Abstract
High-resolution melting (HRM) analysis is a PCR–based method that can be used as a screening assay to identify SARS-CoV-2 variants. However, conventional HRM assays hardly detect slight melting temperature differences at the A–T to T–A transversion. As the N501Y substitution results from A–T to T–A transversion in A23063, few or no studies have shown that a conventional HRM assay can identify N501Y variants. This study successfully developed an HRM assay for identifying the N501Y mutation. Two HRM assays were used in the N501 site because the discrimination results were affected by the virus copy numbers. One is a conventional HRM assay (detectable at 103–106 copies/mL) and the other is a modified HRM assay by adding the wild-type fragment (detectable at 105–1010 copies/mL). Using viral RNAs from cultured variants (Alpha, Beta, and Gamma), a modified HRM assay correctly identified three N501Y variants because of high-copy-number RNAs in those viral samples. The sensitivity and specificity of the N501Y assay were 93.3% and 100%, respectively, based on 209 clinical samples (105 for N501; 104 for N501Y). These results suggest that our HRM-based assay is a powerful tool for rapidly identifying various SARS-CoV-2 variants.
Keywords: SARS-CoV-2, High-resolution melting, N501Y, A23063T, Class 4 single nucleotide polymorphisms, Rapid identification
1. Introduction
The outbreak of the coronavirus disease 2019 (COVID-19) was caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and spread worldwide (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020, Wu et al., 2020, Zhou et al., 2020). Although the rate of SARS-CoV-2 genomic mutation is lower than that of other RNA viruses, many variants have appeared worldwide. Since late 2020, the World Health Organization (WHO) has classified variants of concern (VOCs) and variants of interest according to their risk to public health. COVID-19 specimens were sequenced with the entire viral genome using next-generation sequencing (NGS) and each variant was classified (Harilal et al., 2020). Some developed countries maintain the NGS approach to surveillance for monitoring virus evolution. The NGS approach is essential for tracking SARS-CoV-2 variants and preventing the spread of high-risk variants. However, as NGS analysis is costly and time-consuming, not all specimens can be sequenced in the viral whole genome. A possible solution to the disadvantages of NGS is the combined use of PCR-based screening assays.
In 2020, three VOCs, Alpha from the United Kingdom, Beta from South Africa, and Gamma from Brazil, occurred and spread worldwide. Those VOCs possess some characteristic amino-acid substitutions in the spike receptor-binding domain (RBD) as follows: Alpha has N501Y; Beta has K417N, E484K, and N501Y; and Gamma has K417T, E484K, and N501Y. Common among those variants is the N501Y substitution, which is a key mutation for increased viral infectivity. Several previous studies have indicated that PCR-based screening assays with specific probes could detect the N501Y mutation (Banada et al., 2021, Chaintoutis et al., 2021, Sandoval Torrientes et al., 2021). Indeed, the Japanese government carried out a TaqMan N501Y screening assay to track the Alpha variant.
Our previous studies have shown that high-resolution melting (HRM) analysis, a post-PCR technology based on the melting temperature (Tm) of the amplicon, detected several SARS-CoV-2 mutations (Aoki et al., 2022a, Aoki et al., 2022b, Ferreira et al., 2021). As HRM analysis does not require the design of a specific probe, HRM-based assays can be developed more easily than the TaqMan probe assay, as reported previously (Aoki et al., 2021a). However, this technique must overcome the drawback of the difficulty of detecting class 4 single nucleotide polymorphisms (SNPs) with A–T to T–A complementary transversion using conventional HRM analysis, because of the slight difference in Tm (<0.4 °C) (Cai et al., 2010). Class 4 SNPs are the cause for the A23063T single nucleotide mutation found in the N501Y substitution of SARS-CoV-2. For this reason, HRM-based assays rarely identify the N501Y variant.
