Abstract
The genus Gelidocalamus T. H. Wen, endemic to southern China, is a small but taxonomically problematic genus of Arundinarieae (Poaceae, Bambusoideae). During field work, a population of Gelidocalamus from Zixing, Hunan, was discovered, appearing to be distinct from our previously identified collection. Comparisons of the population of Zixing were performed by using scanning electron microscopy (SEM) and a plastid genome-based phylogeny. Morphologically, it was mostly similar to G.multifolius, but differed by culm leaf erect with densely white pubescence, apical branch sheath much longer than the internodes and foliage leaf larger. Phylogenetically, the new species was well-supported as a sister to the clade of G.multifolius + G.tessellatus, and the above three taxa were clustered in the Shibataea clade (IV) of Arundinarieae. Thus, the new species, formally named as Gelidocalamuszixingensis W.G.Zhang, G.Y.Yang & C.K.Wang, was described and illustrated herein.
Keywords: Gramineae, leaf epidermis, phylogeny, SEM, temperate woody bamboo
Introduction
Arundinarieae (Poaceae: Bambusoideae), i.e., accommodating the temperate woody bamboos, including ca. 581 species in 31 genera (Clark and de Oliveira 2018), is widely accepted as a monophyletic tribe (Guo et al. 2021; Gallaher et al. 2022; Huang et al. 2022), and has been one of the main focuses of botanical research due to its significant ecological and economic value (Triplett 2008). It is mainly distributed in the tropical and subtropical mountains of East Asia, central and southern Africa, Madagascar and eastern North America (Keng and Wang 1996; Soreng et al. 2022). Due to complex allopolyploid history and adaptive radiation events, Arundinarieae has evolved complex and diverse morphological characters, e.g., semelauctant and iterauctant inflorescences, pachymorph and leptomorph rhizomes, and growth habits from solitary to multiple branches, which made it a taxonomically complicated group (Li et al. 2006; Vorontsova et al. 2016; Guo et al. 2021).
As a small but taxonomically problematic genus of Arundinarieae, Gelidocalamus T. H. Wen, 1982, containing ca. 11 species (Li et al. 2006; Zhang et al. 2017; Cai et al. 2021), is endemic to southwestern China, and characterized by a set of morphological features including several branches per node, a single foliage leaf on each ultimate branch typically (except G.multifolius B. M. Yang, 1986) (Yang 1986), and semelauctant inflorescence. In addition, the new shoots occurring in autumn to winter are also a key feature of Gelidocalamus (Keng and Wang 1996; Liu et al. 2017; Nie et al. 2018). Members of Gelidocalamus have a relatively narrow distribution in the southern provinces of China and usually occur along ravines under broad-leaved evergreen forest below 1,000 m elevation, except G.monophyllus (Yi et B. M. Yang) B. M. Yang, 1989, distributed at 1250 m (Li et al. 2016; Nie et al. 2018). However, some “spring-shoot” species (as opposed to some others that produce shoots in the autumn-winter period), e.g., G.rutilans Wen, 1983, G.subsolidus W. T. Lin & Z. J. Feng, 1990, G.solidus C. D. Chu & C. S. Chao, 1984, and G.longiinternodus W. T. Wen & S. C. Chen, 1986, complicate the delimitation of the genus.
Molecular studies of the tribe Arundinarieae have indicated that the conventionally circumscribed Gelidocalamus was polyphyletic, and its “spring-shoot” species were nested with members of Ferrocalamus Hsueh & Keng f., 1982, Shibataea Makino ex Nakai, 1912, Indocalamus Nakai, 1925, and other close relatives (Ma et al. 2017; Guo et al. 2019; Qin et al. 2021). Recently, Guo et al. (2021) investigated Arundinarieae using double digest restriction-site associated DNA (ddRAD) sequences, and revealed that six members of Gelidocalamus formed a monophyletic clade. These taxa (which may be termed the ‘gelido- taxa’) have identical micromorphological characters (i.e., prominent stomata apparatus surrounded by 8–12 short papillae and a dense waxy covering). On the other hand, other “spring-shoot” members were scattered and grouped with other genera. The “gelido-” members of Gelidocalamus are G.stellatus T. H. Wen, 1982, G.tessellatus T. H. Wen & C. C. Chang, 1982, G.annulatus T. H. Wen, 1988, G.latifolius Q. H. Dai & T. Chen, 1985, G.multifolius and G.monophyllus. Two recently reported species G.xunwuensis W. G. Zhang & G. Y. Yang, 2017 and G.fengkaiensis N. H. Xia & Z. Y. Cai, 2021 appear to also be in this group.
During field work in August 2014, a population of Gelidocalamus sp. in Zixing City of Hunan Province, China (25°54'1.75"N, 113°34'9.18"E), was found, and mistakenly identified as G.multifolius due to a somewhat similar morphology. In this study, a detailed comparison among the new species, G.multifolius and G.tessellatus, including characters obtained with scanning electron microscope (SEM) of the foliage leaf epidermis, was made. Moreover, the phylogenetic relationships of the new species including above taxa and allied genera were reassessed based on complete chloroplast genomes.
Materials and methods
Field investigation and sample collection
Mature bamboo leaves were collected from the individuals of the type localities: G. sp from Zixing, Lianping Township of Zixing City in Hunan; G.stellatus, Xiazhuang of Jinggang Mountain in Jiangxi; G.tessellatus, Maolan of Libo County in Guizhou; G.multifolius, Jiuyi Mountain of Ningyuan County in Hunan. Foliage leaves were fixed with the FAA (acetic acid: formalin: ultrapure water: ethanol = 1:2:3:14), and some dried in silica-gel for storage. All voucher specimens were deposited in the herbarium of the College of Forestry, Jiangxi Agricultural University, China (JXAU).
