Abstract
Multidrug-resistant Plasmodium falciparum parasites are a major threat to public health in intertropical regions. Understanding the mechanistic basis, origins, and spread of resistance can inform strategies to mitigate its impact and reduce the global burden of malaria. The recent emergence in Africa of partial resistance to artemisinins, the core component of first-line combination therapies, is particularly concerning. Here, we review recent advances in elucidating the mechanistic basis of artemisinin resistance, driven primarily by point mutations in P. falciparum Kelch13, a key regulator of hemoglobin endocytosis and parasite response to artemisinin-induced stress. We also review resistance to partner drugs, including piperaquine and mefloquine, highlighting a key role for plasmepsins 2/3 and the drug and solute transporters P. falciparum chloroquine-resistance transporter and P. falciparum multidrug-resistance protein-1.
Artemisinin-based combination therapies
Malaria continues to be a major global health problem, with an estimated 241 million cases and 627,000 deaths in 2020 [1]. Plasmodium falciparum, the most virulent causative species, accounts for ~98% of cases. Encouragingly, substantial reductions in the disease burden have been achieved over the past two decades, due in part to the implementation of artemisinin (ART)-based combination therapies (ACTs) as the first-line treatment for uncomplicated P. falciparum malaria in endemic countries worldwide.
ART and its derivatives are highly potent, fast-acting antimalarials that can reduce P. falciparum biomass by up to 10 000-fold every 48-hour asexual blood-stage cycle [2]. A three-day regimen of ART monotherapy is associated with high levels of parasite recrudescence due to a short plasma half-life (typically < 1–2 hr), thus necessitating the use of longer-lasting partner drugs in combination therapies. Partner drugs require a different mode of action, or mechanism of resistance, to reduce the chances of multidrug-resistance selection. The World Health Organization currently recommends six first-line ACTs: artemether-lumefantrine (AL), artesunate-amodiaquine (ASAQ), artesunate-mefloquine (AS-MQ), artesunate-sulfadoxine-pyrimethamine (AS-SP), dihydroartemisinin-piperaquine (DHA-PPQ) and artesunate-pyronaridine (AS-PND) [1]. AS-MQ and DHA-PPQ have been the predominant ACTs used in SE Asia, while AL and ASAQ are the main ACTs used in Africa and account for ~98% of doses delivered worldwide (Table 1).
Table 1.
Artemisinin-based combination therapies recommended by the WHO as first-line treatments for uncomplicated P. falciparum malaria.
| ART derivative | Partner drug | Abbreviation | World use | Regions where first-line | ||||
|---|---|---|---|---|---|---|---|---|
| Artemether | Lumefantrine | AL | 74% | AFRO | AMRO | SEARO | EMRO | WPRO |
| Artesunate | Amodiaquine | ASAQ | 24% | AFRO | ||||
| Dihydroartemisinin | Piperaquine | DHA-PPQ | 1% | AFRO | SEARO | WPRO | ||
| Artesunate | Pyronaridine | AS-PND | 0.1% | SEARO | WPRO | |||
| Artesunate | Mefloquine | AS-MQ | 1% | AMRO | SEARO | WPRO | ||
Artesunate-sulfadoxine-pyrimethamine (AS-SP) is also a recommended first-line ACT in EMRO and SEARO. WHO regions: AFRO, African Region; AMRO, Region of the Americas; SEARO, South-East Asia Region; EMRO, Eastern Mediterranean Region; WPRO, Western Pacific Region.
One of the biggest roadblocks to malaria control is the ability of P. falciparum to rapidly evolve antimalarial resistance. ART-resistant P. falciparum emerged over a decade ago in the Greater Mekong Subregion (GMS), traditionally a hotbed for development of antimalarial resistance. Clinically, ART partial resistance is defined as delayed parasite clearance following artesunate monotherapy or ACT, and can be observed as a parasite-clearance half-life > 5 hr, or parasites microscopically evident on day 3 [3]. Initially, delayed clearance did not result in higher rates of treatment failure when artesunate monotherapy was followed with an effective partner drug [4]. Increased parasite exposure to partner drugs, however, led to the emergence of partner-drug resistance, notably to PPQ [5–9]. The resulting high rates of ACT treatment failure hindered control efforts in the GMS, leading to urgent calls for malaria elimination from this region to prevent the spread of multidrug-resistant parasites [10]. Malaria rates in this region have decreased substantially in recent years, and the greatest current concern is the independent emergence of ART partial resistance in sub-Saharan Africa, where 95% of malaria cases and deaths occur [1]. If ART partial resistance spreads across Africa and precedes partner drug and ACT failure, as witnessed in the GMS, the consequences could be devastating.
Here, we review recent research on resistance to ACTs and their components, including the multifaceted mechanisms underlying ART partial resistance, its geographic spread, resistance to DHA-PPQ in the GMS, and genetic modulators of partner-drug susceptibility.
Artemisinin mode of action and mechanism of resistance
ART is activated by reduced heme iron (Fe2+ heme), which mediates cleavage of the endoperoxide bridge and results in the production of free-radical species (possibly carbon-centered [11]). These free radicals can alkylate heme, proteins, lipids, DNA, and other parasite biomolecules [12]. Widespread cell damage triggers the unfolded protein response through activation of protein kinase 4, leading to phosphorylation of eIF2α and subsequent inhibition of protein translation. This damage also leads to a buildup of polyubiquitinated proteins that are tagged for parasite proteasome-mediated degradation. This proteotoxic stress appears to be further ex-acerbated by ART-mediated inhibition of proteasome function, with studies showing synergy between ART derivatives and proteasome inhibitors [13–15]. Parasite responses to ART show similarities to heat-shock responses, and parasites unable to mount a heat-shock response display increased ART susceptibility [16,17]. ART derivatives may also inhibit the formation of hemozoin, leading to a buildup of Fe2+ heme [18]. This toxic by-product of parasite protease-mediated hemoglobin degradation is thought to be the primary activator of ART [19]. While disruption of the heme-degradation process decreases parasite sensitivity to ART, a recent study provided evidence that increasing heme levels also selectively antagonized ART’s antimalarial action, suggesting a multifactorial relationship between the heme-detoxification pathway and ART activation [20].
The primary genetic drivers of ART resistance, both in vitro and in vivo, are point mutations in P. falciparum K13 (also known as Kelch13), which reside mostly, although not exclusively, in the beta-propeller domain [21,22]. The causal relationship between several K13 mutations and resistance has been validated in vitro through reverse genetics [23–26]. K13 mutations allow a subset of early ring-stage parasites to survive cell-cycle arrest brought on by ART exposure, enabling those parasites to reinitiate transcription and complete their in-traerythrocytic developmental cycle once ART is no longer present at inhibitory concentrations [27–29•]. Resistance in vitro is routinely defined as > 1% survival of early ring-stage parasites exposed for 6 hr to 700 nM DHA (the primary active metabolite of ART) followed by drug-free culture incubation for a further 66 hr (this assay is referred to as the RSA0–3 h) [30]. These resistant parasites constitute a transcriptionally diverse population that differs in its expression of stress-response genes [31]. As discussed below, the mechanism of resistance appears to involve a complex interplay of K13 protein abundance, hemoglobin endocytosis, and the parasite response to stress.
