Abstract
Background:
Behavior and health are inextricably linked. As a result, continuous wearable sensor data offer the potential to predict clinical measures. However, interruptions in the data collection occur, which create a need for strategic data imputation.
Objective:
The objective of this work is to adapt a data generation algorithm to impute multivariate time series data. This will allow us to create digital behavior markers that can predict clinical health measures.
Methods:
We created a bidirectional time series generative adversarial network to impute missing sensor readings. Values are imputed based on relationships between multiple fields and multiple points in time, for single time points or larger time gaps. From the complete data, digital behavior markers are extracted and are mapped to predicted clinical measures.
Results:
We validate our approach using continuous smartwatch data for n=14 participants. When reconstructing omitted data, we observe an average normalized MAE of 0.0197. We then create machine learning models to predict clinical measures from the reconstructed, complete data with correlations ranging from r=0.1230 to r=0.7623. This work indicates that wearable sensor data collected in the wild can be used to offer insights on a person’s health in natural settings.
Keywords: Time series, data imputation, activity learning, health assessment
1. Background
Assessing and promoting health are challenging tasks even when physicians are readily available because health care providers must make decisions based on a typical 20 minute visit with a patient1, aided by results from often-inconclusive laboratory tests. The ability to provide accurate assessments is particularly timely because as the population ages, older adults will likely outnumber children for the first time in US history2, creating a discrepancy between the number of persons needing care and those capable of providing it. Resulting from this changing dynamic, chronic illness rates and healthcare expenditures are increasing3,4. One health domain that is particularly impacted by the aging population is cognitive health. Early detection of cognitive health changes has been identified as a national priority5,6 because this supports more effective treatment and significantly improves the quality of care while reducing health care costs7,8. However, clinic-based assessment is infeasible for many who live in remote areas or remain in their homes due to imposed restrictions. Furthermore, controlling the symptoms of cognitive decline relies on understanding its many influences, including physiology, psychosocial and physical environments, and routine behavior9.
The tight interplay between health and behavior is well documented in the literature10-12. The maturing design of sensor platforms, pervasive computing, and machine learning techniques offer practical, though not fully realized, methods for understanding the relationship between health and behavior and automatically assessing and predicting health status. We hypothesize that a person’s health can be predicted based on digital behavior markers that are collected from continuous, longitudinal wearable sensor data. Specifically, machine learning methods can be used to map a comprehensive set of digital behavior markers onto predicted values for clinical assessment measures13.
Because we can now collect data on ourselves in an ecologically valid manner, we will harness continuously-collected sensor data to create a personalized behavior profile. Despite recent technology advances, most research does not collect continuous data in realistic settings. Laboratory-driven data collections do not reflect natural behavior; behavior markers should be built based on activities sensed “in the wild”14,15.
One practical issue that limits the ability to create an automated behavior profile from wearable sensor data and assess a person’s health is gaps in the data collection. When data are collected in the wild, without imposed controls that ensure collection compliance and data quality, missing data is a common occurrence. Sensor readings will go missing when there are failures in the sensors, device, communication, or storage mechanisms. In our experiments, we collect data from older adult volunteers in their own homes as they perform normal routines. As a result, there are also frequent large gaps in the data collection (i.e., an hour or more) when the participants fail to wear or charge the devices. While there are common reasons for such missing data, in our work we do not incorporate such domain-specific information into the approach.
2. Objective
In this paper, we describe a generative approach to imputing values for multivariate time series data. Data imputation is a well-established problem with numerous available strategies. What makes the imputation problem particularly unique and challenging for smartwatch-based behavior data is that time series data are not i.i.d. and smartwatch data are multivariate, two aspects that are under-represented in the literature. To address this problem, we consider a generative time series model that preserves temporal dynamics together with inter-feature dynamics. The contributions of this paper are the following. First, we discuss adaptation of generative models to impute multivariate time series data. Second, we illustrate the application of this imputation method to collected smartwatch sensor data. Third, we describe how activity labels are applied to the complete time series and used to create digital behavior markers. Fourth, we define a joint inference method to predict clinical measures from the behavior profile. To validate the methods, we compare the imputation accuracy of our imputation algorithm, called Mink (Missing data Imputation Novel Kit), with baseline methods on sampled smartwatch data. Finally, we evaluate the accuracy of our health prediction methodology when missing data are imputed using Mink.
3. Problem Formulation
Consider the setting where multiple sensors are sampled at a constant rate (in our experiments, this rate is 10Hz). We start by formalizing the sensor data time series and the sensor data imputation task.
Definition 1. A time series data stream is an infinite sequence of elements X = {x1, x2, … xi,…}. The ith element of the series is xi. In the case of a multivariate time series, xi is a d-dimensional vector observed at time stamp i16.
Definition 2. We assume that the sensor data collection is a stationary time series. A stationary time series is a process whose statistical properties are constant over time. Thus:
The mean value function is μt = E(xt) and does not depend on time t.
The auto covariance function γ(s, t) = cov(xs,xt) = E[(xs − μs) (xt − μt)] depends on time stamps s and t only through their time difference, ∣s − t∣.
Definition 3. Missing values in a finite-length subset of a time series X1:T = {x1, .., xT} are represented by a mask matrix M. Each element of is defined for time stamp i and feature dimension j as:
| (1) |
Definition 4. An imputation algorithm IA has access to an incomplete dataset consisting of a time series X and mask matrix M. The goal of the algorithm is to replace all values where with a non-NULL value X and a Gaussian noise vector Z. The resulting time series is denoted as and is defined in Equation 2.
| (2) |
Definition 5. Algorithm IA should minimize total normalized mean absolute error (NMAE). NMAE is based on the standard mean absolute error (MAE) definition:
| (3) |
where represents an imputed value (if ) or the observed value (if ) and represents the actual ground truth value.
The NMAE metric is useful when comparing or combining the mean absolute error of features with different scales. Each MAE term is normalized to [0..1] based on the range of values for the corresponding feature. We evaluate the imputation performance of Mink and baseline methods using NMAE.
4. Related Work
4.1. Related Methods
Imputation of missing values is a well-established area of investigation. Researchers have proposed numerous methods to tackle this problem, including replace by constant and value inference using regression. Multiple methods can also be employed and combined, resulting in multiple imputation17,18. Most of these methods assume that data are independent and identically distributed (i.i.d.). They impute values for each feature separately and frequently do not account for the relationships between features. In the case of time series sensor-based behavior monitoring, the data are not i.i.d. Relationships between variables provide important context for imputing values, values need to be imputed simultaneously for multiple features, and the relationships between values at adjacent points in time need to be considered.
