Abstract
Sudden changes in health care utilization during the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic may have impacted the performance of clinical predictive models that were trained prior to the pandemic. In this study, we evaluated the performance over time of a machine learning, electronic health record-based mortality prediction algorithm currently used in clinical practice to identify patients with cancer who may benefit from early advance care planning conversations. We show that during the pandemic period, algorithm identification of high-risk patients had a substantial and sustained decline. Decreases in laboratory utilization during the peak of the pandemic may have contributed to drift. Calibration and overall discrimination did not markedly decline during the pandemic. This argues for careful attention to the performance and retraining of predictive algorithms that use inputs from the pandemic period.
Keywords: SARS-CoV-2, machine learning, algorithm drift, mortality, cancer
INTRODUCTION
Clinical predictive models that rely on electronic health record (EHR) features, such as patient characteristics, encounters, administrative codes, and laboratory values, are increasingly used in health care settings to direct resources to high-risk patients.1 Sudden changes in health care utilization during the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic may have impacted the performance of clinical predictive models.2 Such models are prone to performance changes (“drift”) over time due to changes in (1) the input distribution, including changes in values of input features or patterns of missingness in the inputs due to external causes; (2) changes in the output distribution, including changes in case-mix or risk distribution; or (3) changes in the relationship between inputs and outputs.3,4
OBJECTIVE
To investigate for the possibility of coronavirus disease (COVID)-associated performance drift, we evaluated the performance over time of a machine learning, EHR-based mortality prediction algorithm currently used in clinical practice. Owing to overall decreases in health care utilization, namely outpatient and laboratory encounters, during the pandemic,2 we hypothesized that predicted mortality and true-positive rate (TPR)—the percentage of deceased patients that were predicted as high-risk—would decline during the pandemic.
MATERIALS AND METHODS
Data sources
The study cohort was extracted from Clarity, a database that contains structured data elements of individual EHR data for patients treated at the University of Pennsylvania Health System (UPHS). The EHRs contained patient demographic characteristics, comorbidities, laboratory results, and utilization data. Mortality data were derived from internal administrative data, the EHR, and the Social Security Administration Death Master File, matched to UPHS patients by social security number and date of birth.
Ethics
The University of Pennsylvania Institutional Review Board approved this study with a waiver of informed consent, classifying this study as quality improvement.
Participants
Patients were eligible if they were 18 years or older and had an encounter at 1 of 18 medical or gynecologic oncology clinics within the UPHS between January 2, 2019, and December 31, 2020. All encounters were associated with a mortality risk prediction generated prior to the appointment. Eligible practices included a large tertiary practice, in which clinicians subspecialize in 1 cancer type (eg, lymphoma), and 17 general oncology practices, in which clinicians usually treat multiple cancer types. Benign hematology, genetics, and survivorship visits were excluded. Totally 360 727 encounters were eligible after these exclusions. Encounters with the same contact serial number (CSN) were combined as one distinct encounter including all identified comorbidity conditions. After the combination, 237 336 encounters were included in the analytical sample.
Predictive algorithm
The mortality risks of patients were derived from a gradient boosting machine learning algorithm (GBM) designed to predict 180-day mortality among outpatients with cancer. This model was developed and implemented at UPHS; 559 structured EHR features collected at UPHS were used for training. Since January 2019, this algorithm has been used as part of an intervention to prompt clinicians to initiate serious illness conversations among individuals with ≥10% risk score of 180-day mortality.5 Model features are listed at https://github.com/pennsignals/eol-onc and were derived using demographic, comorbidity, and laboratory values within 180 days prior to the encounter. Risk scores are generated on the Thursday prior to each medical oncology encounter and reflect absolute predicted percentage-point mortality risk. Even during the pandemic period, most appointments were scheduled prior to the previous Thursday, and thus scheduling changes after the prediction was made were rare. All missing variables in the training set were imputed as 0 for count variables and using median imputation (ie, missing values were replaced by the median of all values) for noncount variables. Detailed descriptions of the ML algorithm were described in previous publications.6,7 Clinicians received alerts for up to 6 encounters in the upcoming week with patients whose mortality risk scores were ≥10%, indicating that their patient was high-risk and may be appropriate for a conversation; absolute mortality risk was not presented to the clinician. The overall area under the receiver operator characteristic curve (AUC) of this algorithm was 0.89 (95% CI, 0.88–0.90), and disease-specific AUC ranged from 0.74 to 0.96 in a prospective validation.6
Outcomes
The primary outcome was 180-day mortality from the time of the encounter. Mortality data were derived from the EHR, our internal cancer registry, and the Social Security Administration Death Master File (SSA DMF).
