Abstract
Accurate assessment of treatment response and residual disease is indispensable for the evaluation of cancer treatment efficacy. However, performing tissue biopsies for longitudinal follow-up poses a major challenge in the management of solid tumours like neuroblastoma. In the present study, we evaluated whether circulating miRNAs are suitable to monitor neuroblastoma tumour burden and whether treatment-induced changes of miRNA abundance in the tumour are detectable in serum. We performed small RNA sequencing on longitudinally collected serum samples from mice carrying orthotopic neuroblastoma xenografts that were exposed to treatment with idasanutlin or temsirolimus. We identified 57 serum miRNAs to be differentially expressed upon xenograft tumour manifestation, out of which 21 were also found specifically expressed in the serum of human high-risk neuroblastoma patients. The murine serum levels of these 57 miRNAs correlated with tumour tissue expression and tumour volume, suggesting potential utility for monitoring tumour burden. In addition, we describe serum miRNAs that dynamically respond to p53 activation following treatment of engrafted mice with idasanutlin. We identified idasanutlin-induced serum miRNA expression changes upon one day and 11 days of treatment. By limiting to miRNAs with a tumour-related induction, we put forward hsa-miR-34a-5p as a potential pharmacodynamic biomarker of p53 activation in serum.
Graphical Abstract
Graphical Abstract.
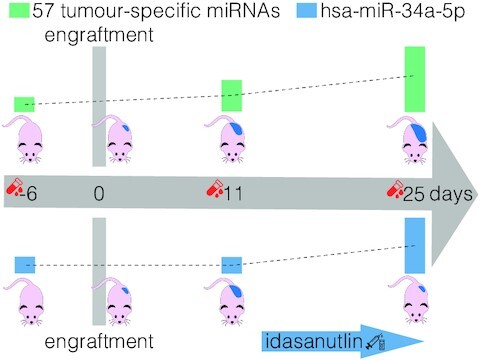
The level of 57 tumour-specific miRNAs reflects tumour burden and hsa-miR-34a-5p is a potential pharmacodynamic biomarker as its abundance in circulation is higher upon effective therapeutic targeting with idasanutlin.
INTRODUCTION
The ongoing improvements in the field of genomic profiling have enabled to study tumour biology in unprecedented detail and to unveil the molecular composition of tumours. However, a hurdle to translate potentially relevant findings into the clinical management of cancer patients is the need for tissue biopsies. This holds especially true for the monitoring of disease activity and the response to treatment over time, as longitudinal invasive sampling is difficult to justify. Novel, non-invasive biomarkers for assessing tumour burden and treatment response are highly desirable for patients with solid tumours like neuroblastoma, a pediatric malignant neoplasm originating from the developing sympathetic nervous system (1). Several non-invasive methodological strategies are under investigation to achieve this goal. Measurement of cell-free DNA for instance, using either a genome-wide approach or quantification restricted to neuroblastoma-associated genes or chromosomal regions, has proven to faithfully recapitulate neuroblastoma tumour biology (2–12).
MicroRNAs, a class of small, non-coding RNAs often found deregulated in numerous cancer types including neuroblastoma (13–16), have recently emerged as promising circulating biomarkers (17–22). In neuroblastoma, reports on miRNAs in circulation are rather scarce. Murray et al. found five miRNAs to be higher expressed in the serum of high-risk neuroblastoma patients, the expression levels of which could be used to distinguish MYCN-amplified neuroblastoma from other neuroblastoma tumours (23). Ramraj et al. performed miRNome profiling by RT-qPCR on serum collected from mouse models of favorable and unfavorable neuroblastoma to identify miRNAs that can distinguish favorable from high-risk neuroblastoma (24). Zeka et al. found nine miRNAs in serum samples strongly associated with tumour volume of murine xenografts and neuroblastoma disease burden and treatment response in metastatic neuroblastoma patients (25). In the present study, we evaluated whether miRNAs are suitable to monitor neuroblastoma tumour burden and whether treatment-induced changes of miRNA abundance in the tumour can be detected in the serum. In order to do so, we performed small RNA sequencing on longitudinally collected serum samples obtained from mice carrying orthotopic neuroblastoma xenografts that were exposed to treatment with idasanutlin (RG7388) or temsirolimus. The low frequency of TP53 mutations counterbalanced by an augmented MDM2 activity renders neuroblastoma highly sensitive to MDM2 antagonists such as idasanutlin (26–32). The rapamycin-derived mTOR-inhibitor temsirolimus has proven therapeutic efficacy in neuroblastoma and is currently under clinical investigation (33,34).
We identified 57 serum miRNAs to be differentially abundant upon xenograft tumour manifestation, out of which 21 were also found specifically expressed in the serum of human high-risk neuroblastoma patients. The murine serum levels of these 57 miRNAs correlated with both tumour tissue expression and tumour volume, suggesting potential utility for monitoring of tumour burden. In addition, we identified idasanutlin-induced serum miRNA expression changes both one day after the start of treatment and upon 11 days of treatment. By limiting to miRNAs with a tumour-related induction, we put forward hsa-miR-34a-5p as a potential pharmacodynamic biomarker of p53 activation in serum.
MATERIALS AND METHODS
Orthotopic xenograft model
Orthotopic neuroblastoma xenografts were generated in four to six week old female athymic immunodeficient NCr nude mice as previously described (35). Briefly, 1 × 106 human luciferase-SH-SY5Y neuroblastoma cells were surgically implanted beneath the renal capsule, towards the superior pole of the kidney. The detailed workflow has been described previously (36). This model closely resembles the growth characteristics of primary adrenal neuroblastoma tumours in humans. Tumour size was determined by luciferase imaging. Mice were peritoneally injected with luciferin and were sedated 15 min later using vaporized isoflurane in an induction chamber and luciferase signal intensity was measured. All animal work was approved by the Baylor College of Medicine Institutional Animal Care and Use Committee (IACUC) protocols (AN-3705 and AN-4810) and carried out in accordance with the relevant guidelines and regulations.
Treatment of mice
Mice were treated with 30 mg/kg idasanutlin (dissolved in hydroxypropylcellulose/Tween-80) or 9 mg/kg temsirolimus (dissolved in PBS/PEG/Tween-20) by oral gavage on a daily basis, five times a week, for a total of 12 days. Idasanutlin was kindly provided by Roche. Temsirolimus was purchased from Sigma Aldrich (PZ0020). After 12 days of treatment, mice were sacrificed and tumours were collected, weighed and snap frozen in liquid nitrogen.
Blood collection and serum preparation
At five different time points—6 days before engraftment, 2 days after engraftment, 11 days after engraftment, 15 days after engraftment and 25 days after engraftment (treatment of the mice was started 14 days after engraftment)—100 μl blood was collected by puncture of the jugular vein using a 4 mm lancet and collected in a BD Vacutainer collection tube with a gel separator. All blood samples were allowed to clot at room temperature for 45 min and were then centrifuged at 2000 × g for 15 min at 4°C using a fixed angle rotor. The supernatant was collected and stored at −80°C. For all serum samples, the degree of hemolysis was determined by measuring levels of free hemoglobin by spectral analysis using the Nanodrop 1000 (ThermoScientific). Absorbance peaks at 414 nm are indicative of free hemoglobin.
Total RNA isolation
For serum samples, RNA was isolated using the miRNeasy serum/plasma kit (Qiagen) according to the manufacturer's instructions. 50 μl of serum was used as input and total RNA was eluted in 12 μl of RNase-free water. For tumour samples, tumour material was denaturated using guanidinium thiocyanate and then lysed using the TissueLyser II (Qiagen) using stainless steel beads (5 mm) two times two minutes at 20 Hz. RNA was isolated using the miRNeasy mini kit (Qiagen), according to the manufacturer's instructions, and total RNA was eluted in 12 μl of RNAse-free water.
tRNA fragment depletion
tRNA halves were depleted from the RNA samples as described previously (37). In short, 12 μl of RNA, 15 μl of 2× hybridization buffer (Supplementary file S1), 1 μl of tRNA halves capture probes (at a final reaction concentration of 0.5 μM for each probe; Supplementary Table S1) and 2 μl of RNase-free water were incubated at 80°C for 2 min to denature the RNA. The mixture was slow-cooled to 22°C (at 0.1°C/min) and placed on ice to allow for efficient hybridization. Dynabeads Myone Streptavidin C1 (Life Technologies) were washed 3 times using a washing buffer (Supplementary file S1). After washing, beads were prepared for RNA manipulation by washing twice with solution A and once with solution B (Supplementary file S1) and suspended in 2× washing buffer to a final concentration of 5 μg/μl. Next, 30 μl of sample was added to 30 μl of beads and the mixture was incubated for 10 minutes at room temperature with gentle mixing. The mixture was then placed on a magnet for 2 min after which the supernatant, containing the depleted RNA, was collected. The depleted RNA was purified by ethanol precipitation and suspended in 7.5 μl RNase-free water.
