Abstract
Objective
To develop an automated deidentification pipeline for radiology reports that detect protected health information (PHI) entities and replaces them with realistic surrogates “hiding in plain sight.”
Materials and Methods
In this retrospective study, 999 chest X-ray and CT reports collected between November 2019 and November 2020 were annotated for PHI at the token level and combined with 3001 X-rays and 2193 medical notes previously labeled, forming a large multi-institutional and cross-domain dataset of 6193 documents. Two radiology test sets, from a known and a new institution, as well as i2b2 2006 and 2014 test sets, served as an evaluation set to estimate model performance and to compare it with previously released deidentification tools. Several PHI detection models were developed based on different training datasets, fine-tuning approaches and data augmentation techniques, and a synthetic PHI generation algorithm. These models were compared using metrics such as precision, recall and F1 score, as well as paired samples Wilcoxon tests.
Results
Our best PHI detection model achieves 97.9 F1 score on radiology reports from a known institution, 99.6 from a new institution, 99.5 on i2b2 2006, and 98.9 on i2b2 2014. On reports from a known institution, it achieves 99.1 recall of detecting the core of each PHI span.
Discussion
Our model outperforms all deidentifiers it was compared to on all test sets as well as human labelers on i2b2 2014 data. It enables accurate and automatic deidentification of radiology reports.
Conclusions
A transformer-based deidentification pipeline can achieve state-of-the-art performance for deidentifying radiology reports and other medical documents.
Keywords: deidentification, radiology, machine learning, NLP, transformer
INTRODUCTION
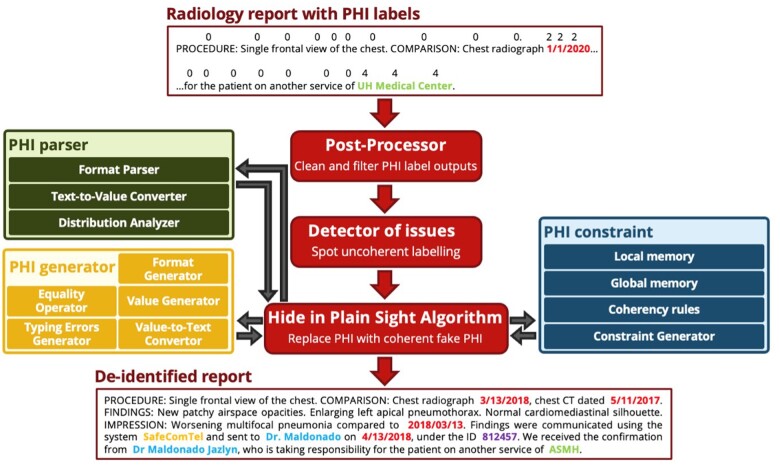
The task of deidentifying medical reports consists of detecting and removing all the protected health information (PHI) from reports, as defined in the Health Insurance Portability and Accountability Act of 1996 (HIPAA) (see Figure 1). HIPAA distinguishes numerous categories of PHI, among which the most frequent are dates, names, locations, identifiers, and phone numbers. The privacy of patients is protected by HIPAA and therefore access to data that includes PHI is limited. Nevertheless, the access to these same data is critical to build machine learning (ML) models capable of solving text-based medical tasks. For this reason, deidentifying medical documents automatically is important to foster the development of ML methods and tools.
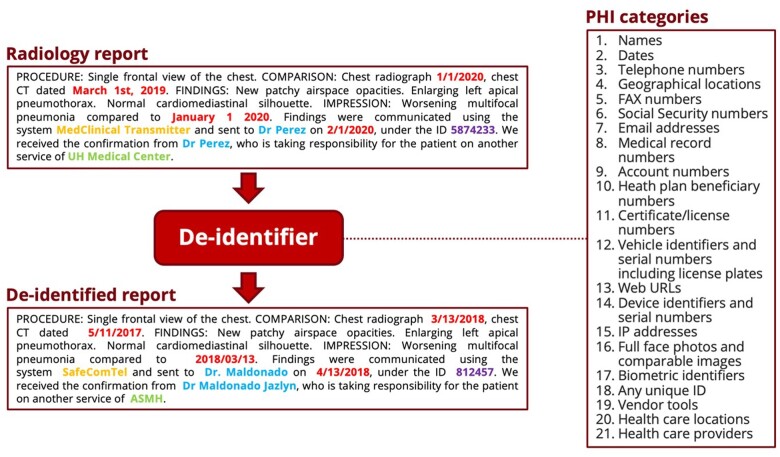
Figure 1.
Overview of the deidentification task: all the PHI in an input radiology report is replaced by coherent synthetic PHI, thereby “hiding in plain sight” any true PHI that might have been missed by the deidentifier. This figure only contains synthetic PHI. PHI: protected health information.
Historically, PHI detection has been automated mostly with rule-based algorithms.1,2 More recent ML approaches tried first to leverage conditional random fields,3,4 then long short-term memory (LSTM) architectures,5,6 and most recently transformers,7 which have achieved state-of-the-art performance across many natural language processing (NLP) tasks.8,9 The development of PHI detection models has been sustained by the release of several datasets, from the i2b2 challenges10,11 to the Physionet1 and Dernoncourt–Lee6 corpora. The removal or substitution of PHI is a less well-studied task, although the “hide in plain sight” approach12 is more robust and promising.
Many previously published algorithms were not designed to deidentify radiology reports. Similarly, public datasets for the deidentification task do not include such reports, limiting the possibility to train models for the radiology domain. Consequently, their results on radiology reports13 are unsatisfactory.
In this study, we propose an automated deidentification pipeline optimized for radiology reports that includes both a PHI detection model and a “hide in plain sight” algorithm that substitutes surrogate synthetic PHI. We apply a training approach for our detection model that optimizes the use of the limited amount of labeled data. In addition, we study its resistance to data shifts, using a variety of training datasets and building a data augmentation technique for this task. We have released our deidentification tool for public use; our model weights are available on Hugging Face at https://huggingface.co/StanfordAIMI/stanford-deidentifier-base; our code is open source and available on GitHub at https://github.com/MIDRC/Stanford_Penn_MIDRC_Deidentifier.