In recent years, researchers have started to investigate the problem of imputation for time series. In addition to the methods mentioned above, other common statistical methods carry forward an observation by copying the value from time t−1 to t, carry backward an observation from t+1 to t, or average the two. These approaches face limitations of the underlying processes being highly dynamic or the existence of a longer sequence of missing values19. Linear or nonlinear regression and forecasting models have been adapted for time series by mapping prior observations t−x .. t−1 (the lag) onto a predicted value for missing time t20,21. Specialized deep network structures are popular as well. Recurrent neural networks are well suited for this task because they retain sequential information in their structure22, although they are typically limited to univariate cases. Researchers have refined this process to combine deep learning with transfer learning for sensor data imputation23, ensuring that the imputed information is customized for each person.
The methods that are most similar to Mink utilize generative adversarial networks (GANs). In this scenario, one agent attempts to impute missing values (the generator) while a second agent attempts to differentiate observed from imputed values (the discriminator). GANs are becoming a standard for imputing i.i.d. data, including multiple imputation24. Yoon et al.25 introduced a GAN data imputer, called GAIN, that boosts performance by supplying a hint vector conditioned on observed values. This approach is effective for i.i.d. data but is not designed to handle the dynamics of time series data. For time series, such generative methods are valuable when large gaps exist in the sequence, because these algorithms will generate long sequences of values. The E2GAN imputer from Luo et al.26 represents recent work to design a GAN structure for time series. This algorithm combines autoencoder-based compression with a recurrent cell to generate time series data. This approach relies on an unsupervised adversarial loss that ensures the discriminator becomes more adept at recognizing imputed data at a rate that parallels the generator’s improved skills at generating imputed values. Another GAN strategy was proposed by Guo et al.27. MTS-GAN incorporates a multichannel convolutional neural network to extract features of each univariate time series, then adds a fully connected network to learn relationships between feature dimensions. In their multivariate time series imputer, MTS-GAN, In this paper, we combine unsupervised approaches found in the earlier methods with a supervised learning component that uses the observed data as an external oracle. This process then utilizes available observations to model the stepwise conditional distributions, resulting in realistic imputed values.
4.2. Applications of Time Series Imputation
To characterize a person’s overall behavior routine, we extract digital behavior markers from sensor data that are automatically labeled with corresponding activity categories. Performing human activity recognition from wearable sensors has become a popular topic for researchers to investigate28,29. Because we can collect continuous data without requiring extra steps for the subject, wearable sensors are a natural choice for assessing health based on sensed behavior. Approaches to activity recognition have considered numerous methods, including decision trees, nearest neighbors, clusters, and ensembles14,30,31, as well as deep networks32-35. Limitations of many of these existing methods are that they focus on basic, repetitive movement types and are often evaluated in laboratory settings. We are interested in extracting markers that reflect a person’s entire behavioral routine, sensed in natural settings. For our experiments in this paper, we pre-trained activity models using techniques that we validated in prior studies36.
Wearable sensor data offer substantial insights into a person’s behavior as well as their health. Typically, prior works analyze a specific behavior such as activity level37,38 or sleep39, with markers that consist of a small set of variables such as step counts or sleep duration40. Some researchers targeted specific sensor-observed behavior markers as a mechanism for assessing the relationship between lifestyle and health. Specifically, Dhana et al.41 quantify healthy behavior as a combination of nonsmoking, physical activity, alcohol consumption, nutrition, and cognitive activities. Individuals who scored higher on this behavior metric had a lower risk of Alzheimer’s dementia. Other researchers have also found that sensor-based behavior patterns are predictive of cognitive health42,43. Li et al.43 found that physical activity was predictive of Alzheimer’s disease, while Aramendi et al.42 predicted cognitive measures of cognitive health and mobility from activity-labeled sensor data.
These studies provide evidence that wearable sensors afford the ability to monitor intervention impact and assess a person’s cognitive health. Within this area of investigation, our proposed approach is unique because we investigate a computational method to monitor and model all a person’s behavior to predict clinical health measures. We utilize the complete set of behavior markers based on both observed and imputed sensor readings to predict multiple measures, then take advantage of the predictive relationship between diverse markers to improve predictive performance. This holistic approach to sensor analysis of behavior and health relies on a method to impute missing values in complex, multivariate time series data.
5. Methods
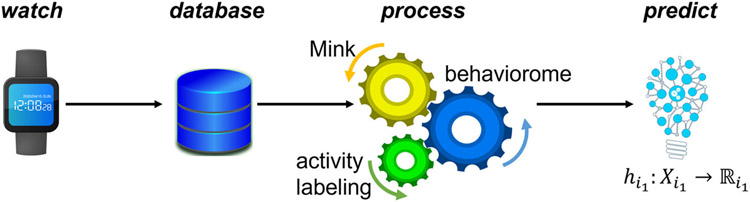
Figure 1 illustrates the steps of our automated sensor-based health assessment process. As the figure shows, the process relies on the ability to accurately impute missing data.
Figure 1.
The process of assessing health from smartwatch data. Data are continuously collected while a participant wears a smartwatch and performs their normal routine. Data are securely stored in a relational database and a processed by imputing missing values (Mink), labeling readings with associated activity labels (activity recognition), and extracting a set of digital behavior markers. A machine learning then maps the behavior profile onto predicted clinical measures.
5.1. Generative Time Series Data Imputation
Our approach to imputing multivariate time series data combines aspects of regression-based sequence prediction, adversarial sequence generation, and time series models. Adapting a definition by Yoon et al.44 for use with multivariate time series data imputation, the goal of Mink is to use training data D to learn a density that best approximates the density of ground-truth data, p(X1:T). The adversarial component of the imputation algorithm attempts to minimize the Jensen-Shannon divergence45 between the estimated and ground-truth densities, shown in Equation 7.
| (7) |
As Yoon et al.44 and Kachuee et al.46 suggest, such adversarial components can be boosted by partnering them with a supervised learning component that learns the temporal relationship between neighboring readings in the sequence. The objective of this component is to minimize the Kullback-Leibler divergence47 between the estimated and true relationship between readings at times t−1 and t, as shown in Equation 8.
| (8) |
Both JS divergence and KL divergence calculate scores that reflect the difference between probability distributions p and . They are both appropriate metrics for this task because they quantify the distance between two data samples based on the corresponding probability distributions. JS divergence is an extension of the KL measure that is symmetric, a property that is needed when comparing estimated and ground-truth data.
