Summary
Background
Environmental surveillance (ES) of a pathogen is crucial for understanding the community load of disease. As an early warning system, ES for SARS-CoV-2 has complemented routine diagnostic surveillance by capturing near real-time virus circulation at a population level.
Methods
In this longitudinal study conducted between January 2022 and June 2022 in 28 sewershed sites in Bengaluru city (∼11 million inhabitants), we quantified weekly SARS-CoV-2 RNA concentrations to track infection dynamics and provide evidence of change in the relative abundance of emerging variants.
Findings
We describe an early warning system using the exponentially weighted moving average control chart and demonstrate how SARS-CoV-2 RNA concentrations in wastewater correlated with clinically diagnosed new COVID-19 cases, with the trends appearing 8–14 days earlier in wastewater than in clinical data. This was further corroborated by showing that the estimated number of infections is strongly correlated with SARS-CoV-2 RNA copies detected in the wastewater. Using a deconvolution matrix, we detected emerging variants of concern up to two months earlier in wastewater samples. In addition, we found a huge diversity in variants detected in wastewater compared to clinical samples. The findings from this study have been discussed regularly with local authorities to inform policy-making decisions.
Interpretation
Our study highlights that quantifying viral titre, correlating it with a known number of cases in the area, and combined with genomic surveillance helps in tracking variants of concern (VOC) over time and space, enabling timely and making informed policy decisions.
Funding
This work has been supported by funding from the Rockefeller Foundation grant to National Centre for Biological Sciences, TIFR) and the Indian Council of Medical Research grant to (FI) Tata Institute for Genetics and Society and Tata Trusts.
Keywords: Environment surveillance, SARS-CoV-2, Genomics, Variants, India, Bengaluru
Research in context.
Evidence before this study
Quantification of SARS-CoV-2 in wastewater by RT-qPCR has been demonstrated as a cost-effective surveillance tool to monitor the pandemic. However, wastewater-based epidemiology of SARS-COV-2 has not been used in making policy decisions in India. Many studies are limited by the lack of information in real-time on the diversity and abundance in SARS-CoV-2 driving the spike in viral load.
Added value of this study
Wastewater-based epidemiology is a powerful approach where epidemiological data must combine with genomic surveillance to gain insights into how the virus populations are changing, and how the virus is evolving. Our real-time analysis shows that genomic surveillance is central to identification of new variants in densely populated cities where tracking pandemic relies heavily on testing symptomatic individuals for the presence of SARS-CoV-2 RNA and counting the positive tests over time.
Implications of all the available evidence
From science to policy perspective, our study is first comprehensive study in India where real-time data was utilised by the municipal government for making policy decisions. Our study highlights that real-time genomic surveillance is the key to understanding the emerging patterns in viral load and variants in the city.
Introduction
Globally, the COVID-19 pandemic (infectious pneumonia caused by Severe Acute Respiratory Syndrome Coronavirus 2: SARS-CoV-2) has brought wastewater-based (sewage) epidemiology (WBE) to the forefront of the health surveillance system. Wastewater testing can provide a parallel and complementary snapshot of community health. WBE has become an integral component of environmental surveillance in more than 60 countries covering over 3000 sites1,2 providing near-real time information on health and community exposure to COVID-19.3 Analysis of sewage identified evidence of SARS-CoV-2 RNA circulating 56 days in advance of the first clinically confirmed case in South America and 91 days in advance of the first clinically confirmed case in Brazil,4 highlighting the crucial role of WBE in disease surveillance. Mostly, detection of SARS-CoV-2 in wastewater was correlated with local COVID-19 incidence, preceding the increase in clinical cases, locally by 1–3 weeks. Furthermore, WBE has also been used to detect regionally prevalent variants of SARS-CoV-2.5 Given its potential to be a relatively widespread, economical, and rapid surveillance tool, WBE provides an excellent opportunity for a developing country like India, where issues such as unprecedented population growth (especially in urban centres), migrating populations, lack of public health systems and lack of integrated health surveillance are a reality.
WBE was first identified as an effective population wide monitoring technique over 40 years ago– with a high capacity for retrospective indication of poliovirus, norovirus, influenza, hepatitis, and measles outbreaks.6 Nonetheless, it has rarely been integrated into the health surveillance system in India except for environmental surveillance of poliovirus (e.g. Global polio eradication initiative).7 Environmental surveillance played a crucial role in poliovirus eradication in India in 2012 and continues to provide vital support in documenting the absence of the polio virus in conjunction with active surveillance of acute flaccid paralysis (AFP) cases.7, 8, 9
In India, tracking of the COVID-19 pandemic relies heavily on testing symptomatic individuals for the presence of SARS-CoV-2 RNA and counting the positive tests over time. With high population density, many SARS-CoV-2 infected persons are likely to be asymptomatic or oligosymptomatic. They are generally not clinically tested, leading to underestimation of COVID-19 trends and prevalence. There is a need for new strategies to track the emergence and spread of SARS-CoV-2 variants. Currently, epidemiological surveillance relies on testing symptomatic cases using the respiratory tract as the principal site for virus replication. However, the virus also replicates in the gastrointestinal tract leading to a high viral load in excreta.10 The Reverse Transcriptase - quantitative PCR (RT-qPCR)-based approach has revealed a strong correlation between SARS-CoV-2 incidence rates and the viral load in wastewater.11, 12, 13 With the continued emergence of new variants, we also need new approaches to identify the diversity between variants that might have escaped individual testing. Alternatively, emerging variants in wastewater suggests that a significant proportion of individuals in the community are infected with it and thus, shedding the virus.
WBE is a powerful approach where epidemiological data must combine with genomic surveillance to gain insights into how the virus populations are changing, and how the virus is evolving. How long does it take for new variants to emerge and spread across cities (major hubs with international airports) and to rural or suburban areas? Wastewater samples are a mixed representative of local lineages circulating in the community. Monitoring SARS-CoV-2 lineages using wastewater has remained challenging due to low-quality reads, fragmented sequences, and the inability to estimate the relative lineage abundance based on patchy variant-defining mutations in a mixed community sample. Furthermore, SARS-CoV-2 lineages classification using pangolin14 and Ultrafast Sample placement on Existing tRees (UShER),15 designed for clinical samples often contain a single dominant variant and tend to underestimate the relative abundances of multiple SARS-CoV-2 lineages in samples with mixtures of viral genomes such as wastewater.
Using longitudinal testing of wastewater and quantitative epidemiological modelling, we aimed to investigate the relationship between wastewater SARS-CoV-2 concentrations, and COVID-19 cases. In addition, we used high throughput sequencing techniques, to quantify the prevalence and genetic diversity including the newly emerging SARS-CoV-2 variants in Bengaluru city (12.9716° N, 77.5946° E, Karnataka, India). Wastewater samples from the inlet of 28 sewage treatment plants covering over 11 million people were monitored for SARS-CoV-2 from August 2021 to June 2022. We used next-generation sequencing of SARS-CoV-2 RNA and modelling of viral concentration from wastewater to explore the dynamics in diversity and abundance of SARS-CoV-2 lineages in Bengaluru city. Our longitudinal study was initiated post second wave (Delta wave) from August 2021 through to the third wave (Omicron wave). Here we present results from January 2022 to June 2022 to show diversity in lineages of SARS-CoV-2 in wastewater and their comparison with the clinical data. This allowed us to test the relationship between an increase in viral concentrations in wastewater with observed variant diversity across spatiotemporal scales. We hypothesise that high variant diversity is expected during a low viral load or low infection rate period. However, there should be a transition in the lineage of the dominant variant at the peak of infection period during high infection rates. In addition, we explore the emergence of new variants that would have escaped identification in clinical surveillance. Furthermore, we evaluate the lag between emerging new lineages in symptomatic individuals and wastewater samples representing community data.
Methods
Wastewater sample collection
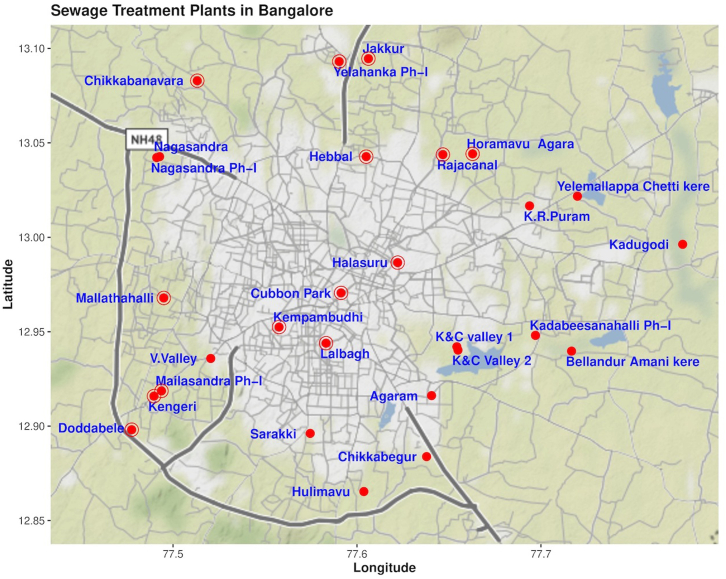
We sampled influent wastewater from 28 sewage treatment plants (STPs) in Bengaluru, India. Grab samples were collected once a week from each STP (January 2022–June 2022) and twice a week from 14 STPs during 0800–1400 h. The details of inflow rate, STP capacity (volume of water), population size is provided in Table 1 and Fig. 1.
Table 1.