K13-propeller mutations have been found in several studies to decrease the abundance of this essential protein, without altering k13 transcription levels [27,32,33••]. This reduction is likely a result of altered protein-folding characteristics and solubility [34] and can differ by background and developmental stage [29•]. Downregulation of K13 protein levels results in decreased sensitivity to ART, and overexpression of either mutant or wild-type K13 resensitizes resistant parasites [33••–36].
K13 was recently localized to peripheral compartments in the parasite plasma membrane as well as vesicular compartments and the endoplasmic reticulum [25•,32,33••,35,37,38]. At the plasma membrane, K13 appears to be concentrated at the neck of hemoglobin-filled cytostomes that traffic the bulk of host hemoglobin from the red blood cell cytosol to the parasite’s lysosome-like digestive vacuole (DV) [32,33••]. Genetic mislocalization of K13 reduces cytostome trafficking and decreases the abundance of hemoglobin-derived peptides [32,33••]. Studies of K13-associated proteins have defined an interactome that includes multiple endocytosis proteins, including AP-2μ and the ubiquitin hydrolase UBP1, both of which have been linked to ART resistance [33••,39,40]. Conditional inactivation of several interactome proteins resulted in reduced ART susceptibility [33••]. These studies indicate a role for K13 in clathrin-independent endocytosis and hemoglobin uptake, suggesting that K13-mediated ring-stage resistance may result from reduced hemoglobin transport to the DV, and subsequent reduction of the drug activator Fe2+ heme.
Mutations in K13 also appear to mediate ART resistance through an increase in the baseline stress response, allowing for more effective mitigation of, or recovery from, the cell damage induced by ART exposure. The precise function of K13 is unknown, however, due to its sequence similarity to a class of Kelch/BTB/POZ ubiquitination adapters, it has been postulated that K13 acts as a ubiquitin ligase adapter that mediates ubiquitin-dependent targeting of proteins for proteasomal degradation [19]. Several analyses have associated K13 mutations with upregulation of chaperones and pathways involved in the unfolded protein response and oxidative stress, and downregulation of ubiquitinating enzymes [25•,27,29•,41]. These factors, in addition to reduced hemoglobin uptake, may explain lower levels of oxidative stress observed in ART-resistant parasites, both at baseline and in response to ART exposure [42]. Increased survival of K13 mutant parasites following ART-induced dormancy may also involve increased baseline phosphorylation of eIF2α and extended ring-stage development [13,32].
Several other interesting phenotypes have been observed in ART-resistant parasites. Enhanced DNA damage repair pathways associated with K13 mutation may aid recovery from ART-induced DNA damage [43]. Mitochondrial proteins are also heavily involved in the ART stress response [44,45], and mutant K13-mediated resistance can be reversed by the mitochondrial electron-transport chain inhibitor atovaquone [29•]. K13 mutant parasites have also been observed to asynchronously reinitiate growth 18–24 hr after removal of DHA pressure, a feature that may be separate from the effect of mutant K13 on restricting hemoglobin endocytosis in ring stages, raising the question of how parasites emerge from their state of ART-induced quiescence [29•].
The degree of protection to ART offered by K13 mutations varies considerably by parasite background, suggesting that secondary determinants contribute to resistance [25•,26,46]. Gene-editing studies recently provided evidence of a greater fitness cost resulting from the introduction of K13 mutations into African strains compared with Asian strains, suggesting that the latter have additional genetic factors that compensate for mutant K13 fitness costs [26]. Mutations in several other proteins have been reported to mediate in vitro resistance to ART, several of which (PI3P, AP-2μ, UBP1, and KIC7) have been colocalized or associated with K13 [33••], and others that are also thought to play a role in vesicular trafficking (coronin, falcipain 2) [19,21,47]. These studies underline the importance of research into K13 and other potential ART-resistance mediators, particularly in light of the recent emergence of resistance in Africa [24••,48–50].
The spread of mutant K13-driven artemisinin partial resistance
ART partial resistance and causative K13 mutations are now widespread across the GMS, which together with partner-drug resistance, has resulted in high ACT failure rates [1]. Over 200 nonsynonymous K13 mutations have been identified in parasite populations globally, of which 12 are validated to confer ART resistance and 10 are associated with resistance (Table 2) [3,21]. The C580Y allele dominates most of the GMS, with the exception of Myanmar where the F466I allele is more common [51•]. The C580Y allele also emerged independently in Guyana and Papua New Guinea, but at low frequency and with no apparent effect as yet on ACT efficacy [52,53].
Table 2.
Molecular markers of antimalarial drug resistance in P. falciparum.
| Class | Drug | Gene (s) | Mutation |
|---|---|---|---|
| 4-aminoquinolines | Chloroquine | pfcrt | Necessary but not sufficient: K76Ta Other mutations include C72S, M74I, N75E, A220S, Q271E, N326S, I356T, and R371I |
| pfmdrl (in combination with mutant pfcrt only) | N86Y, Y184F, S1034C, N1042D, and D1246Y | ||
| Amodiaquine | pfcrt and pfmdrl | PfMDR1 N86Y and PfCRT K76T contribute to decreased parasite susceptibility | |
| Piperaquine | pm2/3 | Increased copy number | |
| pfcrt | T93S, H97Y, F145I, I218F, M343L, C350R, or G353V | ||
| Pyronaridine | Yet to be identified | - | |
| Amino alcohols | Lumefantrine | Yet to be identified | Selects for PfMDR1 N86 and PfCRT K76 |
| Mefloquine | pfmdrl | Increased copy number | |
| Endoperoxide | Artemisinin derivatives | k13 | Validated: P413A, F446I, N458Y, C469Y, M476I, Y493H, R539T, I543T, P553L, R561H, P574L, C580Y, and A675Vb Candidate or associated: P441L, G449A, C469F, A481V, R515K, P527H, N537I/D, G538V, and R622I |
Chloroquine resistance requires at least four mutations that include K76T.
Validated K13 markers have a statistically significant association between the mutation and delayed clearance as well as survival > 1% in the ring-stage survival assay. Candidate or associated markers fulfill one of these conditions to date.
