Abstract
Cancer is a major medical problem worldwide. Due to its high heterogeneity, the use of the same drugs or surgical methods in patients with the same tumor may have different curative effects, leading to the need for more accurate treatment methods for tumors and personalized treatments for patients. The precise treatment of tumors is essential, which renders obtaining an in-depth understanding of the changes that tumors undergo urgent, including changes in their genes, proteins and cancer cell phenotypes, in order to develop targeted treatment strategies for patients. Artificial intelligence (AI) based on big data can extract the hidden patterns, important information, and corresponding knowledge behind the enormous amount of data. For example, the ML and deep learning of subsets of AI can be used to mine the deep-level information in genomics, transcriptomics, proteomics, radiomics, digital pathological images, and other data, which can make clinicians synthetically and comprehensively understand tumors. In addition, AI can find new biomarkers from data to assist tumor screening, detection, diagnosis, treatment and prognosis prediction, so as to providing the best treatment for individual patients and improving their clinical outcomes.
Keywords: artificial intelligence, precision medicine, omics, cancer, medical imaging
Graphical Abstract
Introduction
Cancer is a severe threat to human health with a high mortality and a rising incidence rate (1). Several types of cancer can be cured if they are diagnosed and treated early. However, the treatment of cancer is not ideal at present. Cancer mortality rates remain high and continue to rise, including for prostate, colorectal, and cervical cancer (2). These tumors lack effective screening and treatment methods, resulting in patients not getting timely and effective treatment. Secondly, the heterogeneity of tumors is high, which can create great challenges in their treatment (3). Therefore, new diagnostic and treatment methods that are tailored to individual patients are needed. Precision medicine (PM) is a promising approach that takes individual genetics, environment and lifestyle into account and concentrates on clarifying, diagnosing and treating diseases to create a customized treatment plan for patients through obtaining multi-omics or multi-mode information from individuals (4). Furthermore, artificial intelligence (AI) uses computers or machines to carry out tasks by mimicking or emulating human intelligence, which mainly includes machine learning (ML) and deep learning (DL) (5). AI can process an enormous amount of information to promote the brand-new discovery of PM. AI has shown extraordinary potential in processing, mining and analyzing data and can use the data to develop different models to help achieve PM.
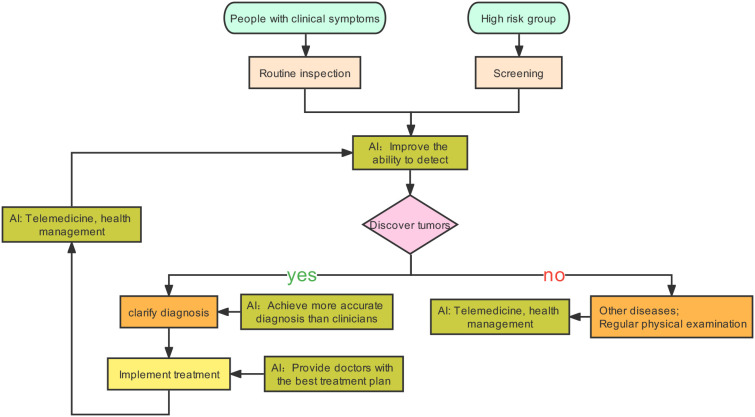
Tumors are generally caught sight of in the following two situations: one is the screening of high-risk groups (6). The other one is the discovery of tumors with clinical manifestations. After the cancer is detected, patients will receive further examinations, such as physical examination, imaging, pathology, and serum tumor markers (6). Based on these results, tumors will be accurately diagnosed, staged, and classified to help the patients benefit from precision treatment. AI can play a part in tumor prevention, screening, diagnosis, treatment, and prognosis prediction (7–10). After AI is injected into the clinical process, it will improve the detection rate of lesions and make the screening method more effective. Secondly, AI can promote the level of diagnosis by helping doctors distinguish between true and false disease progression (7). Finally, AI can calculate the advantages and disadvantages of each treatment scheme and provide the best treatment for patients. In addition, a framework diagram ( Figure 1 ) is added to this article, which shows a series of processes from the discovery of tumor patients to the end of their diagnosis, treatment, and the changes that AI can bring.
Figure 1.
Possible changes caused by AI injection into clinical practice.
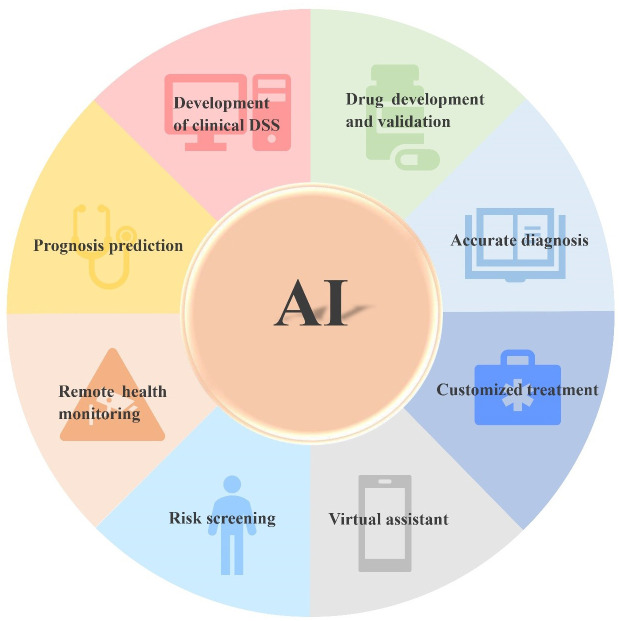
With the development of next-generation sequencing (NGS) technology, omics data, such as genomics, proteomics and transcriptomics, have been accumulated (11). Meanwhile, the massive growth and wide availability of patients’ clinical data such as electronic medical records, clinical trial data, and medical images have led to the era of “big data” (12). The best analysis method is data analysis based on AI, since ML and DL can extract the hidden patterns, important information, and corresponding knowledge behind the data. Based on extracted data, information about the disease is obtained to help clinical analysis. For example, ML and DL can be used to analyze omics data to establish models, generate biomarkers related to diagnosis, classification, and prognosis, provide molecular changes such as DNA, RNA and protein, predict drug efficacy and therapeutic response, and develop targeted drugs (13). Furthermore, as compared with single-omics, multi-omics provides an opportunity to understand the information flow behind a disease (14). Multi-omics integration is crucial to the comprehensive understanding of complex biological processes. Combined with the new longitudinal experimental design, multi-omics can clarify the dynamic relationship between all layers of omics, distinguish the key roles or interactions in system exploitation or complicated phenotypes, clarify the causal relationship and functional mechanisms of complicated diseases, and promote the discovery of PM (15, 16). Quantitative image analysis is a suitable candidate for PM and can assist PM for cancer. ML and DL have been used for quantitatively extracting image features to establish models for diagnosis, monitoring, and predicting recurrence and metastasis, biomarkers and prognosis (17–21). AI can integrate the above data for comprehensive analysis of tumors for the development of a clinical decision support system (DSS) (22). With the continuous improvement of AI algorithms and the improvement of computer software and hardware, AI will mature and will be used more extensively in the medical field ( Figure 2 ) in the future. Therefore, PM for tumors will great evolve.
Figure 2.
Application prospect of AI in tumor.
In the present review, we first introduced the application of AI in omics, and then in pathology and medical imaging, and expanded on how these applications assist PM. Finally, we described the challenges and future directions of AI assisted PM for tumors.
AI-based big data assists PM for cancer
Big data technology mainly includes data analysis, mining, and sharing. It may play a revolutionary role in cancer diagnosis, treatment, prevention, and prognosis, but transforming data into available information to benefit patients is almost at a standstill (23, 24). A major reason for this is that data analysis significantly lags behind data generation (24). The reforms caused by “big data” have affected nearly all aspects of tumor research. For example, the technology can analyze data generated by NGS to discover commonly mutated genes, abnormal gene expression, and biomarkers in tumors for accurate diagnosis and prognosis prediction or to determine the cause of disease and develop targeted drugs for treatment (23, 24). The technology can analyze features that humans can and cannot see in medical images, and mine and filter these features to determine information related to diagnosis, treatment, and prognosis (25, 26). In addition, the technology can analyze patients’ demographic and clinical data, as well as outcome information to predict the factors affecting the prognosis of cancer patients (27). In addition, AI is used to analyze, mine and process tumor-related data, build a health care provider platform based on a significant quantity of tumor-related data, efficiently solve the problem of difficult medical treatment for patients and reduce the waste of unnecessary medical resources (28). Big data reanalysis has been not been sufficiently taken advantage of so far, but we cannot ignore its potential. It can analyze the data in an existing database and provide new insights. For example, Borziak et al. discovered the dedifferentiation markers of liver cancer by using data from existing databases (29). The current big data technology is mainly used in certain fields, such as omics, pathological imaging and medical imaging. However, it does not combine data from multiple fields for data analysis, mining, and sharing, which leads to data not being comprehensively utilized and not meeting clinicians’ and patients’ needs. The challenges in the diagnosis, treatment and monitoring of cancer can be overcome by integrating omics and non-omics data. AI can play an important role in analyzing high-dimensional data-sets with complexity and heterogeneity, especially in multi-omics, intergroup methods and data integration, thus setting forth the cancer molecular mechanism, and identifying new dynamic diagnostic and prognostic biomarkers to provide accurate cancer care (30).
There are certain problems with the current data, such as poor data quality, unstructured databases, inadequate analytics, and lack of delivery (23, 31). Therefore, there is a need for a more authoritative and reliable prospective database. In addition, a longitudinal database is also needed to understand the cancer dynamics of patients in the whole study care continuum (23). Establishing a patient-centered collection of various data-sets will be crucial in the future (32). On this basis, AI-based big data analysis may automatically generate patient diagnosis, personalized treatment plans, and key information for prognostic prediction, thereby helping clinicians provide the best treatment for their patients.
AI assists tumor PM in omics
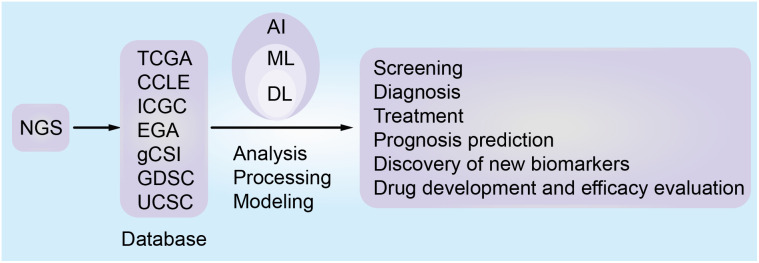
A large amount of data resources ( Table 1 ) generated by NGS can provide key information about tumors. Combining the information with AI will help clarify the etiology and pathogenesis of tumors, and assist the accurate diagnosis, risk stratification, and disease subtype analysis (30, 33). Moreover, AI can identify new therapeutic targets, evaluate the sensitivity and resistance of anticancer drugs, develop new targeted drugs, improve cancer immunotherapy, monitor the recurrence and evolution of the tumor, discover new biomarkers, and predict the prognosis and survival analysis of tumor patients ( Figure 3 ) (34–40). In a few words, AI enables PM for cancer patients, bridging the distance between omics and the clinic. Since NGS produces high-dimensional and complex data, NGS methods for cancer diagnosis usually need higher-dimensional and deeper-seated data coverage to enhance the possibility of detecting a small number of tumor cell mutations and improve the sensitivity and accuracy of AI algorithms (41).
Table 1.