Features
Variables used in the algorithm have been previously published and are provided in Supplementary Table S1.
Statistical analysis
Descriptive analyses compared encounter-level characteristics in the following periods: January 2019–February 2020 (“pre-pandemic”), March–May 2020 (“early pandemic” washout period, representing the early impact of stay-at-home orders and pandemic-related policy changes in hospitals and clinics), and June–December 2020 (“later pandemic”, representing longer-term impact).
Interrupted time series analysis
An interrupted time series (ITS) model was used to evaluate the changes in the absolute 180-day mortality risk scores, the mortality percentage of encounters classified as high-risk, and percentage of laboratory utilization before and after the pandemic, using March–May 2020 as a washout period. The purpose of introducing the washout period in the analysis was to allow any effect of COVID-19 to manifest in patient behaviors as well as clinical practices. The model structure is as follows8:
where represents the time elapsed since the start of the study period with the unit representing the frequency with which observations were taken (ie, month in current study), is a dummy variable indicating the prepandemic period (coded in 0) or the later pandemic period (coded in 1), is the outcome at time t. Monthly average prediction score, or monthly percentage of high-risk encounters, or monthly percentage of laboratory utilization, or monthly calculated TPR were used as the dependent variables in this study. Logit transformation was applied to the outcomes to ensure the predicted trends in later pandemic period ranged within 0 to 1. represents the baseline prediction score at , is interpreted as the change in the outcome associated with a time unit increase (representing the underlying pre-COVID trend), is the level change following the COVID outbreak and indicates the slope change following the COVID outbreak. A Kolmogorov–Smirnov test was performed in SAS to compare the distribution of percentage of high-risk encounters in the before and after COVID period in the sample.
True positive rate comparison
Because we hypothesized that pandemic-related utilization declines would result in underprediction of mortality risk, TPR was the primary performance metric. TPR was calculated based on the total number of predicted high-risk encounters and observed all-cause 180-day mortality from the encounter date, based on the 10% threshold used in practice.5 ITS analysis with logit transformation was performed to evaluate the impact of the pandemic on the model TPR; mortality risk scores from each patient’s first encounter in each month were used to calculate TPR in ITS analyses.9,10
Other performance metrics
In addition to TPR, we compared the following performance metrics between the prepandemic and later pandemic periods: AUC, positive predictive value (PPV), and specificity. Additionally, to compare calibration between the 2 periods, we generated calibration plots and calculated Brier scores,11 a quadratic scoring rule in which the squared differences between actual binary outcomes and predicted probabilities are calculated and lower values indicate higher overall accuracy; As with our TPR calculation, to compare these performance metrics, we used mortality risk scores at a 10% threshold from each patient’s first encounter in each month.
Mechanisms of performance drift
To identify potential mechanisms of performance drift, we first compared patient and encounter characteristics among the prepandemic, early pandemic, and later pandemic periods. We then compared observed versus predicted laboratory utilization using a LASSO model to predict laboratory utilization in the later pandemic period. Prepandemic period data were used to train the model. Predictors included in the model were baseline demographic characteristics and comorbidity conditions. Then the modeled output was applied to the later pandemic period to predict laboratory utilization, including all visit types. Finally, to investigate the potential contributions of greater telemedicine utilization and lower laboratory utilization to performance drift, we examined mortality risk scores associated with and without telemedicine encounters or laboratory visits.
RESULTS
We analyzed mortality risk scores associated with 237 336 in-person and telemedicine medical oncology encounters between January 2019 and December 2020.
