COVID-19 remains a significant driver of healthcare utilisation. Despite widespread immunity from natural infection and vaccination severe disease and adverse outcomes are still observed, albeit at a reduced frequency. Identifying individuals ‘at risk’ of deterioration at the point of presentation is challenging, however vital to inform decision making around the setting, intensity and escalation of care.
The COVID-19 SeverityTM assay (C19-SA) (MeMed, Israel) is a novel near-patient in vitro diagnostic designed for integration into acute care pathways. Using 0.1mL serum the assay quantifies the concentration of three host-released immune proteins (tumour necrosis factor-related apoptosis inducing-ligand, TRAIL; C-reactive protein, CRP; and interferon gamma-induced protein 10, IP-10) and integrates them via a proprietary algorithm1 , 2 in 15 minutes. A numerical score from 0-100 is released to the clinician: 0 representing a low likelihood and 100 a high likelihood of severe outcome.
To date, the C19-SA score has been evaluated in only one unpublished study of 394 patients with COVID-193. Conducted in Germany, Israel and the USA during the first (wild-type) wave of SARS-CoV-2, the score was predictive of severe disease (area under receiver operator curve, AUROC 0.86, 95% confidence interval [CI] 0.81-0.91), displaying superior performance to IL-6 (AUROC 0.77) and CRP (AUROC 0.78) alone. The utility of the assay in the context of modern variants and a population with established immunity is unknown.
Using a similar approach to our prior analysis of serum calprotectin4, we sought to determine the ability of the C19-SA to independently predict a future adverse clinical outcome defined as requirement of non-invasive ventilation (NIV), admission to an intensive care unit (ICU), or death within 4 weeks of presentation to secondary care in patients diagnosed with COVID-19, including assessment of the value of individual components of the composite score (TRAIL, IP-10, CRP).
Between 10th January to 3rd March 2022, we collected remnant serum from the initial blood draw of all patients presenting to the John Radcliffe Hospital, Oxford University NHS Foundation Trust, via the emergency department (ED), ambulatory assessment unit (AAU) or emergency assessment unit (EAU). Samples were then retrospectively analysed on the C19-SA if a positive SARS-CoV-2 PCR was obtained and their presentation was consistent with acute, symptomatic COVID-19 infection (via independent clinical review, see Supplementary Figure 1). Clinicians were blinded to the C19-SA results as well as the individual biomarker concentrations. The Infections in Oxford Research Database (IORD) (Research Ethics Committee and Confidentiality Group Approval: 19/SC/0403, 19/CAG/0144) was used to retrospectively identify individuals and hospital episodes with a C19-SA score to extract relevant demographic, clinical, biochemical and outcome information in an anonymised manner. All analyses were performed in R (version 4.0.2).
In total 52 patients were included. Of these, 6 individuals reached the composite endpoint (3 required NIV, 2 died and 1 individual was admitted onto ICU and subsequently died).
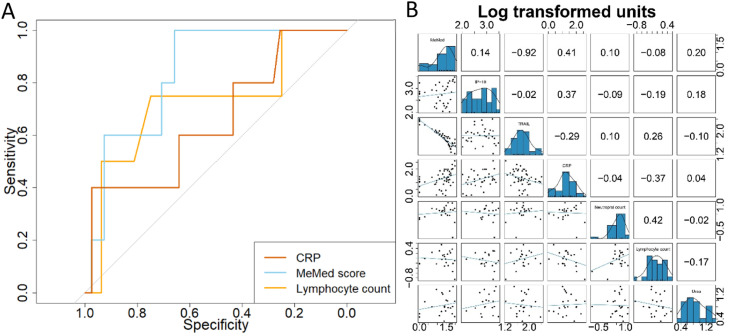
Univariate associations with risk of deterioration were observed for a limited number of variables (Table 1 ). The median value of C19-SA score at presentation in those who deteriorated was 54.5 (IQR 36.8-73), significantly higher than in those who did not (23, IQR 8-42). Oxygen saturation and IP-10 at time of presentation, but not TRAIL, CRP, Glasgow Coma Scale, Body Mass Index, or Charlson comorbidity index were associated with risk of deterioration. The C19-SA score had an AUROC estimate of 0.80 (95% CI 0.63-0.96) compared to 0.71 (0.46-0.96) for CRP (Fig. 1 A). Correlation between the C19-SA score and commonly used biomarkers of disease severity (CRP rho=0.41; lymphocyte count rho=-0.08) was low (Fig. 1B). TRAIL exhibited the strongest correlation with the overall C19-SA score (rho=-0.92).