MATERIALS AND METHODS
Data collection and annotation
This retrospective study used data collected for nonresearch purposes and was approved by our institutional review boards, with a waiver of informed consent. A total of 999 adult male and female radiology reports were collected from multiple hospitals within a single academic health system, University of Pennsylvania Health System (“our Penn corpus”). Reports were randomly sampled from our database of chest X-ray and chest CT reports between November 2019 and November 2020, with an equal proportion of reports from each modality. These reports were annotated by 2 labelers (PJC and CW), with disagreements resolved after discussion with a radiologist who has 9 years of experience (TSC). For each report, the annotations were directly inserted in-line using a spreadsheet, capturing the PHI categories encountered in each radiology report (see Table 1). Even though “Provider names” and “Vendor and software names” are not included on the HIPAA Safe Harbor list of identifiers considered PHI, they are elements that many institutions want removed as part of the deidentification process.
Table 1.
Key characteristics of the datasets used in this study, including number of tokens and spans per PHI category
| PHI category | Our Penn Corpus—999 reports |
Steinkamp Penn Corpus—2501 reports |
Radgraph Stanford Corpus—500 reports |
i2b2 2006—889 reports |
i2b2 2014—1304 reports |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Spans | Tokens | Spans | Tokens | Spans | Tokens | Spans | Tokens | Spans | Tokens | |
| All PHI | 1465 | 7014 | 3428 | 15 207 | 1804 | 9368 | 18 532 | 102 145 | 23 615 | 109 200 |
| Dates | 1245 | 5969 | 2733 | 12 094 | 1138 | 5657 | 7595 | 33 134 | 12 203 | 55 784 |
| Provider names | 150 | 778 | 352 | 1700 | 131 | 1212 | 3612 | 31 385 | 4730 | 26 636 |
| Locations | 18 | 39 | 162 | 598 | 0 | 0 | 2647 | 11 114 | 2253 | 6858 |
| Vendor and software names | 24 | 126 | 84 | 312 | 0 | 0 | 0 | 0 | 206 | 589 |
| IDs | 26 | 89 | 70 | 361 | 527 | 2435 | 3517 | 18 644 | 1505 | 7181 |
| Patient names | 1 | 6 | 21 | 103 | 0 | 0 | 929 | 6236 | 2195 | 9895 |
| Phone numbers | 1 | 7 | 6 | 39 | 8 | 64 | 232 | 1632 | 523 | 2257 |
PHI: protected health information.
We included in our study other datasets previously annotated for the deidentification task: 2501 radiology reports13 from the same institution (“Steinkamp Penn Corpus”) and 500 radiology reports14 from a new institution (“Radgraph Stanford Corpus”), allowing us to measure performance on a new institution. In addition, we incorporated 889 discharge summaries from the 2006 i2b2 challenge11 and 1304 medical notes from the 2014 i2b2 challenge,10 as they are rich in PHI and commonly used for benchmarking deidentifiers (see Figure 2). We manually resolved labeling inconsistencies and did not include other datasets6,12 that were significantly different in format and content.
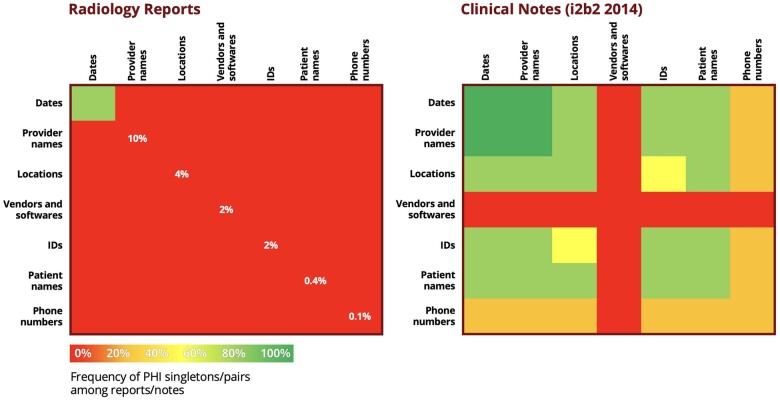
Figure 2.
Comparison of the frequency of PHI categories between radiology reports and clinical notes. Relative to clinical notes, radiology reports are strongly unbalanced and show scarcity of some PHI categories. PHI: protected health information.
As seen in both Table 1 and Figure 2, the distribution of PHI varies significantly across datasets. Radiology reports tend to have a limited amount of PHI, mostly consisting of dates, compared to discharge summaries and medical notes from the i2b2 challenges. In particular, the most critical PHI categories, for example, patient names and IDs, are among the rarest categories in the case of radiology reports. Therefore, building models on top of these reports, capable of achieving enough accuracy on the most critical PHI categories, can be challenging: we investigate the added value brought by training on i2b2 datasets as well, where certain PHI categories are more abundant, with the help of data and fine-tuning methods detailed in the following sections.
PHI detection model
The first step of our automated deidentification pipeline, PHI detection, is handled by a transformer encoder model that has a linear token-level classification head (see Figure 3). We chose this architecture because it achieves better performance on many NLP tasks than older architectures such as LSTMs or conditional random fields. In addition, its subtoken-level tokenizer allows the model to learn proper nouns and other unusual PHI formats.
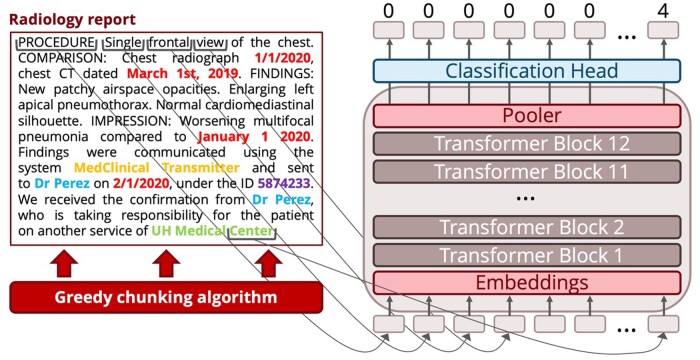
Figure 3.
Model architecture for the PHI detection task. First, the reports are split by a greedy chunking algorithm: chunks are cut between sentences, with no overlap. Then, chunks are fed to the transformer that leverages its attention mechanism to give a hidden representation to each token. A classification head uses these hidden representations to attribute scores, which measure the likelihood that each token belongs to each PHI class. Based on these scores, each token gets classified into its most likely PHI class. This figure only contains synthetic PHI. PHI: protected health information.
For pretraining, we relied on biomedical BERT models and selected PubMedBERT15 based on a preliminary analysis to find the highest performing model before supervised training on PHI data.16
For fine-tuning, we add a greedy chunking algorithm that splits input reports before running the model, which can process no more than 512 tokens at a time. Chunks are cut between sentences, with no overlap. Chunking allowed us to avoid using a recurrent architecture or training a much larger and computationally intensive model such as Big Bird.17 This allows the model to handle the entire report at inference time, optimizing the use of the training data, which frequently has labeled PHI both at the beginning and at the end of each report. Some PHI categories, like dates, are often located at the beginning of the report, while others, like healthcare provider names, are located at the end. The model is then trained with a weighted cross-entropy loss that gives more weight to the PHI tokens.
