Abstract
Growing evidence indicates that long intergenic non-coding RNAs play an important role in cancer progression by affecting gene regulation at the transcriptional and post-transcriptional levels. Recent studies have shown that lincRNA functions as a competitive endogenous RNA, which can interact with and mitigate the function of microRNA. In this study, we investigated the molecular mechanism by which LINC00162 regulates cell proliferation and apoptotic cell death. By analyzing RNA sequencing data, LINC00162 was identified to be a target of heterogeneous nuclear ribonucleoprotein K (hnRNPK). HnRNPK positively regulated LINC00162 expression through p38 mitogen-activated protein kinase. Lowering the level of either hnRNPK or LINC00162 decreased proliferation and colony formation while it increased apoptotic cell death. Small RNA sequencing followed by the ASO pulldown, revealed that LINC00162 interacts directly with miR-485–5p which exhibited tumor-suppressing effects by suppressing cell proliferation and colony formation, and increasing apoptotic cell death. Through the bioinformatic approaches, progestin and adipoQ receptor 4 (PAQR4) was selected as a common target of LINC00162 and miR-485–5p. MiR-485–5p decreased the expression of PAQR4 by directly binding to the 3’UTR of PAQR4 mRNA. Knockdown of hnRNPK and LINC00162 increased the level of functional miR-485–5p, indicating that LINC00162 may compete for miR-485–5p, thereby derepressing PAQR4 expression. Overexpression of either hnRNPK or LINC00162, or inhibition of miR-485–5p, protected cells against etoposide-induced apoptotic death. Our findings demonstrate that a regulatory paradigm implicating hnRNPK, LINC00162, miR-485–5p, and PAQR4 plays an important role in cell proliferation and apoptosis, and is a promising target for cancer therapeutics.
Keywords: LINC00162, hnRNPK, ceRNA, miR-485–5p, PAQR4
1 ǀ. INTRODUCTION
Long non-coding RNAs (lncRNAs) are single-strand RNA transcripts, longer than 200 nucleotides in length, transcribed by RNA polymerase II. Like mRNAs, lncRNAs are 5’ capped, 3’ polyadenylated, and spliced, but do not generally code for protein (Guttman et al., 2009; Iyer et al., 2015). According to their location relative to coding genes, lncRNAs are divided into 5 groups: sense, antisense, bidirectional, intronic, and intergenic lncRNA. Due to their structural diversity, lncRNAs can affect gene expression on many levels, including chromatin remodeling, transcription, translation, and splicing (Schmitz, Grote, & Herrmann, 2016). By influencing the expression of subsets of proteins, lncRNAs participate in many cellular processes, including proliferation, migration, invasion, differentiation, and apoptosis (Aprile, Katopodi, Leucci, & Costa, 2020; M. C. Jiang, Ni, Cui, Wang, & Zhuo, 2019; Panda, Abdelmohsen, & Gorospe, 2017; Schmitt & Chang, 2016).
Accumulating evidence indicates that lncRNAs are expressed abnormally in various cancer cells and exert both oncogenic and tumor-suppressing effects through epigenetic, transcriptional, and post-transcriptional regulation (Deng, Wang, Guo, & Xia, 2016). The cytoplasmic lncRNA mainly functions as a competitive endogenous RNA (ceRNA) of microRNA (miRNA) (Salmena, Poliseno, Tay, Kats, & Pandolfi, 2011). Like many mRNAs, lncRNAs may harbor more than one miRNA recognition elements (MREs) in their sequences and might mitigate the inhibitory function of miRNAs and enhance the expression of subsets of mRNAs. Many lncRNA/miRNA/mRNA networks have been found to regulate cancer cell growth and apoptosis. Examples include lncRNA CCAT2/miR-424/VEGFA (S. L. Sun, Shu, & Tao, 2020), lncRNA XIST/miR-126/IRS1 (Cheng, Luo, & Guo, 2020), lncRNA NEAT1/miR-185–5p/DNMT1 (Yu, Xu, Wu, Wang, & Chen, 2021), lncRNA DLEU1/miR-421/MEF2D (Feng, He, Rao, Diao, & Zhu, 2019), lncRNA PVT1/miR-543/SERPINI1 (Qu, Dai, Guo, Qin, & Liu, 2020), lncRNA UCA1/miR-143/FOSL2 (X. Chen et al., 2020), and LINC00152/miR-612/AKT2 (Cai et al., 2018).
We have investigated the molecular mechanism by which heterogeneous nuclear ribonucleoprotein K (hnRNPK) promotes the malignant properties of cancer. In previous report, two intergenic non-coding RNAs (lincRNAs) were identified as the target of hnRNPK (Lee et al., 2021). LINC00263 strengthens the proliferative and metastatic abilities by sponging miR-148a, thus increasing the expression of calpain 2. We studied here the molecular function of the other lincRNA, LINC00162 which is the most upregulated lincRNA in cutaneous squamous cancer cells, in comparison to normal human epidermal keratinocyte (Luo, Morgan, & Wang, 2016; Piipponen et al., 2016). Since p38 mitogen-activated protein kinase (MAPK) has a significant impact on the expression of LINC00162, it is also known as p38 inhibited cutaneous squamous cell carcinoma associated lincRNA (PICSAR). This study investigates the molecular mechanism by which LINC00162 controls cell proliferation and apoptosis.
2 ǀ. MATERIALS AND METHODS
2.1 ǀ. Cell culture
HeLa, HEK293, U2OS, Caki, 786-O, Beas-2B cells were maintained in DMEM (Hyclone) and H1299, H322 cells were maintained in RPMI (Gibco-BRL). Media were supplemented with 10% FBS (GIBCO-BRL) and 1% antibiotic-antimycotic solution (GIBCO-BRL). Cells were maintained in 37°C in a humidified incubator with 5% CO2.
2.2 ǀ. Chemical treatment
For induction of apoptotic cell death, the cells were treated with etoposide (5 – 20 μM; 341205, Calbiochem) for 48 h. To determine the role of p38 MAPK on the expression of LINC00162, p38 MAPK inhibitor BIRB796 (0.5 or 1 μM; 506172, Calbiochem) was treated into HeLa cells for 24 h. Dimethyl sulfoxide (final concentration = 0.1%) was used for vehicle control.
2.3 ǀ. Plasmid construction, siRNA and miRNA synthesis
Full length LINC00162 (NR_024089.2) was amplified using pfu DNA polymerase (Bioneer) and cloned into a pcDNA3.1 vector. For TAK1 (NM_003188) and PAQR4 (NM_152341) overexpression, the human tagged ORF clone (RC204454 and RC207628, respectively) were purchased (Origene). Small interfering RNAs (siRNAs) directed at HNRNPK mRNA, LINC00162, PAQR4 mRNA, and TAK1 mRNA were synthesized (Bioneer). The sequences of siRNAs are listed in Supplementary table 1. For the overexpression and inhibition of miRNA, a miR-485–5p precursor (PM10837, Ambion) and inhibitor (AM10837, Ambion) were purchased respectively.
2.4 ǀ. siRNA, miRNA and vector transfection
For siRNA and miRNA transfection, an equal number of cells (3.5 × 105 cells/60 mm dish) were plated for overnight and total 100 nM of siRNAs or miRNAs were introduced into cells. For plasmid, designated concentrations of vectors were introduced into cells an equal number of cells (6.0 × 105 cells/60 mm dish). All transfections were performed using Lipofectamine2000 (Invitrogen) in accordance with manufacturers instruction.
2.5 ǀ. Western blot analysis
Transfected cells were lysed in RIPA buffer (10 mM Tris-HCl pH 7.4, 1% NP-40, 1 mM EDTA, 0.1% SDS, 150 mM NaCl) containing complete protease and phosphatase inhibitors (Roche). Equal amounts of protein were separated by SDS–polyacrylamide gel electrophoresis at 80 V for 2.5 h. Gels were transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA) at 15 V for 1.25 h. After blocking with 5% skim milk for 1 h at RT, membranes were incubated with the indicated primary antibody (Supplementary table 3) for overnight at 4°C. Next day, the membranes were washed with TBS-T buffer 3 times for 10 min and incubated with the appropriate secondary antibody (1:10,000) for 1 h at RT. Protein bands were visualized using an enhanced chemiluminescent reagent (1859697, Thermo Scientific). GAPDH was detected as a loading control.
2.6 ǀ. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted using the TRIzol reagent (Invitrogen), in accordance with the manufacturer’s protocol. 1ug of RNAs were used for reverse transcription using the SuperScript III First-Strand Synthesis System (Invitrogen). The level of mRNA was determined by RT-qPCR, using Power SYBR Green PCR Master Mix (Applied Biosystems) and normalized by the level of GAPDH mRNA or 18S RNA. Primer sequences used in this study are listed in Supplementary table 3.
2.7 ǀ. WST-1 Cell viability assay
After 24 h post-transfection, cells (1 × 103 cells) were seeded into each well of 96-well plates. After incubation overnight, the culture media were replaced with 100 μl fresh media, containing 1/10 diluted WST-1 reagent (Takara, Japan) and incubated at 37°C for 30 min. Cell viability was determined by measuring absorbance at 450 mm.
2.8 ǀ. Cell Proliferation and colony forming assay
To access cell proliferation ability, cells (initial cell number = 5 × 104 cells) were resuspended into 6 well plates, and the number of cells was counted every 24 h. For the colony forming assay, an equal number of transfected cells (2 × 102 cells) was seeded into 6-well plates and cell culture media were replaced into fresh in every 3 days. After incubation for 2 weeks, the cell colonies were fixed with 4% paraformaldehyde for 10 min and stained with 0.2% crystal violet for 1.5 h at RT. The clonogenic ability of cells were determined by counting the number of colonies using the Image J program.
2.9 ǀ. Antisense oligonucleotide (ASO) pull-down assay
To search for LINC00162-associated miRNAs, ASO pull-down was performed using non-overlapping biotinylated ASOs recognizing LacZ (four ASOs) and LINC00162 (seven ASOs) (Supplementary figure S4A). Incubation of the whole cell lysates with the biotinylated ASO was followed by coupling with Streptavidin-coupled Dynabeads™ (Invitrogen). RNAs were isolated from the pull-down materials and small RNA sequencing was performed.
2.10 ǀ. Ribonucleoprotein immunoprecipitation (RNP-IP or RIP)
RNP-IP was performed to check the direct interaction between target mRNA and protein. Argonaute-2 (Ago2) antibody was used to check the interaction between target mRNA and miRNA, and hnRNPK antibody was used to check the interaction between hnRNPK with PAQR4 mRNA and LINC00162 (Supplementary table 4). IgG antibody was used in control IP reactions. 120 μg of Dynabeads Protein G (Invitrogen) was coated with 2 μg of each antibody for 24 h at 4°C. Coated beads were incubated with 500 μg of cytoplasmic lysates, which were extracted with polysome extraction buffer (20 mM Tris-HCl pH 7.4, 0.5% NP-40, 100 mM KCl, 5 mM MgCl2) in 4.5 h at 4°C. Samples were washed with 800 μl of NT2 buffer (50 mM Tris-HCl pH 7.4, 0.05% NP-40, 1 mM MgCl2, 150 mM NaCl) for 6 times. Washed samples were Immunoprecipitated samples and treated with 1 U of Turbo DNase I (Invitrogen) at 37°C for 20 min and with 100 μg of protease K at 55°C (Invitrogen) for 20 min. After the RNAs were isolated from antibodies, the levels of lincRNA and target mRNA were determined by RT-qPCR analysis.
