Abstract
While receptor binding is well recognized as a factor in influenza-A virus (IAV) and coronavirus (CoV) host adaptation, the role of viral glycoprotein cleavage has not been studied in detail so far. Interestingly, recent studies suggest that host species may differ in their protease repertoire available for cleavage. Furthermore, it was shown for certain bat-derived CoVs that proteolytic activation provides a critical barrier to infect human cells. Understanding the role of glycoprotein cleavage in different species and how IAV and CoVs adapt to a new protease repertoire may allow evaluating the zoonotic potential and risk posed by these viruses. Here, we summarize the current knowledge on the emergence of a multibasic cleavage site (CS) in the glycoproteins of IAVs and CoVs in different host species. Additionally, we discuss the role of transmembrane serine protease 2 (TMPRSS2) in virus activation and entry and a role of neuropilin-1 in acquisition of a multibasic CS in different hosts.
Current Opinion in Virology 2023, 58:101303
This review comes from a themed issue on Adaptation of viruses to new host
Edited by Silke Stertz and Xander de Haan
For complete overview about the section, refer “Adaptation of viruses to new host (2023)”
https://doi.org/10.1016/j.coviro.2023.101303
1879–6257/© 2023 Elsevier B.V. All rights reserved.
Introduction
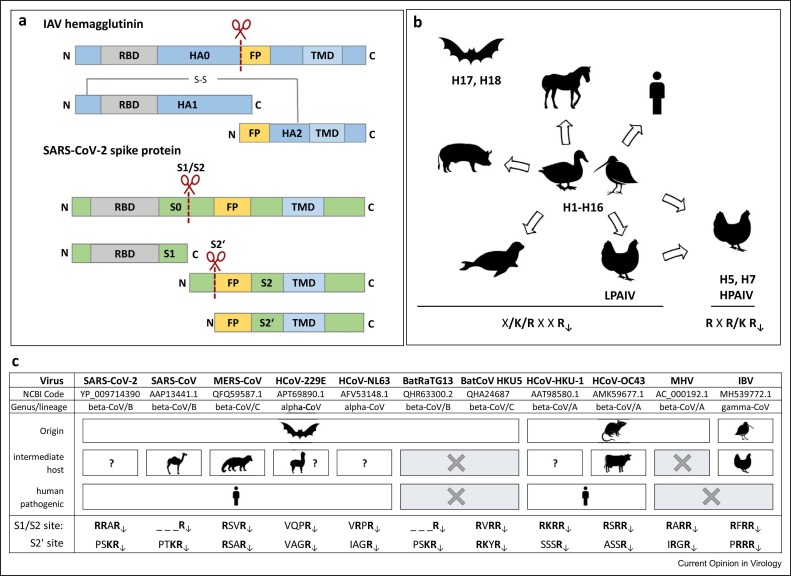
Influenza-A viruses (IAVs) and coronaviruses (CoVs) are pathogens with a broad host spectrum ( Figure 1b, c) and a significant potential for zoonotic transmission. Both IAVs and CoVs are enveloped viruses and possess the major surface glycoproteins hemagglutinin (HA) and spike (S), respectively, that initiate infection by facilitating receptor binding and fusion of viral and cellular membranes 1, 2. HA and S are class-I viral fusion proteins and are synthesized as inactive precursors that must be cleaved post-translationally by a host cell protease to gain their fusion capacity. Cleavage exposes the fusion peptide (FP) and is essential for virus infectivity 2, 3, 4. The IAV HA has to be cleaved at one cleavage site (CS) to be primed for membrane fusion (Figure 1a). Proteolytic cleavage of CoV S has been a puzzling question, but it is now appreciated that CoV S must be proteolytically primed to mediate membrane fusion and several (if not all) CoV S proteins require sequential processing at two sites, S1/S2 and S2′ (Figure 1a, c) 2, 5, 6.
Figure 1.
Cleavage of IAV and CoV fusion proteins and CS motifs common in different host species. (a) IAV HA and CoV S protein are synthesized as precursors and need to be cleaved by host cell proteases to gain their fusion capacity. HA is cleaved at one distinct site immediately upstream of the FP, while S needs to be processed successively at two sites, S1/S2 and S2′, to expose the FP. RBD: receptor-binding domain, TMD: transmembrane domain. (b) Wild aquatic birds are the natural reservoir of IAV subtypes H1–H16 and provide a source of zoonotic transmission to a broad range of avian and mammalian hosts. Genome sequences of subtypes H17 and H18 have only been found in bats. LPAIV and mammalian IAVs possess a monobasic HA CS (R↓). LPAIV can convert into HPAIV via acquisition of a multibasic CS (RXR/KR↓) in poultry. (c) CoV are classified into four genera: alpha, beta, gamma, and delta CoV [75]. Beta-CoV are further divided into four lineages (A–D). CoVs are found in diverse mammalian and avian animal species. Bat and rodent CoV are suggested to serve as sources of alpha- and beta-CoVs, while wild bird CoV are sources of gamma and delta CoVs. The table compares the S1/S2 and S2′ CS motifs of human-pathogenic CoV originating from bats or rodents via potential intermediate hosts with different animal-derived CoV. MHV: murine hepatitis virus.
Single arginine or lysine residues designated as monobasic CS (R/K↓) are cleaved by trypsin-like proteases. Most IAVs, including low pathogenic avian influenza-A viruses (LPAIVs) and human IAVs, possess a monobasic CS. The type-II transmembrane serine protease 2 (TMPRSS2) has been identified as major IAV-activating protease in human airway cells [7]. In contrast, multibasic CSs of the consensus sequence R–X–R/K–R↓ are processed by ubiquitously expressed furin and related proprotein convertases. Highly pathogenic avian influenza-A viruses (HPAIVs) are activated at a multibasic CS, supporting systemic spread in poultry 8, 9.