Lead time of normalised SARS-CoV-2 viral RNA signal weekly SARS-CoV-2 percent positivity.
| Citywide positivity of samples (%) | 55.77% | |||||
| Total size of STP (MLD) | 1142.5 | |||||
| Total size of STP Inflow (MLD) | 926.15 | |||||
| Population size | 11425000 | |||||
| Citywide EWMA using 7-Day Time Lag | ||||||
|
Rate of Change (%) |
LCL–Mean Days |
LCL–Mean Date |
Rate of Change (%) |
Mean–UCL Days |
Mean–UCL Date |
|
| Raw viral load | 20.83% | 14 | June 3-June 17, 2022 | 32.95% | 7 | June 17-June 24, 2022 |
| 34.32% | 21 | May 6-May 27, 2022 | ||||
| Normalised viral load | 9.52% | 14 | June 3-June 17, 2022 | 19.40% | 7 | June 17-June 24, 2022 |
| 17.60% | 21 | May 6-May 27, 2022 | ||||
| Estimated infected individuals | 22.93% | 14 | May 6-May 20, 2022 | 26.81% | 7 | June 17-June 24, 2022 |
| Covid positive cases | 13.22% | 14 | June 3-June 17, 2022 | |||
| Estimated lead time of wastewater to clinical metric = 14 days | ||||||
| Citywide EWMA using 4-Day Time Lag | ||||||
| Raw viral load | 50.50% | 4 | May 18-May 22, 2022 | 60.67% | 4 | June 19-June 23, 2022 |
| 49.22% | 16 | May 30-June 15, 2022 | ||||
| Normalized viral load | 21.85% | 4 | May 18-May 22, 2022 | 30.54% | 4 | June 19-June 23, 2022 |
| 21.06% | 16 | May 30-June 15, 2022 | ||||
| Estimated infected individuals | 22% | 4 | May 10-May 14, 2022 | 44.43% | 4 | June 19-June 23, 2022 |
| 13.92% | 16 | May 30-June 15, 2022 | ||||
| Covid positive cases | 10.21% | 8 | June 3-June 11, 2022 | 7.26% | 12 | June 11-June 23, 2022 |
| Estimated lead time of wastewater to clinical metric = 8 days | ||||||
| Sewershed Site EWMA | ||||||
| Sewershed site (Population) | ||||||
| Kempambudhi (10,000) | 58% positivity | |||||
| 1 MLD; 0.97 MLD Inflow | 26.55% | 7 | June 16-June 23, 2022 | |||
| Lalbagh (15,000) | 61.54% positivity | |||||
| 1.5 MLD | 48.74% | 7 | May 31-June 7, 2022 | |||
| Halasuru (20,000) | 68% positivity | |||||
| 2 MLD; 1.97 MLD Inflow | 179.70% | 14 | April 4-April 19, 2022 | |||
| Cubbon Park (40,000) | 43.48% positivity | |||||
| 4 MLD; 0.97 MLD Inflow | 52.27% | 14 | May 24-June 7, 2022 | |||
| Chikkabanavara (50,000) | 60% positivity | |||||
| 5 MLD; 4.97 MLD Inflow | 110.19% | 21 | May 23- June 13, 2022 | |||
| Mallathahalli (50,000) | 37.50% positivity | |||||
| 5 MLD; 3.76 MLD Inflow | 42.74% | 7 | June 20-June 27, 2022 | |||
| Chikkabegur (50,000) | 60% positivity | |||||
| 5 MLD; 3.57 MLD Inflow | 99.37% | 14 | May 26- June 9, 2022 | 53.69% | 7 | June 16-June 23, 2022 |
| Sarakki (50,000) | 53.58% positivity | |||||
| 5 MLD; 3.29 MLD | 87.35% | 14 | May 26-June 9, 2022 | 118.81% | 7 | June 16-June 23, 2022 |
| Yelahanka Ph-I (100000) | 68.97% positivity | |||||
| 10 MLD; 10.21 MLD Inflow | 38.35% | 7 | May 31-June 7, 2022 | |||
| Hulimavu (100000) | 57.69% positivity | |||||
| 10 MLD; 7.49 MLD Inflow | 82.17% | 14 | May 19-June 2, 2022 | 49.42% | 7 | June 16-June 23, 2022 |
| Yele Mallappa Chetti Kere (150,000) | 65.38% positivity | |||||
| 15 MLD; 14.66 MLD Inflow | 50.09% | 14 | May 25-June 8, 2022 | |||
| Jakkur (150,000) | 43.33% positivity | |||||
| 15 MLD; 15.34 MLD Inflow | 103.75% | 14 | May 27-June 10, 2022 | |||
| K R Puram (200,000)a | 68% positivity | |||||
| 20 MLD; 16.21 MLD Inflow | 32.20% | 14 | May 25-June 8, 2022 | 33.33% | 7 | June 15-June 22, 2022 |
| Nagasandra (200,000) | 45.83% positivity | |||||
| 20 MLD; 13.48 MLD Inflow | 100.52% | 14 | May 23-June 6, 2022 | 61.65% | 7 | June 6- June 13, 2022 |
| Nagasandra Ph-I (200,000) | 36.36% positivity | |||||
| 20 MLD; 10.2 MLD Inflow | 338.96% | 14 | June 13-June 27, 2022 | |||
| Horamavu Agara (200,000) | 58.33% positivity | |||||
| 20 MLD; 18.69 MLD Inflow | 267.42% | 7 | May 25-June 1, 2022 | 83.57% | 7 | June 15-June 22, 2022 |
| Agaram (350000)a | 61.54% positivity | |||||
| 35 MLD; 33.8 MLD Inflow | 75.91% | 21 | May 18-June 8, 2022 | 32.83% | 7 | June 15-June 22, 2022 |
| Rajacanal (400000) | 70% positivity | |||||
| 42 MLD; 42.92 MLD Inflow | 35.53% | 7 | May 3-May 10, 2022 | 4 | ||
| Doddabelee (400000) | 43.48% positivity | |||||
| 40 MLD; 40.46 MLD Inflow | 122.06% | 14 | May 16-May 30, 2022 | |||
| Kadabeesanahalli Ph-I (500000) | 56% positivity | |||||
| 50 MLD; 42.94 MLD Inflow | 52.81% | 14 | June 16-June 30, 2022 | |||
| KC Valley 2 (600000)a | 57.69% positivity | |||||
| 60 MLD; 60.72 MLD Inflow | 26.86% | 7 | May 11-May 18, 2022 | 93.71% | 7 | June 15-June 22, 2022 |
| Kengeri (600000) | 58.33% positivity | |||||
| 60 MLD; 34.29 MLD Inflow | 50.55% | 7 | May 30-June 6, 2022 | |||
| Mailasandra Ph-I (750000) | 57.69% positivity | |||||
| 75 MLD: 64.9 MLD Inflow | 58.32% | 14 | May 23-June 6, 2022 | |||
| Hebbal (1000000) | 66.67% positivity | |||||
| 100 MLD; 78.52 MLD Inflow | 65.69% | April 26-June 7, 2022 | ||||
| Bellandur Amani Kere (900000) | 76% positivity | |||||
| 90 MLD; 94.7 MLD Inflow | 49% | 14 | April 28-May 12, 2022 | 23.38% | 7 | May 12-May 19, 2022 |
| K & C Valley 1 (2480000)a | 73.08% positivity | |||||
| 248 MLD; 219.2 MLD Inflow | 37.10% | 7 | May 25-June 1, 2022 | 33.10% | 7 | June 15- June 22, 2022 |
| V. Valleyb (1,800,000) | 31.25% positivity | |||||
| 180 MLD; 83 MLD Inflow | 77.56% | 21 | June 2-June 23, 2022 | |||
| Kadugodib (60000) | 27.27% positivity | |||||
| 6 MLD; 3.57 MLD Inflow | ||||||
EWMA = Exponentially Weighted Moving Average; MLD = million litres per day.
Sewershed sites showed ‘red alert’ with viral load crossing the upper-class limit (UCL).
Sewershed sites with PCR inhibitors/poor quality data.
Fig. 1.
Location (red dots) of sewage treatment plants sampled in Bengaluru.
Samples were collected in 200 mL plastic bottles, tightly sealed upon collection, and stored at 4 °C in the field. All samples were processed within 24 h at a biosafety level 2 facility following protocol (modified from 11, 12, 13,16, 17, 18, 19) to provide near real-time information on viral concentrations in sewage. Briefly, samples were subjected to heat inactivation and incubated in a 200 mL bottle at 60 °C for 90 min and divided into three replicates. Subsequently, 40 mL of master sample (total 140 mL from 200 mL bottle) was transferred to three 50 mL centrifuge tubes containing 0.9 g NaCl and 4 g polyethylene glycol (PEG; 8000 MW). The PEG and NaCl mixture in the samples was vortexed until it dissolved, and the solution was centrifuged at 11,000 rpm for 30 min at 4 °C. After discarding the supernatant, 600 μL of lysis buffer was added to resuspend the pellets. Finally, the solution was transferred to a centrifuge tube and briefly vortexed to dissolve the pellet. Viral RNA was extracted using Qiagen Viral RNA mini kit protocol following the manufacturer's instructions and 50 μL of elution was stored at −80 °C until subsequent analysis.
Reverse transcriptase-quantitative PCR (RT-qPCR)
Using GenepPath Dx CoViDx One v2.1.1TK-Quantitative multiplex RT-qPCR kit, each sample was screened for the presence of SARS-CoV-2 RNA. The kit targets three viral genes: N-gene, RdRp-gene, and E-gene along with a human control gene (RNAase P gene). Since the kit is designed for a qualitative analysis (positive or negative), COVID-19 Viral Load Calculation Tool (RUO; https://coviquant.genepathdx.com/) was used for the quantifying viral load in the samples. For viral load calculation, three standards provided within the kit were used: high standard = 5000 copies/μL, medium standard = 500 copies/μL and low standard = 50 copies/μL. The online tool uses the Ct values (cut-off 35) of high, medium and low and calculate the unknown in copies/μl of each sample. A negative extraction control and RNA extraction control was analysed with each plate. In order to eliminate the false negative, RT-qPCR was performed on three extracted replicates of each sample and any replicate that generated a result defined as ‘positive’ by the test manufacturer was considered positive. Samples with invalid results were repeated as per the manufacturer's instructions (Supplementary Table S1).
Early warning SARS-CoV-2 detection modelling
By investigating viral RNA concentrations in the wastewater and correlating it with community testing data on a weekly basis, we generated an early warning and real-time map of COVID-19 infection dynamics in the city. We conducted two main analyses using the raw and normalised viral load. At the citywide level, we used consolidated weekly and biweekly data (except invalids) and weekly data for each STP. We calculated the daily viral load, VL (copies/d) for SARS-CoV-2, by normalizing raw viral load, C (copies/mL) to the average daily STP flow, Q(L/d) using Equation (1)19 and P is the population of inhabitants the catchment of the respective sewershed site.
| Equation (1) |
Spatial and temporal dynamics in viral load copies by STPs and at the citywide level
To understand spatial and temporal changes in viral load, we used an Exponentially Weighted Moving Average (EWMA)20 as the monitoring algorithm to detect moderate and persistent shifts in cases. The EWMA is calculated as follow:
Where, is the smoothing parameter with condition and is the mean of the process at time (considered 7-days and 4-days lag; see below).
We used the control limits of the EWMA chart for detecting a mean shift as follow:
Where, is the mean (centreline), is the upper control limit, is the lower control limit, is the process standard deviation, is the width of the boundary of the control and EWMA chart values are computed by using the R package “qcc.”21
The EWMA control chart, as the surveillance algorithm, triggers an alarm of outbreak once the monitoring statistic exceeds the control limit, which is computed based on the design of the control chart. The most significant advantage of the EWMA chart is that it can be used to detect small shifts in the process mean, which is important for early detection and faster response. The EWMA gives the maximum weightage to the most recent observations and exponentially gives less weight to all earlier observations. Since there was no previous wastewater data available from the city and due to the high infectivity of COVID-19, we give more weightage to the most recent observations; therefore, we consider the smoothing parameter , and is the width of the boundary of the control chart (Supplementary Table S2). Similar parameter values have been used for monitoring the COVID-19 outbreak in earlier studies.22, 23 We use the mean line as the threshold, and the ‘early warning’ signal is triggered when the weekly viral load/incidence exceeds the threshold. If predicted data points are below/above the average line, this indicates a lower/higher risk of COVID-19 infection. However, if predicted data points are greater than the projected line (upper control limit), this indicates a ‘red alert’ of infection with COVID-19. The EWMA control chart is robust and can accommodate missing data points (up to 15%). Also, the inconsistency in the data points can be tackled by using the log scales and the above-mentioned smoothing parameters in the EWMA analysis.