Comprehensive omics database resources for building AI models.
| Name | Main features | Web link |
|---|---|---|
| CGHub | Overall data repository; enormous data | https://cghub.ucsc.edu/ |
| TCGA | Comprehensive database; enormous data | https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga |
| CCLE | Comprehensive database; enormous data | https://sites.broadinstitute.org/ccle |
| EGA | Overall data repository; enormous data | https://ega-archive.org/ |
| ICGC | Comprehensive genomics data | https://dcc.icgc.org/ |
| DepMap | High data quality; visualization | https://depmap.org/portal/ |
| SomamiR | Correlation between cancer somatic mutation and miRNA | https://compbio.uthsc.edu/SomamiR/ |
| COSMIC | largest and most comprehensive somatic mutation database; regularly-updated | https://cancer.sanger.ac.uk/cosmic |
| MethyCancer | integrated data of DNA methylation, cancer-related gene, mutation and cancer information | http://methycancer.psych.ac.cn/ |
| CTRP | connecting sensitivity to cancer feature | https://portals.broadinstitute.org/ctrp/ |
| gCSI | Large amount of transcriptomics data | https://pharmacodb.pmgenomics.ca/datasets/4 |
| GDSC | Drug response data; genomics markers of drug sensitivity; update irregularly | https://www.cancerrxgene.org/ |
| NCI60 | Large amount of drug data and genomics data |
https://discover.nci.nih.gov/cellminer/loadDownload.do
https://dtp.cancer.gov/databases_tools/bulk_data.htm |
| canSAR | Comprehensive database; discovery drug | https://cansarblack.icr.ac.uk/ |
| cBioPortal | Large amount of available data | https://www.cbioportal.org/datasets |
| UCSC | Synthetical genomics information | https://genome.ucsc.edu/ |
| dbNSFP | Predictive data | http://bib.oxfordjournals.org/ |
| NONCODE | database dedicated to non-coding RNAs | http://www.noncode.org/ |
| CSD | The positive and negative training sets | http://bib.oxfordjournals.org/ |
| TCIA | A great quantity of medical related image data sets | https://www.tcia.at/home |
| MSKCC | Cancer mutation databases | http://www.cbioportal.org/ |
| ARCHS4 | comprehensive processed mRNA expression data | https://maayanlab.cloud/archs4/ |
Figure 3.
The role of artificial intelligence based on omics database in tumor precision medicine.
AI assists tumor PM in genomics
In recent years, genomics, which relies on nucleotide sequences for data analysis, has become more closely combined with clinical practice (30). The significant accumulation of data has improved the understanding of cancer vulnerability and has enabled us to increasingly anticipate noticeable treatment effects for tumor patients (42). The use of spatial and single-cell genomics may reconstruct the process of tumorigenesis to facilitate a more comprehensive understanding of tumor, decipher the unclear pathogenesis of human beings and develop targeted drugs based on this mechanism (43–47). The combination of ML and genomics data can assist the diagnosis of cancer subtypes, discovering new markers and drug targets, and understanding cancer-driving genes better, which contributes to providing customized treatment for patients (48). For example, Wang et al. developed a compounded deep network model that can diagnose lung cancer subtypes by mixing image-genomics data and can help biomedical professionals determine the potential therapeutic targets by the attention weights of the model (49). In addition, Vanderbilt et al. developed and validated a brand-new approach to identify DNA viruses from corresponding normal or tumor NGS specimens and inquire about virus-tumor type relevance without carrying out extra sequencing. Data on these viruses can provide information for the diagnosis and care of tumor patients. Their study illustrated the function of DNA viruses in the tumor (50). Sudhakar et al. used cancer genomics data and built a pan-cancer model to forecast and identify new driver genes (51). The identification of driving genes can help understand the carcinogenic mechanism and the design of treatment strategies, which has an important biological and clinical significance (52).
AI assists PM for tumors in transcriptomics
Transcriptomics is a powerful means to evaluate all transcripts produced during metabolism (30). Transcriptomics have expanded our knowledge of cancer occurrence and development, tumor microenvironment, and immune-oncology, and can directly determine gene expression levels and analyze the activation of related molecular pathways (53, 54). Transcriptomics is a bridge between genomics and proteomics, mainly involving quantitative reverse-transcription-polymerase chain reaction, microarrays and NGS (RNA-sequencing) (55). Since RNA-sequencing has a higher accuracy in measuring gene expression, it is considered the gold standard for high-throughput gene expression screening (55, 56). Through data mining or more complex mathematical approaches using ML or DL, the features are extracted to facilitate tumor screening and early diagnosis, discover new or previously unknown cancer biomarkers and potential therapeutic targets, as well as drug prioritization, and predict cancer drug sensitivity and prognosis (53–55, 57–62). For example, Warnat-Herresthal et al. found that ML-based transcriptomics can assist in the diagnosis of acute myeloid leukemia (63). Moreover, Ben Azzouz et al. used an ML approach based on transcriptomics data to calculate triple-negative breast cancer subtypes, in order to overcome the barrier of heterogeneity in the treatment of the disease (64). Finally, some ML-based transcriptomics have also been used in the development of prognostic biomarkers for prostate cancer (65), the diagnosis of colorectal cancer (66), and the prediction of immune response (67).
AI assists tumor PM in proteomics
Proteomics can provide comprehensive and quantitative information about proteins in tissues, blood and cell samples (68). Protein expression profiles generated by proteomics and ML-based profile analysis can identify more specific and sensitive protein biomarkers than other single-omics. These biomarkers can diagnose cancer, predict prognosis (69), reveal critical signaling pathways behind disease mechanisms (70, 71), determine new therapeutic targets, evaluate drug therapy efficacy and toxicity (72), and predict therapeutic responses, recurrence, and metastasis (73, 74). Recently, Henry et al. proposed a method of drug ranking using ML to predict drug response using proteomics data, and prioritize drugs in order to identify the most suitable drug for each patient (75). In addition, Federica et al. built a clearer and more transparent DSS to assist in diagnosing high-grade serous ovarian cancer (76). Therefore, AI-based proteomics may play an important role in the accurate diagnosis and treatment of tumors in the future.
Besides the widely used omics data mentioned above, other omics data (metabolomics, immunomics and microbiome data) are also used (77). For instance, disposing of metabolomics data by AI can assist the diagnosis (78, 79), the of treatment response evaluation (80–82), discovery of new biomarkers (83, 84), and determination of patient tolerance (85) and cancer status (invasive or non-invasive) (86). Moreover, the AI model based on immunomics data can forecast the emergency immune characteristics of tumor patients (87).
AI assists tumor PM in multi-omics
Although the current single-omics data can be used for diagnosis, treatment and prediction, they cannot thoroughly and systematically reflect the molecular changes of a tumor (88). Therefore, it is necessary to integrate multi-omics data to comprehensively understand the tumor information and its dynamic development process to screen and accurately diagnose patients, develop tailored treatment strategies, predict prognosis, and monitor recurrence and metastasis (89–92). Some approaches and algorithms of using AI to analyze multi-omics data comprehensively include clustering, factorization, feature transformation, networks-based means and feature extraction (89). These approaches can be used for stratifying medicine, discovering biomarkers (93), pathway analysis, and drug reuse or discovery (89, 94) ( Table 2 ). For example, Ma et al. introduced a new approach that can analyze multi-omics information and related knowledge to reveal the complex relationship between molecular features and clinical characteristics (114). In addition, Wang et al. developed a molecular algorithm for early cancer detection, which is used to confirm malignant cellular tumors according to the spectrum of changes in single-cell copy numbers based on doubtful cells in humoral, resulting in a well-defined cancer diagnosis (115). Furthermore, Olivier B et al. have developed an integrated framework of DL and ML, which can use multi-omics data to accurately predict survival and prognosis (95). Furthermore, except for the above commonly used omics, studies have also focused on linking radiomics with genomics and transcriptomics for accurate diagnosis (116). A multi-task DL framework called OmiEmbed, which can analyze and process several kinds of omics data and simultaneously handle multiple tasks has recently emerged. This disruptive technological breakthrough will significantly promote the development of PM (117). The application of AI to integrate multi-omics data is shown in Table 2 .
Table 2.
Application of AI in the Integration of Multi-omics.
| Clinical application | Data | Model/Algorithm | Performance | References |
|---|---|---|---|---|
| cancer prognosis and survival prediction | RNA-Seq, Methylation, and miRNA | semi-supervised flexible hybrid machine-learning framework | Not applicable | Poirion, O.B., et al. (95) |
| breast cancer subtype identification | mRNA expression, miRNA expression and DNA methylation | deep learning fusion clustering framework | 0.664 | Shuangshuang, L., et al. (91) |
| cancer susceptibility prediction | copy number variations, miRNA expression, and gene expression | multimodal convolutional autoencoder model | 0.9625 | Karim, M.R., et al. (96) |
| identifying Neuroblastoma subtypes | gene expression, copy number alterations, Sequencing Quality Control project | deep learning | 0.74 | Zhang, L., et al. (97) |
| predict the survival of patients with lung cancer | TCGA | unsupervised learning | 0.99 | Takahashi, S., et al. (90) |
| survival stratification of gastric cancer | transcriptomics and epigenomics | bidirectional deep neural networks | 0.76 | Xu, J.M., et al. (98) |
| pan-cancer metastasis prediction | RNA-Seq, microRNA sequencing, and DNA methylation | deep learning | 0.8885 | Albaradei, S., et al. (92) |
| ovarian cancer subtypes identification | mRNA-seq, miRNA-seq, copy number variation, and the clinical information | deep learning | 0.583 | Guo, L. Y., et al. (99) |
| drug repurposing | copy number alteration, DNA methylation, gene expression, pharmacological characteristics for cancer cell lines | deep learning | 0.84 | Wang, Y., et al. (94) |
| predicting lung adenocarcinoma prognostication | mRNA, miRNA, DNA methylation and copy number variations | deep learning | 0.65 | Lee, T.-Y., et al. (100) |
| Diagnostic Classification of Lung Cancer | mRNA expression, miRNA-seq data, and DNA methylation data | deep transfer Learning | 0.824 | Zhu, R., et al. (101) |
| predicting effective therapeutic agents for breast cancer | copy number variations, miRNA, mutation, RNA, protein expression and methylation | deep learning | 0.94 | Khan, D. and S. Shedole (102) |
| predicting survival prognosis for glioma patients | transcription profile, miRNA expression, somatic mutations, copy number variation, DNA methylation, and protein expression | deep learning | 0.990 | Pan, X., et al. (103) |
| Diagnostic classification of cancers | mRNA expression, miRNA-seq, DNA methylation data and clinical information | XGBoost | 0.595-0.872 | Ma, B., et al. (104) |
| identify tumor molecular subtypes | copy number, mRNA, miRNA, DNA methylation and other omics data | consensus clustering and the Gaussian Mixture model | Not applicable | Yang, H., et al. (105) |
| predicting outcome for patients with hepatocellular carcinoma | DNA methylation and mRNA expression data | unsupervised machine-learning | Not applicable | Huang, G. J., et al. (106) |
| predicting the Gleason score levels of prostate cancer and the tumor stage in breast cancer | gene expression, DNA methylation, and copy number alteration | gene similarity network based on uniform manifold approximation and projection and convolutional neural networks | 0.99 | ElKarami, B., et al. (107) |
| patient classification, tumor grade classification, cancer subtype classification | mRNA expression, DNA methylation, and microRNA expression data | Multi-Omics Graph cOnvolutional NETworks | Not applicable | Wang, T. X., et al. (108) |
| cancer prognosis prediction | mRNA, miRNA, DNA methylation, and copy number variation | denoising Autoencoder | Not applicable | Chai, H., et al. (109) |
| cancer subtype classification | gene expression, miRNA expression and DNA methylation data | hierarchical integration deep flexible neural forest framework | 0.885 | Xu, J., et al. (110) |
| Prediction of prognosis of cancer | single nucleotide polymorphism, copy number variant, gene expression, and DNA methylation data | deep learning | 0.67-0.88 | Park, C., et al. (111) |
| tumor Stratification | deoxyribonucleic acid methylation, messenger ribonucleic acid expression data, and protein–protein interactions | Network Embedding; supervised learning; unsupervised clustering algorithm | 0.91 | Li, F., et al. (112) |
| discovery of cancer subtypes | mRNA expression, miRNA expression, DNA methylation, and copy number alterations | end-to-end variational deep learning-based clustering method; Variational Bayes | Not applicable | Rong, Z., et al. (113) |
AI in pathology assists the accurate diagnosis of tumors
Pathological analysis is considered the gold standard of the clinical diagnosis of tumors (118). However, the current shortage of clinical pathologists and their reliance on subjective consciousness for diagnosis leads to low repeatability and unequal diagnostic levels of clinical pathologists, which is not helpful for clinicians’ decision-making with regards to treatment (119). Computational pathology has seen significant developments from the use of improved AI algorithms and computing power. With the use of image analysis of digital pathology, ML and DL, AI has been used to evaluate whole slide imaging (WSI) and produce computer-aided diagnosis systems (CADs), as well as aid cancer prognosis prediction (120–124). At present, the diagnostic ability of the AI-based diagnostic model can be comparable to or even surpass that of experts (125). In combination with human experts, the precision of diagnosis can be even better. It also has the advantages of being less time-consuming, and having a high efficiency and repeatability. Therefore, an increasing number of AI models are being developed to assist clinical pathologists and reduce their workload (120). For example, Ho et al. proposed incorporating AI models into the pathological workflow as the first reader, the second reader, triage, and pre-screening (120, 126, 127).