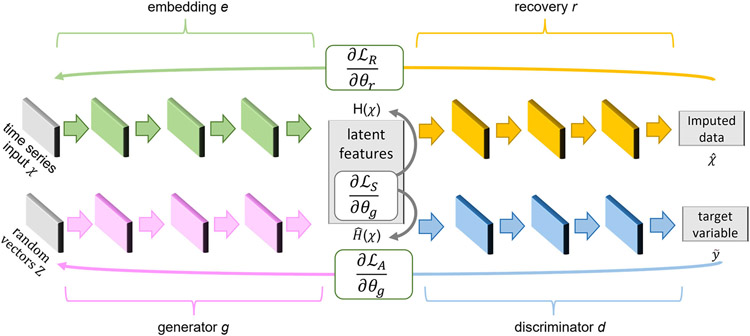
Figure 2 illustrates the architecture of the Mink time series imputation algorithm. As the figure shows, the architecture includes a time series generator g, a time series discriminator d, an embedding function e, and a recovery function r. The method combines a regressive autoencoder (the embedding and recovery elements) with a generative adversarial network (the generator and discriminator components) to optimize the multivariate goals formalized in Equations 7 and 8. The autoencoder components, e and r, are trained together with the generative adversarial network components, g and d, to yield realistic time series values that maintain global properties of the data distribution and temporal relationships between individual readings.
Figure 2.
The Mink time series data imputation architecture. The system processes time series X containing a mixture of observed and missing values and outputs a complete time series with no missing values. To generate realistic data, Mink combines an autoencoder (with embedding function e and recovery function r) and a generative adversarial network (with generator g and discriminator d).
5.1.1. Mink Generative Adversarial Network
Mink employs a generative adversarial network (GAN) to learn realistic time series sequences whose densities emulate those of the real data, as shown in Equation 7. Using the notation from Goodfellow et al.48, a traditional GAN optimizes the value function V(g,d) for generator g and discriminator d, as summarized in Equation 9. In this original formulation, x~pdata draws a sample from the real data distribution and z~pz(z) draws a sample from input Gaussian noise. As the equation expresses, the generator attempts to generate realistic data, the discriminator differentiates real from synthetic data, and the two strengthen each other as they learn.
| (9) |
In the imputation algorithm, the generator creates data in a latent space rather than directly generating time series data. The latent space is defined by the autoencoder, described in the next section. Let ZX represent the vector space from which individual random vectors are sampled. Generator g uses these to create latent vectors in HX. Thus, the generator can be represented as a function g:∏t ZX → ∏t HX. The Mink generator is designed as a stacked recurrent neural network (RNN). All Mink networks utilize a hidden layer of size 24 and a dense final layer that employs a sigmoid activation function. Thus, g generates a synthetic latent vector for time t based on a synthetic latent vector at time t−1, or . The goal of discriminator d is to correctly classify the latent vectors as real data, y, or synthetic data, . Function d is designed as a bidirectional recurrent network with a feedforward output layer.1
The GAN is trained to optimize Equation 9. As a result, the network maximizes the log probability of d correctly discriminating between real and fake samples while at the same time minimizing the log probability of 1−d(g(z)), where d(g(z)) represents the probability that generated data g(z) is real. For our imputation algorithm, we adapt Equation 9 to create an adversarial loss function that trains the GAN. The loss function is shown in Equation 10.
| (10) |
To link these components, the imputation architecture utilizes a third loss function that alternately guides training for the autoencoder and the GAN. This loss function computes a gradient based on the difference between the predicted latent vector at the next time step (the synthetic vector) and the ground truth-derived latent vector at the next time step. This loss function thus reflects the distance between p(Ht∣H1:t−1) and . This stepwise loss is computed as shown in Equation 11.
| (11) |
5.1.2. Autoencoder
To map sample data onto latent features HX, Mink incorporates an autoencoder using an embedding e. A recovery function r then reconstructs data close to the original. To create time series data, the autoencoder captures the temporal relationships between readings, represented as ht = eX (ht−1, xt). The recovery function r is a recurrent network that maps the latent vector back onto the original time series representation, . Mink employs a stacked RNN for both networks where the output for time t only depends on information available at time t−1.
The goal of the architecture’s autoencoder component is to accurately reconstruct the input data from the latent vectors. This component is thus trained using a reconstruction loss that computes the element-wise difference between the original and reconstructed feature values, as shown in Equation 12.
| (12) |
5.1.3. Data Imputation
When training the system, the generator and discriminator functions adversarially optimize , while the autoencoder embedding and recovery functions optimize . Here, parameters α and β balance the loss pairs (we use α=1 and β=10 for our experiments), while parameters θg, θd, θe, and θr govern the generator, discriminator, embedding, and recovery components.
To impute data for missing values conditioned on observed data values, we blend observed data with synthetic data. Mink generates a synthetic data vector that is conditioned on the observed data, the corresponding mask, and a Gaussian noise vector. Missing values in the original data vector X1:T are replaced with their corresponding synthetic component, yielding a complete time series with no missing values.
5.2. Generating a Behavior Profile
Human behavior is one of the biggest drivers of health and wellness49,50. An individual’s activities affect that person, their family, society, and the environment. Health risk behavior is linked to Type 2 diabetes, obesity, heart disease, neurological diseases including ADRDs, and other chronic physical and mental health conditions. For this reason, our overall goal is to model behavior and use machine learning techniques to predict health status from behavior information.
In previous work, theoretical models arose from psychology, sociology, and anthropology to explain the complexities of behavior and the factors that drive it. Until recently, such theories of human behavior and its influences have relied on self-report, which can suffer from retrospective memory limitations51, or experimenter observation, which may introduce confounds and unintended bias52. The maturing of pervasive computing now allows us to collect personal sensor data unobtrusively and continuously. As a result, the field is ripe to create data mining methods to model behavior and predict health.
In our approach, predicting clinical health measures requires five steps. First, we collect continuous sensor data from smartwatches as people perform their normal daily routines. Second, we utilize Mink to impute values for the missing data. Third, we label sensor data streams with corresponding activity labels. Fourth, we extract a set of digital markers. Finally, we use machine learning to map the digital markers onto predicted clinical measures.
5.2.1. Collecting and Labeling Activity Data
To gather behavior-driven sensor data, we designed an app for the Apple Watch to passively and continuously collect sensor data at a constant sampling rate of 10Hz. We currently collect data from the watch accelerometer, gyroscope, and location services. The app periodically queries users to provide ground truth about their current activity and answer in-the-moment questions about their current mood and functionality. During this process, there are frequent gaps in the data collection due to participants failing to charge or wear the smartwatch. As a result, imputation of missing data for one or multiple consecutive time periods is needed before clinical measures can be predicted.
A first step in building a set of digital behavior markers from longitudinal sensor data is labeling data with corresponding activities. Activities represent units of behavior that can be labeled and integrated into the digital behavior markers. While human activity recognition is a popular research topic and many approaches have been proposed32,34,53-59, most approaches operate under controlled laboratory conditions with scripted, movement-based activities60-63. Research has demonstrated a correlation between cognitive health and numerous activities, both simple and complex, that include sleep, work, time out of the home, walking, and socialization64-69. Thus, automatically labeling these activities can improve the ability to assess cognitive and functional health.