The performance of the model was optimized using the methods in Figure 4: data augmentation with the “hide in plain sight” algorithm; ULMFiT fine-tuning methods18,19; hyperparameter optimization with a Tree of Parzen estimator20; and distributed training with scheduling for quick exploration of the hyperparameter space. As part of the ULMFiT methods, the model was trained using discriminative learning rate, which consists of varying the learning speed across layers to avoid catastrophic forgetting phenomena (eg, for the embedding layers). These learning rates were all scheduled with a 1-cycle slanted triangular strategy, supposed to quickly converge to an optimum region of the parameters space before refining the weights. In addition, a final decay of the learning rates was added at the very end of the training, and the scheduling approach used for the learning rates was applied to the momentums as well. Aside these learning rate and momentum strategies, the model training relied on an unfreezing approach that consists of only training the head for the first epoch and then fine-tuning the entirety of the transformer for the remaining epochs, helping smooth the training and maximize the knowledge retained.18
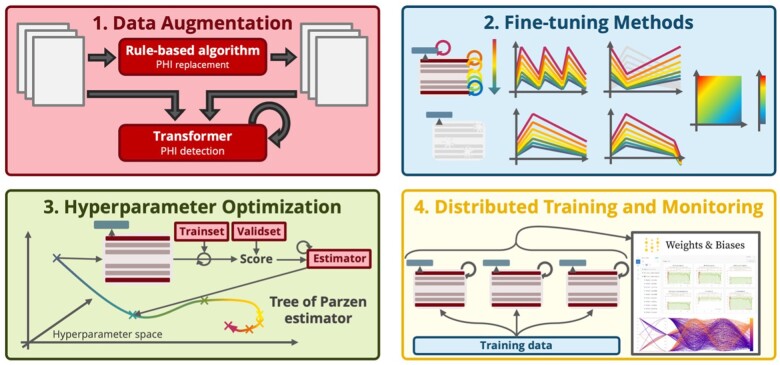
Figure 4.
Model optimization methods, from data augmentation to efficient hyperparameter exploration, are used to make the best use of limited labeled training data. Among the various fine-tuning strategies, discriminative learning rate, 1-cycle slanted triangular scheduling, and the unfreezing approach18 all contribute to enhancing the model performance. In combination with a hyperparameter optimization algorithm, which gets distributed for faster exploration of the hyperparameter space, these allow the deidentifier models to be better trained under the data and compute constraint regimes.
PHI replacement with synthetic PHI “hiding in plain sight”
The second step of our automated deidentification pipeline removes the detected PHI tokens and replaces them with synthetic PHI (Figure 5). This complements the first step by obscuring the few missed PHI entities and serves as a data augmentation tool to diversify the PHI seen in each category.
Figure 5.
Our rule-based synthetic PHI algorithm incorporates modules for each PHI category that can easily be augmented with new modules that handle new types of PHI. For each PHI category, it relies on a parser, a constraint, and a generator. The parser infers the input distributions of content and format. These distributions are skewed by constraints that, for instance, prevent generation of a PHI token identical to the original PHI and maintain the relative order and format of dates in each report. Finally, following these distributions, a rule-based generator replaces each true PHI span with a synthetic one. We elected not to use a neural-based generator to avoid the risk of outputting training data. This figure only contains synthetic PHI. PHI: protected health information.
This rule-based model includes first a postprocessor that cleans the labels of the output, correcting errors that are detectable with hard-coded rules, such as overlapping PHI spans or incorrectly classified stop words. Then, the “hide in plain sight” algorithm removes and replaces the PHI in a manner that could better resist adversarial attacks21 by relying on a stochastic approach that builds both a content and format distribution for each PHI category, based on the parsed input PHI entities. The content and formats are adjusted by in-document and cross-document constraints, which maintain the coherency of dates or healthcare workers and locations, based on short- and long-term memory. These memory units are specific to each PHI category and store each generated PHI, so that the redundancy of PHI within and between reports in the original data can be mimicked (eg, a hospital name common to many reports in the original data will be replaced by a synthetic hospital name, stored and then repeated across many reports in the deidentified data): short-term memory allows to handle report-level redundancy, whereas long-term memory acts at the dataset-level. Using the content and format distributions, fake PHI entities are produced based on token generators (eg, phone numbers, IDs, dates) or public databases (eg, provider and patient names, hospital names, vendor names), such as Census 2000, and all can be randomly altered by typing errors.
Not only is this “hide in plain sight” algorithm used at inference time, to replace the PHI detected by the ML model, but also at training time, as a data augmentation tool. It can be run on the training data and the associated ground-truth labels, therefore producing a new version of the training data, with different PHI entities. Combined with the original training data, this creates a data-augmented training set with more diversified PHI content. In particular, PHI categories that have few entities (eg, patient names) or lack diversity (eg, vendors or hospitals) are enriched through this process: the amount of PHI in each category is doubled, or can be multiplied several times if duplicate PHI in the original training data is counted only once.
Statistical analysis
Train and validation splits were stratified by label, to account for the strongly unbalanced data. Our models were evaluated on 4 hold-out test sets:
Steinkamp Penn test set13: 1023 radiology reports from the same institution as the training data.
Radgraph Stanford test set14: 500 radiology reports from a new institution, to evaluate how well the algorithm performs on data from a different institution.
i2b2 2006 test set11: 220 discharge summaries representing both a format and content shift.
i2b2 2014 test set10: 514 medical notes different both in format and content from previous sets and serving as a common benchmark for deidentification tasks.
Our main criterion is the performance on Steinkamp Penn test set, which is large enough to include rare PHI categories. It contains 6383 PHI tokens, including 5028 dates, 760 provider names, 229 locations, 117 vendor and software names, 164 identifiers, 63 patient names and 25 phone numbers.
We report F1 score, precision, and recall per category and for all PHI. We provide 95% confidence intervals using a bootstrap percentile method with 1000 bootstrap samples.
We use a paired samples Wilcoxon test to measure improvement between different models with P < 0.05 as the measure of significance and Bonferroni correction whenever necessary.