Micromorphological observations of foliage leaf epidermis
After cleaning in the ultrasonic cleaner CPX2800H-C (Branson, USA), the middle portion of foliage leaf (5×5 mm) was dried at room temperature, mounted on stubs, and coated with gold sputtering. Using a scanning electron microscope S-4800 (Hitachi, Japan), leaf epidermal characters were observed and photographed. Terminology for epidermal appendages and leaf blades follows previous studies (Ellis 1979; Ellis et al. 2009; Zhang et al. 2014; Leandro et al. 2019).
Sequencing, assembly and annotation
Total genomic DNA was isolated from foliage leaves dried over silica-gel by a modified CTAB method (Murray and Thompson 1980). Illumina paired-end (2×150 bp) libraries were constructed and sequenced at Novogene Bioinformatics Technology Co. Ltd. (Beijing, China), and ca. 6 GB raw data for each sample was acquired. To improve assembly accuracy, FastQC 0.11.9 (Andrews 2016) and Fastp 0.12.4 (Chen et al. 2018) were used to filter out unpaired and low-depth reads by using default parameters. Complete chloroplast genomes were assembled using the software GetOrganelle 1.7.4 (Jin et al. 2018) with a range of k-mers of 45, 65, 85, 105 and 121, and the filtered reads were transferred to Bandage (Wick et al. 2015) for chloroplast genome scaffolds connection. Then, chloroplast genome sequences were annotated by using CPGAVAS2 (Shi et al. 2019) and manually checked in Geneious 9.1.4 (Kearse et al. 2012), and illustration of the newly sequenced plastome was drawn in the software Chloroplot with default settings (Zheng et al. 2020).
Phylogenetic analysis
To determine the position of the new species, phylogenetic analyses using maximum likelihood (ML) and Bayesian inference (BI) were performed. Besides G. sp. from Zixing (OP920758) and G.multifolius (OP920759), another 18 complete chloroplast genomes of the tribe Arundinarieae were obtained from the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/). Hsuehochloacalcarea (C. D. Chu & C. S. Chao) D. Z. Li & Y. X. Zhang, 2018 was selected as outgroup (Genbank accession numbers see the Table 1 for details).
Table 1.
Information on the 20 complete chloroplast genomes used in this study.
| Species | GenBank accession |
|---|---|
| Ingroup | |
| Acidosasapurpurea (Hsueh & T.P.Yi) Keng f. | HQ337793 |
| Ampelocalamusactinotrichus (Merr. et Chun) S. L. Chen, T. H. Wen et G. Y. Sheng | MH410123 |
| Arundinariagigantea (Walter) Muhl. | NC_020341 |
| Bergbambostessellata (Nees) Stapleton | NC_036816 |
| Gaoligongshaniamegalothyrsa (Handel-Mazzetti) D. Z. Li | JX513419 |
| Gelidocalamusmultifolius B. M. Yang | OP920759 |
| Gelidocalamustessellatus Wen & C. C. Chang | NC_024719 |
| Gelidocalamuszixingensis W.G.Zhang, G.Y.Yang & C.K.Wang | OP920758 |
| Himalayacalamusgyirongensis (Munro) P. C. Keng | NC_043943 |
| Indocalamussinicus (Hance) Nakai | NC_036819 |
| Ravenochloawilsonii (Rendle) D. Z. Li & Y. X. Zhang | JX513421 |
| Kurunadebilis (Thwaites) Attigala, Kaththr. & L.G.Clark | NC_036822 |
| Oldeaniaalpina (K.Schum.) Stapleton | NC_036813 |
| Phyllostachysedulis (Carriere) J. Houzeau | MW007170 |
| Pleioblastusamarus (Keng) Keng f. | MH988736 |
| Sinoasalongiligulata (McClure) N.H.Xia, Q.M.Qin & J.B.Ni | NC_036825 |
| Shibataeachiangshanensis Wen | NC_036826 |
| Shibataeakumasasa (Zollinger ex Steudel) Makino ex Nakai | KU523578 |
| Thamnocalamusspathiflorus (Trinius) Munro | JX513425 |
| Outgroup | |
| Hsuehochloacalcarea (C.D.Chu & C.S.Chao) D.Z.Li & Y.X.Zhang | KJ496369 |
After alignment with MAFFT 7.450 (Katoh and Standley 2013), Maximum likelihood (ML) analysis was generated by IQ-TREE (Nguyen et al. 2015), bootstrap analyses were performed with 1,000 replications, and the best-fit BIC model GTR + F + I + G4 was defined by ModelFinder (Kalyaanamoorthy et al. 2017). Bayesian inference (BI) was conducted using MrBayes 3.2.6 (Ronquist et al. 2012) with the same model. 20,000,000 generations were run to ensure average standard deviation of split frequencies (ASDFs) < 0.01 with sampling frequency set as 2,000 generations. Discarding the first 25% burn-in samples, the optimized topology was printed.