Activity recognition algorithms map sensor data onto corresponding activity names, applying categorical descriptions to sensed behavior. The input is a sequence of sensor readings et=<t, r1, .., rd> collected at time t. To accommodate real-time recognition, features are extracted from a sliding window that are statistical (e.g., min, max, standard deviation, zero crossings, skewness, kurtosis, signal energy), relational (e.g., multi-dimensional correlation, autocorrelation), temporal (e.g., time of day, day of week), navigational (e.g., heading change rate, stop rate, overall trajectory, distance traveled), personal (e.g., frequented locations, distance from user center), and positional (location type, calculated via reverse geocoding using an open street map). A random forest classifier creates a mapping, h:Xt→yt, from a set of descriptors Xt to the corresponding activity, yt. This approach demonstrated a recognition f1 score of 0.85 for 12 activities from 250 individuals in prior work: chores, eat, entertain, errands, exercise, hobby, hygiene, relax, school, sleep, travel, and work70. We use the pretrained model for the remainder of the experiments described in this paper.
5.2.2. Defining and Extracting Digital Behavior Markers
As data are collected and labeled with activity categories, we extract digital descriptors, or markers, that provide insights into a person’s behavior and predictive power for the person’s health. Continual monitoring of daily behavior offers more and finer-resolution insights than are currently available for physician-based or automated health assessment and intervention design. We compute and compile the digital behavior markers that become a person’s behavior profile70,71. The markers are defined in Table 1 and are gathered for each sensor (existing and new) and activity class at multiple time resolutions (e.g., hourly, daily). Our software to generate these markers is available online72.
Table 1.
Digital behavior markers.
| Type | Daily features |
|---|---|
| Statistical summary of sensor values | Maximum, minimum, sum, mean, median, mean/median absolute value, variance, standard deviation, zero/mean crossings, interquartile range, skewness, kurtosis, SMA, power, autocorrelation, computed over multiple time scales |
| Durations | Time spent on each activity, location type, favorite location |
| Occurrences | Time of day for first and last occurrence of each activity, location type, favorite location |
| Sleep | Daytime and nighttime sleep duration, daytime sleep location, nighttime sleep location, number of nighttime sleep interruptions |
| Mobility | Amount of movement inside and outside home, walking speed, number of steps, reverse geocoded location types visited outside the home, total distance traveled |
| Routine | Entropy of daily routine, number of different daily activities, minimum and maximum inactivity times, daily variance in activity durations, occurrence times, and locations, periodogram-derived circadian and diurnal rhythm 87,88 |
5.2.3. Predicting Clinical Health Measures
In the last step, a regression forest is employed to predict the clinical measures C = {c1, c2, .., cn}. The random forest contains 100 decision tree regressors. The trees are built to a depth of 20 using randomly-selected features, then regressors fit a line to the data that belong to each leaf node. We report predictive performance using Pearson correlation. To collect data, we recruited n=14 older adult participants for this study (9 female, 5 male). The mean age was 70.2 (s.d.=7.5) and number of years of education was 16.4 (s.d.=2.5). Detailed participant information is provided in Table 2. In this sample, 6 participants had cognitive impairment, with objective evidence in the memory domain. This study was reviewed and approved by the Washington State University Institutional Review Board (IRB protocol #14460, approved 05/18/2020). Informed consent was obtained from each participant prior to data collection initiation.
Table 2.
Participant information.
| Participant | Age | Cognitive impairment |
Gender | Education (years) |
Missing data (%) |
|---|---|---|---|---|---|
| 1 | 65 | No | female | 18 | 26.17 |
| 2 | 66 | No | female | 20 | 31.49 |
| 3 | 72 | No | female | 18 | 22.80 |
| 4 | 76 | No | female | 20 | 23.27 |
| 5 | 79 | No | female | 18 | 19.92 |
| 6 | 62 | No | male | 16 | 28.32 |
| 7 | 62 | No | male | 20 | 24.05 |
| 8 | 78 | No | male | 18 | 25.35 |
| 9 | 56 | Yes | female | 12 | 21.01 |
| 10 | 70 | Yes | female | 18 | 24.35 |
| 11 | 72 | Yes | female | 14 | 16.79 |
| 12 | 73 | Yes | female | 14 | 27.32 |
| 13 | 58 | Yes | male | 12 | 48.02 |
| 14 | 68 | Yes | male | 20 | 28.82 |
We collected one month of 10Hz continuous smartwatch sensor data (accelerometer, gyroscope, and location) for the participants. As Table 2 indicates, the collected data had many missing entries, including entire evenings for some participants when the watch was not worn. Additionally, we collected clinical assessment measures for each participant at baseline using traditional neuropsychology tests and self-report. The measures and the constructs they assess are listed in Table 3.
Table 3.
Predicted clinical measures.
| Measure | Assessed construct |
|---|---|
| Telephone Interview of Cognitive Status (TICS) 73 | global cognitive status |
| Rey Auditory Verbal Learning Test (RAVLT) 74 | verbal memory |
| Behavioral assessment of the dysexecutive syndrome (BADS) 75 | executive disinhibition |
| Timed Up and Go (TUG) 76 | mobility |
| Quality of Life scale (QOL) 77 | quality of life |
| Short Form Survey (SF-12) 78 | physical and mental health |
| Prospective and Retrospective Memory Questionnaire (PRMQ) 80 | memory |
| Geriatric Depression Scale (GDS) 81 | depression |
| Generalized Anxiety Disorder (GAD) 82 | anxiety |
| Dysexecutive Questionnaire (DEX) 83 | executive function |
| Instrumental Activities of Daily Living – Compensation Scale (IADL-C) 89 | everyday function |
Because this study was conducted during the COVID-19 pandemic, tests were selected that could be administered remotely. The Telephone Interview for Cognitive Status (TICS)73 is administered remotely over a phone and consists of tasks for the participant to perform including word list learning, counting backward, and finger tapping. Other tests, including RAVLT and BADS, were adapted for administration using video conference software. Rey’s Auditory Verbal Learning Test (RAVLT)74 consists of an oral presentation of two lists for immediate recall to assess verbal memory. The Behavioural Assessment of the Dysexecutive Syndrome (BADS)75 is used to evaluate problems that arise during daily activities due to executive disinhibition. This assessment contains thirteen tasks that focus on functional abilities such as planning, problem solving, and temporal judgment. The Timed Up and Go (TUG) test76 requires the participant to stand up from a chair, walk forward, turn around, and return to the chair, which was administered while being remotely monitored by an experimenter. The score reflects the time taken to complete the task and provides an indicator of mobility as well as cognitive health.