2.11 ǀ. Luciferase reporter assay
To examine whether miR-485–5p binds directly to LINC00162 or the 3’UTR of PAQR4 mRNA, luciferase vectors (pmirGLO dual-luciferase vectors; Promega) containing wild-type or mutant sequences of miR-485–5p MREs were constructed. The structures of vectors are presented in Supplementary figure S5 and S8. Following transfection with control miRNA or pre-miR-485–5p, cells were counted and the equal number of (cell number = 5 × 104 cells) cells were inoculated into 24 well plates. Next day, cells were transfected with 100 ng of either wild-type or mutant luciferase vectors. At 24 h post-transfection, luciferase activity was determined using a Dual-GLO™ Luciferase Assay System (Promega).
2.12 ǀ. Cell Fractionation
The subcellular localization of LINC00162 was assessed by fractionation. The cytosolic fraction was obtained using RSB buffer (10 mM Tris-HCl, pH 7.4, 2.5 mM MgCl2, 100 mM NaCl), containing 4 mg/mL digitonin (ThermoFisher Scientific, USA). The nuclear pellets were washed three times with RSB buffer containing digitonin and lysed with RIPA buffer. The levels of LINC00162 in each fraction were determined by RT-qPCR analysis.
2.13 ǀ. Apoptosis analysis using 2D FACS
Following transfection with siRNAs or vectors, HeLa cells were harvested and suspended with 1× Annexin V binding buffer (BD Pharmigen). An equal number of cells (1 × 105 cells) were stained with 15 μl of a mixture of FITC Annexin V and propidium iodide (556547, BD Pharmingen) for 15 min at RT in the dark. The stained cells were analyzed using a flow cytometer (FACS Verse, BD). The degree of apoptosis was calculated by measuring the proportion of late apoptotic cells.
3 ǀ. RESULTS
3.1 ǀ. Expression of LINC00162 is positively regulated by hnRNPK
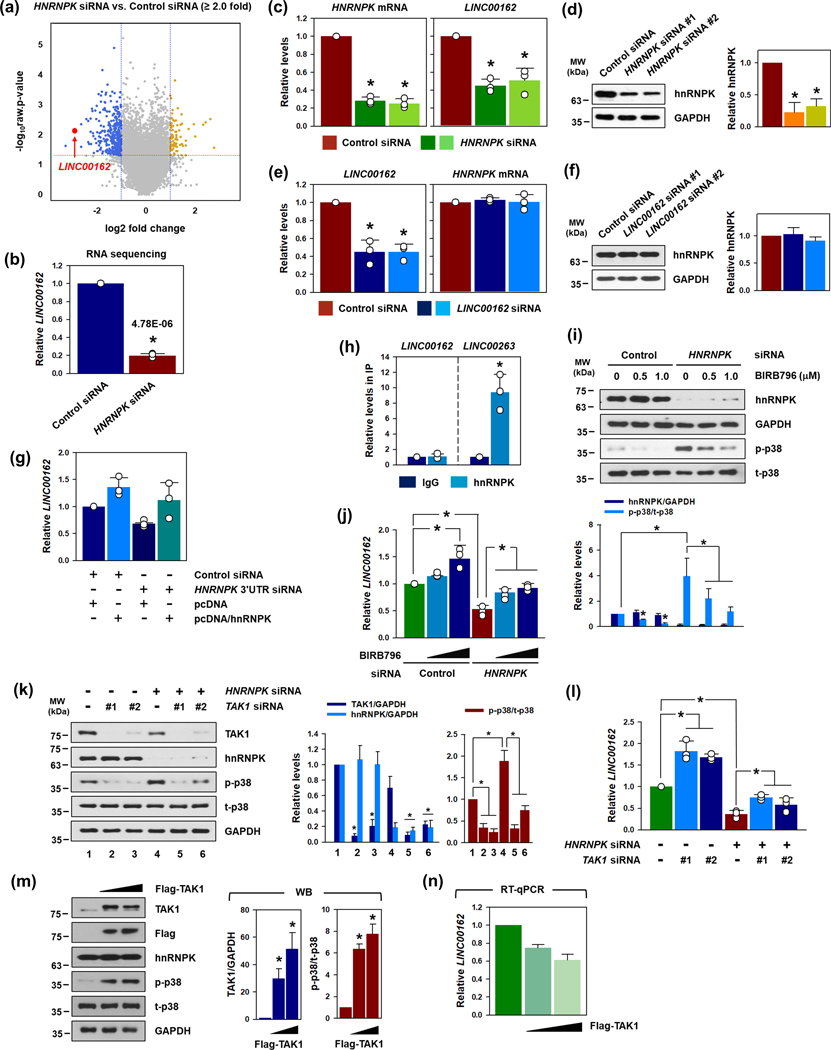
To search for hnRNPK-regulated lincRNAs, RNA sequencing was performed using total RNA isolated from HeLa cells, transfected with either control or hnRNPK siRNA.(Lee et al., 2021) An analysis of the RNA sequencing data (Supplementary figure S1a–S1c) found that the expression of LINC00162 was reduced in hnRNPK-silenced HeLa cells (Figure 1a and 1b). Next, we directly investigated whether hnRNPK reduced the level of LINC00162. The knockdown of hnRNPK by two independent siRNAs efficiently decreased the level of hnRNPK protein and mRNA, inducing a reduction in LINC00162 (Figure 1c and 1d). Conversely, LINC00162-silenced HeLa cells showed a significant decrease in LINC00162, with no change in the hnRNPK protein or mRNA (Figure 1e and 1f). The rescue experiment revealed that knockdown of hnRNPK by a specific siRNA, targeting the 3’UTR of HNRNPK mRNA (Shin et al., 2017), induced a decrease in LINC00162; ectopic hnRNPK restored the expression level of LINC00162 (Figure 1g).
Figure 1. LINC00162 is a hnRNPK-regulated lincRNA.
(a, b) To search for hnRNPK-regulated lincRNA, RNA sequencing was conducted, using total RNA isolated from the control or HNRNPK siRNA. A volcano plot was generated from these data, and LINC00162 was highlighted. (c, d) To verify the RNA sequencing results, HeLa cells were transfected with either the control or HNRNPK siRNAs. The levels of HNRNPK mRNA and LINC00162 were determined by RT-qPCR analysis (c). Western blot analysis was used to check the decrease in hnRNPK protein expression (d). (e-g) To determine whether LINC00162 affected hnRNPK expression levels, HeLa cells were transfected with either the control or two independent LINC00162-targeting siRNAs. The levels of LINC00162 and HNRNPK mRNA were determined by RT-qPCR analysis (e) and the level of hnRNPK protein was checked by Western blot analysis (f). The regulation of LINC00162 by hnRNPK was confirmed through a rescue experiment using the 3’UTR-targeting siRNA and the overexpressing vector of hnRNPK (g). (h) To examine the direct interaction of LINC00162 with hnRNPK, an RIP experiment was conducted using control IgG or hnRNPK antibodies. The levels of LINC00162 and LINC00263 enriched in each IP sample were determined by RT-qPCR analysis. (i, j) HeLa cells were transfected with either the control or HNRNPK siRNA and then treated with various concentrations of BIRB796 (a p38 MAPK inhibitor) for 24 h. The levels of hnRNPK, phospho- and total-p38 MAPK were determined by Western blot analysis (i). Under the conditions above, the level of LINC00162 was determined by RT-qPCR (j). (k) To investigate the role of TAK1 in the activation of p38 MAPK by hnRNPK knockdown, HeLa cells were transfected with HNRNPK siRNA and/or two independent TAK1 siRNAs. The levels of TAK1, hnRNPK, and total/phosphorylated p38 were determined by Western blot. GAPDH was used for loading control. (l) In above condition, the level of LINC00162 was assessed using RT-qPCR. (m, n) The effect of TAK1 overexpression on p38 activation and LINC00162 expression was tested. After 48 h incubation post-transfection of TAK1 overexpression vector, the activation of p38 was examined by Western bot (m). Under the same conditions, the level of LINC00162 was determined by RT-qPCR (n). Western blot data were analyzed using Image J program and student’s t-test, with three independent experiments (* p < 0.05), was used to calculate statistical significance. All data represent mean ± standard deviation (SD).
To define the mechanism by which hnRNPK regulates LINC00162 abundance, direct interactions between hnRNPK and LINC00162 were tested. RIP (RNP-IP) analysis revealed that hnRNPK bound to LINC00263 which is a known hnRNPK-associated lincRNA, but not to LINC00162, suggesting that hnRNPK affects LINC00162 indirectly (Figure 1h). Since the activation of p38 MAPK is known to negatively regulate the expression of LINC00162 (Piipponen et al., 2016), we examined whether the knockdown of hnRNPK influenced p38 MAPK activation. A phospho-kinase proteome profiler experiment confirmed that the knockdown of hnRNPK by two independent siRNAs activated p38 MAPK (Supplementary figure S2a–S2c). The effect of the p38 MAPK inhibitor (BIRB796) was analyzed to confirm that p38 MAPK was involved in regulating LINC00162 levels by hnRNPK. Treating HeLa cells with BIRB796 efficiently decreased the level of phosphorylated p38 MAPK (Figure 1i); inactivating p38 MAPK increased the expression of LINC00162 in both the control and hnRNPK-silenced cells (Figure 1j). Since hnRNPK suppress translationally the expression of transforming growth factor-β-activated kinase 1 (TAK1) (Liepelt et al., 2014), we examined whether TAK1 is involved in the activation of p38 MAPK by knockdown of hnRNPK. The expression of TAK1 was increased in hnRNPK-silenced cells and the knockdown of TAK1 by two independent siRNAs inhibited p38 MAPK activation (Figure 1k). Accordingly, the knockdown of TAK1 resulted in the increase in LINC00162 expression and restored decreased expression of LINC00162 by knockdown of hnRNPK (Figure 1l). On the contrary, the ectopic expression of TAK1 induced the activation of p38 MAPK (Figure 1m). Activation of p38 MAPK by TAK1 overexpression decreased the expression of LINC00162 in a dose-dependent manner (Figure 1n). These results indicate that the knockdown of hnRNPK increased TAK1 expression, leading to the activation of p38. Conclusively, hnRNPK regulates LINC00162 through the TAK1/p38 MAPK-driven processes.