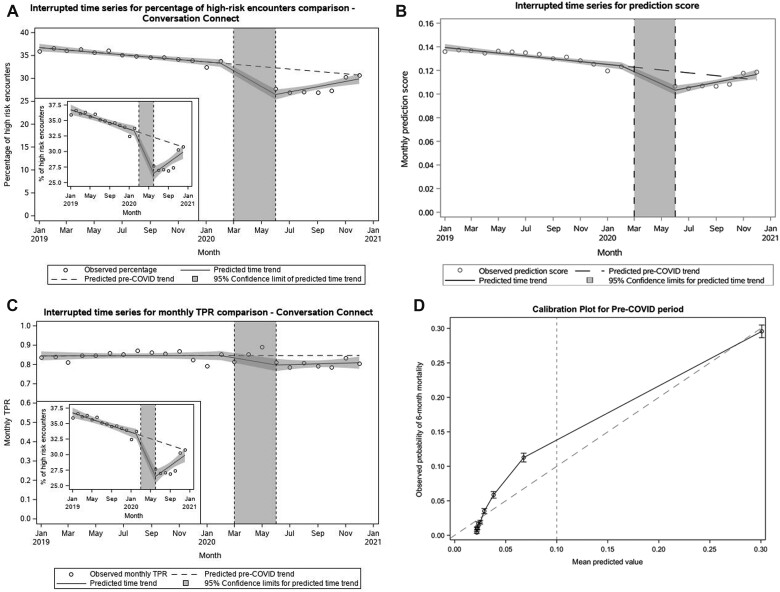
Interrupted time series analyses showed that the later pandemic period was characterized by a 6.8-percentage-point decrease in encounters classified as high-risk (33.3% [prepandemic] vs 26.5% [later-pandemic], P < .001 for level change, Figure 1A). The Kolmogorov–Smirnov test showed that the distributions of percentage of high-risk encounters in the prepandemic and later pandemic periods were different (P < .002). There was a corresponding 2.1 absolute percentage-point decrease in predicted 180-day mortality risk in the later pandemic period (12.4% [prepandemic] vs 10.3% [later pandemic], P < .001 for level change, Figure 1B).
Figure 1.
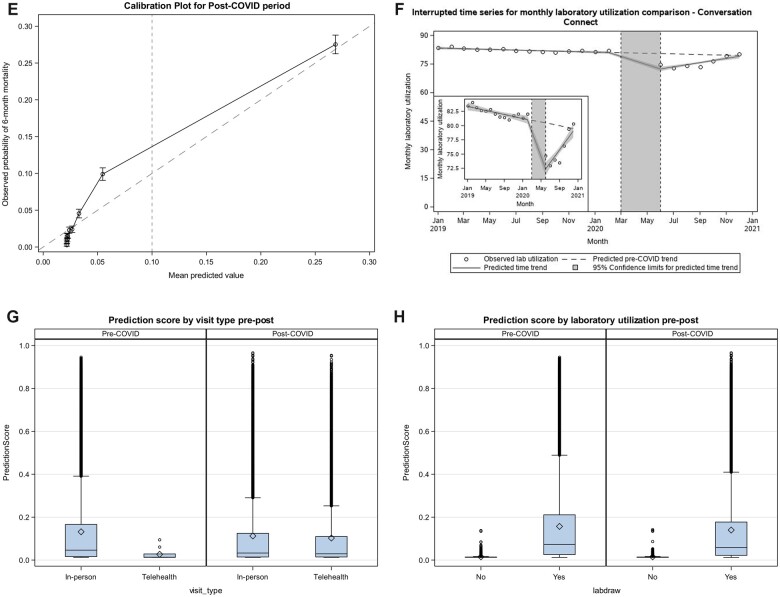
Performance drift in the prediction model during the COVID pandemic. (A–D) Interrupted time series analyses of COVID pandemic impacts on (A) Percentage of high-risk encounters; (B) Absolute predicted 180-day mortality risk; (C) True-positive rate. Circles represent monthly observed averages across all medical oncology encounters. Solid lines reflect time trends in the prepandemic (January 2019–February 2020), early pandemic (March–May 2020), and later pandemic (June–December 2020) periods, with 95% confidence intervals surrounding each time trend in shaded grey. (D, E) Calibration plots for the pre-COVID (D) and post-COVID (E) periods, with a dashed line indicating the clinically relevant threshold used in practice. (F) Interrupted time series analysis of COVID pandemic impact on laboratory utilization. Circles represent monthly observed average percentage of encounters with prior associated laboratory utilization in the prior 180 days across all medical oncology encounters. Solid lines reflect time trends in the prepandemic (January 2019–February 2020), early pandemic (March–May 2020), and later pandemic (June–December 2020) periods, with 95% confidence intervals surrounding each time trend in shaded grey. (G) Prediction score by visit type in pre- versus later pandemic. The mean prediction score