Abstract
Superoxide dismutase (SOD) is a nearly ubiquitous enzyme among organisms that are exposed to oxic environments. The single SOD of Helicobacter pylori, encoded by the sodB gene, has been suspected to be a virulence factor for this pathogenic microaerophile, but mutations in this gene have not been reported previously. We have isolated mutants with interruptions in the sodB gene and have characterized them with respect to their response to oxidative stress and ability to colonize the mouse stomach. The sodB mutants are devoid of SOD activity, based on activity staining in nondenaturing gels and quantitative assays of cell extracts. Though wild-type H. pylori is microaerophilic, the mutants are even more sensitive to O2 for both growth and viability. While the wild-type strain is routinely grown at 12% O2, growth of the mutant strains is severely inhibited at above 5 to 6% O2. The effect of O2 on viability was determined by subjecting nongrowing cells to atmospheric levels of O2 and plating for survivors at 2-h time intervals. Wild-type cell viability dropped by about 1 order of magnitude after 6 h, while viability of the sodB mutant decreased by more than 6 orders of magnitude at the same time point. The mutants are also more sensitive to H2O2, and this sensitivity is exacerbated by increased O2 concentrations. Since oxidative stress has been correlated with DNA damage, the frequency of spontaneous mutation to rifampin resistance was studied. The frequency of mutagenesis of an sodB mutant strain is about 15-fold greater than that of the wild-type strain. In the mouse colonization model, only 1 out of 23 mice inoculated with an SOD-deficient mutant of a mouse-adapted strain became H. pylori positive, while 15 out of 17 mice inoculated with the wild-type strain were shown to harbor the organism. Therefore, SOD is a virulence factor which affects the ability of this organism to colonize the mouse stomach and is important for the growth and survival of H. pylori under conditions of oxidative stress.
The gastric pathogen Helicobacter pylori is a curved, microaerophilic proteobacterium that has been implicated as a causal agent of peptic ulcers and a risk factor for adenocarcinoma. The only known natural hosts of H. pylori are primates, in whose gastric mucosa the organism can persist for years. During such infections, disease symptoms may or may not occur, though gastric inflammation is apparently ubiquitous. The pathogenesis of H. pylori relies on its persistence in surviving a harsh environment, including acidity, peristalsis, and attack by phagocytic cells and their released reactive oxygen species (28).
Using a variety of model systems (reviewed in reference 8), H. pylori virulence factors have been identified that affect the organism's ability to colonize the host. Such factors include motility and urease activity. Urease activity is critical for the establishment of infection (9) and is thought to enable colonization by producing ammonia, which helps neutralize the acidic microenvironment surrounding the organism (28), though this mechanism is not likely to be the only role for urease (10). Motility is required by the bacterium for colonization of the gastric mucosa (11), since H. pylori must move from the acidic environment of the stomach through the mucin, where the pH is higher. Other factors, such as adhesins for recognizing host cell receptors in the gastric epithelium, metabolic enzymes required for growth in vivo, and host defense resistance mechanisms, are likely to be required for colonization, as well (28).
Mechanisms for the detoxification of reactive oxygen species are of particular interest in H. pylori. Despite the fact that this organism is an obligate aerobe, it is unable to grow in atmospheric concentrations of oxygen. Microaerophilic organisms, like H. pylori, are particularly vulnerable to the detrimental effects of oxygen and oxidative stress (20). Nevertheless, they do possess some of the enzymatic machinery needed to eliminate or minimize toxic oxygen-derived products. Organisms that grow in oxic environments must have mechanisms to handle reactive oxygen species (e.g., superoxide anions, peroxides, and hydroxyl radicals) that are by-products of oxygen metabolism (16, 17). In addition to internally generated reactive oxygen species, the successful pathogen must also deal with reactive oxygen species that are generated by phagocytic cells of the host immune response. In H. pylori, genes encoding superoxide dismutase (SOD), catalase, and several putative peroxidases have been identified (1, 31, 33, 37). Of these genes, only katA (catalase) mutants have been characterized (31). Although catalase-defective mutants are no different from the parent in their binding to epithelial cells, this enzyme may be important in detoxification of reactive oxygen species produced by the host immune response. Genes encoding an akyl hydroperoxide reductase, a thiol-specific peroxidase, and other potential detoxification enzymes were identified (6, 28, 37), but mutations in these genes have not been reported or characterized.
Severe toxic effects of superoxide occur when this anion oxidizes the [4Fe-4S] clusters of redox enzymes, such as aconitase, 6-phosphogluconate dehydratase, and fumarase (reviewed in reference 13). This reaction not only disrupts the function of the target enzyme but frees Fe(II), which can bind to the negatively charged DNA and act as a catalyst for hydroxyl radical formation from H2O2 via the Fenton reaction (18, 19, 27). Perhaps the most destructive potential of superoxide is realized by production of the highly reactive hydroxyl radical. When produced in close proximity to DNA, this radical can damage the DNA, inducing mutations. Therefore, an important protective role of SOD, an enzyme that catalyzes the dismutation of superoxide anions (25), is ultimately its prevention of intracellular hydroxyl radical formation.