In this study, we aimed to develop two types of HRM-based assays (a conventional HRM assay; a modified HRM assay by adding the wild-type fragment) to identify the SARS-CoV-2 N501Y mutation. We also investigated the applicability of the current HRM-based assay to the determination of clinical samples.
2. Materials and methods
2.1. Ethical statement
This project was approved by the Research Ethics Committee of Meijo University (approval No. 2020–17–4) and the Aichi Prefectural Institute of Public Health (approval No. 20E-4) and was conducted according to the Infectious Diseases Control Law in Japan.
2.2. Preparation of standard RNA fragments: In vitro T7 transcription
The SARS-CoV-2 sequence was obtained from NCBI (GenBank ID: MN908947), the GISAID database (www.gisaid.org), and the Pango nomenclature system (https://cov-lineages.org/lineages.html). Two RBD DNA fragments (wild-type and N501Y mutant; 600–1000 bp in length) with a 5′ T7 upstream promoter sequence were obtained from Eurofins Genomics K.K. (Tokyo, Japan). In vitro T7 transcription was performed as described previously (Aoki et al., 2021b). The synthesized single-stranded RNA fragments were used as reverse transcriptase (RT)-PCR amplification templates.
2.3. Virus isolation and RNA extraction
The National Institute of Infectious Diseases (Tokyo, Japan) generously provided the Alpha variant (Pango lineage B.1.1.7, hCoV-19/Japan/QK002/2020), Beta variant (Pango lineage B.1.351, hCoV-19/Japan/TY8–612/2021), Gamma variant (Pango lineage P.1, hCoV-19/Japan/TY7–501/2021), and VeroE6/TMPRSS2 cell line. Cells were grown to subconfluence in low-glucose Dulbecco’s modified Eagle’s medium (DMEM; Nihon pharmaceutical, Tokyo, Japan), supplemented with 10% fetal bovine serum (FBS; Sero Scandia, Hellebaek, Denmark), 1 mg/mL G418 (Nacalai Tesque, Kyoto, Japan), and 1 × penicillin–streptomycin (PS; FUJIFILM Wako Pure Chemical, Osaka, Japan) at 37 °C under a humidified 5% CO2 atmosphere. The virus was inoculated after the culture medium was replaced with DMEM supplemented with 2% FBS and 1 × PS. Once the cytopathic effect appeared, the culture medium was harvested and centrifuged, and the supernatants were collected as virus stock. Viral RNA was extracted from the virus stock using spin columns (High Pure Viral RNA Kit; Roche Diagnostics., Basel, Switzerland). All experiments using infectious SARS-CoV-2 were performed in a biosafety level 3 (BSL3) laboratory at Aichi Prefectural Institute of Public Health, according to standard BSL3 guidelines. Purified RNAs were used as RT-PCR amplification templates.
2.4. RT-PCR amplification: First PCR
We performed RT-PCR in a single closed tube using a one step RT-PCR kit (One Step PrimeScript III RT-qPCR Mix, with UNG; Takara Bio Inc., Kusatsu, Japan), according to the manufacturer’s instructions. The primer pairs used were as follows: Outer forward 5′-AATCTTGATTCTAAGGTTGGTGGTAAT-3′ and outer reverse 5′-AACCAAATTAGTAGACTTTTTAGGTCCA-3′. Each DNA fragment was observed as a single, correctly sized band (285 bp). RT-PCR amplification was performed as described previously (Aoki et al., 2021b). After RT-PCR amplification, the reaction mixture was diluted 10,000-fold with water and used as a template for the second PCR and HRM analyses.
2.5. Clinical samples
From March to April 2021, 209 nasopharyngeal swabs or saliva samples were collected from patients suspected of having COVID-19 and those detected with COVID-19 at Aichi Prefectural Institute of Public Health. Viral RNA was extracted from clinical samples using spin columns (QIAamp Viral RNA Mini QIAcube Kit; Qiagen GmbH, Hilden, Germany) and an automated nucleic acid purification system (QIAcube Connect; Qiagen). RT-PCR amplification was performed, and amplicon size was confirmed to be approximately 285 bp as previously reported (Aoki et al., 2021b). The DNA fragments were sequenced (Eurofins Genomics K.K.) and used as a template (with diluted 10,000-fold) for the second PCR and HRM analyses.