3.2 ǀ. LINC00162 regulates cell proliferation and apoptosis
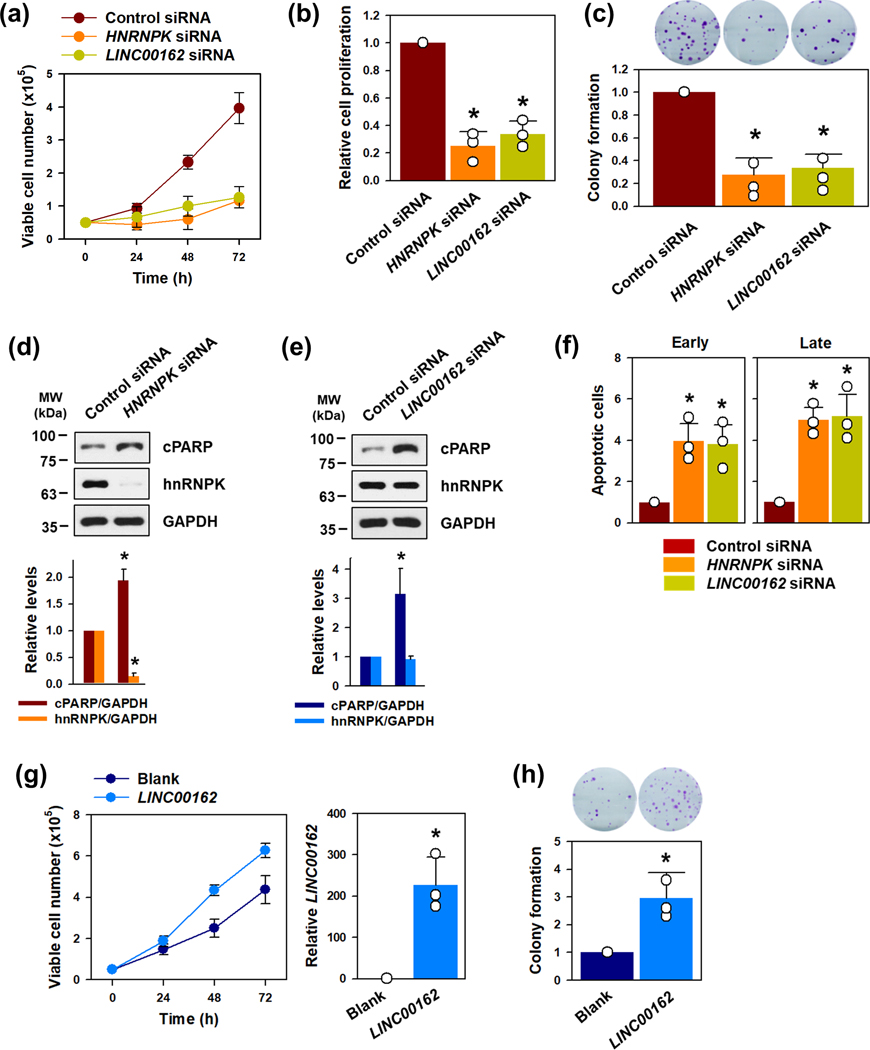
Cell counting, a viability assay, and a colony-forming assay were used to examine the effect of LINC00162 on cell proliferation and apoptosis. The knockdown of either hnRNPK or LINC00162 diminished the cell proliferation rate (Figure 2a and 2b) and inhibited the ability to form colonies (Figure 2c). Moreover, increases in cleaved PARP and in the number of early and late apoptotic cells were observed in cells in which either hnRNPK or LINC00162 were silenced (Figure 2d, 2e, and 2f). The effect of inhibiting cell growth and inducing apoptotic cell death by knockdown of hnRNPK or LINC00162 was verified using two independent siRNAs (Supplementary figure S3a–S3d). Apoptotic cell numbers were determined by a FACS analysis; representative results are shown in Supplementary figure S3e. To test whether LINC00162 accelerates proliferation rate, overexpression vector was manufactured. Introduction of overexpression vector into HeLa cells showed more 150-fold increase in LINC00162. In contrast to the results observed in LINC00162-silenced cells, cell proliferation and colony formation were enhanced by the overexpression of LINC00162 (Figure 2g and 2h, respectively).
Figure 2. LINC00162 regulates cell proliferation and apoptosis.
(a-c) To investigate the role of hnRNPK-regulated LINC00162 in cell proliferation, HeLa cells were transfected with control, hnRNPK, or LINC00162 siRNAs. Cell proliferation was determined by counting the viable cells (a), a WST-1 assay (b), and a colony-forming assay (c). (d-f) Apoptotic cell death by knockdown of hnRNPK or LINC00162 was assessed by checking the level of cleaved PARP (d and e, respectively), and FACS analysis was used to assess the proportion of apoptotic cell death (f). The populations of early and late apoptotic cells were calculated from three independent experiments. (g, h) To examine whether LINC00162 enhanced cell proliferation, cells were transfected with a blank or overexpression vector, the proliferative ability was determined by counting the number of viable cells (g) and colonies (h). Western blot data were analyzed using Image J program and student’s t-test was used to calculate statistical significance through three independent experiments (* p < 0.05). All data represent mean ± standard deviation (SD).
3.3 ǀ. LINC00162 sponges miR-485–5p to regulate cell proliferation and apoptosis
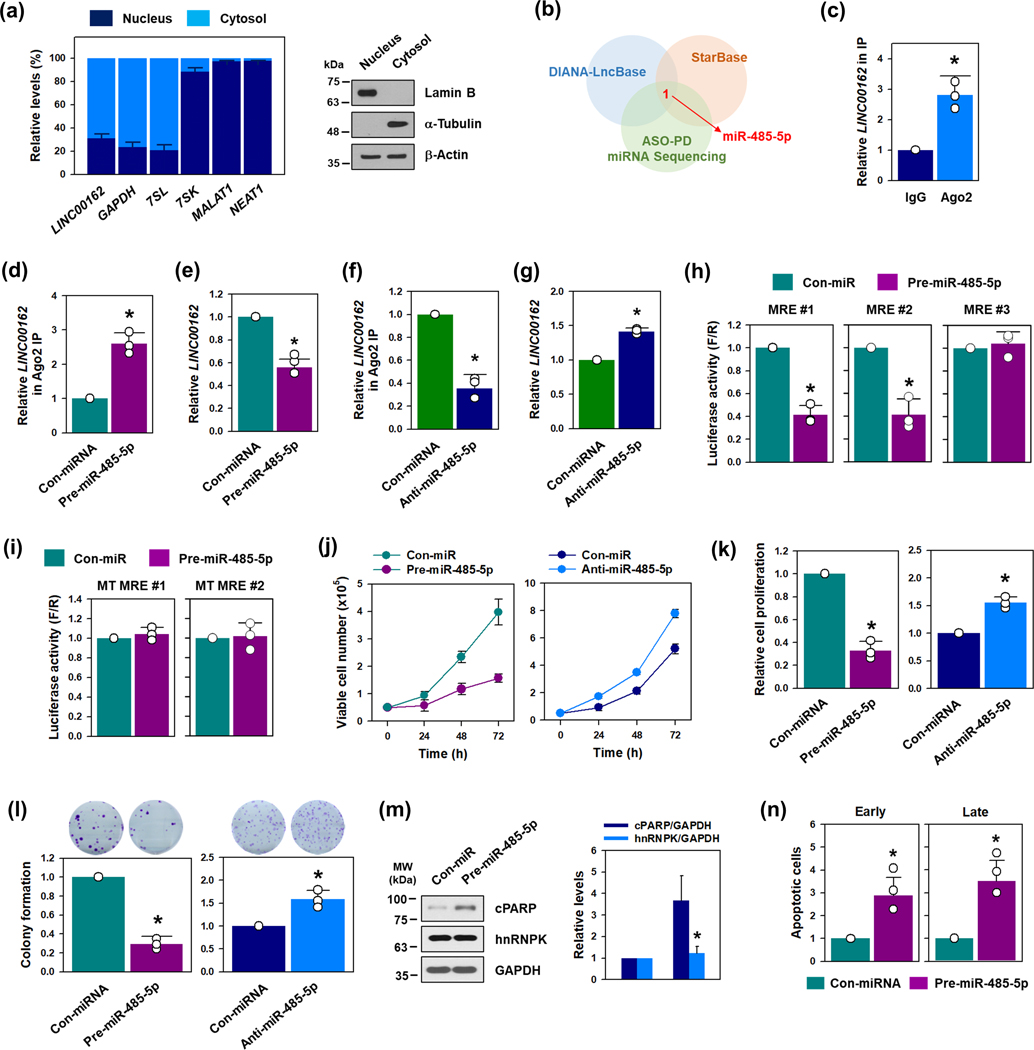
To investigate the action mechanism of LINC00162, the cellular localization of LINC00162 was examined. Whereas lncRNAs NEAT1 and MALAT1 are more abundant in the nucleus, LINC00162 is mainly located in the cytosol, suggesting that LINC00162 may influence the function of miRNAs, as do other cytoplasmic lincRNAs (Figure 3a). An antisense oligonucleotide (ASO) pulldown was conducted to search for LINC00162-bound miRNAs (Supplementary figure S4a). LINC00162-associated miRNAs were screened through miRNA sequencing, using RNA obtained from the ASO pulldown. An analysis of sequencing data found that 24 miRNAs were more enriched in pulldown material when LINC00162 ASO was used than when LacZ ASO was used (Supplementary figure S4b). A comparison of lists of miRNAs predicted to associate with LINC00162 by DIANA-LncBase (www.microrna.gr/LncBase) and StarBase (http://starbase.sysu.edu.ca), and selected through an analysis of miRNA sequencing data, identified only one miRNA, miR-485–5p, at the intersection of these miRNA subsets (Figure 3b). Moreover, there were three miR-485–5p MREs in the sequence of LINC00162 (Supplementary figure S5a). RT-qPCR analysis confirmed that LINC00162 was associated with Ago2 by RIP analysis, supporting the notion that LINC00162 associated with the miRNA-induced silencing complex (miRISC) (Figure 3c). Overexpression of miR-485–5p by transfecting a precursor miRNA increased the enrichment of LINC00162 in miRISC and thus reduced LINC00162 abundance (Figure 3d and 3e). Conversely, inhibition of miR-485–5p by transfecting an antisense miRNA reduced the interaction of Ago2 with LINC00162 and increased LINC00162 abundance in HeLa cells (Figure 3f and 3g). Although reporter luciferase analysis is an artificial tool, since LINC00162 is not a 3′UTR sequence, luciferase vectors derived from pmirGLO containing the three MREs of miR-485–5p were created to test the impact of the miR-485–5p MREs on LINC00162 levels (Supplementary figure S5b). Of the three MREs, only two luciferase activity containing MRE#1 and #2 appear to be functional (Figure 3h). Furthermore, the mutation of the seed sequence in MRE#1 and #2 blocked the inhibitory effect of miR-485–5p (Figure 3i). Based on these results, we propose that miR-485–5p binds to LINC00162 MREs #1 and #2 and lowers LINC00162 levels.
Figure 3. LINC00162 functions as the ceRNA of miR-485–5p.
