientific advisor of Harbinger Health, and also reports employment with Scientific Advisory Board of Zymo Research Inc. P. Sicinski reports Novartis, Genovis, Guidepoint, The Planning Shop, ORIC Pharmaceuticals, Cedilla Therapeutics, Syros Pharmaceuticals, and Exo Therapeutics and also receives research funding from Novartis. A. Toker reports personal fees from JBC outside the submitted work. F. Michor reports grants from Dana-Farber Cancer Institute during the conduct of the study and other support from Harbinger Health, Zephyr AI, and Red Cell Partners outside the submitted work. B.A. Overmoyer reports other support from Incyte Corp. and Dana-Farber Inflammatory Breast Cancer Research Fund, grants from Inflammatory Breast Cancer Research Foundation during the conduct of the study, and other support from Eisai Inc. and Genentech outside the submitted work. K. Polyak reports grants from NCI, Department of Defense, Ludwig Center at Harvard, Susan G. Komen Foundation, and V Foundation during the conduct of the study, grants and personal fees from Novartis, other support from Acrivon Therapeutics, personal fees from Vividion Therapeutics, Aria Pharmaceuticals, and Roche, and personal fees and other support from Scorpion Therapeutics outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
L.E. Stevens: Conceptualization, data curation, investigation, visualization, methodology, writing–original draft. G. Peluffo: Conceptualization, data curation, investigation, methodology, writing–original draft. X. Qiu: Formal analysis. D. Temko: Formal analysis. A. Fassl: Data curation, investigation, methodology, writing–original draft. Z. Li: Formal analysis, visualization. A. Trinh: Formal analysis, visualization, writing–original draft. M. Seehawer: Investigation. B. Jovanović: Investigation. M. Alečković: Investigation. C.M. Wilde: Investigation. R.C. Geck: Formal analysis. S. Shu: Investigation. N.L. Kingston: Data curation, investigation. N.W. Harper: Formal analysis. V. Almendro: Data curation, investigation, visualization, writing–original draft. A.L. Pyke: Investigation. S.B. Egri: Formal analysis, investigation. M. Papanastasiou: Formal analysis, supervision, writing–original draft. K. Clement: Formal analysis. N. Zhou: Formal analysis. S. Walker: Investigation. J. Salas: Investigation. S.Y. Park: Resources. D.A. Frank: Supervision. A. Meissner: Supervision. J.D. Jaffe: Supervision. P. Sicinski: Supervision, funding acquisition. A. Toker: Supervision, funding acquisition. F. Michor: Supervision. H.W. Long: Supervision. B.A. Overmoyer: Resources. K. Polyak: Conceptualization, resources, supervision, funding acquisition, writing–original draft, project administration.
References
- 1. Abraham HG, Xia Y, Mukherjee B, Merajver SD. Incidence and survival of inflammatory breast cancer between 1973 and 2015 in the SEER database. Breast Cancer Res Treat 2021;185:229–38. [DOI] [PubMed] [Google Scholar]
- 2. Garrido-Castro AC, Lin NU, Polyak K. Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment. Cancer Discov 2019;9:176–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bertucci F, Finetti P, Rougemont J, Charafe-Jauffret E, Nasser V, Loriod B, et al. Gene expression profiling for molecular characterization of inflammatory breast cancer and prediction of response to chemotherapy. Cancer Res 2004;64:8558–65. [DOI] [PubMed] [Google Scholar]
- 4. Vermeulen PB, van Golen KL, Dirix LY. Angiogenesis, lymphangiogenesis, growth pattern, and tumor emboli in inflammatory breast cancer: a review of the current knowledge. Cancer 2010;116:2748–54. [DOI] [PubMed] [Google Scholar]
- 5. Kleer CG, Griffith KA, Sabel MS, Gallagher G, van Golen KL, Wu ZF, et al. RhoC-GTPase is a novel tissue biomarker associated with biologically aggressive carcinomas of the breast. Breast Cancer Res Treat 2005;93:101–10. [DOI] [PubMed] [Google Scholar]
- 6. Bertucci F, Finetti P, Birnbaum D, Viens P. Gene expression profiling of inflammatory breast cancer. Cancer 2010;116:2783–93. [DOI] [PubMed] [Google Scholar]
- 7. Woodward WA, Krishnamurthy S, Yamauchi H, El-Zein R, Ogura D, Kitadai E, et al. Genomic and expression analysis of microdissected inflammatory breast cancer. Breast Cancer Res Treat 2013;138:761–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ross JS, Ali SM, Wang K, Khaira D, Palma NA, Chmielecki J, et al. Comprehensive genomic profiling of inflammatory breast cancer cases reveals a high frequency of clinically relevant genomic alterations. Breast Cancer Res Treat 2015;154:155–62. [DOI] [PubMed] [Google Scholar]
- 9. Liang X, Vacher S, Boulai A, Bernard V, Baulande S, Bohec M, et al. Targeted next-generation sequencing identifies clinically relevant somatic mutations in a large cohort of inflammatory breast cancer. Breast Cancer Res 2018;20:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matsuda N, Lim B, Wang Y, Krishnamurthy S, Woodward W, Alvarez RH, et al. Identification of frequent somatic mutations in inflammatory breast cancer. Breast Cancer Res Treat 2017;163:263–72. [DOI] [PubMed] [Google Scholar]