Initial signs of ART partial resistance are now emerging in sub-Saharan Africa. The K13 mutation R561H, which is a validated marker of resistance in the GMS, emerged independently in Rwanda and has rapidly attained a prevalence of 22% in some sites [24••,50]. R561H was significantly associated with day-3 parasitemia [48] and a longer parasite-clearance half-life following AL treatment [50]. Gene editing has shown that R561H can confer a degree of in vitro ART partial resistance similar to that of C580Y and is fitness-neutral [26], indicating that the Rwandan R561H lineage has the potential to predominate in this region.
A recent study from northern Uganda reported an increased prevalence of the K13 mutations C469Y and A675V, attaining 23% and 41%, respectively [54]. Both mutations were associated with delayed parasite clearance following intravenous artesunate monotherapy [49••]. Although A675V demonstrated ex vivo ART resistance [49••], both C469Y and A675V mediated either no significant shift, or only a modest increase in ring-stage survival in a subsequent gene-editing study [46], highlighting the importance of the parasite genetic background in mutant K13-mediated resistance. Continued surveillance across Africa of K13 polymorphisms, parasite-clearance times, and in vitro partial resistance will be extremely important in the coming years.
P. falciparum chloroquine-resistance transporter and P. falciparum multidrug-resistance protein-1 as modulators of susceptibility to artemisinin combination therapies partner drugs
ASAQ and AS-MQ have been used extensively in the GMS, and high failure rates to both have been reported [3,55•]. Artesunate-pyronaridine (AS-PND), the most recently deployed ACT, shows excellent efficacy against DHA-PPQ-resistant infections [56]. AL is the predominant first-line ACT in Africa, and there is no concrete evidence of lumefantrine (LMF) or pyronaridine (PND) resistance. The 4-aminoquinolines amodiaquine (ADQ) and PND act primarily by inhibiting hemozoin formation in the DV, whereas the arylaminoalcohols mefloquine (MFQ) and LMF only partially inhibit hemozoin formation, with their primary targets thought to be located in the cytosol [21,57].
Resistance to ACT partner drugs is mostly associated with point mutations in the P. falciparum chloroquine-resistance transporter (pfcrt) and P. falciparum multidrug-resistance protein-1 (pfmdr1) genes that encode two DV membrane transporters, or amplification of pfmdr1 (Table 2). These mutations often have opposing effects on susceptibilities to partner drugs. Treatment with ASAQ selects for PfCRT K76T, a mutation critically required for chloroquine (CQ) resistance, and PfMDR1 N86Y, a modulator of CQ resistance, while AL treatment selects for the wild-type alleles [58,59]. In vitro, PfMDR1 N86Y reduces susceptibility to ADQ and PND and increases susceptibility to LMF, MFQ, and DHA [60,61]. Additionally, pfmdr1 amplification is the most important determinant of MFQ resistance in Southeast Asia [62,63].
One endogenous function of PfCRT appears to be proton-dependent transport of hemoglobin-derived peptides out of the DV [64,65], while PfMDR1 is predicted to transport solutes into the DV. Wild-type PfMDR1 has been reported to transport diverse pharmacophores, including LMF, MFQ, DHA, PPQ, ADQ, and CQ, and PfMDR1–86Y reduces transport of these compounds [66•]. In contrast, wild-type PfCRT cannot efflux these compounds, and only drug-resistant PfCRT isoforms transport CQ, LMF, and MFQ [66•]. As a result, mutant isoforms of PfCRT and PfMDR1 are thought to decrease accumulation of antimalarials in the DV and increase their concentration in the cytosol, thereby conferring altered drug susceptibility based on the location of the drug targets (Figure 1).
Figure 1.
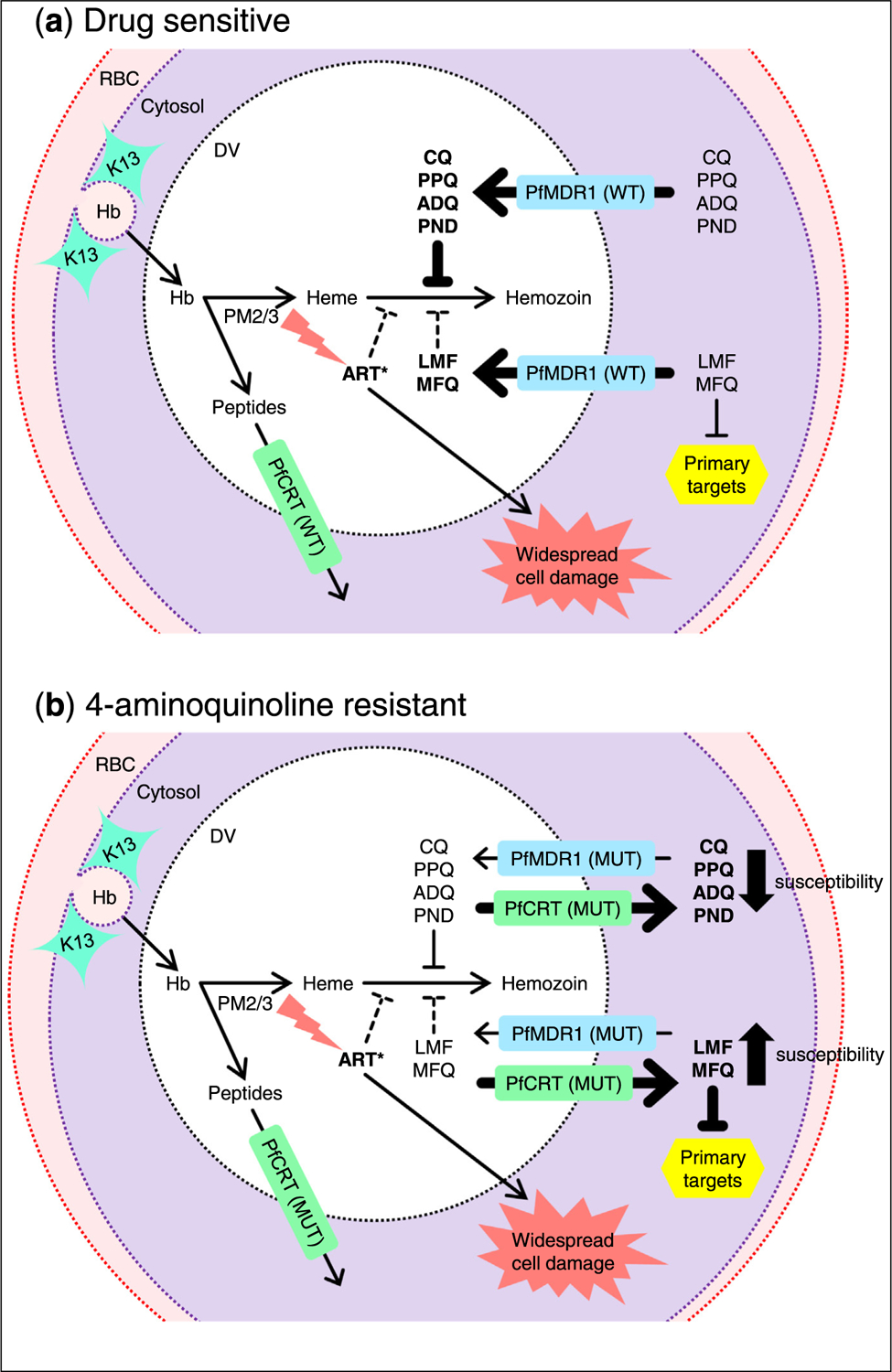
Model of select antimalarial drug modes of action and resistance determinants. (a) K13 is involved in endocytosis of hemoglobin-containing host cytosol and trafficking to the DV, where hemoglobin is degraded by parasite proteases, including plasmepsins 2/3 to release peptides and heme. Fe2+ heme activates ART derivatives via cleavage of their endoperoxide bridge. The 4-aminoquinolines (CQ, PPQ, ADQ, and PND) act primarily by inhibiting biomineralization of toxic heme to inert hemozoin. LMF, MFQ, and ART derivatives partially inhibit hemozoin formation and are thought to act on primary targets in the cytosol. PfMDR1 (WT) transports diverse compounds from the cytosol into the DV. PfCRT (WT) does not mediate drug transport, but transports globin-derived peptide residues from the DV to the parasite cytosol. (b) Mutations in PfMDR1 and PfCRT (MUT) are thought to decrease and confer drug-transport capacity, respectively, reducing drug accumulation in the DV and increasing concentration in the cytosol, resulting in 4-aminoquinoline resistance and increased susceptibility to LMF, MFQ, and ART. Hb, hemoglobin; RBC, red blood cell.
