Abstract
The rhizosphere is an extremely important component of the “one health” scenario by linking the soil microbiome and plants, in which the potential enrichment of antibiotic resistance genes (ARGs) might ultimately flow into the human food chain. Despite the increased occurrence of soil-borne diseases, which can lead to increased use of pesticides and antibiotic-producing biocontrol agents, the understanding of the dynamics of ARG spread in the rhizosphere is largely overlooked. Here, tomato seedlings grown in soils conducive and suppressive to the pathogen Ralstonia solanacearum were selected as a model to investigate ARG spread in the rhizosphere with and without pathogen invasion. Metagenomics data revealed that R. solanacearum invasion increased the density of ARGs and mobile genetic elements (MGEs). Although we found ARGs originating from human pathogenic bacteria in both soils, the enrichment was alleviated in the suppressive soil. In summary, the suppressive soil hindered ARG spread through pathogen suppression and had a lower number of taxa carrying antibiotic resistance.
Keywords: Bacterial pathogen, Invasion, Suppressive soil, Conducive soil, Antibiotic resistance genes (ARGs)
Highlights
-
•
Bacterial pathogen invasion increased the density of ARGs and MGEs within rhizosphere.
-
•
Enriched ARGs were originated from the Invading pathogen and human pathogenic bacteria.
-
•
Suppressive rhizosphere soil could alleviate the ARGs enrichment.
-
•
Lower number of invaded and native taxa carrying ARGs lead to the alleviation.
1. Main
Genes conferring resistance to antibiotics (antibiotic-resistance genes, ARGs) occur naturally [1]. ARGs can be easily exchanged among organisms and surrounding environments [2,3]. For instance, ARGs present in the soil can enter the food chain through rhizosphere dwelling microorganisms [4], the accumulation of which can dramatically increase the health risks of the exposed population. In fact, the acquisition and transfer of ARGs imposes significant harm to human health by generating multi-resistance human pathogens [5]. Therefore, strategies to reduce the amount of ARGs and hinder their spread in soils are crucial.
The accumulation of ARGs in the environment is closely related to artificial interference. Many studies have reported that exogenous contamination, such as the discharge of urban wastewater, manure and sludge from agricultural animals increases ARGs in soil, and the invasion of ARG-rich microorganisms in these wastes is directly linked to the higher human risk [[6], [7], [8]]. In agriculture, intensive cropping and conventional mineral fertilizer application usually increase soil pathogen density [9] and ARG abundance [8,10]. In the presence of mobile genetic elements (MGEs), these ARGs can be easily transferred between microbial community members. However, the subsequent interaction among the coaccumulation factors (pathogens, ARGs and MGEs) around crops remains unclear. After all, soil microbiome under biotic and abiotic stress seem to change their associated microenvironment and consequently leads to the enrichment of ARGs [11,12]. In this context, it is crucial to understand the ARG dynamics of the rhizosphere microbiome of disease-induced soils, as pathogen invasion could promote the spread of ARGs.
We hypothesize that the biotic pressure imposed by pathogens can lead to an increase in ARG-rich taxa, but that the response will depend on the health status of the soil. Conducive soils, which are more susceptible to disturbances and prone to pathogen colonization, would pose a higher risk of antibiotic resistance. The proliferation of ARG-rich pathogens, such as Ralstonia solanacearum which could cause severe bacterial wilt [13], in intensive agriculture, presents an interesting model to access the link between pathogen invasion and ARGs. Here, we experimentally examined how pathogen (R. solanacearum) invasion affects ARGs and MGEs in rhizosphere soil derived from soils collected from two treatments (continuously applied chemical fertilizer or bioorganic fertilizer) at a long-term field experiment site. The soil disease incidence was higher than 60% in the chemical fertilizer treatment and <30% in the bio-organic fertilizer treatment, which led to the designation of conducive (C) and suppressive (S) soils, respectively [14]. After allowing plant root colonization by the S or C microbiome, seedlings were transferred to new pots with germ-free substrate and subsequently inoculated with or without R. solanacearum (Rs). This approach allowed us to identify the potential dynamics of ARGs and MGEs in the rhizosphere, which harbor different suppressiveness levels, under bacterial invasion. The method details are shown in the supplementary file.