RESULTS
Training details
Each of our detection model experiments was optimized across 100 trainings using a tree of Parzen estimator, which explores a hyperparameter space defined for its main characteristics as follows: maximum learning rate between 1e-05 and 5e-04; momentum between 0.8 and 0.95; dropout between 0 and 0.5; weight decay between 1e-06 and 1e-04; and number of epochs from 3 to 5.
Our best model is trained for 1 epoch only with its classification head and learning rate set to 8.5e-05, then over 4 epochs with all its weights and learning rate set to 3e-05. Its weighted loss function uses the same weight, 3, for all PHI categories, and only 1 for non-PHI tokens.
Running all experiments takes between 15 and 75 min per run depending on the size of the training set, totaling 272 h on a single GPU NVIDIA Quadro P5000. Parallelizing across 3 GPUs reduced the duration to a little less than 4 days. As the compute resources are in the Eastern United States (carbon efficiency of 0.37 kg CO2eq/kWh), we estimate 18.1 kg CO2eq of carbon emissions overall.22
Experiments
We train 5 detection models using the same pretraining, PubMedBERT,15 and the same hyperparameter optimization process on various training sets:
Radiology reports only, using our labeled training set along with Steinkamp Penn training set.13
Radiology reports with data augmentation, seeing the impact of our “hide in plain sight” data augmentation method.
i2b2 only, using i2b2 2006 and i2b2 2014 training sets, to study the difference of performance when trained on nonradiology reports and to compare with other deidentifiers based on these datasets.
Radiology reports and i2b2, combining both datasets to evaluate if it results in a gain of performance.
Radiology reports and i2b2 with data augmentation, our largest training set, trying to benefit from all the data from the previous experiments.
Our best model for PHI detection was trained on both radiology reports and i2b2 medical texts and uses our data augmentation method. On the PHI versus non-PHI task, which consists of binarizing the labels and the predictions as either PHI or non-PHI (see “All PHI” in Table 2), it achieves 97.9 F1 score on radiology reports from a known institution, 99.6 F1 score on radiology reports from a new institution, 99.5 F1 score on i2b2 2006, and 98.9 F1 score on i2b2 2014. This task has the advantage of not accounting for misclassifications between different PHI categories. The good performance of our best model on this task underlines its accuracy at detecting spans of PHI, regardless of how it classifies them between the various PHI categories.
Table 2.
Comparison of 5 different training sets, with and without data augmentation, using F1 score (precision, recall)
| PHI category | All PHI | Macro-averaged over PHI categories | Dates | Provider names | Locations | Vendors and softwares | IDs | Patient names | Phone numbers | |
|---|---|---|---|---|---|---|---|---|---|---|
| Steinkamp Penn test set (same institution) | Radiology | 98.3 (98.2, 98.5) | 65.5 (63.9, 67.2) | 99.1 (98.9, 99.2) | 95.1 (91.6, 98.9) | 89.2 (86.5, 92.1) | 81.0 (81.7, 80.3) | 94.0 (88.6, 100) | 0.0 (0.0, 0.0) | 0.0 (0.0, 0.0) |
| Radiology + augmentation | 98.3 (97.8, 98.8) | 92.4 (93.3, 92.0) | 99.0 (98.6, 99.4) | 98.3 (97.8, 98.8) | 90.0 (91.4, 88.6) | 78.8 (71.8, 87.2) | 97.6 (95.3, 100) | 91.5 (98.2, 85.7) | 91.3 (100, 84.0) | |
| i2b2 | 94.9 (93.5, 96.3) | 61.1 (57.9, 70.8) | 98.5 (97.9, 99.2) | 92.1 (91.3, 92.9) | 69.7 (64.1, 76.4) | 0.0 (0.0, 0.0) | 39.0 (31.5, 51.2) | 95.9 (100, 92.1) | 32.8 (20.4, 84.0) | |
| Radiology + i2b2 | 97.5 (97.0, 98.2) | 90.5 (90.3, 90.9) | 98.4 (97.9, 99.0) | 95.9 (93.9, 98.0) | 92.6 (95.4, 90.0) | 76.8 (80.4, 73.5) | 95.9 (92.1, 100) | 95.9 (100, 92.1) | 77.8 (72.4, 84.0) | |
| Radiology + i2b2 + augmentation | 97.9 (98.0, 97.7) | 89.4 (91.5, 88.0) | 98.9 (99.1, 98.6) | 95.6 (92.9, 98.4) | 89.4 (90.6, 88.2) | 65.0 (78.3, 55.6) | 97.3 (95.9, 98.8) | 95.9 (100, 92.1) | 84.0 (84.0, 84.0) | |
| Radgraph Stanford test set (new institution) | Radiology | 99.1 (99.6, 98.6) | 74.3 (74.5, 74.1) | 99.7 (99.8, 99.7) | 99.0 (99.5, 98.5) | None | None | 98.5 (98.8, 98.2) | None | 0.0 (0.0, 0.0) |
| Radiology + augmentation | 99.4 (99.4, 99.5) | 88.0 (99.3, 83.8) | 99.4 (99.1, 99.7) | 99.3 (100, 98.7) | None | None | 98.7 (98.2, 99.3) | None | 54.5 (100, 37.5) | |
| i2b2 | 99.1 (99.8, 98.4) | 95.5 (94.4, 97.2) | 99.8 (99.9, 99.7) | 95.0 (100, 90.5) | None | None | 99.3 (99.8, 98.7) | None | 87.7 (78.0, 100) | |
| Radiology + i2b2 | 99.4 (99.4, 99.3) | 98.7 (98.3, 99.2) | 99.4 (99.1, 99.7) | 99.1 (100, 98.3) | None | None | 99.4 (100, 98.7) | None | 97.0 (94.1, 100) | |
| Radiology + i2b2 + augmentation | 99.6 (99.9, 99.3) | 98.8 (98.4, 99.2) | 99.8 (100, 99.7) | 99.0 (99.9, 98.2) | None | None | 99.2 (99.8, 98.7) | None | 97.0 (94.1, 100) | |
| I2b2 2006 test set | Radiology | 95.6 (98.0, 93.3) | 58.9 (76.2, 58.7) | 97.4 (98.5, 96.2) | 84.7 (84.1, 85.4) | 82.5 (93.0, 74.1) | None | 88.5 (81.9, 96.1) | 0.3 (100, 0.1) | 0.0 (0.0, 0.0) |
| Radiology + augmentation | 96.1 (97.4, 94.8) | 63.9 (80.4, 61.8) | 96.6 (98.2, 95.1) | 83.2 (85.7, 80.9) | 80.1 (95.1, 69.2) | None | 81.5 (70.5, 96.4) | 24.6 (32.6, 19.7) | 17.3 (100, 9.5) | |