H. pylori expresses a dimeric SOD similar to other bacterial iron-cofactored (cytoplasmic) SODs, but the enzyme seems to be associated primarily with the surfaces of the cells (36). The enzyme has been purified, and the gene was cloned and sequenced (33). Recently, the distribution of Fe-SODs among H. pylori strains was studied (4). Different isoforms of the enzyme, apparently representing stable but strain-specific attributes, were identified. Though SOD has been suggested to be a likely virulence factor in H. pylori (6, 28), sodB mutants have not been generated to directly test this suggestion. In Campylobacter coli, which is also a microaerophile with a single SOD gene, sodB mutants were shown to have a decreased ability to colonize the chicken gut and were more sensitive to air for survival, but their growth rates in high-oxygen conditions (in shaken flasks) were unaffected (34).
Mouse colonization assays have been used previously to assay the importance of various genes for stomach colonization in H. pylori (8, 15). To better understand the microaerophily and pathogenesis of H. pylori, we constructed mutant strains with interrupted sodB genes and have characterized them with respect to growth and viability under various O2 conditions, oxidative stress tolerance, and mouse colonization abilities.
MATERIALS AND METHODS
Bacterial strains, growth, and media.
Helicobacter pylori ATCC 43504 was used as the wild-type strain. The Sydney strain, SS1 (22), was used in mouse colonization studies. Other Helicobacter strains were constructed as described below and are listed in Table 1. Cultures were grown at 37°C, under conditions of 5% CO2 and 2 to 12% O2 (varied as described) in a CO2 incubator (Forma Scientific). Blood agar (BA) plates contained Brucella agar (Difco), supplemented with 10% defibrinated sheep blood (Gibson). Where necessary, kanamycin was supplemented at a concentration of 25 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| DH5α | E. coli cloning strain | Lab stock |
| ATCC 43504 | H. pylori Kans Rifs; wild-type strain | Lab stock |
| SS1 | H. pylori Kans Rifs; mouse-adapted strain | D. McGee |
| HPG2 | 43504 sodB::aphA3/SOD− Kanr; aphA3 at NcoI site | This study |
| HPH2 | 43504 sodB::aphA3/SOD− Kanr; aphA3 at SphI site | This study |
| HP0388K | 43504 HP0388::aphA3/Kanr; gene directly downstream of sodB | This study |
| SSG2 | SS1 sodB::aphA3/SOD− Kanr; aphA3 at NcoI site | This study |
| Plasmids | ||
| pBluescript KS(+) | Commercial cloning vector | Stratagene |
| pHP1 | Source of aphA3 cassette | 29 |
| pKSsodB-3 | pKS+ (sodB) | This study |
| pG2 | pKSsodB-3 (aphA3) at unique NcoI site | This study |
| pH2 | pKSsodB-3 (aphA3) at unique SphI site | This study |
| pKS0388 | pKS+ (HP0388); gene directly downstream of sodB | This study |
| pKS388K | pKS0388 (aphA3) at unique NsiI site | This study |
Cloning was performed in Escherichia coli DH5α, grown on Luria-Bertani medium (24) supplemented with either ampicillin (100 μg/ml) or kanamycin (25 μg/ml) as required.
Cloning of the sodB gene of H. pylori ATCC 43504.
To clone the sodB gene from strain ATCC 43504, oligonucleotide primers which flank the gene were designed using the sequence published for strain 26695 (37), with mismatches that introduced restriction endonuclease (EcoRI) sites. The upstream primer had the sequence 5′-GTC TTG AAT TCA ACA AAG AAG-3′, and the downstream primer sequence was 5′-CTA ATT GAA TTC CTA TAT TGC-3′. High-fidelity PCR was performed using Pfu DNA polymerase (Stratagene) as recommended by the manufacturer. Chromosomal DNA from strain ATCC 43504 was used as the template. PCR products were phosphorylated with polynucleotide kinase and cloned into the SmaI site of pBluescript KS(+) (Stratagene) to create pKSsodB-3. Table 1 describes the plasmids used in this study. The sequence of the cloned region was determined by the University of Georgia Molecular Genetics Instrumentation Facility.
Construction of H. pylori mutants.
The kanamycin resistance gene aphA3 from C. coli was excised from the plasmid pHP1 (29) via EcoRI digestion, gel purified (Qiagen), and blunted with T4 DNA polymerase. This fragment was cloned into pKSsodB-3, which had been cut with either NcoI or SphI (both sites are unique and within the coding region of sodB), and blunted with T4 DNA polymerase. To construct sodB mutants, these plasmids, pG2 (aphA3 at the NcoI site) and pH2 (aphA3 at the SphI site), were used to electrotransform H. pylori to the Kanr phenotype by a protocol provided by D. McGee (personal communication). Briefly, H. pylori, grown on BA plates for 48 h, was harvested, washed with 10 ml of cold 15% glycerol–9% sucrose, and resuspended in 500 μl of the same buffer. Fifty-microliter aliquots were held on ice with 2 μl of plasmid (approximately 1 μg) for 10 min and then electroporated (25 μF, 800 Ω, 2.5 kV, 12.5 ms). Mueller-Hinton broth (100 μl) (Difco) was added, and the mixture was then spotted onto BA plates. Plates were incubated at 37°C for 2 days under conditions of 2% oxygen. Growth was scraped from these plates and streaked onto BA plates containing kanamycin, and incubation was continued for 5 to 8 days.