(a) A cellular fractionation assay was used to assess the localization of LINC00162. The levels of LINC00162, NEAT1, MALAT1, 18S, and GAPDH mRNA in each fraction were determined by RT-qPCR. LncRNA NEAT1 and MALAT1 were checked with reference to lncRNAs, which are known to be preferentially expressed in the nucleus. (b) LINC00162-associated miRNAs were screened by comparing the miRNA list obtained from the small RNA sequencing followed by ASO-PD, and predicted by two independent algorithms. (c) The enrichment of LINC00162 in miRISC was assessed by determining its level in the control IgG and Ago2 IP materials. The level of 18S was used for normalization. (d-g) The effect of miR-485–5p on the expression of LINC00162 was examined using Ago2 RNP-IP (d and f) and RT-qPCR analysis (e and g). HeLa cells were transfected with either pre-miR-485–5p (for overexpression: d and e) or anti-miR-485–5p (for inhibition: f and g). The level of LINC00162 in IP materials or the total RNA was determined by RT-qPCR analysis. (h, i) A bioinformatic prediction indicated that three miR-485–5p MREs exist in the sequence of LINC00162. Luciferase vectors containing wild-type (h) or mutant sequences (i) of miR-485–5p MREs were constructed, and luciferase activity was assessed, as described in the Materials and Methods section. (j-n) To assess the effect of miR-485–5p on proliferation and apoptosis, pre-miR-485–5p or anti-miR-485–5p was introduced into HeLa cells for overexpression or inhibition, respectively. Cell proliferation was determined by counting the number of viable cells (j), WST-1 assay (k), and colony forming assay (l). (m, n) In miR-485–5p-overexpresed cells, the apoptotic cell death was assessed by determining the level of cleaved PARP by Western blot (m) and the populations of early and late apoptotic cells were calculated by a FACS analysis (n). Western blot data were analyzed using Image J program and statistical analyses were performed using Student’s t-test in three independent experiments (* p < 0.05). All data represent mean ± standard deviation (SD).
The effect of miR-485–5p on cell proliferation and apoptosis was then assessed (Figure 3j–3l). Overexpression of miR-485–5p by pre-miRNA inhibited cell proliferation and colony formation. Contrarily, inhibition of miR-485–5p by anti-miR enhanced cell proliferation and increased the number of colonies. Moreover, the levels of cleaved PARP were increased by overexpression of miR-485–5p, with no change in hnRNPK expression levels (Figure 3m). FACS analysis revealed that miR-485–5p also increased the proportion of both early and late apoptotic cells (Figure 3n and Supplementary figure S6). Our findings suggest that miR-485–5p is tumor suppressive by lowering LINC00162 abundance.
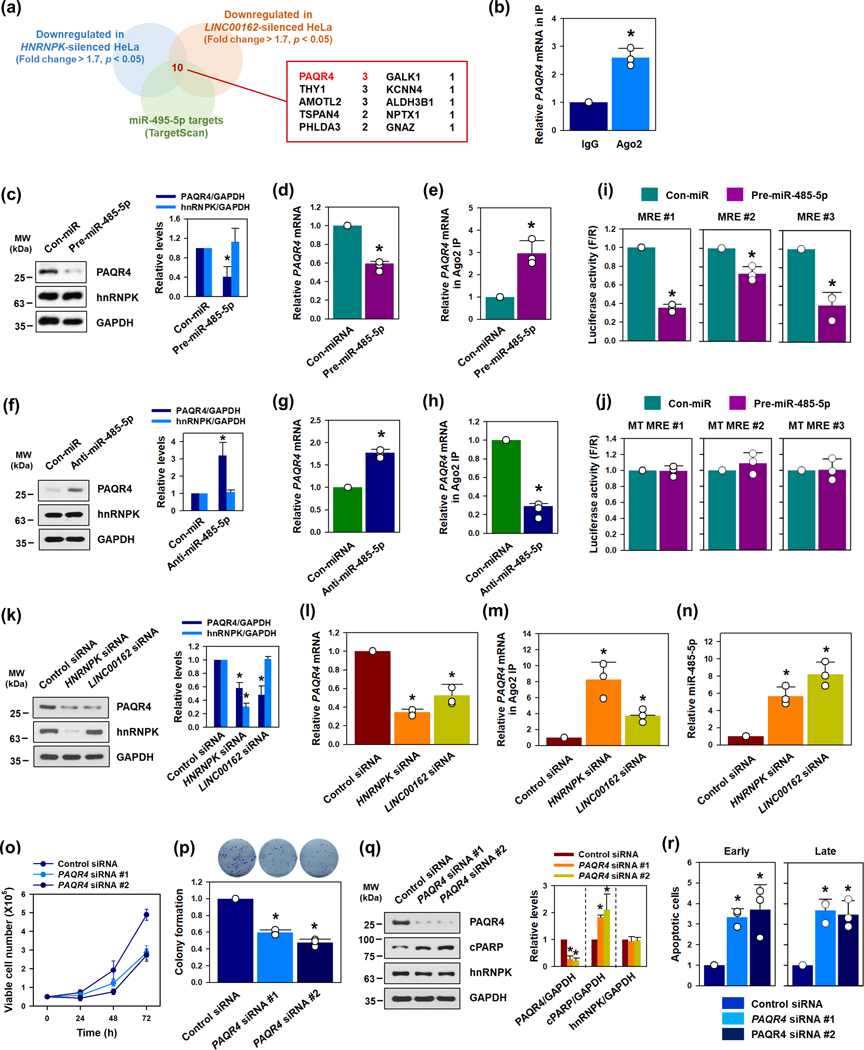
3.4 ǀ. PAQR4 is a target of hnRNPK/LINC00162/miR-485–5p
We compared the mRNAs regulated by silencing hnRNPK and by silencing LINC00162 with those mRNAs predicted to be target genes of miR-485–5p by TargetScan (Figure 4a and Supplementary figure S7a and S7b). Of these, progestin and adipoQ receptor 4 (PAQR4) mRNA was selected for further study because it showed the most significant decrease in hnRNPK- and LINC00162-silenced cells (Supplementary figure S7c), and has three MREs of miR-485–5p (Supplementary figure S8a). Compared to IgG IP, more PAQR4 mRNA was enriched in Ago2 IP, indicating that miRISC is involved in regulating PAQR4 expression (Figure 4b). To determine whether miR-485–5p suppresses PAQR4 expression, pre- and anti-miR-485–5p were used for overexpression and inhibition, respectively. Pre-miR-485–5p significantly decreased the expression level of PAQR4 protein and mRNA (Figure 4c and 4d). Conversely, the inhibition of miR-485–5p increased PAQR4 expression (Figure 4f and 4g).
Figure 4. PAQR4 mRNA is a target of miR-485–5p.
(a) Ten putative target mRNAs in the red box were screened by comparing the lists of downregulated mRNAs obtained from RNA sequencing with a list of predicted target mRNAs of miR-485–5p (TargetScan). (b) To determine whether miRISC was involved in regulating PAQR4, Ago2 RIP analysis was performed. The level of PAQR4 mRNA in IP materials was determined by RT-qPCR. (c-h) HeLa cells were transfected with either control-miR (Con-miR) or pre-miR-485–5p (for miRNA overexpression: c-e), or anti-miR-485–5p (for miRNA inhibition: f-h). The level of PAQR4 protein (c, f) and PAQR4 mRNA (d, g) were determined by Western blot and RT-qPCR analyses, respectively. The association of PAQR4 mRNA with miRISC was examined by RT-qPCR, followed by Ago2 RIP (e, h). (i, j) Bioinformatic prediction revealed three miR-485–5p MREs in the 3’UTR of PAQR4 mRNA. Luciferase vectors containing wild-type (i) or mutant sequences (j) of miR-485–5p MREs, were constructed, and luciferase activity was assessed, as described in the Materials and Methods section. (k-m) To determine whether hnRNPK and LINC00162 could regulate PAQR4, HeLa cells were transfected with either hnRNPK- or LINC00162-targeting siRNA. The levels of PAQR4 protein (k) and PAQR4 mRNA (l) were determined by Western blot and RT-qPCR, respectively. (m) The direct interaction between PAQR4 mRNA and miRISC was assessed through an RIP analysis. (n) Following the knockdown of hnRNPK and LINC00162, an RT-qPCR analysis was used to determine the level of miR-485–5p. (o-r) To assess the role of PAQR4 in cell proliferation and apoptosis, HeLa cells were transfected with control or two independent PAQR4-targeting siRNAs. Cell proliferation was determined by counting the number of viable cells (o) and a colony forming assay (p). Apoptotic cell death was assessed by determining the level of cleaved PARP through Western blot (q). The populations of early and late apoptotic cells were calculated by a FACS analysis (r). Western blot data were analyzed using Image J program and statistical analyses were performed using Student’s t-test in three independent experiments (* p < 0.05). All data represent the means ± standard deviation (SD).
The direct interaction between PAQR4 mRNA and miR-485–5p was assessed by ribonucleoprotein immunoprecipitation (RIP) analysis, using the argonaute 2 (Ago2) antibody (Supplementary figure S9c). Pre-miR-485–5p further enriched PAQR4 mRNA in Ago2 IP, suggesting that miR-485–5p is involved in regulating PAQR4 mRNA through miRISC (Figure 4e). In contrast, the enrichment of PAQR4 mRNA in miRISC was lowered by reducing the level of miR-485–5p (Figure 4h). In addition to the Ago2 RNP-IP assay, luciferase vectors containing a wild-type or mutant sequence of MRE were constructed to verify the direct binding of miR-485–5p (Supplementary figure S8b). As in the case of LINC00162, there were three MREs in the sequence of the 3’UTR of PAQR4 mRNA. The inhibition of luciferase activity by miR-485–5p was observed in all three luciferase vectors (Figure 4i). However, miR-485–5p could not suppress luciferase expression in the case of mutant vectors (Figure 4j). These results indicate that miR-485–5p suppresses PAQR4 expression by directly binding to the 3’UTR of PAQR4 mRNA.
As previously discussed, hnRNPK increases LINC00162 levels and LINC00162 levels are lowered by miR-485–5p. We thus propose that if we reduced hnRNPK or LINC00162, more miR-485–5p would remain available to repress PAQR4 production. In support of this hypothesis, PAQR4 protein and mRNA were downregulated in hnRNPK- or LINC00162-silenced cells (Figure 4k and 4l, respectively; Supplementary figure S9a and S9b). Likewise, knockdown of hnRNPK or LINC00162 also increased the level of PAQR4 mRNA in Ago2 IP (Figure 4m). Moreover, the expression level of miR-485–5p was increased in hnRNPK- and LINC00162-silenced cells (Figure 4n). RIP experiment using hnRNPK antibody indicated that PAQR4 mRNA was not directly associated with hnRNPK (Supplementary figure S9c and S9d). Next, we investigated whether PAQR4 was involved in cell proliferation and apoptosis. The knockdown of PAQR4 by two independent siRNAs retarded cell proliferation and inhibited colony forming ability (Figure 4o and 4p, respectively; Supplementary figure S9e). Interestingly, knockdown of PAQR4 slightly decreased the level of LINC00162 (Supplementary figure S9f). It was assumed that decrease in PAQR4 mRNA gives better accessibility of miR-485–5p to LINC00162. Western blot and FACS analysis indicated that PAQR4-silenced cells showed increased PARP cleavage and a higher proportion of early and late apoptotic cell death compared to the control group (Figure 4q and 4r, respectively; Supplementary figure S9g). The above results lead us to conclude that PAQR4 is positively regulated by hnRNPK and LINC00162, while it is negatively regulated by miR-485–5p; in turn, PAQR4 promotes proliferation and prevents apoptosis.