Aiding diagnosis through ML and DL mainly includes three steps: The first step includes data preprocessing, such as image sharpening, masking and smoothing, image graying and color normalization, data standardization, and data annotation. The second step includes the division of nucleus/tissue. During the third step, models are established for training and verification, diagnosis and prediction (128, 129). For example, the computer-aided diagnosis and prognosis prediction model of WSI based on hematoxylin and eosin staining, can screen, classify and grade tumors (130), and identify micro-metastasis in lymph nodes (25), and microsatellite instability (128). It can also predict the changes at the molecular level (131), the risk of metastasis and recurrence after surgical resection (132) and disease-specific survival (133). Moreover, Armin et al. used a DL model based on digital images of immunohistochemistry to calculate the risk of mortality (134). The role of AI in the digital pathological image is summarized in Figure 4 .
Figure 4.
The role of artificial intelligence based on medical images (digital pathological image and medical imaging) in tumor precision medicine.
Traditional ML methods analyze pathological tissue by manually extracting mainly morphological, textural (135) and spatial features (118). It is easier to understand and explain than DL, and its training sample size is small, especially suitable for the analysis of rare tumor subtypes with a limited sample size. However, manually extracted features have the following limitations (118): They are extracted in an unsupervised way and have nothing to do with the subsequent WSI analysis tasks (136). Only the surface features of the input image can be learned, which is not enough to show the complex features of WSI. It is exceptionally arduous to process multiple WSI images at the same time and the processing speed is slow (137). Compared with ML, DL can automatically extract the features in the image for analysis and can also efficiently process a considerable amount of data (129). The DL model has good scalability (138), but it is easy to overfit, resulting in the low generalization ability of the model. Furthermore, it is characterized by low interpretability and cannot be trusted by clinical pathologists (139). Recently, diagnostic models combining various methods of traditional ML, DL have been developed to integrate their advantages for accurate diagnosis and prediction (129). For example, Sengupta et al. have proposed a novel deep hybrid learning model based on nuclear morphology for accurately diagnosing ovarian cancer (129). It can be safely assumed that in the near future more high-performance prediction models will be developed and enter the clinic to assist clinical pathologists in accurate diagnosis and prognosis prediction, consequently providing accurate and personalized medical care for patients.
AI assists PM for tumors in medical imaging
Imaging is one of the indispensable tools for screening, diagnosis, treatment and follow-ups for several types of tumors. At present, the performance of imaging examination equipment such as thin-layer computed tomography (CT) and multi-parameter magnetic resonance imaging (MRI), is continuously improved, detecting more subtle lesions and producing increasingly complex data. However, this requires more time and effort from radiologists to make a diagnosis, increasing their workload. AI-based data analysis can effectively process huge amounts of data, with the CADs model based on medical images exhibiting high precision and standard (140–142). Introducing AI into clinical practice will help radiologists make diagnostic decisions quickly, accurately, and efficiently, will help focus their energy on advanced decision-making, and promoting accurate medical treatment and personalized treatment for tumors (141, 143). The role of AI in medical imaging is summarized in Figure 4 .
AI assists radiologists in accurately diagnosing tumors
AI has three main tasks in tumor imaging: Detecting, characterizing and monitoring tumors (144). Detection refers to the location of the region of interest in the image. Characterization includes tumor diagnosis, and staging. Monitoring refers to the monitoring of the changes in tumors with time (144). The process of ML-assisted tumor detection and diagnosis is as follows: Image data acquisition, image preprocessing, segmentation of regions of interest, feature selection, establishing the model and carrying out training, verification and testing (145). Among them, feature selection is the most important step, since it is most related to the model’s performance (145). Moreover, DL can automatically extract the feature from the image. Therefore, recent research has increasingly focused on the DL to build “fusion” models for the diagnosis of tumor lesions, including classification, grading and staging, which have been proven to be effective (146–148). For example, Chougrad et al. have built CADs based on deep convolutional neural networks (CNN) to aid radiologists in categorizing breast X-ray masses (149). Moreover, Misra et al. have developed a highly robust DL model for categorizing benign and malignant neoplastic lesions of the breast. The model can improve the accuracy of breast cancer classification by correcting patients whose traditional methods are misclassified (150). Overall, these models can make radiologists more effective in detecting and diagnosing tumors faster, and will likely be popularized and applied to clinical medical treatment soon.
AI assists PM for tumors diagnosed via medical imaging
The choice of treatment depends on the outcome of the diagnosis. For example, if the detected lesion is benign, it can reduce unnecessary surgical resection and other treatments, and provide more targeted medical management for patients. In addition, the use of radiation imaging, a non-invasive diagnostic method, can protect patients from the discomfort caused by biopsy and avoid the risk of implant metastasis in pathological biopsy (151). Moreover, preoperative evaluation of tumor grading prediction using radiology can help select more appropriate treatment options for patients and avoid unnecessary surgery, thereby reducing the patient’s medical burden and avoiding excessive medical treatment (152, 153).
In addition to assisting in accurate diagnosis, AI can also play a significant role in prognosis prediction and treatment of patients. It can predict patient viability based on imaging features and determine the level of treatment needed to achieve optimal survival. The prediction of recurrence, metastasis, surgical margins and therapeutic responses can be used to formulate an optimal therapeutic strategy for individual patients (21, 26).
Accurately identifying and evaluating lesion before an operation can help create appropriate treatment plans for patients and avoid unnecessary treatment measures such as surgery, postoperative radiotherapy, and chemotherapy, which is beneficial to both patients and doctors. For instance, Zhao et al. built a neoplasm grade forecast model of pre-operative G1/2 assessment of nonfunctional pancreatic neuroendocrine tumors by using radiomics to analyze the multi-slice helical CT images (152). In addition, Xie et al. used a CT-based radiomics ML method to distinguish pancreatic mucinous cystic neoplasm from atypical serous cystadenomas prior to surgery (154). The classification and types of tumors are different, and their treatment methods are inconsistent. For example, according to the model established by Zhao and Xie, if the preoperative prediction result is a high-risk or high-grade tumor, it is necessary to strengthen the follow-up treatment, such as postoperative neoadjuvant chemotherapy or radiotherapy (152, 154). Depending on whether the lymph node is metastatic or not, clinicians will choose different treatment options for patients. Therefore, the detection of lymph node metastasis is extremely important. For example, Song et al. established and verified a radiomics nomogram based on dynamic contrast-enhanced MRI, which can predict metastasis of axillary lymph nodes in mastocarcinoma (155). Similarly, Eresen et al. used the radiomics-derived model established by ML to detect metastatic lymph nodes in colorectal cancer patients (156). Predicting preoperative tumor markers and imaging biomarkers can lead to better clinical decision-making and help provide the best treatment for patients. For example, Guo et al. developed LR and LR-SVMSMOTE models based on CT radiomics to predict thyroid cartilage invasion in certain cancers types, such as hypopharyngeal squamous cell carcinoma and laryngeal carcinoma (157). Similarly, Akbari et al. combined the advanced mode analysis and ML method of multi-parameter MRI to provide the prediction space map of tumor invasion and early recurrence possibility to provide more targeted surgical and radiotherapy strategies for tumor patients, aiming to maximize the treatment effect while maintaining neurological function (158).
The DL model based on one of the subsets of AI can assist radiotherapy or oncology doctors in accurately outlining tumor targets, reducing the time doctors take to manually segment images as well as reducing the variation between observers (159–163). The model can predict and verify the therapeutic dose, and allows for the dose prescription to be changed in time to reduce the impact on the surrounding normal tissue, prevent unnecessary radiation, and reduce the occurrence of adverse reactions (164, 165). The model can evaluate the efficacy of radiotherapy and chemotherapy, as well as the therapeutic response, so as to achieve better-personalized prescriptions for patients (166–168). For example, Ermiş et al. have used DL to depict fully automatic brain resection cavity delineation in patients with glioblastoma (169). Likewise, Zhou et al. have developed and tested a three-dimensional DL model capable of predicting the dosage distribution of three-dimensional volume units to carry out intensity-modulated radiation therapy (170). Establishing a dose distribution model prior to treatment helps adjust the dose distribution in advance and reduce the probability of complications from radiotherapy. ML and DL can also assist in post-radiotherapy management, such as distinguishing between the true and false progression of the tumor, radiation necrosis and tumor recurrence, and promoting clinical medical decision-making, thus improving PM (171–174). In addition, the image-based AI model can also assist radiologists in treatment evaluation, including predicting the response of individual cancer patients to chemotherapy or immunotherapy, and monitoring recurrence and metastasis (175–177). Several radiomics-based ML and DL models can predict patient prognosis, such as recrudesce-free and progression-free survival, survival rate, mortality, surgical results, postoperative metastasis and recurrence. According to the prediction results of the model, the corresponding processing is carried out to create a customized treatment scheme for patients and improve the treatment effect and later quality of life. For example, patients with lower overall survival prediction need more intensive treatment. Patients with poor surgical results may want to consider changing the surgical method or choosing non-surgical treatment. Patients with a higher risk of tumor recurrence and metastasis should continue to receive neoadjuvant radiotherapy and chemotherapy (21, 26, 178–180).
Current challenges and future prospects
Although AI is expected to help improve a series of clinical applications against cancer, it does have some challenges and limitations.
One limitation is the lack of standards and imbalance in the data used to build the model (181, 182). These disordered data will lead to the low robustness of the model and be unfit for constructing a DL model with high generalization and precision (129). For example, medical imaging data is generated under different parameters for different devices (5). Digital pathological images are produced by staining with different dyes. The non-standard operation of pathological specimen collection will also affect the quality of pathological images (122). Irregularities in data collection lead to bias. Omics data are also noisy and heterogeneous (183). These data sets, which are generated by different technologies and standards limit the promotion and generalization of AI models, thus limiting their application in clinical practice. In addition, the sample size of training samples and verification samples used to establish the AI model is small, which can easily cause the overfitting of the model (120). Finally, integrating various types of data, such as genomics, transcriptomics, proteomics, metabolomics, immunomics, electronic health records, clinical medical records, pathology and medical images, will help evaluate the tumor comprehensively and develop the best treatment plan for patients (77, 184, 185). Therefore, it is necessary to establish an extensive comprehensive standardized database. However, many types of data have a multi-scale nature, which makes the mechanical connection between data elusive. The biological knowledge of connecting all these variables in a single model is limited, so many data variables will be omitted from the model development process (186). Recently, some studies have combined dynamic modeling and ML to promote the integration of mathematics and clinical oncology. This method can integrate multiple types of data for personalized prediction to assist PM (187). More in-depth research to promote the combination of mechanical modeling and ML approaches is required in the future, so that mathematical oncology can be introduced into clinics. Building deep fusion models such as multi-modality DL is the primary method to develop AI models that can effectively integrate multimodal data information. However, the current method mainly focuses on representation fusion (feature- and decision-level fusion). The main challenge of this method is that the data is highly dimensional, noisy, heterogeneous, and has a small sample size, and there will be data loss during processing (91, 96, 188, 189). Here are some methods to address these barriers: T-distributed stochastic neighborhood embedding, autoencoder, random forest deep feature selection, a stacked autoencoder, gradient descent method, multi-view factorization autoencoder, co-expression network analysis, and regulation techniques (88, 91, 114, 190–193).