SARS-CoV-2 viral load copies in the wastewater can potentially be affected by the incubation period of a particular variant, population susceptibility, and differences in degradation rates in the sewer system. The mean incubation period of the ancestral SARS-CoV-2 variant is 6.4 days,24 the SARS-CoV-2 delta variant is 4.8 days,25, 26 and the omicron variant is 3.6 days.26 To compare the SARS-CoV-2 wastewater signal with COVID-19 cases at the citywide level, we used time lags of 7-days and 4-days to accommodate variable incubation periods of multiple variants seen in the wastewater (see section on variant analysis). Daily reports detailing the number of samples tested and positive for COVID-19 were collected from the BBMP COVID-19 portal (https://apps.bbmpgov.in/Covid19/en/mediabulletin.php). The omicron and delta SARS-CoV-2 variants shed culturable virus more than five days after symptom onset or first positive test,27 and virus in faeces can remain for more than 20 days, in which case prolonged shedding may contribute a significant signal to wastewater.27,28 We modelled EWMA with time lags of 4-days and 7-days. Time-step (4-days and 7-days lag) Pearson's R correlation analyses were performed to evaluate the fit between log-transformed SARS-CoV-2 viral copies in wastewater and reported clinical data. We did not conduct this analysis at the STP level as the clinical testing data for each sewershed catchment size was unavailable. Using citywide clinical testing data gives a biased estimate due to varying inflow and catchment size of each sewershed.
Estimation of infected individuals by STP and citywide
We estimated the number of infected individuals by each STP and citywide. The prevalence of SARS-CoV-2 infection within the catchment of each STP was estimated using the total number of viral RNA copies in wastewater each day, as measured in wastewater by RT-qPCR, and the number of SARS-CoV-2 RNA copies shed in stool by an infected following Ahmed et al.10 Equation (2).
| Equation (2) |
Briefly, SARS-CoV-2 RNA copies/L of wastewater were modelled as point estimates for each date of detection. The daily flow rate of wastewater was calculated for each sewershed using the product of the population of each catchment area and the observed average per capita wastewater rate of 100 L/person/day. The daily stool mass of 128 g was used as per Rose et al.29 and one person shedding SARS-CoV-2 RNA 107copies/g of faeces was used as per Foladori et al.30 Spearman's correlation coefficient was used to calculate the sensitivity of the estimated number of cases and the normalized viral load of each sewershed.
SARS-CoV-2 genome amplification and sequencing
All samples positive with the RTqPCR kit were sequenced at the Next Generation Genomics Facility in National Centre for Biological Sciences, Tata Institute for Fundamental Research, Bengaluru. The libraries were prepared using the Illumina COVIDSeq Test kit (Cat no: 20043675, Illumina Inc, USA). Extracted RNA samples were primed by random hexamers for reverse transcription. The complementary DNA (cDNA) products were amplified using ARTIC V3 primer set targeting the entire SARS-CoV-2 genome and human cDNA targets in two different multiplex PCR reactions. The amplified product was later processed for tagmentation, and adapter ligation using IDT for Illumina Nextera DNA Unique Dual Indexes Sets A–D IDT for Illumina-PCR Indexes Sets 1–4 (384 Indexes, Cat no: 20043137, Illumina Inc, USA). Further enrichment and clean-up were performed as per the manufacturer's instructions.31
Pooled libraries were quantified using a qubit 4.0 fluorometer (Invitrogen, USA), and library sizes were analysed using TapeStation 4200 (Agilent, USA). The libraries were normalised to 2 nM and denatured with 0.1 N NaOH. 8.1 pM of denatured libraries were loaded onto HiSeq Rapid SR flow cell v2 followed by dual indexed Hiseq 2500 - 1 × 50 or custom 1 × 120 cycle workflow as per the manufacturer's instructions (Illumina Inc).
Bioinformatics
Wastewater samples consist of a mixture of variants circulating in a population in contrast with clinical samples which might be infected with a single variant. In the first step, the raw reads were aligned with the reference genome of SARS-CoV-2 (MN908947). The processed reads are aligned with the reference genome by BWA Mem32 and various coverage statistics are taken by SAMtools coverage/bedcov.33 The alignment was used for a single nucleotide variant (SNV) calling using iVar.34 The iVar tool was used to trim the primers and generate a table for each sample with mutation frequency data and an adj p-value (Fisher's test) for altered positions of SARS-CoV-2 from the BAM files. iVar was run with a minimum base quality filter of 20 (default value) using the reference genome of SARS-CoV-2 and the feature file Sars_cov_2.ASM985889v3.101.gff3 from NCBI. For predicting the lineage abundances, a deconvolution matrix was generated using Freyja (https://github.com/andersen-lab/Freyja)35 - a dedicated bioinformatic pipeline for wastewater analysis. From measurements of SNV frequency and sequencing depth at each position in the genome, Freyja returns an estimate of the true lineage abundances in the sample. We cross-validated the efficiency of the deconvolution tool in reliably inferring the mixed samples with clinical data. We first applied the pipeline to clinical data from the study by Bhoyar and colleagues31 and Bengaluru – derived sequences from clinical samples submitted to the GISAID EpiCoV database (hereafter, GISAID; https://www.gisaid.org/). Frejya used the UShER tree with WHO designation and outbreak.info metadata.36
Virus richness and diversity
For community diversity analyses, we used lineage abundances (normalized by the read depth within-sample relative abundances using Freyja) to generate Shannon diversity indices and Bray–Curtis dissimilarity matrices with vegan v2.5-7.37 We also ran Adonis permutational multivariate analysis of variance (PERMANOVA) tests on the distance matrices and performed nonmetric multidimensional scaling on the data and compared the relative abundances of lineages over time with “lmerTest” v3.1-3 using STP as a random effect.38
Role of the funding source
The funders had no role in study design, data collection, data analysis, interpretation, writing of the report.
Results
Longitudinal wastewater sampling can predict SARS-CoV-2 rise 1–2 weeks in advance
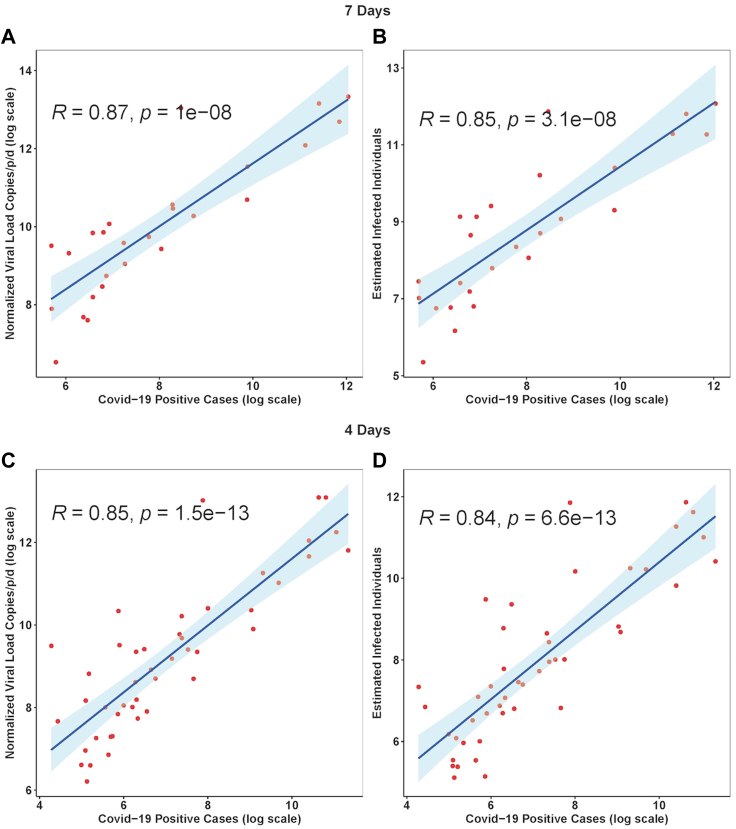
The normalised temporal trend of SARS-CoV-2 RNA in wastewater and new positive cases in Bengaluru city are shown in Fig. 2. Pearson's correlation for citywide viral load showed a marginally stronger correlation with citywide COVID-19 cases with a time lag of 7-days than for 4-days (R2 = 0.87, p = 0.00001; Fig. 3A & B). The sensitivity analysis showed that the estimated number of infections strongly correlates with the log SARS-CoV-2 RNA copies detected in the wastewater (Fig. 3C and D).
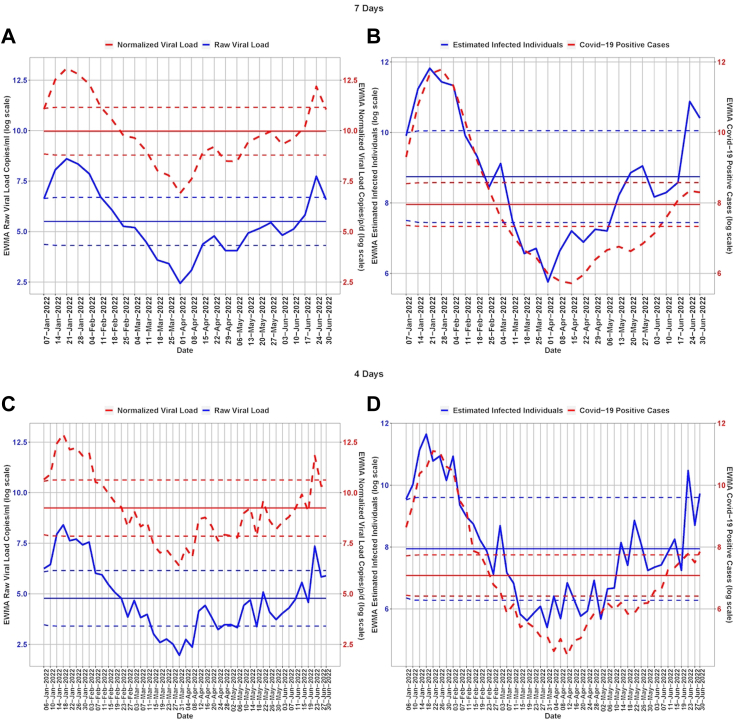
Fig. 2.
Temporal dynamics of normalized viral load in wastewater and COVID-19 cases in Bengaluru. The dark centre red line shows the mean viral load, the dotted line above the mean is the upper control limit and the dotted line below the mean is the lower control limit. The 3rd wave started in late December 2021 with peak in January 2022 and dropped down in February 2022.