An approach similar to that used to isolate the sodB mutants was taken to obtain H. pylori strains with mutations in the downstream gene, known as gene HP0388 in strain 26695 (37). Oligonucleotides that flank the downstream gene (5′-TGT TGA ATT CAA GAC ACT CTG-3′ upstream with the EcoRI site and 5′-CCC ATT TAA ACA GCG CTT CC-3′ downstream with the Eco47III site) were used to amplify the gene, which was then cut with EcoRI and Eco47III and cloned into pBluescript KS(+), which had been cut with EcoRI and HincII, to form pKS0388. Subsequently, HP0388 was interrupted by insertion of the aphA3 cassette into the unique NsiI site to form pKS388K, which was used to transform H. pylori.
Sensitivity of growth to oxygen.
H. pylori strains were streaked for isolated colonies onto BA plates and incubated at 37°C with concentrations of oxygen at 2, 3, 4, 5, 6, 8, or 12%. Growth was scored after 48 h as described in Table 2. Oxygen concentrations were maintained via a Forma 3230 variable oxygen incubator. This instrument continuously measures oxygen levels by thermal conductivity and adjusts to the set concentration by injection of nitrogen gas.
TABLE 2.
Growtha of wild-type and mutant H. pylori strains under various oxygen concentrations
| Strain | Growth under O2 concn (%) of:
|
||||||
|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | 8 | 12 | |
| 43504 | +++ | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| HPG2 | +++ | ++ | + | +/− | +/− | +/− | − |
| HP388K | +++ | NT | NT | NT | NT | NT | ++++ |
Growth was scored after 48 h as follows: −, no growth; +/−, slight growth at the site of heaviest inoculum (see the text); +, significant growth at the site of heaviest inoculum; ++, significant growth and development of isolated, pinpoint colonies; +++, healthy growth with individual colonies up to ∼1 mm; ++++, healthy growth with colonies of >1 mm; NT, not tested.
Sensitivity to oxygen for viability.
To measure the toxicity of oxygen on wild-type and mutant strains of H. pylori, cells grown for 48 h on BA plates at a 2% oxygen concentration were suspended in 150 mM NaCl to an optical density at 600 nm (OD600) of 0.1. Five milliliters of the suspension was stored under conditions of 5% CO2–10% H2–85% N2 or stored in a covered petri plate under normal atmospheric conditions. Both sets of cells were stored at 37°C. Cell populations were measured via plate counts after various times of incubation.
Hydrogen peroxide sensitivity.
Sterile 7.5-mm-pore-size-filter paper disks were applied to BA plates which had been streaked for confluent growth. H2O2, 30% (wt/wt) (20 μl), was applied to each disk, and plates were incubated at 37°C for 48 h before the zones of growth inhibition were measured. Incubation was carried out in an incubator under controlled O2 concentrations which were varied from 2 to 12% partial pressure. The distance from the edge of the disk to the edge of the zone of growth was recorded. No zones of inhibition were observed when H2O was added to the disks instead of H2O2.
Superoxide dismutase activity.
Qualitative activities of cell extracts were assayed in nondenaturing gels by a method based on that of Beauchamp and Fridovich (2). Cells were harvested from BA plates after 48 h of growth, washed in 1.0 ml of lysis buffer (400 mM NaCl, 10 mM Tris [pH 8.0], 10 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM EDTA, 0.1 mM dithiothreitol, 10% [vol/vol] glycerol), and resuspended in 0.5 ml of lysis buffer. Cells were lysed via sonication (two 15-s pulses at an 80% micro-tip maximum with a W-380 Heat Systems-Ultrasonic, Inc., sonicator) on ice. Lysates were cleared by centrifugation at 4°C for 15 min and assayed for protein concentration using a kit (Pierce) based on Coomassie binding. Samples (100 μg) were applied to nondenaturing, 12.5% acrylamide gels and run until the dye front reached the bottom of the gel. Gels were soaked in 0.2% nitroblue tetrazolium for 20 min and then transferred to O2−-generating buffer (100 ml of 36 mM potassium phosphate [pH 7.8], freshly supplemented with 325 mg of N,N,N′,N′-tetramethylethelenediamine and 1 mg of riboflavin) for about 15 min under fluorescent light (10 to 20 cm from a 40-W light) until the gels turned blue and the contrast between the blue background and the achromatic (SOD) bands was easily discerned.
SOD activities were quantified by the method of McCord and Fridovich (25), with minor modifications as described by Maier and Moshiri (23). This method is based on the inhibition of superoxide-mediated cytochrome c reduction by extracts containing SOD. One unit of activity is the amount required to inhibit the reduction of cytochrome c by 50%. Data presented are the averages of three replicates ± the standard deviations.
Determination of spontaneous mutagenesis frequency.