| i2b2 | 99.5 (99.5, 99.5) | 99.1 (99.2, 98.9) | 99.5 (99.2, 99.9) | 99.4 (99.4, 99.4) | 98.0 (98.7, 97.2) | None | 99.8 (99.7, 99.9) | 98.6 (98.3, 99.0) | 99.0 (100, 98.0) | |
| Radiology + i2b2 | 99.4 (99.3, 99.6) | 99.1 (99.3, 99.0) | 99.4 (99.0, 99.9) | 99.1 (98.9, 99.4) | 98.2 (98.7, 97.7) | None | 99.7 (99.5, 99.8) | 99.0 (99.6, 98.4) | 99.4 (100, 98.8) | |
| Radiology + i2b2 + augmentation | 99.5 (99.4, 99.5) | 99.0 (99.0, 99.0) | 99.6 (99.5, 99.8) | 99.0 (98.8, 99.2) | 97.3 (98.2, 96.5) | None | 99.8 (99.7, 99.8) | 99.1 (98.6, 99.6) | 99.3 (99.3, 99.3) | |
| I2b2 2014 test set | Radiology | 93.0 (91.8, 94.3) | 44.6 (40.4, 53.1) | 91.3 (95.4, 87.6) | 78.7 (67.5, 94.2) | 75.2 (70.5, 80.6) | 9.0 (5.5, 24.5) | 58.0 (44.1, 84.8) | 0.0 (0.0, 0.0) | 0.0 (0.0, 0.0) |
| Radiology + augmentation | 93.3 (91.3, 95.4) | 52.4 (62.8, 59.0) | 95.1 (96.7, 93.5) | 80.5 (71.0, 92.9) | 77.5 (75.2, 79.8) | 12.0 (7.6, 28.7) | 68.6 (53.1, 96.8) | 32.0 (69.4, 20.8) | 1.2 (66.7, 0.6) | |
| i2b2 | 99.0 (98.9, 99.0) | 94.0 (96.2, 92.6) | 99.7 (99.6, 99.7) | 97.7 (97.8, 97.6) | 94.6 (92.7, 96.6) | 70.7 (88.1, 59.1) | 98.2 (97.2, 99.2) | 98.1 (98.7, 97.4) | 98.8 (99.2, 98.5) | |
| Radiology + i2b2 | 99.0 (98.6, 99.3) | 91.9 (93.2, 91.0) | 99.6 (99.4, 99.8) | 98.0 (97.0, 98.9) | 93.5 (91.4, 95.6) | 59.3 (70.8, 51.1) | 97.5 (95.9, 99.1) | 97.4 (98.5, 96.2) | 97.8 (99.1, 96.5) | |
| Radiology + i2b2 + augmentation | 98.9 (98.6, 99.3) | 93.9 (96.1, 92.5) | 99.6 (99.6, 99.6) | 97.6 (96.4, 98.8) | 94.3 (92.7, 95.8) | 71.1 (90.3, 58.6) | 98.1 (97.4, 98.8) | 97.4 (97.3, 97.4) | 98.9 (99.0, 98.8) | |
Notes: Each training set is further described in the experiments section. The “All PHI” category corresponds to the PHI versus non-PHI task, where labels and predictions are binarized as either PHI or non-PHI. For each PHI category and test set, the best score is emboldened and underlined, and the second best score only emboldened.
PHI: protected health information.
The results of these experiments are presented in Table 2. Macro-averaging the per-class performances on Steinkamp Penn test set (same institution), the best performing model was trained on radiology report data only, supplemented by data augmentation (P < 0.01). Training on i2b2 only resulted in a loss of more than 3 points of F1 score on the PHI versus non-PHI task (P < 0.01). Training on both i2b2 and radiology reports resulted in a slight reduction in F1 score (P < 0.01), which is mitigated by the use of data augmentation (P < 0.01).
When evaluating on Radgraph Stanford test set, which contained radiology reports from a new institution, the model trained on both radiology reports and i2b2 with data augmentation outperforms models trained only on radiology reports (P < 0.01): an F1 score on all PHI of 99.6 and a recall of 99.3.
Finally, on the i2b2 test sets, models that only saw radiology reports at training time suffer a lower performance, more pronounced on i2b2 2014, losing almost 6 points of F1 score compared to models that saw i2b2 reports at training time (P < 0.01). This loss of performance is limited for dates, the majority category.
Best model performance
Based on these results, we selected as our best model the model trained on both radiology reports and i2b2 texts with data augmentation. As shown in Table 2, that model minimizes the loss of performance on Steinkamp Penn test set while achieving a significant improvement on i2b2 test sets. We believe its more diverse training set, both in terms of document types and PHI entities themselves, makes it more likely to resist data shifts at inference time. Its performance on Radgraph Stanford test set, which outperforms all other models including the ones trained on radiology reports only, supports this hypothesis. This model will be referred to as our “best model” below and as the “stanford-deidentifier-base” when shared on Github and HuggingFace.
On the PHI versus non-PHI task, our best model achieves 97.9 F1 score (with 95% confidence interval [97.1–98.5]) on Steinkamp Penn test set, with 97.7 recall and 98.0 precision. It achieves almost 99 points of F1 score on dates and recall above 98.5 on dates, provider names, and IDs. It suffers a lower performance on vendor and software names, which are less common PHI categories.
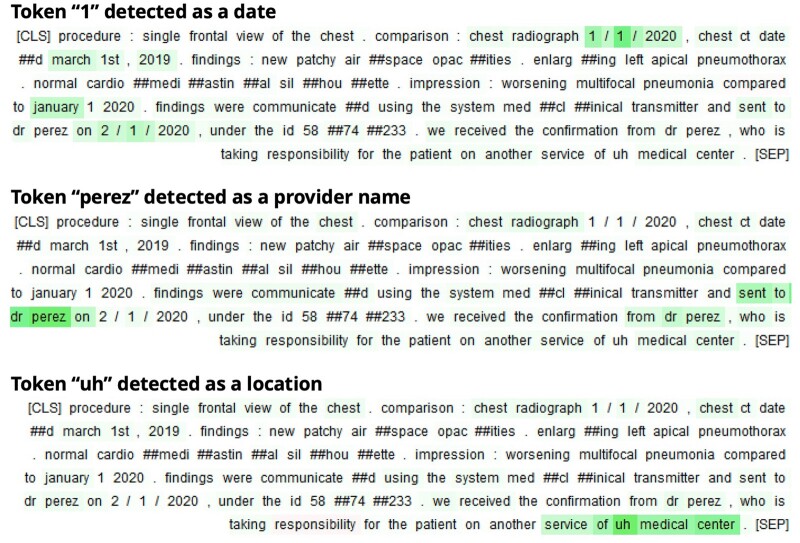
An error analysis shows that most errors are due either to misclassifications between PHI categories or to mislabeling of prefixes or suffixes from PHI spans. For instance, a phone number can start with a “#” token, annotated as PHI but predicted as non-PHI by the model; a professional title, such as “Professor”, could also be missed by the model. Some of these errors could also be considered as annotation errors, stressing the need for coherent and extensive annotation instructions for the deidentification task. When looking at the recall per span regardless of PHI category, our best model detects 99% of the PHI present in radiology reports (see Table 3). This performance is possible by paying attention to both the PHI entities themselves and their context, as seen in Figure 6.