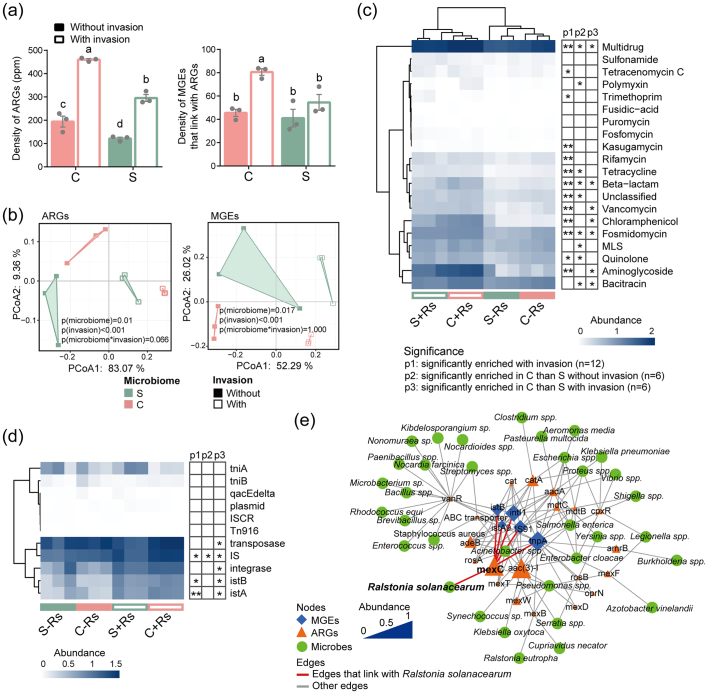
We observed a lower abundance of ARGs in the rhizosphere derived from the suppressive than from the conducive soil, regardless of pathogen invasion. However, R. solanacearum invasion significantly enhanced this value (Fig. 1a). For MGEs, although the whole MGE abundance in the rhizosphere decreased after pathogen invasion (Fig. S1), the abundance of MGEs linked to ARGs increased following R. solanacearum inoculation, and only the conducive rhizosphere showed significant enrichment (Fig. 1a). Moreover, although microbiome origin and pathogen invasion significantly determined the ARG and MGE composition, invasion exerted the most powerful influence (Fig. 1b). Specifically, 13 of the 20 ARG types and 3 of the 11 MGE types significantly increased following pathogen invasion. In addition, the comparison between S and C soils before invasion (S-Rs with C-Rs) revealed that 9 ARG types and 1 MGE type were significantly higher in the conducive rhizosphere. The comparison of the same soils after invasion (S + Rs with C + Rs) showed that 7 ARG and 5 MGE types were significantly higher in the conducive rhizosphere (Fig. 1c, d). The response of aminoglycoside-, chloramphenicol-, and vancomycin-type ARGs and transposase-, integrase-, istA-, and istB-type MGEs following invasion enhanced the difference between the two kinds of rhizosphere microbiomes, which indicates that in comparison to the suppressive rhizosphere, these ARGs and MGEs show more significant responses in the conducive rhizosphere (Fig. 1c, d, and S2a).
Fig. 1.
Abundance and composition of ARGs and MGEs that are linked to ARGs in the rhizosphere. (a) Relative abundance of ARGs and MGEs among the four treatments; (b) PCoA of rhizosphere ARG and MGE composition based on Bray-Curtis distance. The p values were calculated through PERMANOVA based on Bray-Curtis distance; (c) clustered heatmap of ARGs at the type level; (d) clustered heatmap of MGEs at the type level; (e) the network among significantly enriched MGE subtypes and their associated ARGs and the assigned taxonomy of the ARGs. The value of ARG abundance was transformed through log10 transformation. The size of the MGE and ARG nodes was normalized from 0 to 1 based on their abundance. “C”, conducive rhizosphere derived from soil following the long-term application of chemical fertilizer; “S”, suppressive rhizosphere derived from soil following the long-term application of bio-organic fertilizer; “-Rs”, rhizosphere soil without R. solanacearum invasion; “+Rs”, rhizosphere soil after R. solanacearum invasion. Different letters indicate significant differences among the treatments as defined by Tukey's test (p ≤ 0.05). Asterisks indicate significant differences (Mann-Whitney U test, *p ≤ 0.05, **p ≤ 0.01).
Next, we traced dominant ARGs to their contributors. The results showed that almost all ARG contributors in the conducive rhizosphere were more abundant than those in the suppressive rhizosphere following invasion (Fig. S2b). Similar to the abundance of ARGs in R. solanacearum (Fig. S2c), some biocontrol agents, such as Streptomyces and Pseudomonas, and many human conditional pathogens, such as Staphylococcus aureus, Pseudomonas aeruginosa, and Escherichia coli, which contain multidrug-type ARGs, were significantly enriched in the conducive rhizosphere (Fig. S2b). To understand the patterns associated with MGEs following invasion, their associated ARGs, and the potential hosts of the ARGs were visualized as a network (Fig. 1e, S3a, and S3b). The results showed that the pathogen mainly contributed to the multidrug resistance gene mexC, which was potentially connected with various MGEs, such as tnpA, IS91, istB, intl1, and istA9 (Fig. 1e, S3c and S3d). These MGEs were also potentially linked with many ARGs belonging to other microbial hosts, including some of the human conditional pathogens (Fig. 1e).