2.6. HRM analysis: Second PCR
HRM was performed using an HRM reagent (MeltDoctor HRM Master Mix; Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s instructions. The second primer pair was as follows: Forward 5′-TTTCCTTTACAATCATATGGTTTCC-3′ and reverse 5′-GCTGGTGCATGTAGAAGTTCA-3′. Each DNA fragment was observed as a single, correctly sized band (98 bp). PCR amplification was performed as described previously (Aoki et al., 2021b). We performed all reactions in duplicate using a real-time PCR system (LightCycler 96 System; Roche Diagnostics). The HRM curves were analyzed using Gene Scanning Software, version 1.1.0.1320 (Roche Diagnostics) under default settings. We acquired the normalized melting curve and melting peaks (−dF/dT) by setting the premelt and postmelt fluorescence at 100% and 0%, respectively.
3. Results
3.1. HRM analyses of isolated viral RNAs in N501 sites
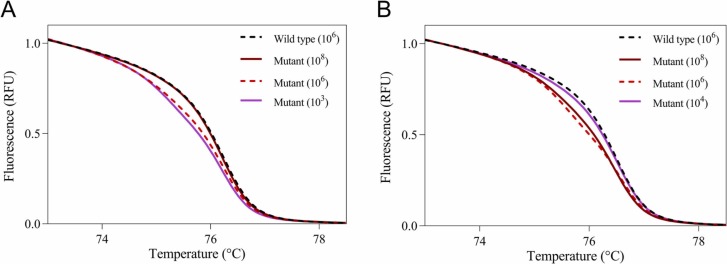
The N501Y mutation was coded by A23063T, meaning a class 4 SNP. The identification of the N501Y variant should be difficult for a conventional HRM analysis. To detect the N501Y mutation, we performed not only a conventional HRM-based assay but also a modified HRM-based assay by adding the wild-type fragment. First, we conducted a conventional HRM-based assay using serial dilutions of wild-type and N501Y RBD fragments. A conventional HRM-based assay could correctly identify the N501Y RBD fragment with a dynamic range of 103–106 copies/mL ( Fig. 1A). As reported previously, the copy number of viruses in the clinical samples ranged from 104–108 copies/mL (Ct values from 15 to 35) (To et al., 2020, Zhu et al., 2020). Thus, only the conventional HRM-based assay will identify a limited number of clinical samples. We then investigated a modified HRM-based assay by adding 106 copies/mL of wild-type fragments in equal amounts with samples. With the addition of the wild-type fragment, the dynamic range of the HRM-based assay was 105–1010 copies/mL (Fig. 1B). Therefore, the HRM-based assay can detect the N501Y mutation with a wide range (103–1010 copies/mL) by combining it with the addition of the wild-type fragment and without.
Fig. 1.
Normalized melting curves of positive control RNAs for the N501 site. Normalized melting curve plots for the N501 site without the addition of the wild-type fragment (A) and with the addition of the wild-type fragment (B) were acquired using standard fragments of 106 copies/mL wild-type (dashed black line), 108 copies/mL N501Y mutant (solid brown line), 106 copies/mL N501Y mutant (dashed red line), and 103 or 104 copies/mL N501Y mutant (solid purple line).