Table 3.
Recall per PHI category for both the simple PHI versus non-PHI task and the same task with at least one token per PHI span needing to be detected
| PHI category | Recall for PHI versus non-PHI | Recall for PHI versus non-PHI with at least one token detected per span |
|---|---|---|
| All PHI | 97.7 | 99.1 |
| Macro-averaged over PHI categories | 93.1 | 96.4 |
| Dates | 98.6 | 99.5 |
| Provider names | 98.4 | 100 |
| Locations | 88.6 | 97.8 |
| Vendors and softwares | 67.5 | 77.8 |
| IDs | 98.8 | 100 |
| Patient names | 100 | 100 |
| Phone numbers | 100 | 100 |
Notes: The first score accounts for misclassifications between PHI categories and the second score for mislabelings of PHI prefixes or suffixes. Scores were computed on Steinkamp Penn test set.
PHI: protected health information.
Figure 6.
Visualization of integrated gradients23 for our model on a radiology report. It highlights the tokens considered by the model to classify a certain entity to a certain PHI class. The model leverages the content of the potential PHI entity, its possible redundancy in the report, and the nearby context to take its decision. This figure only contains synthetic PHI. PHI: protected health information.
Model comparison
Using Steinkamp Penn test set and the results of the associated study,13 we compare our best model to several deidentifiers previously released:
MIST3 trained on 1200 discharge summaries and other medical notes using conditional random fields.
NLM-Scrubber2 built on top of 3093 clinical documents using rules and dictionaries.
Emory HIDE4 developed on 100 pathology reports with conditional random fields.
MIT deid rule-based1 that uses 2434 nursing notes as well as rules and dictionaries.
NeuroNER5,6 created on 2939 medical texts, including i2b2 data, and combining recurrent neural networks and conditional random fields.
The transformer-based MIT deid model,7 trained on 6262 medical notes, including i2b2 datasets.
All ML deidentifiers were retrained on Steinkamp Penn training set (MIST, Emory HIDE, and NeuroNER) except for the transformer-based MIT deid, to test its performance when used directly. Some deidentifiers do not detect some categories and some group categories together, giving them an advantage when looking at specific metrics.
As seen in Table 4, our best model outperforms the best of the others by 4.3 F1 score on PHI versus non-PHI. We notice smaller differences on frequent categories like dates (+1 F1 score) and larger on rare categories (+48.9 F1 score on patients, +8.6 F1 score on provider names).
Table 4.
Comparison of our best model to several publicly available deidentifier models using F1 score (precision, recall), on Steinkamp Penn test set13
| PHI category | MIST | NLM-Scrubber | Emory HIDE | MIT deid rule-based | NeuroNER | MIT deid transformer-based | Our best model (radiology + i2b2 + augmentation) |
|---|---|---|---|---|---|---|---|
| All PHI | 75.5 (94.7, 62.7) | 74.1 (64.1, 87.5) | 92.2 (96.6, 88.2) | 74.0 (81.7, 67.6) | 93.6 (94.5, 92.6) | 78.4 (95.1, 66.7) | 97.9 (98.0, 97.7) |
| Macro- averaged | 61.5 (94.6, 53.7) | 58.8 (51.8, 83.6) | 72.9 (82.1, 66.1) | 28.0 (35.6, 26.0) | 68.6 (75.1, 65.6) | 53.5 (67.9, 48.8) | 89.4 (91.5, 88.0) |
| Dates | 75.1 (97.4, 61.2) | 97.9 (98.3, 97.5) | 96.4 (96.8, 96.0) | 89.0 (96.0, 83.0) | 97.9 (98.4, 97.5) | 83.4 (98.0, 72.6) | 98.9 (99.1, 98.6) |
| Provider names | 80.8 (93.0, 71.4) | None | 86.6 (97.5, 77.9) | None | 87.0 (82.0, 92.6) | 54.3 (84.2, 40.1) | 95.6 (92.9, 98.4) |
| Locations | 79.6 (85.2, 74.7) | None | 83.0 (93.4, 74.7) | 30.8 (51.1, 22.0) | 86.3 (82.0, 77.9) | 45.0 (60.5, 35.8) | 89.4 (90.6, 88.2) |
| Vendors and software | 88.1 (86.7, 89.7) | None | 76.5 (88.6, 67.2) | 6.2 (28.6, 3.4) | 75.9 (82.0, 70.7) | None | 65.0 (78.3, 55.6) |
| IDs | 11.1 (100, 5.9) | None | 90.6 (98.1, 84.1) | 0 (0, 0) | 84.8 (81.1, 88.9) | 55.3 (76.3, 43.3) | 97.3 (95.9, 98.8) |
| Patient names | 19.0 (100, 10.5) | 45.4 (37.3, 57.8) | 0 (0, 0) | 42.1 (37.9, 47.5) | 48.0 (100, 31.6) | None | 95.9 (100, 92.1) |
| Phone numbers | 77.0 (100, 62.5) | 33.0 (19.9, 95.6) | 76.9 (100, 62.5) | 0 (0, 0) | 0 (0, 0) | 29.5 (20.6, 52.0) | 84.0 (84.0, 84.0) |
Notes: Certain cases are left with “None” values, as the corresponding model is not capable of detecting the PHI category. Rule-based models could not be retrained and suffered from differences in what was considered PHI in the original study, which sometimes excluded years or name titles from being labeled as PHI. Our best model was trained on both radiology reports and i2b2 notes with our data augmentation approach. The “All PHI” category corresponds to the PHI versus non-PHI task, where labels and predictions are binarized as either PHI or non-PHI. For each PHI category, the best score is emboldened and underlined.
PHI: protected health information.