The collateral drug sensitivity of pfcrt and pfmdr1 alleles is reflected in changes of allele frequencies in response to replacement of first-line therapies. The prevalence of multicopy pfmdr1 in the GMS decreased dramatically upon withdrawal of AS-MQ [67,68]. The prevalence of pfcrt-76T and pfmdr1-86Y, which increased in Sub-Saharan Africa under CQ pressure, decreased dramatically from 2004 to 2018 in response to widespread uptake of AL [54,69,70]. Worryingly, ex vivo parasites recently collected from patients in Eastern Uganda had a small but significant decrease in susceptibility to LMF, which was accompanied by a significant increase in the prevalence of pfmdr1-N86 to > 99% [71•]. This suggests an increasing prevalence of LMF-tolerant parasites, which is concerning, given the recent emergence of ART partial resistance in east Africa.
Piperaquine resistance associated with multiple plasmepsins 2/3 and novel mutations in P. falciparum chloroquine-resistance transporter
DHA-PPQ replaced AS-MQ as the first-line ACT in Cambodia in the midst of emerging ART resistance, and within a few years, high treatment-failure rates were reported [3]. Multiple genome-wide association studies identified amplification of plasmepsins 2 and 3 (pm2/3) and novel point mutations in pfcrt as being associated with clinical and in vitro PPQ resistance (Table 2) [21]. Multicopy pm2/3 was associated with increased parasite survival at elevated PPQ concentrations [72]. However, pm2 overexpression alone failed to confer in vitro PPQ resistance [73]. Conversely, gene editing validated that novel PfCRT point mutations can drive high-grade in vitro PPQ resistance, including on a pm2/3 single-copy background [74]. Longitudinal surveillance revealed a rapid increase in the prevalence of these PfCRT mutations following DHA-PPQ implementation, with > 98% of parasites harboring these mutations by 2017 [67]. Outside the GMS, a novel PfCRT variant causal for PPQ resistance emerged independently in French Guiana [75]. In China, emergence of novel PfCRT haplotypes conferring in vitro PPQ resistance may explain early PPQ clinical failures in this region [76].
PfCRT mutations that confer PPQ resistance can partially or fully reverse CQ and ADQ resistance based on the isoform on which they arise [74,76,77]. PPQ, a 4-aminoquinoline, exerts its antimalarial action by inhibiting heme detoxification in the DV [77]. Studies of PfCRT-mediated drug transport demonstrate that CQ-resistant isoforms, which have a large transport capacity for CQ, do not transport PPQ [78]. Conversely, PPQ-resistant isoforms efflux PPQ and reduce CQ transportation, leading to increased CQ accumulation in the DV [76].
Conclusions and future outlook
Recent years have seen a tremendous increase in research into the molecular basis of antimalarial drug resistance in P. falciparum parasites, complementing parallel efforts to identify new drug targets and candidate medicines. These efforts provide hope that within the next decade, we can substantially decrease the burden of disease, especially in high-transmission settings in Africa. However, the recent detection in eastern Africa of mutant K13 parasites is particularly worrying, as it may lead to partner-drug resistance and ACT- treatment failures, as earlier occurred in the GMS in south-east Asia. To date, AL remains broadly effective across Africa. Detailed monitoring for resistance, both genotypically and measuring parasite susceptibility, is essential. Genetic crosses to map genetic determinants of resistance, and gene-editing methods to establish causality, provide powerful tools to characterize resistance [79,80]. Mathematical modeling also provides a key approach to optimize treatment and control measures at a national and subnational level [81–83]. Several new approaches are also being actively explored, including triple ACTs, or drug-rotation strategies with multiple ACTs, which could exploit the opposing selective pressures that partner drugs place on PfCRT and PfMDR1 as mediators of parasite susceptibility [2,84•]. These efforts extend to chemoprevention measures such as seasonal malaria chemoprevention or intermittent preventive treatments, whose efficacy can also be optimized through a detailed understanding of the genetic and molecular basis of antimalarial resistance [85]. The exceptional coordination between scientists in academia and industry, supported by multiple organizations including the Bill & Melinda Gates Foundation, the Medicines for Malaria Venture, the World Health Organization, and governmental agencies and science-based funders, is a key foundation for increasing efforts to regain the upper hand against malaria and dramatically reduce its global impact.
Acknowledgements
This work was supported by the National Institutes of Health [R01 AI109023, AI124678, AI050234] and the Bill & Melinda Gates Foundation, Seattle, WA [INV-006599].
Footnotes
Conflict of interest statement
Authors declare no conflicts of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest.
- 1.World Health Organization. World malaria report 2021. https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2021.
- 2.Dhorda M, Amaratunga C, Dondorp AM: Artemisinin and multidrug-resistant Plasmodium falciparum - a threat for malaria control and elimination. Curr Opin Infect Dis 2021, 34:432–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Report on antimalarial drug efficacy, resistance and response: 10 years of surveillance (2010–2019); 2020. https://www.who.int/publications/i/item/9789240012813.