Fig. 3.
Viral concentrations correlate with daily new Covid-19 cases with a 4-d and 7-d time lag in Bengaluru city. The Blue solid line is the linear regression fitting. Light blue area: 95% confidence interval from standard error of the fitting.
We used EWMA of RNA concentration in wastewater (smoothing parameter = 0.7, ) and new clinical cases (smoothing parameter = 0.7, ) to show the qualitative trend in the city. Evidently, there were two major outbreaks- January 2022 and June 2022. The EWMA captured the increased viral load and associated estimated infection during the peak of the SARS-CoV-2 wave in January 2022. The EWMA showed an increased viral trend in June as a ‘red alert’ and an associated estimated number of infections at least 14 days and 8 days in advance using time-lags of 7 days and 4-days, respectively (Table 1, Supplementary Table S3). SARS-CoV-2 was detected throughout the period with a low viral load of 6.38 copies/person/day (log scale) from March 27, 2022–April 8, 2022. While the pattern of viral load mirrored the clinical data, COVID-19 positive cases appeared to remain underreported in the city. The estimated number of infected cases remained high indicating viral detection in wastewater in the early stage (below LCL) of the local outbreak (Fig. 2). Our EWMA analysis showed that using a time lag of 7 days, the increased estimated infected individuals trend reached red alert levels (UCL ∼ 23,106 cases/week) whereas COVID-19 cases were near the mean (threshold value ∼2847 cases/week). Using a time lag of 4 days, the estimated infected individuals reached the red alert (UCL ∼ 14,803 cases/4 days) whereas COVID-19 cases were near the mean and were rising to reach the red alert (UCL ∼2310 cases/4 days).
The viral trend showed an increase from 15th April 2022 which remained below the early warning stage, with a consistent 30–70% rise in the following weeks reaching the ‘early warning’ stage by June 17, 2022 (Table 1).
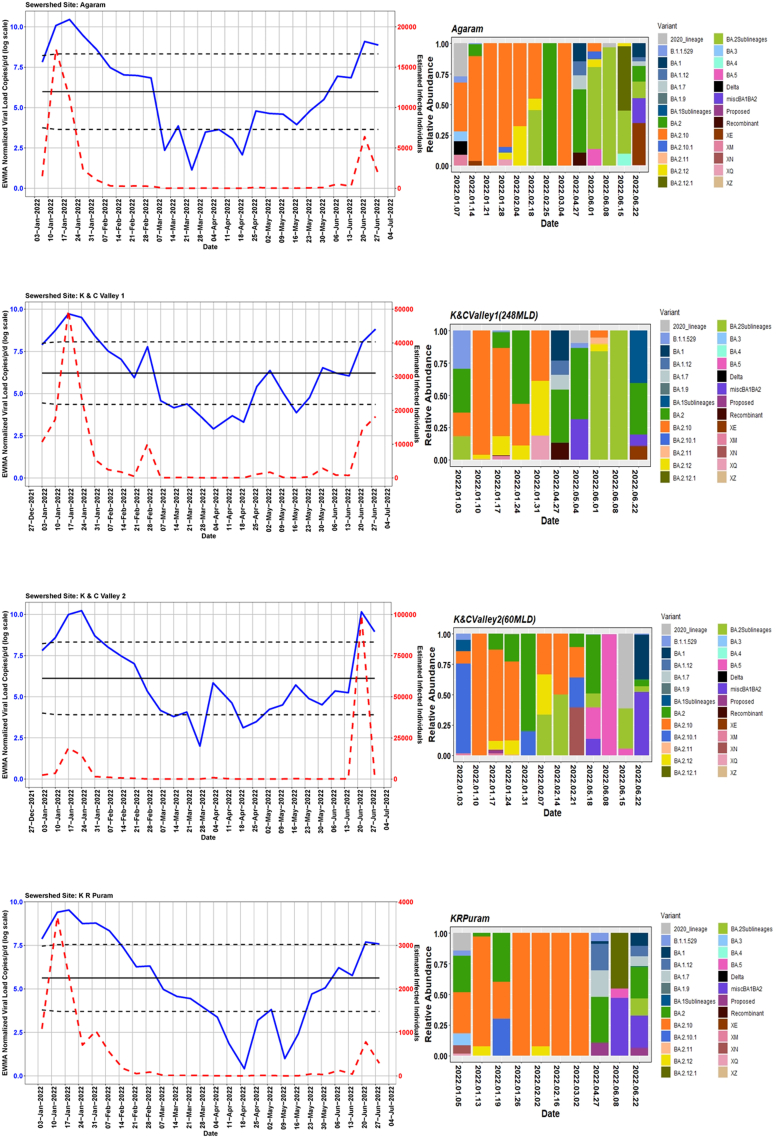
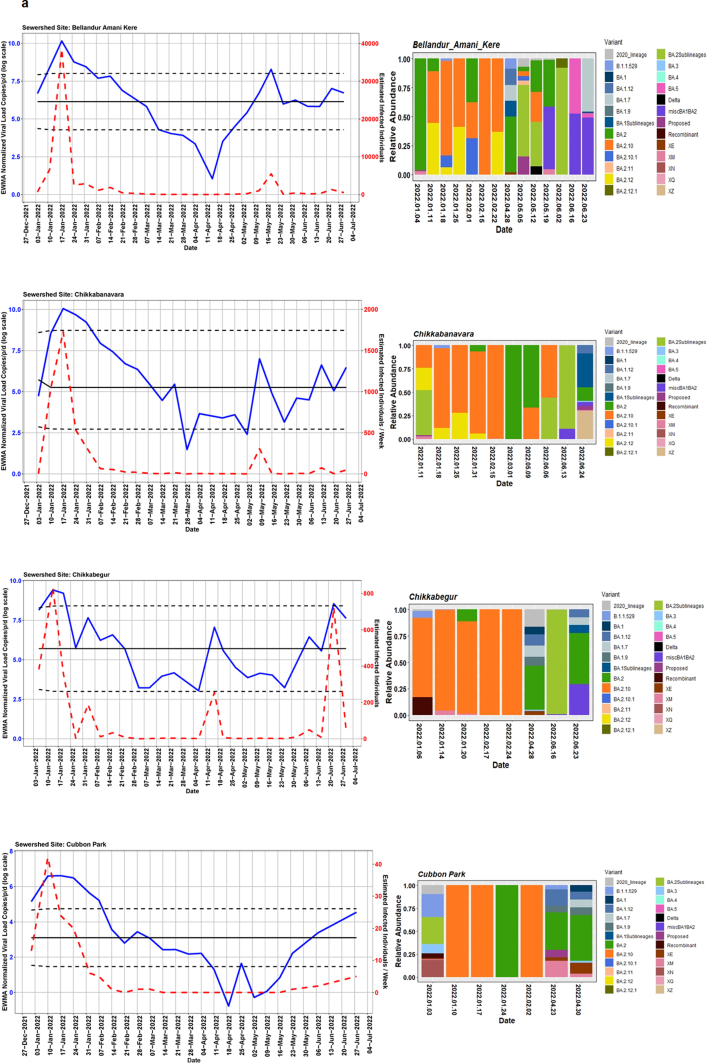
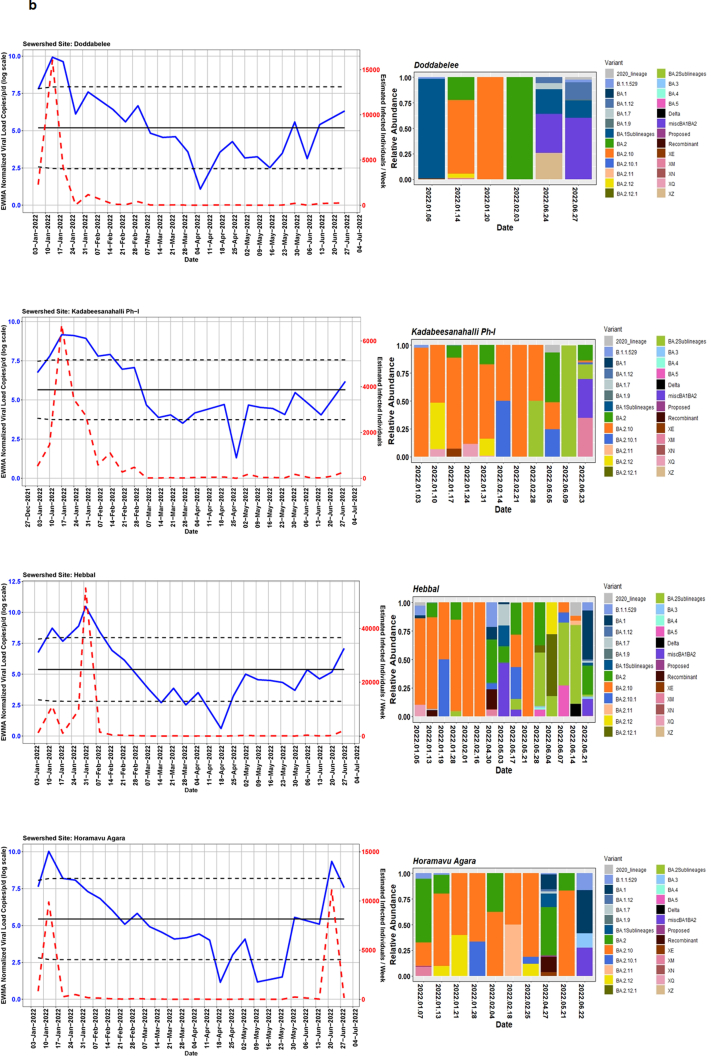
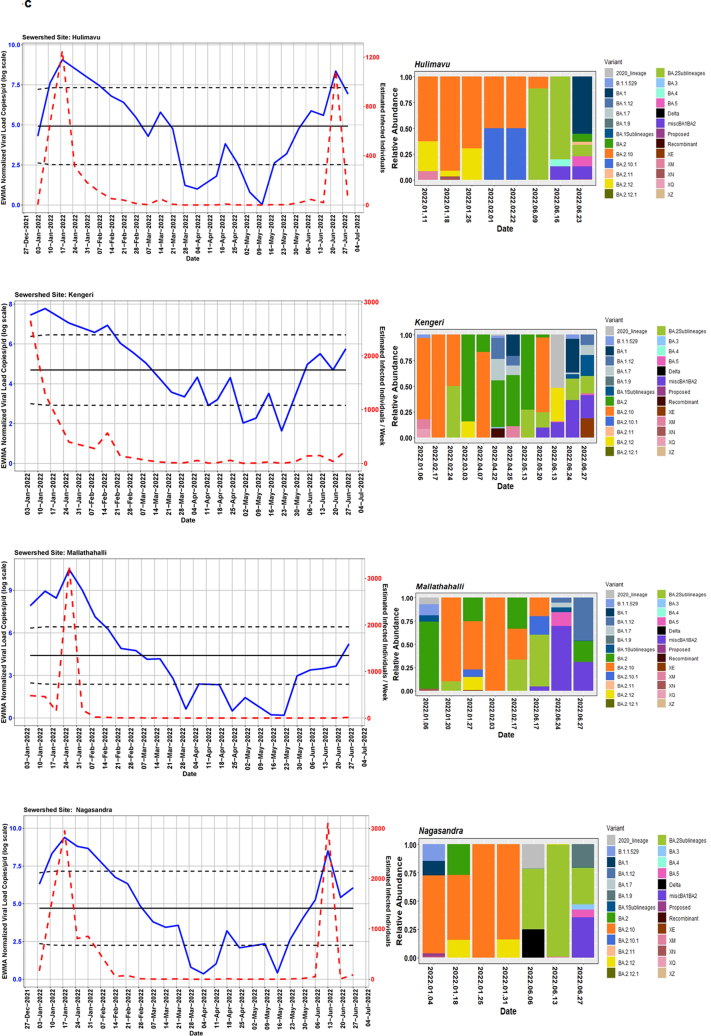
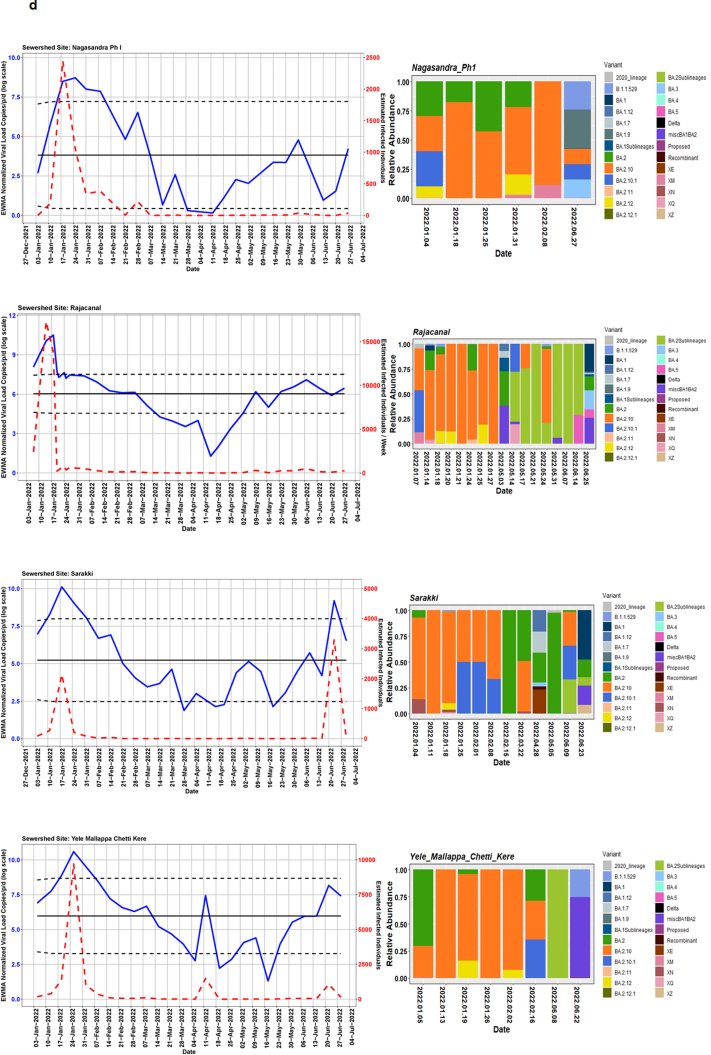