Results
Morphological comparison
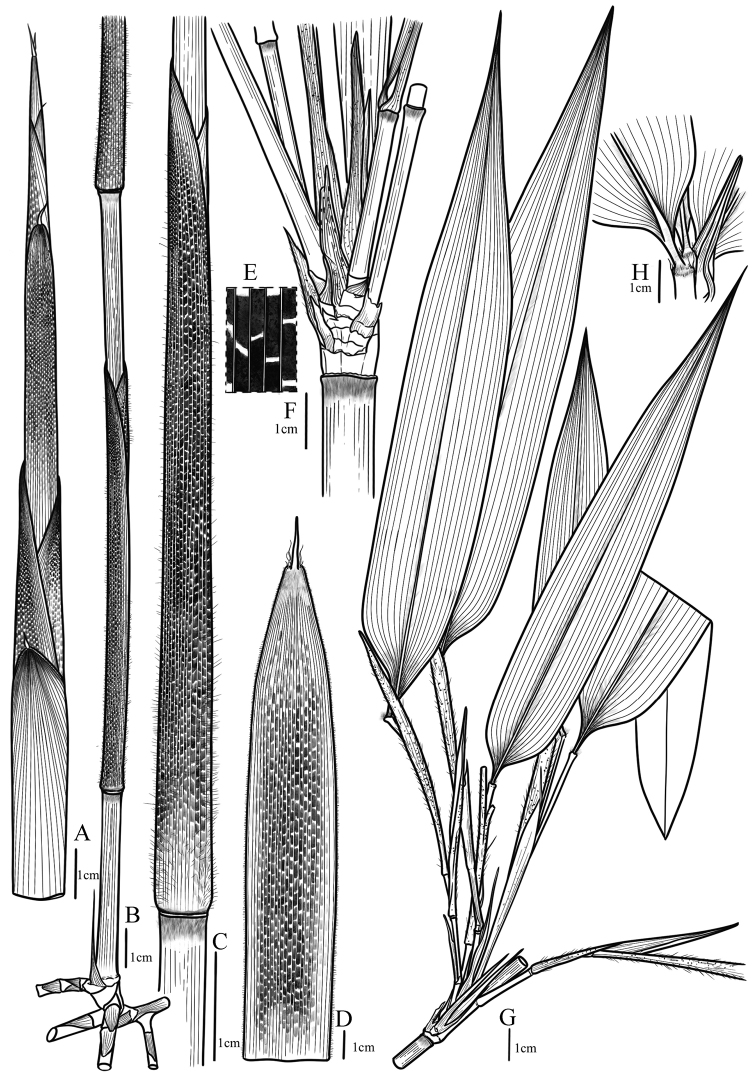
Compared to G.tessellatus, G.zixingensis was mostly similar to G.multifolius in the habit, the morphological characters and new shoots sprouting season, but can be distinguished by the following morphological characters: (a) a ring of white-gray (vs. yellow-brown) appressed pubescence below each culm node; (b) culm leaf sheaths densely white pubescent (vs. glabrous), with brown patches (vs. pale brown), and erect (vs. reflexed) culm sheath blades; (d) branch leaf sheaths setose (vs. glabrous) and much longer, (>3cm) (vs. slightly longer, < 1cm or as long) as the internodes; foliage leaf blades mesophyll (vs. notophyll). (see Table 2, Fig. 1 for details).
Table 2.
Morphological comparison among G.zixingensis, G.multifolius and G.tessellatus.
| Characters | G.zixingensis | G.multifolius | G.tessellatus |
|---|---|---|---|
| Culm | glabrous, each node with a ring of white-gray appressed trichomes below. | glabrous, each node with a ring of yellow-brown appressed trichomes below. | sparsely setae, each node with a ring of golden appressed trichomes below. |
| Culm leaf | sheath pubescent with brown patches, sparsely setae near the base, margin sparsely ciliate; oral setae 2–4 pairs; blade erect. | sheath glabrous with pale brown patches, margin sparsely ciliate; oral setae 3–6 pairs; blade reflexed. | sheath glabrous with purple patches, black setae, margin densely ciliate; oral setae 2–3 pairs; blade erect or reflexed. |
| Branch | sheath papery, densely setae, without black spots, apical branch sheath longer ca. 3 cm than that of the internode. | sheath leathery, with black spots, pubescent, apical branch sheath longer 0.5–1cm than that of the internode. | sheath leathery, without black spots, sparsely setae, apical branch sheath equally or shorter than that of the internode. |
| Foliage leaf | oral setae 1–3 pairs; blade mesophyll, 23–32×3.2–4.9 cm, lateral veins 6–8 pairs | oral setae weak or absent; blade notophyll, 8–14×1.5–2.5 cm, lateral veins 4–6 pairs | oral setae weak or absent; blade mesophyll, 17–35×3.7–5.4 cm, lateral veins 5–6 pairs |
Figure 1.
Comparison of key characters among G.zixingensis, G.multifolius and G.tessellatus. The corresponding scales are at the bottom right of the figures.