in the prepandemic period among the in-person visits was 0.13 (IQR: 0.02–0.17), while among telehealth visits, the mean prediction score was 0.03 (IQR: 0.01–0.03). In the later pandemic period, the mean prediction score among in-person visits was 0.11 (IQR: 0.01–0.12), while among telehealth visits, the mean prediction score was 0.12 (0.02–0.14). (H) Prediction score by laboratory utilization in pre- versus later pandemic. The mean prediction score in the prepandemic period among the encounters with laboratory utilization was 0.16 (IQR: 0.03–0.21), while among the encounters without any laboratory utilization, the mean prediction score was 0.015 (IQR: 0.013–0.014). In the later pandemic period, the mean prediction score among the encounters with labs was 0.14 (IQR: 0.02–0.18), among the encounters without labs was 0.015 (0.012–0.014).
Figure 1.
Continued
The TPR was 81.2%, 75.7%, and 76.2% in February, June, and December 2020. An ITS analysis with logit transformation was performed to evaluate the impact of the pandemic on the model TPR. The onset of the pandemic was associated with an absolute 7.0-percentage-point decrease in TPR (80.9% [prepandemic] vs 73.9% [later pandemic], P = .0203 for the level change) (Figure 1C).
Other than TPR, other performance metrics were similar in the pre- and later pandemic periods (Table 1). Calibration was also similar in the pre- and later-pandemic periods (Figures 1D and E).
Table 1.
Comparison of other performance metrics in the pre- and later-pandemic periods
| Prepandemic | Later-pandemic | |
|---|---|---|
| Mean prediction score | 0.103 | 0.088 |
| TPR | 0.809 | 0.739 |
| AUC | 0.849 | 0.844 |
| Brier score | 0.046 | 0.042 |
| Specificity | 0.762 | 0.807 |
| PPV | 0.168 | 0.175 |
AUC: area under the receiver operator characteristic curve; PPV: positive predictive value; TPR: true-positive rate.
Age, race, average patient encounters per month, insurance type, comorbidity, laboratory values, and overall mortality were similar across the 3 time periods (Table 2).
Table 2.