Small RNA sequencing
For small RNA library preparation, we used the TruSeq small RNA library preparation kit v2 (Illumina) following manufacturer's instructions with small modifications. After PCR amplification, quality of libraries was assessed using a high sensitivity DNA kit on a Bioanalyzer (Agilent) according to manufacturer's instructions. Size selection was performed using 3% agarose dye-free marker H cassettes on a Pippin Prep (Sage Science) following manufacturer's instructions with a specified collection size range of 125–153 bp. Libraries were further purified and concentrated by ethanol precipitation, resuspended in 10 μl of 10 mM Tris–HCl (pH 8.5) and quantified using qPCR (see further). Based on the qPCR results, equimolar library pools were prepared, quality was assessed as described above and the library was further diluted to 4 nM using 10 mM Tris–HCl (pH 8.5). The pooled library was then sequenced at a final concentration of 1.2 pM on a NextSeq 500 using high output v2 kits (single-end, 75 cycles, Illumina). Raw sequencing data is available in the European Genome-Phenome archive (EGAS00001006678).
For RT-qPCR quantification of sequencing libraries to load equimolar concentrations, 1 μl of EtOH purified library was diluted 1:100 000. 2.5 μl of SsoAdvanced universal SYBR green supermix (Bio-Rad Laboratories) and 0.25 μl of each primer (5 μM) (forward primer: AATGATACGGCGACCACCGA; reverse primer: CAAGCAGAAGACGGCATACGA) were combined with 2 μl of diluted library. Reactions were set up in duplicate and performed in a LightCycler 480 (Roche) using the following protocol: enzyme activation at 95°C for 15 min, followed by 44 cycles of 95°C for 5 s, 60°C for 30 s and 72°C for 1 s.
Sequencing data analysis
For the quantification of small RNAs, a dedicated small RNA seq pipeline of Biogazelle (now a CellCarta company) was used. Adaptor trimming was performed using Cutadapt v1.8.1. Reads shorter than 15 bp and those in which no adaptor was found, were discarded. For quality control the FASTX-Toolkit (v0.0.14) was used, a minimum quality score of 20 in at least 80% of bases was applied as a cutoff. The reads were mapped with Bowtie (v1.1.2) without allowing any mismatches. Mapped reads were annotated by matching genomic coordinates of each read with genomic locations of miRNAs (obtained from miRBase, v21) and other small RNAs (obtained from UCSC (human: GRCh37/hg19; mouse: GRCm38/mm10) and Ensembl, v84). As publically available alternatives, we advise the use of miRDeep2 and miRExpress. Further data analysis was performed using RStudio (v0.99.486). Samples with fewer than 0.5 million mapped miRNA reads were omitted. Normalization of miRNA counts and differential expression analysis was performed using the R package DESeq2 (v1.8.2). Species specificity was determined based on the transcript sequence. In the case of perfect sequence conservation between the human and murine transcript, the transcript is annotated with both the human and murine mature miRNA name. Note that the presence of both perfectly conserved and species-specific transcripts of the same mature miRNA can result in multiple annotations.
Filtering hemolysis-associated miRNAs and samples
To identify miRNAs that correlate with hemolysis we determined Pearson's correlation coefficient for the abundance of all miRNA transcripts with the optical density at 414 nm (OD414). A correlation cut-off was determined based on the average correlation coefficient of human-specific miRNAs (mean hsa coefficient + 3 × stdev hsa coefficient), as these transcripts are not expected to be influenced by hemolysis in a murine model system. All miRNA transcripts above the correlation cut-off (n = 165) were excluded from differential expression analyses (Supplementary Table S2). In addition, all serum samples with an OD414 value above 1.5 (n = 4) were excluded for further analysis (Supplementary Table S3).
RT-qPCR of human serum samples
Serum pools were prepared by combining 40 μl of five individual serum samples, as described previously (25). Total RNA was isolated using the miRNeasy serum/plasma kit (Qiagen) according to the manufacturer's instructions. 200 μl of serum was used as input and total RNA was eluted in 12 μl of RNase-free water. cDNA synthesis was performed using the miScript II RT kit (Qiagen) following manufacturer's instructions with 1.5 μl of RNA as input. cDNA was pre-amplified using the miScript PreAMP PCR kit (Qiagen) following manufacturer's instruction with 10 amplification cycles. For each reaction 5 μl of 11-fold diluted cDNA was combined with 7 μl of RNAse-free water, 5 μl of 5× miScript PreAmp Buffer, 2 μl of HotStarTaq DNA Polymerase, 1 μl of miScript PreAmp Universal Primer (5 μM) and 5 μl of miScript PreAmp Primer Mix (5 μM). Whole miRNome RT-qPCR was performed using miScript miRNA PCR arrays (Qiagen) and the miScript SYBR Green PCR kit (Qiagen). For each reaction 2 μl of 22-fold diluted pre-amplified cDNA was combined with 1 μl of RNase-free water, 5 μl of 2× QuantiTect SYBR Green PCR Master Mix, 1 μl of 10× Universal Primer (5 μM). Data were normalized using the modified global mean, as implemented in qbase+ qPCR data-analysis software (Biogazelle), and non-detects imputed. All human work was approved by the Ethical Committee of Ghent University Hospital (EC UZG 2012/035).
RESULTS
Differential abundance analysis reveals circulating miRNAs released by tumour cells
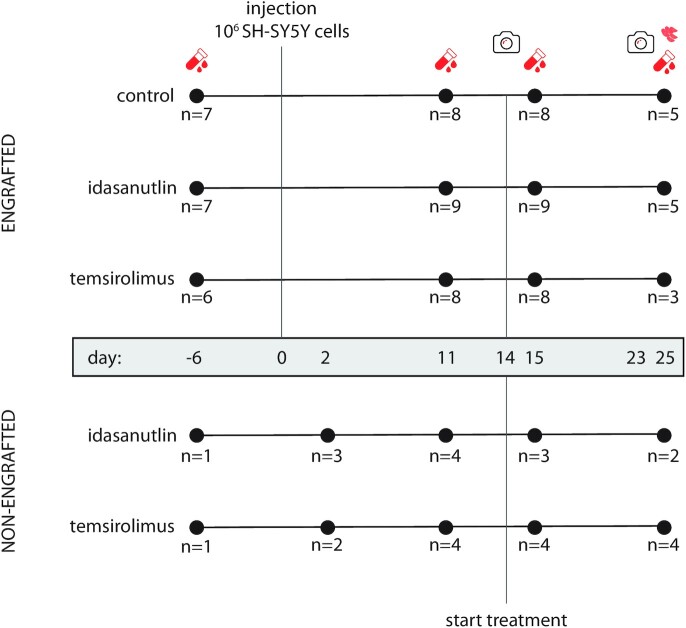
In order to identify circulating miRNAs that can be associated with tumour burden, we performed small RNA sequencing on serum samples of nude mice (n = 25) 6 days before and 11 days after orthotopic engraftment of human SH-SY5Y cells. Concurrently collected serum samples of mice (n = 8) not injected with neuroblastoma tumour cells were used as a negative control (Figure 1). When comparing the abundance profiles before and after tumour cell injection, we found a total of 94 differentially expressed (DE) miRNAs (fold change (FC) >2; P <0.05, Wald test, corrected for multiple testing using Benjamin–Hochberg) (Figure 2A) while in the non-engrafted group we detected six DE miRNAs (Supplementary Table S4A). When comparing serum miRNA abundance between the tumour bearing mice and the non-engrafted mice on both assessed time points, we detected 12 DE miRNA and 61 DE miRNAs (Figure 2B) (Supplementary Table S4B) before and after engraftment respectively. Of note, the serum samples collected on day 2 in the non-engrafted group are grouped with the samples collected on day −6 (Figure 1).