The transformer-based MIT deid algorithm is outperformed both on Steinkamp Penn test set (+19.5 F1 score, P < 0.01) and i2b2 2014 (+0.3 F1 score).
DISCUSSION
Our best model provides state-of-the-art performance on radiology report deidentification as well as several benchmark medical text deidentification tasks. Our algorithm is supplemented by a “hide in plain sight” PHI synthesizer that not only makes any remaining PHI difficult to detect but also serves as a data augmentation technique. Our data augmentation technique had a significant impact on deidentification performance. A model trained with this approach only on radiology reports performs well on most uncommon PHI categories, including patient names and phone numbers. For other relatively rare PHI categories, such as provider names and IDs, data augmentation allows an improvement of more than 3 points of F1 score on Steinkamp Penn test set. The results on Radgraph Stanford test set show that data augmentation is more effective than supplementing the training data with PHI from a different distribution, underlining that the model relies on context and structure in addition to the content of the PHI itself.
Supplementing the training with i2b2 data further improved model performance on reports from a new institution.
These combined improvements led our model to improve upon previously released deidentifiers in almost all PHI categories except vendor and software names, the most difficult categories. The greatest improvements were achieved for provider and patient names, which are highly sensitive PHI categories.
The MIT transformer deidentifier, the most similar model in terms of architecture, was outperformed by our best model on Steinkamp Penn test set and the i2b2 2014 test set. Our model trained only on i2b2, which was used for the MIT model training as well, outperformed it on i2b2 2014 test set and Steinkamp Penn test set, showing the added value of our fine-tuning methods to both maximize performance on i2b2 data and improve model robustness on other domains. Our model also achieves 93.9 macro-averaged F1 score on i2b2 2014, improving upon the 84.8 score for humans on the same categories.10
Recall is the most important metric for deidentification tasks, as any missed PHI may cause harm. Precision also matters to preserve the integrity of the reports for downstream tasks. Our model achieves recall of 99.1% on Steinkamp Penn test set when detecting at least 1 token per span, while maintaining precision of 98.0%. The remaining errors come either from data labeling inconsistencies, misclassifications between PHI categories, misclassifications of prefixes/suffixes of a PHI span, or true errors.
On the highly sensitive categories of PHI (ie, patient names, identifiers, and phone numbers), our model achieves recall of 99%, the only errors being misclassified prefixes/suffixes from correctly classified PHI spans. Only vendor and software names, the least sensitive, but most difficult PHI category to detect, suffered a lower performance: 59% recall on i2b2 2014. In comparison, an inter-annotator agreement study10 on the same dataset showed that human labelers achieved only 52% recall on vendor and software names. The difficulty of correctly classifying this PHI category comes from both its definition, covering a broad range of tool names that can be proper or common nouns, and its scarcity and lack of diversity. For this reason, we included in the released deidentifier command-line tool an option to specify institution-specific vendor and software names, enforcing their detection in the reports that are being deidentified. An institution-specific rule- and dictionary-based model could be used to supplement this model.
Looking at all the cross-domain experiments, it can be noticed that a transformer-based model, along several fine-tuning and hyperparameter optimization methods, leads to optimal performance on each domain when trained on the same domain, be it radiology reports or i2b2 medical notes. Nevertheless, these domain-specific models lose significant performance when tested on a different domain, between –4 and –5 F1 score on the PHI versus non-PHI task. Including training data from the other domain leads to minimizing the loss of performance on both domains compared to the best in-domain models, between 0 and –0.8 F1 score, what can be further reduced with the use of data augmentation. In addition, these cross-domain trained models perform best on data from a new institution and are more robust, as illustrated on the Radgraph Stanford test set. These experiments underline that unifying various deidentification datasets to train a single deidentifier model maintain performance on each domain (within 0.5 F1 score when using data augmentation), while improving model robustness on new institutions, which is of particular interest if these institutions have no training data available. In the setting where training data can be obtained at an institution, we suggest to first build a test set where the performance of our model can be assessed, before taking a decision on directly using it or re-training it first. If re-training is needed, our experiments show that robustness can be maximized and in-domain performance maintained (within 0.5 F1 score) by fine-tuning the model on in-domain data along data from various deidentification corpora. In this case, the annotation scheme must take into account potential labeling inconsistencies, and either conform to the other deidentification datasets or review and update their labels.
One limitation of our work is that we did not determine the precise ratio of data-augmented reports to original reports, nor the ratio of out-of-domain reports (i2b2) to in-domain reports (radiology reports) that would provide the best performance. In addition, we have not yet compared our model with commercially available tools. Finally, it is not yet possible for others to use our radiology report datasets, including benchmarking their models on Steinkamp Penn test set. We plan to address these limitations with future work.
In summary, we have developed and evaluated an automated deidentification pipeline that includes a transformer-based detection model and a “hide in plain sight” rule-based PHI synthesis algorithm. Leveraging the second step of our pipeline as a data augmentation technique and employing out-of-domain reports and an optimized fine-tuning algorithm, our model achieves state-of-the-art performance not only on radiology reports, but also other types of medical notes such as discharge summaries. Our model weights and code are publicly available.
FUNDING
This research is part of MIDRC (The Medical Imaging Data Resource Center) and was made possible by the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health under contracts 75N92020C00008 and 75N92020C00021. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Grant funding: NIH; Independence Blue Cross; RSNA.
AUTHOR CONTRIBUTIONS
Guarantors of integrity of entire study, PJC, TSC, CPL; study concepts/study design or data acquisition or data analysis/interpretation, all authors; manuscript drafting or manuscript revision for important intellectual content, all authors; approval of final version of submitted manuscript, all authors; agrees to ensure any questions related to the work are appropriately resolved, all authors; literature research, PJC; experimental studies, PJC; statistical analysis, PJC; and manuscript editing, all authors.
ACKNOWLEDGMENTS
We would like to acknowledge Stephanie Bogdan for her help in managing the project all along, as well as Jin Long for his assistance in selecting statistical tests. We would also like to acknowledge Nouha Manai from CentraleSupelec and Paris-Saclay University for her help in proofreading and correcting this manuscript.