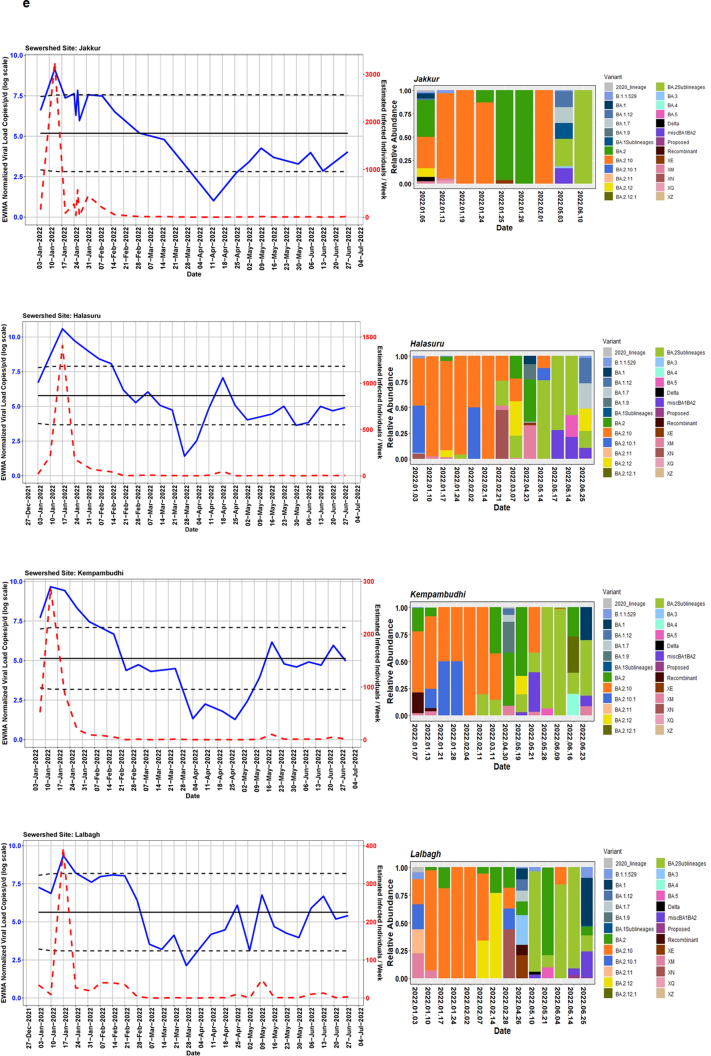
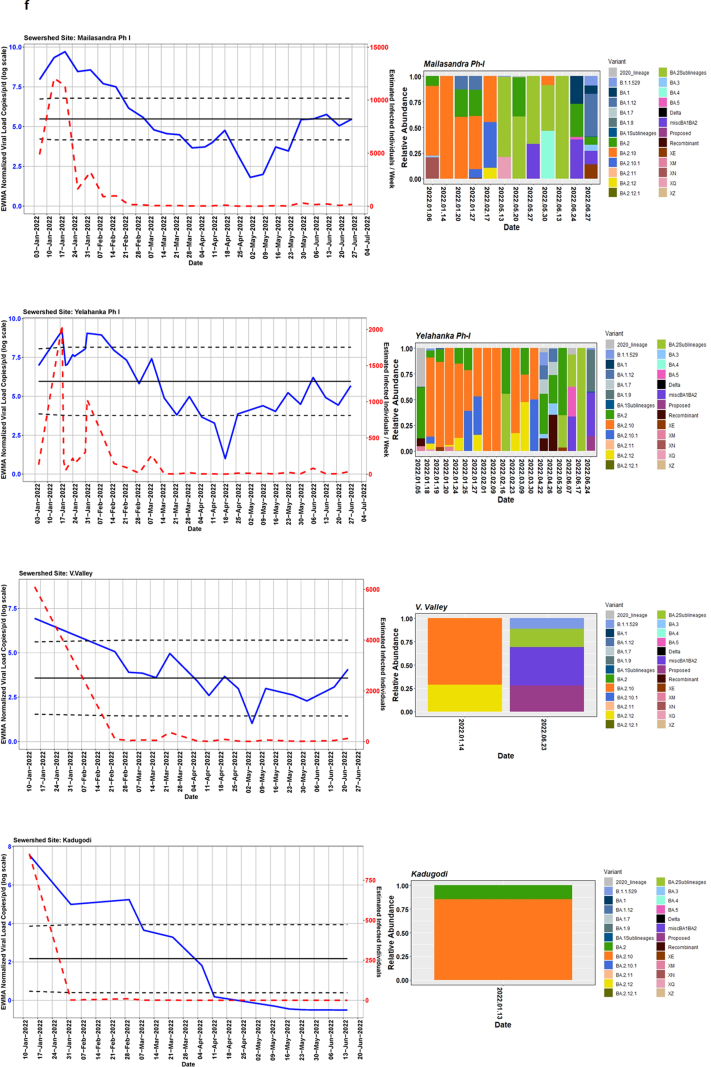
We detected comparable levels and variable SARS-CoV-2 infection dynamics for sewage treatment plants (STPs; Fig. 4). All STPs showed viral signals throughout the study period. However, 10 STPs consistently showed high positivity (Table 1, Supplementary Table S4). The EWMA varied for each STP driven by the capacity, inflow rate and catchment area. Four sewershed sites- Agaram, KR Puram, KC Valley1 and KC Valley2 showed ‘red alert’ with the EWMA viral load higher than the projected line (upper control limit) (Fig. 4). Seventeen sewershed sites; Chikkabegur, Chikkabanvara, Cubbon Park, Doddabelee, Hebbal, Horamavu Agara, Kengeri, Mallathahalli, Rajacanal, V. Valley, Sarakki, Hulimavi, Yele Mallappa Chetti Kere, Nagasandra, Nagasandra-Ph-1, Kadabeesanahalli Ph-1, and Bellandur Amani Kere showed early warning signal with viral load projected above the mean line (Fig. S1 a-f). The EWMA mirrored estimated infected individuals for the catchment area of each STP and mainly reflected with an increase in estimated infected individuals with an increasing viral trend.
Fig. 4.
The exponentially weighted moving average charts for sewershed sites showing ‘red alert’ and the relative abundance of variants by date of collection.
SARS-CoV-2 abundance and diversity in wastewater
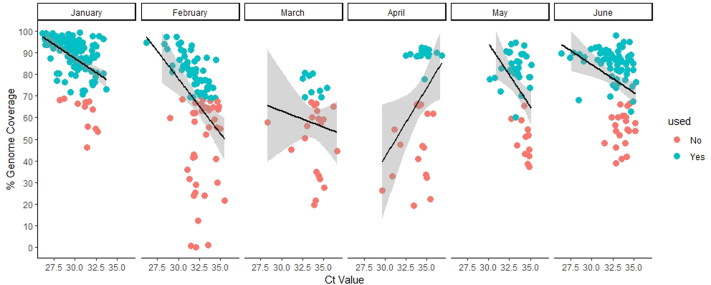
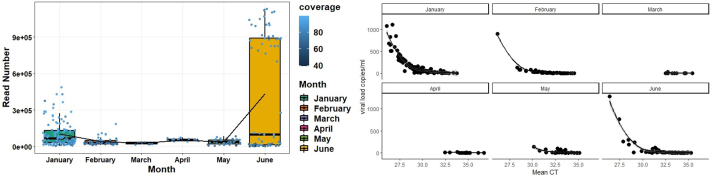
We analysed 878 wastewater samples during Jan 1, 2022–June 30, 2022. To compare wastewater genomic surveillance with clinical surveillance, we sequenced 422 SARS-CoV-2 positive wastewater samples. The genome coverage ranged between 1% and 99%. The CT values showed a negative correlation with viral load copies (r = −0.69, P < 0.0001) and genome coverage (r = −0.44, P < 0.0001) (Fig. S2; Supplementary Table S5). Therefore, we considered samples (n = 304) above 70% genome coverage for subsequent analyses. The low viral copies between 28th March to 8th April 2022 are reflected in our sequencing success; hence, there is a gap in sequencing data despite continuous sampling. The read abundance was not significantly (P = 0.74) different across month (Fig. S3).