- 11. Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer 2012;12:323–34. [DOI] [PubMed] [Google Scholar]
- 12. Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell 2007;11:259–73. [DOI] [PubMed] [Google Scholar]
- 13. Almendro V, Cheng YK, Randles A, Itzkovitz S, Marusyk A, Ametller E, et al. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Rep 2014;6:514–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Park SY, Lee HE, Li H, Shipitsin M, Gelman R, Polyak K, et al. Heterogeneity for stem cell–related markers according to tumor subtype and histologic stage in breast cancer. Clin Cancer Res 2010;16:876–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44+ CD24–stem cell–like breast cancer cells in human tumors. J Clin Invest 2011;121:2723–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Madoux F, Koenig M, Sessions H, Nelson E, Mercer BA, Cameron M, et al. Modulators of STAT transcription factors for the targeted therapy of cancer (STAT3 inhibitors). Probe Reports from the NIH Molecular Libraries Program. Bethesda, MD;2010. [PubMed] [Google Scholar]
- 17. Bieche I, Lerebours F, Tozlu S, Espie M, Marty M, Lidereau R, et al. Molecular profiling of inflammatory breast cancer: identification of a poor-prognosis gene expression signature. Clin Cancer Res 2004;10:6789–95. [DOI] [PubMed] [Google Scholar]
- 18. Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American society of clinical oncology/college of American pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch Pathol Lab Med 2010;134:907–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ianevski A, Giri AK, Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res 2020;48:W488–W93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011;332:687–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qin Q, Mei S, Wu Q, Sun H, Li L, Taing L, et al. ChiLin: a comprehensive ChIP-seq and DNase-seq quality control and analysis pipeline. BMC Bioinf 2016;17:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Anders S, Pyl PT, Huber W. HTSeq—a python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hinohara K, Wu HJ, Sebastien V, McDonald TO, Igarashi KJ, Yamamoto KN, et al. KDM5 histone demethylase activity links cellular transcriptomic heterogeneity to therapeutic resistance. Cancer Cell 2019;35:330–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shu S, Wu HJ, Ge JY, Zeid R, Harris IS, Jovanovic B, et al. Synthetic lethal and resistance interactions with BET bromodomain inhibitors in triple-negative breast cancer. Mol Cell 2020;78:1096–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lun AT, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol 2016;17:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 2018;36:421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Creech AL, Taylor JE, Maier VK, Wu X, Feeney CM, Udeshi ND, et al. Building the connectivity map of epigenetics: chromatin profiling by quantitative targeted mass spectrometry. Methods 2015;72:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Magbanua MJ, Wolf DM, Yau C, Davis SE, Crothers J, Au A, et al. Serial expression analysis of breast tumors during neoadjuvant chemotherapy reveals changes in cell cycle and immune pathways associated with recurrence and response. Breast Cancer Res 2015;17:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Boersma BJ, Reimers M, Yi M, Ludwig JA, Luke BT, Stephens RM, et al. A stromal gene signature associated with inflammatory breast cancer. Int J Cancer 2008;122:1324–32. [DOI] [PubMed] [Google Scholar]
- 34. Iwamoto T, Bianchini G, Qi Y, Cristofanilli M, Lucci A, Woodward WA, et al. Different gene expressions are associated with the different molecular subtypes of inflammatory breast cancer. Breast Cancer Res Treat 2011;125:785–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang X, Terfve C, Rose JC, Markowetz F. HTSanalyzeR: an R/Bioconductor package for integrated network analysis of high-throughput screens. Bioinformatics 2011;27:879–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F, et al. Deriving the consequences of genomic variants with the ensembl API and SNP effect predictor. Bioinformatics 2010;26:2069–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 2004;5:557–72. [DOI] [PubMed] [Google Scholar]