The next set of assessments are questionnaires that were delivered and answered over video communication. These include the Quality of Life (QOL) scale77 that measures the domains of material and physical well-being, relationships, social activities, personal development, and recreation; the short form 12 (SF-12) survey78 that asks questions assessing general health, physical well-being, vitality, social functioning, emotions, and mental health (we separate this into two scores corresponding to the physical and mental health components)79; the Prospective and Retrospective Memory Questionnaire (PRMQ)80 that contains questions about prospective (looking into the future) and retrospective (looking into the past) memory slips in everyday life; the Geriatric Depression Scale (GDS)81 in which participants answer questions in reference to how they felt over the past week to measure depressive symptoms; the seven-item version of the Generalized Anxiety Disorder questionnaire (GAD-7)82 that asks participants how often during the last two weeks they were bothered by specific anxiety symptoms; and the Dysexecutive Functioning Questionnaire (DEX)83 that assesses multiple cognitive-behavioral problems such as sustaining attention, inhibiting inappropriate behaviors, or switching between multiple problem-solving strategies.
The last questionnaire, the Instrumental Activities of Daily Living – Compensation (IADL-C) scale84, was recently designed to assess the functional domains of money and self-management, home-based daily tasks, travel and event memory, and social skills. Unlike earlier assessments, the IADL-C scale is sensitive to the use of compensatory strategies in performing daily tasks that help to overcome memory limitations. The tests and questionnaires provide insights on different, though overlapping, aspects of cognitive health. We hypothesize that cognitive health state is reflected in behavior patterns and thus behavior markers can be used to predict these health assessment scores.
6. Results
We evaluate sensor-based health assessment in two steps. First, we evaluate the performance of our time series imputation method using complete sets of data. Second, we evaluate the performance of our complete method in predicting clinical health measures.
6.1. Evaluation of Data Imputation
We evaluate the accuracy of time series data imputation using NMAE. The participants and days are selected on the criterion that data are complete between 8:00am and 10:00pm on the corresponding date, to provide ground truth for evaluation. In our study, 5 participants collected data that meet this constraint for at least 15 days. We therefore evaluate the imputation of accelerometer and gyroscope sensor values for 15 days of data for 5 participants. In the case of approaches that require model training, we utilize 12 days of data for training for each participant. For all cases, we utilize 3 days of data for testing. The results are thus averaged over 15 days of continuous sensor readings. For each day, we extract a portion of the data, impute the missing values, and compare it with the ground truth. We vary the percentage of missing entries (10%, 20%, or 30%) and the size of the missing data gap (1 second, 1 minute, 1 hour, 12 hours). We randomly select the beginning of each missing data sequence and average results over three random selections.
Table 4 summarizes the results, comparing Mink with several baseline methods. The carry forward, carry backward, bidirectional carry, and MTS-GAN baselines are described in Section 4.1. We also include a two-layer neural network with 100 hidden nodes, a rectified linear activation function, and a learning rate of 0.001 as a baseline regressor. The results in Table 4 are computed based using 12 days of data for training and the following 3 days of data for testing data. Results are averaged over 5 participants, 4 alternative gap sizes, and 3 runs with different random seeds. We employ a paired t-test to determine the statistical significance of the difference in performance between each baseline method and Mink. As the results indicate, the adversarial network provides realistic values, even when a large chunk of consecutive readings is missing. Additionally, this approach outperforms the baseline methods for time series imputation. Because the imputation results are promising for Mink, we next employ the Mink GAN to impute values used for creating the digital behavior markers and inferring clinical measures.
Table 4.
NMAE of imputation methods, each averaged over 5 participants, 4 gap sizes, and 3 random trials. * = the difference in performance is statistically significant (p<.05).
| Carry forward |
Carry backward |
Bidirect carry |
Neural network |
MTS GAN |
KNN (k=3) |
Mink | |
|---|---|---|---|---|---|---|---|
| 10% | 0.0184* | 0.0195* | 0.0180* | 0.0251* | 0.3050* | 0.1381* | 0.0169 |
| 20% | 0.0244* | 0.0245* | 0.0235* | 0.0265 | 0.2944* | 0.1907* | 0.0220 |
| 30% | 0.0225* | 0.0237* | 0.0224* | 0.0290* | 0.2949* | 0.2032* | 0.0207 |
| Average | 0.0216* | 0.0224* | 0.0211* | 0.0268* | 0.2981* | 0.1773* | 0.0197 |
6.2. Evaluation of Clinical Measure Prediction
Finally, we evaluate the predictive performance of the clinical measures for both Mink-based imputation, imputation using constant values, and imputation using bidirectional carry (the second highest-performing imputation method in the previous experiment). For this experiment, we predict the precise numeric score for each assessment measure. Because each measure uses a different score range, we employ the approach used in other studies to evaluate performance by computing correlation between predicted and ground truth scores38,42. The results of this experiment are based on leave-one-subject-out testing and are summarized in Table 5. We observe small correlation for GDS (r=0.1230), GAD (r=0.2125), and QOL (r=0.2436), moderate correlation for RAVLT (r=0.3328), PRMQ (r=0.3984), IADL-C (r=0.4095), DEX (r=0.4401), TICS (r=0.4762), and BADS (r=0.5292), and large correlation for TUG (r=0.6119), SF-12 Physical (r=0.6133), and SF-Mental (r=0.7623). The mean of the correlation values is 0.4294. The generative imputation method employed by Mink does result in an improvement in the mean of the correlation values (0.4294, in comparison with 0.4096 for bidirectional carry and 0.3071 for replace-by-constant).
Table 5.
Pearson correlation of clinical measures using baseline and Mink imputation methods.
| Measure | Constant | Bidirectional carry |
Mink |
|---|---|---|---|
| TICS | 0.6185 | 0.6711 | 0.4762 |
| RAVLT | 0.0743 | 0.2502 | 0.3328 |
| BADS | 0.4818 | 0.4487 | 0.5292 |
| TUG | 0.5076 | 0.4138 | 0.6119 |
| QOL | 0.0172 | 0.1055 | 0.2436 |
| SF-12 Physical | 0.4831 | 0.7084 | 0.6133 |
| SF-12 Mental | 0.5347 | 0.6162 | 0.7623 |
| PRMQ | 0.0006 | 0. 5080 | 0.3984 |
| GDS | 0.1079 | 0.3927 | 0.1230 |
| GAD | 0.4574 | 0.1260 | 0.2125 |
| DEX | 0.1902 | 0.1859 | 0.4401 |
| IADL-C | 0.2115 | 0.4892 | 0.4095 |
| Mean (SD) | 0.3071 (0.2175) | 0.4096 (0.1972) | 0.4294 (0.1771) |
7. Discussion and Conclusions
The long-term goal of this work is to automate health assessment from sensor-observed longitudinal behavior data. In this paper, we address a significant obstacle to this goal by designing a method to impute missing values in the time series data. The results indicate that a generative architecture can be employed for this process. Considering both temporal and between-feature relationships is valuable for such multivariate sensor readings. The results indicate that the generative imputation method outperforms straightforward baseline methods. Additionally, the resulting behavior markers are predictively correlated with collected clinical measures.