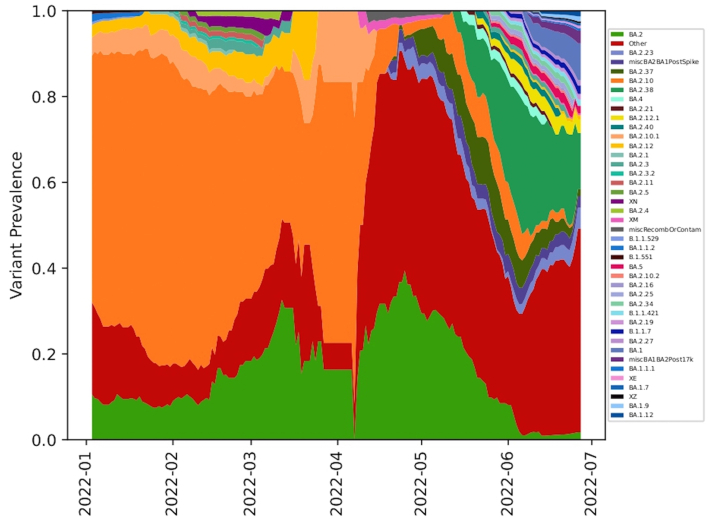
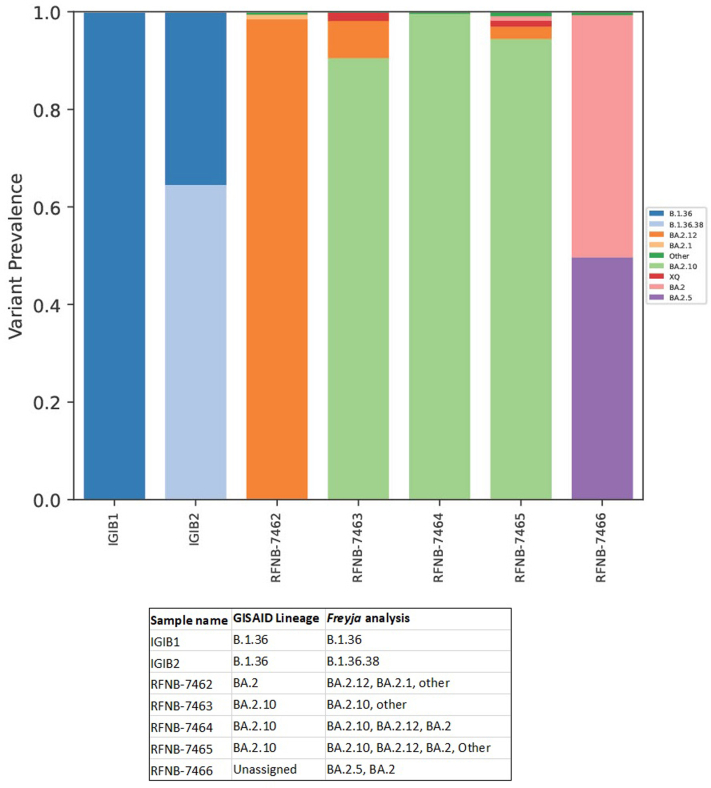
Using Freyja, we analysed relative abundance and diversity in SARS-CoV-2 lineages. Our validation results for clinical samples showed the expected lineages. In addition, a mixture of variants that were not identified using traditional pipeline (see Fig. S4). These clinical samples were reported infected with only one variant, however, we found mutations associated with other variant suggesting mixed infections. Our follow-up analysis using Freyja showed a total abundance of 1220 lineages representing 152 distinct lineages in wastewater from January to June 2022 (Fig. 5). Of these seven lineages from omicron family were in dominant. We found BA.2.10 was the most dominant lineage (14.83%), followed by BA.2 (10.49%), B.1.1529 (5.1%), BA.2.12 (5%) and BA.2.10.1 (3.1%). We found signature mutations of two recombinant lineages of BA.1.1 and BA.2 - XM (3.9%) and XQ and XE (1%).
Fig. 5.
Relative abundance of SARS-CoV-2 lineages in wastewater by date of collection in Bangalore.
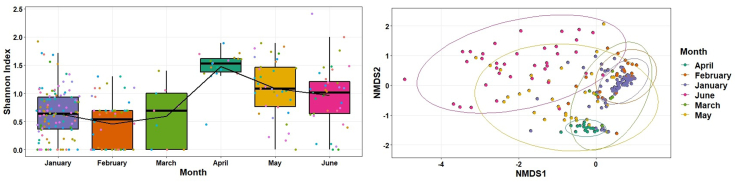
We analysed the Shannon indices to understand overall alpha diversity in SARS-CoV-2 which was significantly different between months (F[5, 253 ] = 19.6, P < 0.001) and then used Tukey's honestly significant difference (HSD) post hoc pairwise comparison testing to show that only January–April, February–April, June–April, March–April, June–February, May–February, June–January and May–January alpha diversities were different from each other (adjusted P value [Padj] < 0.05) (Fig. 6). In contrast, the alpha diversity was not significantly different between STP (F [27, 201] = 0.65, P = 0.90). We also compared the Bray–Curtis dissimilarities of the samples with Adonis and found that overall beta diversity values by month were significantly different (R2 = 0.27, P < 0.001) but not by STP (R2 = 0.09, P = 0.93).
Fig. 6.
Shannon diversity (alpha diversity) in lineages across months and non-metric multidimensional scaling (NMDS) ordination of the Bray–curtis dissimilarities in lineages by month.
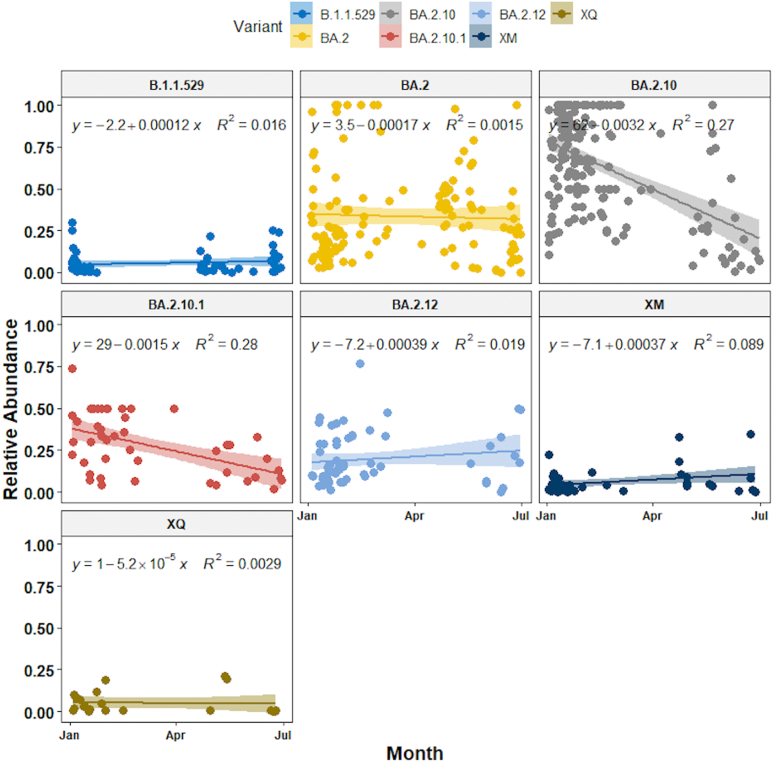
Finally, we used the linear-mixed effect model to show that the relative abundance of seven dominant lineages (VOC) showed change over time - XM (recombinant lineage of BA.1.1 and BA.2) showed an increasing trend whereas BA.2.10, BA.2.10.1 decreased from January to June 2022. BA.2 lineage remained dominant in May along with ‘other’ lineages found in low proportions (<1%). B.1.1.529, BA.2 BA.2.12 and XQ showed no significant change in their relative abundance (Fig. 7). BA.2.12 was the dominant lineage during the third wave in January with high viral load copies/ml.
Fig. 7.
Relative abundance of dominant SARS-CoV-2 lineages showed a variable trend between January 2022 to June 2022.
Comparison between wastewater and clinical data
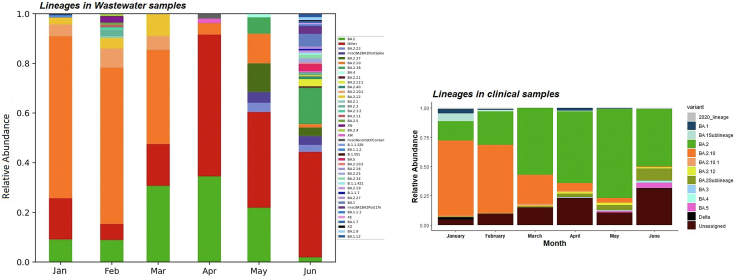
We analysed 13,478 SARS-CoV-2 genomes submitted on GISAID from January to June (January; 7827, February; 3364, March; 364, April; 229, May; 642, June; 1052) from Bengaluru. Of these, 71 were distinct lineages that followed a similar pattern as wastewater samples- BA.2.10 was the dominant lineage with 51.65% of the genomes, 26.90% of the genomes were assigned to BA.2 and ‘unassigned’ (recombinant lineages) constituted 8.73% of the diversity.