- 4.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, et al. : Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 2009, 361:455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saunders DL, Vanachayangkul P, Lon C: Dihydroartemisinin-piperaquine failure in Cambodia. N Engl J Med 2014, 371:484–485. [DOI] [PubMed] [Google Scholar]
- 6.Spring MD, Lin JT, Manning JE, Vanachayangkul P, Somethy S, Bun R, Se Y, Chann S, Ittiverakul M, Sia-ngam P, et al. : Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: an observational cohort study. Lancet Infect Dis 2015, 15:683–691. [DOI] [PubMed] [Google Scholar]
- 7.Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, Almagro-Garcia J, Neal AT, Sreng S, Suon S, et al. : Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis 2017, 17:164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witkowski B, Duru V, Khim N, Ross LS, Saintpierre B, Beghain J, Chy S, Kim S, Ke S, Kloeung N, et al. : A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect Dis 2017, 17:174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Pluijm RW, Imwong M, Chau NH, Hoa NT, Thuy-Nhien NT, Thanh NV, Jittamala P, Hanboonkunupakarn B, Chutasmit K, Saelow C, et al. : Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: a prospective clinical, pharmacological, and genetic study. Lancet Infect Dis 2019, 19:952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Pluijm RW, Amaratunga C, Dhorda M, Dondorp AM: Triple artemisinin-based combination therapies for malaria - a new paradigm? Trends Parasitol 2021, 37:15–24. [DOI] [PubMed] [Google Scholar]
- 11.Posner GH, Oh CH, Wang D, Gerena L, Milhous WK, Meshnick SR, Asawamahasadka W: Mechanism-based design, synthesis, and in vitro antimalarial testing of new 4-methylated trioxanes structurally related to artemisinin: the importance of a carbon-centered radical for antimalarial activity. J Med Chem 1994, 37:1256–1258. [DOI] [PubMed] [Google Scholar]
- 12.Heller LE, Roepe PD: Artemisinin-based antimalarial drug therapy: molecular pharmacology and evolving resistance. Trop Med Infect Dis 2019, 4:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang M, Gallego-Delgado J, Fernandez-Arias C, Waters NC, Rodriguez A, Tsuji M, Wek RC, Nussenzweig V, Sullivan WJ Jr: Inhibiting the Plasmodium eIF2α kinase PK4 prevents artemisinin-induced latency. Cell Host Microbe 2017, 22:766–776 e764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bridgford JL, Xie SC, Cobbold SA, Pasaje CFA, Herrmann S, Yang T, Gillett DL, Dick LR, Ralph SA, Dogovski C, et al. : Artemisinin kills malaria parasites by damaging proteins and inhibiting the proteasome. Nat Commun 2018, 9:3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stokes BH, Yoo E, Murithi JM, Luth MR, Afanasyev P, da Fonseca PCA, Winzeler EA, Ng CL, Bogyo M, Fidock DA: Covalent Plasmodium falciparum-selective proteasome inhibitors exhibit a low propensity for generating resistance in vitro and synergize with multiple antimalarial agents. PLoS Pathog 2019, 15:e1007722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Wang C, Oberstaller J, Thomas P, Otto TD, Casandra D, Boyapalle S, Adapa SR, Xu S, Button-Simons K, et al. : The apicoplast link to fever-survival and artemisinin-resistance in the malaria parasite. Nat Commun 2021, 12:4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tinto-Font E, Michel-Todo L, Russell TJ, Casas-Vila N, Conway DJ, Bozdech Z, Llinas M, Cortes A: A heat-shock response regulated by the PfAP2-HS transcription factor protects human malaria parasites from febrile temperatures. Nat Microbiol 2021, 6:1163–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribbiso KA, Heller LE, Taye A, Julian E, Willems AV, Roepe PD: Artemisinin-based drugs target the Plasmodium falciparum heme detoxification pathway. Antimicrob Agents Chemother 2021, 65:e02137–02120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie SC, Ralph SA, Tilley L: K13, the cytostome, and artemisinin resistance. Trends Parasitol 2020, 36:533–544. [DOI] [PubMed] [Google Scholar]
- 20.Zhu P, Zhou B: The antagonizing role of heme in the antimalarial function of artemisinin: elevating intracellular free heme negatively impacts artemisinin activity in Plasmodium falciparum. Molecules 2022, 27:1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siddiqui FA, Liang X, Cui L: Plasmodium falciparum resistance to ACTs: emergence, mechanisms, and outlook. Int J Parasitol Drugs Drug Resist 2021, 16:102–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paloque L, Coppee R, Stokes BH, Gnadig NF, Niare K, Augereau JM, Fidock DA, Clain J, Benoit-Vical F: Mutation in the Plasmodium falciparum BTB/POZ domain of K13 protein confers artemisinin resistance. Antimicrob Agents Chemother 2022, 66:e0132021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, et al. : K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 2015, 347:428–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.••.Uwimana A, Legrand E, Stokes BH, Ndikumana JM, Warsame M, Umulisa N, Ngamije D, Munyaneza T, Mazarati JB, Munguti K, et al. : Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat Med 2020, 26:1602–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]; Provided evidence of the emergence of the K13 R561H mutation in Rwanda. When introduced into a Dd2 background, the mutations R561H and P574L were shown to significantly increase ring-stage survival, with R561H conferring comparable levels of resistance to C580Y. Phylogenetic analysis of R561H parasites suggested de novo emergence of this mutation.
- 25.•.Siddiqui FA, Boonhok R, Cabrera M, Mbenda HGN, Wang M, Min H, Liang X, Qin J, Zhu X, Miao J, et al. : Role of Plasmodium falciparum Kelch 13 protein mutations in P. falciparum populations from Northeastern Myanmar in mediating artemisinin resistance. mBio 2020, 11:e01134–01119. [DOI] [PMC free article] [PubMed] [Google Scholar]; Authors utilized K13 antibodies to localize the protein to punctate structures that partially overlapped the endoplasmic reticulum. This study also evaluated the in vitro ART response phenotype of four K13 mutations identified on the China-Myanmar border and identified N458Y as causal for resistance. Genetic reversion of field isolates with various K13 mutations to the wild-type allele increased their susceptibility to DHA. K13 mutations were also shown to vary significantly in their fitness cost.