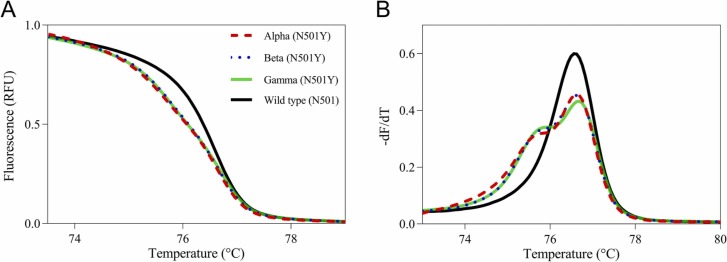
We next investigated the N501Y assay using isolated viral RNAs. By adding the wild-type fragment, the HRM-based assay correctly classified all three variants as N501Y mutants ( Fig. 2). The conventional HRM-based assay could not identify three variants (data not shown) because the isolated viral RNAs were high copy numbers (Ct value <15: more than 108 copies/mL).
Fig. 2.
Normalized melting curves and melting peaks of isolated viral RNAs for the N501 site with the addition of the wild-type fragment. Normalized melting curve plots (A) and melting peak plots (B) for the N501 site with the addition of the wild-type fragment were acquired using isolated viral RNAs of the Alpha (dashed red line), Beta (dotted blue line), and Gamma (solid green line) variants and the wild-type positive control (solid black line).
3.2. HRM analyses of clinical samples in the N501 site
Finally, we confirmed the applicability of the N501Y assay with or without the addition of the wild-type fragment using clinical samples. Sanger sequencing identified 209 samples as the N501 samples (A23063; 105 samples) and N501Y mutants (A23063T; 104 samples). All 105 N501 samples were correctly classified as N501 using the HRM assay both with and without the addition of the wild-type fragment ( Table 1). A total of 97 N501Y samples could be classified as N501Y mutants under at least one condition (with and/or without adding the wild-type fragment). Neither N501Y assay could identify seven of the N501Y samples (Ct values from 25 to 30) as the N501Y mutant. Therefore, we calculated the sensitivity and specificity of the current N501Y assay as 93.3% (97/104) and 100% (105/105), respectively.
Table 1.
Results of the high-resolution melting analyses at the N501 site using clinical samples.
| Without the addition of the wild-type fragment (conventional HRM-based assay) | ||
|---|---|---|
| HRM result | Sanger sequence |
|
| N501 (n = 105) | N501Y (n = 104) | |
| N501 | 105 | 53 |
| N501Y | 0 | 39 |
| Unidentified | 0 | 12 |
| With the addition of the wild-type fragment (modified HRM-based assay) | ||
|---|---|---|
| HRM result | Sanger sequence | |
| N501 (n = 105) | N501Y (n = 104) | |
| N501 | 105 | 10 |
| N501Y | 0 | 78 |
| Unidentified | 0 | 16 |
| Results of the combined assays | ||
|---|---|---|
| N501 (n = 105) | N501Y (n = 104) | |
| Both assays positive | 105 | 21 |
| Either assay positive | 0 | 76 |
| Both assays negative | 0 | 7 |
4. Discussion
The Alpha, Beta, and Gamma variants are the first SARS-CoV-2 variants classified by the WHO as VOCs. Three variants share the N501Y substitution, one of the highest-risk mutations that increases viral infectivity. However, a conventional HRM-based assay can hardly identify the A23063T coding N501Y. This study is the first report regarding the development of an HRM-based assay for detecting the N501Y mutation using the addition of a wild-type fragment. To enhance the discrimination power of the A23063T (N501Y) mutation, we added a small amount of the wild-type fragment to the reaction tube as used previously to identify class 4 SNPs (Liew et al., 2004, Matsumoto et al., 2016). By adding the wild-type fragment, a portion of the mutant amplicon creates a duplex with the wild-type amplicon, leading to a decrease in the Tm value. The sensitivity and specificity of the N501Y assay were 93.3% and 100%, respectively, indicating that our assay may be useful in identifying N501Y variants. In addition, we developed an HRM-based assay for K417N/T and E484K mutations (Figs. S1 and S2). In the K417 sites, this assay can distinguish three variants, such as Alpha, Beta, and Gamma, using a single assay. In the K417 site, all 209 clinical samples were classified as K417 by this assay (data not shown). This result suggests that every N501Y variant in this study was the Alpha variant.