Figure 1.
Schematic representation of the study. Numbers represent the amount of mice at each time point for each condition after deduction of samples with high degree of hemolysis and samples with <0.5 million mapped miRNAs. Red collection tubes represent blood collections, red tumour cells represent the collection of tumour material, cameras represent luciferase imaging.
Figure 2.
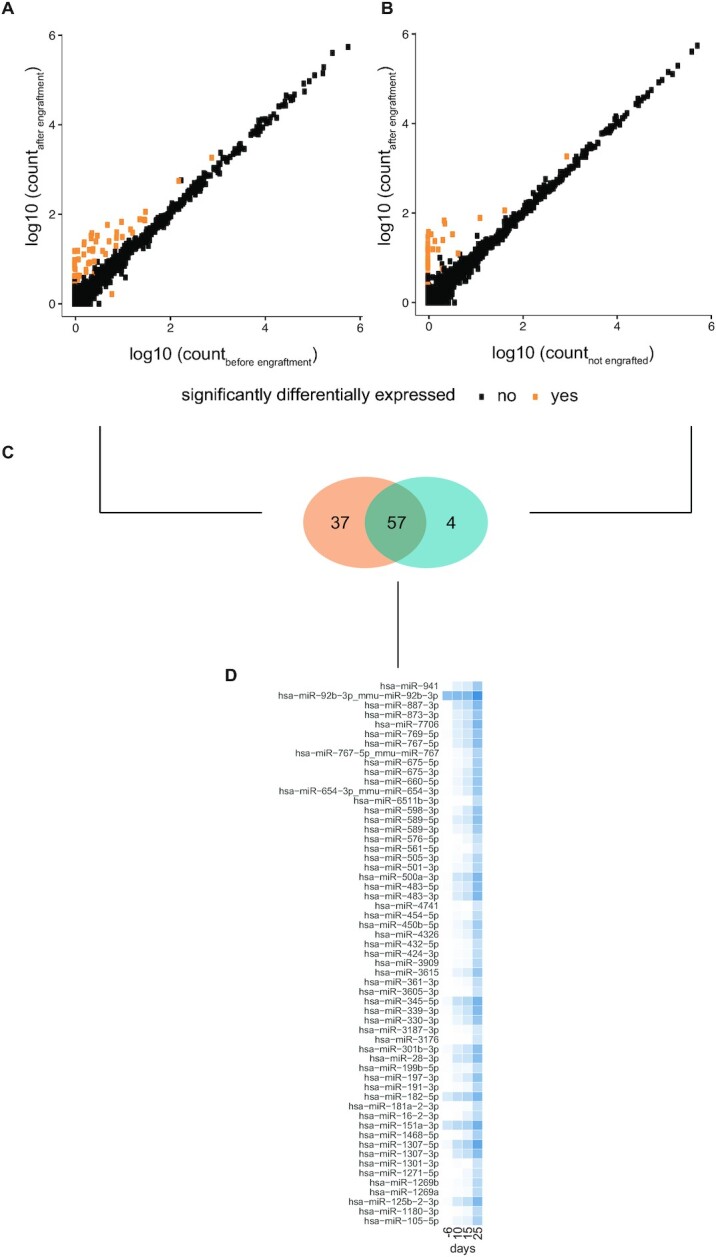
miRNAs differentially expressed in serum of mice carrying orthotopic xenograft tumours of human SH-SY5Y cells. (A) miRNA expression levels in the serum of mice 6 days before and 10 days after engraftment with 106 human SH-SY5Y cells. 94 miRNAs (in orange) were found differentially expressed between the two conditions (P < 0.05 and fold change (FC) >2, Wald test, corrected for multiple testing using Benjamin–Hochberg). (B) miRNA expression levels in the serum of mice 11 days after engraftment with 106 human SH-SY5Y cells and non-engrafted, control mice. 61 miRNAs (in orange) were found differentially expressed between the two conditions (P < 0.05 and FC > 2, Wald test, corrected for multiple testing using Benjamin–Hochberg). (C) Overlap between the two groups of differentially expressed miRNAs. 57 miRNAs are differentially expressed in both comparisons. (D) Heatmap showing an increasing log2 fold change in serum miRNA expression between mice carrying orthotopic xenografts and non-engrafted mice over time. Time points: −6 = 6 days before engraftment; 11 = 11 days after engraftment; 15 = 15 days after engraftment; 25 = 25 days after engraftment.
As we expect tumour-associated miRNAs to be both differentially abundant after engraftment between non-engrafted and engrafted mice, we determined the overlap between the DE miRNAs of these respective comparisons. This resulted in a set of 57 miRNAs that are convincingly altered as a result of tumour manifestation (Figure 2C, Supplementary Table S4C). As expected, we find this set of miRNAs to be heavily enriched for human-specific miRNAs (P < 2.2 × 10–16, Pearson's chi-squared test). Out of the 57 miRNAs, 54 miRNAs have a human specific sequence and three miRNAs have a sequence conserved between mice and humans. Considering human-specific miRNAs must have a tumour-cell origin, this must hold true for these DE miRNAs as well.
In addition, we performed small RNA sequencing on the serum of the same mice, collected at later time points, being 15 days (tumour-bearing mice: n = 8; tumour-free mice: n = 7) and 25 days (tumour-bearing mice: n = 5; tumour-free mice: n = 6) after engraftment (Figure 1). When we consider the expression profiles of the 57 DE miRNAs in all evaluated time points, we observe a gradual increase in expression in engrafted mice which was not observed in non-engrafted mice, evidenced by an increase in fold change between tumour-bearing and non-engrafted mice over time (Figure 2D). While we must note that the non-engrafted mice at these time points have been treated with either idasanutlin or temsirolimus, we do not expect these miRNAs to be influenced by the treatment as virtually all of them are human-specific (and thus absent in non-engrafted mice).
Serum miRNA abundance recapitulates tumour burden
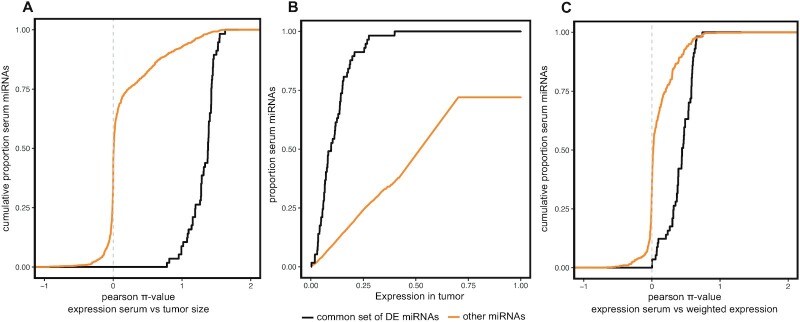
As we observed a gradual increase in serum abundance of the 57 DE miRNAs over time in tumour-carrying mice, we speculated that the serum expression levels of these miRNAs would correlate with tumour size. Tumour size was measured by luciferase imaging at two different time points (14 days and 23 days post tumour cell injection respectively; Figure 1). Pearson's π-values ( = Pearson's ρ × (–log10(P-value)); P-value corrected for multiple testing using Benjamin–Hochberg) between miRNA expression levels in serum (measured 15 days and 25 days after tumour cell injection) and the size of the paired tumour (measured by luciferase imaging 14 days and 23 days post tumour cell injection) were calculated. We found all of the 57 DE miRNAs to significantly correlate with tumour size (Pearson's ρ > 0.3 and adj. P < 0.05) and that these 57 miRNAs correlate significantly better with tumour size than other serum miRNAs (P < 2.2 × 10−16, two-sample Kolmogorov–Smirnov test) (Figure 3A, Supplementary Table S5). We found 334 miRNAs to be correlated with tumour size, out of which 278 are human-specific, 51 are conserved between mouse and human and 5 are mouse-specific (Supplementary Table S5). As we expect the 57 DE serum miRNAs to be also expressed in tumour tissue, we performed small RNA sequencing on end-point tumour samples (n = 13) (Figure 1). When we rank all detected miRNAs according to their mean expression in the tumour samples, we find that the 57 DE serum miRNAs are amongst the top expressed genes in the tumour (Figure 3B). This indicates that miRNAs with a high expression in the tumour are more likely to be detected as differentially expressed in serum. Next, we compared the expression of these miRNAs in paired tumour-serum samples to determine how faithfully serum expression recapitulates expression in the tumour. In order to do so, we calculated the Pearson π-value between miRNA expression in serum collected 25 days post tumour cell injection and the weighted miRNA expression (tumour miRNA expression x tumour size) in the paired tumour for all miRNAs detected in serum. We found that the 57 DE miRNAs correlated significantly stronger with weighted tumour expression than other miRNAs (P < 2.2 × 10–16, two-sample Kolmogorov–Smirnov test) (Figure 3C) (Supplementary Table S6).