- 44. Geck RC, Foley JR, Murray Stewart T, Asara JM, Casero RA Jr, Toker A, et al. Inhibition of the polyamine synthesis enzyme ornithine decarboxylase sensitizes triple-negative breast cancer cells to cytotoxic chemotherapy. J Biol Chem 2020;295:6263–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 2018;46:W486–W94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Quintas-Cardama A, Vaddi K, Liu P, Manshouri T, Li J, Scherle PA, et al. Preclinical characterization of the selective JAK1/2 inhibitor INCB018424: therapeutic implications for the treatment of myeloproliferative neoplasms. Blood 2010;115:3109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov 2014;4:232–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Balko JM, Schwarz LJ, Luo N, Estrada MV, Giltnane JM, Davila-Gonzalez D, et al. Triple-negative breast cancers with amplification of JAK2 at the 9p24 locus demonstrate JAK2-specific dependence. Sci Transl Med 2016;8:334ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nikolsky Y, Nikolskaya T, Bugrim A. Biological networks and analysis of experimental data in drug discovery. Drug Discov Today 2005;10:653–62. [DOI] [PubMed] [Google Scholar]
- 50. Hollern DP, Swiatnicki MR, Andrechek ER. Histological subtypes of mouse mammary tumors reveal conserved relationships to human cancers. PLoS Genet 2018;14:e1007135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang S, Sun H, Ma J, Zang C, Wang C, Wang J, et al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc 2013;8:2502–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ladilov Y, Appukuttan A. Role of soluble adenylyl cyclase in cell death and growth. Biochim Biophys Acta 2014;1842:2646–55. [DOI] [PubMed] [Google Scholar]
- 53. Xie F, Li BX, Kassenbrock A, Xue C, Wang X, Qian DZ, et al. Identification of a potent inhibitor of CREB-mediated gene transcription with efficacious in vivo anticancer activity. J Med Chem 2015;58:5075–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol 2012;30:858–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Teeuwssen M, Fodde R. Cell heterogeneity and phenotypic plasticity in metastasis formation: the case of colon cancer. Cancers (Basel) 2019;11:1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Almendro V, Kim HJ, Cheng YK, Gonen M, Itzkovitz S, Argani P, et al. Genetic and phenotypic diversity in breast tumor metastases. Cancer Res 2014;74:1338–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Drygin D, Ho CB, Omori M, Bliesath J, Proffitt C, Rice R, et al. Protein kinase CK2 modulates IL-6 expression in inflammatory breast cancer. Biochem Biophys Res Commun 2011;415:163–7. [DOI] [PubMed] [Google Scholar]
- 58. Santoni M, Miccini F, Cimadamore A, Piva F, Massari F, Cheng L, et al. An update on investigational therapies that target STAT3 for the treatment of cancer. Expert Opin Investig Drugs 2021;30:245–51. [DOI] [PubMed] [Google Scholar]
- 59. Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst 2008;100:672–9. [DOI] [PubMed] [Google Scholar]
- 60. Opyrchal M, Salisbury JL, Iankov I, Goetz MP, McCubrey J, Gambino MW, et al. Inhibition of Cdk2 kinase activity selectively targets the CD44(+)/CD24(-)/low stem-like subpopulation and restores chemosensitivity of SUM149PT triple-negative breast cancer cells. Int J Oncol 2014;45:1193–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Deng S, Wang C, Wang Y, Xu Y, Li X, Johnson NA, et al. Ectopic JAK-STAT activation enables the transition to a stem-like and multilineage state conferring AR-targeted therapy resistance. Nat Cancer 2022;3:1071–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chan JM, Zaidi S, Love JR, Zhao JL, Setty M, Wadosky KM, et al. Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science 2022;377:1180–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Murthy KS, Zhou H, Makhlouf GM. PKA-dependent activation of PDE3A and PDE4 and inhibition of adenylyl cyclase V/VI in smooth muscle. Am J Physiol Cell Physiol 2002;282:C508–17. [DOI] [PubMed] [Google Scholar]
- 64. Zhang H, Kong Q, Wang J, Jiang Y, Hua H. Complex roles of cAMP-PKA-CREB signaling in cancer. Exp Hematol Oncol 2020;9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Takahashi M, Terwilliger R, Lane C, Mezes PS, Conti M, Duman RS, et al. Chronic antidepressant administration increases the expression of cAMP-specific phosphodiesterase 4A and 4B isoforms. J Neurosci 1999;19:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Owen KL, Brockwell NK, Parker BS. JAK-STAT signaling: a double-edged sword of immune regulation and cancer progression. Cancers (Basel) 2019;11:2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Khan MW, Saadalla A, Ewida AH, Al-Katranji K, Al-Saoudi G, Giaccone ZT, et al. The STAT3 inhibitor pyrimethamine displays anti-cancer and immune stimulatory effects in murine models of breast cancer. Cancer Immunol Immunother 2018;67:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Table S5
Supplementary Table S6
Supplementary Table S7
Supplementary Table S8
Supplementary Table S9
Data Availability Statement
The RNA-seq and ChIP-seq datasets have been deposited to Gene Expression Ominbus (GEO) with the accession number GSE163397. The original mass spectra and the protein sequence database used for searches have been deposited in the public proteomics repository MassIVE (http://massive.ucsd.edu) and are accessible at ftp://massive.ucsd.edu/MSV000090647/.