In addition, data from patients are governed by privacy laws (194). The lack of supervision of these data may lead to breaching patient privacy rules; therefore, appropriate intervention and the improvement of laws and regulations are required. Certainly, studies have focused on solving the privacy problem with regards to patient data. Under the same performance, the privacy vulnerability is reduced by vocabulary selection means (194). At the same time, fusing these data should comply with the principles of medical ethics.
Another limitation is that AI algorithms have been regarded as “black boxes” (139), since the process of their output results is unknown and unexplained, which makes clinicians have low trust in AI and a low willingness to introduce it into the clinical workflow (181). Developing a knowledge-embedded DL model for multi-dimensional data fusion is a hopeful means for this problem (195). An increasing number of studies on AI interpretability and transparency are being conducted. The research aims to make AI transparent and interpretable so that its results can be convincing and easy to introduce into the clinical (128). Traditional ML and DL have their advantages and disadvantages, prompting more research on hybrid learning methods. The current research results show that the hybrid learning model exhibits a better performance, better interpretability, higher transparency and more accurate prediction (129, 148). Although AI has shown the ability to surpass people, since it cannot produce 100% correct results, doctors’ participation is still required for the final diagnosis and treatment decisions (183). Future research will focus on improving the interpretability and performance of AI, because it is an important step for AI to realize clinical application (31).
AI model may be necessary to carry out clinical experiments similar to clinical drug trials because when AI models are initially applied to clinical practice, unexpected clinical conditions will inevitably occur. Only through continuous practice can we better find problems and solve them to improve AI models. However, as AI differs from drugs, its clinical trial plan should also be distinguished from drug clinical trials.
AI can be deployed before, during, and after diagnosis, which respectively stands for cancer prevention, screening, diagnosis, and treatment. For example, before diagnosis, AI can be combined with gene detection, endoscopic examination, and other technology to predict the risk of disease occurrence earlier and carry out risk management for patients to reduce the possibility of disease occurrence (196). Augmented or virtual reality can simulate experiences to improve patient compliance (165). During diagnosis, AI can roughly ask patients for relevant information and process it. Secondly, AI can analyze medical image, blood biochemistry, and other clinical overall data to automatically generate a diagnosis report and a variety of feasible as well as optimal treatment methods. Furthermore, after diagnosis, AI can assist clinicians cut down the damage and maximizing the benefits for patients in surgery, radiotherapy, and chemotherapy. The deployment of AI in clinical practice will improve the efficiency of clinicians, reduce the possibility of clinical errors, improve the medical status in areas with low medical levels, and reduce unnecessary procedures, interventions, and medical costs. In a word, patients and doctors will benefit from AI to achieve a win-win situation.
Most people believe that AI cannot replace doctors (7). AI is an assistant in clinical practice, so the final decision must be made by doctors; the responsibility should also be borne by doctors. However, the clinician cannot control AI because it can make self-development and its development process illegible. Therefore, doctors should not be fully responsible for AI errors. Despite that, when using AI, clinicians should not lose their ability to doubt AI to make accurate diagnoses and treatments and develops the doctor-patient relationship in a sound direction.
Conclusion
AI has shown promising results in certain fields of oncology, including tumor screening, detection, diagnosis, treatment, and prognosis prediction. With the progress of AI, the improvement of computer performance, and the explosive growth of various data, new learning methods, such as the hybrid learning method, will continue to emerge, further improving the overall performance of the model, such as efficient data analysis and accurate prediction. The recent model generated by the ML and DL that can analyze various data sets will also improve the prospects of PM. In conclusion, AI-assisted PM can help detect, diagnose and treat cancer early, as well as assist in the selection of the best treatment scheme, consequently improving the prognosis of patients and improving their treatment results.
Author contributions
JL collected the related papers and was a significant contributor to writing the manuscript. XL, YG, and SH made figures and tables. PR, WW, WL and LZ revised the article. PR, WW, WL and LZ initiated the study and revised the manuscript. All authors contributed to the article and approved the submitted version.
Abbreviations
AI, artificial intelligence; ML, machine learning; DL, deep learning; PM, precision medicine; DSS, decision support system; NGS, next-generation sequencing; WSI, whole slide imaging; CADs, computer-aided diagnosis system; CT, computed tomography; MRI, magnetic resonance imaging; CNN, convolutional neural networks; US, ultrasound; PET/CT, positron emission tomography/computed tomography; DLR, deep learning radiomics; CGHub, Cancer Genomics Hub; TCGA, The Cancer Genome Atlas; CCLE, Cancer Cell Line Encyclopedia; ICGC, International Cancer Genome Consortium; EGA, European Genome-phenome Archive; COSMIC, Catalogue Of Somatic Mutations In Cancer; SomamiR, Somatic mutations altering microRNA-ceRNA interactions; CTRP, Cancer Therapeutics Response Portal; gCSI, The Genentech Cell Line Screening Initiative; GDSC, Genomics of Drug Sensitivity in Cancer; NCI, National Cancer Institute; DepMap, Dependency Map; TCIA, The Cancer Immunome Database.
Funding
This work was supported by China’s National Natural Science Foundation [grant numbers 81972837, 82003203, 82071986, 81771827, 81971721], and the Natural Science Foundation of Hunan Province [grant numbers 2021JJ31011, 2021JJ41058, 2021RC4017].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer (2013) 132(5):1133–45. doi: 10.1002/ijc.27711 [DOI] [PubMed] [Google Scholar]
- 2. Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends–an update. Cancer Epidemiol Biomarkers Prev (2016) 25(1):16–27. doi: 10.1158/1055-9965.EPI-15-0578 [DOI] [PubMed] [Google Scholar]
- 3. Consortium, I.T.P.-C.A.o.W.G . Pan-cancer analysis of whole genomes. Nature (2020) 578(7793):82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. MacEachern SJ, Forkert ND. Machine learning for precision medicine. Genome (2021) 64(4):416–25. doi: 10.1139/gen-2020-0131 [DOI] [PubMed] [Google Scholar]
- 5. Gore JC. Artificial intelligence in medical imaging. Magn Reson Imaging (2020) 68:A1–4. doi: 10.1016/j.mri.2019.12.006 [DOI] [PubMed] [Google Scholar]
- 6. Zhou J, Sun H, Wang Z, Cong W, Wang J, Zeng M, et al. Guidelines for the diagnosis and treatment of hepatocellular carcinoma (2019 edition). Liver Cancer (2020) 9(6):682–720. doi: 10.1159/000509424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Le Berre C, Sandborn WJ, Aridhi S, Devignes M-D, Fournier L, Smail-Tabbone M, et al. Application of artificial intelligence to gastroenterology and hepatology. Gastroenterology (2020) 158(1):76–+. doi: 10.1053/j.gastro.2019.08.058 [DOI] [PubMed] [Google Scholar]
- 8. Senan EM, Jadhav ME, Rassem TH, Aljaloud AS, Mohammed BA, Al-Mekhlafi ZG. Early diagnosis of brain tumour MRI images using hybrid techniques between deep and machine learning. Comput Math Methods Med (2022) 2022. doi: 10.1155/2022/8330833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rajput G, Agrawal S, Biyani K, Vishvakarma SK. Early breast cancer diagnosis using cogent activation function-based deep learning implementation on screened mammograms. Int J Imaging Syst Technol (2022) 32(4):1101–18. doi: 10.1002/ima.22701 [DOI] [Google Scholar]
- 10. Qiu H, Ding S, Liu J, Wang L, Wang X. Applications of artificial intelligence in screening, diagnosis, treatment, and prognosis of colorectal cancer. Curr Oncol (2022) 29(3):1773–95. doi: 10.3390/curroncol29030146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaczmarek E, Pyman B, Nanayakkara J, Tuschl T, Tyryshkin K, Renwick N, et al. Discriminating neoplastic from nonneoplastic tissues using an miRNA-based deep cancer classifier. Am J Pathol (2022) 192(2):344–52. doi: 10.1016/j.ajpath.2021.10.012 [DOI] [PubMed] [Google Scholar]
- 12. Hwang K-T. Clinical databases for breast cancer research. In: Noh D-Y, Han W, Toi M, editors. Translational research in breast cancer. Singapore: Springer Singapore; (2021). p. 493–509. [DOI] [PubMed] [Google Scholar]
- 13. Picard M, Scott-Boyer MP, Bodein A, Perin O, Droit A. Integration strategies of multi-omics data for machine learning analysis. Comput Struct Biotechnol J (2021) 19:3735–46. doi: 10.1016/j.csbj.2021.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol (2017) 18(1):83. doi: 10.1186/s13059-017-1215-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bodein A, Scott-Boyer M-P, Perin O, Lê Cao K-A, Droit A. Interpretation of network-based integration from multi-omics longitudinal data. Nucleic Acids Res (2022) 50(5):e27–7. doi: 10.1093/nar/gkab1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Song M, Greenbaum J, Luttrell J, Zhou WH, Wu C, Shen H, et al. A review of integrative imputation for multi-omics datasets. Front Genet (2020) 11. doi: 10.3389/fgene.2020.570255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol (2017) 14(12):749–62. doi: 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 18. Ibrahim A, Primakov S, Beuque M, Woodruff HC, Halilaj I, Wu G, et al. Radiomics for precision medicine: Current challenges, future prospects, and the proposal of a new framework. Methods (2021) 188:20–9. doi: 10.1016/j.ymeth.2020.05.022 [DOI] [PubMed] [Google Scholar]