Figure 3.
Serum miRNA expression correlates with tumour size. (A) Cumulative fraction of serum miRNAs by Pearson π-value (= Pearson's ρ × (–log10(P-value)); P-value corrected for multiple testing using Benjamin–Hochberg) of the correlation between miRNA serum expression and tumour size (measured by luciferase imaging, 14 days and 23 days after tumour cell injection). The 57 differentially expressed miRNAs (black) correlate significantly better with tumour size than other serum miRNAs (orange) (P < 2.2 × 10–16, Kolmogorov–Smirnov test). (B) Cumulative fraction of serum miRNAs ranked by expression in the tumour. The 57 differentially expressed serum miRNAs (black) are much higher expressed in the tumour than other serum miRNAs (orange) (P < 2.2 × 10–16, Kolmogorov–Smirnov test). All 57 serum miRNAs are among the top expressed tumour miRNAs, with half of these serum miRNAs among the top 10% expressed tumour miRNAs. (C) Cumulative fraction of serum miRNAs by Pearson π-value of the correlation between miRNA serum expression and weighted tumour expression (tumour size × tumour expression). The 57 differentially expressed miRNAs (black) correlate significantly better with weighted tumour expression than other serum miRNAs (orange) (P < 2.2 × 10–16, Kolmogorov–Smirnov test).
DE miRNAs in serum of human neuroblastoma patients
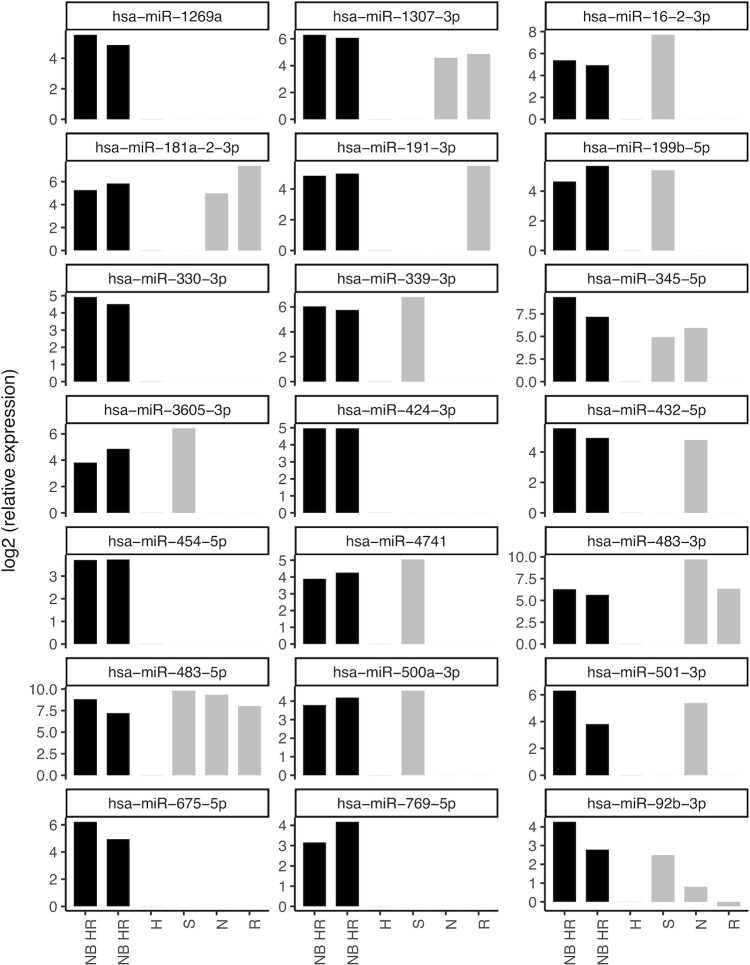
Ideally, liquid biopsy biomarkers for tumour burden should have a low (or absent) basal expression in healthy children. We therefore determined and compared the expression levels of the 57 DE miRNAs in two serum pools of high-risk neuroblastoma patients and one serum pool of healthy children by RT-qPCR. In addition, to evaluate whether these miRNAs are neuroblastoma or cancer specific, we included serum pools collected from children suffering from sarcoma, rhabdomyosarcoma or nephroblastoma. We considered a miRNA to be upregulated in neuroblastoma if the fold change between neuroblastoma and healthy expression was higher than 4 for all neuroblastoma serum pools. Doing so, we found 21 of the 57 DE miRNAs to be upregulated in serum of human neuroblastoma. Some of these miRNAs are putative neuroblastoma specific serum biomarkers (hsa-miR-1269a, hsa-miR-330-3p, hsa-miR-424-3p, hsa-miR-769-5p), while others may be general cancer serum biomarkers (hsa-miR-483-5p) (Figure 4). Details on the serum pooling and clinical characteristics of the patients included in this study were described previously (25).
Figure 4.
miRNA abundance in human serum. miRNA expression levels in serum pools of high-risk neuroblastoma patients (NB HR), healthy children (H), sarcoma patients (S), nephroblastoma patients (N) and rabdomyosarcoma patients (R) of miRNAs both differentially expressed in the serum of mice carrying orthotopic neuroblastoma xenografts and upregulated in both serum pools of high-risk neuroblastoma patients as compared to healthy serum.
Circulating miRNAs reflect idasanutlin treatment
As we have demonstrated that tumoural miRNA abundance is, to a certain degree, reflected in the serum, we wondered whether treatment-induced changes in serum miRNA abundance recapitulate expression changes in the tumour as well. Small RNA sequencing was performed on serum collected from tumour-bearing mice 3 days before, 1 day after and 11 days after start of treatment with 30 mg/kg/day idasanutlin, 9 mg/kg/day temsirolimus or vehicle control (n = 9, 9 and 5 for idasanutlin respectively; n = 8, 8 and 3 for temsirolimus respectively; n = 8, 8 and 5 for control respectively; see Figure 1). To identify miRNAs with a differential treatment effect, we performed Wald tests on the difference of the control/treatment expression ratio 3 days before treatment and 1 day and 11 days after start of treatment.
After 1 day of idasanutlin treatment, we identified 50 DE miRNAs and after 11 days of treatment 20 DE miRNAs (fold change (FC) >2; P <0.05, Wald test, corrected for multiple testing using Benjamin–Hochberg) (Supplementary Table S7). We did not find any DE miRNAs upon temsirolimus treatment, in line with a previous report that did not identify any miRNA expression alterations upon temsirolimus monotherapy in melanoma (38). Next, we filtered out murine-specific miRNAs, as they are unlikely to reflect expression changes inside the tumour and only kept miRNAs that have a human annotation. We further filtered results to only keep miRNAs with a significant expression difference both between control and treated samples and before and after treatment (Wald test, P < 0.05, corrected for multiple testing using Benjamin–Hochberg). This resulted in a total of 31 DE miRNAs (29 up, 2 down) after 1 day of treatment and six DE miRNAs (three up, three down) after 11 days of treatment (Figure 5). It may be possible that the smaller number of detected DE miRNAs after 11 days of treatment (compared to 1 day after treatment) is due to less statistical power as a result of a smaller number of mice in each group.