Priming of CoV S is more complex compared with IAV HA but also more flexible and two CSs appear to offer more possibilities for additional proteases. CoVs show a high variety in CS motifs with different combinations of mono-, di-, and multibasic motifs at the S1/S2 and S2‘ sites (Figure 1c) [2]. It is believed that mono- (and most likely dibasic) motifs are cleaved by TMPRSS2 and related proteases, whereas multibasic CSs are processed by furin. However, lack of expression of TMPRSS2 and other appropriate trypsin-like proteases in cell cultures enables CoVs to use an alternative entry route via the late endosome facilitating S cleavage by endosomal cathepsins 10, 11. Additionally, metalloproteases, including ADAM10, ADAM17, MMP-2, and MMP-9, have been shown to activate severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) S in vitro 11, 12, 13, 14.
Not much attention was drawn on the role of viral glycoprotein cleavage in adaptation of IAVs and CoVs to a new host until recently. In general, it is assumed that orthologous proteases support virus activation in different host species and, therefore, no adaptation is necessary. TMPRSS2 from chicken, duck, swine, and nonhuman primates has been shown to support proteolytic activation of IAV in vitro 15•, 16, 17, 18. However, expression levels and tissue distribution as well as substrate specificity of TMPRSS2 have not been investigated in these species and its role in IAV activation remains to be demonstrated. Even less is known about furin orthologs in different species. It is believed that the important physiological role of furin leads to a highly conserved expression and activity in vertebrates and, therefore, also in virus activation. Studies in fruit bat cells, however, revealed that differences in subcellular localization or activity of furin orthologs may exist [19]. With the emergence of SARS-CoV-2, it has become more recognized that viral glycoprotein cleavage may play a role in CoV transmission and adaptation to new host species, primarily due to the fact that SARS-CoV-2 has acquired a multibasic S1/S2 CS that is not found in the closely related SARS-CoV and BatRaTG13 (Figure 1c) 20, 21.
In this review, we focus on the current knowledge on the role of TMPRSS2 in IAV and CoV activation and beyond, the prevalence and emergence of a multibasic CS in IAV and CoVs in different host species, and neuropilin-1 (NRP1) as a host factor that may be involved in the emergence of a multibasic CS and tissue spreading.
Transmembrane serine protease 2: virus activation and beyond
TMPRSS2 was identified as IAV HA-cleaving protease in 2006 and since then has been shown to activate the fusion proteins of a broad number of respiratory viruses, including influenza-B virus, human metapneumovirus, and various CoVs at monobasic CSs [4]. Lack of TMPRSS2 expression prevents multicycle replication and pathogenesis of IAVs as well as SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), and SARS-CoV-2 in mice 22, 23, 24, 25. This demonstrates that TMPRSS2 provides a promising drug target for therapeutic treatment [26]. Interestingly, multicycle replication of a chimeric MERS-CoV bearing the S protein of a MERS-like virus from Ugandan bats required addition of exogenous trypsin to human Caco-2 cells, whereas MERS-CoV replicated trypsin independent in the cells, most likely due to S cleavage by TMPRSS2. The data indicated that S cleavage may be a critical barrier that needs to be overcome by CoVs to infect a new host [27].
Recent studies suggest that human and murine lung may differ in their protease repertoire available for HA cleavage. Activation of certain human H3N2 IAVs was independent of TMPRSS2 expression in mice, whereas TMPRSS2 was crucial for virus activation in primary human airway cells 6, 23, 24, 28. This may be due to expression of a larger number of appropriate proteases in murine lung and hence cleavage of H3 by a mouse-specific protease. The mouse degradome (complete set of proteases present in an organism) consists of more protease genes (672) compared with human (588) and chicken (460) degradomes [29]. Furthermore, differences in the substrate specificity of orthologous proteases from human and mouse have been described and might contribute to the observed differences in H3 activation 28, 30. Even though mice are not a natural host for IAV, the broader repertoire of trypsin-like proteases could play a role in CoV evolution in rodents.
TMPRSS2 was identified as major activating protease of LPAIV HA of almost all subtypes in human airway cells, suggesting that the transmission of IAV from avian species to humans does not require adaptation to a new protease repertoire [15]. Interestingly, infection studies in murine lung explants showed that the protease dependency can differ between human- and avian-derived HAs. While a virus with human-derived H3 replicated in lung explants of both TMPRSS2-deficient mice and wild-type animals, a virus containing a duck-derived H3 was not able to replicate in lung explants lacking TMPRSS2 expression [15]. However, this has not been analyzed for other HA subtypes. Further investigations are needed to conclude whether avian-derived HAs rely more on TMPRSS2 in proteolytic activation compared with human-derived HAs and if so, whether this affects avian IAV adaptation to new host species.
Some IAV HA subtypes such as H3, H4, LPAIV H5, H9, and H14 possess a basic amino acid in position P4 of the CS motif (K/R–X–X–R↓) that may facilitate activation by additional proteases. The type-II transmembrane serine protease matriptase/ST14 that is broadly expressed among epithelial tissues preferentially cleaves substrates at a R–X–X–R motif 31, 32. Accordingly, H9 with R–S–S–R but not V–S–S–R CS was cleaved by matriptase in addition to TMPRSS2 in vitro and virus activation by matriptase was associated with replication of H9N2 IAV in primary chicken embryo kidney cells [31].
Cleavage of IAV HA by TMPRSS2 takes place intracellularly before release of progeny virus from the infected cell [33]. In contrast, priming of CoV S by TMPRSS2 occurs upon virus entry and is believed to facilitate virus-membrane fusion at or close to the plasma membrane (early entry) 34, 35. In the absence of TMPRSS2, SARS-CoV-2 is taken up by endocytosis and entry occurs via fusion in late endosomes upon S2′ site cleavage by cathepsins (late entry) 10, 11. Using the early entry route may allow CoVs to avoid endosomal restriction by interferon-induced transmembrane (IFITM) proteins, which block viral membrane fusion by preventing hemifusion 36, 37. However, both antiviral and proviral effects have been described for IFITMs in CoV replication 37, 38, 39, 40•. Interestingly, TMPRSS2 has been shown to be beneficial for the virus in either way. Thus, in addition to avoiding endosomal restriction by IFITMs, TMPRSS2 expression was shown to switch IFITM3 activities at the plasma membrane toward enhancement of SARS-CoV-2 infection via yet unknown mechanisms [40].