The frequency of mutagenesis was estimated by a protocol based on that of Sisson et al. (35). Wild-type or mutant cells that had been grown on BA plates at 2% O2 were suspended in phosphate-buffered saline (PBS) at an OD600 of ≈25 (approximately 1010 CFU/ml). One-hundred-microliter samples (neat and 10−1 dilutions) were plated on BA plates containing 2, 5, or 10 μg of rifampin/ml. Dilutions of each suspension were used for total viable cell determinations. All plates were incubated in 2% O2. Mutation frequencies were calculated as the number of Rifr colonies/108 viable cells.
Mouse colonization studies.
Mouse colonization studies were carried out with 5-to-6-week-old, female C57BL/6J mice from Jackson Laboratories. H. pylori SS1 (22) (a gift from D. McGee) and sodB mutants in this strain were each passaged on plates the same number of times (three times for experiment 1 and nine times for experiment 2) for each experiment. Mice were starved for 2 h prior to oral gavage with 100 μl of bacterial suspension. Suspensions were prepared by scraping 48-h growth from BA plates or BA plates with kanamycin into PBS at an OD600 of 1.7 (∼109 CFU/ml). The head space above these suspensions was sparged with Ar gas to maintain low oxygen exposure during the time interval (approximately 15 min) between harvest and gavage. After 20 (experiment 1) or 21 (experiment 2) days, the mice were starved for 3 h and sacrificed, and the stomachs were excised. Stomachs were cut into four pieces each and homogenized in 5 ml of Ar-sparged PBS in 10-ml Dounce hand homogenizers. Samples (100 μl) were plated in duplicate, both neat and at 10−1 dilutions. Plates were incubated for 5 to 7 days at 37°C in a 2% O2 partial pressure atmosphere before colonies were counted.
RESULTS
Cloning and sequencing of the sodB gene of H. pylori ATCC 43504.
The nucleotide and deduced amino acid sequences reported for sodB genes of different H. pylori strains vary slightly (1, 4, 36, 37). To compare the sequences for our wild-type strain, ATCC 43504, with those of other strains, we amplified the gene from chromosomal DNA via high-fidelity PCR using oligonucleotides complementary to strain 26695. The resulting fragment was cloned into pBluescript KS(+) (Stratagene) and sequenced. Compared with the sequence of 26695, we found 21 nucleotide differences within the coding region (data not shown), which confer only three amino acid changes. Compared to the amino acid sequence for ATCC 43504 reported by Bereswill et al. (4), the predicted primary amino acid structure for our strain 43504 differs by one amino acid. We found an aspartic acid encoded at position 54; Bereswill et al. (4) found an alanine at this position. Our predicted protein sequence is identical to that of strains 60190 (36) and 151 (4). Both of these strains belong to the B isoform group of Fe-SODs, described by Bereswill et al. (4).
Inactivation of sodB and the gene directly downstream, HP0388, in H. pylori.
To inactivate sodB, the kanamycin resistance determinant from C. coli was cloned into either the unique NcoI site or the unique SphI site of the sodB gene carried on the pBluescript KS(+) vector. The resulting plasmids were used to transform H. pylori via electroporation. Homologous recombination resulted in kanamycin-resistant, SOD-deficient strains. Transconjugants were incubated for 7 to 8 days at 2% O2 before small colonies could be recovered. All attempts to recover mutants with incubation at 5 or 12% concentrations of O2 failed. PCR amplification of the sodB gene from each the of mutant strains confirmed the insertion of DNA consistent with the presence of the aphA3 gene. HP0388 was inactivated in a similar fashion except that the aphA3 cassette was inserted into the NsiI site of that gene.
Superoxide dismutase activity.
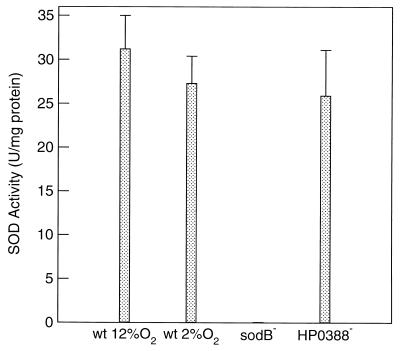
SOD activity was assayed in cell extracts both qualitatively on nondenaturing gels and quantitatively to compare specific activities in extracts from cells grown under two different oxygen stresses. In agreement with the results of Spiegelhalder et al. (36), only a single band of SOD activity was detected for wild-type H. pylori cells (Fig. 1). This band was absent in both sodB mutants (Fig. 1) but present in the HP0388 (downstream gene) mutant (data not shown). Quantitative assays of SOD activity, based on inhibition of cytochrome c reduction, revealed similar levels of activity in wild-type cell extracts whether the organism was grown at 2 or 12%-partial-pressure oxygen (Fig. 2). The sodB mutant had no detectable activity, and cells with interruptions in the downstream gene (HP0388) had levels comparable to that of the wild-type strain.
FIG. 1.

Superoxide dismutase activity of H. pylori cleared cell extracts in nondenaturing, activity-staining gels. Each lane was loaded with ∼100 mg of protein from either wild-type cells (A), HPG2 sodB cells (B), or HPH2 sodB cells (C).
FIG. 2.
Quantitative assay of superoxide dismutase activity in cleared cell extracts of H. pylori strains. O2 concentrations during growth were as indicated for wild-type cells (2 and 12%). The sodB mutant was grown in 2% O2, and the HP0388 mutant was grown in 12% O2. Error bars represent the standard deviations of results for three replicate experiments.