While Mink outperforms a baseline imputation method for clinical measure prediction, the results are not consistent across all clinical measures. One explanation for this finding is that the variance in the generated values can result in larger differences from true values than constant values. While the errors do not occur as often as with the baseline methods, the magnitude of the error may mislead the regression forest. This possibility that the GAN may generate out-of-range values is a limitation of the current approach and can be addressed in future versions of the algorithm.
We also observe that the predictive performance is lower overall for measures with a smaller variance in the collected data. This is due in part to the limited sample size and need for greater diversity in the data. This additional study limitation will be addressed in the future by recruiting a larger population that represents diversity in age, demographics, and health conditions.
In the current work, we assume that the time and duration of missing readings are random values. In practical settings, the missing values may be related to patient physical conditions (e.g., an illness or trip during which the person does not wear the device) or external conditions (e.g., a power outage that prevents the device battery from fully charging). Future enhancements can include modeling such conditions and utilizing the information to improve the design and evaluation of imputation.
Mink successfully outperformed baseline methods in our experiments, but there is room for improvement. We hypothesize that obtaining observational data from a larger and more diversity set of complete days will improve GAN performance and will test this hypothesis in future studies. Additionally, while a generative adversarial network offers an effective way to generate a sequence of missing sensor readings, they are known to suffer from possible mode collapse. As a result, the trained network may generate only a small number of distinct types of readings. While the generated values are realistic, they may lack the variability that exists in the real data. Researchers have investigated strategies to reduce mode collapse85,86. A future step of our work may include addressing this limitation by adapting these strategies for use in time series data.
Acknowledgements
The authors would like to thank Maureen Schmitter-Edgecombe and Justin Frow for their help with data collection. This work is supported in part by National Institutes of Health grant R41EB029774.
Footnotes
Mink code is available online at https://github.com/WSU-CASAS/MINK.
References
- 1.Elflein J. Amount of time U.S. primary care physicians spent with each patient as of 2018. Statista. Published 2019. Accessed June 13, 2020. https://www.statista.com/statistics/250219/us-physicians-opinion-about-their-compensation/ [Google Scholar]
- 2.Iriondo J, Jordan J. Older People Projected to Outnumber Children for First Time in U.S. History.; 2018. https://www.census.gov/newsroom/press-releases/2018/cb18-41-population-projections.html [Google Scholar]
- 3.Center for Medicare and Medicaid Services. NHE Fact Sheet. Center for Medicare and Medicaid Services. Published online 2018. https://www.cms.gov/Research-Statistics-Data-and-Systems/Statistics-Trends-and-Reports/NationalHealthExpendData/NHE-Fact-Sheet [Google Scholar]
- 4.Administration On Aging. Aging statistics. ACL. Published online 2018. https://acl.gov/aging-and-disability-in-america/data-and-research/profile-older-americans [Google Scholar]
- 5.Office of The Assistant Secretary for Planning and Evaluation. National Plan to Address Alzheimer’s Disease: 2018. ASPE. Published 2019. https://aspe.hhs.gov/national-plans-address-alzheimers-disease [Google Scholar]
- 6.Fowler NR, Head KJ, Perkins AJ, et al. Examining the benefits and harms of Alzheimer’s disease screening for family members of older adults: study protocol for a randomized controlled trial. Trials. 2020;21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herman WH, Ye W, Griffin SJ, Simmons RK, Davies MJ, Khunti K. Early detection and treatment of Type 2 diabetes reduce cardiovascular morbidity and mortality: A simulation of the results of the Anglo-Danish-Dutch study of intensive treatment in people with screen-detected diabetes in primary care. Diabetes Care. Published online 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akl A, Snoek J, Mihailidis A. Unobtrusive detection of mild cognitive impairment in older adults through home monitoring. IEEE Journal of Biomedical and Health Informatics. 2017;21(2):339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spruijt-Metz D. Etiology, treatment and prevention of obesity in childhood and adolescence: A decade in review. Journal of Research in Adolescence. 2011;21(1):129–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee MK, Oh J. Health-related quality of life in older adults: Its association with health literacy, self-efficacy, social support, and health-promoting behavior. Healthcare. 2020;8(4):407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson BW, Pettitt A, Flannery JE, Allen NB. Rapid assessment of psychological and epidemiological correlates of COVID-19 concern, financial strain, and health-related behavior change in a large online sample. PLoS ONE. 2020;15(11):e0241990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betsinger TK, DeWitte SN. Toward a bioarchaeology of urbanization: Demography, health, and behavior in cities in the past. American Journal of Physical Anthropology. 2021;175(S72):79–118. [DOI] [PubMed] [Google Scholar]
- 13.Gorman J. “Ome,” the sound of the scientific universe expanding. The New York Times. https://www.nytimes.com/2012/05/04/science/it-started-with-genome-omes-proliferate-in-science.html?_r=1. Published 2012. [Google Scholar]
- 14.Asim Y, Azam MA. Context-aware human activity recognition (CAHAR) in-the-wild using smartphone accelerometer. IEEE Sensors. 2020;8:4361–4371. [Google Scholar]
- 15.Vaizman Y, Ellis K, Lanckriet G. Recognizing detailed human context in the wild from smartphones and smartwatches. IEEE Pervasive Computing. 2017;16(4):62–74. [Google Scholar]
- 16.Tran DH. Automated change detection and reactive clustering in multivariate streaming data. In: IEEE-RIVF International Conference on Computing and Communication Technologies. ; 2019:1–6. [Google Scholar]
- 17.Sterne JAC, White IR, Carlin JB, et al. Multiple imputation for missing data in epidemiological and clinical research: Potential and pitfalls. BMJ. 2009;338:b2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Austin PC, White IR, Lee DS, van Buuren S. Missing data in clinical research: A tutorial on multiple imputation. Canadian Journal of Cardiology. 2021;37(9):1322–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lachin JM. Fallacies of last observation carried forward analyses. Clinical Trials. 2016;13(2):161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bokde N, Alvarez FM, Beck MW, Kulat K. A novel imputation methodology for time series based on pattern sequence forecasting. Pattern Recognition Letters. 2018;116:88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang C, Wang C. Time series data imputation: A survey on deep learning approaches. arXiv. 2020;2011.11347. [Google Scholar]