Rather unexpectedly, the SARS-CoV-2 Omicron variants BA.1 and BA.2 were found to be less efficiently activated by TMPRSS2 compared with other variants in vitro and seem to favor an endosomal entry via cathepsins 41•, 42, 43. Altered TMPRSS2 usage is believed to contribute to the change in tissue tropism of Omicron that replicates well in human nasal cells, but demonstrates significantly less replication in human lung cells compared with the Delta variant 41•, 42. Replication of Omicron BA.1 in nasal and lung cells of TMPRSS2-knockout mice, however, was significantly reduced compared with wild-type animals, indicating that TMPRSS2 is involved in virus activation in mice [44]. Whether or not there are discrepancies in utilizing TMPRSS2 for Omicron S priming in humans and mice remains open. Interestingly, Omicron lineage BA.5 shows increased TMPRSS2 usage when compared with BA.1 and BA.2 [45]. Thus, one may speculate that switching from the TMPRSS2-facilitated entry to a cathepsin-dependent entry route was disadvantageous (in respect of efficient S priming or avoiding restriction by IFITMs or both) and ongoing evolution of Omicron shows a reversion back to TMPRSS2 usage for S priming.
Origin of the multibasic cleavage site in coronaviruses
A multibasic CS is not necessarily linked to enhanced pathogenicity. Two out of four human coronaviruses (HCoVs) associated with common cold contain a multibasic CS at the S1/S2 junction, while the other two HCoVs but also SARS-CoV that causes severe disease possess two monobasic CSs (Figure 1c). Feline enteric coronaviruses (FECVs) possess a multibasic S1/S2 CS and can chronically infect cats for long periods. Feline infectious peritonitis (FIP) viruses that cause deadly FIP arise from FECVs by mutation. Interestingly, among other mutations, amino acid substitutions at the S1/S2 CS modifying the furin cleavage motif, are observed upon development of FIP [46]. The avian infectious bronchitis virus (IBV) harbors a multibasic S1/S2 CS and some strains possess a tribasic S2′ CS motif. IBV causes a highly contagious respiratory disease in chickens. Mortality is usually low, but can vary depending on the strain [47].
The fact that SARS-CoV-2 acquired a multibasic S1/S2 CS due to insertion of four amino acids started a debate of whether the emergence of a furin-cleavable CS appears to be specific for different host species. Bats and rodents are a major natural reservoir of CoV and a source of zoonotic transmission to other host species. While a multibasic S1/S2 CS is common in rodent-derived CoVs (78%), it is rare in bat-derived CoVs (6%) [48]. SARS-CoV-2 probably originated from bats via a potential, yet unknown, intermediate host and probably involving recombination events between different CoVs 25, 38, 49. Thus, its multibasic S1/S2 CS represents rather an exception for a bat-derived CoV.
Importantly, the multibasic CS of SARS-CoV-2 was identified as a critical determinant of virus transmission in ferrets. Loss of the multibasic S1/S2 CS attenuated SARS-CoV-2 and prevented transmission [50]. In agreement with that, deletions of the S1/S2 CS have been shown to arise naturally only at very low levels, and all variants of concern have retained the R–R–A–R motif at the S1/S2 junction 26, 50••. The underlying molecular mechanism remains to be determined. S1/S2 cleavage by furin occurs during egress and may allow for near-complete cleavage and thus efficient receptor binding upon infection of a new host cell. Notably, S1/S2 cleavage is required for angiotensin-converting enzyme 2 (ACE2) binding [51]. ACE2 binding, in turn, enables exposure of the S2′ site and subsequent cleavage by TMPRSS2 [52]. Of note, the P–R–R–A–R↓ motif is suboptimal for furin cleavage due to alanine in P2 position. Several SARS-CoV-2 variants acquired mutations (P681R/H) at P5 predicted to enhance furin cleavage 50••, 53. Whether this represents ongoing adaptation to humans warrants further investigation.
Interestingly, serial infection of Vero cells leads to a loss of the multibasic S1/S2 CS already at low passage numbers, indicating that the multibasic CS has a selective disadvantage for the virus in Vero cells. Loss of the multibasic CS is associated with lack of TMPRSS2 expression. Although it appears illogical that absence of the protease that cleaves at the S2′ site results in mutations at the S1/S2 site, a recent study suggested that it might support more efficient cleavage of the S1/S2 site by cathepsins upon endosomal uptake of the virus 54, 55. Intriguingly, passaging of the Vero-adapted SARS-CoV-2 with mutated S1/S2 CS resulted in prompt reversion to the original multibasic sequence in human Calu-3 airway cells or Caco-2 colon carcinoma cells both expressing TMPRSS2 [55]. The data indicate that SARS-CoV-2 is not critically dependent on TMPRSS2 and furin but able to adapt to another host protease repertoire. Nevertheless, furin and TMPRSS2 seem to be the best option in human cells.
The role for cathepsins in CoV S activation in natural infections is still under debate. Although cathepsins seem to be not crucial for CoV activation in human cells, they may play an important role in virus activation in other host species. Interestingly, a number of bat-derived viruses (e.g. Hendra virus, Nipah virus, and Ebola virus) utilize endosomal cathepsins during their replication cycle 56, 57. A cathepsin-L orthologous protease has been shown to support Hendra virus F-protein activation in fruit bat cells [19]. Additionally, furin-like proteases were described in the cells and shown to support proteolytic activation of parainfluenza virus 5. However, differences in the response of fruit bat cells versus Vero cells to a potent furin inhibitor indicated that subtle differences in subcellular localization or activity of furin may exist between different mammalian species [19]. Whether expression or activity of cathepsins and furin orthologs drives the evolution of specific CS motifs in bats remains unknown, but should be investigated in future studies.