We next constructed co-occurrence networks to examine the interaction between the rhizosphere microbiome and ARGs (Fig. 2). Importantly, Ralstonia was abundant following invasion and was significantly positively correlated with ARG density (Fig. 2b), which indicates that pathogen invasion affected ARG enrichment. More specifically, the network of interactions between bacteria and ARGs was simpler in the absence of invasion (Fig. 2a) but increased in complexity in the presence of the pathogen (Fig. 2b). Most of these resistance genes are multidrug-resistant, followed by aminoglycoside, chloramphenicol, fosmidomycin, and macrolide-resistant (Fig. S2a). Some bacteria, such as Sphingomonas, Pseudoxanthomonas, and Stenotrophomonas, were significantly enriched in the suppressive rhizosphere and negatively correlated with ARGs. In contrast, the bacteria enriched in the conducive rhizosphere showed the opposite trend (Fig. 2b). Finally, an in vitro test demonstrated that the antibiotic resistance of these taxa (Pseudoxanthomonas spp. and Sphingomonas spp.) was significantly less enriched in the suppressive rhizosphere than the potentially responsive ARG contributors (Staphylococcus aureus and Pseudomonas spp.) that were enriched in the conducive rhizosphere (Fig. 2c, S2, and Table S1).
Fig. 2.
Potential interaction between bacteria and ARGs. (a) Networks based on the samples without R. solanacearum invasion; (b) networks based on the samples with R. solanacearum invasion. The abundances of ARGs and bacteria were standardized to a scale of 0–1. The red circles represent ARG subtypes, and the blue circles represent bacterial genera. The pink lines represent the positive correlations between ARGs and bacteria, and the blue lines represent the negative correlations between ARGs and bacteria. Only Spearman correlations with r > 0.06 and p < 0.05 were reserved. The solid line circles and the dashed lines represent the significantly enriched bacteria in the suppressive and conducive rhizospheres, respectively. (c) MIC comparisons between responsive taxa groups with potentially rich and poor ARGs. The taxa group of A (potentially enriched ARG-rich taxa under invasion) contains Staphylococcus aureus and Pseudomonas spp., and B (potentially enriched ARG-poor taxa in the suppressive rhizosphere under invasion) contains Pseudoxanthomonas spp. and Sphingomonas spp. All the MIC values were transformed using the formula (Log2x) + 2, where x indicates the MIC value. The p values were calculated through two-sample Mann-Whitney U tests. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Intensive agricultural cropping contributes to the buildup of pathogenic microorganisms, further adding biotic pressure to the plants and their associated microbiomes, directly influencing “one health” [15]. To compete with pathogen spread, the production of antibiotics by the resident microbiome could significantly increase the level of antibiotics in soil, potentially contributing to the diversity and abundance of ARGs [11,16]. In this scenario, this selective pressure could result in soil enriched with antibiotic resistant human pathogenic bacteria [12], substantially increasing the health risk to humans. However, soil microbial communities with different levels of suppression may have distinct responses following invasion by exogenous microbes [17,18]. Thus, sustainable agricultural practices that promote a decrease in plant pathogen densities can contribute to the “one health” concept by alleviating the risk of ARG spread while promoting plant growth and reducing the environmental footprint of agricultural production.
Our results support the overlooked potential link between pathogen incidence and the increase in antibiotic resistance spread risk. In general, R. solanacearum invasion increased ARG and MGE abundances in both rhizospheres, indicating that the biotic pressure imposed by the pathogen boosted the abundance of ARG-rich taxa. However, ARG enrichment was relatively moderate in the suppressive rhizosphere soil, which occurs in soil following long-term (bio)-organic fertilization, confirming that the response depends on the health status of the soil. It is widely known that soil suppressiveness can be primed by organic amendments through microbiome manipulation [14,19], which can also decrease the soil ARG level [20]. Our previous study showed that rhizosphere soil derived from suppressive soil featured more Sphingomonadaceae and Xanthomonadaceae than that from conducive soil. And these suppressive indicator taxa were also showed the ability to suppress bacterial wilt in rhizosphere [14]. Interestingly, the results here suggest that ARG-poor taxa such as Sphingomonas and Pseudoxanthomonas, belonging to Sphingomonadaceae and Xanthomonadaceae, respectively, were stimulated by pathogen invasion in suppressive soils. In suppressive rhizosphere soil, these species can control bacterial wilt, contributing to lower pathogen density while potentially alleviating the ARG horizontal transfer risk. In contrast, conducive soils, which are more susceptible to disturbances and prone to pathogen colonization, pose a higher risk of ARG. Therefore, we hypothesize that the high suppression induced by organic management is a promising agricultural mode in plant-associated disease control.