Growth under various oxygen concentrations.
The ability of H. pylori mutants to grow under various oxygen concentrations was determined by comparing growth on BA plates in an oxygen-controlled environment to that of the isogenic wild-type strain (Table 2). When incubated at 2% partial-pressure O2, growth of all strains was similar. However, at increasing O2 concentrations, the sodB mutant exhibited a decreased ability to grow. Above 3 to 4% O2, the mutant failed to form colonies on plates and only minimal growth was seen around the point of heaviest inoculum (initial streak on the plates). In contrast, the wild-type strain grew well at all O2 levels (2 to 12%), although it showed slightly greater growth in an O2 concentration of 12% partial pressure than at 2% O2. Growth of the HP0388 mutant was no different from that of the wild type at either 2 or 12% partial pressure O2.
Oxygen tolerance of nongrowing cells.
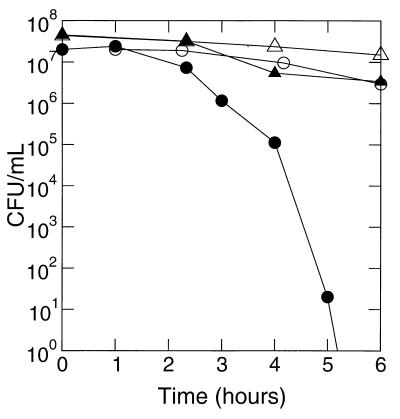
To determine whether the impaired growth of the sodB mutant at higher O2 levels was due to oxygen-dependent cell death, we measured the ability of nongrowing cells to survive periods of oxygen exposure (Fig. 3). A slight drop in viability was noted over the first few hours for both wild-type and mutant strains. However, after a 6-h period in an air-exposed container, the population of the wild-type cells decreased by about 1 order of magnitude (from 107 to 106 CFU/ml), while the mutant population decreased by more than 6 orders of magnitude. The decrease in wild type viability in air was not due to air exposure, since cells incubated in an anoxic environment showed a similar decrease in viability. Between hours 2 and 6, the mutant strain's viability in air rapidly decreased compared with that of cells incubated in an anoxic environment. The mutant population decreased to only about 20 CFU/ml after 5 h of incubation in the air atmosphere, and in three separate experiments no colonies were recovered after 6 h of incubation in air.
FIG. 3.
Survival of nongrowing H. pylori cells under atmospheric oxygen and anoxic environments. Wild-type (triangles) or sodB mutant (circles) cells grown under 2% O2 were suspended in 150 mM NaCl and incubated in either atmospheric oxygen (closed symbols) or without oxygen (open symbols). Samples were removed at the times indicated on the x axis and used for plate counts in a 2% O2 environment.
Susceptibility to hydrogen peroxide.
Decreased SOD activity has been shown to increase sensitivity to hydrogen peroxide for E. coli (5). To test whether H. pylori sodB mutants are more susceptible to this oxidative stress, filter paper disks were applied to plates streaked for confluent growth. Hydrogen peroxide was added to the disks, and the plates were incubated under various oxygen concentrations. Susceptibilities to hydrogen peroxide were measured as zones of inhibition of growth around the disks (Table 3). At each oxygen concentration, the zone of inhibition was considerably larger for the sodB mutants than for the wild type. Additionally, the zone size was affected by the O2 concentration in the incubation atmosphere for both strains. The highest concentrations of O2 (up to 12%) caused the largest zones of H2O2-dependent growth inhibition.
TABLE 3.
Hydrogen peroxide sensitivitiesa of H. pylori strains at various oxygen concentrations
| Strain | Growth inhibition zone (mm) at O2 concn (%) of:
|
|||
|---|---|---|---|---|
| 2 | 3 | 4 | 5 | |
| 43504 | 0 | 3 | 5 | 5 |
| HPG2 | 10 | 17 | 19 | NDb |
Sensitivity assayed as a zone of inhibition of growth, measured from the edge of the disk (containing 20 μl of 30% H2O2) to the edge of growth.
Not determined; growth was too light to accurately determine a zone of inhibition.
Determination of mutation frequency.
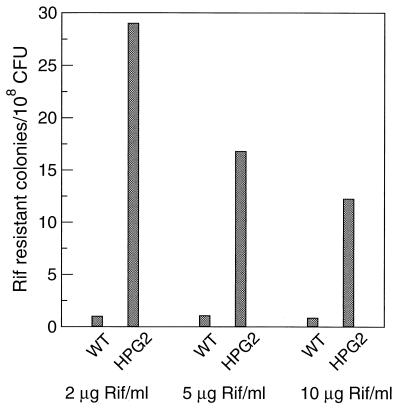
To test whether mutations in sodB increase DNA damage in H. pylori, the mutation frequency was assayed by quantifying the appearance of spontaneous forward mutation to rifampin resistance (Rifr). In four experiments, in which the rifampin concentration ranged from 2 to 10 μg/ml, the wild-type strain exhibited mutation frequencies ranging from 0.84 to 1.69 Rif colonies per 108 viable cells (Fig. 4). The SOD-deficient mutant, HPG2, gave frequencies ranging from 12.3 to 29.0 mutants per 108 viable cells among the four experiments. In each experiment, the mutation frequency of the sodB mutant strain was between 14.0 and 17.4 times higher than that of the wild type.