Figure 5.
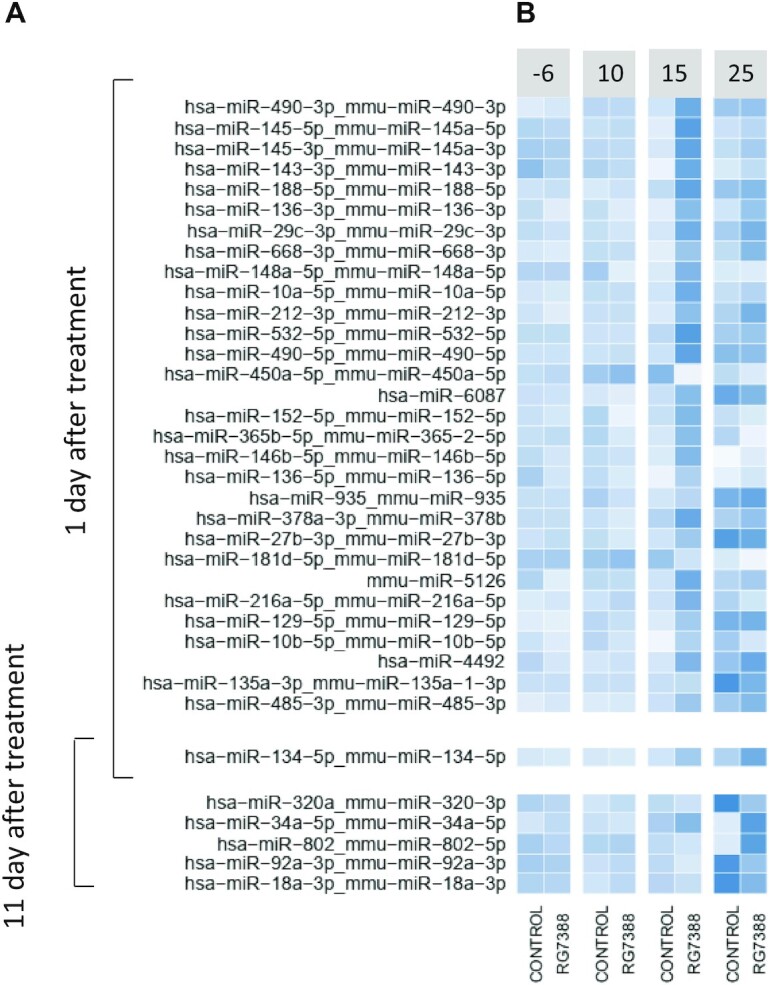
Serum miRNAs responsive to idasanutlin treatment. (A) Differentially expressed miRNAs in serum of mice carrying orthotopic xenograft tumours of SH-SY5Y cells after 1 or 11 days of treatment with idasanutlin (30 mg/kg per day), temsirolimus (9 mg/kg/day) or vehicle control (P < 0.05 and FC > 2, Wald test, corrected for multiple testing using Benjamin–Hochberg). (B) Scaled log2 expression levels of idasanutlin-responsive miRNAs in serum of mice treated with idasanutlin or vehicle control. Time points: −6 = 6 days before engraftment; 10 = 10 days after engraftment and 3 days before treatment; 15 = 15 days after engraftment and 1 day after start treatment; 25 = 25 days after engraftment and 11 days after start treatment.
When examining the expression profiles of these miRNAs in time, we can distinguish some distinctive expression patterns (Figure 5). Amongst the top DE miRNAs after 1 day of idasanutlin treatment are miRNAs of the known p53-regulated miR-143/145 cluster (39,40). Interestingly, this cluster of miRNAs is strongly induced 1 day after treatment but is no longer found differentially expressed 11 days after treatment. On the other hand, after 11 days of idasanutlin treatment, one of the DE miRNAs is miR-34a-5p, a key p53 effector miRNA (41,42).
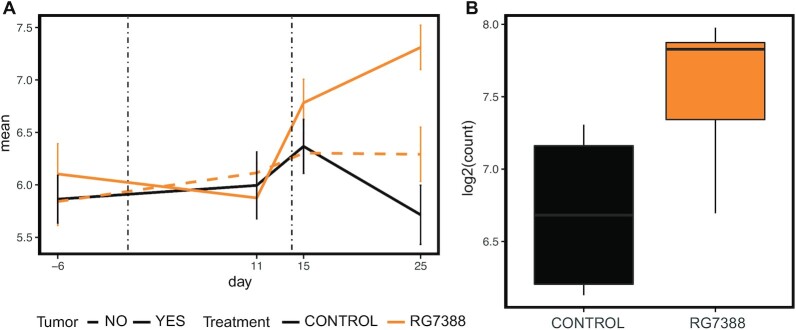
To further ensure a tumour-driven expression change, we compared miRNA expression levels in end-point tumour samples collected from mice treated with idasanutlin (n = 5) and mice treated with vehicle-control (n = 5). We find one miRNA to be differentially expressed in serum after 11 days of treatment (fold change (FC) >1.5; P <0.05, Wald test, corrected for multiple testing using Benjamin–Hochberg) and also upregulated in the tumour of the respective idasanutlin-treated mice (fold change (FC) > 1.5; P < 0.05, Wald test): hsa-miR-34a-5p/mmu-miR-34a-5p (Figure 6). Interestingly, hsa-miR-34a-5p/mmu-miR-34a-5p was also found upregulated after 1 day of treatment although not significantly (P = 0.59). These findings encourage further investigation of hsa-miR-34a-5p/mmu-miR-34a-5p as circulating pharmacodynamic biomarker for p53 activation in neuroblastoma.
Figure 6.
Serum and tumour expression levels of miR-34a-5p. (A) Serum expression levels of miR-34a-5p at different time points before and after treatment with idasanutlin (orange) or vehicle control (black). Serum was collected 6 days before engraftment (−6), 11 days after engraftment (11), one day after start of treatment (15) and 11 days after start of treatment (25). Solid lines represent mice carrying orthotopic xenograft tumours of SH-SY5Y cells; dashed lines represent non-engrafted, tumour-free mice. Error bars represent the standard error of the mean. (B) Boxplots of the expression levels of miR-34a-5p in tumours collected from mice treated for 11 days with idasanutlin or vehicle control.
DISCUSSION
Novel methods to monitor neuroblastoma progression and response to treatment are highly desired. To this purpose, we longitudinally assessed miRNA abundance in the serum of mice carrying orthotopic xenografts of neuroblastoma and exposed to treatment regimens of idasanutlin and temsirolimus, two clinically relevant small molecule drugs.
Regarding miRNAs associated with tumour burden, we found 57 serum miRNAs to be differentially expressed 10 days after tumour engraftment. Serum abundance of these miRNAs was found to strongly correlate with tumour size, with these miRNAs to be among the top expressed in the tumour, and to also correlate positively with tumoural expression. When evaluating the expression of these miRNAs in the serum of human neuroblastoma patients, we found 21 of these miRNAs to be higher expressed in serum from high-risk neuroblastoma patients compared to children without cancer. A detailed discussion concerning the importance of each of these miRNAs is beyond the scope of this manuscript; we do however like to mention that several of these miRNAs have been described in the context of neuroblastoma. hsa-miR-16-2-3p was found upregulated in MYCN-amplified neuroblastoma tumours as compared to neuroblastoma without MYCN amplification (43). Hsa-miR-92b-3p, hsa-miR-1307-3p, hsa-miR-330-3p and hsa-miR-345-5p for instance are known target genes of the MYCN oncogene often found overexpressed in high-risk neuroblastoma (44,45). Consistent with our observations, miR-191-3p, miR-345-5p, miR-92b-3p, miR-339-3p, miR-483-3p and miR-483-5p have all been reported upregulated in metastatic neuroblastoma compared to primary neuroblastoma (46). Of note, detection of tumoural miRNAs may depend on the injected cell line, as some microRNAs define distinct human neuroblastoma cell types, such as the adrenergic (e.g. SH-SY5Y) or mesenchymal cells (47). It is currently unknown whether the differentially abundant miRNAs in our study are specific to adrenergic-type neuroblastoma cells or rather reflect the generic neuroblastoma transcriptome irrespective of the cell state. It is of interest to further study this given the link between the mesenchymal cell phenotype and treatment resistance and relapse (48,49). An important issue regarding the use of circulating miRNAs as blood-based biomarkers concerns the cell of origin. As we used small RNA sequencing to quantify miRNA expression levels, we were able to accurately distinguish between murine and human miRNA sequences. As such, we found the majority of upregulated miRNAs to be human-specific, suggesting they are most likely derived from tumour cells. Whether they are actively released by the tumour or enter circulation as a result of breakdown of apoptotic or necrotic tumour cells remains unclear. Unfortunately miRNA profiling of pure extracellular vesicles or exosomes, actively released by tumour cells, is not yet feasible on minute amounts of serum or plasma, as such an approach could allow to more selectively detect tumour-secreted miRNAs.