Neuropilin-1 and the multibasic cleavage site
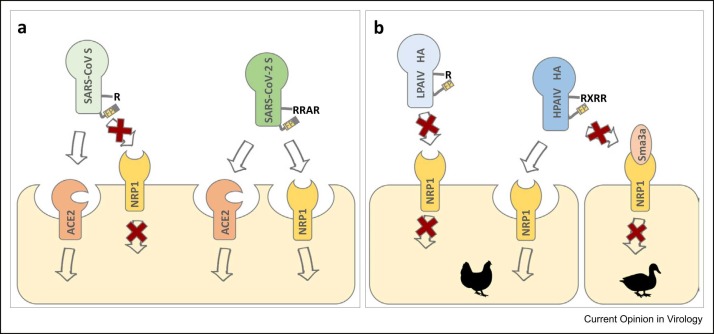
Some viruses that require proteolytic activation of their envelope proteins by furin such as the herpesvirus Epstein–Barr virus or human T-cell lymphotropic virus 1 (HTLV-1) and HTLV-2 have been shown to utilize NRP1 as an additional entry factor [58]. NRP1 is a cell surface receptor with disseminated expression that plays important roles in growth factor signaling, vascular angiogenesis, axonal guidance, and immune function 59, 60. Intriguingly, NRP1 binds peptides with a multibasic sequence (R/K–X–X–R/K) at the C-terminal end (C-end rule) and executes cellular uptake via a mechanism similar to micropinocytosis 59, 61, 62. The C-terminus of the cleaved S1 subunit of SARS-CoV-2 also conforms to the C-end rule and can therefore interact with NRP1 ( Figure 2a). Correspondingly, NRP1 was shown to enhance entry of SARS-CoV-2 into human cells but not that of a mutant virus lacking the multibasic CS 63•, 64•. Virus entry via NRP1 may compensate for the relatively low expression levels of ACE2 in the human respiratory tract and may facilitate infection of multiple organs and tissues, including neurons and endothelial cells 64•, 65•, 66. NRP1, not ACE2, was shown to mediate astrocyte infection by SARS-CoV-2 in brain organoids [65]. Additionally, binding of S1 to NRP1 was predicted to stimulate separation of S1 and S2 and thereby may increase virus infectivity [67]. Cells isolated from human bronchoalveolar lavage fluid from COVID-19 patients showed that NRP1 was upregulated in SARS-CoV-2-infected cells [64]. Mutations at P5 and P7 position of the S1/S2 CS of SARS-CoV-2 variants, namely P681H (Alpha), P681R (Delta), or N679K + P681H (Omicron) enhance the overall basicity of the C-terminus of S1 and may affect NRP1 binding. Based on an in silico molecular docking analysis, Omicron S but not Delta S shows increased binding to NRP1 in comparison to Wuhan S [68].
Figure 2.
NRP1 acts as an alternative entry receptor for SARS-CoV-2 and HPAIV. (a) Both SARS-CoV and SARS-CoV-2 utilize ACE2 as entry receptor. Following S1/S2 cleavage, the multibasic motif at the C-terminus of S1 conforms to the C-end rule and is bound by NRP1 and taken up by the cell. Utilizing NRP1 as additional receptor may enhance entry into human respiratory cells and expand the tissue tropism of SARS-CoV-2. The monobasic motif of the C-terminus of SARS-CoV S1 does not facilitate binding to NRP1. (b) The multibasic motif at the C-terminus of HA1 of a HPAIV provides a NRP1 substrate, whereas HA1 of a LPAIV does not. The higher expression level of the natural NRP1 ligand Sma3a in duck-derived endothelial cells may block binding of HPAIV HA1 to NRP1 and prevent viral uptake, while lower endogenous expression of Sma3a in chicken endothelial cells does not compete with HPAIV HA binding to NRP1 and therefore promotes virus entry. Efficient uptake of HPAIV HA in chicken endothelial cells might support HPAIV genesis in poultry (L.E. Steele et al., abstract The Eighth ESWI Influenza Conference, Virtual Edition, 4–7 December 2021).
HA cleavage has been identified as the prime determinant of avian IAV pathogenicity in poultry for a long time. HPAIVs emerge from LPAIVs by acquisition of a multibasic CS due to insertion of multiple basic amino acids at the CS. Natural HPAIVs are restricted to subtypes H5 and H7 69, 70. Proteolytic activation by furin supports systemic spread of infection with often-fatal outcome. Replication of LPAIVs, on the other hand, is confined to epithelial cells of the respiratory and intestinal tract due to the restricted expression of appropriate trypsin-like proteases. HPAIVs mostly infect endothelial cells in chickens, whereas they are still epitheliotropic in ducks [71]. It is generally assumed that the restricted expression of trypsin-like HA-cleaving proteases serves as primary positive selection pressure for LPAIV to HPAIV conversion. Interestingly, HPAIV are rarely isolated from wild aquatic birds and are believed to emerge after introduction into poultry 70, 72, indicating that a multibasic HA CS provides an advantage in poultry that is not relevant in waterfowl. The C-terminus of the cleaved HA1 subunit of HPAIV is also qualified as a NRP1 substrate, while HA1 of LPAIV is not (Figure 2b). A recent study by Steele and Short suggests that NRP1 might be a trigger for HPAIV genesis in poultry (L.E. Steele et al., abstract The Eighth ESWI Influenza Conference, Virtual Edition, 4–7 December 2021). The data by Steele et al. indicate that a higher endogenous expression level of the natural NRP1 ligand semaphorin 3a (Sema3a) in duck versus chicken endothelial cells blocks binding and uptake of cleaved HA in duck cells, whereas HA is taken up efficiently in chicken-derived endothelial cells (Figure 2b). Thus, efficient binding of HPAIV HA1 to NRP1 on endothelial cells supporting systemic spread of infection occurs only in chicken but not in duck. This might provide the positive selection pressure for LPAIV to HPAIV conversion in chicken. In contrast to chickens, vascular tropism is not a prominent feature of HPAIV pathogenesis in mammals including humans 71, 73. It will thus be of interest to investigate NPR1-dependent uptake of HPAIV HA into mammalian vascular endothelial cells. Notably, carboxypeptidase B has been shown to remove basic amino acids from the C-terminus of HA1 upon cleavage of HA0 into HA1 and HA2 7, 74. Carboxypeptidase B also eliminates the multibasic amino acid motif of the HA1 subunit of fowl plague virus, although not quantitatively, and therefore might interfere with recognition of HA1 as a NRP1 substrate. However, the biological significance of carboxypeptidase trimming of HA1 (and cleaved envelope proteins of other viruses) remains to be investigated in more detail.
Overall, these findings underline that a multibasic CS may not only be beneficial in facilitating virus activation by ubiquitously expressed furin, but also in promoting virus entry into a wide range of tissues and vascular spreading by using NPR1.