Baseline characteristics of medical oncology encounters
| Overall | Prepandemica | Early pandemica | Later pandemica | |
|---|---|---|---|---|
| Number of encounters, N (%) | 237 336 (100.00) | 141 969 (59.82) | 27 582 (11.62) | 67 785 (28.56) |
| Number of unique patients, N (%) | 34 666 (100.00) | 27 609 (79.64) | 12 702 (36.64) | 21 961 (63.35) |
| Monthly encounter rate (total patient visits per month) (SD) | 1.47 (0.88) | 1.50 (0.94) | 1.45 (0.82) | 1.42 (0.78) |
| Prediction score per encounter, mean (SD) | 0.12 (0.18) | 0.13 (0.18) | 0.12 (0.17) | 0.11 (0.17) |
| Telemedicine | 0.11 (0.16) | 0.03 (0.03) | 0.12 (0.16) | 0.10 (0.16) |
| In-person | 0.13 (0.18) | 0.13 (0.18) | 0.12 (0.17) | 0.11 (0.17) |
| No preceding labs | 0.02 (0.01) | 0.02 (0.01) | 0.01 (0.01) | 0.02 (0.01) |
| Preceding labs | 0.15 (0.19) | 0.16 (0.19) | 0.14 (0.18) | 0.14 (0.18) |
| Observed 6-month mortality rate (%) | 7.0 | 7.4 | 6.3 | 6.4 |
| Age, mean (SD) | 61.92 (13.86) | 61.91 (13.74) | 61.95 (13.81) | 61.94 (14.12) |
| Age group, N (%) | ||||
| <55 years old | 62 605 (26.38) | 37 305 (26.28) | 7295 (26.45) | 18 005 (26.56) |
| 55–64 years old | 64 806 (27.31) | 39 407 (27.76) | 7419 (26.90) | 17 980 (26.53) |
| 65–74 years old | 72 553 (30.57) | 43 041 (30.32) | 8587 (31.13) | 20 925 (30.87) |
| 75 and over | 37 372 (15.75) | 22 216 (15.65) | 4281 (15.52) | 10 875 (16.04) |
| Marital status, N (%) | ||||
| Married | 155 676 (65.59) | 92 860 (65.41) | 18 306 (66.37) | 44 510 (65.66) |
| Not married | 81 612 (34.39) | 49 087 (34.58) | 9269 (33.61) | 23 256 (34.31) |
| Missing | 48 (0.02) | 22 (0.02) | 7 (0.03) | 19 (0.03) |
| Race, N (%) | ||||
| Non-Hispanic White | 178 794 (75.33) | 107 305 (75.58) | 20 735 (75.18) | 50 754 (74.87) |
| Hispanic Latino/White | 2834 (1.19) | 1690 (1.19) | 328 (1.19) | 816 (1.20) |
| Non-Hispanic Black | 36 438 (15.35) | 21 691 (15.28) | 4179 (15.15) | 10 568 (15.59) |
| Hispanic Latino/Black | 1072 (0.45) | 667 (0.47) | 119 (0.43) | 286 (0.42) |
| Other | 13 262 (5.59) | 7873 (5.55) | 1632 (5.92) | 3757 (5.54) |
| Unknown | 4936 (2.08) | 2743 (1.93) | 589 (2.14) | 1604 (2.37) |
| Insurance, N (%) | ||||
| Commercial | 26 748 (11.27) | 15 767 (11.11) | 3059 (11.09) | 7922 (11.69) |
| Medicaid | 12 589 (5.30) | 7530 (5.30) | 1412 (5.12) | 3647 (5.38) |
| Medicare | 193 417 (81.50) | 115 766 (81.54) | 22 592 (81.91) | 55 059 (81.23) |
| Self-pay | 943 (0.40) | 515 (0.36) | 142 (0.51) | 286 (0.42) |
| Missing | 3639 (1.53) | 2391 (1.68) | 377 (1.37) | 871 (1.28) |
| Visit type, N (%) | ||||
| In-person | 213 943 (90.14) | 141 958 (99.99) | 22 069 (80.01) | 49 916 (73.64) |
| Telehealth | 23 393 (9.86) | 11 (0.01) | 5513 (19.99) | 17 869 (26.36) |
| Elixhauser comorbidity count, mean (SD) | 2.34 (1.89) | 2.36 (1.88) | 2.33 (1.87) | 2.30 (1.93) |
| Encounters with no preceding laboratory values, N (%) | 46 942 (19.8) | 25 119 (17.7) | 5469 (19.8) | 16 354 (24.1) |
| Lab value, mean (SD) | ||||
| Albumin | 4.34 (22.00) | 4.33 (23.07) | 4.31 (20.10) | 4.38 (20.36) |
| Hemoglobin | 12.10 (1.75) | 12.09 (1.76) | 12.13 (1.72) | 12.10 (1.73) |
| Calcium | 9.17 (0.49) | 9.18 (0.51) | 9.16 (0.46) | 9.17 (0.45) |
| White blood cell | 7.21 (9.08) | 7.16 (8.15) | 7.13 (8.35) | 7.34 (11.00) |
| Total bilirubin | 0.60 (0.62) | 0.61 (0.62) | 0.58 (0.52) | 0.61 (0.68) |
| Alkaline phosphatase | 91.27 (77.04) | 92.47 (81.25) | 90.85 (70.47) | 88.93 (70.20) |
Prepandemic: January 2019–February 2020. Early pandemic: March–May 2020, representing the early impact of stay-at-home orders and pandemic-related policy changes in hospitals and clinics. Later pandemic: June–December 2020, representing longer-term impact.