Besides serum miRNAs associated with tumour burden, an important aim of this study was to identify serum miRNAs that are responsive to treatment. Non-invasive assessment of drug-induced molecular pathway activation could have clinical utility for patient follow-up. In clinical trials of RG7112, an idasanutlin predecessor, the measurement of serum levels of MIC-1, a secreted p53-inducible protein, has successfully been used in estimating p53 activation (50–52). Given p53 is a known modulator of miRNA expression (40,53,54), assessing p53 activation trough circulating miRNA abundance seemed a plausible scenario. Here, we describe for the first time serum miRNAs that dynamically respond to p53 activation following treatment of engrafted mice with idasanutlin. After only 24 hours of treatment we were able to detect significant induction of expression for 31 miRNAs, including known p53 transcriptional targets such as the miR-143/miR-145 cluster. After 11 days of treatment, we detected six DE miRNAs, including miR-34a-5p, another bona fide p53 response mediator. By comparing treatment-induced changes in serum expression in mice carrying tumours to changes in tumour-free mice and by associating expression changes in serum with changes in the tumour we restricted our analyses to miRNAs of which abundance changes in serum are likely to reflect expression changes in the tumour. This resulted in one miRNA that could potentially function as biomarker for p53 activation, hsa-miR-34a-5p. miR-34a-5p is a known p53-regulated miRNA with potent anti-tumour effects. This miRNA is often found lower expressed in unfavourable neuroblastoma and it has been reported that in neuroblastoma targeted activation of p53 can lead to a potent induction of miR-34a expression in vitro (41,55,56). Interestingly, targeted delivery of miR-34a using anti-GD2 coated nanoparticles has potent anti-tumour effects in vivo in neuroblastoma (57). Whether the same holds true for innately circulating miR-34a-5p, or by extension other p53-regulated miRNAs in circulation, would form an interesting subject of further investigation.
In conclusion, we identified circulating miRNAs that are associated with both human neuroblastoma and murine neuroblastoma xenograft tumour burden and uncovered one miRNA that could potentially be used as non-invasive biomarker for p53 pathway activation. Our findings demonstrate that it is feasible to monitor both tumour burden and treatment response by measuring the levels of circulating miRNAs in serum and that expression changes in the tumour are reflected in serum. The identification of treatment-induced alterations of circulating tumour-related miRNAs is an unprecedented finding that holds promise for liquid biopsies as a tool for miRNA-based monitoring of treatment response in cancer patients.
DATA AVAILABILITY
The dataset supporting the conclusions of this article is available at the European Genome-phenome Archive (EGA – accession ID EGAS00001006678).
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the support by UGent Concerted Research Action BOF-GOA, Stichting tegen Kanker and the Hercules foundation. We gratefully acknowledge Biogazelle for providing access to its small RNA sequencing data analysis pipeline and UGent for the use of the supercomputing infrastructure HPC. Funding for open access charge: BOF-GOA [BOF.GOA.2022.0003.05]. Funding resources did not influence the study design and results.
Contributor Information
Alan Van Goethem, OncoRNALab, Cancer Research Institute Ghent (CRIG), Ghent, Belgium; Department of Biomolecular Medicine, Ghent University, Ghent, Belgium.
Jill Deleu, OncoRNALab, Cancer Research Institute Ghent (CRIG), Ghent, Belgium; Department of Biomolecular Medicine, Ghent University, Ghent, Belgium.
Nurten Yigit, OncoRNALab, Cancer Research Institute Ghent (CRIG), Ghent, Belgium; Department of Biomolecular Medicine, Ghent University, Ghent, Belgium.
Celine Everaert, OncoRNALab, Cancer Research Institute Ghent (CRIG), Ghent, Belgium; Department of Biomolecular Medicine, Ghent University, Ghent, Belgium.
Myrthala Moreno-Smith, Department of Pediatrics, Section of Hematology-Oncology, Texas Children's Cancer Center, Baylor College of Medicine, Houston, TX, USA.
Sanjeev A Vasudevan, Department of Pediatrics, Section of Hematology-Oncology, Texas Children's Cancer Center, Baylor College of Medicine, Houston, TX, USA.
Fjoralba Zeka, OncoRNALab, Cancer Research Institute Ghent (CRIG), Ghent, Belgium; Department of Biomolecular Medicine, Ghent University, Ghent, Belgium.
Fleur Demuynck, OncoRNALab, Cancer Research Institute Ghent (CRIG), Ghent, Belgium; Department of Biomolecular Medicine, Ghent University, Ghent, Belgium.
Eveline Barbieri, Department of Pediatrics, Section of Hematology-Oncology, Texas Children's Cancer Center, Baylor College of Medicine, Houston, TX, USA.
Frank Speleman, Department of Biomolecular Medicine, Ghent University, Ghent, Belgium; PPOL, Cancer Research Institute Ghent (CRIG), Ghent, Belgium.
Pieter Mestdagh, OncoRNALab, Cancer Research Institute Ghent (CRIG), Ghent, Belgium; Department of Biomolecular Medicine, Ghent University, Ghent, Belgium.
Jason Shohet, Department of Pediatrics, Division of Pediatric Hematology-Oncology, University of Massachusetts Chan Medical School, Worcester, MA, USA.
Jo Vandesompele, OncoRNALab, Cancer Research Institute Ghent (CRIG), Ghent, Belgium; Department of Biomolecular Medicine, Ghent University, Ghent, Belgium.
Tom Van Maerken, OncoRNALab, Cancer Research Institute Ghent (CRIG), Ghent, Belgium; Department of Biomolecular Medicine, Ghent University, Ghent, Belgium; Department of Laboratory Medicine, AZ Groeninge, Kortrijk, Belgium.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Cancer Online.
FUNDING
National Cancer Plan of the Belgian State [Action 29 to A.V.G.]; Kom op tegen Kanker (Stand up to Cancer, the Flemish Cancer Society); Bijzonder Onderzoeksfonds [BOF22/CDV/077]; Research Foundation – Flanders (FWO) [G0B2820N to J.V., LIQUIDHOPE TRANSCAN-2 project, 1803115N/1510813N to T.V.M., 1S07416N to C.E.].
Conflict of interest statement. J.V. is co-founder of Biogazelle, now a CellCarta company, providing human biofluid exRNA sequencing as a global CRO.