- 19. Refaee T, Wu G, Ibrahim A, Halilaj I, Leijenaar RTH, Rogers W, et al. The emerging role of radiomics in COPD and lung cancer. Respiration (2020) 99(2):99–107. doi: 10.1159/000505429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology (2015) 278(2):563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Preuss K, Thach N, Liang X, Baine M, Chen J, Zhang C, et al. Using quantitative imaging for personalized medicine in pancreatic cancer: A review of radiomics and deep learning applications. Cancers (2022) 14(7). doi: 10.3390/cancers14071654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu Q, Zhai JC, Huo CQ, Li Y, Dong XJ, Li DF, et al. OncoPDSS: an evidence-based clinical decision support system for oncology pharmacotherapy at the individual level. BMC Cancer (2020) 20(1):740. doi: 10.1186/s12885-020-07221-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barker AD, Lee JS. Translating "Big data" in oncology for clinical benefit: Progress or paralysis. Cancer Res (2022) 82(11):2072–5. doi: 10.1158/0008-5472.CAN-22-0100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Malone ER, et al. Molecular profiling for precision cancer therapies. Genome Med (2020) 12(1):8. doi: 10.1186/s13073-019-0703-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chuang W-Y, Chen C-C, Yu W-H, Yeh C-J, Chang S-H, Ueng S-H, et al. Identification of nodal micrometastasis in colorectal cancer using deep learning on annotation-free whole-slide images. Modern Pathol (2021) 34(10):1901–11. doi: 10.1038/s41379-021-00838-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luo H-S, Chen Y-Y, Huang W-Z, Wu S-X, Huang S-F, Xu H-Y, et al. Development and validation of a radiomics-based model to predict local progression-free survival after chemo-radiotherapy in patients with esophageal squamous cell cancer. Radiat Oncol (2021) 16(1):201. doi: 10.1186/s13014-021-01925-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu W, Yang J, Li D, Huang Q, Zhao F, Feng X, et al. Competitive risk analysis of prognosis in patients with cecum cancer: A population-based study. Cancer Control (2021) 28:1073274821989316. doi: 10.1177/1073274821989316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng J, Gao ZJ, Pu LX, He MJ, Fan JP, Wang S, et al. Analysis of tumor disease patterns based on medical big data. J Med Imaging Health Inf (2021) 11(2):478–86. doi: 10.1166/jmihi.2021.3306 [DOI] [Google Scholar]
- 29. Borziak K, Finkelstein J. Utilizing shared big data to identify liver cancer dedifferentiation markers. Stud Health Technol Inform (2022) 289:73–6. doi: 10.3233/SHTI210862 [DOI] [PubMed] [Google Scholar]
- 30. Patel SK, George B, Rai V. Artificial intelligence to decode cancer mechanism: Beyond patient stratification for precision oncology. Front Pharmacol (2020) 11. doi: 10.3389/fphar.2020.01177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kondylakis H, Ciarrocchi E, Cerda-Alberich L, Chouvarda I, Fromont LA, Garcia-Aznar JM, et al. Position of the AI for health imaging (AI4HI) network on metadata models for imaging biobanks. Eur Radiol Exp (2022) 6(1). doi: 10.1186/s41747-022-00281-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marti-Bonmati L, Alberich-Bayarri A, Ladenstein R, Blanquer I, Segrelles JD, Cerda-Alberich L, et al. PRIMAGE project: predictive in silico multiscale analytics to support childhood cancer personalised evaluation empowered by imaging biomarkers. Eur Radiol Exp (2020) 4(1):22. doi: 10.1186/s41747-020-00150-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Song H, Ruan C, Xu Y, Xu T, Fan R, Jiang T, et al. Survival stratification for colorectal cancer via multi-omics integration using an autoencoder-based model. Exp Biol Med (2022) 247(11):898–909. doi: 10.1177/15353702211065010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trubicka J, Grajkowska W, Dembowska-Baginska B. Molecular markers of pediatric solid tumors-diagnosis, optimizing treatments, and determining susceptibility: Current state and future directions. Cells (2022) 11(7):1238. doi: 10.3390/cells11071238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kawazu M, Ueno T, Saeki K, Sax N, Togashi Y, Kanaseki T, et al. HLA class I analysis provides insight into the genetic and epigenetic background of immune evasion in colorectal cancer with high microsatellite instability. Gastroenterology (2022) 162(3):799–812. doi: 10.1053/j.gastro.2021.10.010 [DOI] [PubMed] [Google Scholar]
- 36. Xu F, Chen JH, Huang DH. Pan-cancer analysis identifies FAM49B as an immune-related prognostic maker for hepatocellular carcinoma. J Cancer (2022) 13(1):278–89. doi: 10.7150/jca.65421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson BE, Creason AL, Stommel JM, Keck JM, Parmar S, Betts CB, et al. An omic and multidimensional spatial atlas from serial biopsies of an evolving metastatic breast cancer. Cell Rep Med (2022) 3(2):100525. doi: 10.1016/j.xcrm.2022.100525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Soundararajan M, Eswaran J. Atypical GTPases as drug targets. Anticancer Agents Med Chem (2012) 12(1):19–28. doi: 10.2174/187152012798764705 [DOI] [PubMed] [Google Scholar]
- 39. Kucab JE, Zou XQ, Morganella S, Joel M, Nanda AS, Nagy E, et al. A compendium of mutational signatures of environmental agents. Cell (2019) 177(4):821–+. doi: 10.1016/j.cell.2019.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van de Geer WS, Hoogstrate Y, Draaisma K, Robe PA, Bins S, Mathijssen RHJ, et al. Landscape of driver gene events, biomarkers, and druggable targets identified by whole-genome sequencing of glioblastomas. Neuro-oncology Adv (2022) 4(1):vdab177–vdab177. doi: 10.1093/noajnl/vdab177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Griffith M, Miller CA, Griffith OL, Krysiak K, Skidmore ZL, Ramu A, et al. Optimizing cancer genome sequencing and analysis. Cell Syst (2015) 1(3):210–23. doi: 10.1016/j.cels.2015.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Berger MF, Mardis ER. The emerging clinical relevance of genomics in cancer medicine. Nat Rev Clin Oncol (2018) 15(6):353–65. doi: 10.1038/s41571-018-0002-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gaublomme JT, Li B, McCabe C, Knecht A, Yang Y, Drokhlyansky E, et al. Nuclei multiplexing with barcoded antibodies for single-nucleus genomics. Nat Commun (2019) 10(1):2907. doi: 10.1038/s41467-019-10756-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods (2014) 11(4):417–22. doi: 10.1038/nmeth.2869 [DOI] [PubMed] [Google Scholar]
- 45. Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell (2015) 161(5):1202–14. doi: 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moffitt JR, Hao J, Wang G, Chen KH, Babcock HP, Zhuang X, et al. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc Natl Acad Sci U.S.A. (2016) 113(39):11046–51. doi: 10.1073/pnas.1612826113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stoeckius M, Zheng S, Houck-Loomis B, Hao S, Yeung BZ, Mauck WM. Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol (2018) 19(1):224. doi: 10.1186/s13059-018-1603-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang S, Cai N, Pacheco PP, Narrandes S, Wang Y, Xu W, et al. Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genomics Proteomics (2018) 15(1):41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang X, Yu G, Yan Z, Wan L, Wang W, Lizhen LCC. Lung cancer subtype diagnosis by fusing image-genomics data and hybrid deep networks. IEEE/ACM Trans Comput Biol Bioinf (2021) p:1–1. doi: 10.1109/TCBB.2021.3132292 [DOI] [PubMed] [Google Scholar]
- 50. Vanderbilt CM, Bowman AS, Middha S, Petrova-Drus K, Tang Y-W, Chen X, et al. Defining novel DNA virus-tumor associations and genomic correlates using prospective clinical Tumor/Normal matched sequencing data. J Mol Diagnostics (2022). doi: 10.1016/j.jmoldx.2022.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sudhakar M, Rengaswamy R, Raman K. Novel ratio-metric features enable the identification of new driver genes across cancer types. Sci Rep (2022) 12(1):5. doi: 10.1038/s41598-021-04015-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu HH, Hua X, Shi JX, Chatterjee N, Zhu B. Utilizing patient information to identify subtype heterogeneity of cancer driver genes. Stat Methods Med Res (2022) 31(3):510–9. doi: 10.1177/09622802211055854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Buzdin A, Sorokin M, Garazha A, Sekacheva M, Kim E, Zhukov N, et al. Molecular pathway activation - new type of biomarkers for tumor morphology and personalized selection of target drugs. Semin Cancer Biol (2018) 53:110–24. doi: 10.1016/j.semcancer.2018.06.003 [DOI] [PubMed] [Google Scholar]
- 54. Supplitt S, Karpinski P, Sasiadek M, Laczmanska I. Current achievements and applications of transcriptomics in personalized cancer medicine. Int J Mol Sci (2021) 22(3). doi: 10.3390/ijms22031422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Buzdin A, Sorokin M, Garazha A, Glusker A, Aleshin A, Poddubskaya E, et al. RNA Sequencing for research and diagnostics in clinical oncology. Semin Cancer Biol (2020) 60:311–23. doi: 10.1016/j.semcancer.2019.07.010 [DOI] [PubMed] [Google Scholar]
- 56. Consortium SM-I. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the sequencing quality control consortium. Nat Biotechnol (2014) 32(9):903–14. doi: 10.1038/nbt.2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ma L, Liang Z, Zhou H, Qu L. Applications of RNA indexes for precision oncology in breast cancer. Genomics Proteomics Bioinf (2018) 16(2):108–19. doi: 10.1016/j.gpb.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Clarke PA, te Poele R, Workman P. Gene expression microarray technologies in the development of new therapeutic agents. Eur J Cancer (2004) 40(17):2560–91. doi: 10.1016/j.ejca.2004.07.024 [DOI] [PubMed] [Google Scholar]
- 59. Abdul Aziz NA, Mokhtar NM, Harun R, Mollah MM, Mohamed Rose I, Sagap I, et al. A 19-gene expression signature as a predictor of survival in colorectal cancer. BMC Med Genomics (2016) 9(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Montani F, Marzi MJ, Dezi F, Dama E, Carletti RM, Bonizzi G, et al. miR-test: a blood test for lung cancer early detection. J Natl Cancer Inst (2015) 107(6):djv063. doi: 10.1093/jnci/djv063 [DOI] [PubMed] [Google Scholar]
- 61. Nadal E, Truini A, Nakata A, Lin J, Reddy RM, Chang AC, et al. A novel serum 4-microRNA signature for lung cancer detection. Sci Rep (2015) 5:12464. doi: 10.1038/srep12464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hira ZM, Gillies DF. A review of feature selection and feature extraction methods applied on microarray data. Adv Bioinf (2015) 2015:198363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Warnat-Herresthal S, Perrakis K, Taschler B, Becker M, Bassler K, Beyer M, et al. Scalable prediction of acute myeloid leukemia using high-dimensional machine learning and blood transcriptomics. iScience (2020) 23(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ben Azzouz F, Michel B, Lasla H, Gouraud W, Francois AF, Girka F, et al. Development of an absolute assignment predictor for triple-negative breast cancer subtyping using machine learning approaches. Comput Biol Med (2021) 129. [DOI] [PubMed] [Google Scholar]
- 65. Alkhateeb A, Rezaeian I, Singireddy S, Cavallo-Medved D, Porter LA, Rueda L. Transcriptomics signature from next-generation sequencing data reveals new transcriptomic biomarkers related to prostate cancer. Cancer Inf (2019) 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Long NP, Park S, Anh NH, Nghi TD, Yoon SJ, Park JH, et al. High-throughput omics and statistical learning integration for the discovery and validation of novel diagnostic signatures in colorectal cancer. Int J Mol Sci (2019) 20(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Torang A, Gupta P, Klinke DJ. An elastic-net logistic regression approach to generate classifiers and gene signatures for types of immune cells and T helper cell subsets. BMC Bioinf (2019) 20(1):433. doi: 10.1186/s12859-019-2994-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Masuda T, Mori A, Ito S, Ohtsuki S. Quantitative and targeted proteomics-based identification and validation of drug efficacy biomarkers. Drug Metab Pharmacokinet (2021) 36:100361. doi: 10.1016/j.dmpk.2020.09.006 [DOI] [PubMed] [Google Scholar]