- 22.Cao W, Wang D, Li J, Zhou H, Li L, Li Y. BRITS: Bidirectional recurrent imputation for time series. In: Neural Information Processing Systems. ; 2018:6776–6786. [Google Scholar]
- 23.Wu X, Mattingly S, Mirjafari S, Huang C, Chawla NV. Personalized imputation on wearable sensory time series via knowledge transfer. In: ACM International Conference on Information and Knowledge Management. ; 2020:1625–1634. [Google Scholar]
- 24.Yoon S, Sull S. GAMIN: Generative adversarial multiple imputation network for highly missing data. In: IEEE/CVF Conference on Computer Vision and Pattern Recognition. ; 2020:8456–8464. [Google Scholar]
- 25.Yoon J, Jordon J, van der Schaar M. GAIN: Missing data imputation using generative adversarial networks. In: International Conference on Machine Learning. ; 2018. [Google Scholar]
- 26.Luo Y, Zhang Y, Cai X, Yuan X. E2gan: end-to-end generative adversarial network for multivariate time series imputation. In: International Joint Conference on Artificial Intelligence. ; 2019:3094–3100. [Google Scholar]
- 27.Guo Z, Wan Y, Ye H. A data imputation method for multivariate time series based on generative adversarial network. Neurocomputing. 2019;360(185–197). [Google Scholar]
- 28.Chen K, Zhang D, Yao L, Guo B, Yu Z, Liu Y. Deep learning for sensor-based human activity recognition: Overview, challenges and opportunities. Journal of the ACM. 2020;37(4):111. [Google Scholar]
- 29.Bulling A, Blanke U, Schiele B. A tutorial on human activity recognition using body-worn inertial sensors. ACM Computing Surveys. 2014;46(3):107–140. [Google Scholar]
- 30.Tian Y, Zhang J, Chen L, Geng Y, Wang X. Selective ensemble based on extreme learning machine for sensor-based human activity recognition. Sensors. 2019;19(16):3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nazabal A, Garcia-Moreno P, Artes-Rodriguez A, Ghahramani Z. Human activity recognition by combining a small number of classifiers. IEEE Journal of Biomedical and Health Informatics. 2016;20(5):1342–1351. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Chen Y, Hao S, Peng X, Hu L. Deep learning for sensor-based activity recognition: A survey. Pattern Recognition Letters. 2019;119:3–11. [Google Scholar]
- 33.Hammerla NY, Halloran S, Ploetz T. Deep, convolutional, and recurrent models for human activity recognition using wearables. In: International Joint Conference on Artificial Intelligence. ; 2016. [Google Scholar]
- 34.Ploetz T, Guan Y. Deep learning for human activity recognition in mobile computing. Computer. 2018;51(5):50–59. [Google Scholar]
- 35.Guan Y, Ploetz T. Ensembles of deep LSTM leaners for activity recognition using wearables. Proceedings of the ACM on Interactive, Mobile, Wearable and Ubiquitous Technologies. Published online 2017:11. [Google Scholar]
- 36.Culman C, Aminikhanghahi S, Cook DJ. Easing power consumption of wearable activity monitoring with change point detection. IEEE Transactions on Mobile Computing. Published online 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kankanhalli A, Saxena M, Wadhwa B. Combined interventions for physical activity, sleep, and diet using smartphone apps: A scoping literature review. International Journal of Medical Informatics. 2019;123:54–67. [DOI] [PubMed] [Google Scholar]
- 38.Sprint G, Cook DJ. Unsupervised detection and analysis of changes in everyday physical activity data. Journal of Biomedical Informatics. Published online 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Zambotti M, Cellini N, Goldstone A, Colrain IM, Baker FC. Wearable sleep technology in clinical and research settings. Medicine and Science in Sports and Exercise. 2019;51(7):1538–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang R, Wang W, DaSilva A, et al. Tracking depression dynamics in college students using mobile phone and wearable sensing. Proceedings of the ACM on Interactive, Mobile, Wearable and Ubiquitous Technologies. 2018;2(1):1–26. [Google Scholar]
- 41.Dhana K, Evans DA, Rajan KB, Bennett DA, Morris MC. Healthy lifestyle and the risk of Alzheimer dementia: Findings from 2 longitudinal studies. Neurology. 2020;95(4):e374–e383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alberdi Aramendi A, Weakley A, Schmitter-Edgecombe M, et al. Smart home-based prediction of multi-domain symptoms related to Alzheimer’s Disease. IEEE Journal of Biomedical and Health Informatics. 2018;22(5):1720–1731. doi: 10.1109/JBHI.2018.2798062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Rong Y, Meng H, Lu Z, Kwok T, Cheng H. TATC: Predicting Alzheimer’s disease with actigraphy data. In: ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ; 2018:509–518. [Google Scholar]
- 44.Yoon J, Jarrett Daniel, van der Schaar M. Time-series generative adversarial networks. In: Conference on Neural Information Processing Systems. ; 2019. [Google Scholar]
- 45.Menendez ML, Pardo JA, Pardo L, Pardo MC. The Jensen-Shannon divergence. Journal of the Franklin Institute. 1997;334(2):307–318. [Google Scholar]
- 46.Kachuee M, Karkkainen K, Goldstein O, Darabi S, Sarrafzadeh M. Generative imputation and stochastic prediction. IEEE Transactions on Pattern Analysis and Machine Intelligence. Published online 2021. [DOI] [PubMed] [Google Scholar]
- 47.van Erven T, Harremos P. Renyi divergence and Kullback-Leibler divergence. IEEE Transactions on Information Theory. 2014;60(7):3797–3820. [Google Scholar]
- 48.Goodfellow IJ, Pouget-Abadie J, Mirza M, et al. Generative adversarial networks. Advances in Neural Information Processing Systems. 2014;27:1–9. [Google Scholar]
- 49.Marteau TM, Hollands GJ, Fletcher PC. Changing human behavior to prevent disease: The importance of targeting automatic processes. Science. 2012;337:1492–1495. [DOI] [PubMed] [Google Scholar]
- 50.U.S. Department of Health and Human Services. Healthy People 2020.; 2015. [DOI] [PubMed] [Google Scholar]
- 51.Tourangeau R, Rips LJ, Rasinski K. The Psychology of Survey Response. Cambridge University Press; 2000. [Google Scholar]
- 52.Palmer MG, Johnson CM. Experimenter presence in human behavior analytic laboratory studies: Confound it? Behavior Analysis: Research and Practice. 2019;19(4):303–314. [Google Scholar]
- 53.Li H, Abowd GD, Ploetz T. On specialized window lengths and detector based human activity recognition. In: ACM International Symposium on Wearable Computers. ; 2018:67–71. [Google Scholar]
- 54.Aminikhanghahi S, Cook DJ. Enhancing activity recognition using CPD-based activity segmentation. Pervasive and Mobile Computing. 2019;53(75–89). [Google Scholar]
- 55.Wan J, Li M, O’Grady M, Gu X, Alawlaqi M, O’Hare G. Time-bounded activity recognition for ambient assisted living. IEEE Transactions on Emerging Topics in Computing. Published online 2018. [Google Scholar]
- 56.Du Y, Lim Y, Tan Y. A novel human activity recognition and prediction in smart home based on interaction. Sensors. 2019;19:4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bharti P, De D, Chellappan S, Das SK. HuMAn: Complex activity recognition with multi-modal multi-positional body sensing. IEEE Transactions on Mobile Computing. 2019;18(4):857–870. [Google Scholar]