Compared to the prepandemic period, the early and later pandemic periods had higher proportions of telemedicine encounters (0.01% [prepandemic] vs 20.0% [early pandemic] vs 26.4% [later pandemic]) and encounters with no preceding laboratory draws (17.7% [prepandemic] vs 19.8% [early pandemic] vs 24.1% [later pandemic]). In the ITS analysis controlling for prepandemic trends, the pandemic was associated with an absolute 8.9-percentage-point decrease (81.2% vs 72.3%, P < .0001 for level change) in laboratory utilization associated with the pandemic (Figure 1F). In the later pandemic period, telemedicine encounters (mean predicted risk score 10.3%) and encounters with no preceding laboratory draws (mean predicted risk score 1.5%) were associated with lower predicted risk score than in-person encounters (mean predicted risk score 11.2%, P < .001) or encounters with preceding laboratory draws (mean predicted risk score 14.0%, P < .001), respectively (Figure 1G and H).
DISCUSSION
During the SARS-CoV-2 pandemic period, the performance of a machine learning prognostic algorithm used to prompt serious illness conversations in clinical practice declined substantially, with a 7.0-percentage-point decrease in true-positive rate. The algorithm underidentified patients at high predicted risk of mortality in the initial months of the pandemic period. Towards the end of 2020, prediction scores began to return to prepandemic baselines, presumably due to resumption of routine health care utilization. However, model TPR remained continually below baseline throughout 2020. Declines in encounters associated with laboratory draws and increases in telemedicine utilization—both potentially spurred by pandemic-related stay-at-home orders and patient fears of exposure in health care settings—may have contributed to lower performance. Laboratory utilization likely had a disproportionate impact, as counts of laboratory encounters were included as features of the models in addition to the average and variation in actual laboratory values. Decreased utilization of laboratories in the later pandemic period—rather than changes in lab values themselves—appeared to contribute to decline in performance.
Performance drift of predictive algorithms has been conceptually described in non-COVID settings where characteristics of the input or output distribution differ.3,4 This is one of the first studies to show algorithm performance drift due to SARS-CoV-2 pandemic-related shifts in the input distribution; this drift extended well into 2020. It is unlikely that natural decreases in algorithm performance explain the performance drift. First, our ITS model showed a significant level change after April 2020 that was well below the natural trend in the prepandemic period. Prediction scores dropped sharply after the initial SARS-CoV-2 period, which would be atypical for natural algorithm performance drift. Second, demographic, clinical severity, and cancer-specific characteristics remained largely similar across time periods. Actual mortality in this population also remained consistent, although causes of death (including a higher proportion COVID-related deaths) likely changed, causing a calibration shift. Indeed, a major contributor to algorithm performance declines during the pandemic was a shift in underlying utilization patterns, resulting in a database shift by decreasing the counts of laboratory encounters.
A similar study published in 2021 found that predictive model alerts increased by 43% during the pandemic, even though this model was not trained in the SARS-CoV-2 era.12 Cancellation of elective surgeries and higher than average patient acuity in the underlying patients contributed to this. Our study adds to this and similar literature by suggesting that changes in the frequency of alerts caused by pandemic-related utilization shifts may be associated with decreased accuracy overall. Predictive or risk-adjustment algorithms that use inputs from EHR or claims data should be interpreted with caution, as pandemic-related decreases in utilization may impact performance. Health systems, payers, and clinicians should consider retraining EHR- or claims-based predictive algorithms in the postpandemic era. Alternatively, as calibration did not markedly change in the pre- versus postpandemic periods, health systems could consider setting different thresholds for clinical predictive models in the later and postpandemic periods to account for shifts in predicted risk distribution.