REFERENCES
- 1. Bosse K.R., Maris J.M. Advances in the translational genomics of neuroblastoma: from improving risk stratification and revealing novel biology to identifying actionable genomic alterations. Cancer. 2016; 122:20–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Combaret V., Audoynaud C., Iacono I., Favrot M.-C., Schell M., Bergeron C., Puisieux A. Circulating MYCN DNA as a tumor-specific marker in neuroblastoma patients. Cancer Res. 2002; 62:3646–3648. [PubMed] [Google Scholar]
- 3. Combaret V., Iacono I., Bellini A., Bréjon S., Bernard V., Marabelle A., Coze C., Pierron G., Lapouble E., Schleiermacher G. et al. Detection of tumor ALK status in neuroblastoma patients using peripheral blood. Cancer Med. 2015; 4:540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chicard M., Boyault S., Colmet Daage L., Richer W., Gentien D., Pierron G., Lapouble E., Bellini A., Clement N., Iacono I. et al. Genomic copy number profiling using circulating free tumor DNA highlights heterogeneity in neuroblastoma. Clin. Cancer Res. 2016; 22:5564–5573. [DOI] [PubMed] [Google Scholar]
- 5. Van Roy N., Van Der Linden M., Menten B., Dheedene A., Vandeputte C., Van Dorpe J., Laureys G., Renard M., Sante T., Lammens T. et al. Shallow whole genome sequencing on circulating cell-free DNA allows reliable noninvasive copy-number profiling in neuroblastoma patients. Clin. Cancer Res. 2017; 23:6305–6315. [DOI] [PubMed] [Google Scholar]
- 6. Wang X., Wang L., Su Y., Yue Z., Xing T., Zhao W., Zhao Q., Duan C., Huang C., Zhang D. et al. Plasma cell-free DNA quantification is highly correlated to tumor burden in children with neuroblastoma. Cancer Med. 2018; 7:3022–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yagyu S., Iehara T., Tanaka S., Gotoh T., Misawa-Furihata A., Sugimoto T., London W.B., Hogarty M.D., Teramukai S., Nakagawara A. et al. Serum-based quantification of MYCN gene amplification in young patients with neuroblastoma: potential utility as a surrogate biomarker for neuroblastoma. PLoS One. 2016; 11:e0161039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lodrini M., Sprüssel A., Astrahantseff K., Tiburtius D., Konschak R., Lode H.N., Fischer M., Keilholz U., Eggert A., Deubzer H.E. Using droplet digital PCR to analyze MYCN and ALK copy number in plasma from patients with neuroblastoma. Oncotarget. 2017; 8:85234–85251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chicard M., Colmet-Daage L., Clement N., Danzon A., Bohec M., Bernard V., Baulande S., Bellini A., Deveau P., Pierron G. et al. Whole-exome sequencing of cell-free DNA reveals temporo-spatial heterogeneity and identifies treatment-resistant clones in neuroblastoma. Clin. Cancer Res. 2018; 24:939–949. [DOI] [PubMed] [Google Scholar]
- 10. van Zogchel L.M.J., van Wezel E.M., van Wijk J., Stutterheim J., Bruins W.S.C., Zappeij-Kannegieter L., Slager T.J.E., Schumacher-Kuckelkorn R., Verly I.R.N., van der Schoot C.E. et al. Hypermethylated RASSF1A as circulating tumor DNA marker for disease monitoring in neuroblastoma. JCO Precis. Oncol. 2020; 4:291–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lodrini M., Graef J., Thole-Kliesch T.M., Astrahantseff K., Sprussel A., Grimaldi M., Peitz C., Linke R.B., Hollander J.F., Lankes E. et al. Targeted analysis of cell-free circulating tumor DNA is suitable for early relapse and actionable target detection in patients with neuroblastoma. Clin. Cancer Res. 2022; 28:1809–1820. [DOI] [PubMed] [Google Scholar]
- 12. Bosse K.R., Giudice A.M., Lane M.V., McIntyre B., Schürch P.M., Pascual-Pasto G., Buongervino S.N., Suresh S., Fitzsimmons A., Hyman A. et al. Serial profiling of circulating tumor DNA identifies dynamic evolution of clinically actionable genomic alterations in high-risk neuroblastoma. Cancer Discov. 2022; 12:2800–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen Y., Stallings R.L. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007; 67:976–983. [DOI] [PubMed] [Google Scholar]
- 15. Mestdagh P., Fredlund E., Pattyn F., Schulte J.H., Muth D., Vermeulen J., Kumps C., Schlierf S., De Preter K., Van Roy N. et al. MYCN/c-MYC-induced microRNAs repress coding gene networks associated with poor outcome in MYCN/c-MYC-activated tumors. Oncogene. 2010; 29:1394–1404. [DOI] [PubMed] [Google Scholar]
- 16. De Preter K., Mestdagh P., Vermeulen J., Zeka F., Naranjo A., Bray I., Castel V., Chen C., Drozynska E., Eggert A. et al. miRNA expression profiling enables risk stratification in archived and fresh neuroblastoma tumor samples. Clin. Cancer Res. 2011; 17:7684–7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O’Briant K.C., Allen A. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jin Y., Wong Y.S., Goh B.K.P., Chan C.Y., Cheow P.C., Chow P.K.H., Lim T.K.H., Goh G.B.B., Krishnamoorthy T.L., Kumar R. et al. Circulating microRNAs as potential diagnostic and prognostic biomarkers in hepatocellular carcinoma. Sci. Rep. 2019; 9:10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lawrie C.H., Gal S., Dunlop H.M., Pushkaran B., Liggins A.P., Pulford K., Banham A.H., Pezzella F., Boultwood J., Wainscoat J.S. et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008; 141:672–675. [DOI] [PubMed] [Google Scholar]
- 20. Giglio S., De Nunzio C., Cirombella R., Stoppacciaro A., Faruq O., Volinia S., Baldassarre G., Tubaro A., Ishii H., Croce C.M. et al. A preliminary study of micro-RNAs as minimally invasive biomarkers for the diagnosis of prostate cancer patients. J. Exp. Clin. Cancer Res. 2021; 40:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khan I.A., Rashid S., Singh N., Rashid S., Singh V., Gunjan D., Das P., Dash N.R., Pandey R.M., Chauhan S.S. et al. Panel of serum miRNAs as potential non-invasive biomarkers for pancreatic ductal adenocarcinoma. Sci. Rep. 2021; 11:2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Souza K.C.B., Evangelista A.F., Leal L.F., Souza C.P., Vieira R.A., Causin R.L., Neuber A.C., Pessoa D.P., Passos G.A.S., Reis R.M.V. et al. Identification of cell-free circulating microRNAs for the detection of early breast cancer and molecular subtyping. J. Oncol. 2019; 2019:8393769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Murray M.J., Raby K.L., Saini H.K., Bailey S., Wool S.V., Tunnacliffe J.M., Enright A.J., Nicholson J.C., Coleman N. Solid tumors of childhood display specific serum microRNA profiles. Cancer Epidemiol. Biomark. Prev. 2015; 24:350–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ramraj S.K., Aravindan S., Somasundaram D.B., Herman T.S., Natarajan M., Aravindan N. Serum-circulating miRNAs predict neuroblastoma progression in mouse model of high-risk metastatic disease. Oncotarget. 2016; 7:18605–18619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zeka F., Decock A., Van Goethem A., Vanderheyden K., Demuynck F., Lammens T., Helsmoortel H.H., Vermeulen J., Noguera R., Berbegall A.P. et al. Circulating microRNA biomarkers for metastatic disease in neuroblastoma patients. JCI Insight. 2018; 3:e97021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carr-Wilkinson J., O’Toole K., Wood K.M., Challen C.C., Baker A.G., Board J.R., Evans L., Cole M., Cheung N.K.V., Boos J. et al. High frequency of p53/MDM2/p14ARF pathway abnormalities in relapsed neuroblastoma. Clin. Cancer Res. 2010; 16:1108–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pugh T.J., Morozova O., Attiyeh E.F., Asgharzadeh S., Wei J.S., Auclair D., Carter S.L., Cibulskis K., Hanna M., Kiezun A. et al. The genetic landscape of high-risk neuroblastoma. Nat. Genet. 2013; 45:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van Maerken T., Vandesompele J., Rihani A., De Paepe A., Speleman F. Escape from p53-mediated tumor surveillance in neuroblastoma: switching off the p14ARF-MDM2-p53 axis. Cell Death Differ. 2009; 16:1563–1572. [DOI] [PubMed] [Google Scholar]