- 69. Khan MZI, Tam SY, Law HKW. Advances in high throughput proteomics profiling in establishing potential biomarkers for gastrointestinal cancer. CELLS (2022) 11(6):973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li N, Long Y, Fan X, Liu H, Li C, Chen L, et al. Proteomic analysis of differentially expressed proteins in hepatitis b virus-related hepatocellular carcinoma tissues. J Exp Clin Cancer Res (2009) 28(1):122. doi: 10.1186/1756-9966-28-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Murata M, Matsuzaki K, Yoshida K, Sekimoto G, Tahashi Y, Mori S, et al. Hepatitis b virus X protein shifts human hepatic transforming growth factor (TGF)-beta signaling from tumor suppression to oncogenesis in early chronic hepatitis b. Hepatology (2009) 49(4):1203–17. doi: 10.1002/hep.22765 [DOI] [PubMed] [Google Scholar]
- 72. Wu Z, Doondeea JB, Gholami AM, Janning MC, Lemeer S, Kramer K, et al. Quantitative chemical proteomics reveals new potential drug targets in head and neck cancer. Mol Cell Proteomics (2011) 10(12):M111.011635. doi: 10.1074/mcp.M111.011635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nambu M, Masuda T, Ito S, Kato K, Kojima T, Daiko H, et al. Leucine-rich alpha-2-Glycoprotein 1 in serum is a possible biomarker to predict response to preoperative chemoradiotherapy for esophageal cancer. Biol Pharm Bull (2019) 42(10):1766–71. doi: 10.1248/bpb.b19-00395 [DOI] [PubMed] [Google Scholar]
- 74. Huang X, Zeng Y, Xing X, Zeng J, Gao Y, Cai Z, et al. Quantitative proteomics analysis of early recurrence/metastasis of huge hepatocellular carcinoma following radical resection. Proteome Sci (2014) 12:22. doi: 10.1186/1477-5956-12-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gerdes H, Casado P, Dokal A, Hijazi M, Akhtar N, Osuntola R, et al. Drug ranking using machine learning systematically predicts the efficacy of anti-cancer drugs. Nat Commun (2021) 12(1). doi: 10.1038/s41467-021-22170-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Farinella F, Merone M, Bacco L, Capirchio A, Ciccozzi M, Caligiore D, et al. Machine learning analysis of high-grade serous ovarian cancer proteomic dataset reveals novel candidate biomarkers. Sci Rep (2022) 12(1):3041. doi: 10.1038/s41598-022-06788-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ntzioni E, Chouvarda I. Combining machine learning and network analysis pipelines: The case of microbiome and metabolomics data in colorectal cancer. Stud Health Technol Inf (2022) 289:489–90. doi: 10.3233/SHTI210965 [DOI] [PubMed] [Google Scholar]
- 78. Gupta A, Sagar G, Siddiqui Z, Rao KVS, Nayak S, Saquib N, et al. A non-invasive method for concurrent detection of early-stage women-specific cancers. Sci Rep (2022) 12(1):2301. doi: 10.1038/s41598-022-06274-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Prade VM, Sun N, Shen J, Feuchtinger A, Kunzke T, Buck A, et al. The synergism of spatial metabolomics and morphometry improves machine learning-based renal tumour subtype classification. Clin Trans Med (2022) 12(2). doi: 10.1002/ctm2.666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shen J, Sun N, Zens P, Kunzke T, Buck A, Prade VM, et al. Spatial metabolomics for evaluating response to neoadjuvant therapy in non-small cell lung cancer patients. Cancer Commun (London England) (2022) 42(6):517–35. doi: 10.1002/cac2.12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Liang T-L, Li R-Z, Mai C-T, Guan X-X, Li J-X, Wang X-R, et al. A method establishment and comparison of in vivo lung cancer model development platforms for evaluation of tumour metabolism and pharmaceutical efficacy. Phytomedicine (2022) 96:153831. doi: 10.1016/j.phymed.2021.153831 [DOI] [PubMed] [Google Scholar]
- 82. Miller HA, Yin X, Smith SA, Hu X, Zhang X, Yan J, et al. Evaluation of disease staging and chemotherapeutic response in non-small cell lung cancer from patient tumor-derived metabolomic data. Lung Cancer (2021) 156:20–30. doi: 10.1016/j.lungcan.2021.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Evangelista EB, Kwee SA, Sato MM, Wang L, Rettenmeier C, Xie G, et al. Phospholipids are a potentially important source of tissue biomarkers for hepatocellular carcinoma: Results of a pilot study involving targeted metabolomics. Diagnostics (2019) 9(4):167. doi: 10.3390/diagnostics9040167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Xie Y, Meng W-Y, Li R-Z, Wang Y-W, Qian X, Chan C, et al. Early lung cancer diagnostic biomarker discovery by machine learning methods. Trans Oncol (2021) 14(1):100907. doi: 10.1016/j.tranon.2020.100907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Dykstra MA, Switzer N, Eisner R, Tso V, Foshaug R, Ismond K, et al. Urine metabolomics as a predictor of patient tolerance and response to adjuvant chemotherapy in colorectal cancer. Mol Clin Oncol (2017) 7(5):767–70. doi: 10.3892/mco.2017.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bifarin OO, Gaul DA, Sah S, Arnold RS, Ogan K, Master VA, et al. Machine learning-enabled renal cell carcinoma status prediction using multiplatform urine-based metabolomics. J Proteome Res (2021) 20(7):3629–41. doi: 10.1021/acs.jproteome.1c00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Biswas N, Chakrabarti S. Artificial intelligence (AI)-based systems biology approaches in multi-omics data analysis of cancer. Front Oncol (2020) 10. doi: 10.3389/fonc.2020.588221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tong L, Wu H, Wang MD. Integrating multi-omics data by learning modality invariant representations for improved prediction of overall survival of cancer. Methods (2021) 189:74–85. doi: 10.1016/j.ymeth.2020.07.008 [DOI] [PubMed] [Google Scholar]
- 89. Nicora G, Vitali F, Dagliati A, Geifman N, Bellazzi R. Integrated multi-omics analyses in oncology: A review of machine learning methods and tools. Front Oncol (2020) 10. doi: 10.3389/fonc.2020.01030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Takahashi S, Asada K, Takasawa K, Shimoyama R, Sakai A, Bolatkan A, et al. Predicting deep learning based multi-omics parallel integration survival subtypes in lung cancer using reverse phase protein array data. Biomolecules (2020) 10(10):1460. doi: 10.3390/biom10101460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shuangshuang L, et al. (2020). A deep learning fusion clustering framework for breast cancer subtypes identification by integrating multi-omics data, in: 2020 5TH International Conference on Mechanical, Control and Computer Engineering (ICMCCE 2020), . pp. 1710–4. [Google Scholar]
- 92. Albaradei S, Napolitano F, Thafar MA, Gojobori T, Essack M, Gao X. MetaCancer: A deep learning-based pan-cancer metastasis prediction model developed using multi-omics data. Comput Struct Biotechnol J (2021) 19:4404–11. doi: 10.1016/j.csbj.2021.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lewis JE, Kemp ML. Integration of machine learning and genome-scale metabolic modeling identifies multi-omics biomarkers for radiation resistance. Nat Commun (2021) 12(1):2700. doi: 10.1038/s41467-021-22989-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wang Y, Yang YX, Chen SL, Wang JG. DeepDRK: a deep learning framework for drug repurposing through kernel-based multi-omics integration. Briefings Bioinf (2021) 22(5):bbab048. doi: 10.1093/bib/bbab048 [DOI] [PubMed] [Google Scholar]
- 95. Poirion OB, Jing Z, Chaudhary K, Huang SJ, Garmire LX. DeepProg: an ensemble of deep-learning and machine-learning models for prognosis prediction using multi-omics data. Genome Med (2021) 13(1). doi: 10.1186/s13073-021-00930-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Karim MR, Islam T, Lange C, Rebholz-Schuhmann D, Decker S. Adversary-aware multimodal neural networks for cancer susceptibility prediction from multiomics data. IEEE Access (2022) 10:54386–409. doi: 10.1109/ACCESS.2022.3175816 [DOI] [Google Scholar]
- 97. Zhang L, Lv CK, Jin YQ, Cheng GQ, Fu YB, Yuan DS, et al. Deep learning-based multi-omics data integration reveals two prognostic subtypes in high-risk neuroblastoma. Front Genet (2018) 9. doi: 10.3389/fgene.2018.00477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Xu JM, Xu BH, Li YP, Su ZJ, Yao YP. Unsupervised learning of cross-modal mappings in multi-omics data for survival stratification of gastric cancer. Future Oncol (2021) 18(2):215–30. [DOI] [PubMed] [Google Scholar]
- 99. Guo LY, Wu AH, Wang YX, Zhang LP, Chai H, Liang XF. Deep learning-based ovarian cancer subtypes identification using multi-omics data. Biodata Min (2020) 13(1):10. doi: 10.1186/s13040-020-00222-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lee T-Y, Huang K-Y, Chuang C-H, Lee C-Y, Chang T-H. Incorporating deep learning and multi-omics autoencoding for analysis of lung adenocarcinoma prognostication. Comput Biol Chem (2020) 87:107277. doi: 10.1016/j.compbiolchem.2020.107277 [DOI] [PubMed] [Google Scholar]
- 101. Zhu R, Dai LY, Liu JX, Guo Y. Diagnostic classification of lung cancer using deep transfer learning technology and multi-omics data. Chin J Electron (2021) 30(5):843–52. [Google Scholar]
- 102. Khan D, Shedole S. Leveraging deep learning techniques and integrated omics data for tailored treatment of breast cancer. J Personalized Med (2022) 12(5). doi: 10.3390/jpm12050674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Pan X, Burgman B, Wu E, Huang JH, Sahni N, Stephen Yi S, et al. I-modern: Integrated multi-omics network model identifies potential therapeutic targets in glioma by deep learning with interpretability. Comput Struct Biotechnol J (2022) 20:3511–21. doi: 10.1016/j.csbj.2022.06.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ma B, Meng F, Yan G, Yan H, Chai B, Song F. Diagnostic classification of cancers using extreme gradient boosting algorithm and multi-omics data. Comput Biol Med (2020) 121:103761. doi: 10.1016/j.compbiomed.2020.103761 [DOI] [PubMed] [Google Scholar]
- 105. Yang H, Chen R, Li D, Wang Z. Subtype-GAN: a deep learning approach for integrative cancer subtyping of multi-omics data. Bioinformatics (2021) 37(16):2231–7. doi: 10.1093/bioinformatics/btab109 [DOI] [PubMed] [Google Scholar]
- 106. Huang GJ, Wang C, Fu X. Bidirectional deep neural networks to integrate RNA and DNA data for predicting outcome for patients with hepatocellular carcinoma. Future Oncol (2021) 17(33):4481–95. doi: 10.2217/fon-2021-0659 [DOI] [PubMed] [Google Scholar]
- 107. ElKarami B, Alkhateeb A, Qattous H, Alshomali L, Shahrrava B. Multi-omics data integration model based on UMAP embedding and convolutional neural network. Cancer Inf (2022) 21:11769351221124205. doi: 10.1177/11769351221124205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wang TX, Shao W, Huang Z, Tang HX, Zhang J, Ding ZM, et al. MOGONET integrates multi-omics data using graph convolutional networks allowing patient classification and biomarker identification. Nat Commun (2021) 12(1):3445. doi: 10.1038/s41467-021-23774-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chai H, Zhou X, Zhang Z, Rao J, Zhao H, Yang Y, et al. Integrating multi-omics data through deep learning for accurate cancer prognosis prediction. Comput Biol Med (2021) 134:104481. doi: 10.1016/j.compbiomed.2021.104481 [DOI] [PubMed] [Google Scholar]
- 110. Xu J, Wu P, Chen YH, Meng QF, Dawood H, Dawood H. A hierarchical integration deep flexible neural forest framework for cancer subtype classification by integrating multi-omics data. BMC Bioinf (2019) 20(1):527. doi: 10.1186/s12859-019-3116-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Park C, Oh I, Choi J, Ko S, Ahn J. Improved prediction of cancer outcome using graph-embedded generative adversarial networks. IEEE Access (2021) 9:20076–88. doi: 10.1109/ACCESS.2021.3054894 [DOI] [Google Scholar]