- 58.Kwon M-C, You H, Kim J, Choi S. Classification of various daily activities using convolution neural network and smartwatch. In: IEEE International Conference on Big Data. ; 2018. [Google Scholar]
- 59.Nweke HF, Teh YW, Al-Garadi MA, Alo UR. Deep learning algorithms for human activity recognition using mobile and wearable sensor networks: State of the art and research challenges. Expert Systems with Applications. 2018;105:233–261. [Google Scholar]
- 60.Anguita D, Ghio A, Oneto L, Parra X, Reyes-Ortiz JL. A public domain dataset for human activity recognition using smartphones. In: European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning. ; 2013. [Google Scholar]
- 61.Stisen A, Blunck H, Bhattacharya S, et al. Smart devices are different: Assessing and mitigating mobile sensing heterogeneities for activity recognition. In: ACM Conference on Embedded Networked Sensor Systems. ; 2015:127–140. [Google Scholar]
- 62.Kwapisz JR, Weiss GM, Moore SA. Activity recognition using cell phone accelerometers. In: International Workshop on Knowledge Discovery from Sensor Data. ; 2010. [Google Scholar]
- 63.Lockhart JW, Weiss GM, Xue JC, Gallagher ST, Grosner AB, Pulickal TT. Design considerations for the Wisdm smart phone-based sensor mining architecture. In: International Workshop on Knowledge Discovery from Sensor Data. ; 2011:25–33. [Google Scholar]
- 64.Cook DJ, Schmitter-Edgecombe M, Jonsson L, Morant AV. Technology-enabled assessment of functional health. IEEE Reviews in Biomedical Engineering. 2018;12:319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dodge HH, Mattek NC, Austin D, Hayes TL, Kaye JA. In-home walking speeds and variability trajectories associated with mild cognitive impairment. Neurology. 2012;78(24):1946–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaye J, Mattek N, Dodge HH, et al. Unobtrusive measurement of daily computer use to detect mild cognitive impairment. Alzheimer’s and Dementia. 2014;10(1):10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petersen J, Austin D, Mattek N, Kaye J. Time out-of-home and cognitive, physical, and emotional wellbeing of older adults: A longitudinal mixed effects model. PLoS ONE. Published online 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petersen J, Larimer N, Kaye JA, Pavel M, Hayes TL. SVM to detect the presence of visitors in a smart home environment. In: International Conference of the IEEE Engineering in Medicine and Biology Society. ; 2012:5850–5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cook DJ. Sensors in support of aging-in-place: The good, the bad, and the opportunities. In: National Academies Workshop on Mobile Technology for Adaptive Aging. ; 2019. [Google Scholar]
- 70.Cook D, Schmitter-Edgecombe M. Fusing ambient and mobile sensor features into a behaviorome for predicting clinical health scores. IEEE Access. 2021;2:65033–65043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmitter-Edgecombe M, Sumida CA, Cook DJ. Bridging the gap between performance-based assessment and self-reported everyday functioning: An ecological momentary assessment approach. The Clinical Neuropsychologist. 2020;34(4):678–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.WSU CASAS. Tools. Published 2021. http://casas.wsu.edu/tools/ [Google Scholar]
- 73.Fong TG, Fearing MA, Jons RN, et al. The Telephone Interview for Cognitive Status: Creating a crosswalk with the Mini-Mental State Exam. Alzheimer’s and Dementia. 2009;5(6):492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peaker A, Stewart LE. Rey’s auditory verbal learning test – A review. In: Crawford JR, Parker DM, eds. Developments in Clinical and Experimental Neuropsychology. Plenum Press; 1989. [Google Scholar]
- 75.Wilson BA, Alderman N, Burgess PW, Emslie H, Evans JJ. Behavioural Assessment of the Dysexecutive Syndrome. Thames Valley Test Company; 1996. [Google Scholar]
- 76.Sprint G, Cook D, Weeks D. Towards automating clinical assessments: A survey of the Timed Up and Go (TUG). Biomedical Engineering, IEEE Reviews in. 2015;8:64–77. doi: 10.1109/RBME.2015.2390646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burckhardt CS, Answerson KL. The Quality of Life Scale (QOLS): Reliability, validity, and utilization. Health Quality of Life Outcomes. 2003;1:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huo T, Guo Y, Shenkman E, Muller K. Assessing the reliability of the short form 12 (SF-12) health survey in adults with mental health conditions: a report from the wellness incentive and navigation (WIN) study. Health Quality of Life Outcomes. 2018;16:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ware JE, Koskinski M, Keller SD. SF-12: How to Score the SF-12 Physical and Mental Health Summary Scores.; 1995. [Google Scholar]
- 80.Smith G, Della Sala S, Logie RH, Maylor EA. Prospective and retrospective memory in normal aging and dementia: A questionnaire study. Memory. 2000;8:311–321. [DOI] [PubMed] [Google Scholar]
- 81.Sheikh JI, Yesavage JA. Geriatric Depression Scale (GDS): Recent evidence and development of a shorter version. Clinical Gerontologist. 1986;5:165–173. [Google Scholar]
- 82.Spitzer RL, Kroenke K, Williams JB, Lowe B. A brief measure for assessing generalized anxiety disorder: the GAD-7. Archives of Internal Medicine. 2006;166(10):1092–1097. [DOI] [PubMed] [Google Scholar]
- 83.Gerstorf D, Siedlecki KL, Tucker-Drob EM, Salthouse TA. Executive dysfunctions across adulthood: Measurement properties and correlates of the DEX self-report questionnaire. Neuropsychology, Development, and Cognition Section B, Aging, Neuropsychology and Cognition. 2008;15(4):424–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schmitter-Edgecombe M, Parsey C, Lamb R. Development and Psychometric Properties of the Instrumental Activities of Daily Living: Compensation Scale. Archives of Clinical Neuropsychology. 2014;29(8):776–792. doi: 10.1093/arclin/acu053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu S, Zhang K, Xiao C, Huang JZ, Li MJ, Onizuka M. HSGAN: Reducing mode collapse in GANs by the latent code distance of homogeneous samples. Computer Vision and Image Understanding. 2022;214:103314. [Google Scholar]
- 86.Zuo Z, Zhao L, Li A, et al. Dual distribution matching GAN. Neurocomputing. 2022;478:37–48. [Google Scholar]
- 87.Williams JA, Cook DJ. Forecasting behavior in smart homes based on sleep and wake patterns. Technology and Health Care. 2017;25(1). doi: 10.3233/THC-161255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang W, Harari GM, Wang R, et al. Sensing behavioral change over time: Using within-person variability features from mobile sensing to predict personality traits. Proceedings of the ACM on Interactive, Mobile, Wearable and Ubiquitous Technologies. 2018;2(3):141. [Google Scholar]
- 89.Schmitter-Edgecombe M, Parsey CM, Lamb R. Development and psychometric properties of the instrumental activities of daily living – compensation scale (IADL-C). Neuropsychology. Published online 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]