- 26.Stokes BH, Dhingra SK, Rubiano K, Mok S, Straimer J, Gnädig NF, Deni I, Schindler KA, Bath JR, Ward KE, et al. : Plasmodium falciparum K13 mutations in Africa and Asia impact artemisinin resistance and parasite fitness. Elife 2021, 10:e66277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mok S, Ashley EA, Ferreira PE, Zhu L, Lin Z, Yeo T, Chotivanich K, Imwong M, Pukrittayakamee S, Dhorda M, et al. : Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science 2015, 347:431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett MP, Kyle DE, Sibley LD, Radke JB, Tarleton RL: Protozoan persister-like cells and drug treatment failure. Nat Rev Microbiol 2019, 17:607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.•.Mok S, Stokes BH, Gnädig NF, Ross LS, Yeo T, Amaratunga C, Allman E, Solyakov L, Bottrill AR, Tripathi J, et al. : Artemisinin-resistant K13 mutations rewire Plasmodium falciparum’s intra-erythrocytic metabolic program to enhance survival. Nat Commun 2021, 12:530. [DOI] [PMC free article] [PubMed] [Google Scholar]; Systems-based quantitative transcriptomics, proteomics, and metabolomics profiling of isogenic K13-edited parasites, revealing impacts of mutant K13 on parasite cell-cycle periodicity, the unfolded protein response, protein degradation, peptide levels, vesicular trafficking, and mitochondrial metabolism. Evokes the possibility that mitochondrial sensing might contribute to drug-induced quiescence and subsequent reinitiation of ART-resistant parasite growth.
- 30.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, Lim P, Mao S, Sopha C, Sam B, et al. : Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis 2013, 13:1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nötzel C, Kafsack BFC: PfD123 modulates K13-mediated survival and recovery after artemisinin exposure. bioRxiv 2022, (2022.2001.2027.476788). [Google Scholar]
- 32.Yang T, Yeoh LM, Tutor MV, Dixon MW, McMillan PJ, Xie SC, Bridgford JL, Gillett DL, Duffy MF, Ralph SA, et al. : Decreased K13 abundance reduces hemoglobin catabolism and proteotoxic stress, underpinning artemisinin resistance. Cell Rep 2019, 29:2917–2928 e2915. [DOI] [PubMed] [Google Scholar]
- 33.••.Birnbaum J, Scharf S, Schmidt S, Jonscher E, Hoeijmakers WAM, Flemming S, Toenhake CG, Schmitt M, Sabitzki R, Bergmann B, et al. : A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science 2020, 367:51–59. [DOI] [PubMed] [Google Scholar]; A seminal article on the mechanism of mutant K13-mediated ART resistance, which employed a BioID approach to identify K13-associated proteins, termed the K13 interactome. Inactivation of several of these proteins was shown to significantly reduce hemoglobin transport to the DV and confer ART resistance. This work established an elegant model for resistance through modulation of clathrin-independent hemoglobin endocytosis.
- 34.Schumann R, Bischoff E, Klaus S, Möhring S, Flock J, Keller S, Remans K, Ganter M, Deponte M: Protein abundance and folding rather than the redox state of Kelch13 determine the artemisinin susceptibility of Plasmodium falciparum. Redox Biol 2021, 48:102177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gnädig NF, Stokes BH, Edwards RL, Kalantarov GF, Heimsch KC, Kuderjavy M, Crane A, Lee MCS, Straimer J, Becker K, et al. : Insights into the intracellular localization, protein associations and artemisinin resistance properties of Plasmodium falciparum K13. PLoS Pathog 2020, 16:e1008482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang X, Boonhok R, Siddiqui FA, Xiao B, Li X, Qin J, Min H, Jiang L, Cui L, Miao J: A leak-free inducible CRISPRi/a system for gene functional studies in Plasmodium falciparum. Microbiol Spectr 2022, 10:e0278221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birnbaum J, Flemming S, Reichard N, Soares AB, Mesén-Ramírez P, Jonscher E, Bergmann B, Spielmann T: A genetic system to study Plasmodium falciparum protein function. Nat Methods 2017, 14:450–456. [DOI] [PubMed] [Google Scholar]
- 38.Bhattacharjee S, Coppens I, Mbengue A, Suresh N, Ghorbal M, Slouka Z, Safeukui I, Tang HY, Speicher DW, Stahelin RV, et al. : Remodeling of the malaria parasite and host human red cell by vesicle amplification that induces artemisinin resistance. Blood 2018, 131:1234–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henrici RC, Edwards RL, Zoltner M, van Schalkwyk DA, Hart MN, Mohring F, Moon RW, Nofal SD, Patel A, Flueck C, et al. : The Plasmodium falciparum artemisinin susceptibility-associated AP-2 adaptin μ subunit is clathrin independent and essential for schizont maturation. mBio 2020, 11:e02918–e02919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simwela NV, Hughes KR, Roberts AB, Rennie MT, Barrett MP, Waters AP: Experimentally engineered mutations in a ubiquitin hydrolase, UBP-1, modulate in vivo susceptibility to artemisinin and chloroquine in Plasmodium berghei. Antimicrob Agents Chemother 2020, 64:e02484–02419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rocamora F, Zhu L, Liong KY, Dondorp A, Miotto O, Mok S, Bozdech Z: Oxidative stress and protein damage responses mediate artemisinin resistance in malaria parasites. PLoS Pathog 2018, 14:e1006930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egwu CO, Pério P, Augereau JM, Tsamesidis I, Benoit-Vical F, Reybier K: Resistance to artemisinin in falciparum malaria parasites: a redox-mediated phenomenon. Free Radic Biol Med 2022, 179:317–327. [DOI] [PubMed] [Google Scholar]
- 43.Xiong A, Prakash P, Gao X, Chew M, Tay IJJ, Woodrow CJ, Engelward BP, Han J, Preiser PR: K13-mediated reduced susceptibility to artemisinin in Plasmodium falciparum is overlaid on a trait of enhanced DNA damage repair. Cell Rep 2020, 32:107996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paloque L, Ramadani AP, Mercereau-Puijalon O, Augereau JM, Benoit-Vical F: Plasmodium falciparum: multifaceted resistance to artemisinins. Malar J 2016, 15:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Connelly SV, Manzella-Lapeira J, Levine ZC, Brzostowski J, Krymskaya L, Rahman RS, Ellis AC, Amin SN, Sa JM, Wellems TE: Restructured mitochondrial-nuclear interaction in Plasmodium falciparum dormancy and persister survival after artemisinin exposure. mBio 2021, 12:e0075321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stokes BH, Ward KE, Fidock DA: Evidence of artemisinin-resistant malaria in Africa. N Engl J Med 2022, 386:1385–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma AI, Shin SH, Bopp S, Volkman SK, Hartl DL, Wirth DF: Genetic background and PfKelch13 affect artemisinin susceptibility of PfCoronin mutants in Plasmodium falciparum. PLoS Genet 2020, 16:e1009266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uwimana A, Umulisa N, Venkatesan M, Svigel SS, Zhou Z, Munyaneza T, Habimana RM, Rucogoza A, Moriarty LF, Sandford R, et al. : Association of Plasmodium falciparum kelch13 R561H genotypes with delayed parasite clearance in Rwanda: an open-label, single-arm, multicentre, therapeutic efficacy study. Lancet Infect Dis 2021, 21:1120–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.••.Balikagala B, Fukuda N, Ikeda M, Katuro OT, Tachibana SI, Yamauchi M, Opio W, Emoto S, Anywar DA, Kimura E, et al. : Evidence of artemisinin-resistant malaria in Africa. N Engl J Med 2021, 385:1163–1171. [DOI] [PubMed] [Google Scholar]; This study associated the K13 mutations A675V and C469Y with delayed parasite clearance following artesunate monotherapy in northern Uganda. Ex vivo assessment confirmed a significant increase in the survival of DHA-exposed early ring stages in isolates carrying the A675V mutation. Genotyping of resistant parasites suggested the local emergence of these mutations.