Micromorphological comparison of abaxial foliage leaf epidermis
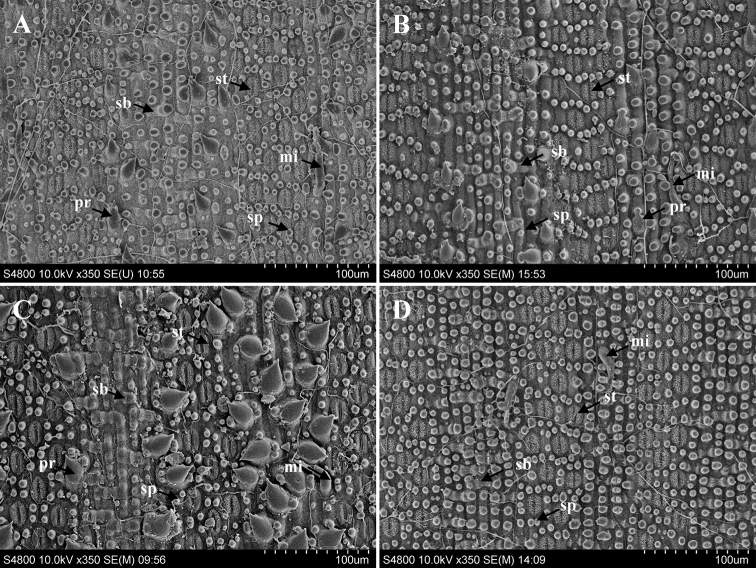
Epidermal traits of the foliage leaf, e.g., short papillae, microhairs, silica bodies and prickles, can be clearly identified under the scanning electron microscope (Fig. 2). Main characters shared by the four selected taxa were: (a) exposed stomatal apparatus, densely covered with white wax and surrounded by 8–10 short papillae; (b) bicellular microhairs, of which the apical one was withered; (c) saddle-shaped silica body, mainly distributed between veins (Table 3). Prickles were sparsely distributed between the veins in G.zixingensis and G.multifolius, and more densely distributed in the G.tessellatus, while no prickles were observed in G.stellatus. Besides, the number of stomatal rows was different, e.g., 3 in G.tessellatus, 3 or 4 in G.stellatus, 4 in G.multifolius, but 5 in G.zixingensis.
Figure 2.
SEM images of the abaxial foliage leaf epidermis AG.zixingensis (Zixing, Hunan, China) BG.multifolius (Ningyuan, Hunan, China) CG.tessellatus (Libo, Guizhou, China) DG.stellatus (Jinggang Mountain, Jiangxi, China). Abbreviations: mi, microhairs; pr, prickles; sb, silica bodies; sp, short papillae; st, stomatal apparatuses.
Table 3.
Micromorphological comparison among four taxa in the study.
| Characters | G.zixingensis | G.multifolius | G.tessellatus | G.stellatus |
|---|---|---|---|---|
| Stomatal apparatus | 5 rows distributed between the veins | 4 rows distributed between the veins | 3 rows distributed between the veins; | 3 or 4 rows distributed between the veins |
| Papillae | 8–10 surrounded the stomatal apparatus | |||
| Microhair | bicellular, apical cell withered | |||
| Prickle | sparsely distributed on the veins | sparsely distributed on the veins | relatively densely distributed on the veins | absent |
| Silica body | saddle-shaped | |||
Phylogenetic analyses based on complete chloroplast genomes
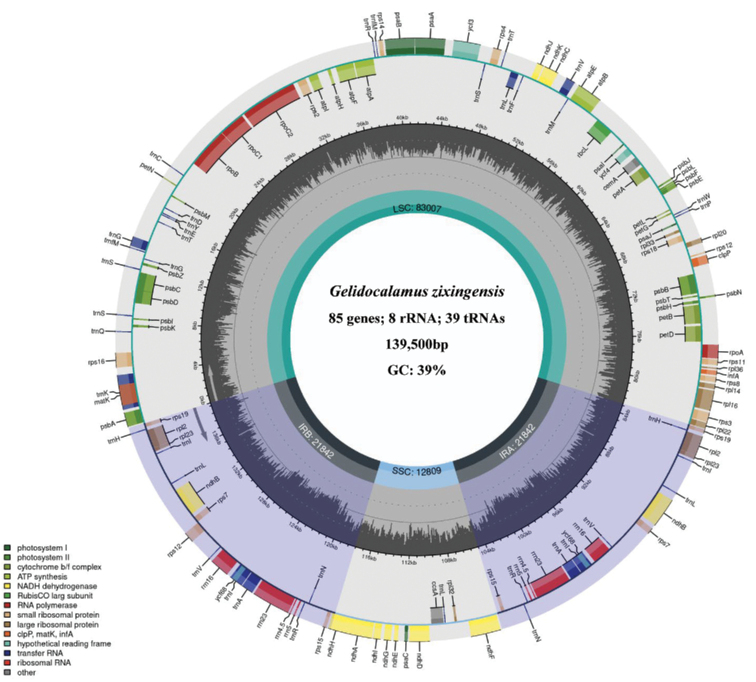
The complete chloroplast genome of Gelidocalamuszixingensis was 139,500 bp in length, comprising a large single copy (LSC) region of 83,007 bp, a small single copy (SSC) region of 12,809 bp and two inverted repeat (IR) regions of 21,842 bp, and its GC content was 39%. The chloroplast genome contained 132 genes, including 85 protein-coding genes, 39 transfer RNAs and 8 ribosomal RNAs (Fig. 3), and the total length of the aligned plastid matrix data was 143,738 bp.
Figure 3.
Complete chloroplast genome map of the Gelidocalamuszixingensis.
Compared to that in G.zixingensis, the total length of chloroplast genome of G.multifolius and G.tessellatus was longer (>200bp), and the differences were mainly in the LSC region (Table 4). Moreover, in comparing the chloroplast genomic variant loci of the three species, a total of 123 SNPs (Single-Nucleotide Polymorphism) and 44 INDELs (Insertion-Deletion) were identified, of which 92 SNPs (74.7%) and 40 INDELs (90.9%) were located in the LSC region.
Table 4.