3.5 ǀ. A regulatory paradigm implicating hnRNPK, LINC00162, miR-485–5p, and PAQR4 protects cancer cells against etoposide-induced apoptotic cell death
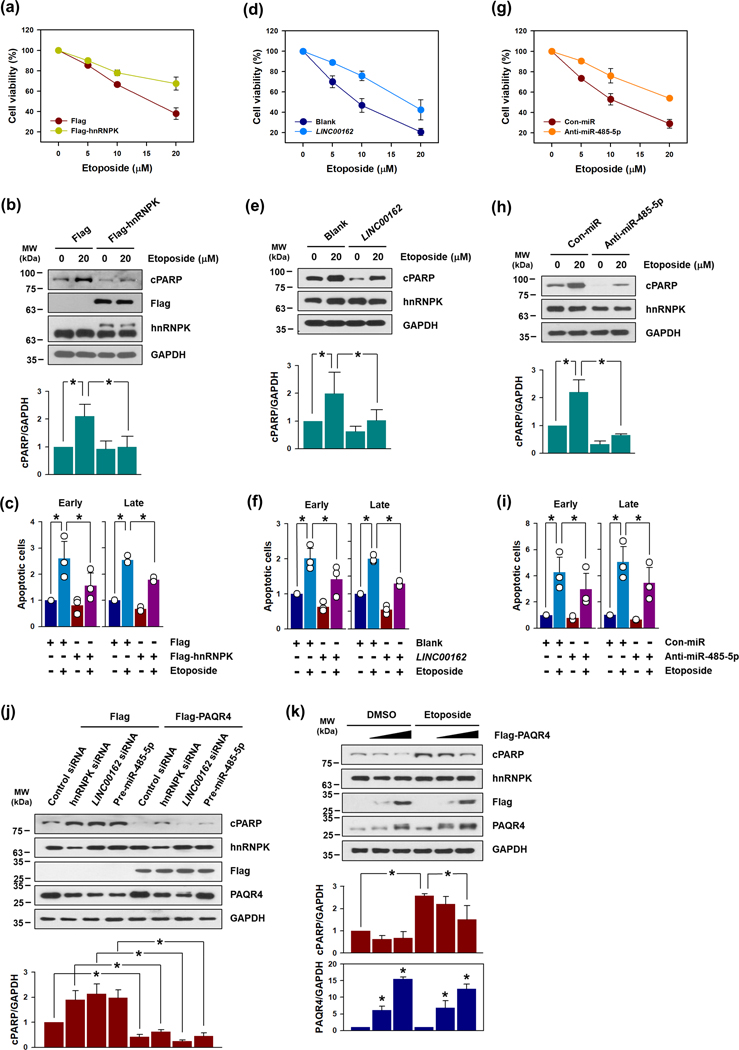
As inducers of apoptosis, DNA-damaging agents such as etoposide and cisplatin are widely used in the treatment of solid cancers.(Norbury & Zhivotovsky, 2004) As LINC00162 was found to regulate apoptotic cell death, we examined whether the levels of miR-485–5p, as regulated by hnRNPK and LINC00162 might protect HeLa cells against etoposide-induced cell death. Treatment of HeLa cells with etoposide induced cell death in a dose-dependent manner (Supplementary figure S10a). We examined the protective effects of hnRNPK or LINC00162 overexpression against etoposide-induced apoptotic cell death. Following transfection with hnRNPK- or LINC00162-overexpressing vectors, HeLa cells were treated with various concentrations of etoposide. The overexpression of hnRNPK and LINC00162 was found to relieve etoposide-induced cytotoxicity (Figures 5a and 5d, respectively). According to Western blot results (Figure 5b and 5e) and FACS analyses (Figure 5c and 5f; Supplementary figure S10b and S10c), the overexpression of either hnRNPK or LINC00162 resulted in a significant reduction in apoptotic cell death. The inhibition of miR-485–5p also lessened etoposide-induced cytotoxicity (Figure 5g) and inhibited apoptotic cell death (Figure 5h and 5i; Supplementary figure S10d). Accordingly, our findings indicate that this regulatory paradigm involving hnRNPK, LINC00162, and miR-485–5p is closely implicated in apoptotic processes and protects cells against apoptotic cell death by a DNA-damaging agent.
Figure 5. Overexpression of hnRNPK or LINC00162, and antagonization of miR-485–5p protected HeLa cells against etoposide-induced apoptotic cell death.
The anti-apoptotic effects of the overexpression of hnRNPK and LINC00162, and the inhibition of miR-485–5p were examined. (a, d, and g) Following transfection with either indicated overexpression vectors or anti-miR-485–5p, HeLa cells were treated with various concentrations of etoposide for 72 h and cell viability was determined using a WST-1 assay. (b, e, and h) After 48 h post-transfection, the cells were treated with DMSO or 20 μM etoposide for 24 h. The levels of cleaved PARP and hnRNPK were determined by Western blot analysis. GAPDH was used for loading control. (c, f, and i) The proportion of apoptotic cell death (early and late) in transfected cells was measured via a FACS analysis, as described above. (j) (k) Western blot data were analyzed using Image J program and statistical analyses were performed using Student’s t-test in three independent experiments (* p < 0.05). All data represent the means ± standard deviation (SD).
3.6 ǀ. miR-485–5p is a key molecule of the oncogenic function of LINC00162
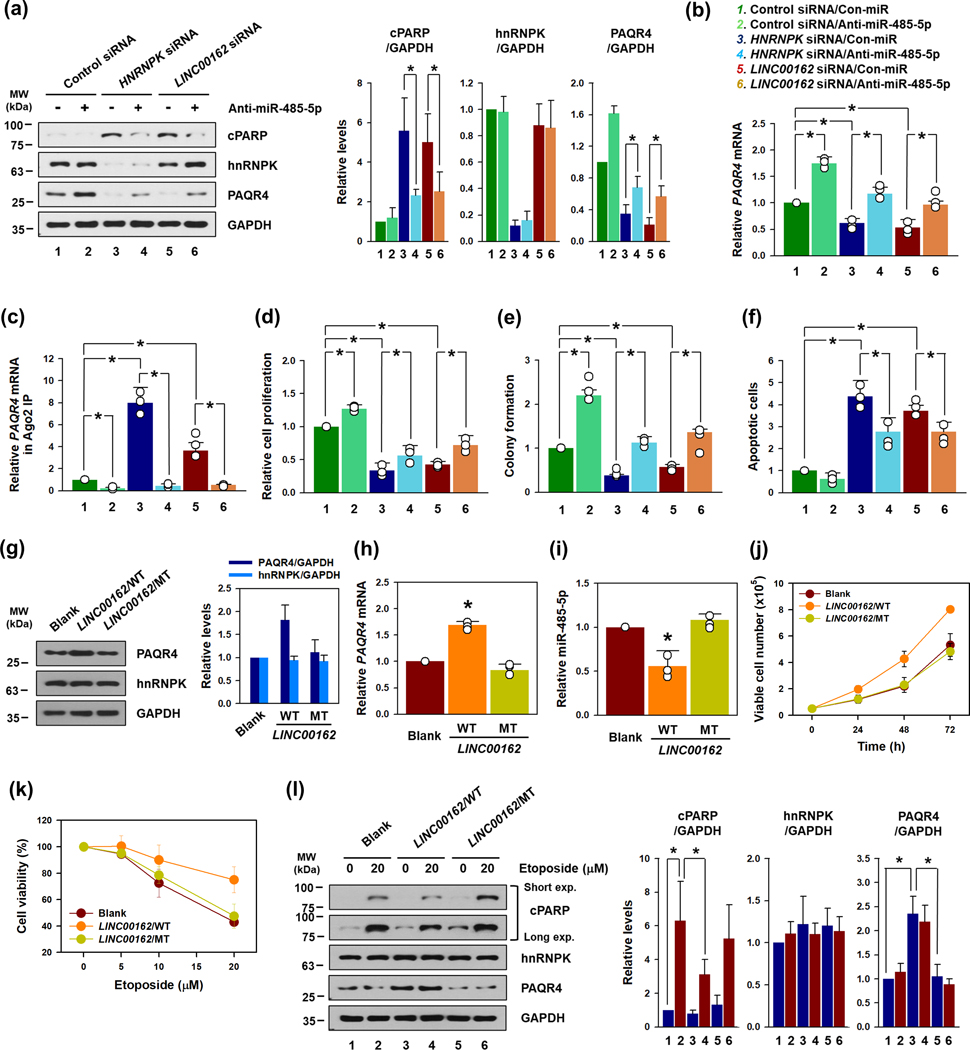
To verify that miR-485–5p is required for the oncogenic function of LINC00162, we investigated whether inhibiting miR-485–5p could reverse the effects of LINC00162. As observed earlier, the knockdown of hnRNPK or LINC00162 decreased PAQR4 expression. However, antagonization of miR-485–5p by anti-miRNA led to a recovery in the expression of PAQR4 protein (Figure 6a) and PAQR4 mRNA (Figure 6b) through a knockdown of hnRNPK or LINC00162. The Ago2 RIP data showed that the increased association of PAQR4 mRNA with miRISC in LINC00162-silenced cells was blocked by the inhibition of miR-485–5p (Figure 6c). Accordingly, PAQR4 expression, cell proliferation (Figure 6d) and colony formation (Figure 6e and Supplementary figure S11a) were recovered by inhibition of miR-485–5p. In addition, the proportion of apoptotic cells increased by the knockdown of hnRNPK and LINC00162 was reversed by anti-miR-485–5p (Figure 6f and Supplementary figure S11b).
Figure 6. Antagonizing miR-485–5p recovered cell proliferation and inhibited apoptotic cell death through a knockdown of hnRNPK and LINC00162.
To investigate whether miR-485–5p was responsible for the oncogenic functions of hnRNPK and LINC00162, siRNAs targeting hnRNPK or LINC00263 were introduced into HeLa cells with either control miRNA or anti-miR-485–5p. The levels of PAQR4 protein and PAQR4 mRNA were determined by Western blot (a) and RT-qPCR (b) analyses. (c) Ago2 RIP analysis was carried out to prove that inhibition of miR-485–5p decreased the enrichment of PAQR4 mRNA in miRISC. (d-f) In same conditions, cellular proliferation (d), colony formation (e), and apoptotic cell death (f) were also examined, as described in the Materials and Methods section. (g-j) To block miR-485–5p-decoying activity of LINC00162, we constructed mutated LINC00162 vector that could not sequester miR-485–5p. The levels of PAQR4 protein and mRNA were determined by Western blot (g) and RT-qPCR (h) analyses, respectively. (i) The effect of WT and MT LINC00162 on the expression of miR-485–4p was assessed by RT-qPCR. (j) Following the transfection of HeLa cells with either wild type (WT) or mutated (MT) LINC00162 vector, cell proliferation was examined by counting the number of viable cells. (k) Following transfection with either WT or MT LINC00162 vector, HeLa cells were treated with various concentrations of etoposide for 72 h and cell viability was determined using a WST-1 assay. (l) After 48 h post-transfection as described above, the cells were treated with DMSO or 20 μM etoposide for 24 h. The levels of cleaved PARP, hnRNPK, and PAQR4 were determined by Western blot analysis. GAPDH was used for loading control. Western blot data were analyzed using Image J program and statistical analyses were performed using Student’s t-test in three independent experiments (* p < 0.05). All data represent the means ± standard deviation (SD).