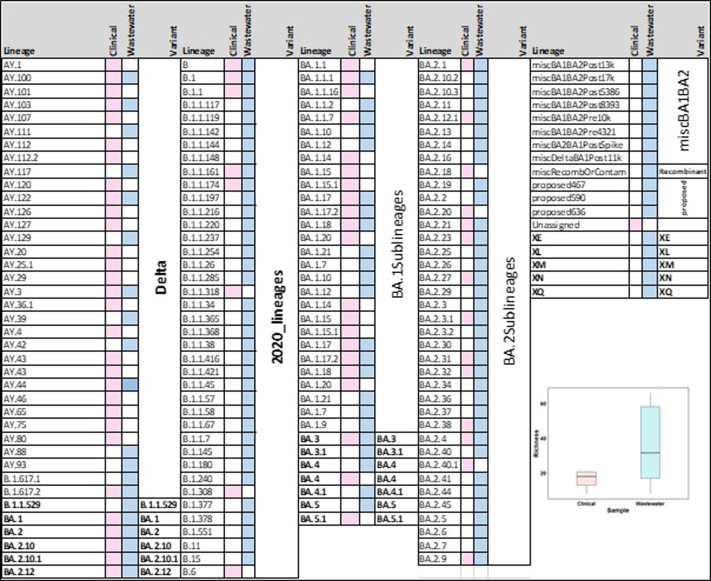
To test whether wastewater genomic surveillance can detect changes in lineage abundance circulating at the community level, we compared dominant Variant of Concern (VOC; variants associated with high transmissibility or immune evasion) detection rates between clinical and wastewater sequenced data. We found that the general trend in lineage abundance remained very comparable to the dynamics between wastewater and clinical data. Furthermore, the wastewater genomic surveillance consistently recorded mutations associated with ‘other’ lineages in low frequency (<1%), which were otherwise not seen in the clinical samples. There was a huge mismatch in lineage diversity which was significantly higher in wastewater than recorded in clinical samples (Mann–Whitney U test, P < 0.001; Fig. S5).
To understand if wastewater sampling can help in the early detection of emerging variants in the city, we compared the first detection of VOC in wastewater with the collection dates of clinical samples sequenced as part of genomic surveillance in Bengaluru. In contrast, the exact collection dates for many samples were not mentioned in the database, which prevented us from conducting a temporal comparison across several lineages. Nonetheless, we compared the month of the emergence of VOC in clinical samples from GISAID. For example, BA.2.10.1, BA.2.12 were detected two months prior in wastewater in January 2022 to the first detection in clinical samples in March 2022 (Fig. 8).
Fig. 8.
Aggregated relative abundance of SARS-CoV-2 variants by month in wastewater in the left panel analysed using Freyja and in clinical genomic surveillance in the right panel. Lineages retrieved in less than 1% relative abundance are aggregated as ‘Other’. BA.1Sublineages are aggregate of lineages less than 1% − BA.1.1, BA.1.1.1, BA.1.1.16, BA.1.1.7, BA.1.14, BA.1.15, BA.1.15.1, BA.1.17, BA.1.17.2, BA.1.18, BA.1.20. BA.2Sublineages are aggregate of lineages less than 1% − BA.12.1, BA.2.18, BA.2.20, BA.2.21, BA.2.23, BA.2.27, BA.2.3, BA.2.3.1, BA.2.31, BA.2.32, BA.2.38, BA.2.4, BA.2.40.1, BA.2.9. See Fig. S5 for details.
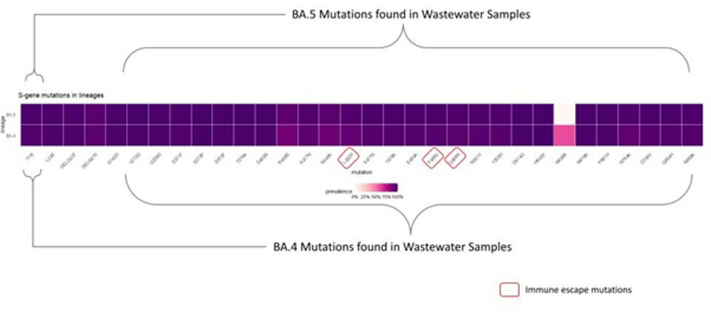
Until June end, there were 12 genomes of BA.4, and 58 genomes of BA.5 were submitted on GISAID. The first clinical sample of BA.5 was collected on May 11, 2022 and sequenced in wastewater sample collected on May 18, 2022 from two STPs (Lalbagh and KC Valley2). BA.5 was detected on two consecutive sampling on June 8, and June 15, 2022 from KC Valley2. Similarly, BA.4. was first isolated on May 24, 2022 in clinical samples and first found in wastewater on May 28, 2022 from Hebbal sewershed site and May 30, 2022 from Mailasandra subsequently sequenced in a sample collected on June 16, 2022 in Hulimavu. In total, 73 spike mutations were recorded in BA.4 and BA.5 in the wastewater sample (Fig. S6). The increases in lineage detection frequency for VOC - ‘unassigned’ (recombinant lineages probably equates to XM, XE, XN in wastewater) showed an increasing trend whereas BA.2.10 and BA.2 declined with time.
Our study shows a localised increase in variant diversity and a spike in VOC abundance -BA.1, BA.1 sub lineages, BA.2 sub lineages, and prevalence of ancient SARS-CoV-2 lineages recorded in 2020 and Delta lineages across STPs. In addition, the lineage richness was higher in the ‘red alert’ stage of an emerging wave than during the ‘wave’ phase (see Fig. 2). Both wastewater and clinical datasets showed low viral richness during March–April 2022. However, there was a huge surge in lineage diversity in June 2022 which corresponded with increasing viral load both at citywide as well as STPs. This further highlight that the increasing trend was contributed by a shift from BA.2.10 to many BA.2 sub-lineages rather than one dominating lineage at the community level. BA.2.10 was the most dominant lineage seen in both wastewater and clinical samples during the peak of the third wave in January 2022 (Fig. 5).
Discussion
Our longitudinal study provides an in-depth analysis of the SARS-CoV-2 viral concentration and how this relates to the lineage dynamics in wastewater capturing data from more than 11 million people in Bengaluru city. The strong correlation between wastewater viral concentrations and daily reports of new clinically confirmed COVID-19 cases (Fig. 3) – combined with the time lag between the wastewater signal and clinical data – suggests that newly infected individuals contribute significant viral loads to the wastewater. This further suggests that most of this shedding may occur early in infection (asymptomatic phase), prior to the individual seeking healthcare and being tested.39 Therefore, wastewater surveillance can be used as a complementary tool to clinical testing to predict trends in new COVID-19 cases. SARS-CoV-2 concentrations in wastewater began to increase exponentially in late March 2022 which led to a mask mandate by the April 26 2022. This period coincided with the opening of schools and an increase in public movement with a reduced remote working. Wastewater surveillance revealed the largest number of positive cases likely to be Mahadevapura zone, East Zone, and Bommanahalli. By June 7, 2022 (https://timesofindia.indiatimes.com/city/bengaluru/mahadevapura-has-highest-viral-load-sewage-analysis/articleshow/92117008.cms), there was renewed mask mandate which corresponded with a surge in viral load in four sewersheds. We note that inadequate testing, changing criteria of testing and population seeking home tests over RTPCR could introduce uncertainty to the reported cases. Nonetheless, the time-lag used for modelling fits the data and signifies the timing of infection that wastewater signals may reflect shedding dynamics early in infection.
Each STP showed a variable viral dynamic which could be primarily driven by individual shedding rates, viral stability in wastewater, and flow rates of the influent. Our sensitivity analysis indicated that the estimated number of infected individuals at a citywide level was four folds higher than reported cases. This information has been crucial in enhancing testing in targeted locations for the early detection of asymptomatic cases. One of the limitations of our analysis is the lack of clinical testing data from catchment areas of each sewershed site which could help correlate with viral load to devise an effective and economical strategy to track the timing, location, and magnitude of SARS-CoV-2 activity outbreaks. Two sewershed sites, Kadugodi, and V. Valley, had technical challenge in obtaining high quality data and sequences from samples with low viral load and elevated levels of PCR inhibitors (Fig. S1f). SARS-CoV-2 is sensitive to environmental degradation and (temperature, pH, chemicals), processing procedures of STP which can affect the persistence of detectable genetic material in wastewater.40
Wastewater-based genomic surveillance of SARS-CoV-2 has been a valuable resource in identification of novel mutations and early emergence of new variants.41 Wastewater-based genomic data is often fragmented and does not capture all variant-defining mutations on a single genome. Nonetheless, with an increase in detection of multiple variant-associated mutations or mutations not shared by other known variants, we classify these lineages as variant-like upon following Freyja which used the UShER tree with WHO designation and outbreak.info metadata.36 Our wastewater samples contained diverse SARS-CoV-2 lineages which showed significant difference across months but not by sewershed sites suggesting that there was no geographical variation in viral composition in the city. Furthermore, we found low proportion of lineages which were not reported in the clinical data. This discrepancy highlights that clinical testing and sequencing as per the national guidelines, only symptomatic individuals are tested. There were lineages only found in clinical samples but not seen in the wastewater which probably suggests that a very low number of individuals harboured those variants. This further signifies that dominant VOC infecting a large proportion of individuals exhibited similar trends in the wastewater. Furthermore, the clinical data is analysed using a traditional pipeline which assigns a single variant based on the dominant mutations in the sample. We used a pipeline designed to calculate proportions of VOC from the environmental samples that is a highly reproducible computational analysis pipeline with comprehensive reports. Our validation of the pipeline using clinical samples showed mutations associated with mixed lineages in clinical samples (Fig. S1) which probably highlights that the diversity in clinical data might be underestimated and the need to consider alternate approaches.
Given that we sequenced only 2% of wastewater samples relative to the sequenced clinical samples, the emergence of variants in wastewater followed a trend that matched with other countries. For example, the rise of BA.5 is occurring at the same time as the decline in BA.2 in several countries. We detected BA.2.10.1, BA.2.12 two months prior in wastewater in January 2022 to the first detection in clinical samples in March 2022 in Bengaluru whereas BA.4 and BA.5 were detected in 4–7 days late in wastewater. The emergence of a variant in the wastewater implies that a significant proportion of individuals in the community are infected with that variant and shedding the virus. Wastewater testing can provide a less biased snapshot of viral diversity and community health.42 Whereas a late detection in the clinical sample could happen due to limited or biased testing, sequencing43 or a large proportion of individuals were asymptomatic or home testing upon COVID-19 symptoms. Clinical samples were sequenced from a selected hospital which is not representative of the Bengaluru population. Karthikeyan and colleagues35 observed varying periods of VOC lineage detection relative to clinical genomic surveillance, which was attributed to different virus shedding characteristics across lineages.44
While we saw a temporal trend in variants, there was no geographical structure in lineages across sewershed sites. Nonetheless, we noticed a similar pattern in viral load and shift from BA.2.10 to BA.2 sub-lineages-we observed a strong positive association between the local VOC frequency and estimated number of infected individuals which were consistent with the increased transmissibility of this VOC identified.
Our study highlights that quantifying viral titre, correlating with the known number of cases in the area combined with genomic surveillance helps in tracking of VOCs over time and space and helps inform policy-making decisions to control new outbreaks. Several WBE initiatives for SARS-CoV-2 monitoring were established worldwide, and currently, the COVIDpoops19 initiative1 lists 128 dashboards and our data is displayed as part of the Bengaluru pandemic response initiative. The findings from this study were discussed regularly with Bruhat Bengaluru Mahanagara Palike (BBMP) and Bangalore Water Supply and Sewerage Board (BWSSB) to inform policy-making decisions. Our approach can support establishing WBE for monitoring and early-warning system for detecting pathogens beyond SARS-CoV-2.