Concluding remarks
There are many open questions about the evolution and benefits of distinct CS motifs and protease usage in different IAV and CoV host species. Recent studies suggest that host differences may exist in the virus-activating protease repertoire. Whether these variations play a role in virus activation and provide a barrier that needs to be overcome by IAV and CoV in host adaptation remains to be further investigated. Importantly, viral glycoprotein cleavage has been recognized as a factor that is not only essential for virus infectivity but furthermore may promote virus infection via facilitating binding to additional receptors or by allowing the virus to avoid cellular restriction factors.
Understanding the role of glycoprotein cleavage and proteases involved in different host species may allow to evaluate the zoonotic potential and risk posed by IAVs and CoVs. Much attention is devoted to bats and rodents in determining the role of viral glycoprotein processing in host adaptation of zoonotic viruses, particularly CoVs. However, bats and rodents harbor a number of other viruses and comparative analyses of proteolytic activation of different viruses in these hosts might unravel basic mechanisms that drive the evolution of distinct CSs. Moreover, birds are an important host reservoir for IAV and CoV and should be included in future studies.
Conflict of interest statement
The authors declare no competing conflicts of interest.
Acknowledgements
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) SFB 1021 (project B07) and by the LOEWE Center DRUID (project D1).
Data Availability
No data were used for the research described in the article.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
-
•
of special interest
-
••
of outstanding interest
- 1.Skehel J.J., Wiley D.C. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. 〈http://www.ncbi.nlm.nih.gov/pubmed/10966468〉 [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann M., Hofmann-Winkler H., Pöhlmann S. In: Activation of Viruses by Host Proteases. Böttcher-Friebertshäuser E., Garten W., Klenk H.D., editors. Springer International Publishing; 2018. Priming time: how cellular proteases arm coronavirus spike proteins; pp. 71–98. [Google Scholar]
- 3.Huang R., Wahn K., Klenk H.-D., Rott R. Fusion between cell membrane and liposomes containing the glycoproteins of influenza virus. Virology. 1980;104:294–302. doi: 10.1016/0042-6822(80)90334-7. [DOI] [PubMed] [Google Scholar]
- 4.Böttcher-Friebertshäuser E. In: Activation of Viruses by Host Proteases. Böttcher-Friebertshäuser E., Garten W., Klenk H.D., editors. Springer International Publishing; 2018. Membrane-anchored serine proteases: host cell factors in proteolytic activation of viral glycoproteins; pp. 153–203. [Google Scholar]
- 5.Haan C.A.M., de, Haijema B.J., Schellen P., Wichgers Schreur P., te Lintelo E., et al. Cleavage of group 1 coronavirus spike proteins: how furin cleavage is traded off against heparan sulfate binding upon cell culture adaptation. J Virol. 2008;82:6078–6083. doi: 10.1128/JVI.00074-08. 〈http://www.ncbi.nlm.nih.gov/pubmed/18400867〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Limburg H., Harbig A., Bestle D., Stein D.A., Moulton H.M., et al. TMPRSS2 is the major activating protease of influenza A virus in primary human airway cells and influenza B virus in human type II pneumocytes. J Virol. 2019;93 doi: 10.1128/JVI.00649-19. 〈http://www.ncbi.nlm.nih.gov/pubmed/31391268〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garten W., Bosch F.X., Linder D., Rott R., Klenk H.-D. Proteolytic activation of the influenza virus hemagglutinin: the structure of the cleavage site and the enzymes involved in cleavage. Virology. 1981;115:361–374. doi: 10.1016/0042-6822(81)90117-3. [DOI] [PubMed] [Google Scholar]
- 8.Stieneke-Gröber A., Vey M., Angliker H., Shaw E., Thomas G., et al. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 1992;11:2407–2414. doi: 10.1002/j.1460-2075.1992.tb05305.x. 〈http://www.ncbi.nlm.nih.gov/pubmed/1628614〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. 〈http://www.ncbi.nlm.nih.gov/pubmed/21325420〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., et al. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. 〈http://www.ncbi.nlm.nih.gov/pubmed/16081529〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jocher G., Grass V., Tschirner S.K., Riepler L., Breimann S., et al. ADAM10 and ADAM17 promote SARS-CoV-2 cell entry and spike protein-mediated lung cell fusion. EMBO Rep. 2022;23 doi: 10.15252/embr.202154305. 〈http://www.ncbi.nlm.nih.gov/pubmed/35527514〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto M., Gohda J., Kobayashi A., Tomita K., Hirayama Y., et al. Metalloproteinase-dependent and TMPRSS2-independent cell surface entry pathway of SARS-CoV-2 requires the furin cleavage site and the S2 domain of spike protein. mBio. 2022;13 doi: 10.1128/mbio.00519-22. 〈http://www.ncbi.nlm.nih.gov/pubmed/35708281〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harte J.V., Wakerlin S.L., Lindsay A.J., McCarthy J.V., Coleman-Vaughan C. Metalloprotease-dependent S2'-activation promotes cell-cell fusion and syncytiation of SARS-CoV-2. Viruses. 2022;14:2094. doi: 10.3390/v14102094. 〈http://www.ncbi.nlm.nih.gov/pubmed/36298651〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benlarbi M., Laroche G., Fink C., Fu K., Mulloy R.P., et al. Identification and differential usage of a host metalloproteinase entry pathway by SARS-CoV-2 Delta and Omicron. iScience. 2022;25 doi: 10.1016/j.isci.2022.105316. 〈http://www.ncbi.nlm.nih.gov/pubmed/36254158〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Bestle D., Limburg H., Kruhl D., Harbig A., Stein D.A., et al. Hemagglutinins of Avian influenza viruses are proteolytically activated by TMPRSS2 in human and murine airway cells. J Virol. 2021;95 doi: 10.1128/JVI.00906-21. 〈http://www.ncbi.nlm.nih.gov/pubmed/34319155〉 [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that TMPRSS2 is the major activating protease of almost all IAV HA subtypes in human and murine airway cells. Additionally, the study shows that duck TMPRSS2 is a potential IAV-activating protease in waterfowl.