Along with inhibition of miR-485–5p, the mutated LINC00162 vector was constructed to prove that sponging miR-485–5p by LINC00162 is required for its oncogenic effects. Since miR-485–5p binds to MRE#1 and #2 of LINC00162 (Figure 3h), we constructed mutated LINC00162 to prevent miR-485–5p from binding to both MREs (Supplementary figure S12). The effects of wild type (WT) and mutated (MT) LINC00162 on the expression of PAQR4 and miR-485–5p were assessed by Western blot and RT-qPCR. As expected, the expression level of PAQR4 protein (Figure 6g) and mRNA (Figure 6h) was increased by WT LINC00162, but not by MT. In the case of WT, the expression of miR-485–5p was increased, while in the case of MT, there was no change (Figure 6i). The comparison of cell growth rates showed an increase in WT but no difference from control in MT (Figure 6j). As observed earlier, overexpression of WT LINC00162 protect cells against etoposide-induced apoptotic cell death. However, HeLa cells transfected with MT LINC00162 did not show any protective effect (Figure 6k). Western blot analysis revealed that HeLa cells transfected with WT LINC00162 showed lower amount of cleaved PARP compared to control cells. However, in case of MT LINC00162, PARP cleavage occurred similar to control cells (Figure 6l). These results further connect the effects of miR-485–5p downregulating PAQR4 and inducing apoptotic cell death with the knockdown of LINC00162.
3.7 ǀ. Overexpression of PAQR4 reversed the tumor-suppressing effects of miR-485–5p.
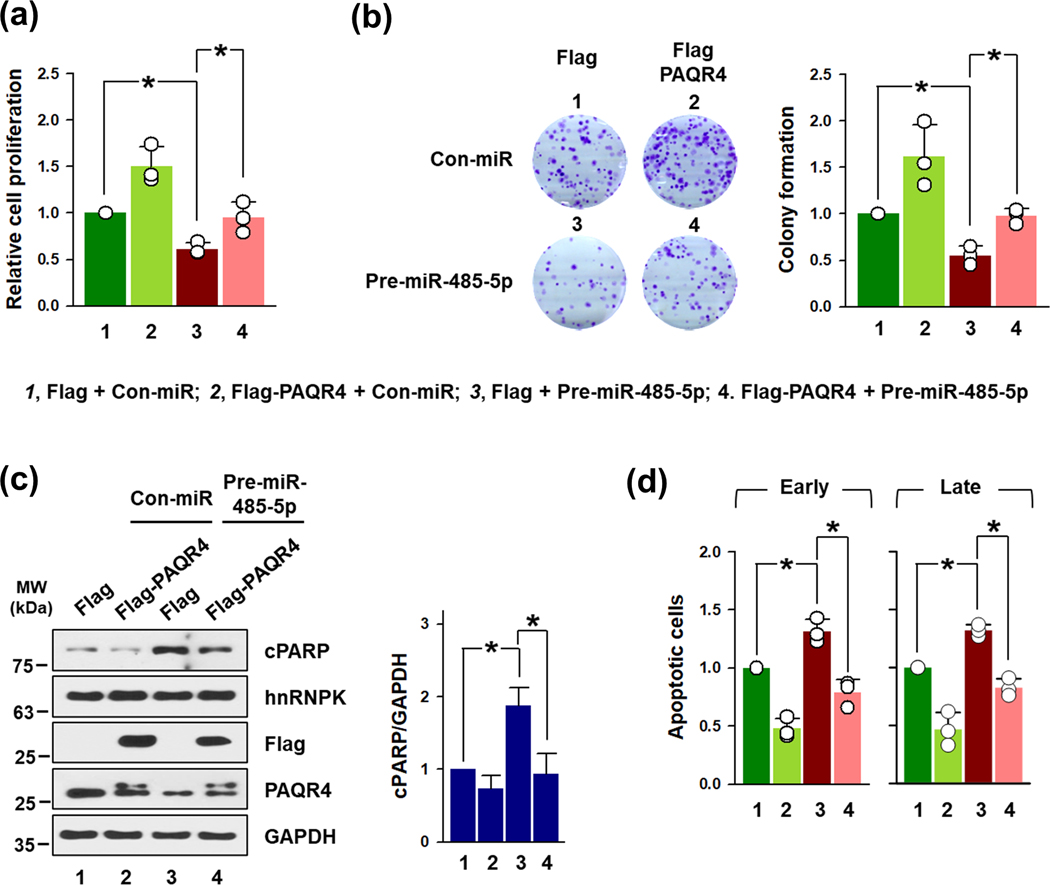
To examine that PAQR4 is involved in the tumor-suppressing effects of miR-485–5p, rescue experiments were conducted. HeLa cells were cotransfected with Flag-PAQR4 and pre-miR-485–5p and cellular proliferation was measured by WST1 assay. As observed in previous results, cellular proliferation was increased by PAQR4 overexpression and decreased by miR-485–5p (Figure 7a). In addition, PAQR4 overexpression showed the effect of recovering the reduction in cell growth by miR-485–5p. These results were observed in the same way in colon-forming assay. The decrease in number of colonies by miR-485–5p was recovered by PAQR4 expression (Figure 7b). Next, we tested whether overexpression of PAQR4 inhibits apoptotic cell death by miR-485–5p by Western blot and FACS experiments. PAQR4 overexpression lowered the increased level of cleaved PARP by miR-485–5p (Figure 7c) and also decreased the population of early and late apoptotic cells those were induced by miR-485–5p (Figure 7d). These results demonstrate that the tumor-suppressing effects of miR-485–5p is due to its suppressive effect on PAQR4.
Figure 7. PAQR4 reversed miR-485–5p-elicited growth retardation and apoptotic cell death.
To examine whether ectopic expression of PAQR4 reversed the effects of miR-485–5p, HeLa cells were cotransfected with pre-miR-485–5p and PAQR4 vector. Cell proliferation was measured by WST1 assay (a) and colony forming assay (b) as described in above. (c) The levels of cleaved PARP, hnRNPK, Flag-PAQR4, and endogenous PAQR4 were determined by Western blot analysis. GAPDH was used for loading control. Western blot data were analyzed using Image J program and statistical analyses were performed using Student’s t-test in three independent experiments (* p < 0.05). (d) The proportion of apoptotic cell death (early and late) in transfected cells was measured via a FACS analysis, as described above. All data represent the means ± standard deviation (SD).
3.8 ǀ. The oncogenic hnRNPK/LINC00162/miR-485–5p/PAQR4 axis was applicable to various types of cancer.
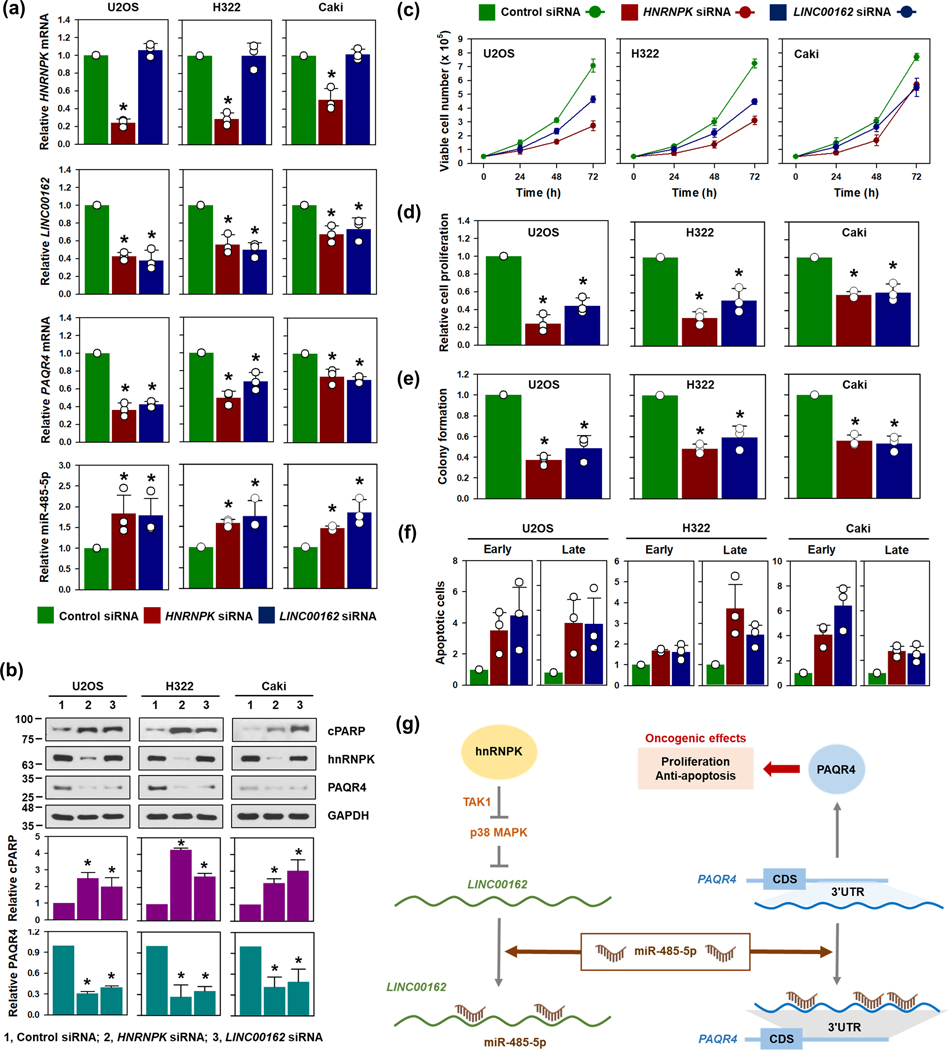
The levels of LINC00162, miR-485–5p, ad PAQR4 mRNA were compared between normal and cancer cells: for lung cancer, H1299 and H322 compared to Beas2B (Supplementary figure S13a); for renal cancer, 789-O and Caki compared to HEK293 (Supplementary figure S13b). In both cancer types, LINC00162 and PAQR4 mRNA showed higher expression in cancer cells compared to normal cells, while miR-485–5p showed less expressed in cancer cells.
To make our findings solid, the oncogenic function of hnRNPK/LINC00162/miR-485–5p/PAQR4 axis was verified in U2OS (osteosarcoma), H322 (lung adenocarcinoma), and Caki (Clear cell carcinoma) cells. In all tested cells, decrease in LINC00162 was observed in hnRNPK-silenced cells. However, knockdown of LINC00162 did not affect the expression of hnRNPK (Figure 8a and 8b). Knockdown of hnRNPK or LINC00162 reduced the level of PAQR4 mRNA by increasing miR-485–5p. Similar to the results observed in HeLa cells, decrease in hnRNPK and LINC00162 inhibited cell growth and colony forming ability, and caused apoptotic cell death (Figure 8c–8f; Supplementary figure S13c and S13d). In conclusion, our findings are not specific to cell-type, suggesting it is generally applicable to various cancer cells.