- 50.Straimer J, Gandhi P, Renner KC, Schmitt EK: High prevalence of Plasmodium falciparum K13 mutations in Rwanda is associated with slow parasite clearance after treatment with artemether-lumefantrine. J Infect Dis 2021, 225:1411–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.•.Imwong M, Dhorda M, Myo Tun K, Thu AM, Phyo AP, Proux S, Suwannasin K, Kunasol C, Srisutham S, Duanguppama J, et al. : Molecular epidemiology of resistance to antimalarial drugs in the Greater Mekong subregion: an observational study. Lancet Infect Dis 2020, 20:1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]; Comprehensive molecular epidemiology study describing the evolution and spread of antimalarial drug resistance across the GMS from 2007 to 2018. Details the emergence and establishment of separate dominant ART-resistant lineages in Myanmar (with K13 F446I) and the eastern GMS (K13 C580Y), with no evidence for the spread of PPQ resistance between the two regions.
- 52.Mathieu LC, Cox H, Early AM, Mok S, Lazrek Y, Paquet JC, Ade MP, Lucchi NW, Grant Q, Udhayakumar V, et al. : Local emergence in Amazonia of Plasmodium falciparum k13 C580Y mutants associated with in vitro artemisinin resistance. Elife 2020, 9:e51015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miotto O, Sekihara M, Tachibana SI, Yamauchi M, Pearson RD, Amato R, Gonçalves S, Mehra S, Noviyanti R, Marfurt J, et al. : Emergence of artemisinin-resistant Plasmodium falciparum with kelch13 C580Y mutations on the island of New Guinea. PLoS Pathog 2020, 16:e1009133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asua V, Conrad MD, Aydemir O, Duvalsaint M, Legac J, Duarte E, Tumwebaze P, Chin DM, Cooper RA, Yeka A, et al. : Changing prevalence of potential mediators of aminoquinoline, antifolate, and artemisinin resistance across Uganda. J Infect Dis 2021, 223:985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.•.Mairet-Khedim M, Leang R, Marmai C, Khim N, Kim S, Ke S, Kauy C, Kloeung N, Eam R, Chy S, et al. : Clinical and in vitro resistance of Plasmodium falciparum to artesunate-amodiaquine in Cambodia. Clin Infect Dis 2021, 73:406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]; Report of clinical resistance to artesunate-amodiaquine in Cambodia, which was significantly associated with in vitro resistance in a novel amodiaquine parasite survival assay. Resistance was not associated with previous molecular markers of reduced ADQ susceptibility (PfMDR1 86Y and PfCRT 76T).
- 56.Manh ND, Thanh NV, Quang HH, Van NTT, San NN, Phong NC, Birrell GW, Edstein MD, Edgel KA, Martin NJ, et al. : Pyronaridine-artesunate (Pyramax) for treatment of artemisinin- and piperaquine-resistant Plasmodium falciparum in the central highlands of Vietnam. Antimicrob Agents Chemother 2021, 65:e0027621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong W, Bai XC, Sleebs BE, Triglia T, Brown A, Thompson JK, Jackson KE, Hanssen E, Marapana DS, Fernandez IS, et al. : Mefloquine targets the Plasmodium falciparum 80S ribosome to inhibit protein synthesis. Nat Microbiol 2017, 2:17031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maiga H, Grivoyannis A, Sagara I, Traore K, Traore OB, Tolo Y, Traore A, Bamadio A, Traore ZI, Sanogo K, et al. : Selection of pfcrt K76 and pfmdr1 N86 coding alleles after uncomplicated malaria treatment by artemether-lumefantrine in Mali. Int J Mol Sci 2021, 22:6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beavogui AH, Diawara EY, Cherif MS, Delamou A, Diallo N, Traore A, Millimouno P, Camara D, Sylla MM, Toure AA, et al. : Selection of PfCRT 76T and PfMDR1 86Y mutant Plasmodium falciparum after treatment of uncomplicated malaria with artesunate-amodiaquine in Republic of Guinea. J Parasitol 2021, 107:778–782. [DOI] [PubMed] [Google Scholar]
- 60.Veiga MI, Dhingra SK, Henrich PP, Straimer J, Gnädig N, Uhlemann AC, Martin RE, Lehane AM, Fidock DA: Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun 2016, 7:11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Madamet M, Briolant S, Amalvict R, Benoit N, Bouchiba H, Cren J, Pradines B: The Plasmodium falciparum chloroquine resistance transporter is associated with the ex vivo P. falciparum African parasite response to pyronaridine. Parasit Vectors 2016, 9:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, et al. : Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet 2004, 364:438–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calçada C, Silva M, Baptista V, Thathy V, Silva-Pedrosa R, Granja D, Ferreira PE, Gil JP, Fidock DA, Veiga MI: Expansion of a specific Plasmodium falciparum PfMDR1 haplotype in Southeast Asia with increased substrate transport. mBio 2020, 11:e02093–02020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shafik SH, Cobbold SA, Barkat K, Richards SN, Lancaster NS, Llinás M, Hogg SJ, Summers RL, McConville MJ, Martin RE: The natural function of the malaria parasite’s chloroquine resistance transporter. Nat Commun 2020, 11:3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanchez CP, Manson EDT, Moliner Cubel S, Mandel L, Weidt SK, Barrett MP, Lanzer M: The knock-down of the chloroquine resistance transporter PfCRT is linked to oligopeptide handling in Plasmodium falciparum. Microbiol Spectr 2022, Jul 18:65, 10.1128/spectrum.01101-22 In press e0110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.•.Shafik SH, Richards SN, Corry B, Martin RE: Mechanistic basis for multidrug resistance and collateral drug sensitivity conferred to the malaria parasite by polymorphisms in PfMDR1 and PfCRT. PLoS Biol 2022, 20:e3001616. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors provide evidence, using a Xenopus oocyte expression system, that wild-type allele PfMDR1 transports DHA and several ACT partner drugs and that mutant isoforms that augment CQ resistance have reduced transport. CQ-resistant PfCRT isoforms were found to transport CQ as well as LMF and MFQ (the latter two displayed a 10-fold lower capacity compared with CQ; PPQ was not tested).