Antimicrobial resistomes in the soil can be absorbed by plants and then migrate into the food chain, serving as an additional risk to human health [4]. Therefore, reducing the amount of ARGs in agricultural soil, especially in plant-associated soil, is particularly important for human health. We propose a conceptual model for ARG spread following biotic stress based on our results. First, the functional microorganisms in suppressive soil limit the migration and colonization of R. solanacearum and directly inhibit the horizontal and vertical transfer of its ARGs; second, the resident response of the ARG-poor microbiome reduces the space of microorganisms with a high ARG microbiome, thus indirectly limiting the abundance of ARGs in indigenous communities, and reducing the risk to human health (Fig. 3).
Fig. 3.
Conceptual model of ARG spread under biotic disturbance caused by pathogen invasions.
The results of this study highlight the intricate yet unknown dynamics of the soil ARG spread mode that is influenced by the rhizosphere microbiome with different suppressiveness levels. We postulate that the invading pathogen leads to a shift in microbiome composition and activation of ARG-rich functional taxa to restrict biotic stress and inhibit pathogen propagation. Specific ARG-poor taxa that respond to pathogen invasions in suppressive soil may be a promising solution for this scenario. Notably, more work in soil-borne disease control should consider ARGs when estimating the “one health” contribution to humans in future studies.
Ethics statement
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required for this specific study.
Funding statement
This work was supported by the Natural Science Foundation of Jiangsu Province, China (BK20200562), the Fundamental Research Funds for the Central Universities (XUEKEN2022004), the National Natural Science Foundation of China (42090065, 32002132, 41977044), the Guidance Foundation, the Hainan Institute of Nanjing Agricultural University (NAUSY-MS10), the China Postdoctoral Science Foundation (2020M671520), and the Priority Academic Program Development of the Jiangsu Higher Education Institutions (PAPD).
CRediT authorship contribution statement
Yuchan Li: Visualization, Methodology, Validation, Writing – original draft. Xuhui Deng: Conceptualization, Methodology, Writing – original draft, Writing – review & editing, Funding acquisition. Na Zhang: Methodology, Investigation, Data curation. Zongzhuan Shen: Methodology, Writing – review & editing. Rong Li: Conceptualization, Writing – review & editing, Funding acquisition. Qirong Shen: Conceptualization, Funding acquisition. Joana Falcao Salles: Writing – review & editing.
Declaration of Competing Interest
The authors have no conflicting interests related to this manuscript.
Acknowledgments
Not available.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.onehlt.2023.100481.
Contributor Information
Xuhui Deng, Email: dengxuhui@njau.edu.cn.
Rong Li, Email: lirong@njau.edu.cn.
Appendix A. Supplementary data
Supplementary material
Data availability
The raw sequence data for the metagenomics of all samples were submitted to the NCBI Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/) with the accession number SRP319842.