The TaqMan probe assay is the most popular tool for detecting SNPs with specific probes. As the arrangement of the probe affects the assay’s specificity, optimizing the probe location and length is necessary to develop an SNP genotyping assay. In this study, we developed the HRM-based N501Y assay by adding the wild-type fragment. As the HRM-based assay can be constructed without designing a specific probe, the development of this assay requires less time. Thus, our HRM-based assay is more simple and less costly than a probe-based assay, such as the TaqMan probe assay. The addition of 106 copies/mL of wild-type fragments may also be applicable for other SARS-CoV-2 mutations coded by the A–T exchange, such as L452Q (coded by T22917A), though it is required for the optimization and validation of the concentration of the wild-type fragment added.
Although this HRM-based assay has some advantages over other screening assays, it also has limitations for identifying SARS-CoV-2 variants. First, we validated our HRM assay for the N501Y mutation using clinical samples in a confined area of Japan. To verify the usability of our N501Y assay, more diverse samples must be analyzed. Second, the concentration of wild-type fragments added was investigated only at 106 copies/mL. Third, this assay needs a two-step PCR protocol to improve the detection limit.
In conclusion, we developed HRM-based assays to determine the presence of N501Y SARS-CoV-2 mutation. This study demonstrates that the HRM assay can detect the N501Y mutation without the need for designing a specific probe. Thus, our HRM-based assay is an easier and less costly tool for identifying the SARS-CoV-2 N501Y mutation than probe-based assays, such as the TaqMan probe. Although this assay can be a useful screening tool for identifying SARS-CoV-2 variants, further studies are required to overcome this study’s limitations.
Funding
This work was supported in part by the Daiko Foundation and Meijo University Research Project for Countermeasures Against COVID-19.
Authorship statement
All the authors participated in the interpretation of the results. A.A. designed and conducted the HRM analyses and wrote the original draft of the manuscript. H.A. and M.I. conducted virus isolation and RNA purification and edited the manuscript. Y.M. performed sequence analyses. Y.O. planned the study and edited the manuscript. K.S., M.K., M.K., K.O., T.S., and H.J. planned the study and supervised the project. All the authors critically reviewed and approved the final version of the manuscript.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgments
The authors would like to thank the staff of the Laboratory of Virology, Department of Microbiology and Medical Zoology, Aichi Prefectural Institute of Public Health, who performed COVID-19 PCR testing and RNA purification from clinical samples.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jviromet.2023.114678.
Appendix A. Supplementary material
Supplementary material
.
References
- Aoki A., Adachi H., Mori Y., Ito M., Sato K., Okuda K., Sakakibara T., Okamoto Y., Jinno H. Discrimination of SARS-CoV-2 Omicron Sublineages BA.1 and BA.2 Using a High-Resolution Melting-Based Assay: a Pilot Study. Microbiol Spectr. 2022;10 doi: 10.1128/spectrum.01367-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki A., Adachi H., Mori Y., Ito M., Sato K., Okuda K., Sakakibara T., Okamoto Y., Jinno H. A rapid screening assay for L452R and T478K spike mutations in SARS-CoV-2 Delta variant using high-resolution melting analysis. J. Toxicol. Sci. 2021;46:471–476. doi: 10.2131/jts.46.471. [DOI] [PubMed] [Google Scholar]