While some studies have shown evidence of changes in scores from predictive models,12 ours is the first to examine changes in algorithm performance during the pandemic, measured by the decrease in the TPR. Our study argues for considering retraining models with training data during the lengthy COVID pandemic period, given likely utilization shifts that may induce longer-term changes in performance. Even in nonpandemic scenarios, regular retraining of predictive algorithms may be useful to address gradual changes in algorithm performance. For models where the pandemic resulted in underidentification of high-risk patients, setting lower risk thresholds may be an alternative solution. Shocks such as the pandemic should also prompt more frequent diagnostic checks on performance. Pandemic-related performance shifts are also likely to affect other performance models, including risk stratification and risk-adjustment algorithms used by large payers that utilize data during the pandemic period for inputs.13 Alternatively, reinforcement learning systems,14 shift-stable algorithms, or adaptive continuous learning methods that continuously monitor and correct shift can enhance clinical applicability by accounting for changes in the underlying dataset.
Limitations of this study include potentially limited generalizability because it includes a single health system. However, our study includes 18 diverse medical oncology practices spanning 2 states. We were also unable to account for unmeasured contributors to changes in underlying risk. The distribution of other unmeasured markers of patient acuity, including performance status and cancer burden, may have contributed to fewer alerts during the pandemic. While we relied on a combination of cancer registry data, the Social Security Administration Death Master File, and institutional data to measure the outcome of death, and while overall rates of death remained consistent throughout the study period, it is possible that we did not capture all deaths during the later pandemic period. Indeed, it is known that the SSA DMF and EHR data likely underestimate death rates compared with gold-standard sources such as the Centers for Disease Control’s National Death Index (NDI).15,16 However NDI data are infrequently updated and were not present for either the pre- and later-pandemic periods in this study; therefore, any potential bias underestimating death was assumed to be consistent throughout the time period. Furthermore, because fewer high-risk alerts were generated in the later pandemic period, it is likely that an increase in deaths would have resulted in a further decrease in the TPR of the algorithm. Additionally, our analysis of mechanisms of drift relied on descriptive analyses. Other methods such as neural networks may point to learning features of the input data, including combinations of features, that would lend more insight into drift mechanisms. Finally, we did not have access to the cause of death in our dataset. We did not observe that the pandemic period nor receipt of algorithm-based nudges were associated with meaningfully higher mortality rates. However, it is possible that subtle changes in mortality rates associated with alerts or changes in the distributions of causes of death during the pandemic may have also contributed to the performance drift observed in this study.
CONCLUSION
For a mortality prediction model used to identify individuals who need timely advance care planning, performance and identification of high-risk patients substantially declined for a sustained period during the SARS-CoV-2 pandemic period, driven partially by decreases in laboratory utilization during the peak of the pandemic. This argues for careful attention to the performance and retraining of predictive algorithms that use inputs from the pandemic period.
FUNDING
This work was supported by grants from the National Cancer Institute to RBP (K08-CA-263541) and JC (R01-HL138306, UL1-TR001878).
AUTHOR CONTRIBUTIONS
ASN and JC contributed equally as co-senior authors.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Colleen Brensinger for analytic support and Jay Fein for assistance with manuscript preparation.
CONFLICT OF INTEREST STATEMENT
Dr Parikh reports receiving grants from Humana, the National Institutes of Health, Prostate Cancer Foundation, National Palliative Care Research Center, Conquer Cancer Foundation, and Veterans Administration; personal fees and equity from GNS Healthcare, Inc. and Onc.AI; personal fees from Cancer Study Group, Thyme Care, Humana, and Nanology; honorarium from Flatiron, Inc. and Medscape; and serving on the board (unpaid) of the Coalition to Transform Advanced Care, all outside the submitted work.
Contributor Information
Ravi B Parikh, Department of Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA; Department of Medical Ethics and Health Policy, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA; Corporal Michael J. Crescenz VA Medical Center, Philadelphia, Pennsylvania, USA.