Table 1.
Characteristics of patients with a MeMed COVID-19 Severity™ assay score available at presentation to AAU, EAU or ED during January to March 2022. Univariate P-values were calculated using the Mann-Whitney-U test for continuous variables and Fisher's Exact Test for nominal variables.
| Characteristic | COVID-19 diagnosis (n=52) | COVID-19 with no outcome (n=46) | COVID-19 with outcome (n=6) | Evidence of difference between outcome groups (P) |
|---|---|---|---|---|
| Demographics | ||||
| Age at presentation in years | 66 (50-80) | 64 (48-80) | 68 (64-85) | NS |
| Female sex | 23 (44) | 19 (41) | 4 (67) | NS |
| BMI | 28 (24-32) | 28 (24-31) | 35 (26-36) | NS |
| Charlson comorbidity index | 3 (0-13) | 0 (0-12) | 6 (4-15) | NS |
| Index of multiple deprivation | 16 (8-21) | 16 (8-22) | 17 (15-18) | NS |
| Clinical measures at presentation | ||||
| Supplemental oxygen number | 14 (27) | 11 (24) | 3 (50) | NS |
| Respiratory rate | 19 (18-22) | 19 (18-21) | 21 (18-24) | NS |
| Oxygen saturation | 97 (96-99) | 98 (96-99) | 95 (93-95) | ⁎⁎ |
| Glasgow coma scale | 15 (15-15) | 15 (15-15) | 15 (15-15) | NS |
| Outcomes | ||||
| Death | 3 (6) | - | 3 (50) | NA |
| Requiring NIV | 3 (6) | - | 3 (50) | NA |
| ICU Admission | 1 (2) | - | 1 (17) | NA |
| Biomarkers | ||||
| Neutrophil count (x10*9/L) | 4.9 (2.6-6.4) | 4.8 (2.6-6.4) | 4.0 (2.0-7.5) | NS |
| Lymphocytes count (x10*9/L) | 0.9 (0.6-1.6) | 1.1 (0.6-1.7) | 0.6 (0.4-0.8) | NS |
| Urea (mmol/L) | 6.3 (4.1-10.1) | 6.0 (3.7-10.2) | 6.4 (6.1-9.8) | NS |
| C-reactive protein (mg/L) | 18.7 (10.3-48.4) | 17.4 (7.8-40.0) | 61.7 (17.9-102.9) | NS |
| MeMed score, (units) | 26.0 (9.5-46.3) | 23.0 (8.0-42.0) | 54.5 (36.8-73.0) | * |
| IP-10, (pg/ml) | 656.5 (235.0-1273.8) | 522.0 (235.0-908.0) | 1696.0 (1199.0-2028.0) | * |
| TRAIL, (mg/L) | 57.0 (39.0-84.5) | 58.0 (39.0-86.5) | 54.5 (40.8-62.3) | NS |
Continuous variables are represented with median and IQR, nominal variables with frequency and column percentage (of valid cases).
NS: non-significant
P < 0.05.
P < 0.01.
NA: not applicable.
Fig. 1.
Biomarkers to predict the risk of deterioration of patients presenting to ambulatory care and A&E with COVID-19. A) The receiver operating curve (ROC) distributions for two biomarkers and the MeMed COVID-19 Severity™ assay measured in 52 patients with confirmed COVID-19. ROC analyses were undertaken using the pROC package and the comparison of the paired ROC statistics was calculated using DeLong's test5. B) The Spearman rank rho correlation between the log10-transformed distributions of the predominant biomarkers associated with deterioration in COVID-19, measured in 52 patients with confirmed COVID-19.
In multivariate analysis, the addition of the C19-SA score to a basic risk of deterioration model (age+sex) led to an incremental increase in predictive value, however this could be replicated via substitution with the lymphocyte count (Supplementary Table 1). Employing the C19-SA score in the full risk model, as opposed to CRP alone or the lymphocyte count, again afforded only limited improvement (C-statistic 0.96 (95% CI 0.96-1) vs 0.91 [0.89-1] and 0.92 [0.88-1] respectively).
We conclude that in this cohort of patients presenting to a tertiary hospital during the Omicron surge of COVID-19 in the UK, the MeMed C19-SA score alone could predict deterioration, as defined by a composite endpoint. In multivariate risk models it however demonstrated limited additive predictive value over traditional and routinely available biomarkers including CRP and the lymphocyte count.