CONFLICT OF INTEREST STATEMENT
Personal financial interests: Board of directors and shareholder, Bunkerhill Health; Option holder, whiterabbit.ai; Advisor and option holder, Galileo CDS; Advisor and option holder, Sirona Medical; Advisor and option holder, Adra; Advisor and option holder, Kheiron; Advisor, Sixth Street; Chair, SIIM Board of Directors; Member at Large, Board of Directors of the Pennsylvania Radiological Society; Member at Large, Board of Directors of the Philadelphia Roentgen Ray Society; Member at Large, Board of Directors of the Association of University Radiologists (term just ended in June); Honoraria, Sectra (webinars); Honoraria, British Journal of Radiology (section editor); Speaker honorarium, Icahn School of Medicine (conference speaker); Speaker honorarium, MGH (conference speaker); Co-founder, River Records. Recent grant and gift support paid to academic institutions involved: Carestream; Clairity; GE Healthcare; Google Cloud; IBM; IDEXX; Hospital Israelita Albert Einstein; Kheiron; Lambda; Lunit; Microsoft; Nightingale Open Science; Nines; Philips; Subtle Medical; VinBrain; Whiterabbit.ai; Lowenstein Foundation; Gordon and Betty Moore Foundation; Paustenbach Fund.
Contributor Information
Pierre J Chambon, Department of Radiology, Stanford University, Stanford, California, USA; Department of Applied Mathematics and Engineering, Paris-Saclay University, Ecole Centrale Paris, Paris, France.
Christopher Wu, Department of Radiology, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Jackson M Steinkamp, Department of Radiology, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Jason Adleberg, Department of Radiology, Mount Sinai Health System, New York, New York, USA.
Tessa S Cook, Department of Radiology, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Curtis P Langlotz, Department of Radiology, Stanford University, Stanford, California, USA.
DATA AVAILABILITY
The datasets used for this analysis involve i2b2 data, which is available at https://portal.dbmi.hms.harvard.edu/, and data from University of Pennsylvania and Stanford University that may be available upon reasonable request. Please contact the corresponding author, Pierre J. Chambon, for details.
REFERENCES
- 1. Neamatullah I, Douglass M, Lehman L, et al. Automated de-identification of free-text medical records. BMC Med Inform Decis Mak 2008; 8: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kayaalp M, Browne AC, Dodd ZA, Sagan P, McDonald CJ.. De-identification of address, date, and alphanumeric identifiers in narrative clinical reports AMIA Annu Symp Proc 2014;2014:767–76. [PMC free article] [PubMed] [Google Scholar]
- 3. Aberdeen J, Bayer S, Yeniterzi R, et al. The MITRE Identification Scrubber Toolkit: design, training, and assessment. Int J Med Inform 2010; 79 (12): 849–59. [DOI] [PubMed] [Google Scholar]
- 4. Gardner J, Xiong L.. An integrated framework for de-identifying unstructured medical data. Data Knowl Eng 2009; 68 (12): 1441–51. [Google Scholar]
- 5. Dernoncourt F, Lee J, Szolovits P.. NeuroNER: an easy-to-use program for named-entity recognition based on neural networks. In: conference on empirical methods in natural language processing: system demonstrations. Copenhagen, Denmark: Association for Computational Linguistics; 2017: 97–102. [Google Scholar]
- 6. Dernoncourt F, Lee J, Uzuner O, Szolovits P.. De-identification of patient notes with recurrent neural networks. J Am Med Inform Assoc 2017; 24 (3): 596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson A, Bulgarelli L, Pollard TJ.. Deidentification of free-text medical records using pre-trained bidirectional transformers. Proc ACM Conf Health Inference Learn (2020) 2020; 2020: 214–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. Advances in neural information processing systems 30 [published online ahead of print 2017]. arXiv preprint arXiv: 1706.03762.
- 9. Devlin J, Chang M, Lee K, Toutanova K. Bert: pre-training of deep bidirectional transformers for language understanding [published online ahead of print 2018]. arXiv preprint arXiv:1810.04805.
- 10. Stubbs A, Uzuner Ö.. Annotating longitudinal clinical narratives for de-identification: the 2014 i2b2/UTHealth corpus. J Biomed Inform 2015; 58 Suppl: S20–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Uzuner O, Luo Y, Szolovits P.. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc 2007. Sep–Oct; 14 (5): 550–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carrell D, Malin B, Aberdeen J, et al. Hiding in plain sight: use of realistic surrogates to reduce exposure of protected health information in clinical text. J Am Med Inform Assoc 2013. Mar-Apr; 20 (2): 342–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Steinkamp J, Pomeranz T, Adleberg J, et al. Evaluation of automated public de-identification tools on a corpus of radiology reports. Radiol Artif Intell 2020; 2 (6): e190137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jain S, Agrawal A, Saporta A, et al. RadGraph: extracting clinical entities and relations from radiology reports [published online ahead of print 2021]. arXiv:2106.14463.
- 15. Tinn R, Cheng H, Gu Y, et al. Fine-tuning large neural language models for biomedical natural language processing [published online ahead of print 2021]. arXiv:2112.07869. [DOI] [PMC free article] [PubMed]
- 16. Gu Y, Tinn R, Cheng H, et al. Domain-specific language model pretraining for biomedical natural language processing. ACM Trans Comput Healthc (HEALTH) 2022; 3 (1): 1–23. [Google Scholar]
- 17. Zaheer M, Guruganesh G, Dubey K, et al. Big bird: transformers for longer sequences. Adv Neural Inf Process Syst 2020; 33: 17283–97. [Google Scholar]
- 18. Howard J, Ruder S. Universal language model fine-tuning for text classification [published online ahead of print 2018]. arXiv:1801.06146.
- 19. Chambon P, , CookT, , Langlotz C.. Improved fine-tuning of in-domain transformer model for inferring COVID-19 presence in multi-institutional radiology reports. J Digit Imaging2022; 1–14. doi:10.1007/s10278-022-00714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bergstra J, Bardenet R, Bengio Y, Kégl B. Algorithms for hyper-parameter optimization. In: proceedings of the 24th international conference on neural information processing systems (NIPS'11). Red Hook, NY: Curran Associates Inc.; 2011: 2546–54.
- 21. Carrell D, Cronkite D, Li M, et al. The machine giveth and the machine taketh away: a parrot attack on clinical text deidentified with hiding in plain sight. J Am Med Inform Assoc 2019; 26: 1536–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lacoste A, Luccioni A, Schmidt V, Dandres T. Quantifying the carbon emissions of machine learning [published online ahead of print 2019]. arXiv:1910.09700.
- 23. Sundararajan M, Taly A, Yan Q. Axiomatic attribution for deep networks. In: international conference on machine learning. PMLR; 2017; 3319–28. arXiv preprint arXiv: 1703.01365.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used for this analysis involve i2b2 data, which is available at https://portal.dbmi.hms.harvard.edu/, and data from University of Pennsylvania and Stanford University that may be available upon reasonable request. Please contact the corresponding author, Pierre J. Chambon, for details.