FIG. 4.
Spontaneous rifampicin resistance in wild-type and sodB H. pylori. Resistance was assayed by colony counts at three concentrations of rifampicin (as indicated) after incubation at 2% O2 for 48 h. The data shown for the 5-μg/ml Rif level is the mean for two separate experiments. Results for the 2- and 10-μg/ml Rif levels are from single experiments.
Mouse colonization of sodB mutants.
SOD has been implicated as a virulence factor in other organisms, including Campylobacter coli, where SOD was shown to be a colonization factor (34). To determine whether a deficiency in SOD activity is important for H. pylori colonization, the relative abilities of wild-type and sodB-deficient strains to colonize the mouse stomach were evaluated in two separate experiments. In each of the two experiments, with mice inoculated with the mouse-adapted strain SS1, H. pylori was recovered from the stomachs of 15 out of 17 animals (Table 4). However, only one of 23 mice that were inoculated with SSG2 (sodB mutant) was found to harbor the strain 3 weeks after oral administration. Colonies isolated from this mouse stomach were confirmed to be the sodB mutant strain based on their kanamycin resistance and their inability to grow at 12% O2. Both the wild-type and mutant strains were replated an identical number of times after the initial isolation of the mouse-colonizing strain. This step was done to reduce the possibility that repeated subculturing of the mutant led to the decrease in mouse infectivity that was observed. Also, the decrease in mutant colonization is not due to O2 exposure of the stomach isolates, since the stomachs were homogenized in low-O2 conditions, and the cells plated from the stomach homogenate dilutions were rapidly transported to the incubator containing 2% O2.
TABLE 4.
Mouse colonization abilitiesa of wild-type strains and sodB mutant strains
| Experiment no. | Colonization of stomachs by:
|
|
|---|---|---|
| Wild type | sodB mutant | |
| 1 | 7/8 | 1/8 |
| 2 | 8/9 | 0/15 |
Reported as number of mouse stomachs from which H. pylori was recovered/total number of stomachs assayed. Colonization for the wild type ranged from 1.7 × 103 to 8.9 × 105 viable cells/g of stomach. The mutant colonization (one stomach from experiment 1) was 1.7 × 103 viable cells/g of stomach, but the other stomachs that had been inoculated with the sodB mutant strain had no colonies on the plates, even from those receiving 0.1 ml plated directly from the 5-ml stomach homogenate solution.
DISCUSSION
SOD activity appears to be a common trait among organisms that live in and consume oxygen (26). This activity is a primary defense against the ubiquitous respiratory production of superoxide (17). In combination with hydrogen peroxide, another partially reduced O2 by-product of respiration (16), superoxide gives rise to highly reactive hydroxyl radicals that can damage nearby macromolecules like DNA or membrane lipids. Because microaerophilic organisms may be especially sensitive to the reactive forms of oxygen (20), and because this oxygen sensitivity is likely to be especially important in pathogenic organisms that must confront the oxidative attack of the host immune system, we wished to investigate the properties of H. pylori that are deficient in SOD activity. We demonstrate that sodB mutants of this microaerophile are even more sensitive than the parent strain to oxygen and oxygen metabolites and that they are deficient in colonization of the mouse stomach.
The growth ability of the sodB mutant was decreased relative to that of the wild-type strain at O2 levels above 2%. We observed some slight growth of the mutant in incubators held at O2 levels between 3 and 8%, although no isolated colonies appeared under these conditions. This observation suggests the presence of low-O2 microenvironments around the point of heaviest inoculation. Similar low-O2 microenvironments surrounding cells grown under different O2 concentrations may help to explain why we saw no difference in SOD activity levels between cells harvested from growth at 2 or 12% O2. Alternatively, SOD activities may be constitutive or regulated by factors that were not varied in our experiments.
We saw an increase in hydrogen peroxide sensitivity as O2 levels were increased for both the wild-type and sodB strains. For each concentration, the mutant was more sensitive than the wild type. This oxygen-dependent peroxide sensitivity and hypersensitivity of the mutant are consistent with observations with E. coli, for which a major toxic effect of superoxide is its generation of catalysts for hydroxyl radical formation from hydrogen peroxide (19).
In contrast to E. coli, which has three known SODs (3), H. pylori has only one, an iron-cofactored species (36). Fe-SODs are normally associated with the cytoplasm (14) and are ordinarily involved in protection against internally generated superoxide. The SOD of H. pylori seems to be associated with the cell surface (36), consistent with a role in protection against exogenously produced superoxide like that incurred during an oxidative burst via a host inflammatory response. Use of an iron-cofactored SOD for protection from exogenous superoxide has been noted for C. jejuni, for which interruptions in sodB decreased the rate of survival inside host cells (32).