Comparison of complete chloroplast genomes of three taxa in the study.
| Characters | G.zixingensis | G.multifolius | G.tessellatus |
|---|---|---|---|
| Total length | 139,500 | 139,745 | 139,712 |
| LSC region | 83,007 | 83,252 | 83,220 |
| SSC region | 12,809 | 12,809 | 12,808 |
| IR region | 21,842 | 21,842 | 21,842 |
| Total genes | 132 | 132 | 132 |
| CDS | 85 | 85 | 85 |
| tRNA | 39 | 39 | 39 |
| rRNA | 8 | 8 | 8 |
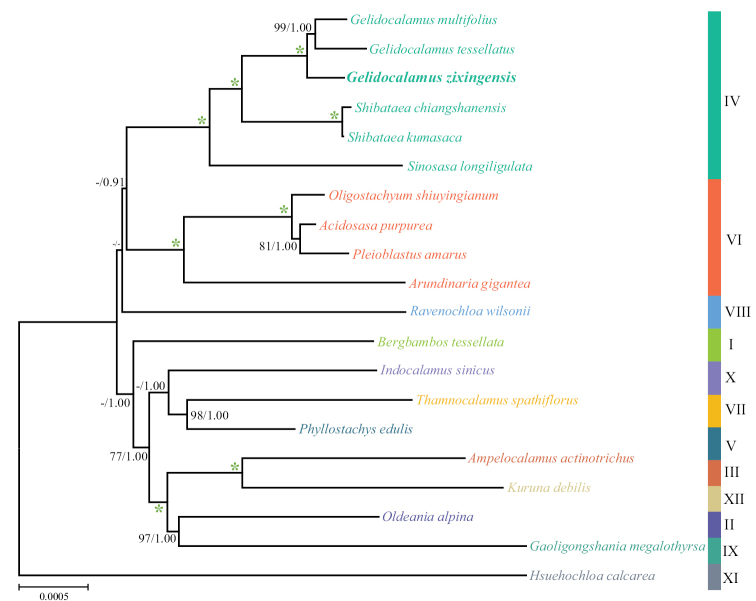
The majority-rule consensus tree with both maximum likelihood (ML) and Bayesian inference (BI) analyses was shown in Fig. 4. Arundinarieae is well-supported as a monophyletic entity, finely divided into 12 lineages (I–XII). There is high support for the G.zixingensis being a sister to the G.multifolius + G.tessellatus clade (bootstrap value of 100% in ML analysis and posterior probability of 1.0 in BI analysis), and the above 3 species were clustered with members of Shibataea e.g., S.chiangshanensis and S.kumasaca, member of Sinosasa, e.g., S.longiligulata, to form the IV clade (bootstrap value of 100% in ML analysis and posterior probability of 1.0 in BI analysis).
Figure 4.
Phylogenetic consensus tree of the Gelidocalamuszixingensis based on plastid genome dataset with maximum likelihood and Bayesian analyses. Only bootstrap values (BS) ≥ 75% and posterior probabilities (PP)≥0.75 are indicated at each node, otherwise dashes. The green asterisk indicates support of 100% BS and 1.00 PP. The letters represent the major chloroplast marker-based clade (I-XII) in which the selected taxa are located.
Taxonomic treatment
. Gelidocalamus zixingensis
W.G.Zhang, G.Y.Yang & C.K.Wang sp. nov.
51FE44B8-4B5B-5DCE-8D99-DEED3EC11BA6
urn:lsid:ipni.org:names:77311677-1
Figure 5.
GelidocalamuszixingensisA habitat B new shoot C, J branch and its leaf sheath D, F, G, H culm and its leaf sheath E transection of culm and pith-cavity I leaf Sheath.
Figure 6.
GelidocalamuszixingensisA new shoot B leptomorph rhizomes and culm C–E culm and its sheath, brown patches F–H branch and its sheaths, leaf and leaf sheath.
Diagnosis.
The new species is morphologically similar to G.multifolius, but differs by having densely white pubescence (vs. glabrous) on the culm leaf sheaths, culm leaf blades erect (vs. reflexed); apical branch sheaths much longer (vs. slightly longer or equilong) than the internodes; foliage leaf blades mesophyll (vs. notophyll).
Type.
China. Hunan, Zixing County, Lianping Township, Chengkang Village, under the forest, 25°54'1.75"N, 113°34'9.18"E, elev. ca. 594 m, 18 Oct. 2015, W.G. Zhang et al. LPC031 (holotype: JXAU!)
Description.
Rhizomes leptomorph. Culms 1.7–4.2 m, 3.5–10 mm in diameter; erect, apically slightly nodding; internodes initially covered with white pubescence, ca. 14–35 cm long, wall 0.6–1.9 mm thick; each node with a ring of white-gray appressed pubescence below sheath scar; branching intravaginal, arising from 5th node above ground, ca. 4–11 (16) branches per node; branches equal or subequal, ca. 5–30 cm long. Culm leaves sheaths persistent, 12–19 cm, culm leaf sheath abaxially with brown patches, densely white pubescent and sparsely setose near the base; culm leaf blade erect, linear-lanceolate, 0.5–2 cm long, 2 mm wide, apex acuminate, base blunt or truncate, ca.1/3 as wide as sheath apex, oral setae 2–4 on each side of the sheath apex, ca. 4 mm long; auricles absent; ligule truncate, ca. 0.5 mm high, scabrous. Branch sheath papery, white setose, without black spots, margins ciliate; sub-apical branch sheath ca. 3 cm beyond the internode. Foliage leaves usually solitary on ultimate branches; ligule truncate, ca. 1 mm, scabrous; auricles absent; oral setae 1–3 pairs straight or curved; leaf blade broadly lanceolate, usually 23.4–32.5×3.2–4.9 cm, lateral veins 6–8 pairs, abaxial surface basally pubescent, base cuneate and asymmetrical, margins serrulate and slightly revolute near base.