References
- 1.Dcosta V.M., King C.E., Kalan L., Morar M., Sung W.W.L., Schwarz C., Froese D., Zazula G., Calmels F., Debruyne R., Golding G.B., Poinar H.N., Wright G.D. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 2.Allen H.K., Donato J., Wang H.H., Cloud-Hansen K.A., Davies J., Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 3.Pehrsson E.C., Tsukayama P., Patel S., Mejía-Bautista M., Sosa-Soto G., Navarrete K.M., Calderon M., Cabrera L., Hoyos-Arango W., Bertoli M.T., Berg D.E., Gilman R.H., Dantas G. Interconnected microbiomes and resistomes in low-income human habitats. Nature. 2016;533:212–216. doi: 10.1038/nature17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Q.L., Cui H.L., Su J.Q., Penuelas J., Zhu Y.G. Antibiotic resistomes in plant microbiomes. Trends Plant Sci. 2019;24:530–541. doi: 10.1016/j.tplants.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Zhu Y.G., Zhao Y., Zhu D., Gillings M., Penuelas J., Ok Y.S., Capon A., Banwart S. Soil biota, antimicrobial resistance and planetary health. Environ. Int. 2019;131 doi: 10.1016/j.envint.2019.105059. [DOI] [PubMed] [Google Scholar]
- 6.Berglund B., Fick J., Lindgren P.E. Urban wastewater effluent increases antibiotic resistance gene concentrations in a receiving northern European river. Environ. Toxicol. Chem. 2015;34:192–196. doi: 10.1002/etc.2784. [DOI] [PubMed] [Google Scholar]
- 7.Su J.Q., Wei B., Ou-Yang W.Y., Huang F.Y., Zhao Y., Xu H.J., Zhu Y.G. Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ. Sci. Technol. 2015;49:7356–7363. doi: 10.1021/acs.est.5b01012. [DOI] [PubMed] [Google Scholar]
- 8.Dungan R.S., Strausbaugh C.A., Leytem A.B. Survey of selected antibiotic resistance genes in agricultural and non-agricultural soils in south-Central Idaho. FEMS Microbiol. Ecol. 2019;95:71. doi: 10.1093/femsec/fiz071. [DOI] [PubMed] [Google Scholar]
- 9.Xiong W., Zhao Q., Zhao J., Xun W., Li R., Zhang R., Wu H., Shen Q. Different continuous cropping spans significantly affect microbial community membership and structure in a Vanilla-grown soil as revealed by deep pyrosequencing. Microb. Ecol. 2015;70:209–218. doi: 10.1007/s00248-014-0516-0. [DOI] [PubMed] [Google Scholar]
- 10.Sun S., Lu C., Liu J., Williams M.A., Yang Z., Gao Y., Hu X. Antibiotic resistance gene abundance and bacterial community structure in soils altered by ammonium and nitrate concentrations. Soil Biol. Biochem. 2020;149 doi: 10.1016/j.soilbio.2020.107965. [DOI] [Google Scholar]
- 11.Fang H., Wang H., Cai L., Yu Y. Prevalence of antibiotic resistance genes and bacterial pathogens in long-term manured greenhouse soils as revealed by metagenomic survey. Environ. Sci. Technol. 2015;49:1095–1104. doi: 10.1021/es504157v. [DOI] [PubMed] [Google Scholar]
- 12.Brandl M.T. Plant lesions promote the rapid multiplication of Escherichia coli O157:H7 on postharvest lettuce. Appl. Environ. Microbiol. 2008;74:5285–5289. doi: 10.1128/AEM.01073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown D.G., Swanson J.K., Allen C. Two host-induced Ralstonia solanacearum genes, acrA and dinF, encode multidrug efflux pumps and contribute to bacterial wilt virulence. Appl. Environ. Microbiol. 2007;73:2777–2786. doi: 10.1128/AEM.00984-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng X., Zhang N., Li Y., Zhu C., Qu B., Liu H., Li R., Bai Y., Shen Q., Falcao Salles J. Bio-organic soil amendment promotes the suppression of Ralstonia solanacearum by inducing changes in the functionality and composition of rhizosphere bacterial communities. New Phytol. 2022;235:1558–1574. doi: 10.1111/nph.18221. [DOI] [PubMed] [Google Scholar]
- 15.Li X., Ding C., Zhang T., Wang X. Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 2014;72:11–18. doi: 10.1016/j.soilbio.2014.01.019. [DOI] [Google Scholar]
- 16.Haas D., Défago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005;3:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 17.Xing J., Jia X., Wang H., Ma B., Falcão Salles J., Xu J. The legacy of bacterial invasions on soil native communities. Environ. Microbiol. 2021;23:669–681. doi: 10.1111/1462-2920.15086. [DOI] [PubMed] [Google Scholar]
- 18.Chapelle E., Mendes R., Bakker P.A.H., Raaijmakers J.M. Fungal invasion of the rhizosphere microbiome. ISME J. 2016;10:265–268. doi: 10.1038/ismej.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonanomi G., Lorito M., Vinale F., Woo S.L. Organic amendments, beneficial microbes, and soil microbiota: toward a unified framework for disease suppression. Annu. Rev. Phytopathol. 2018;56:1–20. doi: 10.1146/annurev-phyto-080615-100046. [DOI] [PubMed] [Google Scholar]
- 20.Wang J., Gu J., Wang X., Song Z., Dai X., Guo H., Yu J., Zhao W., Lei L. Enhanced removal of antibiotic resistance genes and mobile genetic elements during swine manure composting inoculated with mature compost. J. Hazard. Mater. 2021;411 doi: 10.1016/j.jhazmat.2021.125135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The raw sequence data for the metagenomics of all samples were submitted to the NCBI Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/) with the accession number SRP319842.