- 67.Shrestha B, Shah Z, Morgan AP, Saingam P, Chaisatit C, Chaorattanakawee S, Praditpol C, Boonyalai N, Lertsethtakarn P, Wojnarski M, et al. : Distribution and temporal dynamics of Plasmodium falciparum chloroquine resistance transporter mutations associated with piperaquine resistance in northern Cambodia. J Infect Dis 2021, 224:1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Imwong M, Suwannasin K, Srisutham S, Vongpromek R, Promnarate C, Saejeng A, Phyo AP, Proux S, Pongvongsa T, Chea N, et al. : Evolution of multidrug resistance in Plasmodium falciparum: a longitudinal study of genetic resistance markers in the Greater Mekong Subregion. Antimicrob Agents Chemother 2021, 65:e0112121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ehrlich HY, Bei AK, Weinberger DM, Warren JL, Parikh S: Mapping partner drug resistance to guide antimalarial combination therapy policies in sub-Saharan Africa. Proc Natl Acad Sci USA 2021, 118:e2100685118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ehrlich HY, Jones J, Parikh S: Molecular surveillance of antimalarial partner drug resistance in sub-Saharan Africa: a spatial-temporal evidence mapping study. Lancet Microbe 2020, 1:e209–e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.•.Tumwebaze PK, Katairo T, Okitwi M, Byaruhanga O, Orena S, Asua V, Duvalsaint M, Legac J, Chelebieva S, Ceja FG, et al. : Drug susceptibility of Plasmodium falciparum in eastern Uganda: a longitudinal phenotypic and genotypic study. Lancet Microbe 2021, 2:e441–e449. [DOI] [PMC free article] [PubMed] [Google Scholar]; Longitudinal study of P. falciparum isolates collected from Eastern Uganda, which showed sustained susceptibility to ten antimalarial drugs including DHA and ACT partner drugs. Lumefantrine was noted to show a small but significant decrease in susceptibility. Frequencies of pfmdr1 and pfcrt wild-type allele significantly increased in the last decade, demonstrating their selection in response to widespread use of artemether-lumefantrine.
- 72.Bopp S, Magistrado P, Wong W, Schaffner SF, Mukherjee A, Lim P, Dhorda M, Amaratunga C, Woodrow CJ, Ashley EA, et al. : Plasmepsin II-III copy number accounts for bimodal piperaquine resistance among Cambodian Plasmodium falciparum. Nat Commun 2018, 9:1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silva M, Calçada C, Teixeira M, Veiga MI, Ferreira PE: Multigenic architecture of piperaquine resistance trait in Plasmodium falciparum. Lancet Infect Dis 2020, 20:26–27. [DOI] [PubMed] [Google Scholar]
- 74.Ross LS, Dhingra SK, Mok S, Yeo T, Wicht KJ, Kümpornsin K, Takala-Harrison S, Witkowski B, Fairhurst RM, Ariey F, et al. : Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat Commun 2018, 9:3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pelleau S, Moss EL, Dhingra SK, Volney B, Casteras J, Gabryszewski SJ, Volkman SK, Wirth DF, Legrand E, Fidock DA, et al. : Adaptive evolution of malaria parasites in French Guiana: reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc Natl Acad Sci USA 2015, 112:11672–11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Small-Saunders JL, Hagenah LM, Wicht KJ, Dhingra SK, Deni I, Kim J, Vendome J, Gil-Iturbe E, Roepe PD, Mehta M, et al. : Evidence for the early emergence of piperaquine-resistant Plasmodium falciparum malaria and modeling strategies to mitigate resistance. PLoS Pathog 2022, 18:e1010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dhingra SK, Redhi D, Combrinck JM, Yeo T, Okombo J, Henrich PP, Cowell AN, Gupta P, Stegman ML, Hoke JM, et al. : A variant PfCRT isoform can contribute to Plasmodium falciparum resistance to the first-line partner drug piperaquine. mBio 2017, 8:e00303–e00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim J, Tan YZ, Wicht KJ, Erramilli SK, Dhingra SK, Okombo J, Vendome J, Hagenah LM, Giacometti SI, Warren AL, et al. : Structure and drug resistance of the Plasmodium falciparum transporter PfCRT. Nature 2019, 576:315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Button-Simons KA, Kumar S, Carmago N, Haile MT, Jett C, Checkley LA, Kennedy SY, Pinapati RS, Shoue DA, McDew-White M, et al. : The power and promise of genetic mapping from Plasmodium falciparum crosses utilizing human liver-chimeric mice. Commun Biol 2021, 4:734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okombo J, Kanai M, Deni I, Fidock DA: Genomic and genetic approaches to studying antimalarial drug resistance and Plasmodium biology. Trends Parasitol 2021, 37:476–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tun STT, von Seidlein L, Pongvongsa T, Mayxay M, Saralamba S, Kyaw SS, Chanthavilay P, Celhay O, Nguyen TD, Tran TN, et al. : Towards malaria elimination in Savannakhet, Lao PDR: mathematical modelling driven strategy design. Malar J 2017, 16:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Camponovo F, Ockenhouse CF, Lee C, Penny MA: Mass campaigns combining antimalarial drugs and anti-infective vaccines as seasonal interventions for malaria control, elimination and prevention of resurgence: a modelling study. BMC Infect Dis 2019, 19:920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao B, Saralamba S, Lubell Y, White LJ, Dondorp AM, Aguas R: Determinants of MDA impact and designing MDAs towards malaria elimination. Elife 2020, 9:e51773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.•.van der Pluijm RW, Tripura R, Hoglund RM, Pyae Phyo A, Lek D, Ul Islam A, Anvikar AR, Satpathi P, Satpathi S, Behera PK, et al. : Triple artemisinin-based combination therapies versus artemisinin-based combination therapies for uncomplicated Plasmodium falciparum malaria: a multicentre, open-label, randomised clinical trial. Lancet 2020, 395:1345–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]; Results from this large multicenter, open-label, randomized trial provide evidence that dihydroartemisinin-piperaquine plus mefloquine, and artemether-lumefantrine plus amodiaquine as triple ACTs are efficacious, well tolerated, and safe treatments of uncomplicated P. falciparum malaria, including in areas with artemisinin and ACT partner-drug resistance.
- 85.Plowe CV: Malaria chemoprevention and drug resistance: a review of the literature and policy implications. Malar J 2022, 21:104. [DOI] [PMC free article] [PubMed] [Google Scholar]