Our mouse studies indicate that H. pylori colonization is greatly affected by sodB mutations. However, from these experiments we cannot determine whether the decreased viability of the sodB mutants under oxidative conditions prohibits their survival before they can establish residency in the mucosa or whether they are more sensitive and fall prey to the oxidative burst of a host immune response while residing in the mucin. Because we did confirm that one mouse became colonized by strain HPG2, we cannot report that sodB is required for colonization. Due to animal variability in a model system such as this, we assume that atypical conditions which allowed growth and survival of H. pylori existed in this single mouse.
To investigate whether the phenotype of the sodB mutants was due to polar disruption of downstream gene expression, we isolated isogenic mutants with aphA3 cassette interruptions in this gene. HP0388 is the designation for the gene that is 54 bp directly downstream of sodB in strain 26695 (37). The putative gene product has protein sequence homology to a hypothetical protein of unknown function from Haemophilus influenzae (HI0319) and to a putative methyl transferase in E. coli (EC1870) (http://www.tigr.org). The intergenic region, between sodB and HP0388, contains a putative transcriptional terminator, a 38-bp region with a 13-base inverted repeat (with one mismatch), followed by seven thymidine residues over the next nine bases. Unlike mutations in sodB, mutations in HP0388 do not affect SOD activity or growth in 12% O2. Based on these data, we suggest that the phenotypes reported here for sodB mutants are not the result of polar effects. Though a slight possibility exists that the virulence defect is due to this hypothetical product, which is possibly under the same transcriptional control as sodB, the obvious problem of an oxidative stress-related deficiency due to the sodB mutation is the likely cause for the decreased colonization.
In addition to [4Fe-4S] damage and subsequent hydroxyl radical damage, superoxide toxicity may include the reduction or oxidation of other compounds (12) and interaction with nitric oxide (NO) to form peroxynitrite (30). Because NO levels in gastric juice are fairly high (30), superoxide-dependent peroxynitrite toxicity may be an important factor in the defective colonization abilities of sodB mutants. Exposure to NO causes a transient decrease in respiration in E. coli but a sustained inhibition of respiration in H. pylori (30). Nagata et al. (30) have presented evidence that this inhibition is due to peroxynitrite production and is linked to higher superoxide levels in H. pylori. These data are consistent with the idea that sodB mutants of H. pylori may be compromised in respiration, another factor that could affect the ability of this organism to survive in its host.
Known H. pylori virulence factors include a variety of proteins that are involved in its pathogenesis, such as VacA and Cag (21). Another group of virulence factors is clearly important for colonization of H. pylori in the gastric mucosa. These include urease, motility factors (flagellin), and SOD. Because of this organism's microaerophilic nature and the increased levels of reactive oxygen in the infected host (7), we expect that other factors involved in the response to oxidative stress are likely to be required for virulence.
ACKNOWLEDGMENTS
We thank S. Maier for technical assistance and D. McGee for kindly providing strains and plasmids.
This research was supported by U.S. Department of Energy grant DE-FG02-99ER20321 to R.J.M.
REFERENCES
- 1.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 2.Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 3.Benov L, Fridovich I. Escherichia coli expresses a copper- and zinc-containing superoxide dismutase. J Biol Chem. 1994;269:25310–25314. [PubMed] [Google Scholar]
- 4.Bereswill S, Neuner O, Strobel S, Kist M. Identification and molecular analysis of superoxide dismutase isoforms in Helicobacter pylori. FEMS Microbiol Lett. 2000;183:241–245. doi: 10.1111/j.1574-6968.2000.tb08965.x. [DOI] [PubMed] [Google Scholar]
- 5.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doig P, de Jonge B L, Alm R A, Brown E D, Uria-Nickelsen M, Noonan B, Mills S D, Tummino P, Carmel G, Guild B C, Moir D T, Vovis G F, Trust T J. Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol Mol Biol Rev. 1999;63:675–707. doi: 10.1128/mmbr.63.3.675-707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drake I M, Mapstone N P, Schorah C J, White K L M, Chalmers D M, Dixon M F, Axon A T R. Reactive oxygen species activity and lipid peroxidation in Helicobacter pylori associated gastritis: relation to gastric mucosal ascorbic acid concentrations and effect of H. pylori eradication. Gut. 1998;42:768–771. doi: 10.1136/gut.42.6.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaton K A. Animal models of Helicobacter gastritis. Curr Top Microbiol Immunol. 1999;241:123–154. doi: 10.1007/978-3-642-60013-5_8. [DOI] [PubMed] [Google Scholar]
- 9.Eaton K A, Brooks C L, Morgan D R, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. 1991;59:2470–2475. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eaton K A, Krakowka S. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect Immun. 1994;62:3604–3607. doi: 10.1128/iai.62.9.3604-3607.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eaton K A, Suerbaum S, Josenhans C, Krakowka S. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect Immun. 1996;64:2445–2448. doi: 10.1128/iai.64.7.2445-2448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 13.Fridovich I. Superoxide anion radical (O2−), superoxide dismutases, and related matters. J Biol Chem. 1997;272:18515–18517. doi: 10.1074/jbc.272.30.18515. [DOI] [PubMed] [Google Scholar]