- Aoki A., Mori Y., Okamoto Y., Jinno H. Simultaneous screening of SARS-CoV-2 Omicron and Delta variants using high-resolution melting analysis. Biol. Pharm. Bull. 2022;45:394–396. doi: 10.1248/bpb.b21-01081. [DOI] [PubMed] [Google Scholar]
- Aoki A., Mori Y., Okamoto Y., Jinno H. Development of a genotyping platform for SARS-CoV-2 variants using high-resolution melting analysis. J. Infect. Chemother. 2021;27:1336–1341. doi: 10.1016/j.jiac.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banada P., Green R., Banik S., Chopoorian A., Streck D., Jones R., Chakravorty S., Alland D. A simple reverse transcriptase PCR melting-temperature assay to rapidly screen for widely circulating SARS-CoV-2 variants. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.00845-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Yuan Y., Lin Q., Chan P. Allele-specific extension allows base-pair neutral homozygotes to be discriminated by high-resolution melting of small amplicons. Anal. Biochem. 2010;406:29–33. doi: 10.1016/j.ab.2010.06.043. [DOI] [PubMed] [Google Scholar]
- Chaintoutis S.C., Chassalevris T., Tsiolas G., Balaska S., Vlatakis I., Mouchtaropoulou E., Siarkou V.I., Tychala A., Koutsioulis D., Skoura L., Argiriou A., Dovas C.I. A one-step real-time RT-PCR assay for simultaneous typing of SARS-CoV-2 mutations associated with the E484K and N501Y spike protein amino-acid substitutions. J. Virol. Methods. 2021;296 doi: 10.1016/j.jviromet.2021.114242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira B., da Silva-Gomes N.L., Coelho W., da Costa V.D., Carneiro V.C.S., Kader R.L., Amaro M.P., Villar L.M., Miyajima F., Alves-Leon S.V., de Paula V.S., Leon L.A.A., Moreira O.C. Validation of a novel molecular assay to the diagnostic of COVID-19 based on real time PCR with high resolution melting. PLoS One. 2021;16 doi: 10.1371/journal.pone.0260087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harilal D., Ramaswamy S., Loney T., Suwaidi H.A., Khansaheb H., Alkhaja A., Varghese R., Deesi Z., Nowotny N., Alsheikh-Ali A., Abou Tayoun A. SARS-CoV-2 whole genome amplification and sequencing for effective population-based surveillance and control of viral transmission. Clin. Chem. 2020;66:1450–1458. doi: 10.1093/clinchem/hvaa187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew M., Pryor R., Palais R., Meadows C., Erali M., Lyon E., Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin. Chem. 2004;50:1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- Matsumoto N., Mori S., Hasegawa H., Sasaki D., Mori H., Tsuruda K., Imanishi D., Imaizumi Y., Hata T., Kaku N., Kosai K., Uno N., Miyazaki Y., Yanagihara K. Simultaneous screening for JAK2 and calreticulin gene mutations in myeloproliferative neoplasms with high resolution melting. Clin. Chim. Acta. 2016;462:166–173. doi: 10.1016/j.cca.2016.09.023. [DOI] [PubMed] [Google Scholar]
- Sandoval Torrientes M., Castello Abietar C., Boga Riveiro J., Alvarez-Arguelles M.E., Rojo-Alba S., Abreu Salinas F., Costales Gonzalez I., Perez Martinez Z., Martin Rodriguez G., Gomez de Ona J., Coto Garcia E., Melon Garcia S. A novel single nucleotide polymorphism assay for the detection of N501Y SARS-CoV-2 variants. J. Virol. Methods. 2021;294 doi: 10.1016/j.jviromet.2021.114143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., Yip C.C.-Y., Cai J.-P., Chan J.M.-C., Chik T.S.-H., Lau D.P.-L., Choi C.Y.-C., Chen L.-L., Chan W.-M., Chan K.-H., Ip J.D., Ng A.C.-K., Poon R.W.-S., Luo C.-T., Cheng V.C.-C., Chan J.F.-W., Hung I.F.-N., Chen Z., Chen H., Yuen K.-Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Guo J., Xu Y., Chen X. Viral dynamics of SARS-CoV-2 in saliva from infected patients. J. Infect. 2020;81:e48–e50. doi: 10.1016/j.jinf.2020.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material