Phenology.
New shoots in October.
Etymology.
The species epithet refers to the locality of the type specimen: Zixing County, Hunan, China.
Vernacular names.
Zī Xīng Duăn Zhī Zhú (Chinese pronunciation), 资兴短枝竹 (Chinese name).
Distribution and habitat.
To date, this species has only been found under evergreen broad-leaved forest along river banks at 500–600 m in Chengkang Village, Lianping Township, Zixing County. Species growing in the surrounding area include Quercusmyrsinifolia Blume, 1871 (Fagaceae), Araliachinensis L., 1868 (Araliaceae), Euryajaponica Thunb., 1783 (Pentaphylacaceae), and Liriopespicata (Thunb.) Lour., 1790 (Asparagaceae).
Morphological key of the species of Gelidocaldums of China
| 1 | Culm internodes glabrous | 2 |
| – | Culm internodes hairy | 5 |
| 2 | Culm leaf sheaths glabrous | 3 |
| – | Culm leaf sheaths pubescent with sparse setae | Gelidocalamuszixingensis |
| 3 | Culm leaf sheath margins glabrous or one margin ciliate; oral setae absent or small | 4 |
| – | Culm leaf sheath margins ciliate; oral setae 3–6 pairs | Gelidocalamusmultifolius |
| 4 | Oral setae of culm leaves 1–2 pairs, small; branch sheath margins glabrous; branch sheaths with black spots | Gelidocalamusstellatus |
| – | Oral setae of culm leaves none or several; branch sheath margins ciliate at one side, the other side glabrous or apically ciliate; branch sheaths without black spots | Gelidocalamusfengkaiensis |
| 5 | Culm leaf sheaths covered with appressed brown short hairs | 6 |
| – | Culm leaf sheaths covered with white erect thin hairs | 8 |
| 6 | Culms up to 5m tall, more than 1cm in diam | Gelidocalamustessellatus |
| – | Culms less than 4m, less than 1cm in diam | 7 |
| 7 | Culm leaf sheaths with white villus, margins with cilia | Gelidocalamusmonophyllus |
| – | Culm leaf sheaths hairless, margins glabrous | Gelidocalamusxunwuensis |
| 8 | Culm leaf sheath margins densely ciliate, oral setae 1 pair, foliage leaves 1(–2) per ultimate branch, lateral veins 6–9 pairs | Gelidocalamuslatifolius |
| – | Culm leaf sheath margins hairless, oral setae 2–3 pairs, foliage leaves 1–3 per ultimate branch, lateral veins 4–6 pairs | Gelidocalamusannulatus |
Discussion
Morphologically, although its inflorescence is not seen so far, G.zixingensis is undoubtedly a member of the genus Gelidocalamus, because it possesses all key characters of the genus, i.e., leptomorph rhizomes, several branches per node, typically a single foliage leaf on each ultimate branch, semelauctant inflorescence (Wen 1982; Li et al. 2006; Yi et al. 2008). However, it is obviously different from other species of the genus, e.g., conspicuously longer than the internodes and culm leaf densely white pubescent. At first glance, G.zixingensis is similar to G.multifolius in appearance, but can be distinguished by a ring of white-gray appressed pubescence below each node, culm sheaths densely pubescent with brown patches, sub-apical branch sheath much longer than the internode, and a single foliage leaf on each ultimate branch.
Previous studies indicated that leaf epidermal features were of taxonomic significance in Bambusoideae (Soderstrom and Ellis 1988; Yang et al. 2008; Zhang et al. 2014; Leandro et al. 2019). According to papilla form and distribution patterns around the stomatal apparatus of the abaxial leaf epidermis, Gelidocalamus can be classified into at least three types: (a) short papillae, none on the stomatal apparatus, e.g., G.stellatus, G.multifolius, G.tessellatus; (b) elongate or short papillae overarching the stomata, e.g., G.subsolidus, G.solidus; (c) many short papillae, completely covering stomatal apparatus, e.g., G.monophyllus (Wu et al. 2014). Compared to those of the “spring-shoot” taxa, leaf epidermal characters in the “gelido-” members of Gelidocalamus are relatively stable, and can be used as a diagnostic feature. In the present study, epidermal traits of foliage leaf in G.zixingensis were found to be consistent with these of six “gelido-” members in Gelidocalamus (except G.monophyllus, Liu et al. 2017; Nie et al. 2018), and the difference mainly lay in the fact that G.zixingensis had 5 rows of stomatal apparatus.
The tribe Arundinarieae was known for its complex phylogenetic relationships. Despite many previous attempts based on different datasets having been made, intractable problems, such as low resolution or heavily conflicting topologies, still arose (Zhang et al. 2012; Yang et al. 2013; Zeng et al. 2014; Ma et al. 2017). Recently, with the wider sampling of Arundinarieae, Guo et al. (2021) provided a robust phylogenetic tree of the tribe, referred from the ddRAD dataset, which was mostly consistent with the morphological data. In the phylogenetic analysis, only six “gelido-” members formed a monophyletic lineage, although all members of Gelidocalamus belonged to the leptomorph lineage. Together with our present molecular phylogenetic analysis, we confirm that G.zixingensis belongs to the genus Gelidocalamus, and it is closely related to G.multifolius.