- 112. Li F, Sun ZS, Liu JX, Shang JL, Dai LY, Liu XK. NESM: a network embedding method for tumor stratification by integrating multi-omics data. G3-Genes Genomes Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Rong Z, Liu Z, Song J, Cao L, Yu Y, Qiu M, et al. MCluster-VAEs: An end-to-end variational deep learning-based clustering method for subtype discovery using multi-omics data. Comput Biol Med (2022) 150:106085. doi: 10.1016/j.compbiomed.2022.106085 [DOI] [PubMed] [Google Scholar]
- 114. Ma TL, Zhang AD. Integrate multi-omics data with biological interaction networks using multi-view factorization AutoEncoder (MAE). BMC Genomics (2019) 20:944. doi: 10.1186/s12864-019-6285-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wang Z, Zhao Y, Shen X, Zhao Y, Zhang Z, Yin H, et al. Single-cell genomics-based molecular algorithm for early cancer detection. Analytical Chem (2022) 94(5):2607–14. doi: 10.1021/acs.analchem.1c04968 [DOI] [PubMed] [Google Scholar]
- 116. Trivizakis E, Souglakos J, Karantanas A, Marias K. Deep radiotranscriptomics of non-small cell lung carcinoma for assessing molecular and histology subtypes with a data-driven analysis. Diagnostics (2021) 11(12):2383. doi: 10.3390/diagnostics11122383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zhang X, Xing Y, Sun K, Guo Y. OmiEmbed: A unified multi-task deep learning framework for multi-omics data. Cancers (2021) 13(12):3047. doi: 10.3390/cancers13123047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wu Y, Cheng M, Huang S, Pei Z, Zuo Y, Liu J, et al. Recent advances of deep learning for computational histopathology: Principles and applications. Cancers (2022) 14(5):1199. doi: 10.3390/cancers14051199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Robertson S, Azizpour H, Smith K, Hartman J. Digital image analysis in breast pathology–from image processing techniques to artificial intelligence. Trans Res (2018) 194:19–35. doi: 10.1016/j.trsl.2017.10.010 [DOI] [PubMed] [Google Scholar]
- 120. Ho C, Zhao Z, Chen XF, Sauer J, Saraf SA, Jialdasani R, et al. A promising deep learning-assistive algorithm for histopathological screening of colorectal cancer. Sci Rep (2022) 12(1):2222. doi: 10.1038/s41598-022-06264-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Srinidhi CL, Ciga O, Martel AL. Deep neural network models for computational histopathology: A survey. Med Image Anal (2021) 67:101813. doi: 10.1016/j.media.2020.101813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Iizuka O, Kanavati F, Kato K, Rambeau M, Arihiro K, Tsuneki M. Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Sci Rep (2020) 10(1):1504. doi: 10.1038/s41598-020-58467-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kainz P, Pfeiffer M, Urschler M. Segmentation and classification of colon glands with deep convolutional neural networks and total variation regularization. PeerJ (2017) 5:e3874. doi: 10.7717/peerj.3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Lui TKL, Guo CG, Leung WK. Accuracy of artificial intelligence on histology prediction and detection of colorectal polyps: A systematic review and meta-analysis. Gastrointest Endosc (2020) 92(1):11–22.e6. doi: 10.1016/j.gie.2020.02.033 [DOI] [PubMed] [Google Scholar]
- 125. Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. Jama (2017) 318(22):2199–210. doi: 10.1001/jama.2017.14585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Dembrower K, Wåhlin E, Liu Y, Salim M, Smith K, Lindholm P, et al. Effect of artificial intelligence-based triaging of breast cancer screening mammograms on cancer detection and radiologist workload: a retrospective simulation study. Lancet Digit Health (2020) 2(9):e468–74. doi: 10.1016/S2589-7500(20)30185-0 [DOI] [PubMed] [Google Scholar]
- 127. Stenzinger A, Alber M, Allgäuer M, Jurmeister P, Bockmayr M, Budczies J, et al. Artificial intelligence and pathology: From principles to practice and future applications in histomorphology and molecular profiling. Semin Cancer Biol (2022) 84:129–43. doi: 10.1016/j.semcancer.2021.02.011 [DOI] [PubMed] [Google Scholar]
- 128. Su F, Li J, Zhao X, Wang B, Hu Y, Sun Y, et al. Interpretable tumor differentiation grade and microsatellite instability recognition in gastric cancer using deep learning. Lab Invest (2022) 102(6):641–9. doi: 10.1038/s41374-022-00742-6 [DOI] [PubMed] [Google Scholar]
- 129. Sengupta D, Ali SN, Bhattacharya A, Mustafi J, Mukhopadhyay A, Sengupta K. A deep hybrid learning pipeline for accurate diagnosis of ovarian cancer based on nuclear morphology. PloS One (2022) 17(1):e0261181. doi: 10.1371/journal.pone.0261181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Bulten W, Balkenhol M, Belinga JA, Brilhante A, Çakır A, Egevad L, et al. Artificial intelligence assistance significantly improves Gleason grading of prostate biopsies by pathologists. Mod Pathol (2021) 34(3):660–71. doi: 10.1038/s41379-020-0640-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med (2018) 24(10):1559–67. doi: 10.1038/s41591-018-0177-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Ye ZX, Zhang YX, Liang YB, Lang JD, Zhang XL, Zang GL, et al. Cervical cancer metastasis and recurrence risk prediction based on deep convolutional neural network. Curr Bioinf (2022) 17(2):164–73. doi: 10.2174/1574893616666210708143556 [DOI] [Google Scholar]
- 133. Kulkarni PM, Robinson EJ, Sarin Pradhan J, Gartrell-Corrado RD, Rohr BR, Trager MH, et al. Deep learning based on standard H&E images of primary melanoma tumors identifies patients at risk for visceral recurrence and death. Clin Cancer Res (2020) 26(5):1126–34. doi: 10.1158/1078-0432.CCR-19-1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Meier A, Nekolla K, Hewitt LC, Earle S, Yoshikawa T, Oshima T, et al. Hypothesis-free deep survival learning applied to the tumour microenvironment in gastric cancer. J Pathol Clin Res (2020) 6(4):273–82. doi: 10.1002/cjp2.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Peikari M, Gangeh MJ, Zubovits J, Clarke G, Martel AL. Triaging diagnostically relevant regions from pathology whole slides of breast cancer: A texture based approach. IEEE Trans Med Imaging (2016) 35(1):307–15. doi: 10.1109/TMI.2015.2470529 [DOI] [PubMed] [Google Scholar]
- 136. Mao B, Zhang L, Ning P, Ding F, Wu F, Lu G, et al. Preoperative prediction for pathological grade of hepatocellular carcinoma via machine learning-based radiomics. Eur Radiol (2020) 30(12):6924–32. doi: 10.1007/s00330-020-07056-5 [DOI] [PubMed] [Google Scholar]
- 137. Komura D, Ishikawa S. Machine learning approaches for pathologic diagnosis. Virchows Arch (2019) 475(2):131–8. doi: 10.1007/s00428-019-02594-w [DOI] [PubMed] [Google Scholar]
- 138. Fakoor R, Ladhak F, Nazi A, Huber M. Using deep learning to enhance cancer diagnosis and classification. In: Proc Int Conf Mach (2013) 28:3937–49. [Google Scholar]
- 139. Mobadersany P, Yousefi S, Amgad M, Gutman DA, Barnholtz-Sloan JS, Velázquez Vega JE, et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci United States America (2018) 115(13):E2970–9. doi: 10.1073/pnas.1717139115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Al-Antari MA, Han SM, Kim TS. Evaluation of deep learning detection and classification towards computer-aided diagnosis of breast lesions in digital X-ray mammograms. Comput Methods Programs BioMed (2020) 196:105584. doi: 10.1016/j.cmpb.2020.105584 [DOI] [PubMed] [Google Scholar]
- 141. Hussein S, Kandel P, Bolan CW, Wallace MB, Bagci U. Lung and pancreatic tumor characterization in the deep learning era: Novel supervised and unsupervised learning approaches. IEEE Trans Med Imaging (2019) 38(8):1777–87. doi: 10.1109/TMI.2019.2894349 [DOI] [PubMed] [Google Scholar]
- 142. Bertelli E, Mercatelli L, Marzi C, Pachetti E, Baccini M, Barucci A, et al. Machine and deep learning prediction of prostate cancer aggressiveness using multiparametric MRI. Front Oncol (2021) 11:802964. doi: 10.3389/fonc.2021.802964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Ayalew YA, Fante KA, Mohammed MA. Modified U-net for liver cancer segmentation from computed tomography images with a new class balancing method. BMC Biomed Eng (2021) 3(1):4–4. doi: 10.1186/s42490-021-00050-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J Clin (2019) 69(2):127–57. doi: 10.3322/caac.21552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Dhahri H, Al Maghayreh E, Mahmood A, Elkilani W, Faisal Nagi M. Automated breast cancer diagnosis based on machine learning algorithms. J Healthcare Eng (2019) 2019:4253641. doi: 10.1155/2019/4253641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Liu RY, Pan DR, Xu Y, Zeng H, He ZL, Lin JB, et al. A deep learning-machine learning fusion approach for the classification of benign, malignant, and intermediate bone tumors. Eur Radiol (2022) 32(2):1371–83. doi: 10.1007/s00330-021-08195-z [DOI] [PubMed] [Google Scholar]
- 147. Alanazi MF, Ali MU, Hussain SJ, Zafar A, Mohatram M, Irfan M, et al. Brain Tumor/Mass classification framework using magnetic-Resonance-Imaging-Based isolated and developed transfer deep-learning model. Sensors (2022) 22(1):372. doi: 10.3390/s22010372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Zahoor MM, Qureshi SA, Bibi S, Khan SH, Khan A, Ghafoor U, et al. A new deep hybrid boosted and ensemble learning-based brain tumor analysis using MRI. Sensors (2022) 22(7):2726. doi: 10.3390/s22072726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Chougrad H, Zouaki H, Alheyane O. Deep convolutional neural networks for breast cancer screening. Comput Methods Programs BioMed (2018) 157:19–30. doi: 10.1016/j.cmpb.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 150. Misra S, Jeon S, Managuli R, Lee S, Kim G, Yoon C, et al. Bi-modal transfer learning for classifying breast cancers via combined b-mode and ultrasound strain imaging. IEEE Trans Ultrasonics Ferroelectrics Frequency Control (2022) 69(1):222–32. doi: 10.1109/TUFFC.2021.3119251 [DOI] [PubMed] [Google Scholar]
- 151. Brunese L, Mercaldo F, Reginelli A, Santone A. Radiomics for Gleason score detection through deep learning. Sensors (Basel Switzerland) (2020) 20(18):5411. doi: 10.3390/s20185411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Zhao Z, Bian Y, Jiang H, Fang X, Li J, Cao K, et al. CT-radiomic approach to predict G1/2 nonfunctional pancreatic neuroendocrine tumor. Acad Radiol (2020) 27(12):e272–81. doi: 10.1016/j.acra.2020.01.002 [DOI] [PubMed] [Google Scholar]
- 153. Gurgitano M, Angileri SA, Roda GM, Liguori A, Pandolfi M, Lerardi AM, et al. Interventional radiology ex-machina: impact of artificial intelligence on practice. Radiologia Med (2021) 126(7):998–1006. doi: 10.1007/s11547-021-01351-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Xie T, Wang X, Zhang Z, Zhou Z. CT-based radiomics analysis for preoperative diagnosis of pancreatic mucinous cystic neoplasm and atypical serous cystadenomas. Front Oncol (2021) 11:621520. doi: 10.3389/fonc.2021.621520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Song D, Yang F, Zhang Y, Guo Y, Qu Y, Zhang X, et al. Dynamic contrast-enhanced MRI radiomics nomogram for predicting axillary lymph node metastasis in breast cancer. Cancer Imaging (2022) 22(1):17. doi: 10.1186/s40644-022-00450-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Eresen A, Li Y, Yang J, Shangguan J, Velichko Y, Yaghmai V, et al. Preoperative assessment of lymph node metastasis in colon cancer patients using machine learning: a pilot study. Cancer Imaging (2020) 20(1):30. doi: 10.1186/s40644-020-00308-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Guo R, Guo J, Zhang L, Qu X, Dai S, Peng R, et al. CT-based radiomics features in the prediction of thyroid cartilage invasion from laryngeal and hypopharyngeal squamous cell carcinoma. Cancer Imaging (2020) 20(1):81. doi: 10.1186/s40644-020-00359-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Akbari H, Macyszyn L, Da X, Bilello M, Wolf RL, Martinez-Lage M, et al. Imaging surrogates of infiltration obtained via multiparametric imaging pattern analysis predict subsequent location of recurrence of glioblastoma. Neurosurgery (2016) 78(4):572–80. doi: 10.1227/NEU.0000000000001202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Fung NTC, Hung WM, Sze CK, Lee MCH, Ng WT. Automatic segmentation for adaptive planning in nasopharyngeal carcinoma IMRT: Time, geometrical, and dosimetric analysis. Med Dosim (2020) 45(1):60–5. doi: 10.1016/j.meddos.2019.06.002 [DOI] [PubMed] [Google Scholar]