- 29. Van Maerken T., Speleman F., Vermeulen J., Lambertz I., De Clercq S., De Smet E., Yigit N., Coppens V., Philippé J., De Paepe A. et al. Small-molecule MDM2 antagonists as a new therapy concept for neuroblastoma. Cancer Res. 2006; 66:9646–9655. [DOI] [PubMed] [Google Scholar]
- 30. Van Maerken T., Ferdinande L., Taildeman J., Lambertz I., Yigit N., Vercruysse L., Rihani A., Michaelis M., Cinatl J., Cuvelier C.A. et al. Antitumor activity of the selective MDM2 antagonist nutlin-3 against chemoresistant neuroblastoma with wild-type p53. J. Natl. Cancer Inst. 2009; 101:1562–1574. [DOI] [PubMed] [Google Scholar]
- 31. Lakoma A., Barbieri E., Agarwal S., Jackson J., Chen Z., Kim Y., McVay M., Shohet J.M., Kim E.S. The MDM2 small-molecule inhibitor RG7388 leads to potent tumor inhibition in p53 wild-type neuroblastoma. Cell Death Discov. 2015; 1:15026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen L., Rousseau R.F., Middleton S.A., Nichols G.L., Newell D.R., Lunec J., Tweddle D.A. Pre-clinical evaluation of the MDM2-p53 antagonist RG7388 alone and in combination with chemotherapy in neuroblastoma. Oncotarget. 2015; 6:10207–10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen L., Pastorino F., Berry P., Bonner J., Kirk C., Wood K.M., Thomas H.D., Zhao Y., Daga A., Veal G.J. et al. Preclinical evaluation of the first intravenous small molecule MDM2 antagonist alone and in combination with temozolomide in neuroblastoma. Int. J. Cancer. 2019; 144:3146–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnsen J.I., Segerström L., Orrego A., Elfman L., Henriksson M., Kågedal B., Eksborg S., Sveinbjörnsson B., Kogner P. Inhibitors of mammalian target of rapamycin downregulate MYCN protein expression and inhibit neuroblastoma growth in vitro and in vivo. Oncogene. 2008; 27:2910–2922. [DOI] [PubMed] [Google Scholar]
- 35. Agarwal S., Ghosh R., Chen Z., Lakoma A., Gunaratne P.H., Kim E.S., Shohet J.M. Transmembrane adaptor protein PAG1 is a novel tumor suppressor in neuroblastoma. Oncotarget. 2016; 7:24018–24026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patterson D.M., Shohet J.M., Kim E.S. Preclinical models of pediatric solid tumors (neuroblastoma) and their use in drug discovery. Curr. Protoc. Pharmacol. 2011; 52:14.17.1–14.17.18. [DOI] [PubMed] [Google Scholar]
- 37. Van Goethem A., Yigit N., Everaert C., Moreno-Smith M., Mus L.M., Barbieri E., Speleman F., Mestdagh P., Shohet J., Van Maerken T. et al. Depletion of tRNA-halves enables effective small RNA sequencing of low-input murine serum samples. Sci. Rep. 2016; 6:37876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wagenseller A.G., Shada A., D’Auria K.M., Murphy C., Sun D., Molhoek K.R., Papin J.A., Dutta A., Slingluff C.L MicroRNAs induced in melanoma treated with combination targeted therapy of Temsirolimus and Bevacizumab. J. Transl. Med. 2013; 11:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang L., Shi Z.-M., Jiang C.-F., Liu X., Chen Q.-D., Qian X., Li D.-M., Ge X., Wang X.-F., Liu L.-Z. et al. MiR-143 acts as a tumor suppressor by targeting N-RAS and enhances temozolomide-induced apoptosis in glioma. Oncotarget. 2014; 5:5416–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suzuki H.I., Yamagata K., Sugimoto K., Iwamoto T., Kato S., Miyazono K. Modulation of microRNA processing by p53. Nature. 2009; 460:529–533. [DOI] [PubMed] [Google Scholar]
- 41. Cole K.A., Attiyeh E.F., Mosse Y.P., Laquaglia M.J., Diskin S.J., Brodeur G.M., Maris J.M. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol. Cancer Res. 2008; 6:735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., Bentwich Z., Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007; 26:731–743. [DOI] [PubMed] [Google Scholar]
- 43. Megiorni F., Colaiacovo M., Cialfi S., McDowell H.P., Guffanti A., Camero S., Felsani A., Losty P.D., Pizer B., Shukla R. et al. A sketch of known and novel MYCN-associated miRNA networks in neuroblastoma. Oncol. Rep. 2017; 38:3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mestdagh P., Boström A.-K., Impens F., Fredlund E., Van Peer G., de Antonellis P., von Stedingk K., Ghesquière B., Schulte S., Dews M. et al. The miR-17-92 microRNA cluster regulates multiple components of the TGF-β pathway in neuroblastoma. Mol. Cell. 2010; 40:762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hsu C.-L., Chang H.-Y., Chang J.-Y., Huang H.-C., Juan H.-F. Unveiling MYCN regulatory networks in neuroblastoma via integrative analysis of heterogeneous genomics data. Oncotarget. 2016; 7:36293–36310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guo J., Dong Q., Fang Z., Chen X., Lu H., Wang K., Yin Y., Cai X., Zhao N., Chen J. et al. Identification of miRNAs that are associated with tumor metastasis in neuroblastoma. Cancer Biol. Ther. 2010; 9:446–452. [DOI] [PubMed] [Google Scholar]
- 47. Samaraweera L., Grandinetti K.B., Huang R., Spengler B.A., Ross R.A. MicroRNAs define distinct human neuroblastoma cell phenotypes and regulate their differentiation and tumorigenicity. 2014; 14:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boeva V., Louis-Brennetot C., Peltier A., Durand S., Pierre-Eugène C., Raynal V., Etchevers H.C., Thomas S., Lermine A., Daudigeos-Dubus E. et al. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat. Genet. 2017; 49:1408–1413. [DOI] [PubMed] [Google Scholar]
- 49. van Groningen T., Koster J., Valentijn L.J., Zwijnenburg D.A., Akogul N., Hasselt N.E., Broekmans M., Haneveld F., Nowakowska N.E., Bras J. et al. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet. 2017; 49:1261–1266. [DOI] [PubMed] [Google Scholar]
- 50. Patnaik A., Tolcher A., Beeram M., Nemunaitis J., Weiss G.J., Bhalla K., Agrawal M., Nichols G., Middleton S., Beryozkina A. et al. Clinical pharmacology characterization of RG7112, an MDM2 antagonist, in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2015; 76:587–595. [DOI] [PubMed] [Google Scholar]
- 51. Ray-Coquard I., Blay J.-Y., Italiano A., Le Cesne A., Penel N., Zhi J., Heil F., Rueger R., Graves B., Ding M. et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol. 2012; 13:1133–1140. [DOI] [PubMed] [Google Scholar]
- 52. Yang H., Filipovic Z., Brown D., Breit S.N., Vassilev L.T. Macrophage inhibitory cytokine-1: a novel biomarker for p53 pathway activation. Mol. Cancer Ther. 2003; 2:1023–1029. [PubMed] [Google Scholar]
- 53. Yamakuchi M., Lotterman C.D., Bao C., Hruban R.H., Karim B., Mendell J.T., Huso D., Lowenstein C.J. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:6334–6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Georges S.A., Biery M.C., Kim S.-Y., Schelter J.M., Guo J., Chang A.N., Jackson A.L., Carleton M.O., Linsley P.S., Cleary M.A. et al. Coordinated regulation of cell cycle transcripts by p53-inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008; 68:10105–10112. [DOI] [PubMed] [Google Scholar]
- 55. Welch C., Chen Y., Stallings R.L. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007; 26:5017–5022. [DOI] [PubMed] [Google Scholar]
- 56. Rihani A., Van Goethem A., Ongenaert M., De Brouwer S., Volders P.-J., Agarwal S., De Preter K., Mestdagh P., Shohet J., Speleman F. et al. Genome wide expression profiling of p53 regulated miRNAs in neuroblastoma. 2015; 5:9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tivnan A., Orr W.S., Gubala V., Nooney R., Williams D.E., McDonagh C., Prenter S., Harvey H., Domingo-Fernández R., Bray I.M. et al. Inhibition of neuroblastoma tumor growth by targeted delivery of microRNA-34a using anti-disialoganglioside GD2 coated nanoparticles. 2012; 7:e38129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset supporting the conclusions of this article is available at the European Genome-phenome Archive (EGA – accession ID EGAS00001006678).