- 16.Peitsch C., Klenk H.-D., Garten W., Böttcher-Friebertshäuser E. Activation of influenza A viruses by host proteases from swine airway epithelium. J Virol. 2014;88:282–291. doi: 10.1128/JVI.01635-13. 〈http://www.ncbi.nlm.nih.gov/pubmed/24155384〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertram S., Heurich A., Lavender H., Gierer S., Danisch S., et al. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035876. 〈http://www.ncbi.nlm.nih.gov/pubmed/22558251〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zmora P., Molau-Blazejewska P., Bertram S., Walendy-Gnirß K., Nehlmeier I., et al. Non-human primate orthologues of TMPRSS2 cleave and activate the influenza virus hemagglutinin. PLoS One. 2017;12 doi: 10.1371/journal.pone.0176597. 〈http://www.ncbi.nlm.nih.gov/pubmed/28493964〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Najjar F., Lampe L., Baker M.L., Wang L.-F., Dutch R.E. Analysis of cathepsin and furin proteolytic enzymes involved in viral fusion protein activation in cells of the bat reservoir host. PLoS One. 2015;10 doi: 10.1371/journal.pone.0115736. 〈http://www.ncbi.nlm.nih.gov/pubmed/25706132〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bestle D., Heindl M.R., Limburg H., van Lam van T., Pilgram O., et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance. 2020;3 doi: 10.26508/lsa.202000786. 〈http://www.ncbi.nlm.nih.gov/pubmed/32703818〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78:779–784.e5. doi: 10.1016/J.MOLCEL.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., et al. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. 2019;93 doi: 10.1128/JVI.01815-18. 〈http://www.ncbi.nlm.nih.gov/pubmed/30626688〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatesuer B., Bertram S., Mehnert N., Bahgat M.M., Nelson P.S., et al. Tmprss2 is essential for influenza H1N1 virus pathogenesis in mice. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003774. 〈http://www.ncbi.nlm.nih.gov/pubmed/24348248〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarnow C., Engels G., Arendt A., Schwalm F., Sediri H., et al. TMPRSS2 is a host factor that is essential for pneumotropism and pathogenicity of H7N9 influenza A virus in mice. J Virol. 2014;88:4744–4751. doi: 10.1128/JVI.03799-13. 〈http://www.ncbi.nlm.nih.gov/pubmed/24522916〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li F., Han M., Dai P., Xu W., He J., et al. Distinct mechanisms for TMPRSS2 expression explain organ-specific inhibition of SARS-CoV-2 infection by enzalutamide. Nat Commun. 2021;12 doi: 10.1038/s41467-021-21171-x. 〈http://www.ncbi.nlm.nih.gov/pubmed/33558541〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller C., Böttcher-Friebertshäuser E., Lohoff M. TMPRSS2, a novel host-directed drug target against SARS-CoV-2. Signal Transduct Target Ther. 2022;7:251. doi: 10.1038/s41392-022-01084-x. 〈http://www.ncbi.nlm.nih.gov/pubmed/35871159〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Menachery V.D., Dinnon K.H., Yount B.L., McAnarney E.T., Gralinski L.E., et al. Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. J Virol. 2020;94 doi: 10.1128/JVI.01774-19. 〈http://www.ncbi.nlm.nih.gov/pubmed/31801868〉 [DOI] [PMC free article] [PubMed] [Google Scholar]; The data of this study indicate that proteolytic cleavage of the S protein is the primary infection barrier for two bat-derived group 2C CoVs in human cell cultures.
- 28.Harbig A., Mernberger M., Bittel L., Pleschka S., Schughart K., et al. Transcriptome profiling and protease inhibition experiments identify proteases that activate H3N2 influenza A and influenza B viruses in murine airways. J Biol Chem. 2020;295:11388–11407. doi: 10.1074/jbc.RA120.012635. 〈http://www.ncbi.nlm.nih.gov/pubmed/32303635〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quesada V., Velasco G., Puente X.S., Warren W.C., López-Otín C. Comparative genomic analysis of the zebra finch degradome provides new insights into evolution of proteases in birds and mammals. BMC Genom. 2010;11 doi: 10.1186/1471-2164-11-220. 〈http://www.ncbi.nlm.nih.gov/pubmed/20359326〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magnen M., Elsässer B.M., Zbodakova O., Kasparek P., Gueugnon F., et al. Kallikrein-related peptidase 5 and seasonal influenza viruses, limitations of the experimental models for activating proteases. Biol Chem. 2018;399:1053–1064. doi: 10.1515/hsz-2017-0340. 〈http://www.ncbi.nlm.nih.gov/pubmed/29883316〉 [DOI] [PubMed] [Google Scholar]
- 31.Baron J., Tarnow C., Mayoli-Nüssle D., Schilling E., Meyer D., et al. Matriptase, HAT, and TMPRSS2 activate the hemagglutinin of H9N2 influenza A viruses. J Virol. 2013;87:1811–1820. doi: 10.1128/JVI.02320-12. 〈http://www.ncbi.nlm.nih.gov/pubmed/23192872〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oberst M.D., Singh B., Ozdemirli M., Dickson R.B., Johnson M.D., et al. Characterization of matriptase expression in normal human tissues. J Histochem Cytochem. 2003;51:1017–1025. doi: 10.1177/002215540305100805. 〈http://www.ncbi.nlm.nih.gov/pubmed/12871983〉 [DOI] [PubMed] [Google Scholar]
- 33.Böttcher-Friebertshäuser E., Freuer C., Sielaff F., Schmidt S., Eickmann M., et al. Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J Virol. 2010;84:5605–5614. doi: 10.1128/JVI.00140-10. 〈http://www.ncbi.nlm.nih.gov/pubmed/20237084〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park J.-E., Li K., Barlan A., Fehr A.R., Perlman S., et al. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci USA. 2016;113:12262–12267. doi: 10.1073/pnas.1608147113. 〈http://www.ncbi.nlm.nih.gov/pubmed/27791014〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koch J., Uckeley Z.M., Doldan P., Stanifer M., Boulant S., et al. TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells. EMBO J. 2021;40 doi: 10.15252/embj.2021107821. 〈http://www.ncbi.nlm.nih.gov/pubmed/34159616〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li K., Markosyan R.M., Zheng Y.-M., Golfetto O., Bungart B., et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003124. 〈http://www.ncbi.nlm.nih.gov/pubmed/23358889〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wrensch F., Winkler M., Pöhlmann S. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses. 2014;6:3683–3698. doi: 10.3390/v6093683. 〈http://www.ncbi.nlm.nih.gov/pubmed/25256397〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J., Cui W., Tian B. The potential intermediate hosts for SARS-CoV-2. Front Microbiol. 2020;11 doi: 10.3389/FMICB.2020.580137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prelli Bozzo C., Nchioua R., Volcic M., Koepke L., Krüger J., et al. IFITM proteins promote SARS-CoV-2 infection and are targets for virus inhibition in vitro. Nat Commun. 2021;12:4584. doi: 10.1038/s41467-021-24817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Shi G., Kenney A.D., Kudryashova E., Zani A., Zhang L., et al. Opposing activities of IFITM proteins in SARS-CoV-2 infection. EMBO J. 2021;40 doi: 10.15252/embj.2020106501. 〈http://www.ncbi.nlm.nih.gov/pubmed/33270927〉 [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uncovers new pro- and antiviral mechanisms of IFITM3 and shows that TMPRSS2 expression can shift the balance of IFITM3 activities toward enhancement of infection.