We developed an early warning system, and its performance was demonstrated by detecting and tracking the SARS- COV-2 infections and variant diversity in the local community and city levels from January 2022 to June 2022. The EWMA chart was able to capture the historical COVID-19 infection patterns and distinguish between endemic situations and the outbreak patterns. Real-time genomic surveillance is the key to understanding the emerging patterns in viral load and variants in the city as it helps to develop a pandemic plan as well as be prepared for future pandemics.
Contributors
FI, UR and VS: designed the study; SL: conducted EWMA modelling, SG, AN, SJ: processed the samples, conducted RNA extractions and RT-qPCR; DS: library preparation and sequencing; ND and KP: conducted bioinformatic analysis. RM and FI secured funding. FI analysed the data and wrote the manuscript. All authors approved the final version of the manuscript.
Data sharing statement
All raw wastewater sequencing data will be available via the NCBI Sequence Read Archive under the BioProject ID PRJNA916712. Consensus sequences from clinical surveillance are all available on GISAID. Freyja is hosted publicly on github (https://github.com/andersen-lab/Freyja) and is available under a BSD-2-Clause License (https://doi.org/10.5281/zenodo.6585067, version 1.3.7). Freyja is accessible as a package via bioconda (https://bioconda.github.io/recipes/freyja/README.html).
Editor note
The Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
Declaration of interests
None.
Acknowledgments
This work has been supported by funding from the Rockefeller Foundation grant to National Centre for Biological Sciences, TIFR) and the Indian Council of Medical Research grant to (FI) Tata Institute for Genetics and Society and Tata Trusts. We sincerely thank Bengaluru Water Supply and Sewerage Board for providing access to the sewershed sites. We are grateful to NCBS sequencing facility for facilitating the sequencing of wastewater samples. Finally, we thank Dr. Giridhar Babu for constructive feedback and constant support throughout this study.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.lansea.2023.100151.
Appendix A. Supplementary data
Supplementary Fig. S1_a.
Supplementary Fig. S1_b.
Supplementary Fig. S1_c.
Supplementary Fig. S1_d.
Supplementary Fig. S1_e.
Supplementary Fig. S1_f.
Supplementary Fig. S2.
Supplementary Fig. S3.
Supplementary Fig. S4.
Supplementary Fig. S5.
Supplementary Fig. S6.
References
- 1.COVIDPoops19 Summary of global SARS-CoV-2 wastewater monitoring efforts by UC Merced researchers. Univ Calif Merced. 2022 https://ucmerced.maps.arcgis.com/apps/dashboards/c778145ea5bb4daeb58d31afee389082 URL596. [Google Scholar]
- 2.Naughton C.C., Roman F.A., Grace Alvarado A.F., et al. Show us the data: global COVID-19 wastewater monitoring efforts, equity, and gaps. medRxiv. 2021;712 doi: 10.1101/2021.03.14.21253564. 2021.03.14.21253564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguiar-Oliveira M.L., Campos A., Matos A.R., et al. Wastewater-based epidemiology (WBE) and viral detection in polluted surface water: a valuable tool for COVID-19 surveillance-A brief review. Int J Environ Res Publ Health. 2020;10(17):9251. doi: 10.3390/ijerph17249251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caicedo-Ochoa Y., Rebellón-Sánchez D.E., Peñaloza-Rallón M., et al. Effective reproductive number estimation for initial stage of COVID-19 pandemic in Latin American countries. Int J Infect Dis. 2020;95:316–318. doi: 10.1016/j.ijid.2020.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crits-Christoph A., Kantor R.S., Olm M.R., et al. Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. mBio. 2021;12(No.1) doi: 10.1128/mBio.02703-20. e027033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edition 4th Guidelines for drinking-water quality WHO chronicle. vol. 38. 2011. pp. 104–108. [PubMed] [Google Scholar]
- 7.Deshpande J.M., Shetty S.J., Siddiqui Z.A., et al. Environmental surveillance system to track wild poliovirus transmission. Appl Environ Microbiol. 2003;69:2919–2927. doi: 10.1128/AEM.69.5.2919-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hovi T., Stenvik M., Partanen H., et al. Poliovirus surveillance by examining sewage specimens. Quantitative recovery of virus after introduction into sewerage at remote upstream location. Epidemiol Infect. 2001;127:101–106. doi: 10.1017/s0950268801005787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asghar H., Diop O.M., Weldegebriel G., et al. Environmental surveillance for polioviruses in the global polio eradication initiative. J Infect Dis. 2014;210(Suppl. 1):S294–S303. doi: 10.1093/infdis/jiu384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahmed W., Angel N., Edson J., et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medema G., Been F., Heijnen L., et al. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr Opin Environ Sci Health. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peccia J., Zulli A., Brackney D.E., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La Rosa G., Iaconelli M., Mancini P., et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rambaut A., Holmes E.C., O’Toole Á., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turakhia Y., Thornlow B., Hinrichs A.S., et al. Ultrafast Sample placement on Existing tRees (UShER) enables real-time phylogenetics for the SARS-CoV-2 pandemic. Nat Genet. 2021;53(6):809–816. doi: 10.1038/s41588-021-00862-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sapula S.A., Whittall J.J., Pandopulos A.J., et al. An optimized and robust PEG precipitation method for detection of SARS-CoV-2 in wastewater. Sci Total Environ. 2021;785 doi: 10.1016/j.scitotenv.2021.147270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zdenkova K., Bartackova J., Cermakova E., et al. Monitoring COVID-19 spread in Prague local neighborhoods based on the presence of SARS-CoV-2 RNA in wastewater collected throughout the sewer network. medRxiv. 2021 doi: 10.1016/j.watres.2022.118343. 2021.7.28.21261272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang T., Breitbart M., Lee W.H., et al. RNA viral Community in Human Feces: prevalence of plant pathogenic viruses. PLoS Biol. 2005;4 doi: 10.1371/journal.pbio.0040003. Article e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isaksoon F., Lundy L., Hedström A., et al. Evaluating the use of alternative normalization approaches on SARS-CoV-2 concentrations in wastewater: Experiences from two catchments in Northern Sweden. Environments. 2022;9:39. doi: 10.3390/environments9030039. [DOI] [Google Scholar]
- 20.Roberts S.W. Control chart tests based on geometric moving averages. Technometrics. 1959;1:239–250. [Google Scholar]
- 21.Scrucca L. qcc: an R package for quality control charting and statistical process control. dim (pistonrings) 2004;1(200):3. [Google Scholar]
- 22.Yupaporn A., Rapin S. EWMA control chart based on its first hitting time and coronavirus alert levels for monitoring symmetric COVID-19 cases. Asia Pacific J Trop Med. 2021;14(8):364. [Google Scholar]
- 23.Bagarella G., Maistrello M., Minoja M., et al. Early detection of SARS-CoV-2 epidemicwaves: lessons from the syndromic surveillance in Lombardy, Italy. Int. J. Environ. Res. Public Health. 2022;19 doi: 10.3390/ijerph191912375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elias C., Sekri A., Leblanc P., et al. The incubation period of COVID-19: a meta-analysis. Int J Infect Dis. 2021;104:708–710. doi: 10.1016/j.ijid.2021.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du Z., Liu C., Wang L., et al. Shorter serial intervals and incubation periods in SARS-CoV-2 variants than the SARS-CoV-2 ancestral strain. J Trav Med. 2022;5:1–3. doi: 10.1093/jtm/taac052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant R., Charmet T., schaeffer L., et al. Impact of SARS-CoV-2 Delta variant on incubation, transmission settings and vaccine effectiveness: results from a nationwide case-control study in France. Lancet Reg Health Europe. 2022;13 doi: 10.1016/j.lanepe.2021.100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boucau J., Marino C., Regan J., et al. Duration of shedding of culturable virus in SARS-CoV-2 Omicron (BA.1) infection. N Engl J Med. 2022;387(3):275–277. doi: 10.1056/NEJMc2202092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wölfel R., Corman V.M., Guggemos W., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020:1–10. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 29.Rose C., Parker A., Jefferson B., et al. The characterization of feces and urine: a review of the literature to inform advanced treatment technology. Crit Rev Environ Sci Technol. 2015;45(17):1827–1879. doi: 10.1080/10643389.2014.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foladori P., Cutrupi F., Segata N., et al. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhoyar R.C., Jain A., Sehgal P., et al. High throughput detection and genetic epidemiology of SARS-CoV-2 using COVIDSeq next-generation sequencing. PLoS One. 2021;16(2) doi: 10.1371/journal.pone.0247115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H. Aligning sequence reads, clone sequences and assembly con∗gs with BWA-MEM. 2014. figshare. Poster. [DOI]
- 33.Li H., Handsaker B., Wysoker A., et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grubaugh N.D., Gangavarapu K., Quick J., et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20:8. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karthikeyan S., Levy J.I., De Hoff P., et al. Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission. Nature. 2022;609:101–108. doi: 10.1038/s41586-022-05049-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsueng G., Mullen J.L., Alkuzweny M., et al. 2021. Outbreak Info Research Library: A Standardized, Searchable Platform To Discover and Explore COVID-19 Resources. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oksanen J., Blanchet F.G., Friendly M., et al. vegan: community ecology package. 2017. https://github.com/vegandevs/vegan
- 38.Kuznetsova A., Brockhoff P.B., Christensen R.H.B. lmerTest package: tests in linear mixed effects models. J Stat Software. 2017;82(13):1–26. [Google Scholar]
- 39.Wu F., Zhang J., Xiao A., et al. vol. 5. 2020. (SARS-CoV-2 Titers in wastewater are higher than expected from clinically confirmed cases mSystems). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohapatra S., Gayathri Menon N., Mohapatra G., et al. The novel SARS-CoV-2 pandemic: possible environmental transmission, detection, persistence and fate during wastewater and water treatment. Sci Total Environ. 2021;765 doi: 10.1016/j.scitotenv.2020.142746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dharmadhikari T., Rajput V., Yadav Rakeshkumar, et al. High throughput sequencing based direct detection of SARS-CoV-2 fragments in wastewater in Pune, West India. Sci Total Environ. 2022;807 doi: 10.1016/j.scitotenv.2021.151038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Randazzo W., Truchado P., Cuevas-Ferrando E., et al. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brito A.F., Semenova E., Dudas G., et al. Global disparities in SARS-CoV-2 genomic surveillance. medRxiv. 2021 doi: 10.1101/2021.08.21.21262393. [Preprint] 2021.08.21.21262393 PMID: 34462754; PMCID: PMC8404891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singanayagam A., Hakki S., Dunning J., et al. Community transmission and viral load kinetics of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study. Lancet Infect Dis. 2022;22:183–195. doi: 10.1016/S1473-3099(21)00648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.