Figure 8. The oncogenic hnRNPK/LINC00162/miR-485–5p/PAQR4 axis was applicable to various types of cancer.
(a-f) To make our findings solid, the oncogenic function of hnRNPK/LINC00162/miR-485–5p/PAQR4 axis was verified in U2OS, H322, and Caki cells. Cells were transfected with hnRNPK or LINC00162 siRNA for 48 h. The expression levels of HNRNPK mRNA, LINC00162, PAQR4 mRNA, and miR-485–5p were determined by RT-qPCR (a) and Western blot (b). Cell proliferation was measured by counting the number of viable cells (c), WST-1 assay (d), and colony formation (e). The population of early and late apoptotic cell death was calculated by a FACS analysis (f). (g) A schematic of the proposed action mechanism of above regulatory molecules. Please refer to the main text for a detailed description.
Based on the results of this work, we suggest a schematic model to summarize the action mechanism of oncogenic LINC00162, as shown in Figure 8g. Briefly, hnRNPK regulates LINC00162 levels in a p38 MAPK-dependent manner. Predominantly residing in the cytosol, LINC00162 may compete for miR-485–5p, thereby derepressing PAQR4 production.
4 ǀ. DISCUSSION
LincRNAs are intergenic lncRNAs and have been found to participate in cancer phenotypes including proliferation, apoptosis, and metastasis (Aprile et al., 2020; Dhamija & Diederichs, 2016; N. Jiang, Zhang, Gu, Li, & Shang, 2021). With increasing evidence that lincRNAs are abnormally expressed in various cancers, they have become a potential target for cancer treatment. In this study, we discovered a hnRNPK-regulated oncogenic lincRNA, LINC00162. It was reported that LINC00162 enhances the migratory ability of squamous carcinoma by downregulating integrin α2β1 and α5β1 (Piipponen, Heino, Kahari, & Nissinen, 2018); it also accelerates the proliferation of pancreatic ductal adenocarcinoma (Lu et al., 2020). LINC00162 is highly abundant and closely associated with the proliferation of bladder cancer cells (X. Wang et al., 2020); it can negatively regulate the expression of its neighboring gene PTTG1IP by binding to THRAP3, a transcription-related protein.
The mechanisms by which lncRNAs affect cancer-associated genes often depend on their subcellular localization (Batista & Chang, 2013; L. L. Chen, 2016). In the nucleus, lncRNAs modulate gene transcription by forming ribonucleoprotein complexes or by recruiting chromatin remodeling complexes as scaffolds or guides. Alongside their roles in the nucleus, lncRNAs also regulate target gene expression by associating with miRNAs, mRNAs, and proteins. For abundant lncRNAs, a competing endogenous RNA (ceRNA) hypothesis proposes that RNAs with same MREs can compete for the same miRNAs, indirectly affecting the stability and translation of mRNAs. The optimal effect of ceRNA occurs when the quantities of miRNA and ceRNA are nearly equal (Kumar et al., 2014; Mukherji et al., 2011; Tay, Karreth, & Pandolfi, 2014). The activity of ceRNA also depends on its cellular localization and the number of MREs within it (Ala et al., 2013). LINC00162 is known to promote proliferation, migration, and invasion of fibroblast-like synoviocytes by decoying miR-4701–5p in rheumatoid arthritis (Bi et al., 2019). It is also reported that LINC00162 was elevated in cisplatin-resistant cutaneous squamous cell carcinoma (D. Wang, Zhou, Yin, & Zhou, 2020). Knockdown of LINC00162 inhibited cisplatin resistance and malignant phenotypes including proliferation, migration, and invasion by interfering the inhibitory function of miR-485–5p on REV3L expression. In hepatocellular carcinoma, LINC00162 activates PI3K/Akt/mTOR pathway by sponging EIF6-targeting miR-588 (Liu, Mo, et al., 2020). Our findings reveal that LINC00162 functions as a sponge of miR-485–5. Several lncRNAs are reported to function as sponge of miR-485–5p (Bao et al., 2019; Liu, Yang, et al., 2020; Tu et al., 2019; Y. Wang et al., 2018; Zhang, Hu, Zhou, & Gao, 2018). In hepatocellular cell carcinoma, lncRNA DSCR8 activates the Wnt/β-catenin pathway by decoying FZD7-targeting miR-485–5p (Y. Wang et al., 2018). While LINC00460 is oncogenic by decoying miR-485–5p and increasing the expression of p21-activated kinase 1 (Tu et al., 2019). LINC00467 also has oncogenic functions, as it enhances cellular proliferation and diminishes apoptotic cell death through miR-485–5p/DPAGT1 (Liu, Yang, et al., 2020). In addition to lincRNAs, the tumor-suppressing properties of miR-485–5p are interrupted by antisense transcripts, which have a miRNA recognition element (MRE) of miR-485–5p. FLVCR1-AS1 and FOXD2-AS1 promote malignant properties, including proliferation, migration, and invasion, by decoying miR-485–5p in cholangiocarcinoma and papillary thyroid cancer, respectively (Bao et al., 2019; Y. Zhang et al., 2018).
As previously reported, we found that hnRNPK upregulates LINC00263 by directly binding and enhancing its stability (Lee et al., 2021). Although LINC00162 is also regulated by hnRNPK, no molecular interaction was observed, indicating that hnRNPK indirectly regulates LINC00162 levels. It is well-known that hnRNPK can affect various cellular signaling pathways (Z. Wang et al., 2020). For example, the transformation of growth factor-beta-activated kinase 1 (TAK1) is translationally regulated by hnRNPK through interaction with the 3’UTR of TAK1 mRNA (Liepelt et al., 2014; X. L. Sun et al., 2019). The knockdown of hnRNPK increases the expression of TAK1 and thus, activates p38 MAPK. We found that the decrease in LINC00162 levels by the knockdown of hnRNPK was reversed through treatment with a p38 MAPK inhibitor, suggesting that hnRNPK regulates LINC00162 in a p38-dependent manner. In addition to the regulation of LINC00162 through TAK1/p38 MAPK, the functional relationship between hnRNPK and LINC00162 is thought to be very diverse. For example, cellular signaling is also a remarkable mechanism whereby LINC00162 regulates hnRNPK. According to a previous report, LINC00162 can regulate the activation of the GSK3β through PI3K/Akt (Fan et al., 2015). It is known that GSK3β regulates the localization of hnRNPK by inducing phosphorylation of hnRNPK, suggesting that LINC00162 may influence the oncogenic function of hnRNPK. It was recently reported that hnRNPK also interacts with SINE-derived nuclear RNA localization (SIRLOIN)-containing lncRNAs and mediates nuclear enrichment. It is anticipated when abnormal expression of hnRNPK occurs, the cellular localization of SIRLOIN-containing RNAs in the nucleus and cytoplasm might be affected (Zhu, Wang, & Xu, 2019). Although we haven’t expanded our investigation to the role of hnRNPK in the localization of LINC00162, there might be several possible links between hnRNPK-regulated LINC00162 and SIRLOIN.
We demonstrate here that LINC00162 increased PAQR4 expression by sponging miR-485–5p. Since there are three MREs in LINC00162 and PAQR4 mRNA sequences, miR-485–5p was expected to strongly regulate both of them. Since hnRNPK is reported to regulate the expression of target genes at transcriptional level, we can’t rule out the possibility that hnRNPK controls the transcription of PAQR4 by directly affecting transcriptional machinery or transcriptional factors. The possibility of hnRNPK directly controlling the expression of PAQR4 should also be considered. In general, hnRNPK increases the stability or translation of target mRNA. Since PAQR4 mRNA was lowered by knockdown of hnRNPK, the translational regulation can be neglected. To confirm this possibility, we examined the direct interaction between hnRNPK and PAQR4 mRNA through RIP experiment. Although PAQR4 mRNA was predicted to harbor the binding motif of hnRNPK, it was not enriched in hnRNPK IP (Supplementary figure S9d), indicating that hnRNPK affects PAQR4 expression in an indirect way. PAQR4 is reported to play a critical role in the regulation of various cellular processes, including cancer malignancy (L. Wang et al., 2017; Ye, Gao, Guo, Zhang, & Jiang, 2020; H. Zhang et al., 2018). Although it is highly expressed in cancers, the function of PAQR4 is not fully understood. PAQR4 is significantly overexpressed in prostate cancer and positively correlated with poor prognosis. In breast cancer, PAQR4 exhibits a tumorigenic effect by stabilizing cyclin-dependent kinase 4 (H. Zhang et al., 2018) and activating the PI3K/Akt pathway (Ye et al., 2020) but additional pathways are likely implicated.
We conclude that LINC00162, a novel hnRNPK-regulated lincRNA, exhibits oncogenic effects, including accelerated proliferation and anti-apoptosis, by being a target of miR-485–5p and reducing the levels of functional miR-485–5p in the cell. In turn, thus allows PAQR4 levels to rise. Taken together, the components of this regulatory paradigm-hnRNPK, LINC00162, miR-485–5p, and PAQR4-may represent promising targets for cancer therapy.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by a grant from the Basic Research Program (HHK, NRF-2021R1F1A1063119) through the National Research Foundation (NRF), funded by the Korean Ministry of Science and ICT, and by grants from the Future Medicine 20–30 Project (HHK, SMX1210751) funded by Samsung Medical Center, and the Global Ph.D. Fellowship Program (WJL, NRF-2017H1A2A1045644) through the National Research Foundation of Korea (NRF), funded by the Ministry of Education. PRP and MG were funded by the NIA IRP, NIH.