Supplementary Material
Acknowledgements
We are grateful to Chun-Ling Long (Zhejiang Normal University), Ting Kong (Jiangxi Matou Mountain National Nature Reserve), Wei-Jian Li (Nanchang Business College, Jiangxi Agricultural University), Yu-Guang Liu (Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences) for their work in field surveys and sampling. This study was financially supported by the National Natural Science Foundation of China [31960335, 31260043] and Key R & D Planned Projects of Jiangxi Province [20192BBF60015].
Citation
Wang C-K, Guo R, Guo C-C, Yang G-Y, Zhang W-G (2023) Gelidocalamus zixingensis (Poaceae, Bambusoideae, Arundinarieae), a new species from southern China revealed by morphological and molecular evidence. PhytoKeys 218: 29–45. https://doi.org/10.3897/phytokeys.218.96849
Funding Statement
National Natural Science Foundation of China [31960335, 31260043] and Key R & D Planned Projects of Jiangxi Province [20192BBF60015]
Contributor Information
Guang-Yao Yang, Email: yanggy2004@126.com.
Wen-Gen Zhang, Email: wgzhang@jxau.edu.cn.
Supplementary materials
Gelidocalamuszixingensis complete chloroplast genome sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Cheng-Kun Wang, Rong Guo, Chun-Ce Guo, Guang-Yao Yang, Wen-Gen Zhang
Data type
Phylogenetic.
References
- Andrews S. (2016) FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Cai ZY, Zhou XX, Wong KM, Xia NH. (2021) Gelidocalamusfengkaiensis (Poaceae: Bambusoideae), a new bamboo species from Guangdong, China, with an analysis of branch development in relation to flowering. Botanical Studies (Taipei, Taiwan) 62(1): 12. 10.1186/s40529-021-00319-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. (2018) fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics (Oxford, England) 34(17): i884–i890. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed]
- Clark LG, de Oliveira RP. (2018) Diversity and evolution of the New World bamboos (Poaceae: Bambusoideae: Bambuseae, Olyreae). 11th World Bamboo Congress, Xalapa, Mexico: Proceedings, pt. 2. Plymouth: MA: World Bamboo Organization, 35–47.
- Ellis RP. (1979) A procedure for standardizing comparative leaf anatomy in the Poaceae. II. The epidermis as seen in surface view. Bothalia 12(4): 641–671. 10.4102/abc.v12i4.1441 [DOI] [Google Scholar]
- Ellis B, Daly D, Hickey L, Johnson K, Mitchell J, Wilf P, Wing S. (2009) Manual of Leaf Architecture. Cornell University Press, New York, 24–25. 10.1079/9781845935849.0000 [DOI]
- Gallaher TJ, Peterson PM, Soreng RJ, Zuloaga FO, Li DZ, Clark LG, Tyrrell CD, Welker CAD, Kellogg EA, Teisher JK. (2022) Grasses through space and time: An overview of the biogeographical and macroevolutionary history of Poaceae. Journal of Systematics and Evolution 60(3): 522–569. 10.1111/jse.12857 [DOI] [Google Scholar]
- Guo C, Guo ZH, Li DZ. (2019) Phylogenomic analyses reveal intractable evolutionary history of a temperate bamboo genus (Poaceae: Bambusoideae). Plant Diversity 41(4): 213–219. 10.1016/j.pld.2019.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Ma PF, Yang GQ, Ye XY, Guo Y, Liu JX, Liu YL, Eaton DAR, Guo ZH, Li DZ. (2021) Parallel ddRAD and Genome Skimming Analyses Reveal a Radiative and Reticulate Evolutionary History of the Temperate Bamboos. Systematic Biology 70(4): 756–773. 10.1093/sysbio/syaa076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhang L, Columbus JT, Hu Y, Zhao Y, Tang L, Guo Z, Chen W, McKain M, Bartlett M, Huang CH, Li DZ, Ge S, Ma H. (2022) A well-supported nuclear phylogeny of Poaceae and implications for the evolution of C4 photosynthesis. Molecular Plant 15(4): 755–777. 10.1016/j.molp.2022.01.015 [DOI] [PubMed] [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. (2018) GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv: 256479. 10.1101/256479 [DOI]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. (2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14(6): 587–589. 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (Oxford, England) 28(12): 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keng PC, Wang ZP. (1996) Gramineae. Flora of China, vol. 9(1). Science Press, Beijing, 622–630.
- Leandro TD, Scatena VL, Clark LG. (2019) Comparative leaf blade anatomy and micromorphology in the systematics and phylogeny of Bambusoideae (Poaceae: Poales). Botanical Journal of the Linnean Society 192(1): 165–183. 10.1093/botlinnean/boz074 [DOI] [Google Scholar]
- Li DZ, Wang ZP, Zhu ZD, Xia NH, Jia LZ, Guo ZH, Yang GY, Stapleton CMA. (2006) Bambuseae (Poaceae). In: Wu ZY, Raven PH, Hong DY. (Eds) Flora of China.vol. 22 Science Press and Missouri Botanical Garden Press, Beijing and St. Louis, 7–180.