- 160. Lustberg T, van Soest J, Gooding M, Peressutti D, Aljabar P, van der Stoep J, et al. Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol (2018) 126(2):312–7. doi: 10.1016/j.radonc.2017.11.012 [DOI] [PubMed] [Google Scholar]
- 161. Elguindi S, Zelefsky MJ, Jiang J, Veeraraghavan H, Deasy JO, Hunt MA, et al. Deep learning-based auto-segmentation of targets and organs-at-risk for magnetic resonance imaging only planning of prostate radiotherapy. Phys Imaging Radiat Oncol (2019) 12:80–6. doi: 10.1016/j.phro.2019.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Schouten JPE, Noteboom S, Martens RM, Mes SW, Leemans CR, de Graaf P, et al. Automatic segmentation of head and neck primary tumors on MRI using a multi-view CNN. Cancer Imaging (2022) 22(1):8. doi: 10.1186/s40644-022-00445-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Lin L, Dou Q, Jin YM, Zhou GQ, Tang YQ, Chen WL, et al. Deep learning for automated contouring of primary tumor volumes by MRI for nasopharyngeal carcinoma. Radiology (2019) 291(3):677–86. doi: 10.1148/radiol.2019182012 [DOI] [PubMed] [Google Scholar]
- 164. Zhang J, Cheng Z, Fan Z, Zhang Q, Zhang X, Yang R, et al. A feasibility study for in vivo treatment verification of IMRT using Monte Carlo dose calculation and deep learning-based modelling of EPID detector response. Radiat Oncol (2022) 17(1):31. doi: 10.1186/s13014-022-01999-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Auloge P, Cazzato RL, Ramamurthy N, de Marini P, Rousseau C, Garnon J, et al. Augmented reality and artificial intelligence-based navigation during percutaneous vertebroplasty: a pilot randomised clinical trial. Eur Spine J (2020) 29(7):1580–9. doi: 10.1007/s00586-019-06054-6 [DOI] [PubMed] [Google Scholar]
- 166. Tomaszewski MR, Latifi K, Boyer E, Palm RF, El Naqa I, Moros EG, et al. Delta radiomics analysis of magnetic resonance guided radiotherapy imaging data can enable treatment response prediction in pancreatic cancer. Radiat Oncol (2021) 16(1):237. doi: 10.1186/s13014-021-01957-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Bousabarah K, Blanck O, Temming S, Wilhelm M-L, Hoevels M, Baus WW, et al. Radiomics for prediction of radiation-induced lung injury and oncologic outcome after robotic stereotactic body radiotherapy of lung cancer: results from two independent institutions. Radiat Oncol (2021) 16(1):74. doi: 10.1186/s13014-021-01805-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Zhang DH, Duan YC, Guo J, Wang YW, Yang Y, Li ZH, et al. Using multi-scale convolutional neural network based on multi-instance learning to predict the efficacy of neoadjuvant chemoradiotherapy for rectal cancer. IEEE J Trans Eng Health Med (2022) 10. doi: 10.1109/JTEHM.2022.3156851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Ermiş E, Jungo A, Poel R, Blatti-Moreno M, Meier R, Knecht U, et al. Fully automated brain resection cavity delineation for radiation target volume definition in glioblastoma patients using deep learning. Radiat Oncol (2020) 15(1):100. doi: 10.1186/s13014-020-01553-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Zhou J, Peng Z, Song Y, Chang Y, Pei X, Sheng L, et al. A method of using deep learning to predict three-dimensional dose distributions for intensity-modulated radiotherapy of rectal cancer. J Appl Clin Med Phys (2020) 21(5):26–37. doi: 10.1002/acm2.12849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Peng L, Parekh V, Huang P, Lin DD, Sheikh K, Baker B, et al. Distinguishing true progression from radionecrosis after stereotactic radiation therapy for brain metastases with machine learning and radiomics. Int J Radiat Oncol Biol Phys (2018) 102(4):1236–43. doi: 10.1016/j.ijrobp.2018.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Zhang Z, Yang J, Ho A, Jiang W, Logan J, Wang X, et al. A predictive model for distinguishing radiation necrosis from tumour progression after gamma knife radiosurgery based on radiomic features from MR images. Eur Radiol (2018) 28(6):2255–63. doi: 10.1007/s00330-017-5154-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Shaver MM, et al. Optimizing neuro-oncology imaging: A review of deep learning approaches for glioma imaging. Cancers (2019) 11(6):829. doi: 10.3390/cancers11060829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Wang K, Qiao Z, Zhao X, Li X, Wang X, Wu T, et al. Individualized discrimination of tumor recurrence from radiation necrosis in glioma patients using an integrated radiomics-based model. Eur J Nucl Med Mol Imaging (2020) 47(6):1400–11. doi: 10.1007/s00259-019-04604-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Cha KH, Hadjiiski L, Chan HP, Weizer AZ, Alva A, Cohan RH, et al. Bladder cancer treatment response assessment in CT using radiomics with deep-learning. Sci Rep (2017) 7(1):8738. doi: 10.1038/s41598-017-09315-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Gong J, Bao X, Wang T, Liu JY, Peng WJ, Shi JY, et al. A short-term follow-up CT based radiomics approach to predict response to immunotherapy in advanced non-small-cell lung cancer. Oncoimmunology (2022) 11(1):2028962. doi: 10.1080/2162402X.2022.2028962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Bhardwaj D, Dasgupta A, DiCenzo D, Brade S, Fatima K, Quiaoit K, et al. Early changes in quantitative ultrasound imaging parameters during neoadjuvant chemotherapy to predict recurrence in patients with locally advanced breast cancer. Cancers (2022) 14(5):1247. doi: 10.3390/cancers14051247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Liu G, Poon M, Zapala MA, Temple WC, Vo KT, Matthay KK, et al. Incorporating radiomics into machine learning models to predict outcomes of neuroblastoma. J Digital Imaging (2022) 35(3):605–12. doi: 10.1007/s10278-022-00607-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Cheng SH, Cheng YJ, Jin ZY, Xue HD. Unresectable pancreatic ductal adenocarcinoma: Role of CT quantitative imaging biomarkers for predicting outcomes of patients treated with chemotherapy. Eur J Radiol (2019) 113:188–97. doi: 10.1016/j.ejrad.2019.02.009 [DOI] [PubMed] [Google Scholar]
- 180. Parr E, Du Q, Zhang C, Lin C, Kamal A, McAlister J, et al. Radiomics-based outcome prediction for pancreatic cancer following stereotactic body radiotherapy. Cancers (Basel) (2020) 12(4):1051. doi: 10.3390/cancers12041051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Schilling KG, Landman BA. AI In MRI: A case for grassroots deep learning. Magn Reson Imaging (2019) 64:1–3. doi: 10.1016/j.mri.2019.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Li BJ, Wang TY, Nabavi S, Assoc Comp M. (2021). Cancer molecular subtype classification by graph convolutional networks on multi-omics data, in: 12TH ACM Conference on Bioinformatics, Computational Biology, and Health Informatics (ACM-BCB 2021), . [Google Scholar]
- 183. Wu WT, Li YJ, Feng AZ, Li L, Huang T, Xu AD, et al. Data mining in clinical big data: the frequently used databases, steps, and methodological models. Mil Med Res (2021) 8(1):44. doi: 10.1186/s40779-021-00338-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Yu KH, Berry GJ, Rubin DL, Ré C, Altman RB, Snyder M. Association of omics features with histopathology patterns in lung adenocarcinoma. Cell Syst (2017) 5(6):620–627.e3. doi: 10.1016/j.cels.2017.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. James TA, Fan B. ASO author reflections: Using tumor genomics to predict axillary response to chemotherapy in breast cancer. Ann Surg Oncol (2021) 28(3):1326–7. doi: 10.1245/s10434-020-09455-w [DOI] [PubMed] [Google Scholar]
- 186. Hatzikirou H. Combining dynamic modeling with machine learning can be the key for the integration of mathematical and clinical oncology: Comment on “Improving cancer treatments via dynamical biophysical models” by m. kuznetsov, j. clairambault, v. volpert. Phys Life Rev (2022) 40:1–2. doi: 10.1016/j.plrev.2022.01.002 [DOI] [PubMed] [Google Scholar]
- 187. Kuznetsov M, Clairambault J, Volpert V. Improving cancer treatments via dynamical biophysical models. Phys Life Rev (2021) 39:1–48. doi: 10.1016/j.plrev.2021.10.001 [DOI] [PubMed] [Google Scholar]
- 188. Chen RJ, Lu MY, Wang J, Williamson DFK, Rodig SJ, Lindeman NI, et al. Pathomic fusion: An integrated framework for fusing histopathology and genomic features for cancer diagnosis and prognosis. IEEE Trans Med Imaging (2022) 41(4):757–70. doi: 10.1109/TMI.2020.3021387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Sun D, Wang M, Li A. A multimodal deep neural network for human breast cancer prognosis prediction by integrating multi-dimensional data. IEEE/ACM Trans Comput Biol Bioinform (2019) 16(3):841–50. doi: 10.1109/TCBB.2018.2806438 [DOI] [PubMed] [Google Scholar]
- 190. Huang Z, Zhan X, Xiang S, Johnson TS, Helm B, Yu CY, et al. SALMON: Survival analysis learning with multi-omics neural networks on breast cancer. Front Genet (2019) 10:166. doi: 10.3389/fgene.2019.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Khan AN, Ihalage AA, Ma Y, Liu B, Liu Y, Hao Y. Deep learning framework for subject-independent emotion detection using wireless signals. PloS One (2021) 16(2):e0242946. doi: 10.1371/journal.pone.0242946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Davatzikos C. Machine learning in neuroimaging: Progress and challenges. Neuroimage (2019) 197:652–6. doi: 10.1016/j.neuroimage.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Hu Y, Zhao L, Li Z, Dong X, Xu T, Zhao Y. Classifying the multi-omics data of gastric cancer using a deep feature selection method. Expert Syst Appl (2022) 200:116813. doi: 10.1016/j.eswa.2022.116813 [DOI] [Google Scholar]
- 194. Yoon HJ, Stanley C, Christian JB, Klasky HB, Blanchard AE, Durbin EB, et al. Optimal vocabulary selection approaches for privacy-preserving deep NLP model training for information extraction and cancer epidemiology. Cancer Biomarkers (2022) 33(2):185–98. doi: 10.3233/CBM-210306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Leng DJ, Zheng LY, Wen YQ, Zhang YH, Wu LL, Wang J, et al. A benchmark study of deep learning-based multi-omics data fusion methods for cancer. Genome Biol (2022) 23(1):171. doi: 10.1186/s13059-022-02739-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Huang LM, Yang WJ, Huang ZY, Tang CW, Li J. Artificial intelligence technique in detection of early esophageal cancer. World J Gastroenterol (2020) 26(39):5959–69. doi: 10.3748/wjg.v26.i39.5959 [DOI] [PMC free article] [PubMed] [Google Scholar]