- 41•.Meng B., Abdullahi A., Ferreira I.A.T.M., Goonawardane N., Saito A., et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. 〈http://www.ncbi.nlm.nih.gov/pubmed/35104837〉 [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that Omicron S is less efficiently cleaved by furin and TMPRSS2 and prefers virus entry via endosomes and S cleavage by endosomal cathepsins.
- 42.Zhao H., Lu L., Peng Z., Chen L.-L., Meng X., et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect. 2022;11:277–283. doi: 10.1080/22221751.2021.2023329. 〈http://www.ncbi.nlm.nih.gov/pubmed/34951565〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willett B.J., Grove J., MacLean O.A., Wilkie C., Lorenzo G. de, et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat Microbiol. 2022;7:1161–1179. doi: 10.1038/s41564-022-01143-7. 〈http://www.ncbi.nlm.nih.gov/pubmed/35798890〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44•.Iwata-Yoshikawa N., Kakizaki M., Shiwa-Sudo N., Okura T., Tahara M., et al. Essential role of TMPRSS2 in SARS-CoV-2 infection in murine airways. Nat Commun. 2022;13 doi: 10.1038/s41467-022-33911-8. 〈http://www.ncbi.nlm.nih.gov/pubmed/36243815〉 [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study the authors show that the Omicron variant requires TMPRSS2 expression for efficient virus replication in mice in vivo and is therefore not an exception in TMPRSS2 usage in mice.
- 45••.Aggarwal A., Akerman A., Milogiannakis V., Silva M.R., Walker G., et al. SARS-CoV-2 Omicron BA.5: evolving tropism and evasion of potent humoral responses and resistance to clinical immunotherapeutics relative to viral variants of concern. EBioMedicine. 2022;84 doi: 10.1016/j.ebiom.2022.104270. 〈http://www.ncbi.nlm.nih.gov/pubmed/36130476〉 [DOI] [PMC free article] [PubMed] [Google Scholar]; The data of the study reveal the reversion of the Omicron BA.5 variant in tropism back to using TMPRSS2 for S priming instead of favoring the endosomal entry pathway.
- 46.Licitra B.N., Millet J.K., Regan A.D., Hamilton B.S., Rinaldi V.D., et al. Mutation in spike protein cleavage site and pathogenesis of feline coronavirus. Emerg Infect Dis. 2013;19:1066–1073. doi: 10.3201/eid1907.121094. 〈http://www.ncbi.nlm.nih.gov/pubmed/23763835〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackwood M.W., Wit S. In: Diseases of Poultry. Swayne D.E., Boulianne M., Logue C.M., McDougald L.R., Nair V., et al., editors. Wiley; 2020. Infectious bronchitis; pp. 167–188. [Google Scholar]
- 48••.Stout A.E., Millet J.K., Stanhope M.J., Whittaker G.R. Furin cleavage sites in the spike proteins of bat and rodent coronaviruses: implications for virus evolution and zoonotic transfer from rodent species. One Health. 2021;13 doi: 10.1016/j.onehlt.2021.100282. 〈http://www.ncbi.nlm.nih.gov/pubmed/34179330〉 [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provides a comprehensive analysis of furin CSs found in rodent CoV and shows that a multibasic S1/S2 CS in S is common in rodent CoV, whereas it is typically not present in bat CoV.
- 49.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. 〈http://www.ncbi.nlm.nih.gov/pubmed/32015507〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Peacock T.P., Goldhill D.H., Zhou J., Baillon L., Frise R., et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol. 2021;6:899–909. doi: 10.1038/s41564-021-00908-w. 〈http://www.ncbi.nlm.nih.gov/pubmed/33907312〉 [DOI] [PubMed] [Google Scholar]; This study reveals that the furin CS is an important determinant of SARS-CoV-2 transmission. Analysis of a large number of SARS-CoV-2 sequences isolated from COVID-19 patients showed very low frequencies of naturally occurring mutants with deletions at the furin CS.