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Ala U, Karreth FA, Bosia C, Pagnani A, Taulli R, Leopold V, . . . Pandolfi PP (2013). Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc Natl Acad Sci U S A, 110(18), 7154–7159. doi: 10.1073/pnas.1222509110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprile M, Katopodi V, Leucci E, & Costa V. (2020). LncRNAs in Cancer: From garbage to Junk. Cancers (Basel), 12(11). doi: 10.3390/cancers12113220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W, Cao F, Ni S, Yang J, Li H, Su Z, & Zhao B. (2019). lncRNA FLVCR1-AS1 regulates cell proliferation, migration and invasion by sponging miR-485–5p in human cholangiocarcinoma. Oncol Lett, 18(3), 2240–2247. doi: 10.3892/ol.2019.10577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, & Chang HY (2013). Long noncoding RNAs: cellular address codes in development and disease. Cell, 152(6), 1298–1307. doi: 10.1016/j.cell.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Guo XH, Mo BY, Wang ML, Luo XQ, Chen YX, . . . Zheng SG (2019). LncRNA PICSAR promotes cell proliferation, migration and invasion of fibroblast-like synoviocytes by sponging miRNA-4701–5p in rheumatoid arthritis. EBioMedicine, 50, 408–420. doi: 10.1016/j.ebiom.2019.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhang J, Wu P, Yang W, Ye Q, Chen Q, & Jiang C. (2018). Blocking LINC00152 suppresses glioblastoma malignancy by impairing mesenchymal phenotype through the miR-612/AKT2/NF-kappaB pathway. J Neurooncol, 140(2), 225–236. doi: 10.1007/s11060-018-2951-0 [DOI] [PubMed] [Google Scholar]
- Chen LL (2016). Linking Long Noncoding RNA Localization and Function. Trends Biochem Sci, 41(9), 761–772. doi: 10.1016/j.tibs.2016.07.003 [DOI] [PubMed] [Google Scholar]
- Chen X, Wang Z, Tong F, Dong X, Wu G, & Zhang R. (2020). lncRNA UCA1 Promotes Gefitinib Resistance as a ceRNA to Target FOSL2 by Sponging miR-143 in Non-small Cell Lung Cancer. Mol Ther Nucleic Acids, 19, 643–653. doi: 10.1016/j.omtn.2019.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cheng Z, Luo C, & Guo Z. (2020). LncRNA-XIST/microRNA-126 sponge mediates cell proliferation and glucose metabolism through the IRS1/PI3K/Akt pathway in glioma. J Cell Biochem, 121(3), 2170–2183. doi: 10.1002/jcb.29440 [DOI] [PubMed] [Google Scholar]
- Deng K, Wang H, Guo X, & Xia J. (2016). The cross talk between long, non-coding RNAs and microRNAs in gastric cancer. Acta Biochim Biophys Sin (Shanghai), 48(2), 111–116. doi: 10.1093/abbs/gmv120 [DOI] [PubMed] [Google Scholar]
- Dhamija S, & Diederichs S. (2016). From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer, 139(2), 269–280. doi: 10.1002/ijc.30039 [DOI] [PubMed] [Google Scholar]
- Fan X, Xiong H, Wei J, Gao X, Feng Y, Liu X, . . . Liu, L. (2015). Cytoplasmic hnRNPK interacts with GSK3beta and is essential for the osteoclast differentiation. Sci Rep, 5, 17732. doi: 10.1038/srep17732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, He M, Rao M, Diao J, & Zhu Y. (2019). Long noncoding RNA DLEU1 aggravates glioma progression via the miR-421/MEF2D axis. Onco Targets Ther, 12, 5405–5414. doi: 10.2147/OTT.S207542 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, . . . Lander ES (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature, 458(7235), 223–227. doi: 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, . . . Chinnaiyan AM (2015). The landscape of long noncoding RNAs in the human transcriptome. Nat Genet, 47(3), 199–208. doi: 10.1038/ng.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang MC, Ni JJ, Cui WY, Wang BY, & Zhuo W. (2019). Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res, 9(7), 1354–1366. [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Zhang X, Gu X, Li X, & Shang L. (2021). Progress in understanding the role of lncRNA in programmed cell death. Cell Death Discov, 7(1), 30. doi: 10.1038/s41420-021-00407-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Armenteros-Monterroso E, East P, Chakravorty P, Matthews N, Winslow MM, & Downward J. (2014). HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature, 505(7482), 212–217. doi: 10.1038/nature12785 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lee WJ, Shin CH, Ji H, Jeong SD, Park MS, Won HH, . . . Kim HH (2021). hnRNPK-regulated LINC00263 promotes malignant phenotypes through miR-147a/CAPN2. Cell Death Dis, 12(4), 290. doi: 10.1038/s41419-021-03575-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepelt A, Mossanen JC, Denecke B, Heymann F, De Santis R, Tacke F, . . . Ostareck-Lederer A. (2014). Translation control of TAK1 mRNA by hnRNP K modulates LPS-induced macrophage activation. RNA, 20(6), 899–911. doi: 10.1261/rna.042788.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Mo H, Sun L, Wang L, Chen T, Yao B, . . . Yang N. (2020). Long noncoding RNA PICSAR/miR-588/EIF6 axis regulates tumorigenesis of hepatocellular carcinoma by activating PI3K/AKT/mTOR signaling pathway. Cancer Sci, 111(11), 4118–4128. doi: 10.1111/cas.14631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yang S, Chen X, Dong S, Zhou S, & Xu S. (2020). LncRNA LINC00467 acted as an oncogene in esophageal squamous cell carcinoma by accelerating cell proliferation and preventing cell apoptosis via the miR-485–5p/DPAGT1 axis. J Gastroenterol Hepatol. doi: 10.1111/jgh.15201 [DOI] [PubMed] [Google Scholar]
- Lu Y, Wu M, Fu J, Sun Y, Furukawa K, Ling J, . . . Chiao PJ (2020). The overexpression of long intergenic ncRNA00162 induced by RelA/p65 promotes growth of pancreatic ductal adenocarcinoma. Cell Prolif, 53(5), e12805. doi: 10.1111/cpr.12805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Morgan SL, & Wang KC (2016). PICSAR: Long Noncoding RNA in Cutaneous Squamous Cell Carcinoma. J Invest Dermatol, 136(8), 1541–1542. doi: 10.1016/j.jid.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, & van Oudenaarden A. (2011). MicroRNAs can generate thresholds in target gene expression. Nat Genet, 43(9), 854–859. doi: 10.1038/ng.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norbury CJ, & Zhivotovsky B. (2004). DNA damage-induced apoptosis. Oncogene, 23(16), 2797–2808. doi: 10.1038/sj.onc.1207532 [DOI] [PubMed] [Google Scholar]
- Panda AC, Abdelmohsen K, & Gorospe M. (2017). SASP regulation by noncoding RNA. Mech Ageing Dev, 168, 37–43. doi: 10.1016/j.mad.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piipponen M, Heino J, Kahari VM, & Nissinen L. (2018). Long non-coding RNA PICSAR decreases adhesion and promotes migration of squamous carcinoma cells by downregulating alpha2beta1 and alpha5beta1 integrin expression. Biol Open, 7(11). doi: 10.1242/bio.037044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piipponen M, Nissinen L, Farshchian M, Riihila P, Kivisaari A, Kallajoki M, . . . Kahari VM (2016). Long Noncoding RNA PICSAR Promotes Growth of Cutaneous Squamous Cell Carcinoma by Regulating ERK1/2 Activity. J Invest Dermatol, 136(8), 1701–1710. doi: 10.1016/j.jid.2016.03.028 [DOI] [PubMed] [Google Scholar]
- Qu C, Dai C, Guo Y, Qin R, & Liu J. (2020). Long non-coding RNA PVT1-mediated miR-543/SERPINI1 axis plays a key role in the regulatory mechanism of ovarian cancer. Biosci Rep, 40(6). doi: 10.1042/BSR20200800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, & Pandolfi PP (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell, 146(3), 353–358. doi: 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AM, & Chang HY (2016). Long Noncoding RNAs in Cancer Pathways. Cancer Cell, 29(4), 452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz SU, Grote P, & Herrmann BG (2016). Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci, 73(13), 2491–2509. doi: 10.1007/s00018-016-2174-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin CH, Lee H, Kim HR, Choi KH, Joung JG, & Kim HH (2017). Regulation of PLK1 through competition between hnRNPK, miR-149–3p and miR-193b-5p. Cell Death Differ, 24(11), 1861–1871. doi: 10.1038/cdd.2017.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SL, Shu YG, & Tao MY (2020). LncRNA CCAT2 promotes angiogenesis in glioma through activation of VEGFA signalling by sponging miR-424. Mol Cell Biochem, 468(1–2), 69–82. doi: 10.1007/s11010-020-03712-y [DOI] [PubMed] [Google Scholar]
- Sun XL, Wang ZL, Wu Q, Jin SQ, Yao J, & Cheng H. (2019). LncRNA RMST activates TAK1-mediated NF-kappaB signaling and promotes activation of microglial cells via competitively binding with hnRNPK. IUBMB Life, 71(11), 1785–1793. doi: 10.1002/iub.2125 [DOI] [PubMed] [Google Scholar]
- Tay Y, Karreth FA, & Pandolfi PP (2014). Aberrant ceRNA activity drives lung cancer. Cell Res, 24(3), 259–260. doi: 10.1038/cr.2014.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J, Zhao Z, Xu M, Chen M, Weng Q, & Ji J. (2019). LINC00460 promotes hepatocellular carcinoma development through sponging miR-485–5p to up-regulate PAK1. Biomed Pharmacother, 118, 109213. doi: 10.1016/j.biopha.2019.109213 [DOI] [PubMed] [Google Scholar]
- Wang D, Zhou X, Yin J, & Zhou Y. (2020). Lnc-PICSAR contributes to cisplatin resistance by miR-485–5p/REV3L axis in cutaneous squamous cell carcinoma. Open Life Sci, 15(1), 488–500. doi: 10.1515/biol-2020-0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang R, You X, Zhang H, Wei S, Cheng T, . . . Chen Y. (2017). The steady-state level of CDK4 protein is regulated by antagonistic actions between PAQR4 and SKP2 and involved in tumorigenesis. J Mol Cell Biol, 9(5), 409–421. doi: 10.1093/jmcb/mjx028 [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang R, Wu S, Shen L, Ke M, Ouyang Y, . . . Nan A. (2020). Super-Enhancer LncRNA LINC00162 Promotes Progression of Bladder Cancer. iScience, 23(12), 101857. doi: 10.1016/j.isci.2020.101857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sun L, Wang L, Liu Z, Li Q, Yao B, . . . Liu Q. (2018). Long non-coding RNA DSCR8 acts as a molecular sponge for miR-485–5p to activate Wnt/beta-catenin signal pathway in hepatocellular carcinoma. Cell Death Dis, 9(9), 851. doi: 10.1038/s41419-018-0937-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Qiu H, He J, Liu L, Xue W, Fox A, . . . Xu J. (2020). The emerging roles of hnRNPK. J Cell Physiol, 235(3), 1995–2008. doi: 10.1002/jcp.29186 [DOI] [PubMed] [Google Scholar]
- Ye J, Gao M, Guo X, Zhang H, & Jiang F. (2020). Breviscapine suppresses the growth and metastasis of prostate cancer through regulating PAQR4-mediated PI3K/Akt pathway. Biomed Pharmacother, 127, 110223. doi: 10.1016/j.biopha.2020.110223 [DOI] [PubMed] [Google Scholar]
- Yu H, Xu A, Wu B, Wang M, & Chen Z. (2021). Long noncoding RNA NEAT1 promotes progression of glioma as a ceRNA by sponging miR-185–5p to stimulate DNMT1/mTOR signaling. J Cell Physiol, 236(1), 121–130. doi: 10.1002/jcp.29644 [DOI] [PubMed] [Google Scholar]
- Zhang H, Han R, Ling ZQ, Zhang F, Hou Y, You X, . . . Chen Y. (2018). PAQR4 has a tumorigenic effect in human breast cancers in association with reduced CDK4 degradation. Carcinogenesis, 39(3), 439–446. doi: 10.1093/carcin/bgx143 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hu J, Zhou W, & Gao H. (2018). LncRNA FOXD2-AS1 accelerates the papillary thyroid cancer progression through regulating the miR-485–5p/KLK7 axis. J Cell Biochem. doi: 10.1002/jcb.28072 [DOI] [PubMed] [Google Scholar]
- Zhu S, Wang Z, & Xu J. (2019). Connecting Versatile lncRNAs with Heterogeneous Nuclear Ribonucleoprotein K and Pathogenic Disorders. Trends Biochem Sci, 44(9), 733–736. doi: 10.1016/j.tibs.2019.06.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.