Yichen Zhang, Department of Medical Ethics and Health Policy, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Likhitha Kolla, Department of Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA; Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Corey Chivers, Department of Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Katherine R Courtright, Department of Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Jingsan Zhu, Department of Medical Ethics and Health Policy, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Amol S Navathe, Department of Medicine, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA; Department of Medical Ethics and Health Policy, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA; Corporal Michael J. Crescenz VA Medical Center, Philadelphia, Pennsylvania, USA.
Jinbo Chen, Department of Biostatistics, Epidemiology and Informatics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
DATA AVAILABILITY
The data that support the findings of this study are available on request. Information about the prediction algorithm can be found here: https://github.com/pennsignals/eol-onc.
REFERENCES
- 1. Paxton C, Niculescu-Mizil A, Saria S.. Developing predictive models using electronic medical records: challenges and pitfalls. AMIA Annu Symp Proc 2013; 2013: 1109–15. [PMC free article] [PubMed] [Google Scholar]
- 2. Moynihan R, Sanders S, Michaleff ZA, et al. Impact of COVID-19 pandemic on utilisation of healthcare services: a systematic review. BMJ Open 2021; 11 (3): e045343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davis SE, Lasko TA, Chen G, Siew ED, Matheny ME.. Calibration drift in regression and machine learning models for acute kidney injury. J Am Med Inform Assoc 2017; 24 (6): 1052–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wong A, Otles E, Donnelly JP, et al. External validation of a widely implemented proprietary sepsis prediction model in hospitalized patients. JAMA Intern Med 2021; 181 (8): 1065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Manz CR, Parikh RB, Small DS, et al. Effect of integrating machine learning mortality estimates with behavioral nudges to clinicians on serious illness conversations among patients with cancer: a stepped-wedge cluster randomized clinical trial. JAMA Oncol 2020; 6 (12): e204759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Manz CR, Chen J, Liu M, et al. Validation of a machine learning algorithm to predict 180-day mortality for outpatients with cancer. JAMA Oncol 2020; 6 (11): 1723–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Parikh RB, Manz C, Chivers C, et al. Machine learning approaches to predict 6-month mortality among patients with cancer. JAMA Netw Open 2019; 2 (10): e1915997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bernal JL, Cummins S, Gasparrini A.. Interrupted time series regression for the evaluation of public health interventions: a tutorial. Int J Epidemiol 2017; 46 (1): 348–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Efron B, Tibshirani RJ.. An Introduction to the Bootstrap. Boca Raton, FL: CRC Press; 1994. [Google Scholar]
- 10. Davison AC, Hinkley DV.. Bootstrap Methods and Their Application. Cambridge: Cambridge University Press; 1997. [Google Scholar]
- 11. Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 2010; 21 (1): 128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wong A, Cao J, Lyons PG, et al. Quantification of sepsis model alerts in 24 US hospitals before and during the COVID-19 pandemic. JAMA Netw Open 2021; 4 (11): e2135286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luo Y, Wunderink RG, Lloyd-Jones D.. Proactive vs reactive machine learning in health care: lessons from the COVID-19 pandemic. JAMA 2022; 327 (7): 623–4. [DOI] [PubMed] [Google Scholar]
- 14. Bastani H, Drakopoulos K, Gupta V, et al. Efficient and targeted COVID-19 border testing via reinforcement learning. Nature 2021; 599 (7883): 108–13. [DOI] [PubMed] [Google Scholar]
- 15. Navar AM, Peterson ED, Steen DL, et al. Evaluation of mortality data from the social security administration death master file for clinical research. JAMA Cardiol 2019; 4 (4): 375–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gensheimer MF, Narasimhan B, Henry AS, Wood DJ, Rubin DL.. Accuracy of electronic medical record follow-up data for estimating the survival time of patients with cancer. JCO Clin Cancer Inform 2022; 6: e2200019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on request. Information about the prediction algorithm can be found here: https://github.com/pennsignals/eol-onc.