- 51.Wrobel A.G., Benton D.J., Xu P., Roustan C., Martin S.R., et al. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat Struct Mol Biol. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. 〈http://www.ncbi.nlm.nih.gov/pubmed/32647346〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu S., Zheng X., Zhou B., Li J., Chen M., et al. SARS-CoV-2 spike engagement of ACE2 primes S2' site cleavage and fusion initiation. Proc Natl Acad Sci USA. 2022;119 doi: 10.1073/pnas.2111199119. 〈http://www.ncbi.nlm.nih.gov/pubmed/34930824〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lubinski B., Fernandes M.H.V., Frazier L., Tang T., Daniel S., et al. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience. 2022;25 doi: 10.1016/j.isci.2021.103589. 〈http://www.ncbi.nlm.nih.gov/pubmed/34909610〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sasaki M., Uemura K., Sato A., Toba S., Sanaki T., et al. SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009233. 〈http://www.ncbi.nlm.nih.gov/pubmed/33476327〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaudhry M.Z., Eschke K., Hoffmann M., Grashoff M., Abassi L., et al. Rapid SARS-CoV-2 adaptation to available cellular proteases. J Virol. 2022;96 doi: 10.1128/jvi.02186-21. 〈http://www.ncbi.nlm.nih.gov/pubmed/35019723〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. 〈http://www.ncbi.nlm.nih.gov/pubmed/15831716〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pager C.T., Dutch R.E. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J Virol. 2005;79:12714–12720. doi: 10.1128/JVI.79.20.12714-12720.2005. 〈http://www.ncbi.nlm.nih.gov/pubmed/16188974〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Balistreri G., Yamauchi Y., Teesalu T. A widespread viral entry mechanism: the C-end Rule motif-neuropilin receptor interaction. Proc Natl Acad Sci USA. 2021;118 doi: 10.1073/pnas.2112457118. 〈http://www.ncbi.nlm.nih.gov/pubmed/34772761〉 [DOI] [PMC free article] [PubMed] [Google Scholar]; This review proposes that binding of viral envelope proteins to neuropilins plays an underappreciated and prevalent role in the transmission and pathogenesis of a number of viruses.
- 59.Guo H.-F., Vander Kooi C.W. Neuropilin functions as an essential cell surface receptor. J Biol Chem. 2015;290:29120–29126. doi: 10.1074/jbc.R115.687327. 〈http://www.ncbi.nlm.nih.gov/pubmed/26451046〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mayi B.S., Leibowitz J.A., Woods A.T., Ammon K.A., Liu A.E., et al. The role of neuropilin-1 in COVID-19. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009153. 〈http://www.ncbi.nlm.nih.gov/pubmed/33395426〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teesalu T., Sugahara K.N., Kotamraju V.R., Ruoslahti E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc Natl Acad Sci USA. 2009;106:16157–16162. doi: 10.1073/pnas.0908201106. 〈http://www.ncbi.nlm.nih.gov/pubmed/19805273〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pang H.-B., Braun G.B., Friman T., Aza-Blanc P., Ruidiaz M.E., et al. An endocytosis pathway initiated through neuropilin-1 and regulated by nutrient availability. Nat Commun. 2014;5:4904. doi: 10.1038/ncomms5904. 〈http://www.ncbi.nlm.nih.gov/pubmed/25277522〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63•.Daly J.L., Simonetti B., Klein K., Chen K.-E., Williamson M.K., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. 〈http://www.ncbi.nlm.nih.gov/pubmed/33082294〉 [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors found that the furin-cleaved S1 fragment of the SARS-CoV-2 S protein binds directly to cell surface NRP1. Blocking this interaction with a small-molecule inhibitor or monoclonal antibodies reduced viral infection in cell culture.
- 64•.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. 〈http://www.ncbi.nlm.nih.gov/pubmed/33082293〉 [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that NRP1, which is known to bind furin-cleaved substrates, potentiates SARS-CoV-2 infectivity.
- 65•.Kong W., Montano M., Corley M.J., Helmy E., Kobayashi H., et al. Neuropilin-1 mediates SARS-CoV-2 infection of astrocytes in brain organoids, inducing inflammation leading to dysfunction and death of neurons. mBio. 2022;13 doi: 10.1128/mbio.02308-22. 〈http://www.ncbi.nlm.nih.gov/pubmed/36314791〉 [DOI] [PMC free article] [PubMed] [Google Scholar]; The study demonstrates that NPR1 functions as primary receptor for SARS-CoV-2 entry into astrocytes resulting in neuronal dysfunction and death.
- 66.Hikmet F., Méar L., Edvinsson Å., Micke P., Uhlén M., et al. The protein expression profile of ACE2 in human tissues. Mol Syst Biol. 2020;16 doi: 10.15252/msb.20209610. 〈http://www.ncbi.nlm.nih.gov/pubmed/32715618〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Z.-L., Buck M. Neuropilin-1 assists SARS-CoV-2 infection by stimulating the separation of Spike protein S1 and S2. Biophys J. 2021;120:2828–2837. doi: 10.1016/j.bpj.2021.05.026. 〈http://www.ncbi.nlm.nih.gov/pubmed/34087218〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baindara P., Roy D., Mandal S.M., Schrum A.G. Conservation and enhanced binding of SARS-CoV-2 Omicron spike protein to coreceptor neuropilin-1 predicted by docking analysis. Infect Dis Rep. 2022;14:243–249. doi: 10.3390/idr14020029. 〈http://www.ncbi.nlm.nih.gov/pubmed/35447881〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abdelwhab E.-S.M., Veits J., Mettenleiter T.C. Genetic changes that accompanied shifts of low pathogenic avian influenza viruses toward higher pathogenicity in poultry. Virulence. 2013;4:441–452. doi: 10.4161/viru.25710. 〈http://www.ncbi.nlm.nih.gov/pubmed/23863606〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alexander D.J. An overview of the epidemiology of avian influenza. Vaccine. 2007;25:5637–5644. doi: 10.1016/J.VACCINE.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 71.Short K.R., Veldhuis Kroeze E.J.B., Reperant L.A., Richard M., Kuiken T. Influenza virus and endothelial cells: a species specific relationship. Front Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00653. 〈http://www.ncbi.nlm.nih.gov/pubmed/25520707〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vijaykrishna D., Bahl J., Riley S., Duan L., Zhang J.X., et al. Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000161. 〈http://www.ncbi.nlm.nih.gov/pubmed/18818732〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reperant L.A., Rimmelzwaan G.F., Kuiken T. Avian influenza viruses in mammals. Rev Sci Tech. 2009;28:137–159. doi: 10.20506/rst.28.1.1876. 〈http://www.ncbi.nlm.nih.gov/pubmed/19618623〉 [DOI] [PubMed] [Google Scholar]
- 74.Garten W., Linder D., Rott R., Klenk H.-D. The cleavage site of the hemagglutinin of fowl plague virus. Virology. 1982;122:186–190. doi: 10.1016/0042-6822(82)90387-7. [DOI] [PubMed] [Google Scholar]
- 75.Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. 〈http://www.ncbi.nlm.nih.gov/pubmed/22278237〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data were used for the research described in the article.