- 14.Fridovich I. Oxygen toxicity: a radical explanation. J Exp Biol. 1998;201:1203–1209. doi: 10.1242/jeb.201.8.1203. [DOI] [PubMed] [Google Scholar]
- 15.Ghiara P, Marchetti M, Blaser M J, Tummuru M K R, Cover T L, Segal E D, Tompkins L S, Rappuoli R. Role of Helicobacter pylori virulence factors vacuolating cytotoxin, CagA, and urease in a mouse model of disease. Infect Immun. 1995;63:4154–4160. doi: 10.1128/iai.63.10.4154-4160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez-Flecha B, Demple B. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J Biol Chem. 1995;270:13681–13687. doi: 10.1074/jbc.270.23.13681. [DOI] [PubMed] [Google Scholar]
- 17.Imlay J A, Fridovich I. Assay of metabolic superoxide production in Escherichia coli. J Biol Chem. 1991;266:6957–6965. [PubMed] [Google Scholar]
- 18.Keyer K, Gort A S, Imlay J A. Superoxide and the production of oxidative DNA damage. J Bacteriol. 1995;177:6782–6790. doi: 10.1128/jb.177.23.6782-6790.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keyer K, Imlay J A. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krieg N R, Hoffman P S. Microaerophily and oxygen toxicity. Annu Rev Microbiol. 1986;40:107–130. doi: 10.1146/annurev.mi.40.100186.000543. [DOI] [PubMed] [Google Scholar]
- 21.Labigne A, de Reuse H. Determinants of Helicobacter pylori pathogenicity. Infect Agents Dis. 1996;5:191–202. [PubMed] [Google Scholar]
- 22.Lee A, O'Rourke J, De Ungria M C, Robertson B, Daskalopoulos G, Dixon M F. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–1397. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- 23.Maier R J, Moshiri F. Role of the Azotobacter vinlandii nitrogenase-protective Shethna protein in preventing oxygen-mediated cell death. J Bacteriol. 2000;182:3854–3857. doi: 10.1128/jb.182.13.3854-3857.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 25.McCord J M, Fridovich I. Superoxide dismutase. An enzymic function of erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 26.McCord J M, Keele B B, Jr, Fridovich I. An enzyme-based theory of obligate anaerobiosis: the physiological function of superoxide dismutase. Proc Natl Acad Sci USA. 1971;68:1024–1027. doi: 10.1073/pnas.68.5.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCormick M L, Buettner G R, Britigan B E. Endogenous superoxide dismutase levels regulate iron-dependent hydroxyl radical formation in Escherichia coli exposed to hydrogen peroxide. J Bacteriol. 1998;180:622–625. doi: 10.1128/jb.180.3.622-625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGee D J, Mobley H L. Mechanisms of Helicobacter pylori infection: bacterial factors. Curr Top Microbiol Immunol. 1999;241:155–180. doi: 10.1007/978-3-642-60013-5_9. [DOI] [PubMed] [Google Scholar]
- 29.McGee D J, Radcliff F J, Mendz G L, Ferrero R L, Mobley H L. Helicobacter pylori rocF is required for arginase activity and acid protection in vitro but is not essential for colonization of mice or for urease activity. J Bacteriol. 1999;181:7314–7322. doi: 10.1128/jb.181.23.7314-7322.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagata K, Yu H, Nishikawa M, Kashiba M, Nakamura A, Sato E F, Tamura T, Inoue M. Helicobacter pylori generates superoxide radicals and modulates nitric oxide metabolism. J Biol Chem. 1998;273:14071–14073. doi: 10.1074/jbc.273.23.14071. [DOI] [PubMed] [Google Scholar]
- 31.Odenbreit S, Wieland B, Haas R. Cloning and genetic characterization of Helicobacter pylori catalase and construction of a catalase-deficient mutant strain. J Bacteriol. 1996;178:6960–6967. doi: 10.1128/jb.178.23.6960-6967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pesci E C, Cottle D L, Picket C L. Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect Immun. 1994;62:2687–2694. doi: 10.1128/iai.62.7.2687-2694.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pesci E C, Picket C L. Genetic organization and enzymatic activity of a superoxide dismutase from the microaerophilic human pathogen Helicobacter pylori. Gene. 1994;143:111–116. doi: 10.1016/0378-1119(94)90614-9. [DOI] [PubMed] [Google Scholar]
- 34.Purdy D, Cawthraw S, Dickinson J H, Newell D G, Park S F. Generation of a superoxide dismutase (SOD)-deficient mutant of Campylobacter coli: evidence for the significance of SOD in campylobacter survival and colonization. Appl Environ Microbiol. 1999;65:2540–2546. doi: 10.1128/aem.65.6.2540-2546.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sisson G, Jeong J Y, Goodwin A, Bryden L, Rossler N, Lim-Morrison S, Raudonikiene A, Berg D E, Hoffman P S. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and Escherichia coli containing a cloned H. pylori rdxA+ (nitroreductase) gene. J Bacteriol. 2000;182:5091–5096. doi: 10.1128/jb.182.18.5091-5096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spiegelhalder C, Gerstenecker B, Kersten A, Schiltz E, Kist M. Purification of Helicobacter pylori superoxide dismutase and cloning and sequencing of the gene. Infect Immun. 1993;61:5315–5325. doi: 10.1128/iai.61.12.5315-5325.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]