Our findings agree with a previous evaluation of the C19-SA3, although suggest a lower overall predictive ability (AUROC 0.8 vs 0.86). This may reflect our smaller sample size, more diverse cohort including those not admitted to hospital, or the reduced frequency in adverse outcomes with modern variants and prevalent immunity. The data is also in accordance with studies investigating the C19-SA's component biomarkers. Elevated IP-10 has been reported in patients with COVID-19 and is associated with ICU admission and mortality, whereas TRAIL exhibits an inverse correlation between length of hospital and ICU stay6 , 7. Alternative studies have demonstrated the predictive capability of IP-10 and TRAIL at day 3 of hospitalisation, however not on presentation8. Whilst we observed significant elevation in IP-10 in patients experiencing an adverse outcome (1696 pg/mL vs 522 pg/mL) but no difference in TRAIL (54.4 mg/L vs 58 mg/L), the high correlation between TRAIL and the overall C19-SA suggests a meaningful contribution.
Given its size, our study is at risk of type II error and further work evaluating the C19-SA is required to clarify its place and value in different care settings. Whilst the double-blind design minimises risk of selection bias and has previously demonstrated the ability to observe statistically significant associations between biomarkers and clinical outcomes4, serum may not have been available from all eligible patients (no sample, insufficient volume). We believe this was a random effect and does not introduce systematic bias. Finally, it should be noted that the data relates to patients infected with currently circulating variants, includes those attending lower acuity services (e.g. same day emergency care) and patients not requiring hospitalisation.
Declaration of Competing Interest
This was an investigator-led study and MeMed had no control over the study design, data analysis or interpretation. MeMed reviewed the study design, result and interpretation and made no changes prior to submission. Reagents for MeMed COVID-19 Severity™ measures were provided by MeMed, no financial support was received aside from this. All authors declare no conflicts of interest.
Acknowledgement
We thank the healthcare professionals for contributing to this study. We thank the people of Oxfordshire who contribute to the Infections in Oxfordshire Research Database and Jack Cregan for managing the data extracts.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2023.01.017.
Appendix. Supplementary materials
References
- 1.Oved K, Cohen A, Boico O, Navon R, Friedman T, Etshtein L, et al. A novel host-proteome signature for distinguishing between acute bacterial and viral infections. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hainrichson M, Avni N, Eden E, Feigin P, Gelman A, Halabi S, et al. A point-of-need platform for rapid measurement of a host-protein score that differentiates bacterial from viral infection: Analytical evaluation. Clin Biochem. 2022 Apr 26 doi: 10.1016/j.clinbiochem.2022.04.012. S0009-9120(22)00115-1. [DOI] [PubMed] [Google Scholar]
- 3.Mastboim NS, Angel A, Shaham O, Ber TI, Navon R, Simon E, et al. 2021. An immune-protein signature combining TRAIL, IP-10 and CRP for accurate prediction of severe COVID-19 outcome. medRxiv doi: 10.1101/2021.06.27.21259196. [DOI] [PMC free article] [PubMed]
- 4.Mentzer AJ, James T, Yongya M, Cox S, Paddon K, Shine B, et al. Serum calprotectin is not an independent predictor of severe COVID-19 in ambulatory adult patients. J Infect. 2022 Feb;84(2):e27–e29. doi: 10.1016/j.jinf.2021.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011 Mar 17;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tegethoff SA, Danziger G, Kühn D, Kimmer C, Adams T, Heintz L, et al. TNF-related apoptosis-inducing ligand, interferon gamma-induced protein 10, and C-reactive protein in predicting the progression of SARS-CoV-2 infection: a prospective cohort study. Int J Infect Dis. 2022 May 25;122:178–187. doi: 10.1016/j.ijid.2022.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lev S, Gottesman T, Sahaf Levin G, Lederfein D, Berkov E, Diker D, et al. Observational cohort study of IP-10’s potential as a biomarker to aid in inflammation regulation within a clinical decision support protocol for patients with severe COVID-19. PLoS One. 2021;16(1) doi: 10.1371/journal.pone.0245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sophonsri A, Le D, Lou M, Ny P, Minejima E, Chambliss AB, et al. Temporal dynamics of host immune response associated with disease severity and time to recovery in patients hospitalized for COVID-19. Critical Care Explor. 2022;4(9):e0760. doi: 10.1097/CCE.0000000000000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.