- Li WJ, Zhang WG, Tang M, Ji CF, Yang GY. (2016) The Specimen Collection Situation and Species Distribution of Gelidocalamus. Journal of Bamboo Research 35: 1–7. [Google Scholar]
- Liu YG, Li WJ, Tang M, Yang GY, Zhang WG. (2017) Taxonomic re-evaluation of some Gelidocalamus (Poaceae: Bambusoideae) taxa from Southeast China. Phytotaxa 299(1): 111–117. 10.11646/phytotaxa.299.1.9 [DOI] [Google Scholar]
- Ma PF, Vorontsova MS, Nanjarisoa OP, Razanatsoa J, Guo ZH, Haevermans T, Li DZ. (2017) Negative correlation between rates of molecular evolution and flowering cycles in temperate woody bamboos revealed by plastid phylogenomics. BMC Plant Biology 17(1): 260. 10.1186/s12870-017-1199-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8(19): 4321–4326. 10.1093/nar/8.19.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. (2015) IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Molecular Biology and Evolution 32(1): 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie TJ, Li WJ, Ji XN, Liu YG, Li ZY, Yang GY, Zhang WG. (2018) Re-evaluation of the taxonomy of Gelidocalamusstellatus (Poaceae: Bambusoideae) and its infraspecific taxa from southern China. Phytotaxa 356(3): 215–225. 10.11646/phytotaxa.356.3.3 [DOI] [Google Scholar]
- Qin QM, Tong YH, Zheng XR, Ni JB, Xia NH. (2021) Sinosasa (Poaceae: Bambusoideae), a new genus from China. Taxon 70(1): 27–47. 10.1002/tax.12422 [DOI] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. (2019) CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Research 47(W1): W65–W73. 10.1093/nar/gkz345 [DOI] [PMC free article] [PubMed]
- Soderstrom TR, Ellis RP. (1988) The woody bamboos (poaceae: bambuseae) of sri lanka: a morphological-anatomical study. Smithsonian Contributions to Botany 72(72): 1–75. 10.5479/si.0081024X.72 [DOI] [Google Scholar]
- Soreng RJ, Peterson PM, Zuloaga FO, Romaschenko K, Clark LG, Teisher JK, Gillespie LJ, Barberá P, Welker CAD, Kellogg EA, Li DZ, Davidse G. (2022) A worldwide phylogenetic classification of the Poaceae (Gramineae) III: An update. Journal of Systematics and Evolution 60(3): 476–521. 10.1111/jse.12847 [DOI] [Google Scholar]
- Triplett JK. (2008) Phylogenetic relationships among the temperate bamboos (Poaceae: Bambusoideae) with an emphasis on Arundinaria and allies. PhD Thesis, Iowa State University, America.
- Vorontsova M, Clark LG, Dransfield J, Govaerts R, Baker WJ. (2016) World Checklist of Bamboos and Rattans. Science Press, Beijing.
- Wen TH. (1982) A new genus and some new species of Bambusoideae from China. Journal of Bamboo Research 1(1): 20–45. [Google Scholar]
- Wick RR, Schultz MB, Zobel J, Holt KE. (2015) Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics (Oxford, England) 31(20): 3350–3352. 10.1093/bioinformatics/btv383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Long CL, Yu F, Ji CF, Wang X, Yang GY. (2014) Leaf Micromorphology of Gelidocalamus Wen and Its Taxonomical Significance. Zhiwu Kexue Xuebao 32: 211–215. [Google Scholar]
- Yang BM. (1986) Three new species of bamboo native of Hunan. Nature Science Journal of Hunan Normal University 3: 89–94. [Google Scholar]
- Yang HQ, Wang H, Li DZ. (2008) Comparative morphology of the foliage leaf epidermis, with emphasis on papillae characters, in key taxa of woody bamboos of the Asian tropics (Poaceae: Bambusoideae). Botanical Journal of the Linnean Society 156(3): 411–423. 10.1111/j.1095-8339.2007.00736.x [DOI] [Google Scholar]
- Yang HM, Zhang YX, Yang JB, Li DZ. (2013) The monophyly of Chimonocalamus and conflicting gene trees in Arundinarieae (Poaceae: Bambusoideae) inferred from four plastid and two nuclear markers. Molecular Phylogenetics and Evolution 68(2): 340–356. 10.1016/j.ympev.2013.04.002 [DOI] [PubMed] [Google Scholar]
- Yi TP, Shi JY, Ma LS, Wang HT, Yang L. (2008) Iconographia Bambusoidearum Sinicarum. Science Press, Beijing, 214–215.
- Zeng L, Zhang Q, Sun R, Kong H, Zhang N, Ma H. (2014) Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nature Communications 5(1): 4956. 10.1038/ncomms5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YX, Zeng CX, Li DZ. (2012) Complex evolution in Arundinarieae (Poaceae: Bambusoideae): Incongruence between plastid and nuclear GBSSI gene phylogenies. Molecular Phylogenetics and Evolution 63(3): 777–797. 10.1016/j.ympev.2012.02.023 [DOI] [PubMed] [Google Scholar]
- Zhang YX, Zeng CX, Li DZ. (2014) Scanning electron microscopy of the leaf epidermis in Arundinarieae (Poaceae: Bambusoideae): evolutionary implications of selected micromorphological features. Botanical Journal of the Linnean Society 176(1): 46–65. 10.1111/boj.12192 [DOI] [Google Scholar]
- Zhang WG, Ji XN, Liu YG, Li WJ, Yang GY. (2017) Gelidocalamusxunwuensis (Poaceae, Bambusoideae), a new species from southeastern Jiangxi, China. PhytoKeys 85: 59–67. 10.3897/phytokeys.85.13804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Poczai P, Hyvonen J, Tang J, Amiryousefi A. (2020) Chloroplot: An online program for the versatile plotting of organelle genomes. Frontiers in Genetics 11: 576124. 10.3389/fgene.2020.576124 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gelidocalamuszixingensis complete chloroplast genome sequences
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Cheng-Kun Wang, Rong Guo, Chun-Ce Guo, Guang-Yao Yang, Wen-Gen Zhang
Data type
Phylogenetic.